Summary
The Nostoc sp. strain CCCryo 231-06 is a cyanobacterial strain capable of surviving under extreme conditions and thus is of great interest for the astrobiology community. The knowledge of its complete genome sequence would serve as a guide for further studies. However, a major concern has been placed on the effects of contamination on the quality of sequencing data without a reference genome. Here, we report the use of microfluidic technology combined with single cell sequencing and de novo assembly to minimize the contamination and recover the complete genome of the Nostoc strain CCCryo 231-06 with high quality. 100% of the whole genome was recovered with all contaminants removed and a strongly supported phylogenetic tree. The data reported can be useful for comparative genomics for phylogenetic and taxonomic studies. The method used in this work can be applied to studies that require high-quality assemblies of genomes of unknown microorganisms.
Subject areas: Microbiology, Microbial genomics, Space sciences, Astrobiology
Graphical abstract
Highlights
-
•
This work uses a microfluidic platform for Nostoc single cell sequencing
-
•
This technology provides minimal contamination in single cell sequencing
-
•
Complete genome of the Nostoc strain CCCryo 231-06 was recovered with high quality
Microbiology; Microbial genomics; Space sciences; Astrobiology
Introduction
Nostoc is a genus of filamentous cyanobacteria common in both terrestrial and aquatic habitats. Colonies of Nostoc species have been found in extreme environmental conditions, from freezing and dry Antarctic valleys and the Arctic to hot desert soils and springs (Dodds et al., 1995; Potts, 2000). The successful survival of Nostoc species in these habitats is largely due to their ability to recover their metabolic activities within days after rehydration even though they have been dessicated for years. Therefore, Nostoc species have been of great interest for the astrobiology community and the search of life on Mars mainly because of their dessication and freezing resistance and ultraviolet (UV) tolerance (Wang et al., 2010; Kimura et al., 2017a, 2017b). For example, dried colonies of Nostoc sp. HK-01 were found alive after long-term high vacuum and UV exposure and were able to grow on analogous Martian soil, and thus were proposed for experimentation under space environmental conditions (Kimura et al., 2014). Nostoc sp. strain CCCryo 231-06, isolated from permafrost soil and rock substrates in Victoria Land, Antarctica, was selected and used as a microbial species for the Biology and Mars Experiment (BIOMEX) on the International Space Station (ISS)(De Vera et al., 2019). As a preferred model species in astrobiology and extraterrestrial planetary research (Kimura et al., 2014; Tomita-Yokotani et al., 2013), the availability of the complete genome sequence of this Nostoc sp. will lead to a better understanding of this group of photosynthesizing and oxygen producing microorganisms and can serve as a guide for further studies. In addition, the knowledge of the complete genome enables global approaches to biological functions in extremophiles and understanding of their survival and resistance mechanisms and sheds new light on the molecular processes conferring extraordinary resistance to environmental stressors.
Whole genome sequencing (WGS) technology and bioinformatic tools play a significant role in obtaining the complete genome sequences of unknown or unculturable microbial species (Quainoo et al., 2017; Buermans and Den Dunnen, 2014). However, one of the increasing concerns has focused on the effects of contamination on the quality of sequencing data as it can impact the genetic findings and the fidelity of the genome sequences of species with no available reference genome (Low et al., 2019). The sources of contamination can be unwanted cells in the population or extracellular DNA in the sample, reagents, or instruments. To improve the quality of sequencing data and lead to meaningful scientific findings, efforts have been directed to identify or minimize contaminants in WGS applications. Bioinformatic tools have been developed to recognize and remove contaminants in the sequencing data (Olson et al., 2017; Lu and Salzberg, 2018). Microfluidic platforms are becoming popular for WGS applications due to their unique ability to handle nanoliters of fluid in a controlled manner, allowing for nanoscale reactions with minimal contamination (Kim et al., 2017; Liu and Walther-Antonio, 2017; Blainey and Quake, 2011). However, the challenge still remains especially when the contamination stems from closely related species.
To further address this challenge, single-cell whole genome sequencing (SC-WGS) using microfluidic technologies is starting to gain attention (Liu et al., 2019a; Hosokawa et al., 2017; Landry et al., 2013). The key steps in SC-WGS include single cell isolation, lysis, and amplification of femto to picograms of total DNA to reach the quantity sufficient for library preparation and sequencing. Multiple displacement amplification (MDA) (Dean et al., 2001) has been a popular option for single-cell whole genome amplification (SC-WGA) in microfluidic platforms (Marcy et al., 2007; Zare and Kim, 2010; Binga et al., 2008). It is based on φ29 DNA polymerase and random primers to replicate template DNA with high fidelity and lower error rates following relatively simple procedures compatible with microfluidic platforms (Motley et al., 2014; De Bourcy et al., 2014; Chen et al., 2014).
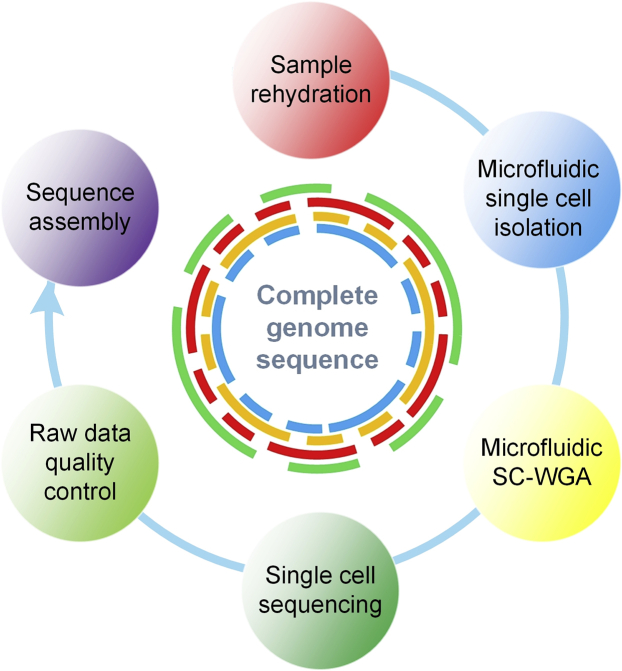
In this work, we perform SC-WGS on the Nostoc sp. strain CCCryo 231-06 using microfluidic technology to obtain its complete genome sequence with minimal contamination. The microfluidic platform is shown in Figure 1 and the overall workflow is shown in Figure 2. We chose this Nostoc strain because this species was recently selected and used as a microbial species for the BIOMEX experiment on the ISS, which could lead to findings on the limits of life on Mars and provide further understanding of the kind of species that are likely to survive in Mars-like habitats. The complete genome sequence of strain CCCryo 231-06 would lead to a more complete understanding of the genetic features of this species and provide reference genome data for comparative genomic analyses of evolution among different Nostoc species. In addition, the demonstrated methodology can potentially impact the study of other microorganisms that are of astrobiological significance, such as Phormidium and Leptolyngbya, the other two dominant cyanobacterial species in the mat communities alongside Nostoc that can survive extreme temperatures and UV in the Arctic and Antarctic (Vincent et al., 2004; Quesada and Vincent, 2012).
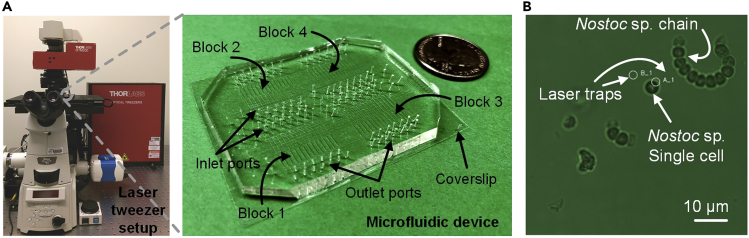
Figure 1.
Optofluidic platform overview
(A) An optofluidic platform consists of a microscope, laser tweezers and a microfluidic device consists of 4 identical reaction blocks for high-throughput SC-WGA.
(B) The use of laser tweezers to trap single cells of Nostoc strain CCCryo 231-06. Laser trap A_1 was turned on and trapped a single cell in the sample suspension; laser trap B_1 was turned off.
Figure 2.
An overview of the workflow of SC-WGS for Nostoc strain CCCryo 231-06
Results and discussion
Sequencing data and genome properties
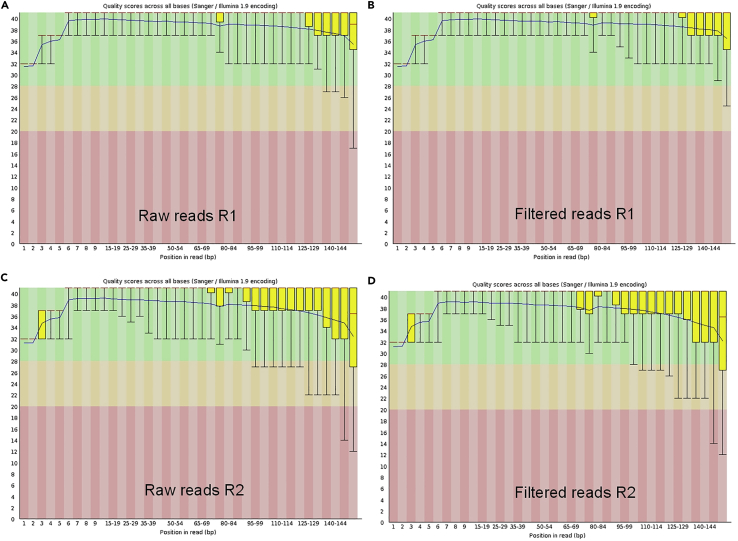
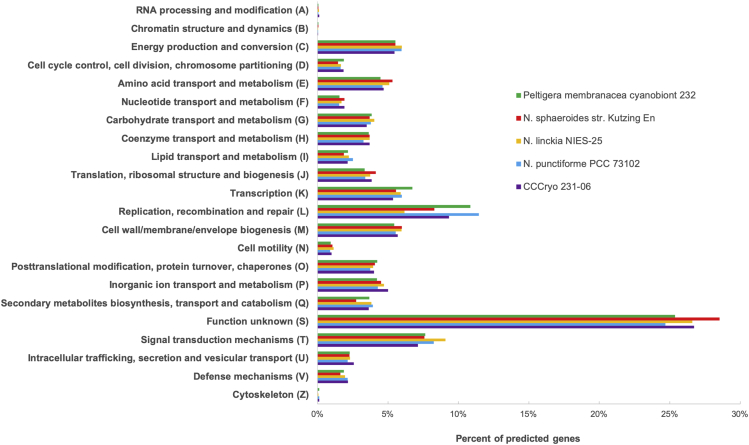
A complete list of samples, exposure conditions and number of single cells sequenced is summarized in Table 1. To ensure high quality of the de novo assembly, raw reads were filtered for final assembly. The draft genome consists of 721 contigs containing a mean length of 7,079,046 bp and a GC content of 41.75%. The filtering has led to higher quality sequencing reads for de novo assembly with N50 of 22,738 bp and L50 of 94 bp (Figure 3). A summary of the statistics before and after filtering is presented in Table 2. Nostoc sp. CCCryo 231-06 genome statistics is shown in Table 3. The de novo assembled genome of the Nostoc strain CCCryo 231-06 is 7.8 Mbp in size with ∼750 fragments. The recovered ribosomal RNA (rRNA) gene sequence matches the deposited 16S rRNA gene sequence of Nostoc strain CCCryo 231-06. Several contaminants were detected in the sequencing data as shown in Figure 4; these contaminants were removed prior to subsequent analysis. The genome recovery completeness was evaluated using CheckM and essentially reached 100% completeness without known contamination. We also compared the genome of Nostoc strain CCCryo 231-06 with its close relatives (Peltigera membranacea, Nostoc sphaeroides, Nostoc linckia, and Nostoc punctiforme) based on Clusters of Orthologous Groups of proteins (COGs) database (Figure 5). Each COG functional categories represent a family of orthologous protein-coding genes. Of the 7,939 total genes in the Nostoc CCCryo 231-06, 99.4% are protein-encoding genes, 57.5% of the genes are assigned to functions, and 61.9% of the genes are assigned to COGs. No significant differences in gene category usage were apparent among the genomes of these species. Supplemental functional analysis to provide a more granular, gene-specific view was performed on the same group of Nostoc species using OrthoFinder (Emms and Kelly, 2019). 82% of genes in the gene pool across the five species appeared in an orthogroup, with only 4.1% of them in species-specific orthogroups. 45% of orthogroups contained genes from all species. Of the 7891 protein-coding genes in Nostoc CCCryo 231-06, 81% appeared in multispecies orthogroups (Table S1).
Table 1.
A list of samples of the Nostoc sp. strain CCCryo 231-06 exposed to different conditions and the number of single cells sequenced
| Sample No. | Sample name | Exposure condition | Number of single cells | Cultured at CCCryoa as |
|---|---|---|---|---|
| 1 | 231-06 Control | 4 | 231-06 | |
| 2 | 1-1-t-05: 231-06, BG11 | Flight, BIOMEX, UV, space | 4 | 231-06_C1 |
| 3 | 1-1-t-06: 231-06, Lunar | 4 | 231-06_D1 | |
| 4 | 1-1-b-05: 231-06, BG11 | Flight, BIOMEX, dark, space | 4 | 231-06_C2 |
| 5 | 1-1-b-06: 231-06, Lunar | 4 | 231-06_D2 | |
| 6 | 2-1-t-05: 231-06, S-MRS | Flight, BIOMEX, UV, Mars | 4 | 231-06_C4 |
| 7 | 2-1-t-06: 231-06, P-MRS | 3 | 231-06_D4 | |
| 8 | 2-1-b-05: 231-06, S-MRS | Flight, BIOMEX, dark, Mars | 4 | 231-06_C5 |
| 9 | 2-1-b-06: 231-06, P-MRS | 4 | 231-06_D5 | |
| 10 | 1-1-t-05: 231-06, BG11 | Ground, BIOMEX, UV, space | 4 | - |
| 11 | 1-1-t-06: 231-06, Lunar | 4 | - | |
| 12 | 2-1-t-05: 231-06, S-MRS | Ground, BIOMEX, UV, Mars | 4 | - |
| 13 | 2-1-t-06: 231-06, P-MRS | 1 | - |
CCCryo database is available at <www.cccryo.fraunhofer.de>
Figure 3.
Quality control of the sequencing data
(A) Raw data reads R1-based sequence quality.
(B) Filtered reads R1-based sequence quality.
(C) Raw data reads R2-based sequence quality.
(D) Filtered reads R2-based sequence quality.
Table 2.
Sequencing data filtering quality control
| Before filtering | After filtering | |
|---|---|---|
| Mean length | 151 bp | 137 bp |
| Total reads | 454.38 M | 447.57 M |
| Total bases | 68.61 G | 61.38 G |
| Q20 bases | 96.53% | 97.05% |
| Q30 bases | 91.67% | 92.39% |
Table 3.
Nostoc sp. CCCryo 231-06 genome statistics
| Attribute | Value | % of total |
|---|---|---|
| Genome size (bp) | 7,079,046 | |
| DNA G+C (bp) | 41.75 | |
| Total contigs | 721 | |
| Contig N50 length (bp) | 22,738 | |
| Contig L50 count | 94 | |
| Estimated essential completeness (%) | 100 | |
| Estimated contig contamination (%) | 0 | |
| Total genes | 7,939 | |
| Protein encoding genes | 7,891 | 99.4% |
| rRNA gnes | 4 | 0.0005% |
| tRNA genes | 44 | 0.0055% |
| Genes with function prediction | 4,566 | 57.5% |
| Genes assigned to COGs | 4,911 | 62.9% |
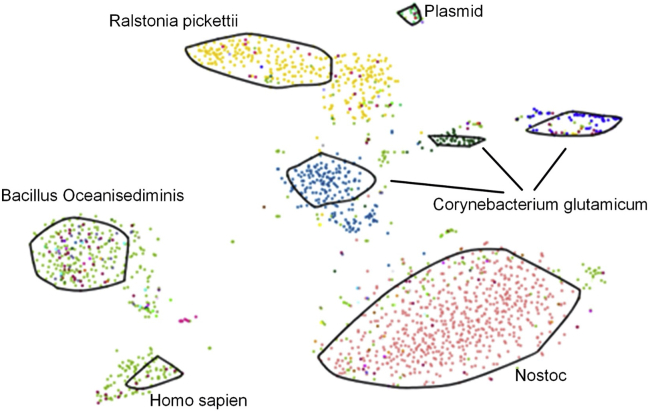
Figure 4.
Representation of metagenome contigs for manual binning
Highlighted is the bin for the Nostoc strain CCCryo 231-06. Other organisms are present in the sequence data, being members of the culture or environmental contaminants.
Figure 5.
A COG usage bar chart shows the percent of predicted genes of Nostoc strain CCCryo 231-06 and four close relatives
No significant differences in gene category usage were apparent among the genomes of these species.
Contamination analysis
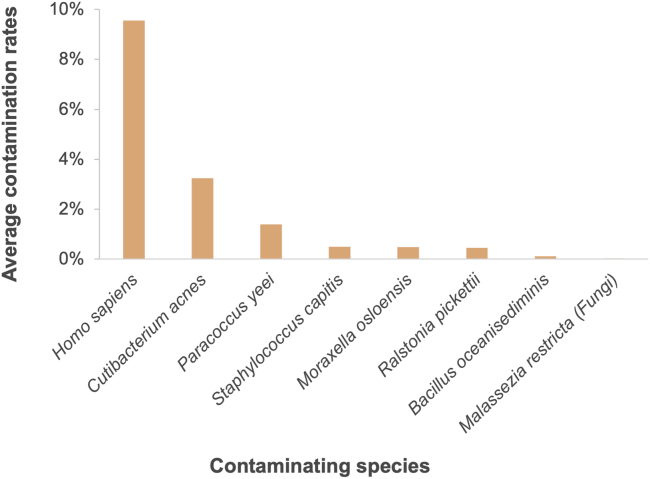
Contaminants detected in the sequenced reads are presented in Figure 6. Homo sapiens reads are the most prominent contamination detected among the sequenced reads, and this non-specific amplification of human DNA has been reported as a common contaminant in whole genome amplification and remains a challenge (Hammond et al., 2016; Walker et al., 2020). Among the contaminating microorganisms, nearly half are frequent colonizers of human skin while the rest are typically found in nature. Cutibacterium acnes, Staphylococcus capitis, Moraxella osloensis, and Malassezia restricta (Fungi) are more common on human skin (Andersson et al., 2019; Platsidaki and Dessinioti, 2018; Saunders et al., 2012, Zaidel et al., Hadano et al., 2012); among these, Cutibacterium acnes showed the most prominent reads as it is one of the most common bacterial species on human skin flora and also an opportunistic pathogen that can promote acne vulgaris (Andersson et al., 2019; Platsidaki and Dessinioti, 2018). These contaminants can be introduced during any stage of the entire process including handling, experimentation, sample transfer, library preparation, and sequencing; they can also stem from equipment, reagents, and supplies. On the other hand, Ralstonia pickettii, Paracoccus yeei, and Bacillus oceanisediminis are typically found in nature environments including soil, brines, rivers, lakes, and marine environments (Coenye et al., 2003; Koskinen et al., 2017; Zhang et al., 2010) and thus are more likely to be sourced from the original culture or exist in the extracellular polysaccharide matrix of the Nostoc strain (Huo et al., 2021). Overall, because this platform provides visual confirmation of single cell prior to cell lysis and DNA amplification, the contamination rate is significantly lowered compared with the sequencing of colonies of Nostoc species (contamination rate ranging from 80% to <5%)(Huo et al., 2021; Hirose et al., 2021). In this work, identified contaminants were removed from the sequenced reads prior to the genome sequence assembly of Nostoc strain CCCryo 231-06.
Figure 6.
A bar chart shows contaminants detected within amplified Nostoc sp. CCCryo 231-06 samples
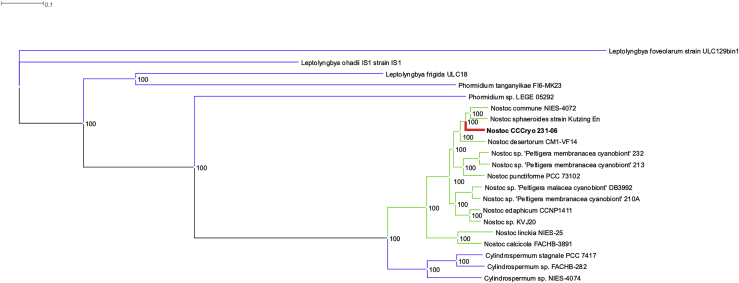
Phylogenetic tree
A phylogenetic analysis was carried out using the amino acid sequences of the protein-encoded genes. The phylogenetic tree was generated using PATRIC Phylogenetic Tree Building Service to show the position of strain CCCryo 231-06 relative to other members of the genus Nostoc (Figure 7). The concatenated tree has a bootstrap value of 100 at each node, indicating high support and reproducibility of these nodes. Nostoc sphaeroides strain Kutzing En is the closest relative of Nostoc strain CCCryo 231-06. Cylindrospermum, Phormidium, and Leptolyngbya were used as outgroup members.
Figure 7.
A protein-based phylogenetic tree highlighting the position of the Nostoc strain CCCryo 231-06 relative to the neighboring species and other members of Nostoc. Cylindrospermum spp., Leptolyngbya spp., and Phormidium spp. are the outgroup genomes
Bootstrap values are shown at each subtree.
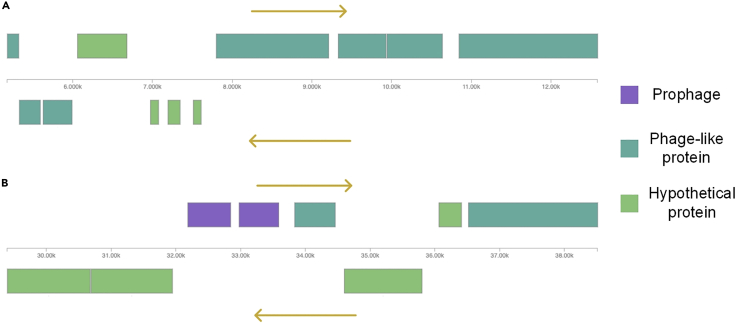
Prophage identification
Two putative prophages were identified in the genome of the Nostoc strain CCCryo 231-06 using the PHASTER tool. The prophage shown in Figure 8A is most similar to a putative prophage belonging to N. punctiforme PCC 73102 (91% BLAST identity), whereas the prophage shown in Figure 8B has similarity to a putative prophage belonging to N. commune HK-02 (93% BLAST identity), which is suggestive of horizontal transmission among Nostoc spp. The BLAST searches were performed against the nt database of NCBI.
Figure 8.
Two putative prophages identified in the genome of Nostoc strain CCCryo 231-06
(A) A prophage similar to a putative prophage belonging to N. punctiforme PCC 73102 (91% BLAST identity).
(B) A prophage similar to a putative prohage belonging to N. commune HK-02 (93% BLAST identity).
Limitations and alternative strategies
This platform is ideal for capturing round-shaped cells of 2–15 μm in diameter, the direct trapping of a filament of microorganism can be challenging. The laser tweezers used in this work are inadequate to separate single Nostoc cells from the gelatinous filaments. Laser-capture microdissection is an advanced tool that can dissect single cells from a filamentous species with high precision for the subsequent single cell isolation; however, it would require the dissection of a critical mass of single cells for the subsequent processing in microfluidic devices. Alternatively, it is possible to use more common methods such as sonication, vortexing, and micropestling to obtain single cells in suspension prior to introducing it into the microfluidic device. The same approach can be applied to other filamentous cyanobacteria cells such as Phormidium and Leptolyngbya.
This approach offers high single-cell confidence and minimal contamination but requires time and labor-intensive efforts to ensure the purity of the isolated cells especially in environmental or biological samples that are often complex in composition, thus limiting the use of this technology to small-scale studies (<50 cells). To perform large-scale studies, droplet microfluidic technology (Teh et al., 2008; Sohrabi et al., 2020) is an ideal tool due to its ability to generate thousands of microscale droplets within seconds to encapsulate single cells in an automated manner. As the cell encapsulation process in droplet microfluidics is random, on one hand, it can present cell selections with reduced bias; on the other hand, the number of target cell captured within the droplets follows a Poisson distribution rather than confirmed single cells (Lagus and Edd, 2013). One way to enhance single-cell capture rate within droplets is to adjust the original cell concentration so that >75% of the droplets contain no cells, and ∼22% contain single cells. Moreover, the droplet-based process can also capture undesired cells (e.g., contaminants) in the sample. Integrating sample pre-purification steps such as filtration and deterministic lateral displacement strategy (Hochstetter et al., 2020; McGrath et al., 2014) can enhance the usability of droplet microfluidic technology to large-scale single cell sequencing of cyanobacteria. In short, each platform offers a unique set of advantages and limitations; the choice of the tools largely relies on the needs of the particular research study.
Conclusions
This study represents the use of microfluidic technologies and single-cell whole genome sequencing for the complete construction of the genome of strain CCCryo 231-06 of Nostoc, a member of the Cyanobacteria. The resulting assembled genome essentially reached completeness with high quality for further analysis. Furthermore, this is the first genome description of the Nostoc strain CCCryo 231-06, which may guide future research aimed at characterizing the role of specific genes and pathways of this strain. Ultimately, this methodology can be applied to various microbial single-cell studies that require the sequencing and assembly of unknown or unculturable species with high quality for scientific discovery. With the rapidly evolving micro/nanotechnologies, we envision that multiple technological paradigms can be integrated into a single device for high-throughput single cell sequencing without compromising purity.
Limitations of study
The single cell isolation platform used in this work is operated manually and is thus better suited for small-scale experiments that serve as a proof of concept of the application of the microfluidic-based single cell technology.
STAR★Methods
Key resource table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| The antarctic strain CCCryo 231-06 (= UTEX EE21; CCMEE 391) of the cyanobacterium Nostoc sp. | Biobank CCCryo at Branch Bioanalytics and Bioprocesses of the Fraunhofer Institute for Cell Therapy and Immunology (IZI-BB), Potsdam, Germany | CCCryo 231-06 |
| Chemicals, peptides, and recombinant proteins | ||
| Pluronic F127 | Sigma Aldrich | Cat# P2443; CAS: 9003-11-6 |
| PBS | Fisher Scientific | Cat# BP243820 |
| PDMS | Momentive | RTV615 |
| UltraPure™ 0.5M EDTA, pH 8.0 | ThermoFisher | Cat# 15575020 |
| Ready-Lyse™ Lysozyme Solution | Lucigen | Cat# R1804M |
| Dithiothreitol (DTT) | Bio-rad | Cat# 1610611 |
| Low-EDTA TE (1X), pH 8.0 | VWR | Cat# 10128-588 |
| NEBNext® Multiplex Oligos for Illumina® | New England Biolabs | Cat# E7335S |
| SPRIselect Beads | Beckman Coulter | Cat# B23318 |
| Deposited data | ||
| Genome sequence | This paper | NCBI, accession# PRJNA721463, the locus tag prefix is KBT16. |
| Experimental models: Organisms/strains | ||
| The antarctic strain CCCryo 231-06 (= UTEX EE21; CCMEE 391) of the cyanobacterium Nostoc sp. | Biobank CCCryo at Branch Bioanalytics and Bioprocesses of the Fraunhofer Institute for Cell Therapy and Immunology (IZI-BB), Potsdam, Germany | CCCryo 231-06 |
| Software and algorithms | ||
| Samtools | Schneider et al. (2012) | http://samtools.sourceforge.net/ |
| BioBloomTools | Chu et al. (2014) | https://mybiosoftware.com/tag/biobloomtools |
| Atropos | Didion et al. (2017) | https://bio.tools/atropos |
| BBTools | https://sourceforge.net/projects/bbmap/, 2019 | https://sourceforge.net/projects/bbmap/ |
| MEGAHIT | Li et al. (2015) | https://bio.tools/megahit |
| BusyBee | Laczny et al. (2017) | https://ccb-microbe.cs.uni-saarland.de/busybee |
| CheckM | Parks et al. (2015) | https://github.com/Ecogenomics/CheckM |
| GTDBtk tool | Parks et al. (2018) | https://github.com/Ecogenomics/GTDBTk |
| Prokka | Seemann et al., 2014 | https://github.com/tseemann/prokka |
| eggNOG mapper | Huerta-Cepas al., 2017 | https://github.com/eggnogdb/eggnog-mapper |
| PATRIC | Davis et al. (2020) | https://www.patricbrc.org/ |
| RAxML | Stamatakis et al., 2014 | https://cme.h-its.org/exelixis/web/software/raxml/ |
| Dendroscope 3 | Huson et al., 2012 | https://uni-tuebingen.de/ |
| PHASTER | Arndt et al. (2016) | https://phaster.ca/ |
| Other | ||
| REPLI-g Single Cell Kit | Qiagen | Cat# 150,343 |
| NEBNext Ultra II DNA Library Prep Kit for Illumina | New England Biolabs | Cat# E7103 |
| HiSeq 3000/4000 PE Cluster Kit | Illumina | Cat# PE-410-1001 |
| HiSeq 3000/4000 SBS Kit (300 cycles) | Illumina | Cat# FC-410-1003 |
Resource availability
All resources are commericially available and the sources can be found in the key resource table.
Lead contact
Further information should be directed to the lead contact, Marina Walther-Antonio (waltherantonio.marina@mayo.edu).
Materials availability
This study did not generate new or unique reagents or materials.
Method details
Cell preparation
The antarctic strain CCCryo 231-06 (= UTEX EE21; CCMEE 391) of the cyanobacterium Nostoc sp. was obtained from the Culture Collection of Cryophilic Algae (CCCryo) at the Branch Bioanalytics and Bioprocesses of the Fraunhofer Institute for Cell Therapy and Immunology (IZI-BB) in Potsdam, Germany. They were collected, cultured, and maintained in cooperation with the German Aerospace Center (DLR) Berlin. The samples were received in desiccated form. 200 μL of sample diluent (0.08% Pluronic F127 (Sigma Aldrich) in Phosphate Buffer Saline (PBS)) was added to the sample followed by mild micro-pestling for 30 s to resuspend the cells. The final concentration reached ∼10 cells/μL to facilitate single-cell trapping.
Microfluidic experimental setup
This work was performed in our optofluidic platform at Mayo Clinic (Rochester, MN)(Liu et al., 2018, 2019b) with a customized microfluidic device for high-throughput SC-WGA. Briefly, this platform consists of a microscope (Nikon Eclipse), optical tweezers (Thorlabs) and a custom-built Polydimethylsiloxane (PDMS) microfluidic device with 4 identical reactions blocks that contains 14 parallel reaction lines in each block (Figure 1A). Each reaction line has sets of valves that allow for the creation of isolated microchambers. The number of microchambers in each reaction line corresponds to the number of reagents that needs to be sequentially added to perform the SC-WGA reactions. The sample inlets of the microfluidic devices was designed in a way as to minimize cross-contamination of samples exposed to different conditions. The major advantages of using optical tweezers to isolate single cells from a population include high target single cell confidence, providing a way to visually ensure that only one single cell is trapped into a microchamber and thus maintaining minimal possibility of sequencing contaminating cells unintendedly. A representative image of using laser tweezers to trap a single Nostoc cell is shown in Figure 1B.
Each reaction block in the microfluidic device was designed to accommodate 13 single cells of strain CCCryo 231-06 exposed to different conditions and one negative control (sterile phosphate buffered saline (PBS)). Therefore, a total of 4 reaction blocks should support the SC-WGA of 4 single cells from each of the 13 samples. However, it was increasingly difficult to identify Nostoc cells in 2 two samples, and only three cells and one single cell from these two samples were sequenced respectively. A complete list of samples, exposure conditions and number of single cells sequenced is summarized in Table 1.
Microfluidic-based SC-WGA
The general workflow of SC-WGA in a microfluidic chip is shown in Figure 2. Prior to introducing the cells into the microfluidic device, the sample channel in the microfluidic device was primed with chip diluent (0.04% Pluronic F127 in phosphate buffered saline (PBS)) for 30 min to prevent the cells from sticking to the PDMS surface. The prepared cell suspension was then introduced into the device, and single cells of the Nostoc strain CCCryo 231-06 were trapped and transported into microchambers by optical tweezers. In the sample, there were micron-sized floating sphere-like and irregular-shaped objects which can be heterotrophic bacterial cells or debris clusters that can house various contaminants. To best ensure the purity of single cells for downstream amplification, we visually verified that the isolated cells were not attached to visible contaminants. Moreover, contaminants that inadvertently entered the cell isolation chambers were trapped and moved out of the chambers using laser tweezers prior to the lysis step.
Lysis and genome amplification procedures of the Nostoc sp. cells followed our optimized protocol for bacterial single cell lysis and whole genome amplification in a microfluidic device (Liu et al., 2018). Briefly, this experiment used Qiagen REPLI-g Single Cell Kit with optimized lysis conditions. 3 cycles of heat-shock was performed by alternately placing the microfluidic chip on a 65°C hotplate and a −20°C cold block for 2 min each. A custom-made lysis buffer containing 0.5 mM of EDTA, 200 U/μL of Ready-Lyse lysozyme (Epicenter), 200 mM Dithiothreitol (DTT) (BioRad) was introduced into the microchambers and the microfluidic device was incubated on a hotplate at 37°C for 2 h. D2 lysis and DNA denaturing buffer suppied in the Single Cell Kit was introduced and incubated on a hotplate at 37°C for 1 h, followed by the addition of the neutralizing buffer to terminate DNA denaturation at room temperature. The polymerase was added into the reaction chambers, and the device was placed on a hotplate at 32°C for 16 h. The amplification was terminated by incubating the microfluidic device at 65°C for 3 min and cooled on ice. The amplified DNA was collected from the outlet ports of the device through gel-loading pipet tips and transferred into 96 microwell plates for downstream processing. All the supplies and reagents were filtered (0.2 μm), autoclaved or UV-sterilized, except for the DNA polymerase.
Library construction and sequencing
DNA was normalized to 10 ng in 50 μL of low TE buffer and sheared in a microTUBE AFA fiber plate using a Covaris LE220 instrument (Covaris, Woburn, MA, USA) to acquire fragments of approximately 200 bp. After shearing, libraries were prepared using NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, USA) and dual-indexed using NEBNext Multiplex Oligos (New England Biolabs) according to the manufacturer’s instructions. SPRIselect Beads (Beckman Coulter, Indianapolis, IN, USA) were used for all library clean-up steps. Libraries were sequenced generating 150 bp paired end reads using an Illumina HiSeq 4000 (Illumina, San Diego, CA, USA).
Preprocessing of sequenced reads
We converted the bam files to fastq files using SAMtools (Schneider et al., 2012). To enhance the quality and reliability of sequencing reads for downstream analysis, we trimmed sequencing adapters and trimmed low-quality bases using Atropos version 1.1.19 (Didion et al., 2017; Martin, 2011). We also removed putative contaminants including environmental bacteria, fungi and human-relevant contaminants using BioBloomTools version 2.1.1 (Chu et al., 2014).
De novo genome assembly
One of the major challenges of bacterial SC-WGS is that the whole genome of a target single cell can only be partially recovered. However, with sufficient number of single cells (regardless of different experimental conditions), it is possible to co-assemble a consensus Nostoc genome to near completion. Therefore, in this work, we co-assembled using all 48 samples targeting the Nostoc isolate. We emphasize the “consensus” aspect of this recovered sequence, thus the sequence will not completely match the genotype present after each exposure condition. As a clarification, to compensate for the observation that the genome recovered from each exposure condition is not fully recovered, we combined the sequencing data from all conditions, and achieved an essentially complete, consensus representation, as measured by the presence of a full complement of essential single-copy genes expected for Bacteria. The assumption is that there is only a few point mutations differentiating the genotype for each experimental condition, and are otherwise near-clonal in nature. This co-assembled consensus genome was to be used as a reference for the variant calling procedures and a template for gene/function annotation to inform the significance of the detected variants. In this case, we determined that consensus reference would be an acceptable substitute for the purpose of identifiying interested variants and offering insights into the functions present in Nostoc sp. CCCryo 231-06. Findings pertaining to Nostoc sp. CCCryo 231-06 exposure to different conditions will be presented in a separate publication.
Specifically, we combined the reads from all samples, and to offset the uneven coverage of sequenced reads introduced by the whole-genome amplification process, we digitally normalized the read coverage using the BBNorm tool from the BBTools suite version 38.26 (https://sourceforge.net/projects/bbmap/, 2019) to a target coverage of 100X. We then took the combined, digitally-normalized reads and used the MEGAHIT de novo metagenomic assembler version 1.1.3 (Li et al., 2015) using the “meta-sensitive” preset. To reconstruct the complete genome of this Nostoc strain we needed to separate its contigs from the contigs from other organisms (members of its community and putative contaminant sequences). We used the BusyBee tool to identify and select the bin of our target organism (Laczny et al., 2017). We then assessed the initial quality of the recovered genome including the completeness and contamination based on a set of normally single copy gene markers using checkM version 1.0.13 (Parks et al., 2015), and then refined the bin using the refine tool following the procedure outlined in Parks et al. (Parks et al., 2017). Finally, we used the GTDBtk tool version 0.2.2 (Parks et al., 2018) to putatively determine the taxonomic placement of the recovered genome.
Genome annotation
The recovered genome of this Nostoc strain CCCryo 231-06 was annotated using Prokka version 1.13 (Seemann, 2014), and annotation with the categories of Clusters of Orthologous Groups was performed on the genome of this Nostoc strain and other representative species of the Nostoc genus using eggNOG mapper version 2 (Huerta-Cepas et al., 2017, 2018).
Phylogenic tree building
The protein-based phylogenetic tree was built using the Phylogenetic Tree Building Service from PATRIC(Davis et al., 2020). Briefly, the PATRIC Phylogenetic Tree Building Service codon tree method selects a predefined number of PATRIC global protein family (PGFams) for random genes to build an alignment (In this case, the predefined number was 1000 different PGFams). Representative genomes from the Nostoc genus were selected, and the reference genomes of Cylindrospermum stagnale as well as the representative species from the Phormidium and Leptolyngbya genera were used as the outgroup taxa. Briefly, the PATRIC Phylogenetic Tree Building Service tool utilizes PATRIC PGFams as homology groups and single-copy genes for protein alignment via RAxML (Stamatakis, 2014). The Codon Tree option was selected with gene number 1000. Tree visualization was performed in Dendroscope V3 (Huson and Scornavacca, 2012). All 1000 genes were shown in Figure S1.
Identification of putative prophages
The assembled consensus genome was submitted to the PHASTER prophage identification tool (Arndt et al., 2016), which returns putative prophage region coordinates and annotates the genes found in them. The proposed prophages were also used to search the NCBI nt database using BLAST, to find genomes where these prophages are also present, and to quantify identity to these genomes.
Acknowledgments
This research was funded by the following sources. Marina Walther-Antonio acknowledges The Ivan Bowen Family Foundation and CTSA Grant Number KL2 TR002379 from the National Center for Advancing Translational Sciences (NCATS). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. Dirk Schulze-Makuch and Jean-Pierre de Vera acknowledge support from ERC Advanced Grant “HOME” (# 339231), and in addition, Jean-Pierre de Vera acknowledges support from ESA for the BIOMEX project (ESA-ILSRA 2009-0834) and DLR for the DLR-FuW-Project BIOMEX (2474128). Charles S. Cockell was supported by the Science and Technology Facilities Council (STFC), Grant No. ST/M001261/1. Mickael Baqué acknowledges the support of the Deutsche Forschungsgemeinschaft (DFG – German Research Foundation) for the project “Raman Biosignatures for Astrobiology Research” (RaBioFAM; project number: 426601242). In addition, we thank the Microbiome Program and the Center for Individualized Medicine, Mayo Clinic for their support, and Dr. Alexander Revzin at Mayo Clinic for granting us the access to his microfabrication facilities.
Author contributions
Experimental methodology: Y.L. and W.M.A.; Bioinformatic methodology: P.J.; Formal analysis: P.J., Y.L., and W.H.; Library optimization, preparation, and sequencing: S.M.; Funding acquisition and project supervision: D.S.M., J.P.d.V., C.C., T.L., M.B., and M.W.A.; NGS optimization and sequencing supervision, B.E. and J.J.; Writing–original draft, Y.L. and P.J.; Writing–review & editing, Y.L., W.H., D.S.M., J.P.d.V., C.C., T.L., M.B., and M.W.A.
Declaration of interests
The authors declare no competing interests.
Published: May 20, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104291.
Supplemental information
Data and code availability
-
•
The complete genome sequence has been deposited at NCBI and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This work does not involve original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Andersson T., Ertürk Bergdahl G., Saleh K., Magnúsdóttir H., Stødkilde K., Andersen C.B.F., Lundqvist K., Jensen A., Brüggemann H., Lood R. Common skin bacteria protect their host from oxidative stress through secreted antioxidant RoxP. Sci. Rep. 2019;9:3596. doi: 10.1038/s41598-019-40471-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt D., Grant J.R., Marcu A., Sajed T., Pon A., Liang Y., Wishart D.S. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binga E.K., Lasken R.S., Neufeld J.D. Something from (almost) nothing: the impact of multiple displacement amplification on microbial ecology. ISME J. 2008;2:233–241. doi: 10.1038/ismej.2008.10. [DOI] [PubMed] [Google Scholar]
- Blainey P.C., Quake S.R. Digital MDA for enumeration of total nucleic acid contamination. Nucleic Acids Res. 2011;39:e19. doi: 10.1093/nar/gkq1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buermans H., Den Dunnen J. Next generation sequencing technology: advances and applications. Biochim. Biophys. Acta (BBA)-Molecular Basis Dis. 2014;1842:1932–1941. doi: 10.1016/j.bbadis.2014.06.015. [DOI] [PubMed] [Google Scholar]
- Chen M., Song P., Zou D., Hu X., Zhao S., Gao S., Ling F. Comparison of multiple displacement amplification (MDA) and multiple annealing and looping-based amplification cycles (MALBAC) in single-cell sequencing. PLoS One. 2014;9 doi: 10.1371/journal.pone.0114520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J., Sadeghi S., Raymond A., Jackman S.D., Nip K.M., Mar R., Mohamadi H., Butterfield Y.S., Robertson A.G., Birol I. BioBloom tools: fast, accurate and memory-efficient host species sequence screening using bloom filters. Bioinformatics. 2014;30:3402–3404. doi: 10.1093/bioinformatics/btu558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenye T., Goris J., De Vos P., Vandamme P., Lipuma J.J. Classification of Ralstonia pickettii-like isolates from the environment and clinical samples as Ralstonia insidiosa sp. nov. Int. J. Syst. Evol. Microbiol. 2003;53:1075–1080. doi: 10.1099/ijs.0.02555-0. [DOI] [PubMed] [Google Scholar]
- Davis J.J., Wattam A.R., Aziz R.K., Brettin T., Butler R., Butler R.M., Chlenski P., Conrad N., Dickerman A., Dietrich E.M., et al. The PATRIC Bioinformatics Resource Center: expanding data and analysis capabilities. Nucleic Acids Res. 2020;48:D606–D612. doi: 10.1093/nar/gkz943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bourcy C.F., De Vlaminck I., Kanbar J.N., Wang J., Gawad C., Quake S.R. A quantitative comparison of single-cell whole genome amplification methods. PLoS One. 2014;9 doi: 10.1371/journal.pone.0105585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vera J.-P., Alawi M., Backhaus T., Baqué M., Billi D., Böttger U., Berger T., Bohmeier M., Cockell C., Demets R., et al. Limits of life and the habitability of Mars: the ESA space experiment BIOMEX on the ISS. Astrobiology. 2019;19:145–157. doi: 10.1089/ast.2018.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean F.B., Nelson J.R., Giesler T.L., Lasken R.S. Rapid amplification of plasmid and phage DNA using phi29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didion J.P., Martin M., Collins F.S. Atropos: specific, sensitive, and speedy trimming of sequencing reads. PeerJ. 2017;5 doi: 10.7717/peerj.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds W.K., Gudder D.A., Mollenhauer D. The ecology of Nostoc. J. Phycol. 1995;31:2–18. doi: 10.1111/j.0022-3646.1995.00002.x. [DOI] [Google Scholar]
- Emms D.M., Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadano Y., Ito K., Suzuki J., Kawamura I., Kurai H., Ohkusu K. Moraxella osloensis: an unusual cause of central venous catheter infection in a cancer patient. Int. J. Gen. Med. 2012;5:875. doi: 10.2147/ijgm.s36919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond M., Homa F., Andersson-Svahn H., Ettema T.J.G., Joensson H.N. Picodroplet partitioned whole genome amplification of low biomass samples preserves genomic diversity for metagenomic analysis. Microbiome. 2016;4:52. doi: 10.1186/s40168-016-0197-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y., Ohtsubo Y., Misawa N., Yonekawa C., Nagao N., Shimura Y., Fujisawa T., Kanesaki Y., Katoh H., Katayama M., et al. Genome sequencing of the NIES Cyanobacteria collection with a focus on the heterocyst-forming clade. DNA Res. 2021;28:dsab024. doi: 10.1093/dnares/dsab024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstetter A., Vernekar R., Austin R.H., Becker H., Beech J.P., Fedosov D.A., Gompper G., Kim S.-C., Smith J.T., Stolovitzky G., et al. Deterministic lateral displacement: challenges and perspectives. ACS Nano. 2020;14:10784–10795. doi: 10.1021/acsnano.0c05186. [DOI] [PubMed] [Google Scholar]
- Hosokawa M., Nishikawa Y., Kogawa M., Takeyama H. Massively parallel whole genome amplification for single-cell sequencing using droplet microfluidics. Sci. Rep. 2017;7:5199. doi: 10.1038/s41598-017-05436-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J., Forslund K., Coelho L.P., Szklarczyk D., Jensen L.J., Von Mering C., Bork P. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 2017;34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J., Szklarczyk D., Heller D., Hernández-Plaza A., Forslund S.K., Cook H., Mende D.R., Letunic I., Rattei T., Jensen L.J., et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2018;47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo D., Li H., Cai F., Guo X., Qiao Z., Wang W., Yu G., Li R. Genome evolution of filamentous cyanobacterium Nostoc species: from facultative symbiosis to free living. Microorganisms. 2021;9:2015. doi: 10.3390/microorganisms9102015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D.H., Scornavacca C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- Kim S., De Jonghe J., Kulesa A.B., Feldman D., Vatanen T., Bhattacharyya R.P., Berdy B., Gomez J., Nolan J., Epstein S., Blainey P.C. High-throughput automated microfluidic sample preparation for accurate microbial genomics. Nat. Commun. 2017;8:13919. doi: 10.1038/ncomms13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Arai M., Katoh H., Ajioka R., Baba K.I., Sato S., Tomita-Yokotani K. 44th International Conference on Environmental Systems. 2014. Utilization of a cyanobacterium, Nostoc sp. HK-01, under the space environment. [Google Scholar]
- Kimura S., Inoue K., Katoh H., Ichikawa S., Tomita-Yokotani K. 47th International Conference on Environmental Systems. 2017. Tolerance and growth of a terrestrial cyanobacterium, Nostoc sp. HK-01 under harsh environments. [Google Scholar]
- Kimura S., Tomita-Yokotani K., Katoh H., Sato S., Ohmori M. Complete life cycle and heat tolerance of dry colonies of a terrestrial cyanobacterium, Nostoc sp. HK-01. Biol. Sci. Space. 2017;31:1–8. doi: 10.2187/bss.31.1. [DOI] [Google Scholar]
- Koskinen K., Rettberg P., Pukall R., Auerbach A., Wink L., Barczyk S., Perras A., Mahnert A., Margheritis D., Kminek G., Moissl-Eichinger C. Microbial biodiversity assessment of the European Space Agency’s ExoMars 2016 mission. Microbiome. 2017;5:143. doi: 10.1186/s40168-017-0358-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laczny C.C., Kiefer C., Galata V., Fehlmann T., Backes C., Keller A. BusyBee Web: metagenomic data analysis by bootstrapped supervised binning and annotation. Nucleic Acids Res. 2017;45:W171–W179. doi: 10.1093/nar/gkx348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagus T.P., Edd J.F. A review of the theory, methods and recent applications of high-throughput single-cell droplet microfluidics. J. Phys. D: Appl. Phys. 2013;46:114005. doi: 10.1088/0022-3727/46/11/114005. [DOI] [Google Scholar]
- Landry Z.C., Giovanonni S.J., Quake S.R., Blainey P.C. Optofluidic cell selection from complex microbial communities for single-genome analysis. Methods Enzymol. 2013:61–90. doi: 10.1016/b978-0-12-407863-5.00004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Liu C.-M., Luo R., Sadakane K., Lam T.-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- Liu Y., Jeraldo P., Jang J.S., Eckloff B., Jen J., Walther-Antonio M. Bacterial single cell whole transcriptome amplification in microfluidic platform shows putative gene expression heterogeneity. Anal. Chem. 2019;91:8036–8044. doi: 10.1021/acs.analchem.8b04773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schulze-Makuch D., De Vera J.-P., Cockell C., Leya T., Baqué M., Walther-Antonio M. The development of an effective bacterial single-cell lysis method suitable for whole genome amplification in microfluidic platforms. Micromachines. 2018;9:367. doi: 10.3390/mi9080367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Walther-Antonio M. Microfluidics: a new tool for microbial single cell analyses in human microbiome studies. Biomicrofluidics. 2017;11 doi: 10.1063/1.5002681. [DOI] [Google Scholar]
- Liu Y., Yao J., Walther-Antonio M. Whole genome amplification of single epithelial cells dissociated from snap-frozen tissue samples in microfluidic platform. Biomicrofluidics. 2019;13 doi: 10.1063/1.5090235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low A.J., Koziol A.G., Manninger P.A., Blais B., Carrillo C.D. ConFindr: rapid detection of intraspecies and cross-species contamination in bacterial whole-genome sequence data. PeerJ. 2019;7 doi: 10.7717/peerj.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Salzberg S.L. Removing contaminants from databases of draft genomes. PLoS Comput. Biol. 2018;14 doi: 10.1371/journal.pcbi.1006277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcy Y., Ishoey T., Lasken R.S., Stockwell T.B., Walenz B.P., Halpern A.L., Beeson K.Y., Goldberg S.M.D., Quake S.R. Nanoliter reactors improve multiple displacement amplification of genomes from single cells. PLoS Genet. 2007;3:e155. doi: 10.1371/journal.pgen.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- McGrath J., Jimenez M., Bridle H. Deterministic lateral displacement for particle separation: a review. Lab. A Chip. 2014;14:4139–4158. doi: 10.1039/c4lc00939h. [DOI] [PubMed] [Google Scholar]
- Motley S.T., Picuri J.M., Crowder C.D., Minich J.J., Hofstadler S.A., Eshoo M.W. Improved multiple displacement amplification (iMDA) and ultraclean reagents. BMC Genomics. 2014;15:443. doi: 10.1186/1471-2164-15-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson N.D., Zook J.M., Morrow J.B., Lin N.J. Challenging a bioinformatic tool’s ability to detect microbial contaminants using in silico whole genome sequencing data. PeerJ. 2017;5 doi: 10.7717/peerj.3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks D.H., Chuvochina M., Waite D.W., Rinke C., Skarshewski A., Chaumeil P.-A., Hugenholtz P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- Parks D.H., Imelfort M., Skennerton C.T., Hugenholtz P., Tyson G.W. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks D.H., Rinke C., Chuvochina M., Chaumeil P.-A., Woodcroft B.J., Evans P.N., Hugenholtz P., Tyson G.W. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2017;2:1533–1542. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- Platsidaki E., Dessinioti C. Recent advances in understanding Propionibacterium acnes (Cutibacterium acnes) in acne. F1000Res. 2018:7. doi: 10.12688/f1000research.15659.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts M. Springer; 2000. Nostoc. The Ecology of Cyanobacteria. [Google Scholar]
- Quainoo S., Coolen J.P.M., Van Hijum S.A., Huynen M.A., Melchers W.J.G., Van Schaik W., Wertheim H.F.L. Whole-genome sequencing of bacterial pathogens: the future of nosocomial outbreak analysis. Clin. Microbiol. Rev. 2017;30:1015–1063. doi: 10.1128/cmr.00016-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada A., Vincent W.F. Ecology of Cyanobacteria II. Springer; 2012. Cyanobacteria in the cryosphere: snow, ice and extreme cold.https://link.springer.com/chapter/10.1007/978-94-007-3855-3_14 [Google Scholar]
- Saunders C.W., Scheynius A., Heitman J. Malassezia fungi are specialized to live on skin and associated with dandruff, eczema, and other skin diseases. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- Sohrabi S., Kassir N., Keshavarz Moraveji M. Droplet microfluidics: fundamentals and its advanced applications. RSC Adv. 2020;10:27560–27574. doi: 10.1039/d0ra04566g. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh S.-Y., Lin R., Hung L.-H., Lee A.P. Droplet microfluidics. Lab. A Chip. 2008;8:198–220. doi: 10.1039/b715524g. [DOI] [PubMed] [Google Scholar]
- Tomita-Yokotani K., Kimura S., Kimura Y., Igarashi Y., Ajioka R., Sato S., Katoh H., Baba K. Dried colony in cyanobacterium, Nostoc sp. HK-01---Several high space environment tolerances for``Tanpopo''Mission. Int. Astrobiol. Workshop. 2013;2013:1033. [Google Scholar]
- Vincent W.F., Mueller D.R., Bonilla S. Ecosystems on ice: the microbial ecology of markham ice shelf in the high arctic. Cryobiology. 2004;48:103–112. doi: 10.1016/j.cryobiol.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Walker S.P., Barrett M., Hogan G., Flores Bueso Y., Claesson M.J., Tangney M. Non-specific amplification of human DNA is a major challenge for 16S rRNA gene sequence analysis. Sci. Rep. 2020;10:16356–16357. doi: 10.1038/s41598-020-73403-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Hao Z., Huang Z., Chen L., Li X., Hu C., Liu Y. Raman spectroscopic analysis of a desert cyanobacterium Nostoc sp. in response to UVB radiation. Astrobiology. 2010;10:783–788. doi: 10.1089/ast.2009.0407. [DOI] [PubMed] [Google Scholar]
- Zaidel, E., Di Toro, D., Kazelián, L., Neme, R. O., Lespada, M. I., Arcondo, F. & Zylberman, M. Staphylococcus Capitis Endocarditis: Living with S. Capitis. 10.19102/icrm.2012.031007 [DOI]
- Zare R.N., Kim S. Microfluidic platforms for single-cell analysis. Annu. Rev. Biomed. Eng. 2010;12:187–201. doi: 10.1146/annurev-bioeng-070909-105238. [DOI] [PubMed] [Google Scholar]
- Zhang J., Wang J., Fang C., Song F., Xin Y., Qu L., Ding K. Bacillus oceanisediminis sp. nov., isolated from marine sediment. Int. J. Syst. Evol. Microbiol. 2010;60:2924–2929. doi: 10.1099/ijs.0.019851-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The complete genome sequence has been deposited at NCBI and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This work does not involve original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.