Abstract
Hypertrophic cardiomyopathy (HCM) is a common inherited heart disease with a prevalence of about 0.2%. HCM is typically caused by mutations in genes encoding sarcomere or sarcomere-associated proteins. Here, we characterized induced pluripotent stem cell (iPSC) lines generated from the peripheral blood mononuclear cells of three HCM patients each carrying c.433C > T, c.610C > T, or c.235C > T mutation in the TNNI3 gene by non-integrated Sendai virus. All of the three lines exhibited normal morphology, expression of pluripotent markers, stable karyotype, and the potential of trilineage differentiation. The cardiomyocytes differentiated from these iPSC lines can serve as useful tools to model HCM in vitro.
1. Resource table
| Unique stem cell lines identifier | 1) SCVIi017-A 2) SCVIi018-A 3) SCVIi019-A |
|---|---|
|
| |
| Alternative name(s) of stem cell lines | |
| Institution | Stanford Cardiovascular Institute, Stanford, CA, US |
| Contact information of distributor | Joseph C. Wu, joewu@stanford.edu |
| Type of cell lines | iPSC |
| Origin | Human |
| Additional origin info required for human ESC or iPSC | Age: 43 (SCVIi017-A), 23 (SCVIi018-A), 23 (SCVIi019-A) Sex: Female (SCVIi017-A), Male (SCVIi018-A), Male (SCVIi019-A) Ethnicity if known: Not Hispanic or Latino (all three lines) |
| Cell Source | Blood |
| Clonality | Clonal |
| Associated disease | Hypertrophic cardiomyopathy (HCM) |
| Gene/locus |
TNNI3 c.433C > T (SCVIi017-A) TNNI3 c.610C > T (SCVIi018-A) TNNI3 c.235C > T (SCVIi019-A) |
| Date archived/stock date | Aug 3rd, 2021 |
| Cell line repository/bank |
https://hpscreg.eu/cell-line/SCVIi017-A
https://hpscreg.eu/cell-line/SCVIi018-A https://hpscreg.eu/cell-line/SCVIi019-A |
| Ethical approval | The generation of the lines was approved by the Administrative Panel on Human Subjects Research (IRB) under IRB #29904 “Derivation of Human Induced Pluripotent Stem Cells (Biorepository)”. |
2. Resource utility
Three induced pluripotent stem cell (iPSC) lines were generated from three hypertrophic cardiomyopathy (HCM) patients each carrying different heterozygous mutation in the TNNI3 gene. These fully characterized iPSC lines can be differentiated into cardiomyocytes to understand the complex pathogenic mechanisms of HCM.
3. Resource details
HCM is a genetic disorder characterized by left ventricular hypertrophy. HCM is predominantly caused by mutations in genes encoding sarcomere or sarcomere-associated proteins (Lan et al., 2013; Marian & Braunwald, 2017; Wu et al., 2019). Thin filaments of the sarcomeres are composed of tropomyosin, troponin and actin (van der Velden & Stienen, 2019). The TNNI3 gene encodes cardiac troponin I (cTnI), a subunit of the troponin complex. Notably, mutations in TNNI3 have been reported in 2%–7% of HCM cases (Mogensen et al., 2004).
In this report, we generated three iPSC lines SCVIi017-A, SCVIi018-A, and SCVIi019-A from three HCM patients each carrying distinct mutation in TNNI3. Peripheral blood mononuclear cells (PBMCs) collected from these patients were reprogrammed into iPSCs using Sendai virus carrying reprogramming factors OCT4, SOX2, KLF4, and c-MYC. All of the three iPSC lines showed typical iPSC morphology (Fig. 1A). High expression levels of pluripotency markers were confirmed by immunofluorescence staining and reverse transcription quantitative polymerase chain reaction (RT-qPCR) (Fig. 1B and 1C). Genetic testing confirmed c.433C > T, c.610C > T and c.235C > T mutations in TNNI3 of SCVIi017-A, SCVIi018-A, and SCVIi019-A, respectively (Fig. 1D). Neither reprogramming nor long-term maintenance compromised the karyotype integrity of these iPSC lines (Fig. 1E). All of the three iPSC lines demonstrated full potential to generate three lineages by expressing endoderm (Sox17 and Foxa2), mesoderm (Brachyury and Tbx6), and ectoderm (Otx2 and Pax6) markers (Fig. 1F). While trace amount of Sendai virus was detectable at early passages of iPSCs, it was absent at passages 24–27 (Fig. 1G). All iPSC clones were tested negative for mycoplasma (Supplementary Fig. 1). A set of 16 polymorphic short tandem repeats (STR) analysis confirmed the identicalness of the three iPSC lines to the patients’ PBMCs (data archived) (Table 1).
Fig. 1.
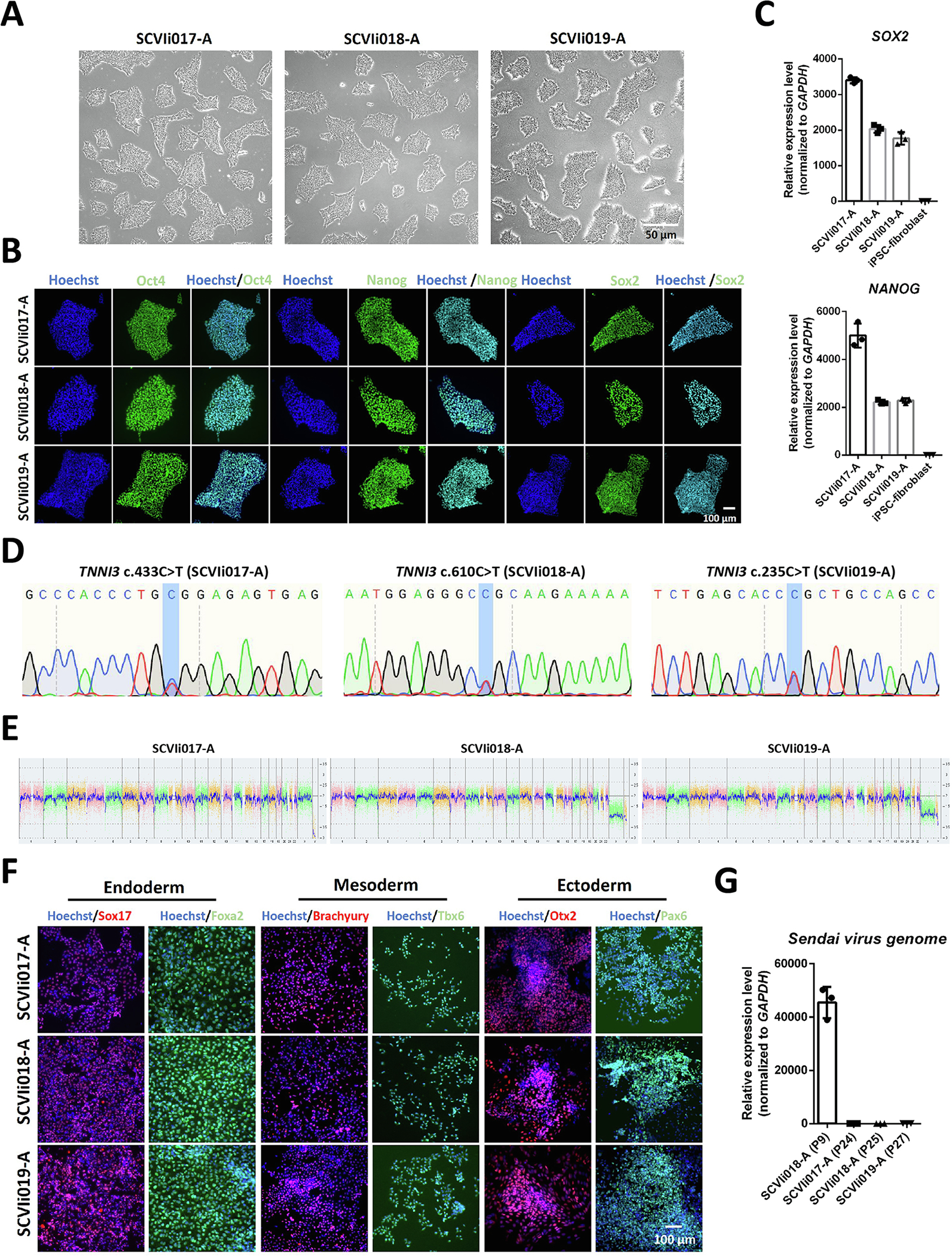
Characterization of iPSC lines derived from hypertrophic cardiomyopathy patients carrying TNNI3 mutations. (A) Brightfield images of the iPSC lines. Scale bar, 50 μm. (B) Immunofluorescent staining images for pluripotency markers OCT4, SOX2, and NANOG. Scale bar, 100 μm. (C) Quantification of NANOG and SOX2 expression by RT-qPCR. IPSC-derived fibroblasts were used as a negative control. (D) Results of Sanger sequencing showing TNNI3 mutations. (E) Results of KaryoStat assay. (F) Immunofluorescent staining images for markers of three germ layers. Scale bar, 100 μm. (G) Quantification of Sendai virus (SEV) expression by RT-qPCR.
Table 1.
Characterization and validation.
| Classification | Test | Result | Data |
|---|---|---|---|
|
| |||
| Morphology | Photography brightfield | Visual record of the line: normal | Fig. 1 panel A |
| Phenotype | Qualitative analysis Immunocytochemistry |
Positive expression of pluripotency markers: Oct3/4, Nanog, Sox2 | Fig. 1 panel B |
| Quantitative analysisRT-qPCR | NANOG and SOX2 are highly expressed | Fig. 1 panel C | |
| Genotype | Whole genome array(KaryoStat™ Assay)Resolution 1–2 Mb | Normal karyotype: 46, XY and 46, XX | Fig. 1 panel E |
| Identity | Microsatellite PCR (mPCR) or STR analysis | N/A 16 loci tested, all matched |
N/A Submitted in archive with journal |
| Mutation analysis (IF APPLICABLE) | Sequencing | Heterozygous Heterozygous Heterozygous |
Fig. 1 panel D |
| Southern blot or WGS | N/A | N/A | |
| Microbiology and virology | Mycoplasma | Mycoplasma testing by luminescence. Negative |
Supplementary Fig. 1 |
| Differentiation potential | Directed differentiation | Positive expression of three germ layer markers by immunocytochemistry | Fig. 1 panel F |
| List of recommended germ layer markers | Expression of these markers has to be demonstrated at mRNA (RT-PCR) or protein (IF) levels, at least 2 markers need to be shown per germ layer | Ectoderm: Pax6, Otx2 Endoderm: Sox17, Foxa2 Mesoderm: Brachyury, Tbx6 |
Fig. 1 panel F |
| Donor screening (OPTIONAL) | HIV 1 + 2 Hepatitis B, Hepatitis C | N/A | N/A |
| Genotype additional info (OPTIONAL) | Blood group genotyping HLA tissue typing |
N/A N/A |
N/A N/A |
4. Materials and methods
4.1. Reprogramming
PBMCs were isolated and collected by gradient centrifugation from the peripheral blood of patients. PBMCs were isolated by Percoll separation (GE Healthcare) and purified by washing with DPBS buffer (Thermo Fisher Scientific). After replating, PBMCs were cultured in PBMC medium containing complete StemPro-34 medium (Thermo Fisher Scientific) supplemented with 100 ng/mL SCF (Peprotech), 100 ng/mL FLT3 (Thermo Fisher Scientific), 20 ng/mL IL-3 (Peprotech), 20 ng/mL IL-6 (Thermo Fisher Scientific), and 20 ng/mL EPO (Thermo Fisher Scientific). PBMCs were reprogrammed to iPSCs by the CytoTune®-iPSC Sendai Reprogramming Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Briefly, transduced PBMCs were resuspended and plated. The StemPro™−34 medium was refreshed every two days. At day 7, the medium was changed to fresh StemMACS™ iPS-Brew XF medium (Miltenyi Biotechnology). Medium was refreshed every other day until day 10–15 post-infection when colonies were ready to be picked. Picked colonies were further expanded and frozen down for downstream applications.
4.2. Cell culture
iPSCs were cultured in StemMACS™ iPS-Brew XF medium in 6-well plates coated with Matrigel (Corning) at a dilution of 1:400 in a humidified incubator at 37 °C with 5% CO2. Medium was changed every other day. iPSCs were passaged at a ratio of 1:6 to 1:12. Y-27632 (10 μM), a potent inhibitor of ROCK1 (Selleck Chemicals), was added in the medium during the first 24 h of cell replating to improve cell survival and attachment.
4.3. Immunofluorescence staining
iPSCs at passages 15–20 and iPSC derivatives were fixed with 4% paraformaldehyde for 15 min at room temperature (RT), permeabilized with 0.3% Triton X-100 (Sigma) for 10 min at RT, and blocked with 3% bovine serum albumin (BSA, Sigma) for 30 min at RT. Then cells were incubated with primary antibodies overnight at 4 °C and fluorescence-conjugated secondary antibodies for 60 min at RT. Cell nuclei were counter stained with Hoechst 33342 (Thermo Fisher Scientific) for 5 min at RT. Images were captured using an inverted fluorescence microscope. The antibody information and dilution ratios are listed in Table 2.
Table 2.
Reagents details.
| Antibodies used for immunocytochemistry/flow-cytometry | ||||
|
| ||||
| Antibody | Dilution | Company Cat # | RRID | |
|
| ||||
| Pluripotency Markers | Rabbit Anti-Nanog | 1:200 | ProteintechCat# 142951-1-AP | RRID: AB_1607719 |
| Pluripotency Markers | Mouse IgG2bκ Anti-Oct-3/4 | 1:200 | Santa CruzBiotechnologyCat# sc-5279 | RRID: AB_628051 |
| Pluripotency Markers | Mouse IgG1κ Anti-Sox2 | 1:200 | Santa CruzBiotechnologyCat# sc-365823 | RRID: AB_10842165 |
| Ectoderm marker | Goat Anti-Otx2 | 1:200 | R&D SystemsCat# 963273 | RRID: AB_2157172 |
| Ectoderm marker | Rabbit Anti-Pax6 | 1:100 | Thermo FisherScientificCat# 42-6600 | RRID: AB_2533534 |
| Endoderm marker | Goat Anti-Sox17 | 1:200 | R&D SystemsCat# 963121 | RRID: AB_355060 |
| Endoderm marker | Rabbit Anti-Foxa2 | 1:250 | Thermo FisherScientificCat# 701698 | RRID: AB_2576439 |
| Mesoderm marker | Goat Anti-Brachyury | 1:200 | R&D SystemsCat# 963427 | RRID: AB_2200235 |
| Mesoderm marker | Rabbit Anti-Tbx6 | 1:200 | Thermo FisherScientificCat# PA5-35102 | RRID: AB_2552412 |
| Secondary antibody | Alexa Fluor 488 Goat Anti-Mouse (H + L) | 1:500 | Thermo FisherScientificCat# A-32723 | RRID: AB_2633275 |
| Secondary antibody | Alexa Fluor 488 Goat Anti-Rabbit (H + L) | 1:500 | Thermo FisherScientificCat# A-32731 | RRID: AB_2633280 |
| Secondary antibody | Alexa Fluor 594 Donkey Anti-Goat (H + L) | 1:500 | Thermo FisherScientificCat# A-11058 | RRID: AB_2534105 |
| Primers | ||||
|
| ||||
| Target | Size of band | Forward/Reverse primer (5′-3′) | ||
| Sendai virus plasmids (qPCR) | Sendai virus genome | 181 bp | Mr04269880_mr | |
| Pluripotency marker (qPCR) | SOX2 | 258 bp | Hs04234836_s1 | |
| Pluripotency marker (qPCR) | NANOG | 327 bp | Hs02387400_g1 | |
| House-keeping gene (qPCR) | GAPDH | 91 bp | Hs02758991_g1 | |
| Genotyping | TNNI3 c.433C > THeterozygous | 525 bp | Forward: CCATGGGTTGGGAAACAGAAAATReverse: GCCTTAGCCCACACTCACCTTCT | |
| Genotyping | TNNI3 c.610C > THeterozygous | 593 bp | Forward: GGAGGGAAGACAGGGATTCTTGAReverse: GTGTGTCCATGTGTCCACCTGTC | |
| Genotyping | TNNI3 c.235C > THeterozygous | 582 bp | Forward: ATCCTTCCTTGCTCCATCTCACCReverse: TGGGTAAGGACAGCCATATTGGA | |
RRID Requirement for antibodies: use http://antibodyregistry.org/ to retrieve RRID for antibodies and include ID in table as shown in examples.
4.4. Trilineage differentiation potential assay
iPSCs at passages 15–20 were differentiated using the STEMdiff™ trilineage differentiation kit (Stemcell Technologies) according to the manufacturer’s instructions. Differentiations were assessed by the expressions of classical lineage markers in each germ layer.
4.5. RT-qPCR
Total RNA was extracted by miRNeasy Micro Kit (Qiagen). RT-qPCR was performed by iScript™ Reverse Transcription Supermix (Bio-rad) according to the manufacturer’s instructions. iPSCs at passages 15–20 were used for the detection of pluripotency markers. iPSCs at passages 24–27, as well as early passage (P9), were used for the detection of Sendai virus genome.
4.6. Karyotyping
A total of 2 × 106 iPSCs were collected from each line between passages 11–15 and analyzed using the KaryoStat™ assay (Thermo Fisher Scientific).
4.7. Short tandem repeat (STR) analysis
Genomic DNAs of PBMCs and iPSCs at passages 15–20 were isolated by QuickExtract™ DNA Extraction Solution (Lucigen). STR analysis was performed using a CLA IdentiFiler™ Direct PCR Amplification Kit (Thermo Fisher Scientific). Capillary electrophoresis was performed on ABI3130xl by the Stanford Protein Nucleic Acid (PAN) Facility.
4.8. Mycoplasma detection
Mycoplasma detection was performed by a MycoAlert™ Detection Kit (Lonza) according to the manufacturer’s instructions.
4.9. DNA sequencing
Genomic DNA was isolated from iPSCs at passages 15–20 using the QuickExtract™ DNA Extraction Solution (Lucigen) and amplified by PCR. Information of the designed primers was listed in Table 2. Purified PCR products were subjected to Sanger sequencing. The presence of TNNI3 mutations was identified by aligning the Sanger sequencing data with wildtype TNNI3 sequence using SnapGene software.
Supplementary Material
Acknowledgment
This work was supported by National Institutes of Health 75N92020D00019, R01 HL126527, R01 HL130020, and P01 HL141084 (JCW).
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scr.2021.102597.
References
- Lan F, Lee AS, Liang P, Sanchez-Freire V, Nguyen PK, Wang L, Han L, Yen M, Wang Y, Sun N, Abilez OJ, Hu S, Ebert AD, Navarrete EG, Simmons CS, Wheeler M, Pruitt B, Lewis R, Yamaguchi Y, Ashley EA, Bers DM, Robbins RC, Longaker MT, Wu JC, 2013. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell 12 (1), 101–113. 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marian AJ, Braunwald E, 2017. Hypertrophic cardiomyopathy: genetics, pathogenesis, clinical manifestations, diagnosis, and therapy. Circ Res 121 (7), 749–770. 10.1161/CIRCRESAHA.117.311059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen J, Murphy RT, Kubo T, Bahl A, Moon JC, Klausen IC, Elliott PM, McKenna WJ, 2004. Frequency and clinical expression of cardiac troponin I mutations in 748 consecutive families with hypertrophic cardiomyopathy. J. Am. Coll. Cardiol 44 (12), 2315–2325. 10.1016/j.jacc.2004.05.088. [DOI] [PubMed] [Google Scholar]
- van der Velden J, Stienen GJM, 2019. Cardiac disorders and pathophysiology of sarcomeric proteins. Physiol. Rev 99 (1), 381–426. 10.1152/physrev.00040.2017. [DOI] [PubMed] [Google Scholar]
- Wu H, Yang H, Rhee JW, Zhang JZ, Lam CK, Sallam K, Chang ACY, Ma N, Lee J, Zhang H, Blau HM, Bers DM, Wu JC, 2019. Modelling diastolic dysfunction in induced pluripotent stem cell-derived cardiomyocytes from hypertrophic cardiomyopathy patients. Eur. Heart J 40 (45), 3685–3695. 10.1093/eurheartj/ehz326. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


