Abstract
Pancreatic ductal adenocarcinoma (PDAC) is the most common neoplastic disease of the pancreas, accounting for more than 90% of all pancreatic malignancies. As a highly lethal malignancy, PDAC is the fourth leading cause of cancer-related deaths worldwide with a 5-year overall survival of less than 8%. The efficacy and outcome of PDAC treatment largely depend on the stage of disease at the time of diagnosis. Surgical resection followed by adjuvant chemotherapy remains the only possibly curative therapy, yet 80–90% of PDAC patients present with non-resectable PDAC stages at the time of clinical presentation. Despite our advancing knowledge of PDAC, the prognosis remains strikingly poor, which is primarily due to the difficulty of diagnosing PDAC at the early stages. Recent advances in glycoproteomics and glycomics based on mass spectrometry have shown that aberrations in protein glycosylation plays a critical role in carcinogenesis, tumor progression, metastasis, chemo-resistance and immuno-response of PDAC and other types of cancers. A growing interest has thus been placed upon protein glycosylation as a potential early detection biomarker for PDAC. We herein take stock of the advancements in the early detection of PDAC that were carried out with mass spectrometry, with special focus on protein glycosylation.
Introduction
PDAC Cancer Statistics
Pancreatic ductal adenocarcinoma (PDAC) is a devastating cancer with poor prognosis and rising incidents. PDAC accounts for more than 90% of all pancreatic malignancies[1], and only 10–20% of PDAC patients are surgically resectable [2]. By 2030, PDAC is predicted to emerge as the second leading cause of cancer-related death in the United States, surpassing breast cancer [3–5]. The major challenges with treating PDAC lie in the difficulty of early detection and the particularly aggressive cancer biology[6]. The PDAC prognosis would be improved significantly when malignant lesions are identified at early stages and resected surgically[7]. Among the top 5 lethal cancers, PDAC is the only type without an early detection strategy[8].
PDAC Pathological Features and Progression
PDAC is an infiltrating epithelial neoplasm with glandular differentiation and sialo-type and sulfated acid mucin production (well and moderately differentiated tumors), which is derived from the pancreatic ductal tree[9]. It is characterized histologically by its highly desmoplastic stroma embedding tubular and ductlike structures, causing ductal obstruction and vascular involvement[10]. PDAC is generally a multinodular and sclerotic solid tumor with poorly-defined margins and a whitish cut surface [9]. Apart from conventional PDAC, other PDAC variants including adenosquamous carcinoma (ASqC), colloid carcinoma, hepatoid carcinoma (HC), medullary carcinoma of the pancreas (MCP), signet ring cell carcinoma (SRCC), undifferentiated carcinoma with osteoclast-like giant cells (UCOGC), and undifferentiated carcinoma (UC) are characterized to have different histologic features and prognostic landscape [10].
It is believed that the pathogenesis of PDAC follows a step-wise progression similar to that of colorectal carccinoma [11, 12], which is characterized by the transition of a normal pancreatic duct to pre-invasive precursor lesions including the most common pancreatic intraepithelial neoplasia (PanIN), ultimately these advanced precursor lesions would develop into an invasive PDAC[10, 11]. The gradual accumulation of genetic mutations such as KRAS predominantly drives the progression of PDAC, with KRAS mutations found in almost all PDAC[13]. Other genetic mutations, including the loss of function in tumor suppressor genes such as CDKN2A, TP53, or SMAD4, the activation of oncogenic Her-2/neu[11, 14], and germline mutations in the genes BRCA1/2, ATM, MLH1, TP53, or CDKN2A[15–17], are also found in PDAC. The aggregation of multiple genomic aberrations in advanced PanIN lesions results in an invasive phenotype and subsequent metastatic disease. The aggregation of multiple genomic aberrations in advanced PanIN lesions results in an invasive phenotype and subsequent metastatic disease.
PDAC Diagnosis
PDAC presents clinical symptoms that are nonspecific and overlap with other conditions such as chronic pancreatitis, therefore a diagnosis for PDAC could only be made with a reasonable level of certainty after further investigations [9]. In addition, the early stages of PDAC are often clinically silent, creating extra challenges for PDAC diagnosis. Currently, the diagnosis and staging of PDAC relies heavily on imaging and cytology diagnostic methods, with 10% of PDAC remains isoattenuating on CT and approximately 80% of the patients diagnosed at advanced inoperable stages [2, 10]. Nevertheless, CT-based diagnoses of PDAC have an overall sensitivity of 89% and specificity of 90%. Since invasive approaches are often associated with delay in diagnosis, non-invasive biomarker could potentially provide a valuable complement. Non-invasive serum biomarkers represent one of the most attractive methods due to the low risk of patients and ease of access [18]. Currently, the serum marker carbohydrate antigen 19–9 (CA 19–9) is the only clinical biomarker established for pancreatic cancer management [19]. CA 19–9 is used extensively for disease monitoring, especially for recurrence assessment after surgery [19]. However, CA 19–9 does not provide adequate accuracy and sensitivity for early detection and diagnosis purposes, since an elevated level of CA 19–9 could indicate either PDAC or benign biliary/pancreatic disease [20]. The advance of proteomic technology based on mass spectrometry has propelled the establishment of various pancreatic cancer biomarker identification methods [21–23] and the discovery of several serum biomarkers to address the specificity and sensitivity issues with CA 19–9[24]. Despite the efforts, none of these biomarkers could surpass the performance of CA 19–9 to be translated into clinical use for the early detection of PDAC[24, 25]. Capitalizing the biological relevance with tumor progression [26] and prevalence of protein glycosylation might be a promising path for PDAC early detection.
Protein Glycosylation and Its Cancer Implication
Protein glycosylation is the one of the most abundant protein modifications, involving proteins comprising ~50% of the human proteome[27]. Protein glycosylation is highly diversified to conform with various biological and physiological functions[28–33]. Protein glycosylation is a multi-step enzymatic process that occurs in the endoplasmic reticulum and Golgi apparatus[34]. N-linked and O-linked glycosylation are the most common protein modifications involving glycan conjugation. N-linked glycans are linked to the amide group of asparagine residues in a consensus Asn-X-Ser/Thr sequence (X could be any amino acid other than proline), with, N-glycans in eukaryotic cells sharing a common core sequence, Manα1–3(Manα1–6)Manβ1–4GlcNAcβ1–4GlcNAcβ1–Asn-X-Ser/Thr [34]. In contrast, O-linked glycans are linked to the hydroxyl group of serine, threonine or tyrosine residues without an obvious motif preference and involving various initiating monosaccharides [34]. Apart from N- and O-linked glycosylation, major components of the extracellular matrix (ECM), such as proteoglycan proteins, including galectins and hyaluronan, also contain abundant sugar moieties, with differential abundance of these ECM-related proteoglycans impacting cell proliferation and migration.
Protein glycosylation can be altered in structure and density (hyper, hypo, or neo-glycosylation) in association with changes in cellular pathways in cancer [35, 36], which was first described 50 years ago [37]. Since then, altered glycosylation patterns have long been considered as hallmarks in various epithelial cancers [38–42], including PDAC. Aberrant protein glycosylation can influence cancer cells proliferation, metastasis, invasion, and interactions within the tumor microenvironment [35]. Glycosylation alterations that are often associated with cancer include fucosylation, sialyation, increased GlcNAc branching of N-glycans and hyper-expression of truncated O-glycans such as Tn and sialyl-Tn (sTn) [43]. These signature alterations in cancer-associated glycosylation may provide novel diagnostic markers and even therapeutic targets.
In general, the carcinogenesis relies on a series of sequential obligatory steps beginning with the detachment from neighboring cells as well as the concomitant degradation and remodeling of the basement membrane (BM), which is the barrier to tumor cells dissemination [44, 45]. Tumor cells acquire the ability to penetrate the surrounding tissues through the disruption of cell-cell junction, which seals the luminal of blood vessels [46, 47]. Consequently, tumor cells start to separate from the primary tumor and enter the systemic circulatory, and migrate to distant organ to form metastatic. A critical step in metastatic process is the epithelial-mesenchymal transition (EMT), where the epithelial cells exhibit altered phenotypes leading to novel functions, characterized by enhanced migratory and invasive potentials. EMT was first observed in normal embryonic development of organogenesis, during which the cells lose the polarity and transform into fibroblast-like cells which express mesenchymal makers [48]. Glycosylation changes were previously reported to occur during TGF-β-induced EMT[49–51]. Using the TGF-β treatment to induce the expression of oncofetal fibronectin (onfN), Freire-de-Lima and colleagues showed a direct correlation of the O-linked glycosylation in regulating EMT process in human prostate epithelial cells. [52]. Similarly, by inducing the expression of mesenchymal EMT genes and by stimulating MMP-2 activity in breast carcinoma, the role of GALNT14 in regulating the cellular proliferation, migration, and invasion has been validated [53]. In our recent study, we observed a dramatic reduction in the migratory phenotype, when cells were endogenously overexpressed with the endoplasmic reticulum N-linked glycosylated enzyme, the Mannosyl-oligosaccharide 1,2-alpha-mannosidase IA (Man1A1). Our finding highlighted the importance of glycosylation and its related enzymes in the process of carcinogenesis and EMT [54].
The cell surface glycans have been proven to play an important role in cancer development and resistant mechanism [43]. We and others have shown the role of core fucosylation in the development of cancer progression and castration resistance mechanisms, melanoma metastasis [55] as well as T-cell exhaustion [56]. Using prostate cancer models, we have shown how core fucosylation on the epidermal growth factor receptor (EGFR) switches the tropism of cancer cells from nuclear receptor signaling (androgen receptor) to the cell surface receptor (EGFR) mechanisms to escape castration-induced cell death [57]. Similarly, cell surface glycans have been implicated in antigen mimicry to avoid immune attacks [58]. The hyper sialyation on the cancer cell surface have been shown to make the cell a primary ligand for sialic acid binding immunoglobulins type lectin (siglecs), which are found on the surface of immune cells [59]. Siglecs would promote immunosuppressive signaling upon bounding to sialylated glycans, thus conferring protection to the cancer cells from NK cells.
Aberrant Protein Glycosylation in PDAC
Protein glycosylation in healthy pancreas acts as a protective and lubricative shield of the pancreatic ducts [60]. In PDAC patients, the changes in protein glycosylation exert a significant effect on tumor transformation and progression, where the normal pancreatic ducts become obstructed and vascularized [10]. PDAC-associated glycosylation abnormalities can extend beyond the pancreatic neoplasms and are often shed into the circulation system. Proteomic analysis of relevant body fluids such as serum, bile fluid, urine, pancreatic juice or cyst fluids, and pancreatic tissues, have revealed glycoprotein-associated alterations that contribute to the carcinogenesis and progression of PDAC [61–68]. These changes include increases in the sialyl Lewis antigens (sLeA and sLeX), an increase in truncated mucin-type O-glycans (Tn and sTn), increased branched and fucosylated N-glycans, upregulation of specific proteoglycans and galectins and increased O-GlcNAcylation [18] (Table 1).
Table 1.
Summary of PDAC-associated glycosylation alterations.
| PDAC-ASSOCIATED GLYCOSYLATION ALTERATION | FEATURES | REFERENCES |
|---|---|---|
|
| ||
| SIALYL LEWIS A | Upregulated | [69] |
| Could be detected by CA 19–9 assay on various proteins including mucins | [60, 70–77] | |
| SIALYL LEWIS X | Upregulated | [78–81] |
| Linked to invasion and metastasis | [82] | |
| Detected on pancreatic cancer-associated proteins | [77, 83–85] | |
| TN AND STN ANTIGENS | Hyper-expressed | [86, 87] |
| Linked to metastasis and poor prognosis | [81, 87, 88] | |
| BRANCHED AND FUCOSYLATED N-GLYCANS | Highly-branched N-glycans are found in pancreatic cancers | [89, 90] |
| Extensive fucosylation | [91–93] | |
| Detected on pancreatic cancer-associated proteins | [94–96] | |
| PROTEOGLYCANS | Overexpressed | [97–99] |
| Linked to cancer progression | [100, 101] | |
| GALECTINS (GLYCAN BINDING PROTEINS) | Overexpressed Galectin-1 and Galectin-3 | [102–105] |
PDAC Glycoproteomics
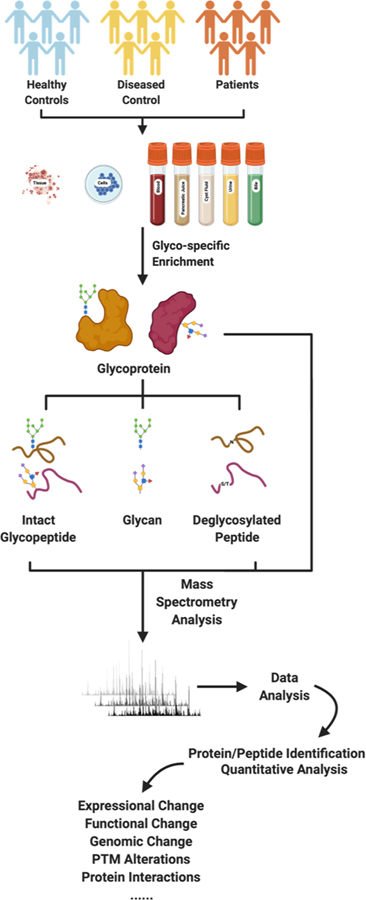
Proteomics has been extensively applied in PDAC studies, ranging from the studies elucidating the mechanism implicated in pancreatic carcinogenesis to the mining of protein biomarkers for PDAC early detection or therapeutic purposes [61–68]. Glycoproteomics, as a branch of proteomics, especially focuses on the proteomic characterization of glycosylated proteins and establishing the correlation between glycoproteomic signatures and biological functions in both healthy and diseased states. Mass spectrometry-based glycoproteomics has been the most powerful and versatile approach available for the characterization of glycoproteome in complex biological samples. A recently published work led by Liwei Cao et al. identified 1,727 N-linked glycosites with significant (adjusted p < 0.01) elevation, 75 N-linked glycoproteins with >2 fold change in PDAC[106]. Of note, 18 out of the 75 N-linked glycoproteins were identified for the first time, with the rest being catalogued in the Pancreatic Cancer Database[107, 108]. Despite the fact that current MS-based glycoproteomics could warrant us with a decent amount of identifications of potential glycoprotein as biomarker candidates, challenges remain for the comprehensive glycoproteomic characterization of a clinical sample. Glycosylation is a highly diversified process with various compositions, structures (linear or branched) and linkages [35]. Subsequently, the heterogeneity generates various substoichiometric modifications to the glycopeptide that decrease the quantities of unique glycoforms [109], which requires better sample enrichment strategies and an increased sensitivity of the mass spectrometry instrumentations. A typical glycoproteomic pipeline consists of sample preparation and enrichment, mass spectrometry (MS) analysis and bioinformatics interpretation [35] (Figure 1).
Figure 1.
A typical pipeline for PDAC glycoproteomic study.
Advances in mass spectrometry-based glycoproteomics
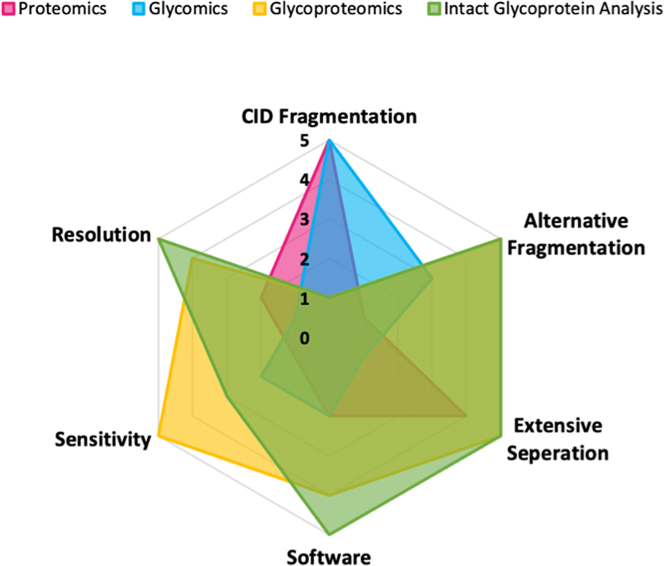
Glycoproteomic is an important branch of proteomics which uses MS as a basic tool for glycoprotein structure elucidation, yet the development of glycoproteomics lags behind compared to other protein modifications due to the plasticity of glycoproteins. Currently, glycoproteomic studies focuses on three different levels, glycan profiling, glycopeptide profiling and glycoprotein profiling [110],with varying instrument requirements (Figure 2) [111]. Due to the lower ionization efficiency of glycans, the sensitivity of the glycomics studies should be slightly higher than that of bottom-up proteomics, and even more so for glycoproteomics studies, where the heterogeneous glycans are subdivided over the peptide backbones. In this context, extensive separation is almost mandatory for glycoproteomics and intact glycoprotein analyses, and less of a requirement in glycomics studies, where information of glycan composition may be obtained by direct MALDI-TOF or MALDI-FTICR analysis of released glycans [111]. In most bottom-up strategies, CID fragmentation is the method of choice. However, CID is generally not sufficient in glycoproteomics and intact glycoprotein analysis unlike that in glycomics analysis. Alternative dissociation techniques such as ETD, EThcD, UVPD, or AI-ETD are needed to provide more comprehensive fragmentation of both the peptide backbone and the glycan in order to identify intact glycopeptides and glycoproteins. As a result of more complex MS spectra containing both the glycans and peptide backbones information, more powerful bioinformatics software is required to assign the MS spectra for structural elucidation. Mass spectrometry resolution (e.g. mass accuracy) is defined as m/Δm, where Δm is the full width at half-maximum. Compared to proteomics, glycomics studies require a higher resolution in order to accurately differentiate glycan peaks from non-glycan contaminant peaks rapidly. A higher resolution is generally required in glycoproteomics studies and even more so in intact glycoprotein analysis, allowing high-confidence identifications and structural elucidation of glycoforms.
Figure 2.
Radar graph showing the instrument requirements for proteomics, glycomics, glycoproteomics, and intact glycoprotein analysis.
Glycomics
Glycomics is the most extensively developed field of glycosylation-associated profiling, focusing on the elucidation of glycan structures via the release of glycan moieties using chemical or enzymatic strategies from glycoproteins and other kinds of sugar-containing macromolecules [112]. Typically, N-linked glycans are released by enzymatic digestion [113], while O-linked glycans are released by β-elimination or chemoenzymatic digestion [114–116]. The released glycans can be directly analyzed by matrix-assisted laser desorption/ionization (MALDI) [117], or via liquid chromatography-electrospray ionization mass spectrometry (LC/ESI-MS) that integrates in-line separation of glycan structures based on their properties[118]. MALDI stands out for the simple sample preparation process with salts and compatibility with non-surfactant contaminants. However, the high degree of vibrational excitation in the ionization process of MALDI may trigger the fragmentation of terminal glycans that are labile, such as sialic acid, sulfate and phosphate group, and subsequent loss of information in the process of data acquisition. ESI greatly reduces the loss of acidic glycan residues during the ionization process, but the LC/MS is more labor intensive due to additional steps to remove the salts and any other contaminants. In addition, sample ionization can result in the loss of more labile moieties, thus additional steps to modify glycan structures can prevent residue loss [119, 120]. Permethylation is a common approach adopted for preserving whole glycan structural information [121]. In addition, permethylation can improve native glycan ionization through the universal conversion of free oxygen-containing nucleophilic groups such as carboxylic acid, hemiketal and hydroxyl groups in glycans. Overall, glycomics offers a high throughput methodology with a low consumption of biological samples, which is important in clinical studies, thus providing rationale for its respective extensive applications cancer biomarker discovery.
Glycopeptide analysis
Glycopeptide analysis is the major component of glycoproteomic-based studies, wherein glycoproteins are subjected to proteolytic digestion by enzymes such as Lys-C and trypsin, with the resulting peptide sample utilized for proteomic analysis. Even though ~50% of proteins are glycosylated [122], glycopeptides represent a minor fraction of the total amount of global peptides. Further confounding glycopeptide stoichiometry, glycoproteins can have variable glycosite occupancy and diverse heterogeneous glycan structures, with an individual glycosite conjugated to a myriad of glycan attachments. The combination of low-abundance, diverse glycan structures and variable fragment patterns of MS signals make glycopeptide detection in global peptides extremely challenging. To overcome these weaknesses in glycopeptide detection, enrichment methods using the physicochemical properties of glycopeptides or integrating chemoenzymatic reactions have been introduced to improve the coverage of glycopeptides and aid glycoproteomic studies [123].
Several common glycopeptide enrichment approaches include lectin affinity binding methods, hydrophilic interaction liquid chromatography (HILIC), hydrazide chemistry methods, titanium dioxide binding methods, and boronic acid binding methods, as well as hybrid methods integrating chemical and enzymatic reactions. Due to the specific recognition of terminal glycan structures on glycopeptides or glycoproteins, lectins are widely used in N-linked or O-linked glycopeptide and glycoprotein enrichment. [124]. As one lectin only selectively bind to a specific terminal glycan structures, this enrichment method brings biases with certain glycans. HILIC based methods rely on the hydrophilic interactions between glycopeptides and matrix [125]. However, the normal HILIC is problematic in terms of low specificity, with co-elution of hydrophilic non-glycopeptides. Several improvements on the stationary phase of HILIC matrices have been developed to help increase the glycopeptide enrichment specificity, including the conjugation with various hydrophilic groups to enhance the hydrogen-bonding and changing to zwitterionic based HILIC (ZIC-HILIC) [126]. Similarly, mixed mode strong anion exchange (MAX) has also been utilized to improve specificity of hydrophilic interactions to enrich glycopeptides [127]. Apart from these non-covalent bonding enrichment methods, there are several strategies where sugar molecules are covalently bonded for the enrichment of glycopeptides, including the hydrazide chemistry method [128], which has been shown to have the highest specificity for glycopeptide enrichment. Hydrazide based oxidation method relies on the hydrazone formation from hydrazide and aldehyde group generated via glycan cis-diol periodate oxidation. The enriched glycopeptides are then selectively released from the bead via N-glycosidase F (PNGase F) digestion at the N-linked glycosylation site. Cleavage with PNGase F will convert the asparagine residue to an aspartic residue, allowing MS identification of N-linked glycosylation site. Further adaptations of hydrazide chemistry method allow for N-linked glycopeptides, O-linked glycopeptides and intact glycopeptides analyses [129]. Titanium dioxide (TiO2) can strongly coordinate with negatively charged oxygen present in residues such as charged sialic acid residues, conferring TiO2 with the ability to enrich sialic acid-containing glycopeptides [130]. However, negative phosphorous residues could also coordinate with TiO2 and results in co-elution of phosphopeptides with sialic acid-containing glycopeptides. Boronic acids chelation with cis-diol on glycans to form cyclic boronate esters is another way to enrich glycopeptides [131]. One drawback of these strategies is that they enrich glycopeptides without preference for particular glycan structures (N-linked or O-linked), thus several chemoenzymatic enrichment strategies were developed to enrich a specific class of glycopeptides of interest. Based on the hydrazide chemistry, N-linked glycans and glycosite-containing peptides (NGAG) can be extracted from solid phase coupled with chemoenzymatic reactions [132]. A similar design was developed recently to specifically enrich and map mucin type O-GalNAcylation at a large-scale [114].
Meanwhile, parallel to the development of sample preparation methodologies, advances in analytical methods also contribute to the improvement of glycopeptide analysis, especially, MS instrumentation approaches and associated data analysis algorithms to interpret the mass spectra. Inherent structural differences between peptide backbones and glycan structures result in varied MS fragmentation efficiency. Collision-induced dissociation (CID) and high energy collision dissociation (HCD) not only fragment peptide backbones, but also glycans, while electron-capture dissociation (ECD) and electron-transfer dissociation (ETD), the peptide backbone is fragmented more readily thus preserving the glycan structure information [133]. Combining multiple fragmentation approaches could potentially help to get information for both glycans and peptide backbones [134]. Photodissociation, especially ultraviolet photodissociation (UVPD) is another fragmentation strategy for the acquisition of intact glycan information [135].
In addition to fragmentation strategy, different data acquisition strategies are also considered in glycoproteomics studies. Data Dependent Acquisition (DDA) is the go-to method when it comes to glycoproteomics, due to its robustness and flexibility in neo-glycan species discovery. In DDA, individual precursor ions would be isolated sequentially in a narrow m/z window, with intensity being the one of the primary selection criteria. This semi-stochastic trait of DDA is beneficial in increasing new discovery rates, however, this selection preference often “shadow” those peptides of low abundance, such as glycopeptides. Therefore, identification and quantification of glycopeptides from DDA data is quite challenging [136]. Compared to DDA, all precursor ions that are detected in the pre-set isolation windows (usually 10–20 m/z) would be subject to co-fragmentation in Data-Independent Acquisition (DIA). This indiscriminative fragmentation of DIA is beneficial for glycoproteomics since DIA provides a broader dynamic range of detected signals, improved identification reproducibility, accurate and sensitive quantification, as well as enhanced protein coverage [137]. As an emerging method in bottom-up glycoproteomics, DIA is still at its infant stage. Despite the attractive strengths of DIA over DDA, the inherent complexity of glycopeptides poses tremendous analytical and bioinformatics challenges.
In addition, new data analysis pipelines are indispensable in deciphering the various glycopeptide structures. While spectral assignments of formerly glycosylated peptides using glyco-enrichment strategies that focus on generation of deglycosylated glycopeptides (i.e. subjected to releasing glycans from glycosylation sites of the glycoproteins) are compatible with established proteomic search pipelines, intact glycopeptide analysis is far more challenging. To this end, more than twenty different software tools were developed to interpret glycopeptide data for identification and quantification as well as to analyze and annotate glycans and glycopeptides [138]: Byonic [139], pGlyco 2.0 [140], GPQuest [141], Integrated GlycoProteome Analyzer (I-GPA) [142], LaCyTools [143], GP Finder [144], gFinder [145], GlycoMaster DB [146], Glycopep grader (GPG) [147], GlycoPep Detector (GPD) [148], GlycoPeptideSearch (GPS) [149], MassyTools[150], GlycoSpectrumScan[151], GlycopeptideGraphMS[152], GlycoFragwork[153], and GlycReSoft [154], as well as the most recent MSFragger-Glyco[155] and O-Pair search[156]. An extensive description of the algorithms, strengths and weaknesses of most of the available glyco-related software is summarized by Haojie Lu [157] and the comparison study is described by Morten Anderson et al [158]. Of note, Software such as GPQuest [141] and pGlyco [159] have the capabilities to analyze the intact glycopeptide raw data, enabling the delineation of the peptide backbone, site of glycosylation, and glycan structure composition. MSFragger-Glyco and O-Pair search represent the most recent developments in intact glycopeptide identification, both algorithms use ion-indexed search strategy, which could make more accurate assignments in less amount of time compared to traditional search algorithms such as the commercially available software, Byonic. To conform with the pressing need to decipher accumulating glycomics and glycoproteomics data, glyco-analysis is moving toward the next stage of more comprehensive and quantitative identification. A handful of software are reported for quantitative purposes: the above mentioned LaCyTools [143] and GlycopeptideGraphMS [152] are both python-based with an improved glycopeptide identification and quantitation, Integrated GlycoProteome Analyzer (I-GPA) [142] enables an automated label-free quantitation, pQuant [160] could quantitate more peptides with accuracy compared to MaxQuant [161]. Bioinformatics in glycopeptide analysis methods have grown substantially in the last decade and will continue to increase, subsequently improving our understanding of glycosylation in the disease state.
Glycoprotein analysis
Glycoprotein analysis is the directly analysis of intact glycoprotein by MS without any digestion, i.e. the “top down” approach [162], [163]. “Top-down” Intact glycoprotein analysis has its intrinsic advantages as this approach captures and identifies the entirety of proteoforms of an individual protein, which allows the direct assessment of modifications on protein backbones from the same protein molecule. In contrast, the “bottom up” approach relies heavily on individual peptide identification, which may potentially lead to the inference problem of “peptide-to-protein” [164]. In addition to the former, modifications that are related to each other (i.e. modifications that co-exist on a specific proteoform) may not be captured by “bottom-up” approach as well. In bacterial glycoproteomic studies, the top-down approach was proven to be a powerful tool to discover new glycans. [165]. In addition, the top-down approach has been routinely utilized for the assessment and characterization of glycoproteins used for therapeutics and monoclonal antibodies [166]. Distinct from bottom-up proteomics separation methods, which tend to use hydrophobic-based separation (e.g. C18 matrices) of peptides, top-down proteomics incorporates multiple in-line separation strategies, including reverse phase liquid chromatography (RPLC), size exclusion chromatography (SEC) and capillary zone electrophoresis (CZE) to reduce the sample heterogeneity, as well as enabling detection of a boarder range of molecular weights [167]. As an emerging new technology, intact glycoprotein analysis has shown great potential in clinical study and biomarker discovery. Despite its powerful potentials and technological advancements, limitations persist for top-down proteomics. For example, the solubilization challenge of hydrophobic proteins such as membrane proteins, the scarcity of optimal sample separation or fractionation methods for glycoproteins, and the limited scalability of top-down method to high molecular-weight proteins, as well as the suppression of low-abundance proteins like glycoproteins, the difficulty to localize labile PTMs accurately during LC–MS/MS, remain to be tackled. Apart from the technical hurdles, additional bioinformatics tools are also needed to process the complex data generated by top-down method [168, 169].
Glycoproteomic profiling of PDAC cells and tissues
In PDAC, the surgically-obtained tumor tissues consist of cancerous lesions interspersed with activated stroma, which can account for up to 90% of the whole resected tumor volume. These activated stromal cells include activated fibroblasts, myofibroblasts, immune cells, neo-endothelial cells and extra cellular matrix (ECM) components [68]. Proteomic characterization of the whole tissue or purified cancer cells would not only provide insight into the differential protein profiles of these various cell types, but also reveal the cross-talk between cancer cells and the surrounding tumor microenvironment.
In PDAC tissues, an overall increase of N-linked glycosylation was observed compared to healthy pancreas[94]. In addition, this N-glycosylation occupancy change was also implicated in pathways associated with pancreatic cancer, such as TGF-β, TNF, and NF-kappa-B pathways, one of the major carriers of CA 19–9 antigen, MUC5AC, individual glycoproteins including carcinoembryonic antigen-related cell adhesion molecule 5 (CEACAM5), insulin-like growth factor binding protein 3 (IGFBP3), galectin-3-binding protein (LGALS3BP), cathepsin D (CTSD), as well as integrins and CD antigens (including the pancreatic cancer stem-like cells marker CD44) [94, 170]. In-depth mapping of these PDAC-associated glycoproteins revealed that the occupancy change in N-linked glycosylation was specific to not only protein but also glycosylation site. What is worth noting is that the increase of N-linked glycosylation of many of these glycoproteins was also observed in chronic pancreatic tissues, which makes the PDAC glycoproteomic changes even more confounding [35]. As the common precursor for PDAC, pancreatic intraepithelial neoplasia (PanIN), especially the pre-invasive PanIN3 is believed to be the most clinically relevant stage for early detection of curable pancreatic neoplasia [68]. Glycoproteomic profiling of PanIN 3 using iTRAQ and ICAT techniques found several dysregulated glycoproteins, including the ECM components laminin beta 1 (LAMB1) and decorin (DCN), and other proteins such as 14–3-3 theta(YWHAQ), galectin-1 (LGALS1, glycan binding), vimentin (VIM) and actinin (ACTN) in PanIN 3 [171]. In addition, the quantitative proteomics analysis of whole pancreatic cancer tissues had previously revealed that the stromal-epithelial interaction is critical for carcinogenesis [172].
Glycoproteomic techniques were also applied to profile the glycoproteome of isolated PDAC-associated cells, including neoplastic cells, stromal cells and stem cells. Cell surface proteins are largely comprised of transmembrane glycoproteins, such as receptor tyrosine kinases (RTKs), which play an indispensable role in cell signaling, trafficking and cell-cell interactions [35]. N-linked glycosylation alterations in these cell-surface receptors, including epithelial growth factor receptor (EGFR), integrins, and TGF β receptor (TGFβR), were found to influence their functionality, which has implications in carcinogenesis and cancer progression [96, 173–175]. Apart from antibody-based enrichment of cell surface glycoproteins, studies utilizing cell surface capturing (CSC) technology[176] or biorthogonal chemical reporter[177] were carried out to characterize N-linked glycopeptides enriched from the surface of pancreatic cancer cells, revealing the overexpression of CD109[178] and ecto-50-nucleotidase[179]. Another study based on multi-lectin affinity chromatography and nano-LC MS/MS investigated the differentially expressed glycoproteins between the malignant phenotype of pancreatic cancer stem-like CD24+/CD44+ cells and CD24-/CD44+ cells, where the hyper-expression of CD24 was positively linked to late-stage pancreatic adenocarcinomas and the expression of CD13 was negatively linked to late-stage pancreatic adenocarcinomas[180].
Proteomic profiling of protein glycosylation abnormalities in body fluids
Detection regimes based on body fluids are more preferable due to the facts that they are considered less invasive and more accessible. Moreover, PDAC-associated molecular events and signaling can induce protein alterations in various cellular pathways at different levels. These protein alterations, are structurally or quantitatively reflected at the proteome level and may be detectable in PDAC-associated body fluids[64], which would provide valuable clinical information related to disease pathology and severity. In this context, the proteomic characterization of abnormal protein glycosylation patterns in body fluids associated with PDAC may present meaningful opportunities for detecting PDAC at early stages. Depending on the nature of the body fluids (including serum/plasma, pancreatic juice, pancreatic cyst fluids, urine, and bile) and its proximity to the pancreatic neoplastic lesions, can influence the resulting proteomic profiles [64, 181]. In general, serum/plasma represents the proteome of the circulating system, which consists of various functional blood proteins and proteins shed from different tissues, while urine can be viewed as an ultrafiltrate of plasma. Pancreatic juice can be considered a proximal fluid, and is secreted directly into the pancreatic ducts containing many secreted enzymes. Similarly, pancreatic cyst fluid may include mucins and other proteins of interest derived from tissues. Finally, bile could be considered relevant a source for PDAC-associated diagnostics since pancreatic diseases could cause biliary stenosis[64, 181], and may also leak proteins that could be leverage for disease information.
Serum/plasma
The detection and measurement of CA 19–9 antigen in PDAC-associated body fluids is the only FDA approved biomarker monitoring for PDAC, but it falls short in terms of specificity and sensitivity[19]. Instead of analyzing the whole fluid and delineating differential abundance of protein components, it is reasonable to focus on glycosylation alterations of certain proteins involved in neoplastic progression. In fact, CA 19–9 test detects the abnormalities associated with sialyl Lewis antigen of mucins and other carriers of CA 19–9, such as MUC1, MUC5AC and MUC16. Although the individual measurement of CA 19–9 glycan on MUC1, MUC5AC and MUC16 did not improve the performance of the test significantly, it was found that the combined measurement of standard CA 19–9 test and the measurement of CA 19–9 glycan levels on MUC5AC and MUC16 did enhance the discrimination of malignant from benign pancreatic disease[75]. Other distinct glycosylation alterations of MUC1 and MUC5AC including Tn antigen, fucosylation and Lewis antigens were also observed in pancreatic cancer serum [91].
Apart from CA 19–9, other protein glycosylation abnormalities associated with pancreatic cancer were also observed in patient sera. One glycomic study observed an increase in N-linked glycosylation branching as well as hyper-fucosylation and hyper-sialylation in the sera from pancreatic cancer patients[90]. The glycosylation of another pancreas-associated protein – serum ribonuclease 1 (RNASE1), exhibited an overall 40% increase in core fucosylation in the sera of pancreatic cancer patients[93]. Investigation on other major acute-phase proteins (APP) in sera from pancreatic cancer and chronic pancreatitis patients, including alpha-1-acid glycoprotein (AGP1 or ORM1), haptoglobin (HP), fetuin-A (AHSG), α−1-antitrypsin (SERPINA1) and transferrin (TF), revealed an increased level of sialyl Lewis X as well as branching, which could be associated to inflammatory response[182]. In addition, an increase of core fucosylation was also observed on AGP1 and HP in the sera of advanced pancreatic cancer patients, which could be relevant to cancer and may serve as pancreatic cancer signature[182]. The diagnostic potential of fucosylated HP was later validated in several studies, it is possible to detect fucosylated HP as an indicator for advanced pancreatic cancer[183, 184]. However, fucosylated HP could also be detected in other diseases such as hepatocellular carcinoma, liver cirrhosis, gastric cancer, and colorectal cancer[183], and the detection of fucosylated HP alone does not provide satisfactory accuracy for pancreatic cancer diagnosis[184].
An elevated level of other glycoproteins were identified in the sera of pancreatic cancer patients using two-dimensional gel electrophoresis (2DE) and MS, such as fibrinogen γ (FGG) [185], apolipoprotein E (apoE)[186, 187], alpha-1-antichymotrypsin (α1AC)[186], leucine-rich alpha-2-glycoprotein (LRG)[188, 189], and inter-alpha-trypsin inhibitor (IαI)[186]. In comparison to healthy controls and chronic pancreatitis serum samples, another study identified and evaluated several differentially expressed glycoproteins in PDAC serum samples, including tissue inhibitor of metalloproteinase-1 (TIMP1), intercellular adhesion molecule 1 (ICAM1), zinc-alpha 2-glycoprotein 1 (AZGP1), lactoferrin (LF), thrombospondin-1 (THBS1) [190], and lipopolysaccharide-binding protein (LBP) [23]. It was demonstrated that a combination of TIMP1 and ICAM1 could outperform CA 19–9 in terms of the accuracy of distinguishing patients from the healthy and chronic pancreatitis controls, and that AZGP1 could be the biomarker candidate for chronic pancreatitis [23]. Later on, thrombospondin-2 (THBS2) in complement to CA 19–9 was also found to have an improved discrimination capability for PDAC [191]As another accessible body fluid in clinical testing, urine is also a valuable source for disease detection. A proteomics analysis of urine samples collected from patients with PDAC, chronic pancreatitis as well as healthy controls yielded several differential expressed proteins associated with PDAC, including CD59 glycoprotein (CD59) [192]. In particular, these differentially expressed proteins, such as CD90/Thy-1 [193], annexin A10 [194], annexin A2 (ANXA2) [68] and gelsolin (GSN) [68], have been found overexpressed in pancreatic cancer tissues [68], which demonstrated the potential of using the levels of these PDAC-tissue associated proteins in urine and as means for of pancreatic disease detection and discrimination.
Pancreatic juice and pancreatic cyst fluid
Compared to serum/plasma, the collection of pancreatic juice and pancreatic cyst fluid generally requires endoscopy and fine-needle aspiration. Apart from this limitation, pancreatic juice and pancreatic cyst fluid are rich in cancer-specific proteins, providing us with ample material to advance the development of diagnostic biomarkers for PDAC. Due to the advances in proteomics, the proteomic characterizations of pancreatic juice and pancreatic cyst fluid are more comprehensive than ever. Proteomic analyses of pancreatic juice have been conducted in patients with pancreatic adenocarcinoma [195–199], pre-malignant pancreatic neoplasia [200], and pancreatitis [196, 201], as well as in individuals with normal pancreas [202]. Hundreds of proteins such as pancreatic enzymes and other pancreas-associated proteins were identified across different pancreatic juice samples in these studies [64]. Using isotope-code affinity tag (ICAT) technology and tandem MS, several differentially expressed glycoproteins were identified in PDAC samples compared to healthy controls, including kallikrein 1 (KLK1), insulin-like growth factor binding protein 2 (IGFBP2), lithostathine 1 (REG1A, REG1B), pancreatic secretory granule membrane major glycoprotein (GP2), and pancreatic ribonuclease 1 (RNASE1) [195]. In addition, another study identified the overexpression of matrix metalloproteinase-9 (MMP9), and α1-β-glycoprotein (A1BG) in pancreatic juices from PDAC patients. A1BG was also found to be significantly elevated in PDAC patient sera [203], which indicates the fact that cancer-related proteins can often extend beyond the vicinity of cancerous pancreatic tissues.
Apart from PanIN lesions, the most common precursor of PDAC, pancreatic cystic neoplasms also have malignant potentials, such as intraductal papillary mucinous neoplasms (IPMNs) and mucinous cystic neoplasms (MCNs). Moreover, both PanIN 3 and IPMNs/MCNs share many similar molecular features with PDAC, including proteomic changes [181]. One study focusing on the proteomic profiling of pancreatic cyst fluids suggested that the protein family members of amylase, mucins, CEACAMs, and S100 proteins may be candidate biomarkers for the discrimination of pancreatic cysts with malignant potential from benign lesions [204]. Another study identified several hyper-fucosylated glycoproteins such as triacylglycerol lipase and pancreatic α-amylase in pancreatic cyst fluids from IPMNs and MCNs [205]. These proteins are also found to be differentially expressed in PDAC, warranting research into early detection of PDAC at clinically-relevant stages such as PanIN3 and IPMNs/MCNs.
Bile
More than half of pancreatic cancer patients have jaundice as one of the symptoms [20], which is caused by a build-up of bilirubin, a component of bile. Normally, bile is produced by the liver and discharged by biliary ducts into the small intestine to aid lipids digestion. When the bile duct is blocked (biliary stenosis), the accumulating bilirubin in tissues would eventually cause jaundice. Biliary stenosis can be caused by pancreatic diseases, such as the malignant pancreatic adenocarcinoma or chronic pancreatitis [64]. One proteomics study of bile samples from patients with pancreatic adenocarcinoma, cholangiocarcinoma, chronic pancreatitis, and gallstones, identified 127 proteins, including the overexpressed CEACAM6 and MUC1 in pancreatic cancer and cholangiocarcinoma samples [206]. Detecting pancreatic cancer-associated proteins in bile is thus proven to be feasible, however, biliary stenosis may be either due to benign conditions such as chronic pancreatitis or malignant conditions including cholangiocarcinoma and pancreatic adenocarcinoma [206], and the etiological identification of one specific condition remains highly challenging.
Proteomic profiling of protein glycosylation abnormalities on pancreatic cancer-derived exosomes
Exosome are a subset of extracellular vesicles (EVs), with an average size of ~100 nm. Exosomes were first discovered back in 1983 [207], but their role in intercellular communication was only recognized recently. As a compact bioactive cargo with a unique liquid bilayer structure containing nucleic acids, lipids and proteins from donor cells, exosomes could alter the pathophysiological conditions of recipient cells through merging. Accumulating evidence have shown that exosomes play critical roles in cancers, especially tumor-derived exosomes (TEXs) [208]. Hence, exosomes are featured at the forefront of biomarker research in various diseases including pancreatic cancers. Exosomes have been obtained from biofluids such as saliva, blood, urine, pancreatic juice and ascites in PDAC [209]. Abnormalities in glycosylation (Table 1) can be displayed on the cell surfaces, extracellular EVs or proteins directly derived from PDAC cancer cells, making exosomes an exciting candidate for early diagnosis biomarker discovery. Notably, more than 80% of the exosome surface proteins are estimated to be glycosylated [210]. A proteomics study of outer membrane-associated proteins in urine exosomes identified 49 proteins, 25 of which are disease-specific glycoproteins [211]. One of such glycoproteins is olfactomedin-4 (OLFM4), which was reported to be related to the chemo-resistance and poor prognosis in pancreatic cancer [212].
In addition to glycoproteins, proteoglycans are also found on exosomes derived from PDAC. One study found that glypican-1 (GPC1), which is overexpressed in PDAC cancer cells, is also significantly elevated in serum exosomes derived from PDAC patients but not the chronic pancreatitis patients. In fact, it has been proved that GPC1+ exosomes provide superior specificity and sensitivity of PDAC diagnosis to CA 19–9 [213]. Following studies yielded either similar [214–219] or contradictory results, which is not unaccounted for. Glypican-1 is not only found on cancerous tissues but also extensively expressed in brain, gastrointestinal tract, urinary and reproductive systems [220], exosomes produced other than cancerous tissues could confound cancer-derived ones. The specificity of the glypican-1 antibody is not validated stringently, which could easily generate false-positive results [221]. In addition, the exosomes extracted for glypican-1 estimation could be biased [222]. Nonetheless, GPC+ exosomes present wonderful opportunities in PDAC early diagnosis once coupled with more rigorous experimental and statistical validations.
The potential of developing diagnostic markers from exosomes for PDAC is undeniable. However, we are a long way to clinical applications of exosomes as diagnostic tools. First, the cellular and molecular mechanisms of exosomes in PDAC tumorigenesis, progression, metastasis and resistance are unclear. Second, we are in short of effective exosome extraction methods and high-throughput analysis methods, mass spectrometry-based approaches represent the current state-of-art technique. Last but not least, the establishment of an effective biomarker for PDAC early diagnosis (specificity > 99.99%), is challenging due to the low-incidence rate of PDAC in the general population (< 1%) [223], and PDAC exhibit overlapping signatures with other benign conditions.
Concluding remarks and future perspectives
Protein glycosylation is undeniably a major player in the carcinogenesis of pancreatic cancers. Pancreatic cancer-associated changes in protein glycosylation have profound biological ramifications on cellular pathways and signaling at different levels, granting tumor cells with aggressive features. PDAC represents a pancreatic cancer entity of astonishingly high malignancy and poor prognosis, as well as constantly rising patient numbers. It is becoming increasingly pressing that a reliable biomarker is developed for PDAC early-stage diagnosis and disease management. To this end, an overflow of potential biomarkers for PDAC were published over the years, however, their clinical applicability is often considered limited as a result of: expression in benign diseases (for example, pancreatitis) other than PDAC, low predictive value in asymptomatic patients (0.5–0.9%), and varying specificity (70–90%) and sensitivity (68–91%) [224]. The emerging technologies of glycoproteomics, glycomics, and other glycoprotein analytical techniques provide robust tools for the inquisition into the sophistications of protein glycosylation involved in pancreatic cancers. Additionally, there are increasing evidence that disease-associated changes in protein glycosylation is fundamental for the discovery and development of cancer biomarkers [225]. In this context, pancreatic cancer-associated changes in protein glycosylation can undeniably “sweeten the pot” of biomarker discovery.
However, the changes in PDAC glycoproteomes are intricate and can overlap with associated diseases or disease complications such as chronic pancreatitis [226, 227], jaundice [228] and new-onset diabetes (NOD) [229]. Meanwhile, PDAC is considered “uncommon” despite the devastating effects, accounting for only 3% of all cancer cases [230], with a global incidence rate lower than 1% [224]. As a result, the diagnosis of PDAC is often delayed and tricky, which directly leads to poor prognosis and increased diagnoses on inoperable PDAC patients. While proteomic efforts have been made to draw a line between the low-grade precursors (PanINs, IPMNs/MCNs) and PDAC for the early diagnosis attempts, the distinct mechanism and timeline of progression still require further validation and clinical assessment. In addition to the confounding pathological signatures exist in PDAC, technical challenges still remain in various aspects of biomarker discovery based on glycoproteomics. For example, due to the lack of effective consensus enrichment strategy for O-linked glycopeptides, the glycoproteome information of O-linked glycosylation is lacking, which poses further challenges for bioinformatics analysis of O-glycoproteome. Moreover, the less abundant protein glycosylation could not be detected due to the sensitivity limit of current mass spectrometry instrumentations. Consequently, comprehensive protein glycosylation mapping of complex samples is yet to be achieved. Nonetheless, the role of glycosylation in pancreas-related diseases is of emerging interest, many strategies have been demonstrated to target protein glycosylation for improved diagnostic and therapeutic biomarkers. To date, despite the current technology hurdles, significant findings have laid groundwork in the field of glycoproteomics and provide a compelling rationale for targeting protein glycosylation for future biomarker discoveries.
Despite the temptation to discover more potential biomarkers for the early detection of PDAC, the deployment of current FDA-approved CA 19–9 in parallel with other biomarkers such as carcinoembryonic antigen (CEA), or a serum biomarker panel, could amend for the unsatisfactory sensitivity and specificity while using CA 19–9 solely [231]. Another way of achieving better diagnosis power is to combine CA 19–9 with the cost-effective CT/MRI, which is potentially the most accurate method in detecting recurrent pancreatic carcinoma [232, 233]. In addition to proteomics and glycoproteomics candidates, an agglomerate effort of genomics, transcriptomics, proteomics and metabolomics could present us with new insights, also new challenges in developing novel diagnostic and therapeutic tools.
On top of this quest for “perfect” diagnostic biomarker of cancers including PDAC, it is worth noting that biomarkers don’t always tell a “full” story. Discovery of a biomarker that might be associated with an increased cancer risk doesn’t necessarily mean a patient will get cancer or has cancer. Most cancers have multiple biomarkers while some have no identifiable biomarkers. Among these identifiable biomarkers, some could potentially be “driving” the cancer onset and progression. Targeting such biomarkers may be a potential treatment option, but does not always lead to a viable one. Additionally, new abnormalities would be constantly generating during cancer growth and immune evasion. Previously established biomarkers might lose diagnostic and therapeutic power. Currently, there is no effective immunotherapy for the treatment of PDAC as recently reviewed by Kanan Panchal et al. [234], mostly due to the stroma-rich environment of PDAC, which is reported to inhibit spontaneous and therapeutically induced antitumor immunity [235]. As the fourth pillar of cancer treatment (surgery, radiation, and chemotherapy are the initial three pillars), targeted immunotherapy holds great promise in curbing PDAC. With additional efforts and knowledge regarding the immunologic factors involved in the tumorigenesis, progression and immune evasion, development of immunotherapeutic that have direct effect on PDAC could be foreseen. Apart from direct targeting, immunotherapy strategies revolving around the reversion of immunosuppressive environment (TME) have also been proposed, they could be concluded by two directions: (1) T-cell pivoting to restore immunosurveillance and (2) myeloid cells redirecting to condition tumors with increased sensitivity to cytotoxic therapies [236]. Establishing an effective immunotherapeutic for PDAC is still more than a stone away, instead, targeted drug delivery of chemotherapeutic drugs utilizing nanoparticle could potentially overcome the stroma-rich barrier present in PDAC. Future considerations of PDAC targeted therapy based on biomarker should be focusing on multiple aspects including gene mutations and DNA damage repair, alerted receptors, TME, stroma-depleting and etc., there is always going to be a dire need for new biomarkers and therapeutic agents. And maybe, glycoprotein is the way to look.
Acknowledgements
This work was supported in part by the National Institutes of Health under grants and contracts of National Cancer Institute, the Early Detection Research Network (EDRN, U01CA152813) and Clinical Proteomics Tumor Analysis Consortium (U24CA160036 and U24CA210985).
Biography

Shao-Yung earned his B.S in Chemical Engineering from National Taiwan University in 2017. He started his graduate studies as a PhD student in September, 2017, in Chemical and Biomolecular Engineering. He joined Dr. Hui Zhang’s lab in 2018, and he is currently working on deciphering the interplays of cell surface proteins via a mass spectrometry approach. He’s also investigating the workflow of intact glycoproteomics to help elucidate the role of protein-glycan structures in biological states and bioprocesses.

My research goals focus on leveraging innovative proteomic approaches to elucidate the underlying molecular mechanisms that drive pathobiology in a variety of oncological malignancies. Integrating total protein measurements with protein post-translational modification information, including glycosylation and phosphorylation occupancy, enables the delineation of disease-specific features that can not be detected using other “omic” methodologies. I have a particular interest in the development of tissue- and extracellular vesicle (EV)-based protein targets that can be utilized for prognostic, diagnostic, and theranostic applications to further the implementation of personalized medicine in the clinical setting.

My research goals focus on using systems biology tools to dissect pathways responsible for the diverse cancer phenotypes that were previously not possible using conventional methods. As a member of the EDRN and CPTAC consortiums, our group is pushing the boundaries to discover novel surrogate glycoprotein biomarkers and therapeutic targets for metastatic castration resistant prostate cancer. My extensive training in field of proteomics, glycoproteomics and translational medicine has enabled me to embark on the road of discoveries for metastatic cancers.

Yuefan Wang obtained his B.S degree in 2009 from Peking University. After undergraduate research with Professors Zhen Yang and Jiahua Chen, he continued to work at the same group, and received his Ph. D. degree in organic chemistry in 2014 from Peking University. He then took a postdoctoral fellow in Professor Jun O Liu’s group of Johns Hopkins University to explore macrocycle induced protein-protein interactions. He is currently working in Professor Hui Zhang’ group of Johns Hopkins University. His research interests focus on protein-protein interactions and single cell study from chemical proteomics approaches, and their application in cancer research.

Yuanwei is an aspiring young scientist at the Center for Biomarker Discovery & Translation, Johns Hopkins University, with research interests spanning glycoproteomics, single-molecule protein sequencing and glycotransferase engineering. She’s dedicated to deciphering the relationship between glycoproteomics and pathological intricacies. Her passion also includes transforming the scientific findings into applications that can make a difference in fields like diagnostics. She graduated as a student of honor from Peking University, and now, as a second-year Ph.D. student at Johns Hopkins, she’s enjoying the process of brainstorming and bouncing off ideas for the scientific investigations at Professor Hui Zhang’s group.

Dr. Hui Zhang pioneered the development of technology for comprehensive analyses of glycoproteins (Nature Biotechnology 2003). This work and a recent work (Nature Biotechnology 2016) have opened up the field of glycoproteomics for characterization of glycoproteins on their glycosylation sites, glycans, and glycosite-specific glycosylation. Recently, her team at Johns Hopkins University completed the proteomic characterization of ovarian cancer, kidney cancer, head and neck cancer, prostate cancer, and pancreatic cancer sponsored by NIH/NCI/CPTAC and NIH/NCI/EDRN. These studies reveal impacts of genomic and transcriptomic alterations on proteins and protein phosphorylation, glycosylation, and acetylation.
References
- [1].American Cancer Society, “American Cancer Society: Cancer Facts and Figures 2021,” Atlanta, Ga: American Cancer Society, pp. 13–15, 2021. [Google Scholar]
- [2].Kleeff J et al. , “Pancreatic cancer,” Nature Reviews Disease Primers, 2016, doi: 10.1038/nrdp.2016.22. [DOI] [PubMed] [Google Scholar]
- [3].Chari ST et al. , “Early Detection of Sporadic Pancreatic Cancer: Summative Review,” Pancreas. 2015. doi: 10.1097/MPA.0000000000000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zins M, Matos C, and Cassinotto C, “Pancreatic adenocarcinoma staging in the era of preoperative chemotherapy and radiation therapy,” Radiology. 2018. doi: 10.1148/radiol.2018171670. [DOI] [PubMed] [Google Scholar]
- [5].Conroy T et al. , “FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer,” New England Journal of Medicine, 2018, doi: 10.1056/nejmoa1809775. [DOI] [PubMed] [Google Scholar]
- [6].Orth M et al. , “Pancreatic ductal adenocarcinoma: Biological hallmarks, current status, and future perspectives of combined modality treatment approaches,” Radiation Oncology. 2019. doi: 10.1186/s13014-019-1345-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sohn TA, Lillemoe KD, Cameron JL, Huang JJ, Pitt HA, and Yeo CJ, “Surgical palliation of unresectable periampullary adenocarcinoma in the 1990s,” Journal of the American College of Surgeons, 1999, doi: 10.1016/S1072-7515(99)00049-6. [DOI] [PubMed] [Google Scholar]
- [8].Chari ST, Sharma A, and Maitra A, “Early detection of sporadic pancreatic ductal adenocarcinoma: Problems, promise, and prospects,” Annals of Internal Medicine, 2020, doi: 10.7326/M19-2336. [DOI] [PubMed] [Google Scholar]
- [9].Luchini C, Capelli P, and Scarpa A, “Pancreatic Ductal Adenocarcinoma and Its Variants,” Surgical Pathology Clinics. 2016. doi: 10.1016/j.path.2016.05.003. [DOI] [PubMed] [Google Scholar]
- [10].Schawkat K, Manning MA, Glickman JN, and Mortele KJ, “Pancreatic ductal adenocarcinoma and its variants: Pearls and perils,” Radiographics, 2020, doi: 10.1148/rg.2020190184. [DOI] [PubMed] [Google Scholar]
- [11].Hruban RH, Goggins M, Parsons J, and Kern SE, “Progression model for pancreatic cancer,” Clinical Cancer Research. 2000. [PubMed] [Google Scholar]
- [12].McGuigan A, Kelly P, Turkington RC, Jones C, Coleman HG, and McCain RS, “Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes,” World Journal of Gastroenterology. 2018. doi: 10.3748/wjg.v24.i43.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Waters AM and Der CJ, “KRAS: The critical driver and therapeutic target for pancreatic cancer,” Cold Spring Harbor Perspectives in Medicine, 2018, doi: 10.1101/cshperspect.a031435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Jones S et al. , “Core signaling pathways in human pancreatic cancers revealed by global genomic analyses,” Science, 2008, doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Easton DF et al. , “Genome-wide association study identifies novel breast cancer susceptibility loci,” Nature, 2007, doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pihlak R, Valle JW, and McNamara MG, “Germline mutations in pancreatic cancer and potential new therapeutic options,” Oncotarget, 2017, doi: 10.18632/oncotarget.17291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hu C et al. , “Association between inherited germline mutations in cancer predisposition genes and risk of pancreatic cancer,” 2018. doi: 10.1001/jama.2018.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Munkley J, “The glycosylation landscape of pancreatic cancer (Review),” Oncology Letters. 2019. doi: 10.3892/ol.2019.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Winter JM, Yeo CJ, and Brody JR, “Diagnostic, prognostic, and predictive biomarkers in pancreatic cancer,” Journal of Surgical Oncology, 2013, doi: 10.1002/jso.23192. [DOI] [PubMed] [Google Scholar]
- [20].Stark A and Eibl G, “Pancreatic Ductal Adenocarcinoma,” Pancreapedia: Exocrine Pancreas Knowledge Base. 2015. doi: 10.3998/panc.2015.14. [DOI] [Google Scholar]
- [21].Wehr AY, Hwang WT, Blair IA, and Yu KH, “Relative quantification of serum proteins from pancreatic ductal adenocarcinoma patients by stable isotope dilution liquid chromatography-mass spectrometry,” 2012. doi: 10.1021/pr201011f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Faca VM et al. , “A mouse to human search for plasma proteome changes associated with pancreatic tumor development,” PLoS Medicine, 2008, doi: 10.1371/journal.pmed.0050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pan S et al. , “Protein alterations associated with pancreatic cancer and chronic pancreatitis found in human plasma using global quantitative proteomics Profiling,” Journal of Proteome Research, 2011, doi: 10.1021/pr101148r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Nie S et al. , “Glycoprotein biomarker panel for pancreatic cancer discovered by quantitative proteomics analysis,” Journal of Proteome Research, 2014, doi: 10.1021/pr400967x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Pereira SP et al. , “Early detection of pancreatic cancer,” The Lancet Gastroenterology and Hepatology. 2020. doi: 10.1016/S2468-1253(19)30416-9. [DOI] [Google Scholar]
- [26].Drake PM et al. , “Sweetening the pot: Adding glycosylation to the biomarker discovery equation,” Clinical Chemistry. 2010. doi: 10.1373/clinchem.2009.136333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gill DJ, Clausen H, and Bard F, “Location, location, location: New insights into O-GalNAc protein glycosylation,” Trends in Cell Biology. 2011. doi: 10.1016/j.tcb.2010.11.004. [DOI] [PubMed] [Google Scholar]
- [28].Haltiwanger RS and Lowe JB, “Role of glycosylation in development,” Annual Review of Biochemistry. 2004. doi: 10.1146/annurev.biochem.73.011303.074043. [DOI] [PubMed] [Google Scholar]
- [29].Helenius A and Aebi M, “Intracellular functions of N-linked glycans,” Science. 2001. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- [30].Ohtsubo K and Marth JD, “Glycosylation in Cellular Mechanisms of Health and Disease,” Cell. 2006. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- [31].Rudd PM, Elliott T, Cresswell P, Wilson IA, and Dwek RA, “Glycosylation and the immune system,” Science. 2001. doi: 10.1126/science.291.5512.2370. [DOI] [PubMed] [Google Scholar]
- [32].Woods RJ, Edge CJ, and Dwek RA, “Protein surface oligosaccharides and protein function,” Nature Structural Biology, 1994, doi: 10.1038/nsb0894-499. [DOI] [PubMed] [Google Scholar]
- [33].Hart GW and Copeland RJ, “Glycomics hits the big time,” Cell, vol. 143, no. 5, pp. 672–676, 2010, doi: 10.1016/j.cell.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].A. C. R. D.; E. J. D.; F. H. H.; S. P.; B. C. R.; H. G. W.; Varki EM and E., Essentials of Glycobiology, 3rd edition. 2015. [Google Scholar]
- [35].Pan S, Brentnall TA, and Chen R, “Glycoproteins and glycoproteomics in pancreatic cancer,” World Journal of Gastroenterology. 2016. doi: 10.3748/wjg.v22.i42.9288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hu Y et al. , “Integrated Proteomic and Glycoproteomic Characterization of Human High-Grade Serous Ovarian Carcinoma,” Cell Reports, vol. 33, no. 3, 2020, doi: 10.1016/j.celrep.2020.108276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Meezan E, Wu HC, Black PH, and Robbins PW, “Comparative Studies on the Carbohydrate-Containing Membrane Components of Normal and Virus-Transformed Mouse Fibroblasts. II. Separation of Glycoproteins and Glycopeptides by Sephadex Chromatography,” Biochemistry, 1969, doi: 10.1021/bi00834a039. [DOI] [PubMed] [Google Scholar]
- [38].Dennis JW, Granovsky M, and Warren CE, “Glycoprotein glycosylation and cancer progression,” Biochimica et Biophysica Acta - General Subjects. 1999. doi: 10.1016/S0304-4165(99)00167-1. [DOI] [PubMed] [Google Scholar]
- [39].Kobata A and Amano J, “Altered glycosylation of proteins produced by malignant cells, and application for the diagnosis and immunotherapy of tumours,” Immunology and Cell Biology, 2005, doi: 10.1111/j.1440-1711.2005.01351.x. [DOI] [PubMed] [Google Scholar]
- [40].Ono M and Hakomori S, “Glycosylation defining cancer cell motility and invasiveness,” 2003. doi: 10.1023/B:GLYC.0000018019.22070.7d. [DOI] [PubMed] [Google Scholar]
- [41].Pinho SS and Reis CA, “Glycosylation in cancer: Mechanisms and clinical implications,” Nature Reviews Cancer. 2015. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- [42].Brooks S et al. , “Altered Glycosylation of Proteins in Cancer: What Is the Potential for New Anti-Tumour Strategies,” Anti-Cancer Agents in Medicinal Chemistry, 2008, doi: 10.2174/187152008783330860. [DOI] [PubMed] [Google Scholar]
- [43].Stowell SR, Ju T, and Cummings RD, “Protein glycosylation in cancer,” Annual Review of Pathology: Mechanisms of Disease, 2015, doi: 10.1146/annurev-pathol-012414-040438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Liotta LA, Steeg PS, and Stetler-Stevenson WG, “Cancer metastasis and angiogenesis: An imbalance of positive and negative regulation,” Cell. 1991. doi: 10.1016/0092-8674(91)90642-C. [DOI] [PubMed] [Google Scholar]
- [45].Guo W and Giancotti FG, “Integrin signalling during tumour progression,” Nature Reviews Molecular Cell Biology, vol. 5, no. 10. pp. 816–826, 2004. doi: 10.1038/nrm1490. [DOI] [PubMed] [Google Scholar]
- [46].M DM and Baluk P, “Significance of blood vessel leakiness in cancer,” Cancer Research, vol. 62, no. 18, pp. 5381–5385, 2002. [PubMed] [Google Scholar]
- [47].Hashizume H et al. , “Openings between defective endothelial cells explain tumor vessel leakiness,” American Journal of Pathology, 2000, doi: 10.1016/S0002-9440(10)65006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Their JP, “Epithelial-mesenchymal transitions in tumor progression,” Nature Reviews Cancer. 2002. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- [49].Lange T, Samatov TR, Tonevitsky AG, and Schumacher U, “Importance of altered glycoprotein-bound N- and O-glycans for epithelial-to-mesenchymal transition and adhesion of cancer cells,” Carbohydrate Research, 2014, doi: 10.1016/j.carres.2014.01.010. [DOI] [PubMed] [Google Scholar]
- [50].Li X, Wang X, Tan Z, Chen S, and Guan F, “Role of glycans in cancer cells undergoing epithelial-mesenchymal transition,” Frontiers in Oncology. 2016. doi: 10.3389/fonc.2016.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zhang J, Ten Dijke P, Wuhrer M, and Zhang T, “Role of glycosylation in TGF-β signaling and epithelial-to-mesenchymal transition in cancer.,” Protein & cell, Jun. 2020, doi: 10.1007/s13238-020-00741-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Freire-de-Lima L et al. , “Involvement of O-glycosylation defining oncofetal fibronectin in epithelial-mesenchymal transition process,” Proceedings of the National Academy of Sciences of the United States of America, 2011, doi: 10.1073/pnas.1115191108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Huanna T et al. , “GALNT14 mediates tumor invasion and migration in breast cancer cell MCF-7,” Molecular Carcinogenesis, 2015, doi: 10.1002/mc.22186. [DOI] [PubMed] [Google Scholar]
- [54].Pan J et al. , “Glycoproteomics-based signatures for tumor subtyping and clinical outcome prediction of high-grade serous ovarian cancer,” Nature Communications, vol. 11, no. 1, pp. 1–13, 2020, doi: 10.1038/s41467-020-19976-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Agrawal P et al. , “A Systems Biology Approach Identifies FUT8 as a Driver of Melanoma Metastasis,” Cancer Cell, 2017, doi: 10.1016/j.ccell.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Okada M et al. , “Blockage of Core Fucosylation Reduces Cell-Surface Expression of PD-1 and Promotes Anti-tumor Immune Responses of T Cells,” Cell Reports, 2017, doi: 10.1016/j.celrep.2017.07.027. [DOI] [PubMed] [Google Scholar]
- [57].Höti N et al. , “A comprehensive analysis of FUT8 overexpressing prostate cancer cells reveals the role of EGFR in castration resistance,” Cancers, 2020, doi: 10.3390/cancers12020468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Nardy AFFR, Freire-de-Lima L, Freire-de-Lima CG, and Morrot A, “The sweet side of immune evasion: Role of Glycans in the Mechanisms of Cancer Progression,” Frontiers in Oncology. 2016. doi: 10.3389/fonc.2016.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Lübbers J, Rodríguez E, and van Kooyk Y, “Modulation of Immune Tolerance via Siglec-Sialic Acid Interactions,” Frontiers in immunology. 2018. doi: 10.3389/fimmu.2018.02807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Moniaux N, Andrianifahanana M, Brand RE, and Batra SK, “Multiple roles of mucins in pancreatic cancer, a lethal and challenging malignancy,” British Journal of Cancer. 2004. doi: 10.1038/sj.bjc.6602163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Aspinall-O’Dea M and Costello E, “The pancreatic cancer proteome - Recent advances and future promise,” Proteomics - Clinical Applications. 2007. doi: 10.1002/prca.200700144. [DOI] [PubMed] [Google Scholar]
- [62].Cecconi D, Palmieri M, and Donadelli M, “Proteomics in pancreatic cancer research,” Proteomics. 2011. doi: 10.1002/pmic.201000401. [DOI] [PubMed] [Google Scholar]
- [63].Coleman O, Henry M, McVey G, Clynes M, Moriarty M, and Meleady P, “Proteomic strategies in the search for novel pancreatic cancer biomarkers and drug targets: Recent advances and clinical impact,” Expert Review of Proteomics. 2016. doi: 10.1586/14789450.2016.1167601. [DOI] [PubMed] [Google Scholar]
- [64].Pan S, Brentnall TA, and Chen R, “Proteomics analysis of bodily fluids in pancreatic cancer,” Proteomics. 2015. doi: 10.1002/pmic.201400476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Pawa N, Wright JM, and Arulampalam THA, “Mass spectrometry based proteomic profiling for pancreatic cancer,” Journal of the Pancreas. 2010. [PubMed] [Google Scholar]
- [66].Sun C, Rosendahl AH, Ansari D, and Andersson R, “Proteome-based biomarkers in pancreatic cancer,” World Journal of Gastroenterology. 2011. doi: 10.3748/wjg.v17.i44.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Tonack S, Aspinall-O’Dea M, Neoptolemos JP, and Costello E, “Pancreatic cancer: Proteomic approaches to a challenging disease,” Pancreatology, 2009, doi: 10.1159/000212083. [DOI] [PubMed] [Google Scholar]
- [68].Pan S, Brentnall TA, Kelly K, and Chen R, “Tissue proteomics in pancreatic cancer study: Discovery, emerging technologies, and challenges,” Proteomics. 2013. doi: 10.1002/pmic.201200319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Schmiegel WH, Greten H, and Thiele HG, “Characterization of CA 19–9 Bearing Mucins as Physiological Exocrine Pancreatic Secretion Products,” Cancer Research, 1986. [PubMed] [Google Scholar]
- [70].Magnani JL et al. , “A monoclonal antibody-defined antigen associated with gastrointestinal cancer is a ganglioside containing sialylated lacto-N-fucopentaose II,” Journal of Biological Chemistry, 1982, doi: 10.1016/S0021-9258(19)45389-1. [DOI] [PubMed] [Google Scholar]
- [71].Magnani JL et al. , “A monosialoganglioside is a monoclonal antibody-defined antigen of colon carcinoma,” Science, 1981, doi: 10.1126/science.7209516. [DOI] [PubMed] [Google Scholar]
- [72].Herlyn M, Sears HF, Steplewski Z, and Koprowski H, “Monoclonal antibody detection of a circulating tumor-associated antigen. I. Presence of antigen in sera of patients with colorectal, gastric, and pancreatic carcinoma,” Journal of Clinical Immunology, 1982, doi: 10.1007/BF00916897. [DOI] [PubMed] [Google Scholar]
- [73].Magnani JL, Ginsburg V, Steplewski Z, and Koprowsk H, “Identification of The Gastrointestinal and Pancreatic Cancer-Associated Antigen Detected by Monoclonal Antibody 19–9 in the Sera of Patients as a Mucin,” Cancer Research, 1983. [PubMed] [Google Scholar]
- [74].Yue T et al. , “Identification of blood-protein carriers of the CA 19–9 antigen and characterization of prevalence in pancreatic diseases,” Proteomics, 2011, doi: 10.1002/pmic.201000827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Yue T et al. , “Enhanced discrimination of malignant from benign pancreatic disease by measuring the CA 19–9 antigen on specific protein carriers,” PLoS ONE, 2011, doi: 10.1371/journal.pone.0029180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Partyka K, Maupin KA, Brand RE, and Haab BB, “Diverse monoclonal antibodies against the CA 19–9 antigen show variation in binding specificity with consequences for clinical interpretation,” Proteomics, 2012, doi: 10.1002/pmic.201100676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Tang H et al. , “Glycans Related to the CA19–9 Antigen Are Increased in Distinct Subsets of Pancreatic Cancers and Improve Diagnostic Accuracy Over CA19–9,” CMGH, 2016, doi: 10.1016/j.jcmgh.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Pour PM, Tempero MM, Takasaki H, Uchida E, Takiyama Y, and Burnett DA, “Expression of Blood Group-related Antigens ABH, Lewis A, Lewis B, Lewis X, Lewis Y, and CA 19–9 in Pancreatic Cancer Cells in Comparison with the Patient’s Blood Group Type,” Cancer Research, 1988. [PubMed] [Google Scholar]
- [79].Singh S et al. , “Upregulation of glycans containing 3′ fucose in a subset of pancreatic cancers uncovered using fusion-tagged lectins,” Journal of Proteome Research, 2015, doi: 10.1021/acs.jproteome.5b00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Tang H et al. , “Glycan motif profiling reveals plasma sialyl-Lewis X elevations in pancreatic cancers that are negative for sialyl-Lewis A,” Molecular and Cellular Proteomics, 2015, doi: 10.1074/mcp.M114.047837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Balmaña M et al. , “Identification of potential pancreatic cancer serum markers: Increased sialyl-Lewis X on ceruloplasmin,” Clinica Chimica Acta, 2015, doi: 10.1016/j.cca.2015.01.007. [DOI] [PubMed] [Google Scholar]
- [82].Takahashi S et al. , “Overexpression of sialyl Lewis x antigen is associated with formation of extratumoral venous invasion and predicts postoperative development of massive hepatic metastasis in cases with pancreatic ductal adenocarcinoma,” Pathobiology, 2001, doi: 10.1159/000048767. [DOI] [PubMed] [Google Scholar]
- [83].hyun Rho J et al. , “Discovery of sialyl Lewis A and Lewis X modified protein cancer biomarkers using high density antibody arrays,” Journal of Proteomics, 2014, doi: 10.1016/j.jprot.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Bossen EH, Borowitz MJ, Levine SJ, and Tuck FL, “Antigens of human pancreatic adenocarcinoma cells defined by murine monoclonal antibodies,” Cancer Research, 1982. [PubMed] [Google Scholar]
- [85].Kawa S et al. , “Epitope analysis of SPan-1 and DUPAN-2 using Synthesized Glycoconjugates Sialyllact-N-Fucopentaose II and Sialyllact-N-Tetraose,” Pancreas, 1994, doi: 10.1097/00006676-199411000-00003. [DOI] [PubMed] [Google Scholar]
- [86].Remmers N et al. , “Aberrant expression of mucin core proteins and O-linked glycans associated with progression of pancreatic cancer,” Clinical Cancer Research, 2013, doi: 10.1158/1078-0432.CCR-12-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Hofmann BT et al. , “COSMC knockdown mediated aberrant O-glycosylation promotes oncogenic properties in pancreatic cancer,” Molecular Cancer, 2015, doi: 10.1186/s12943-015-0386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Radhakrishnan P et al. , “Immature truncated O-glycophenotype of cancer directly induces oncogenic features,” Proceedings of the National Academy of Sciences of the United States of America, 2014, doi: 10.1073/pnas.1406619111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Park HM et al. , “Mass spectrometry-based N-linked glycomic profiling as a means for tracking pancreatic cancer metastasis,” Carbohydrate Research, 2015, doi: 10.1016/j.carres.2015.04.019. [DOI] [PubMed] [Google Scholar]
- [90].Zhao J, Qiu W, Simeone DM, and Lubman DM, “N-linked glycosylation profiling of pancreatic cancer serum using capillary liquid phase separation coupled with mass spectrometric analysis,” Journal of Proteome Research, 2007, doi: 10.1021/pr0604458. [DOI] [PubMed] [Google Scholar]
- [91].Yue T, Goldstein IJ, Hollingsworth MA, Kaul K, Brand RE, and Haab BB, “The prevalence and nature of glycan alterations on specific proteins in pancreatic cancer patients revealed using antibody-lectin sandwich arrays,” Molecular and Cellular Proteomics, 2009, doi: 10.1074/mcp.M900135-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Kamada Y et al. , “Reevaluation of a lectin antibody ELISA kit for measuring fucosylated haptoglobin in various conditions,” Clinica Chimica Acta, 2013, doi: 10.1016/j.cca.2012.12.014. [DOI] [PubMed] [Google Scholar]
- [93].Barrabés S et al. , “Glycosylation of serum ribonuclease 1 indicates a major endothelial origin and reveals an increase in core fucosylation in pancreatic cancer,” Glycobiology, 2007, doi: 10.1093/glycob/cwm002. [DOI] [PubMed] [Google Scholar]
- [94].Pan S et al. , “Quantitative glycoproteomics analysis reveals changes in N-glycosylation level associated with pancreatic ductal adenocarcinoma,” Journal of Proteome Research, 2014, doi: 10.1021/pr4010184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Pan S, Tamura Y, Chen R, May D, McIntosh MW, and Brentnall TA, “Large-scale quantitative glycoproteomics analysis of site-specific glycosylation occupancy,” Molecular BioSystems, 2012, doi: 10.1039/c2mb25268f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Contessa JN, Bhojani MS, Freeze HH, Rehemtulla A, and Lawrence TS, “Inhibition of N-linked glycosylation disrupts receptor tyrosine kinase signaling in tumor cells,” Cancer Research, 2008, doi: 10.1158/0008-5472.CAN-07-6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Weber CK et al. , “Biglycan is overexpressed in pancreatic cancer and induces G1-arrest in pancreatic cancer cell lines,” Gastroenterology, 2001, doi: 10.1053/gast.2001.27222. [DOI] [PubMed] [Google Scholar]
- [98].Conejo JR et al. , “Syndecan-1 expression is up-regulated in pancreatic but not in other gastrointestinal cancers,” International Journal of Cancer, 2000, doi: . [DOI] [PubMed] [Google Scholar]
- [99].Kleeff J et al. , “The cell-surface heparan sulfate proteoglycan glypican-1 regulates growth factor action in pancreatic carcinoma cells and is overexpressed in human pancreatic cancer,” Journal of Clinical Investigation, 1998, doi: 10.1172/JCI4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Whipple CA, Young AL, and Korc M, “A KrasG12D-driven genetic mouse model of pancreatic cancer requires glypican-1 for efficient proliferation and angiogenesis,” Oncogene, 2012, doi: 10.1038/onc.2011.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Aikawa T et al. , “Glypican-1 modulates the angiogenic and metastatic potential of human and mouse cancer cells,” Journal of Clinical Investigation, 2008, doi: 10.1172/JCI32412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Qian D et al. , “Galectin-1-driven upregulation of SDF-1 in pancreatic stellate cells promotes pancreatic cancer metastasis,” Cancer Letters, 2017, doi: 10.1016/j.canlet.2017.03.024. [DOI] [PubMed] [Google Scholar]
- [103].Zhao W et al. , “Galectin-3 Mediates Tumor Cell–Stroma Interactions by Activating Pancreatic Stellate Cells to Produce Cytokines via Integrin Signaling,” Gastroenterology, 2018, doi: 10.1053/j.gastro.2017.12.014. [DOI] [PubMed] [Google Scholar]
- [104].Chen R et al. , “Stromal galectin-1 expression is associated with long-term survival in resectable pancreatic ductal adenocarcinoma,” Cancer Biology and Therapy, 2012, doi: 10.4161/cbt.20842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Chen R et al. , “Proteins associated with pancreatic cancer survival in patients with resectable pancreatic ductal adenocarcinoma,” Laboratory Investigation, 2015, doi: 10.1038/labinvest.2014.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Cao L et al. , “Proteogenomic characterization of pancreatic ductal adenocarcinoma,” Cell, vol. 184, no. 19, pp. 5031–5052.e26, Sep. 2021, doi: 10.1016/J.CELL.2021.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Harsha HC et al. , “A Compendium of Potential Biomarkers of Pancreatic Cancer,” PLOS Medicine, vol. 6, no. 4, p. e1000046, Apr. 2009, doi: 10.1371/JOURNAL.PMED.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].JK T et al. , “Pancreatic Cancer Database: an integrative resource for pancreatic cancer,” Cancer biology & therapy, vol. 15, no. 8, pp. 963–967, 2014, doi: 10.4161/CBT.29188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Woo CM, Iavarone AT, Spiciarich DR, Palaniappan KK, and Bertozzi CR, “Isotope-targeted glycoproteomics (IsoTaG): A mass-independent platform for intact N- and O-glycopeptide discovery and analysis,” Nature Methods, 2015, doi: 10.1038/nmeth.3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Yang Y, Franc V, and Heck AJR, “Glycoproteomics: A Balance between High-Throughput and In-Depth Analysis,” Trends in Biotechnology. 2017. doi: 10.1016/j.tibtech.2017.04.010. [DOI] [PubMed] [Google Scholar]
- [111].Ruhaak LR, Xu G, Li Q, Goonatilleke E, and Lebrilla CB, “Mass Spectrometry Approaches to Glycomic and Glycoproteomic Analyses,” Chemical Reviews, 2018, doi: 10.1021/acs.chemrev.7b00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Zaia J, “Mass spectrometry and glycomics,” OMICS A Journal of Integrative Biology. 2010. doi: 10.1089/omi.2009.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Plummer TH, Elder JH, Alexander S, Phelan AW, and Tarentino AL, “Demonstration of peptide:N-glycosidase F activity in endo-β-N-acetylglucosaminidase F preparations,” Journal of Biological Chemistry, vol. 259, no. 17, pp. 10700–10704, 1984, doi: 10.1016/s0021-9258(18)90568-5. [DOI] [PubMed] [Google Scholar]
- [114].Yang W, Ao M, Hu Y, Li QK, and Zhang H, “Mapping the O-glycoproteome using site-specific extraction of O-linked glycopeptides (EXoO),” Molecular Systems Biology, 2018, doi: 10.15252/msb.20188486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Yang W, Song A, Ao M, Xu Y, and Zhang H, “Large-scale site-specific mapping of the O-GalNAc glycoproteome,” Nature Protocols, 2020, doi: 10.1038/s41596-020-0345-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Yang W, Ao M, Song A, Xu Y, Sokoll L, and Zhang H, “Mass Spectrometric Mapping of Glycoproteins Modified by Tn-Antigen Using Solid-Phase Capture and Enzymatic Release,” Analytical Chemistry, 2020, doi: 10.1021/acs.analchem.0c01564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Harvey DJ, Royle L, Radcliffe CM, Rudd PM, and Dwek RA, “Structural and quantitative analysis of N-linked glycans by matrix-assisted laser desorption ionization and negative ion nanospray mass spectrometry,” Analytical Biochemistry, 2008, doi: 10.1016/j.ab.2008.01.025. [DOI] [PubMed] [Google Scholar]
- [118].Zaia J, “On-line separations combined with MS for analysis of glycosaminoglycans,” Mass Spectrometry Reviews, 2009, doi: 10.1002/mas.20200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Zaia J, “Mass spectrometry of oligosaccharides,” Mass Spectrometry Reviews, 2004, doi: 10.1002/mas.10073. [DOI] [PubMed] [Google Scholar]
- [120].Shah P, Yang S, Sun S, Aiyetan P, Yarema KJ, and Zhang H, “Mass spectrometric analysis of sialylated glycans with use of solid-phase labeling of sialic acids,” Analytical Chemistry, vol. 85, no. 7, pp. 3606–3613, 2013, doi: 10.1021/ac3033867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Hakomori SI, “A rapid permethylation of glycolipid, and polysaccharide catalyzed by methylsulfinyl carbanion in dimethyl sulfoxide,” Journal of Biochemistry. 1964. doi: 10.1093/oxfordjournals.jbchem.a127869. [DOI] [PubMed] [Google Scholar]
- [122].Moremen KW, Tiemeyer M, and Nairn AV, “Vertebrate protein glycosylation: Diversity, synthesis and function,” Nature Reviews Molecular Cell Biology. 2012. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Suttapitugsakul S, Sun F, and Wu R, “Recent Advances in Glycoproteomic Analysis by Mass Spectrometry,” Analytical Chemistry. 2019. doi: 10.1021/acs.analchem.9b04651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Abbott KL and Pierce JM, “Lectin-based glycoproteomic techniques for the enrichment and identification of potential biomarkers,” in Methods in Enzymology, 2010. doi: 10.1016/S0076-6879(10)80020-5. [DOI] [PubMed] [Google Scholar]
- [125].Chen CC, Su WC, Huang BY, Chen YJ, Tai HC, and Obena RP, “Interaction modes and approaches to glycopeptide and glycoprotein enrichment,” Analyst. 2014. doi: 10.1039/c3an01813j. [DOI] [PubMed] [Google Scholar]
- [126].Mysling S, Palmisano G, Hojrup P, and Thaysen-Andersen M, “Utilizing ion-pairing hydrophilic interaction chromatography solid phase extraction for efficient glycopeptide enrichment in glycoproteomics,” Analytical Chemistry, 2010, doi: 10.1021/ac100530w. [DOI] [PubMed] [Google Scholar]
- [127].Yang W et al. , “Comparison of Enrichment Methods for Intact N- and O-Linked Glycopeptides Using Strong Anion Exchange and Hydrophilic Interaction Liquid Chromatography,” Analytical Chemistry, 2017, doi: 10.1021/acs.analchem.7b03641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Zhang H, jun Li X, Martin DB, and Aebersold R, “Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry,” Nature Biotechnology, 2003, doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- [129].Nilsson J et al. , “Enrichment of glycopeptides for glycan structure and attachment site identification,” Nature Methods, 2009, doi: 10.1038/nmeth.1392. [DOI] [PubMed] [Google Scholar]
- [130].Larsen MR, Jensen SS, Jakobsen LA, and Heegaard NHH, “Exploring the sialiome using titanium dioxide chromatography and mass spectrometry,” Molecular and Cellular Proteomics, 2007, doi: 10.1074/mcp.M700086-MCP200. [DOI] [PubMed] [Google Scholar]
- [131].Xiao H, Chen W, Smeekens JM, and Wu R, “An enrichment method based on synergistic and reversible covalent interactions for large-scale analysis of glycoproteins,” Nature Communications, 2018, doi: 10.1038/s41467-018-04081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Sun S et al. , “Comprehensive analysis of protein glycosylation by solid-phase extraction of N-linked glycans and glycosite-containing peptides,” Nature Biotechnology, 2016, doi: 10.1038/nbt.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Zhu Z, Su X, Clark DF, Go EP, and Desaire H, “Characterizing O-Linked glycopeptides by electron transfer dissociation: Fragmentation rules and applications in data analysis,” Analytical Chemistry, 2013, doi: 10.1021/ac401814h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Yu Q et al. , “Electron-Transfer/Higher-Energy Collision Dissociation (EThcD)-Enabled Intact Glycopeptide/Glycoproteome Characterization,” Journal of the American Society for Mass Spectrometry, 2017, doi: 10.1007/s13361-017-1701-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Brodbelt JS, “Photodissociation mass spectrometry: New tools for characterization of biological molecules,” Chemical Society Reviews. 2014. doi: 10.1039/c3cs60444f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].C. D and Z. J, “Methods to improve quantitative glycoprotein coverage from bottom-up LC-MS data,” Mass spectrometry reviews, 2021, doi: 10.1002/MAS.21692. [DOI] [PubMed] [Google Scholar]
- [137].Ye Z and Vakhrushev SY, “The Role of Data-Independent Acquisition for Glycoproteomics,” Mol Cell Proteomics, vol. 20, p. 100042, 2021, doi: 10.1074/mcp.R120.002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Abrahams JL, Taherzadeh G, Jarvas G, Guttman A, Zhou Y, and Campbell MP, “Recent advances in glycoinformatic platforms for glycomics and glycoproteomics,” Current Opinion in Structural Biology. 2020. doi: 10.1016/j.sbi.2019.11.009. [DOI] [PubMed] [Google Scholar]
- [139].Bern M, Kil YJ, and Becker C, “Byonic: Advanced peptide and protein identification software,” Current Protocols in Bioinformatics, vol. 40, no. SUPPL.40, pp. 13.20.1–13.20.14, Dec. 2012, doi: 10.1002/0471250953.bi1320s40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Zhang Y, Zhu J, Yin H, Marrero J, Zhang X-X, and Lubman DM, “ESI–LC–MS Method for Haptoglobin Fucosylation Analysis in Hepatocellular Carcinoma and Liver Cirrhosis,” Journal of Proteome Research, vol. 14, no. 12, pp. 5388–5395, Dec. 2015, doi: 10.1021/acs.jproteome.5b00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Toghi Eshghi S, Shah P, Yang W, Li X, and Zhang H, “GPQuest: A spectral library matching algorithm for site-specific assignment of tandem mass spectra to intact n-glycopeptides,” Analytical Chemistry, 2015, doi: 10.1021/acs.analchem.5b00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Park GW et al. , “Integrated GlycoProteome Analyzer (I-GPA) for Automated Identification and Quantitation of Site-Specific N-Glycosylation,” Scientific Reports, vol. 6, no. February, pp. 1–12, 2016, doi: 10.1038/srep21175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Jansen BC et al. , “LaCyTools: A Targeted Liquid Chromatography–Mass Spectrometry Data Processing Package for Relative Quantitation of Glycopeptides,” Journal of Proteome Research, vol. 15, no. 7, pp. 2198–2210, Jul. 2016, doi: 10.1021/ACS.JPROTEOME.6B00171. [DOI] [PubMed] [Google Scholar]
- [144].Zhu J et al. , “Differential Quantitative Determination of Site-Specific Intact N -Glycopeptides in Serum Haptoglobin between Hepatocellular Carcinoma and Cirrhosis Using LC-EThcD-MS/MS,” Journal of Proteome Research, vol. 18, no. 1, p. acs.jproteome.8b00654, Oct. 2018, doi: 10.1021/acs.jproteome.8b00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Kim JW et al. , “GFinder: A Web-Based Bioinformatics Tool for the Analysis of N-Glycopeptides,” Journal of Proteome Research, vol. 15, no. 11, pp. 4116–4125, 2016, doi: 10.1021/acs.jproteome.6b00772. [DOI] [PubMed] [Google Scholar]
- [146].He L, Xin L, Shan B, Lajoie GA, and Ma B, “GlycoMaster DB: Software to assist the automated identification of N-linked glycopeptides by tandem mass spectrometry,” Journal of Proteome Research, vol. 13, no. 9, pp. 3881–3895, 2014, doi: 10.1021/pr401115y. [DOI] [PubMed] [Google Scholar]
- [147].Woodin CL, Hua D, Maxon M, Rebecchi KR, Go EP, and Desaire H, “GlycoPep grader: A web-based utility for assigning the composition of N-linked glycopeptides,” Analytical Chemistry, vol. 84, no. 11, pp. 4821–4829, 2012, doi: 10.1021/ac300393t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Zhu Z, Hua D, Clark DF, Go EP, and Desaire H, “GlycoPep detector: A tool for assigning mass spectrometry data of N-linked glycopeptides on the basis of their electron transfer dissociation spectra,” Analytical Chemistry, vol. 85, no. 10, pp. 5023–5032, 2013, doi: 10.1021/ac400287n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Chandler KB, Pompach P, Goldman R, and Edwards N, “Exploring site-specific N-glycosylation microheterogeneity of haptoglobin using glycopeptide CID tandem mass spectra and glycan database search,” Journal of Proteome Research, vol. 12, no. 8, pp. 3652–3666, 2013, doi: 10.1021/pr400196s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Jansen BC et al. , “MassyTools: A High-Throughput Targeted Data Processing Tool for Relative Quantitation and Quality Control Developed for Glycomic and Glycoproteomic MALDI-MS,” Journal of Proteome Research, vol. 14, no. 12, pp. 5088–5098, 2015, doi: 10.1021/acs.jproteome.5b00658. [DOI] [PubMed] [Google Scholar]
- [151].Deshpande N, Jensen PH, Packer NH, and Kolarich D, “GlycoSpectrumScan: Fishing glycopeptides from MS spectra of protease digests of human colostrum sIgA,” Journal of Proteome Research, vol. 9, no. 2, pp. 1063–1075, 2010, doi: 10.1021/pr900956x. [DOI] [PubMed] [Google Scholar]
- [152].Choo MS, Wan C, Rudd PM, and Nguyen-Khuong T, “GlycopeptideGraphMS: Improved Glycopeptide Detection and Identification by Exploiting Graph Theoretical Patterns in Mass and Retention Time,” Analytical Chemistry, vol. 91, no. 11, pp. 7236–7244, 2019, doi: 10.1021/acs.analchem.9b00594. [DOI] [PubMed] [Google Scholar]
- [153].Mayampurath A, Yu CY, Song E, Balan J, Mechref Y, and Tang H, “Computational framework for identification of intact glycopeptides in complex samples,” Analytical Chemistry, vol. 86, no. 1, pp. 453–463, 2014, doi: 10.1021/ac402338u. [DOI] [PubMed] [Google Scholar]
- [154].Klein J, Carvalho L, and Zaia J, “Application of network smoothing to glycan LC-MS profiling,” Bioinformatics, vol. 34, no. 20, pp. 3511–3518, 2018, doi: 10.1093/bioinformatics/bty397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Polasky DA, Yu F, Teo GC, and Nesvizhskii AI, “Fast and comprehensive N- and O-glycoproteomics analysis with MSFragger-Glyco,” Nature Methods, vol. 17, no. 11, pp. 1125–1132, Nov. 2020, doi: 10.1038/s41592-020-0967-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Lu L, Riley NM, Shortreed MR, Bertozzi CR, and Smith LM, “O-Pair Search with MetaMorpheus for O-glycopeptide characterization,” Nature Methods, vol. 17, no. 11, pp. 1133–1138, Nov. 2020, doi: 10.1038/s41592-020-00985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Lu H, “Mass spectrometry based glycoproteomics and its clinic application”.
- [158].Kawahara R et al. , “Community Evaluation of Glycoproteomics Informatics Solutions Reveals High-Performance Search Strategies of Serum N- and O-Glycopeptide Data,” bioRxiv, p. 2021.03.14.435332, Sep. 2021, doi: 10.1101/2021.03.14.435332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Zeng WF et al. , “pGlyco: A pipeline for the identification of intact N-glycopeptides by using HCD- and CID-MS/MS and MS3,” Scientific Reports, 2016, doi: 10.1038/srep25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Liu C et al. , “pQuant Improves Quantitation by Keeping out Interfering Signals and Evaluating the Accuracy of Calculated Ratios,” Analytical Chemistry, vol. 86, no. 11, pp. 5286–5294, Jun. 2014, doi: 10.1021/AC404246W. [DOI] [PubMed] [Google Scholar]
- [161].Tyanova S, Temu T, and Cox J, “The MaxQuant computational platform for mass spectrometry-based shotgun proteomics,” Nature Protocols 2016 11:12, vol. 11, no. 12, pp. 2301–2319, Oct. 2016, doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- [162].Chen B, Brown KA, Lin Z, and Ge Y, “Top-Down Proteomics: Ready for Prime Time?,” Analytical Chemistry. 2018. doi: 10.1021/acs.analchem.7b04747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Donnelly DP et al. , “Best practices and benchmarks for intact protein analysis for top-down mass spectrometry,” Nature Methods, 2019, doi: 10.1038/s41592-019-0457-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Huang T, Wang J, Yu W, and He Z, “Protein inference: a review,” Briefings in Bioinformatics, vol. 13, no. 5, pp. 586–614, Sep. 2012, doi: 10.1093/BIB/BBS004. [DOI] [PubMed] [Google Scholar]
- [165].Valguarnera E, Kinsella RL, and Feldman MF, “Sugar and Spice Make Bacteria Not Nice: Protein Glycosylation and Its Influence in Pathogenesis,” Journal of Molecular Biology. 2016. doi: 10.1016/j.jmb.2016.04.013. [DOI] [PubMed] [Google Scholar]
- [166].Srzentić K et al. , “Interlaboratory Study for Characterizing Monoclonal Antibodies by Top-Down and Middle-Down Mass Spectrometry,” Journal of the American Society for Mass Spectrometry, 2020, doi: 10.1021/jasms.0c00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Staub A, Guillarme D, Schappler J, Veuthey JL, and Rudaz S, “Intact protein analysis in the biopharmaceutical field,” Journal of Pharmaceutical and Biomedical Analysis. 2011. doi: 10.1016/j.jpba.2011.01.031. [DOI] [PubMed] [Google Scholar]
- [168].Chen B, Brown KA, Lin Z, and Ge Y, “Top-Down Proteomics: Ready for Prime Time?,” Analytical Chemistry, vol. 90, no. 1, pp. 110–127, Jan. 2017, doi: 10.1021/ACS.ANALCHEM.7B04747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Melby JA et al. , “Novel Strategies to Address the Challenges in Top-Down Proteomics,” Journal of the American Society for Mass Spectrometry, vol. 32, pp. 1278–1294, Jun. 2021, doi: 10.1021/JASMS.1C00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Nigjeh EN, Chen R, Allen-Tamura Y, Brand RE, Brentnall TA, and Pan S, “Spectral library-based glycopeptide analysis–detection of circulating galectin-3 binding protein in pancreatic cancer,” Proteomics - Clinical Applications, 2017, doi: 10.1002/prca.201700064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Pan S et al. , “Quantitative proteomics investigation of pancreatic intraepithelial neoplasia,” Electrophoresis, 2009, doi: 10.1002/elps.200800752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Chen R et al. , “Pancreatic cancer proteome: The proteins that underlie invasion, metastasis, and immunologic escape,” Gastroenterology, 2005, doi: 10.1053/j.gastro.2005.08.001. [DOI] [PubMed] [Google Scholar]
- [173].Purchio AF et al. , “Identification of mannose 6-phosphate in two asparagine-linked sugar chains of recombinant transforming growth factor-β1 precursor,” Journal of Biological Chemistry, 1988, doi: 10.1016/S0021-9258(18)68207-9. [DOI] [PubMed] [Google Scholar]
- [174].Sato C, Kim JH, Abe Y, Saito K, Yokoyama S, and Kohda D, “Characterization of the N-oligosaccharides attached to the atypical Asn- X-Cys sequence of recombinant human epidermal growth factor receptor,” Journal of Biochemistry, 2000, doi: 10.1093/oxfordjournals.jbchem.a022585. [DOI] [PubMed] [Google Scholar]
- [175].Zheng M, Fang H, and Hakomori SI, “Functional role of N-glycosylation in α5β1 integrin receptor. De-N- glycosylation induces dissociation or altered association of α5 and β1 subunits and concomitant loss of fibronectin binding activity,” Journal of Biological Chemistry, 1994, doi: 10.1016/S0021-9258(17)32719-9. [DOI] [PubMed] [Google Scholar]
- [176].Wollscheid B et al. , “Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins,” Nature Biotechnology, 2009, doi: 10.1038/nbt.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Agard NJ and Bertozzi CR, “Chemical approaches to perturb,profile,andperceive glycans,” Accounts of Chemical Research, 2009, doi: 10.1021/ar800267j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].S Haun R and Yang Fan C, “CD109 Overexpression in Pancreatic Cancer Identified by Cell-Surface Glycoprotein Capture,” Journal of Proteomics & Bioinformatics, 2014, doi: 10.4172/jpb.s10-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Haun RS, Quick CM, Siegel ER, Raju I, Mackintosh SG, and Tackett AJ, “Bioorthogonal labeling cell-surface proteins expressed in pancreatic cancer cells to identify potential diagnostic/therapeutic biomarkers,” Cancer Biology and Therapy, 2015, doi: 10.1080/15384047.2015.1071740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Zhu J, He J, Liu Y, Simeone DM, and Lubman DM, “Identification of glycoprotein markers for pancreatic cancer CD24 +CD44 + stem-like cells using nano-LC-MS/MS and tissue microarray,” Journal of Proteome Research, 2012, doi: 10.1021/pr201059g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Pan S, Brentnall TA, and Chen R, “Proteome alterations in pancreatic ductal adenocarcinoma,” Cancer Letters. 2020. doi: 10.1016/j.canlet.2019.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Sarrats A et al. , “Glycosylation of liver acute-phase proteins in pancreatic cancer and chronic pancreatitis,” Proteomics - Clinical Applications, 2010, doi: 10.1002/prca.200900150. [DOI] [PubMed] [Google Scholar]
- [183].Miyoshi E and Nakano M, “Fucosylated haptoglobin is a novel marker for pancreatic cancer: Detailed analyses of oligosaccharide structures,” Proteomics. 2008. doi: 10.1002/pmic.200800046. [DOI] [PubMed] [Google Scholar]
- [184].Miyoshi E, Shinzaki S, Moriwaki K, and Matsumoto H, “Identification of fucosylated haptoglobin as a novel tumor marker for pancreatic cancer and its possible application for a clinical diagnostic test,” in Methods in Enzymology, 2010. doi: 10.1016/S0076-6879(10)78006-X. [DOI] [PubMed] [Google Scholar]
- [185].Bloomston M, Zhou JX, Rosemurgy AS, Frankel W, Muro-Cacho CA, and Yeatman TJ, “Fibrinogen γ overexpression in pancreatic cancer identified by large-scale proteomic analysis of serum samples,” Cancer Research, 2006, doi: 10.1158/0008-5472.CAN-05-3659. [DOI] [PubMed] [Google Scholar]
- [186].Yu KH, Rustgi AK, and Blair IA, “Characterization of proteins in human pancreatic cancer serum using differential gel electrophoresis and tandem mass spectrometry,” Journal of Proteome Research, 2005, doi: 10.1021/pr050174l. [DOI] [PubMed] [Google Scholar]
- [187].Yan L et al. , “Confounding effect of obstructive jaundice in the interpretation of proteomic plasma profiling data for pancreatic cancer,” Journal of Proteome Research, 2009, doi: 10.1021/pr800451h. [DOI] [PubMed] [Google Scholar]
- [188].Deng R, Lu Z, Chen Y, Zhou L, and Lu X, “Plasma proteomic analysis of pancreatic cancer by 2-dimensional gel electrophoresis,” Pancreas, 2007, doi: 10.1097/MPA.0b013e31802f2483. [DOI] [PubMed] [Google Scholar]
- [189].Kakisaka T et al. , “Plasma proteomics of pancreatic cancer patients by multi-dimensional liquid chromatography and two-dimensional difference gel electrophoresis (2D-DIGE): Up-regulation of leucine-rich alpha-2-glycoprotein in pancreatic cancer,” Journal of Chromatography B: Analytical Technologies in the Biomedical and Life Sciences, 2007, doi: 10.1016/j.jchromb.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Jenkinson C et al. , “Decreased serum thrombospondin-1 levels in pancreatic cancer patients up to 24 months prior to clinical diagnosis: Association with diabetes mellitus,” Clinical Cancer Research, vol. 22, no. 7, pp. 1734–1743, 2016, doi: 10.1158/1078-0432.CCR-15-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Kim J et al. , “Detection of early pancreatic ductal adenocarcinoma with thrombospondin-2 and CA19–9 blood markers,” Science Translational Medicine, vol. 9, no. 398, p. eaah5583, Jul. 2017, doi: 10.1126/scitranslmed.aah5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Weeks ME et al. , “Analysis of the urine proteome in patients with pancreatic ductal adenocarcinoma,” Proteomics - Clinical Applications, 2008, doi: 10.1002/prca.200780164. [DOI] [PubMed] [Google Scholar]
- [193].Zhu J, Thakolwiboon S, Liu X, Zhang M, and Lubman DM, “Overexpression of cd90 (thy-1) in pancreatic adenocarcinoma present in the tumor microenvironment,” PLoS ONE, vol. 9, no. 12, pp. 1–20, 2014, doi: 10.1371/journal.pone.0115507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Zhu J, Wu J, Pei X, Tan Z, Shi J, and Lubman DM, “Annexin A10 is a candidate marker associated with the progression of pancreatic precursor lesions to adenocarcinoma,” PLoS ONE, vol. 12, no. 4, pp. 1–19, 2017, doi: 10.1371/journal.pone.0175039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Chen R et al. , “Quantitative proteomic profiling of pancreatic cancer juice,” Proteomics, 2006, doi: 10.1002/pmic.200500702. [DOI] [PubMed] [Google Scholar]
- [196].Chen R et al. , “Comparison of pancreas juice proteins from cancer versus pancreatitis using quantitative proteomic analysis,” Pancreas, 2007, doi: 10.1097/01.mpa.0000240615.20474.fd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Grønborg M et al. , “Comprehensive proteomic analysis of human pancreatic juice,” Journal of Proteome Research, 2004, doi: 10.1021/pr0499085. [DOI] [PubMed] [Google Scholar]
- [198].Lv S et al. , “Transthyretin, identified by proteomics, is overabundant in pancreatic juice from pancreatic carcinoma and originates from pancreatic islets,” Diagnostic Cytopathology, 2011, doi: 10.1002/dc.21484. [DOI] [PubMed] [Google Scholar]
- [199].Makawita S et al. , “Integrated proteomic profiling of cell line conditioned media and pancreatic juice for the identification of pancreatic cancer biomarkers,” Molecular and Cellular Proteomics, 2011, doi: 10.1074/mcp.M111.008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Chen R et al. , “Elevated level of anterior gradient-2 in pancreatic juice from patients with pre-malignant pancreatic neoplasia,” Molecular Cancer, 2010, doi: 10.1186/1476-4598-9-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Paulo JA, Kadiyala V, Banks PA, Steen H, and Conwell DL, “Mass spectrometry-based (GeLC-MS/MS) comparative proteomic analysis of endoscopically (ePFT) collected pancreatic and gastroduodenal fluids,” Clinical and Translational Gastroenterology, 2012, doi: 10.1038/ctg.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Doyle CJ et al. , “The proteome of normal pancreatic juice,” Pancreas, 2012, doi: 10.1097/MPA.0b013e31822862f6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Tonack S et al. , “ITRAQ reveals candidate pancreatic cancer serum biomarkers: Influence of obstructive jaundice on their performance,” British Journal of Cancer, 2013, doi: 10.1038/bjc.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Ke E et al. , “Proteomic Analyses of Pancreatic Cyst Fluids,” Pancreas, 2009, doi: 10.1097/MPA.0b013e318193a08f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Mann BF, Goetz JA, House MG, Schmidt CM, and Novotny MV, “Glycomic and proteomic profiling of pancreatic cyst fluids identifies hyperfucosylated lactosamines on the N-linked glycans of overexpressed glycoproteins,” Molecular and Cellular Proteomics, 2012, doi: 10.1074/mcp.M111.015792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Farina A, Dumonceau JM, Frossard JL, Hadengue A, Hochstrasser DF, and Lescuyer P, “Proteomic analysis of human bile from malignant biliary stenosis induced by pancreatic cancer,” Journal of Proteome Research, 2009, doi: 10.1021/pr8004925. [DOI] [PubMed] [Google Scholar]
- [207].Harding C and Stahl P, “Transferrin recycling in reticulocytes: pH and iron are important determinants of ligand binding and processing,” Biochemical and Biophysical Research Communications, vol. 113, no. 2, pp. 650–658, Jun. 1983, doi: 10.1016/0006-291X(83)91776-X. [DOI] [PubMed] [Google Scholar]
- [208].Dai J et al. , “Exosomes: key players in cancer and potential therapeutic strategy,” Signal Transduction and Targeted Therapy 2020 5:1, vol. 5, no. 1, pp. 1–10, Aug. 2020, doi: 10.1038/s41392-020-00261-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].M X et al. , “Glycoproteomic bioanalysis of exosomes by LC-MS for early diagnosis of pancreatic cancer,” Bioanalysis, vol. 13, no. 11, pp. 861–864, Jun. 2021, doi: 10.4155/BIO-2021-0036. [DOI] [PubMed] [Google Scholar]
- [210].Erozenci LA, Böttger F, Bijnsdorp I. v., and Jimenez CR, “Urinary exosomal proteins as (pan-)cancer biomarkers: insights from the proteome,” FEBS Letters, vol. 593, no. 13, pp. 1580–1597, Jul. 2019, doi: 10.1002/1873-3468.13487. [DOI] [PubMed] [Google Scholar]
- [211].Hildonen S, Skarpen E, Halvorsen TG, and Reubsaet L, “Isolation and mass spectrometry analysis of urinary extraexosomal proteins,” Scientific Reports 2016 6:1, vol. 6, no. 1, pp. 1–14, Nov. 2016, doi: 10.1038/srep36331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Ohkuma R et al. , “High expression of olfactomedin-4 is correlated with chemoresistance and poor prognosis in pancreatic cancer,” PLOS ONE, vol. 15, no. 1, p. e0226707, Jan. 2020, doi: 10.1371/JOURNAL.PONE.0226707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Melo SA et al. , “Glypican-1 identifies cancer exosomes and detects early pancreatic cancer,” Nature 2015 523:7559, vol. 523, no. 7559, pp. 177–182, Jun. 2015, doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Qian JY, Tan YL, Zhang Y, Yang YF, and Li XQ, “Prognostic value of glypican-1 for patients with advanced pancreatic cancer following regional intra-arterial chemotherapy,” Oncology Letters, vol. 16, no. 1, pp. 1253–1258, Jul. 2018, doi: 10.3892/OL.2018.8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Yang KS et al. , “Multiparametric plasma EV profiling facilitates diagnosis of pancreatic malignancy,” Science Translational Medicine, vol. 9, no. 391, May 2017, doi: 10.1126/SCITRANSLMED.AAL3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Li J et al. , “GPC1 exosome and its regulatory miRNAs are specific markers for the detection and target therapy of colorectal cancer,” Journal of Cellular and Molecular Medicine, vol. 21, no. 5, pp. 838–847, May 2017, doi: 10.1111/JCMM.12941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Lewis JM, Vyas AD, Qiu Y, Messer KS, White R, and Heller MJ, “Integrated Analysis of Exosomal Protein Biomarkers on Alternating Current Electrokinetic Chips Enables Rapid Detection of Pancreatic Cancer in Patient Blood,” ACS Nano, vol. 12, no. 4, pp. 3311–3320, Apr. 2018, doi: 10.1021/ACSNANO.7B08199. [DOI] [PubMed] [Google Scholar]
- [218].Campbell DH et al. , “Detection of glypican-1 (GPC-1) expression in urine cell sediments in prostate cancer,” PLoS ONE, vol. 13, no. 4, Apr. 2018, doi: 10.1371/JOURNAL.PONE.0196017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Levin RA et al. , “Development of a reliable assay to measure glypican-1 in plasma and serum reveals circulating glypican-1 as a novel prostate cancer biomarker,” Oncotarget, vol. 9, no. 32, pp. 22359–22367, Apr. 2018, doi: 10.18632/ONCOTARGET.25009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Kim M-S et al. , “A draft map of the human proteome,” Nature 2014 509:7502, vol. 509, no. 7502, pp. 575–581, May 2014, doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Baker M, “Blame it on the antibodies,” Nature, vol. 521, no. 7552, pp. 274–276, May 2015, doi: 10.1038/521274A. [DOI] [PubMed] [Google Scholar]
- [222].Wang S, Qiu Y, and Bai B, “The Expression, Regulation, and Biomarker Potential of Glypican-1 in Cancer,” Frontiers in Oncology, vol. 0, p. 614, Jul. 2019, doi: 10.3389/FONC.2019.00614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].AY X et al. , “Global incidence and mortality of pancreatic diseases: a systematic review, meta-analysis, and meta-regression of population-based cohort studies,” The lancet. Gastroenterology & hepatology, vol. 1, no. 1, pp. 45–55, 2016, doi: 10.1016/S2468-1253(16)30004-8. [DOI] [PubMed] [Google Scholar]
- [224].Hanna-Sawires RG et al. , “Clinical Perspective on Proteomic and Glycomic Biomarkers for Diagnosis, Prognosis, and Prediction of Pancreatic Cancer,” International Journal of Molecular Sciences 2021, Vol. 22, Page 2655, vol. 22, no. 5, p. 2655, Mar. 2021, doi: 10.3390/IJMS22052655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Taniguchi N, Hancock W, Lubman DM, and Rudd PM, “The second golden age of glycomics: From functional glycomics to clinical applications,” Journal of Proteome Research. 2009. doi: 10.1021/pr801057j. [DOI] [PubMed] [Google Scholar]
- [226].Lowenfels AB et al. , “Pancreatitis and the Risk of Pancreatic Cancer,” New England Journal of Medicine, 1993, doi: 10.1056/nejm199305203282001. [DOI] [PubMed] [Google Scholar]
- [227].Malka D et al. , “Risk of pancreatic adenocarcinoma in chronic pancreatitis,” Gut, 2002, doi: 10.1136/gut.51.6.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Hidalgo L et al. , “Obstructive jaundice as a complication of a peptic duodenal ulcer mimicking pancreatic cancer,” Endoscopy, 2010, doi: 10.1055/s-0030-1255785. [DOI] [PubMed] [Google Scholar]
- [229].Pannala R, Basu A, Petersen GM, and Chari ST, “New-onset diabetes: a potential clue to the early diagnosis of pancreatic cancer,” The Lancet Oncology. 2009. doi: 10.1016/S1470-2045(08)70337-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Sah RP et al. , “Phases of Metabolic and Soft Tissue Changes in Months Preceding a Diagnosis of Pancreatic Ductal Adenocarcinoma,” Gastroenterology, 2019, doi: 10.1053/j.gastro.2019.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].S. J et al. , “Identification of Serum Biomarker Panels for the Early Detection of Pancreatic Cancer,” Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology, vol. 28, no. 1, pp. 174–182, Jan. 2019, doi: 10.1158/1055-9965.EPI-18-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].H. J. A et al. , “Accuracy of multi-detector computed tomography, fluorodeoxyglucose positron emission tomography-CT, and CA 19–9 levels in detecting recurrent pancreatic adenocarcinoma,” JOP : Journal of the pancreas, vol. 14, no. 4, pp. 466–468, 2013, doi: 10.6092/1590-8577/1529. [DOI] [PubMed] [Google Scholar]
- [233].Pedrazzoli S, Sperti C, Beltrame V, and Bissoli S, “Accuracy of CA 19–9 and Radiologic Imaging in Detecting Recurrence After Resection for Pancreatic Cancer,” JOP. Journal of the Pancreas, vol. 14, no. 6, pp. 680–681, 2013, Accessed: Sep. 19, 2021. [Online]. Available: https://pancreas.imedpub.com/accuracy-of-ca-199-and-radiologic-imaging-in-detecting-recurrence-after-resection-for-pancreatic-cancer.php?aid=593 [DOI] [PubMed] [Google Scholar]
- [234].Panchal K, Sahoo RK, Gupta U, and Chaurasiya A, “Role of targeted immunotherapy for pancreatic ductal adenocarcinoma (PDAC) treatment: An overview,” International Immunopharmacology, vol. 95, p. 107508, Jun. 2021, doi: 10.1016/J.INTIMP.2021.107508. [DOI] [PubMed] [Google Scholar]
- [235].Balachandran VP, Beatty GL, and Dougan SK, “Broadening the Impact of Immunotherapy to Pancreatic Cancer: Challenges and Opportunities,” Gastroenterology, vol. 156, no. 7, pp. 2056–2072, May 2019, doi: 10.1053/J.GASTRO.2018.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Beatty GL, Eghbali S, and Kim R, “Deploying Immunotherapy in Pancreatic Cancer: Defining Mechanisms of Response and Resistance,” 10.1200/EDBK_175232, no. 37, pp. 267–278, Oct. 2018, doi: 10.1200/EDBK_175232. [DOI] [PubMed] [Google Scholar]