Abstract
Salmonella enteritidis is an important food-borne pathogen. The use of antibiotics is a serious threat to animal and human health, owing to the existence of resistant strains and drug residues. Lactic acid bacteria, as a new alternative to antibiotics, has attracted much attention. In this study, we investigated the antibacterial potential and underlying mechanism of Lactobacillus rhamnosus SQ511 against S. enteritidis ATCC13076. The results revealed that L. rhamnosus SQ511 significantly inhibited S. enteritidis ATCC13076 growth or even caused death. Laser confocal microscopic imaging revealed that the cell-free supernatant (CFS) of L. rhamnosus SQ511 elevated the reactive oxygen species level and bacterial membrane depolarization in S. enteritidis ATCC13076, leading to cell death. Furthermore, L. rhamnosus SQ511 CFS had severely deleterious effects on S. enteritidis ATCC13076, causing membrane destruction and the release of cellular materials. In addition, L. rhamnosus SQ511 CFS significantly reduced the expression of virulence, motility, adhesion, and invasion genes in S. enteritidis ATCC13076 (P < 0.05), and considerably inhibited motility and biofilm formation capacity (P < 0.05). Thus, antimicrobial compounds produced by L. rhamnosus SQ511 strongly inhibited S. enteritidis growth, mobility, biofilm formation, membrane disruption, and reactive oxygen species generation, and regulated virulence-related gene expressions, presenting promising applications as a probiotic agent.
Keywords: Lactobacillus rhamnosus SQ511, Salmonella enteritidis ATCC13076, Antagonistic activity, Antibacterial mechanism, Cell-free supernatant
Introduction
Salmonella spp. are major foodborne pathogens and one of the leading causes of serious diseases. Salmonella-infected animals usually have no clinical symptoms, but their body and meat are the main reservoirs of the pathogen. It is estimated that the number of food poisoning cases in the USA may be as high as 48 million a year, with Salmonella and Campylobacter diseases alone affecting as many as 2 million people a year (Rohde et al. 2016). In USA, more than 70% of human salmonellosis cases are caused by the consumption of contaminated poultry and eggs (Andino and Hanning 2015). Given the surge in the demand for poultry meat and eggs, this association between zoonotic food-borne infections and poultry products is worrisome (Chousalkar et al. 2018). Thus, gut microorganisms are key to the entry of Salmonella spp. into the food chain continuum through contaminated meat and/or eggs (Harvey et al. 2011). Early control measures to reduce and prevent Salmonella infection and colonization are essential to block pathogen contamination on poultry products (Gast et al. 2016) and promote public health (Rajan et al. 2017; Umaraw et al. 2017). Therefore, the microbiological safety of poultry products is a major problem from a public health and economic perspective (Alali and Hofacre 2016).
Excessive use of antibiotics in livestock and the poultry industry, and the development of antibiotic resistance in bacteria have drawn significant attention to the study of probiotics. Probiotics are considered to be the most promising antibiotic substitutes against intestinal pathogen infections (Afolayan et al. 2017) and have been proven to be useful alternatives to growth promoters; thus, evolving into one of the mainstream products for the use of antibiotics in livestock production (Jouany and Morgavi 2007). Lactic acid bacteria (LAB) are the most representative strains of probiotics, and play an important role in the health and production of livestock (Allen et al. 2013) and poultry, owing to their beneficial effects on maintaining gastrointestinal microbial balance, resistance to intestinal pathogens, and ability to support the immune system (Adetoye et al. 2018).
Salmonella enteritidis is one of the most serious food-borne bacterial pathogens in the chicken industry, causing severe public health problems. In this study, we evaluated the potential antibacterial effects and possible underlying mechanism of Lactobacillus rhamnosus SQ511 against S. enteritidis ATCC13076. The results obtained could help to prevent and control S. enteritidis infection and ensure the safety of animal and poultry food products.
Materials and methods
Bacterial culture
Lactobacillus rhamnosus SQ 0511 was isolated from the intestines of healthy broilers by researchers in our laboratory and cultivated in De Man Rogosa Sharpe (MRS, BD, Sparks, MD, USA) medium. S. enteritidis ATCC13076 was obtained from Shanghai Institute of Veterinary Medicine, Chinese Academy of Agricultural Sciences, China, and cultivated in Luria–Bertani (LB, BD, Sparks, MD, USA) medium. All the bacterial cultures were stored in 50% (V/V) glycerol solution at a ratio of 1:1. The two strains were cultivated under constant shaking at 150 rpm and 37 °C for 16–18 h.
Bacterial culture and preparation of cell-free culture supernatants of L. rhamnosus SQ511
The overnight well-grown culture of L. rhamnosus SQ511 was centrifuged at 10,000×g for 10 min at 4 °C. The resultant CFS was filter-sterilized using 0.2-µm filters (Millipore, Bedford, MA, USA), neutralized to pH 6.5 with 1 M NaOH, and used for further experiments (Khan and Kang 2016).
Determination of antibacterial activity of L. rhamnosus SQ511
The ability of L. rhamnosus SQ511 to inhibit pathogen growth was investigated, as described previously with some modifications (Benavides et al. 2016). After overnight culture, L. rhamnosus SQ511 was inoculated into improved MRS liquid medium at a ratio of 1:100 (v/v) and cultured for 18 h at 37 °C under anaerobic conditions. The bacterial culture medium was centrifuged at 10,000×g for 10 min at 4 °C, and the supernatant and bacterial cells were, respectively, collected and filtered. Subsequently, the supernatant was filtered with 0.22-µm filter, diluted with PBS buffer, and suspended to its original concentration.
To determine the antimicrobial activity of L. rhamnosus SQ511, the indicator S. enteritidis ATCC13076 strain was grown in LB broth with concentration adjusted to 107 CFU/mL, and the culture was poured onto pre-prepared nutrient agar plates containing several Oxford cups, which were removed when the agar solidified. Subsequently, L. rhamnosus SQ511 culture, supernatant, resuspended bacterial cells (100 µL), and MRS medium were spotted onto the wells and incubated at 37 °C. After 12 h of incubation, the inhibition zones were determined. The mean diameters of the inhibition zones were estimated, and inhibition halos > 15 mm indicated high inhibitory activity (Benavides et al. 2016). The experiments were repeated thrice independently.
Analysis of the effect of L. rhamnosus SQ511 on S. enteritidis ATCC13076 viability
After the activation of L. rhamnosus SQ511 and S. enteritidis ATCC13076, the bacterial cells were cultured for 20 h with concentrations adjusted to 107 CFU/mL, and the following treatment groups were established: (1) 0.1 mL of L. rhamnosus SQ511 was inoculated into MRS liquid medium along with 0.1 mL of S. enteritidis ATCC10376; (2) 0.1 mL of L. rhamnosus SQ511 was inoculated into MRS liquid medium for 24 h, and then 0.1 mL of S. enteritidis ATCC13076 was added; and (3) 0.1 mL of S. enteritidis ATCC13076 was inoculated into MRS liquid medium for 24 h, and then, 0.1 mL of L. rhamnosus SQ511 was added. All the cultures were incubated at 37 °C. At 24, 48, 72, 96, and 120 h of incubation, the survival of S. enteritidis ATCC13076 and L. rhamnosus SQ511 were measured by plating the culture on TSB and MRS agar plates and enumerating viable cells.
Fluorescence microscopy
Live and dead cells were evaluated by fluorescent staining (Robertson et al. 2019) using LIVE/DEAD®BacLight™ Bacterial Viability Kit (L7007, Sigma, Germany). Overnight-grown S. enteritidis ATCC13076 cells (107 CFU/mL) were treated with or without L. rhamnosus SQ511 CFS (minimum inhibitory concentration (MIC)) for 4 h at 37 °C. After incubation, the cells were harvested by centrifugation, washed with PBS, and stained with SYTO9/PI (1:1) for 10 min. Subsequently, the cells were washed with PBS and visualized under a laser scanning confocal microscope (Olympus, Japan, FV1000). Similarly, to assess the effect of L. rhamnosus SQ511 CFS on reactive oxygen species (ROS) content and membrane potential, H2DCFDA and rhodamine 123 fluorescent staining were performed, respectively. In brief, S. enteritidis ATCC13076 cells (107 cells/mL) were treated with or without CFS of L. rhamnosus SQ511 for 4 h, followed by centrifugation to harvest the cells. After washing with PBS, the cells were stained with H2DCFDA (10 mM) or rhodamine 123 (1 mg/mL) in the dark for 20 min. The cells were thoroughly washed with PBS to remove the dye and observed under a laser confocal microscope. All the experiments were repeated thrice independently and at least three different fields of view were observed for each culture.
Crystal violet uptake assay
Alterations to membrane permeability were detected by crystal violet assays (Devi et al. 2010). Suspensions of S. enteritidis ATCC10376 (107 cells/mL) were prepared in LB broth, and the cells were harvested at 10,000 × g for 5 min. Then, the cells were washed thrice and suspended in PBS. The CFS of L. rhamnosus SQ511 at the MIC was added to the cell suspension and incubated at 37 °C for 4 h. Control samples were prepared in a similar manner without L. rhamnosus SQ511 treatment. After incubation, the cells were harvested at 10,000×g for 5 min, suspended in PBS containing 10 mg/mL crystal violet, incubated for 10 min at 37 °C, and centrifuged at 10,000×g for 5 min. The optical density (OD590) of the supernatant was measured using a spectrophotometer, with the OD value of crystal violet solution (originally used in the assay) considered as 100% excluded. The percentage of crystal violet uptake was calculated as follows: % Dye uptake = 100 – ((OD of the sample/OD value of crystal violet solution) 100).
Cellular materials release
Measurement of release of cellular materials (DNA) from S. enteritidis ATCC10376 cells was conducted by inoculating the mid-log phase culture (107 cells/mL) into NaCl (0.85%) in the presence or absence of L. rhamnosus SQ511 CFS and incubating at 37 °C. After incubation for 0, 30, 60, and 120 min, 1 mL of the culture broth was transferred to a tube and centrifuged at 10,000 rpm. The absorbance of the supernatant was measured at 260 nm using a spectrophotometer, and the results were expressed as OD recorded at each time interval. Simultaneously, the amount of released protein in the presence or absence of L. rhamnosus SQ511 CFS was determined using Bradford reagent (Devi et al. 2010). In brief, 5 mL of overnight-grown cells (107 cells/mL) was harvested at 8000 rpm, washed thoroughly thrice with normal saline (0.85% NaCl), and finally suspended in 1 mL of saline in the presence of L. rhamnosus SQ511 CFS. A control without L. rhamnosus SQ511 CFS treatment was also established under the same experimental conditions. The protein release was quantified at different time points (0, 30, 60, and 120 min).
To confirm the damaging effect of L. rhamnosus SQ511 CFS on S. enteritidis ATCC10376 cell membrane, the supernatant of L. rhamnosus SQ511 CFS-treated bacterial suspension was subjected to SDS-PAGE (Devi et al. 2010). In brief, the bacterial cells were grown to OD 2.0 and then harvested at 10,000 rpm for 5 min at room temperature, and the bacterial pellet was washed twice with PBS (pH 7.4) and resuspended in PBS. Then, L. rhamnosus SQ511 CFS at the MIC was added to the cell suspension and incubated at 37 °C for 6 and 12 h. After treatment, the suspensions were centrifuged at 10,000 rpm for 10 min. Control samples were prepared in a similar manner without L. rhamnosus SQ511 CFS treatment. After exposure to L. rhamnosus SQ511 CFS for 6 and 12 h, the bacterial cells were centrifuged at 10,000×g for 5 min and the supernatant was discarded. The cell pellet was rinsed and resuspended in 0.01 mol/L phosphate buffer (Wang et al. 2015). Before SDS-PAGE analysis, the bacterial cell suspension was disrupted by ultrasonication for 10 min (200 W) and centrifuged for 3 min (10,000×g). Then, the supernatant was collected and the protein concentration was determined (Bradford 1976). Buffer (20 mL containing 250 mmol/L Tris–HCl (pH 6.8), 10% SDS, 0.5% bromophenol blue, 50% glycerin, and 5% β-mercaptoethanol) was added to 80 mL of sample with a protein concentration of approximately 3 mg/mL. The mixture was boiled for 5 min and cooled on ice. Subsequently, 25 mL of the supernatant of each sample was collected for SDS-PAGE analysis. After electrophoresis, the gel was stained with Coomassie brilliant blue R-250 and then decolorized to obtain the separated protein bands.
Scanning electron microscopy
The potential effect of L. rhamnosus SQ511 CFS on the cell morphology of S. enteritidis ATCC10376 was determined using scanning electron microscopy (SEM) (Khan and Kang 2016). S. enteritidis ATCC10376 was incubated with or without L. rhamnosus SQ511 CFS for 4 h, washed thrice with PBS, and centrifuged at 5000 rpm. The cells were collected and further fixed in glutaraldehyde (2.5 g/100 mL) for 2 h. After fixation, dehydration was performed using increasing concentrations of ethanol from 50 to 100%, and the cells were dried with liquid CO2. The dried cells were coated with gold in a sputtering coater and the samples were observed under SEM.
Motility assay
Salmonella enteritidis ATCC13076 isolates were cultured in LB broth in the presence or absence of MIC of L. rhamnosus SQ511 CFS or MRS medium (control) at 37 °C for 14 h. Then, 10 µL of the culture (108 CFU/mL) was added to the center of an LB agar plate (LB medium-containing 0.03% agar) and incubated for 12 h at 37 °C, and the zone of motility was measured (Amalaradjou et al. 2014), and the entire experiment was repeated thrice.
Determination of the effect of L. rhamnosus SQ511 CFS on the biofilm formation ability of S. enteritidis ATCC13076
The pathogenic bacterial cells were inoculated into LB broth and their biofilm formation ability and the effects of L. rhamnosus SQ511 CFS on biofilm formation were determined (Aoudia et al. 2016). In brief, 3 mL of S. enteritidis ATCC13076 (106 CFU/mL) culture was added to a test tube. To determine the effect of L. rhamnosus SQ511 CFS on biofilm formation ability of the pathogen, L. rhamnosus SQ511 CFS at the MIC was added to the test tube with bacterial culture. As a negative control, 3 mL of LB broth was added to blank test tube without bacterial culture. All the tubes were incubated for 48 h at 37 °C. To quantify biofilm formation, the tubes were gently washed thrice with 3 mL of sterile distilled water, and the attached bacteria were fixed with 3 mL of methanol for 15 min. Then, the liquid was removed and the test tubes were air-dried at room temperature for 30–45 min. Subsequently, 3 mL of 2% (v/v) crystal violet solution was added to each test tube and held at ambient temperature for 15 min. The excess stain was removed by placing the test tubes under gently running tap water. Then, 3 mL of ethanol was added to the tubes and slowly agitated for 10 min for destaining, and the OD595 values were obtained and plotted.
Effect of L. rhamnosus SQ511 on S. enteritidis ATCC13076 virulence, motility, and the expression of colonization genes
Salmonella enteritidis ATCC13076 (106 CFU/mL) was grown to early stationary phase in LB broth supplemented with SICs (sub-inhibitory concentrations) of L. rhamnosus SQ511 CFS or MRS medium at 37 °C. The RNA was isolated using the RNeasy Mini Kit, according to the manufacturer’s protocol, and quantified using a Nanodrop. The candidate genes were mainly Salmonella motility genes (flgG, fimD, and prot6E), adhesion and invasion genes (sopB and invH), type III secretory system (T3SS) effector genes (sipA, sipB, pipB, ssaV, orf245, and spvB), and cell membrane and cell wall integrity genes (hflK, lrp, ompR, and tatA). The cDNA was synthesized using the iScript Reverse Transcriptase Kit, and used as a template for Salmonella virulence, motility, and expression assays for colonization genes. Specific primers for candidate genes were selected from previous studies. The primers were custom-synthesized by Sangon Biotech (Shanghai) Co., Ltd. All primer sequences are listed in the study by Shi et al. (2019). The PCR was conducted as follows: 95 °C for 30 s, followed by 40 cycles of denaturation at 95 °C for 5 s, annealing at 58 °C for 20 s, and extension at 72 °C for 20 s. RT-qPCR was performed on StepOnePlus™ platform using SYBR Green assay under standard thermal cycling conditions. Duplicate samples were used for the assay, and the experiment was repeated thrice. The data were normalized to the endogenous control, and comparative quantification (2˗∆∆Ct) was performed to detect changes in the relative gene expression between CFS-treated and untreated control (MRS) samples (He et al. 2010).
Statistical analyses
The results are presented as means ± SEM. GraphPad Prism 5.0 software was employed to perform the calculations. Statistical significance was calculated using Student’s t test.
Results
Antimicrobial activity of L. rhamnosus SQ511
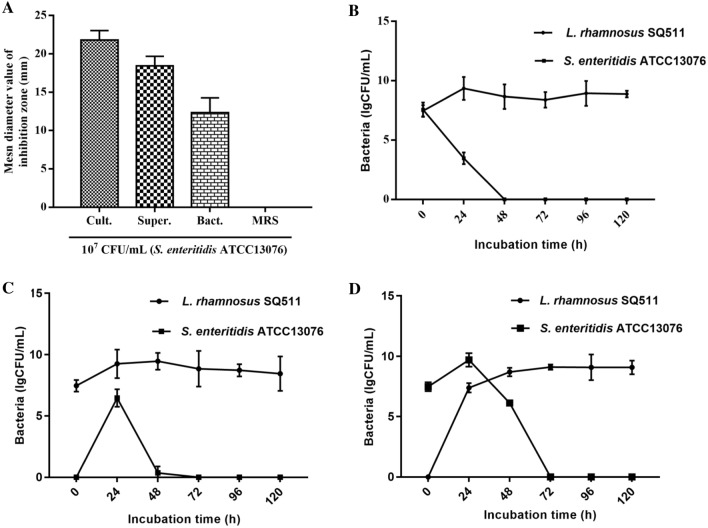
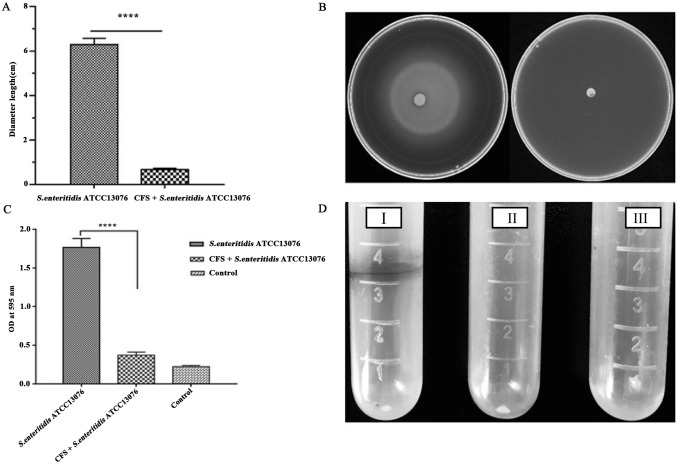
The L. rhamnosus SQ511 culture solution, supernatant, and cells exhibited antimicrobial activity against 107 CFU/mL S. enteritidis ATCC13076, with mean inhibition zone diameters of 21.82, 18.45, and 12.34 mm, respectively (Fig. 1A). Figure 1B–D shows the growth patterns of L. rhamnosus SQ511 and S. enteritidis ATCC13076 in co-culture. The viable counts of each bacterium were determined. When L. rhamnosus SQ511 and S. enteritidis ATCC13076 were inoculated simultaneously and cultured for 48 h, viable S. enteritidis ATCC13076 cells were almost completely inactivated (Fig. 1B). In contrast, when S. enteritidis ATCC13076 was inoculated after 24 h of L. rhamnosus SQ511 inoculation, the survival rate of S. enteritidis ATCC13076 cells at 48 h of co-culture was 0.6 CFU/mL, and almost all S. enteritidis ATCC13076 cells were inactivated at 72 h of culture (Fig. 1C). When S. enteritidis ATCC13076 cells were inoculated 24 h prior to L. rhamnosus SQ511 inoculation, the number of S. enteritidis ATCC13076 cells was 9.7 and 6.13 CFU/mL at 24 and 48 h of culture, respectively; however, co-culture for 72 h inactivated almost all S. enteritidis ATCC13076 cells (Fig. 1D).
Fig. 1.
A Antimicrobial activity of L. rhamnosus SQ511 against S. enteritidis ATCC13076. Mean inhibition zone diameter in mm recorded after 12 h of incubation. Bars represent means ± standard errors (SEs) of three independent experiments. B Strains L. rhamnosus SQ511 and S. enteritidis ATCC13076 were inoculated at the same time and cultured for 120 h. C L. rhamnosus SQ511 was inoculated for 24 h before S. enteritidis ATCC13076 inoculation. D S. enteritidis ATCC13076 was inoculated for 24 h before L. rhamnosus SQ511 inoculation. Data are presented as means ± SEM of three independent experiments
Live/dead bacterial cell evaluation
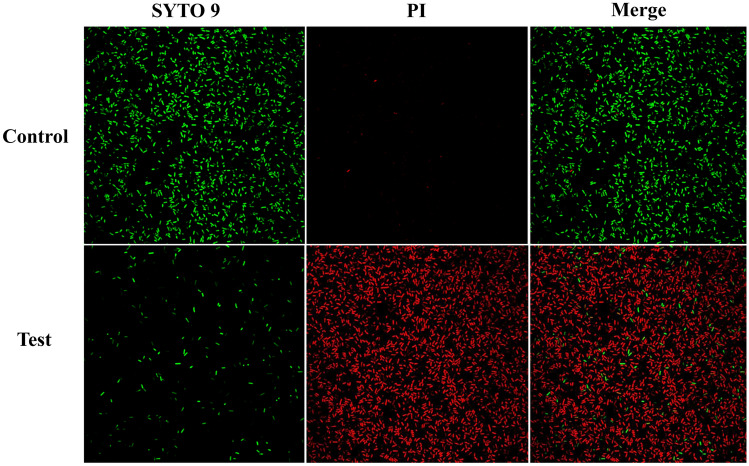
The dead cells were detected by red fluorescence owing to staining of DNA by the fluorescent dye PI, while the live cells were detected by green fluorescence owing to penetration of SYTO9 across the cell wall to stain the nucleic acid. We found high red fluorescence in S. enteritidis ATCC13076 cells treated with L. rhamnosus SQ511 CFS, indicating the bactericidal effect of L. rhamnosus SQ511 on the pathogen (Fig. 2).
Fig. 2.
Microscopic assessment of S. enteritidis ATCC13076 after treatment with CFS of L. rhamnosus SQ511. The bacterial cell death was examined via SYTO9/PI staining and fluorescence microscopy. Green fluorescence indicates live bacteria and red fluorescence denotes dead bacteria
Reduction of membrane potential and ROS generation in S. enteritidis ATCC10376 by L. rhamnosus SQ511
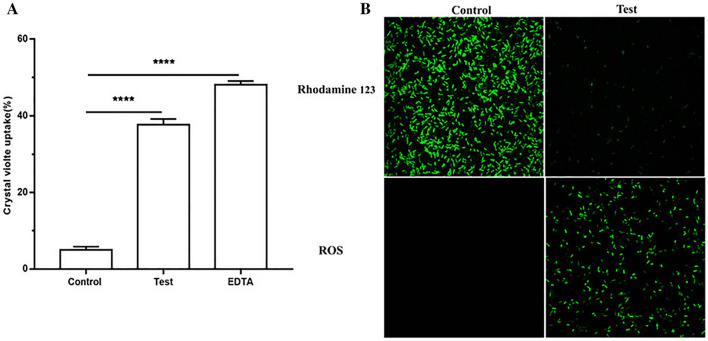
Crystal violet absorption assay was used to determine the membrane permeability of S. enteritidis ATCC13076 cells. The absorbance of crystal violet by S. enteritidis ATCC13076 cells without and with L. rhamnosus SQ511 CFS treatment was 4.987% ± 0.4967% and 37.69% ± 0.8526%, respectively (Fig. 3A). In addition, L. rhamnosus SQ511 CFS-treated S. enteritidis ATCC13076 cells presented reduced fluorescence intensity of rhodamine 123 after 4 h (Fig. 3B), indicating that L. rhamnosus SQ511 had an adverse effect on the bacterial membrane potential. Furthermore, the effect of L. rhamnosus SQ511 CFS on oxidative stress in S. enteritidis ATCC13076 was determined using H2DCFDA staining to detect ROS accumulation (Fig. 3B). In S. enteritidis ATCC13076 cells, ROS are deacetylated by esterases and H2DCFDA staining produces a visible green color under the fluorescence microscope. Treatment of S. enteritidis ATCC13076 cells with L. rhamnosus SQ511 CFS for 4 h increased the ROS levels, indicating that L. rhamnosus SQ511 CFS induced oxidative stress in S. enteritidis ATCC13076 cells.
Fig. 3.
A Crystal violet uptake of S. enteritidis ATCC13076 treated with the CFS of L. rhamnosus SQ511. EDTA (0.25 M) was used as a positive control. The data are represented as means ± SEM of three independent experiments, ****P < 0.0001, when compared with control. B Effect of CFS of L. rhamnosus SQ511 on S. enteritidis ATCC13076 membrane potential and ROS generation detected by H2DCFDA staining. The bacterial cells were incubated with or without CFS of L. rhamnosus SQ511 for 4 h, stained with rhodamine 123 and H2DFFDA, and observed under laser confocal microscopy (×100)
Effect of CFS of L. rhamnosus SQ511 on cellular materials release from S. enteritidis ATCC10376
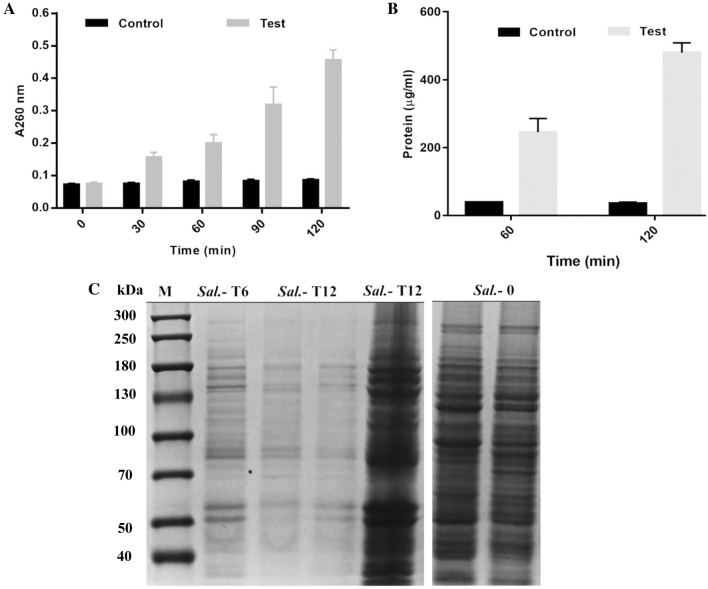
The CFS of L. rhamnosus SQ511 showed potential to disrupt the cell membrane integrity of S. enteritidis ATCC13076, leading to the release of cellular materials. As depicted in Fig. 4A, the CFS of L. rhamnosus SQ511 led to the release of intracellular materials from S. enteritidis ATCC13076 (based on absorbance at 260 nm), whereas the controls showed no release of cellular contents. Simultaneously, protein release from S. enteritidis ATCC13076 cells was detected using Bradford method. Protein release serially increased from 245.7 to 480.1 mg/mL in S. enteritidis ATCC13076 cells treated with L. rhamnosus SQ511 CFS at 6 and 12 h, respectively (Fig. 4B). Subsequently, the protein contents of S. enteritidis ATCC13076 cells exposed to L. rhamnosus SQ511 CFS for 6 and 12 h were analyzed by SDS-PAGE. The protein profiles of S. enteritidis ATCC13076 cells exposed to L. rhamnosus SQ511 CFS significantly differed from those of the control. The protein bands of the control were clearer and denser, while those of the cells treated with L. rhamnosus SQ511 CFS were fainter, with some bands even disappearing (Fig. 4C).
Fig. 4.
A Effects of CFS of L. rhamnosus SQ511 on the release of cellular materials from S. enteritidis ATCC13076 based on absorbance value recorded at 260 nm. B Protein release as a result of L. rhamnosus SQ511 CFS treatment. C SDS-PAGE profiles of proteins of S. enteritidis ATCC13076 treated with or without L. rhamnosus SQ511 CFS. M, molecular weight marker; Sal. Salmonella control, Sal-T Salmonella under treatment
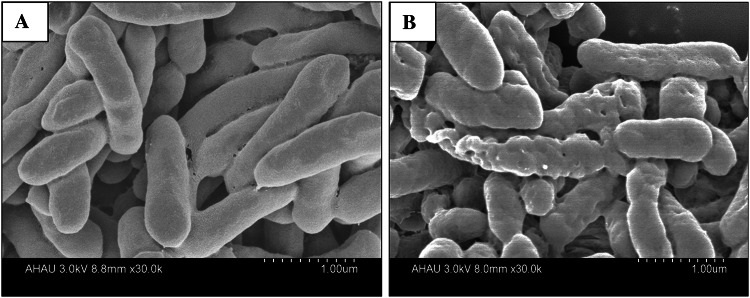
SEM
SEM was conducted to visualize the effect of L. rhamnosus SQ511 CFS on the cell morphology of S. enteritidis ATCC13076 (Fig. 5). The morphological structure of S. enteritidis ATCC13076 cells without L. rhamnosus SQ511 CFS treatment remained intact, with rod-like shape and intact cell surface (Fig. 5A). In contrast, treatment with L. rhamnosus SQ511 CFS shrunk the surface of S. enteritidis ATCC13076 cells, with the appearance of a large number of dimples (Fig. 5B).
Fig. 5.
SEM analysis of S. enteritidis ATCC13076. A Control cells without treatment showing regular and intact morphologies. B Cells treated with the CFS of L. rhamnosus SQ511 presenting distorted cell wall morphology
Effect of L. rhamnosus SQ511 on S. enteritidis ATCC13076 motility
We evaluated the ability of L. rhamnosus SQ511 CFS to inhibit the mobility of S. enteritidis ATCC13076. Treatment with CFS of L. rhamnosus SQ511 significantly (P < 0.05) reduced S. enteritidis ATCC13076 motility, when compared with that noted in the control (untreated). As shown in Fig. 6A, L. rhamnosus SQ511 CFS reduced the zone of motility of S. enteritidis ATCC13076 to 0.6689 ± 0.03391 cm, when compared with that in the control (6.294 ± 0.1629 cm). Figure 6B illustrates the observed reduction in S. enteritidis ATCC13076 motility following treatment with CFS of L. rhamnosus SQ511.
Fig. 6.
Determination of the motility of S. enteritidis ATCC13076 and the influence of L. rhamnosus SQ511 on the biofilm formation ability of S. enteritidis ATCC13076. A Effect of inhibitory concentrations of L. rhamnosus SQ511 supernatants on S. enteritidis ATCC13076 motility. Data are presented as means ± SEM. B Motility assay performed with S. enteritidis ATCC1307 treated with or without the SIC of L. rhamnosus SQ511 supernatant. C Biofilm formation of S. enteritidis ATCC13076 was assessed by crystal violet staining. D Inhibitory activity of L. rhamnosus SQ511 against S. enteritidis ATCC13076 biofilm formation. 1. S. enteritidis ATCC13076; 2. Addition of MIC of L. rhamnosus SQ511 CFS; 3. Control, LB medium
Effect of L. rhamnosus SQ511 on S. enteritidis ATCC13076 biofilm formation
The results obtained revealed the potent inhibitory effect of L. rhamnosus SQ511 CFS on the biofilm formation ability of S. enteritidis ATCC13076 (Fig. 6C), which was confirmed by a significant reduction in OD values (P < 0.05), whereas the control cells (untreated) showed strong biofilm formation ability (Fig. 6D).
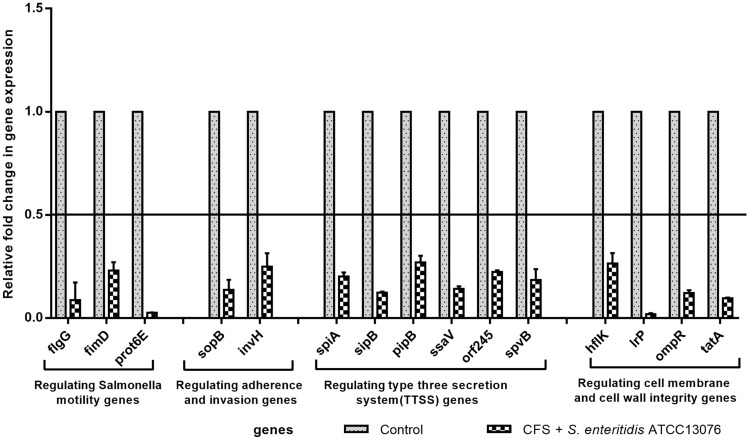
Effect of L. rhamnosus SQ511 on the expression of genes critical for virulence, motility, and adherence of S. enteritidis ATCC13076
To understand the underlying molecular mechanism of the effect of L. rhamnosus SQ511 CFS on S. enteritidis ATCC13076, real-time quantitative PCR was used to evaluate the differential expression of genes associated with virulence, motility, and adherence of the pathogen. When compared with the untreated control, the CFS of L. rhamnosus SQ511 significantly downregulated the expression of motor-related, adhesion, invasion, T3SS, and cell membrane and cell wall integrity genes in S. enteritidis ATCC13076 (P < 0.05; Fig. 7).
Fig. 7.
Effect of L. rhamnosus SQ511 CFS on the expression of motility genes, adhesion genes, and virulence genes of S. enteritidis ATCC13076
Discussion
Although many L. rhamnosus strains have been reported to have broad-spectrum antibacterial properties (Barbieri et al. 2017; Valente et al. 2019), its underlying mechanism of action against pathogenic bacteria is not fully understood. To comprehend the action of L. rhamnosus against pathogens in vitro, we evaluated L. rhamnosus SQ511 isolated from chicken feces from different perspectives for its potential use as probiotics and to determine its possible antibacterial mechanism against S. enteritidis ATCC13076. The study, based on inhibition zone and co-culture experiments, showed that L. rhamnosus SQ511 could inhibit the growth of S. enteritidis ATCC13076, which may be related to the low pH and antimicrobial compounds produced by L. rhamnosus SQ511 (Benavides et al. 2016). Previous studies have shown that both acidic environment and pressure induction in the outer membrane may affect the survival of pathogens (Delley et al. 2015). In the present study, L. rhamnosus SQ511 culture, CFS, and cells showed antagonistic activity against S. enteritidis ATCC13076. These results indicated that the antibacterial activity of L. rhamnosus SQ511 may be related to the bacterium itself, in addition to the metabolic or low pH conditions. It has been reported that an antagonistic effect exists between two different bacteria (Popova et al. 2012). Effective antibacterial compounds must penetrate or destroy the plasma membrane of the pathogen (Ibrahim et al. 2000). In the present study, the CFS of L. rhamnosus SQ511 disrupted S. enteritidis ATCC13076 cell membrane, resulting in the release of cellular contents, as evidenced by the absorbance values measured at 260 nm. Antibacterial agents mainly rupture the bacterial membrane by membrane decomposition and ROS production. In the present study, we observed an increase in crystal violet absorption in S. enteritidis ATCC13076 cells treated with L. rhamnosus SQ511 CFS, which indicated an increase in the membrane leakage, altering the permeability of the cell membrane of the pathogen. In addition, depolarization of S. enteritidis ATCC13076 cell membrane was verified using the fluorescent dye rhodamine 123. During initiation of the mechanisms of cell death, ROS production is known to play a crucial role in commencing cessation of the cell (Khan et al. 2017). In the present study, we confirmed an increase in the level of ROS in S. enteritidis ATCC13076 cells treated with L. rhamnosus SQ511 CFS, which revealed that the S. enteritidis ATCC13076 cells were under high oxidative stress in response to L. rhamnosus SQ511 CFS treatment. Previous studies have indicated that high oxidative stress is closely related to adverse effects on cellular components such as DNA and proteins (Warnes et al. 2012).
SEM can be used to observe the morphological changes and membrane structure of treated cells, and has been employed to examine membranes damaged by the antimicrobial peptide temporin L (Mangoni et al. 2004). In the present study, SEM analysis showed changes in the morphology of S. enteritidis ATCC13076 cells treated with L. rhamnosus SQ511 CFS, and the cell surface shrunk with the appearance of a large number of membrane pores. It has been reported that antimicrobial peptides can disintegrate cell membranes (Killian and von Heijne 2000). Similar morphological changes have also been observed in Escherichia coli (Arakha et al. 2015), and high levels of blistering and dimple formation have been noted in E. coli treated with a cationic antimicrobial peptide (i.e., gramicidin) (Hartmann et al. 2010). However, it is not clear whether membrane damage is the only mode of action of L. rhamnosus SQ511 CFS, and DNA and cellular proteins may also be affected in addition to membranes. Proteins play an important role in the life activities of bacterial cells. In the present study, CFS of L. rhamnosus SQ511 enhanced the leakage of proteins through S. enteritidis ATCC13076 cell membrane, significantly disrupting the cells. When S. enteritidis ATCC13076 was exposed to CFS of L. rhamnosus SQ511 for 120 min, similar to the above-mentioned protein content leakage, the protein content in the supernatant was higher than that of the control group, and the soluble protein SDS-PAGE of S. enteritidis ATCC13076 further confirmed CFS of L. rhamnosus SQ511 has a significant effect on bacterial proteins by disrupting or inhibiting the synthesis of S. enteritidis ATCC13076 resulting in its death. Similar antibacterial mechanism of LAB has also been reported against S. enteritis, E. coli, and Listeria sp. (Wang et al. 2015).
Salmonella typhimurium causes intestinal infection by adhering and invading intestinal epithelial cells, a process involving multiple virulence genes. The adhesion process is controlled by the gene encoding pili, while invasion is controlled by SPI-1. SPI-1 plays a key role in the pathogenesis of Salmonella spp., and encodes a T3SS and related effector proteins (Upadhyaya et al. 2013). When S. typhimurium is affected by the environment, the expression of these virulence genes will also alter, leading to changes in their adhesion and invasion abilities, which affect their pathogenicity. It has been found that probiotics affect the virulence of S. typhimurium. The cell-free fermentation supernatant of Lactobacillus reuteri S5 has been reported to significantly reduce the adhesion and invasion ability of S. enteritidis ATCC13076 in vitro (Shi et al. 2019). Analysis of the bacteriostatic mechanism of probiotics at the genetic level indicated that the changes in adhesion and invasiveness revealed by sopB and invH genes were not comprehensive. Based on the findings of previous research, in the present study, adhesion-related flagellar genes and SPI-1 invasion related genes were examined. The results indicated that L. rhamnosus SQ511 significantly downregulated S. enteritidis ATCC13076 flagellar genes, fimD, flgG, and prot6E, invasion genes, sopB and invH, and effector protein encoding genes. In addition, co-culture of L. rhamnosus SQ511 with S. enteritidis ATCC13076 significantly inhibited the growth of S. enteritidis ATCC13076. These findings indicated that L. rhamnosus SQ511 not only inhibits S. enteritidis ATCC13076 growth, but also reduces the expression of pathogenic genes and correspondingly weakens the ability of the pathogen to adhere and invade the host cells.
The CFS of L. rhamnosus SQ511 significantly inhibited the motility and biofilm formation of S. enteritidis ATCC13076. Previous studies have shown that the decrease in motility of Salmonella spp. may be owing to the structural or functional defects of the bacterial motility apparatus (Bayoumi and Griffiths 2012; Lorkowski et al. 2014). Similarly, the present study also found that the CFS of L. rhamnosus SQ511 inhibited the flagellar gene expression of S. enteritidis ATCC13076. Many pathogenic bacteria have the ability to form biofilms, which can protect the cells from the effects of antibacterial agents (Kim et al. 2013). As the formation of bacterial biofilms poses a major threat to human health, it is of remarkable significance to inhibit or destroy the biofilm formed by pathogenic bacteria to reduce their resistance to antibacterial agents. L. rhamnosus SQ511 was found to inhibit the growth, motility, biofilm formation, and expression of virulence genes of S. enteritidis ATCC13076, which indicate its significant potential for use as a biocontrol agent.
Conclusion
Lactobacillus rhamnosus SQ511 was demonstrated to inhibit the growth, motility, biofilm formation, and expression of adhesion, invasion, and virulence genes of S. enteritidis ATCC13076, as well as to disrupt the integrity of the pathogen’s cell membrane, resulting in the release of intracellular substances (DNA and proteins). The CFS of L. rhamnosus SQ511 had an adverse effect on the electrical potential of S. enteritidis ATCC13076. In addition, this study is the first to determine that L. rhamnosus SQ511 CFS induced oxidative stress involving ROS. Thus, L. rhamnosus SQ511 isolated from chicken feces has the potential for use as a biocontrol agent against S. enteritidis infection.
Acknowledgements
We thank Anhui Key Laboratory for Biodiversity Research and Ecological Protection in Southwest Anhui for its platform support.
Author contributions
LJ: conceived and designed the experiments; JS: performed the experiments; LY and HJ: analyzed the data; LY contributed to reagents/materials/analysis tools; and SS and DQ: wrote the manuscript. All authors have critically read and contributed to the manuscript and approved the final version.
Funding
This work was supported by the Anhui Natural Science Foundation (Grant No. 2108085QC138), Key Research and Development Plan of Anhui Province (Grant No. 202104f06020024), Open Fund Project of Anhui Key Laboratory for Biodiversity Research and Ecological Protection in Southwest Anhui (Grant No. Wy2021001), and National Entrepreneurship Practice Project (Grant No. 202110732044x, S202110372099).
Declarations
Conflict of interest
The authors declare no conflict of interest.
Contributor Information
Shuiqin Shi, Email: shishuiqin0511@163.com.
Li Gong, Email: 158191448@qq.com.
Hao Yu, Email: 3485137216@qq.com.
Guangyu He, Email: 2910604600@qq.com.
Jingjing Zhang, Email: 2953005326@qq.com.
Yu Han, Email: 2687041157@qq.com.
Yannan Liu, Email: 2742783579@qq.com.
Jie Hu, Email: 2423799422@qq.com.
Jinsheng Dong, Email: 2164799751@qq.com.
Jia Liu, Email: 1825443884@qq.com.
Kai Zhao, Email: anhui0511@126.com.
Duoqi Zhou, Email: Duoqizhou@163.com.
References
- Adetoye A, Pinloche E, Adeniyi BA, Ayeni FA. Characterization and anti-salmonella activities of lactic acid bacteria isolated from cattle faeces. BMC Microbiol. 2018 doi: 10.1186/s12866-018-1248-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afolayan AO, Ayeni FA, Ruppitsch W. Antagonistic and quantitative assessment of indigenous lactic acid bacteria in different varieties of Ogi against gastrointestinal pathogens. Pan Afr Med J. 2017;27:22–22. doi: 10.11604/pamj.2017.27.22.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alali WQ, Hofacre CL. Preharvest food safety in broiler chicken production. Microbiol Spectr. 2016;4:4. doi: 10.1128/microbiolspec.PFS-0002-2014. [DOI] [PubMed] [Google Scholar]
- Allen HK, Levine UY, Looft T, Bandrick M, Casey TA. Treatment, promotion, commotion: antibiotic alternatives in food-producing animals. Trends Microbiol. 2013;21(3):114–119. doi: 10.1016/j.tim.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Amalaradjou MAR, Kim KS, Venkitanarayanan K. Sub-inhibitory concentrations of trans-cinnamaldehyde attenuate virulence in Cronobacter sakazakii in vitro. Int J Mol Sci. 2014;15(5):8639–8655. doi: 10.3390/ijms15058639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andino A, Hanning I. Salmonella enterica: survival, colonization, and virulence differences among serovars. Sci World J. 2015 doi: 10.1155/2015/520179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoudia N, Rieu A, Briandet R, Deschamps J, Chluba J, Jego G, Garrido C, Guzzo J. Biofilms of Lactobacillus plantarum and Lactobacillus fermentum: effect on stress responses, antagonistic effects on pathogen growth and immunomodulatory properties. Food Microbiol. 2016;53:51–59. doi: 10.1016/j.fm.2015.04.009. [DOI] [PubMed] [Google Scholar]
- Arakha M, Saleem M, Mallick BC, Jha S. The effects of interfacial potential on antimicrobial propensity of ZnO nanoparticle. Sci Rep. 2015 doi: 10.1038/srep09578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri N, Herrera M, Salva S, Villena J, Alvarez S. Lactobacillus rhamnosus CRL1505 nasal administration improves recovery of T-cell mediated immunity against pneumococcal infection in malnourished mice. Benef Microbes. 2017;8(3):393–405. doi: 10.3920/bm2016.0152. [DOI] [PubMed] [Google Scholar]
- Bayoumi MA, Griffiths MW. In vitro inhibition of expression of virulence genes responsible for colonization and systemic spread of enteric pathogens using Bifidobacterium bifidum secreted molecules. Int J Food Microbiol. 2012;156(3):255–263. doi: 10.1016/j.ijfoodmicro.2012.03.034. [DOI] [PubMed] [Google Scholar]
- Benavides AB, Ulcuango M, Yepez L, Tenea GN. Assessment of the in vitro bioactive properties of lactic acid bacteria isolated from native ecological niches of Ecuador. Rev Argent Microbiol. 2016;48(3):236–244. doi: 10.1016/j.ram.2016.05.003. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Chousalkar K, Gast R, Martelli F, Pande V. Review of egg-related salmonellosis and reduction strategies in United States, Australia, United Kingdom and New Zealand. Crit Rev Microbiol. 2018;44(3):290–303. doi: 10.1080/1040841x.2017.1368998. [DOI] [PubMed] [Google Scholar]
- Delley M, Bruttin A, Richard M, Affolter M, Rezzonico E, Brueck WM. In vitro activity of commercial probiotic Lactobacillus strains against uropathogenic Escherichia coli. Fems Microbiol Lett. 2015 doi: 10.1093/femsle/fnv096. [DOI] [PubMed] [Google Scholar]
- Devi KP, Nisha SA, Sakthivel R, Pandian SK. Eugenol (an essential oil of clove) acts as an antibacterial agent against Salmonella typhi by disrupting the cellular membrane. J Ethnopharmacol. 2010;130(1):107–115. doi: 10.1016/j.jep.2010.04.025. [DOI] [PubMed] [Google Scholar]
- Gast RK, Guraya R, Jones DR, Anderson KE, Karcher DM. Colonization of internal organs by Salmonella enteritidis in experimentally infected laying hens housed in enriched colony cages at different stocking densities. Poult Sci. 2016;95(6):1363–1369. doi: 10.3382/ps/pew037. [DOI] [PubMed] [Google Scholar]
- Hartmann M, Berditsch M, Hawecker J, Ardakani MF, Gerthsen D, Ulrich AS. Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLa as revealed by transmission and scanning electron microscopy. Antimicrob Agents Chemother. 2010;54(8):3132–3142. doi: 10.1128/aac.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey PC, Watson M, Hulme S, Jones MA, Lovell M, Berchieri A, Jr, Young J, Bumstead N, Barrow P. Salmonella enterica Serovar Typhimurium colonizing the lumen of the chicken intestine grows slowly and upregulates a unique set of virulence and metabolism genes. Infect Immun. 2011;79(10):4105–4121. doi: 10.1128/iai.01390-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He GZ, Tian WY, Qian N, Cheng AC, Deng SX. Quantitative studies of the distribution pattern for Salmonella enteritidis in the internal organs of chicken after oral challenge by a real-time PCR. Vet Res Commun. 2010;34(8):669–676. doi: 10.1007/s11259-010-9438-6. [DOI] [PubMed] [Google Scholar]
- Ibrahim HR, Sugimoto Y, Aoki T. Ovotransferrin antimicrobial peptide (OTAP-92) kills bacteria through a membrane damage mechanism. BBA-Gen Subjects. 2000;1523(2–3):196–205. doi: 10.1016/s0304-4165(00)00122-7. [DOI] [PubMed] [Google Scholar]
- Jouany JP, Morgavi DR. Use of 'natural' products as alternatives to antibiotic feed additives in ruminant production. Animal. 2007;1(10):1443–1466. doi: 10.1017/s1751731107000742. [DOI] [PubMed] [Google Scholar]
- Khan I, Kang SC. Probiotic potential of nutritionally improved Lactobacillus plantarum DGK-17 isolated from Kimchi—a traditional Korean fermented food. Food Control. 2016;60:88–94. doi: 10.1016/j.foodcont.2015.07.010. [DOI] [Google Scholar]
- Khan I, Bahuguna A, Kumar P, Bajpai VK, Kang SC. Antimicrobial potential of carvacrol against uropathogenic Escherichia coli via membrane disruption, depolarization, and reactive oxygen species generation. Front Microbiol. 2017 doi: 10.3389/fmicb.2017.02421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian JA, von Heijne G. How proteins adapt to a membrane-water interface. Trends Biochem Sci. 2000;25(9):429–434. doi: 10.1016/s0968-0004(00)01626-1. [DOI] [PubMed] [Google Scholar]
- Kim S, Bang J, Kim H, Beuchat LR, Ryu J-H. Inactivation of Escherichia coli O157:H7 on stainless steel upon exposure to Paenibacillus polymyxa biofilms. Int J Food Microbiol. 2013;167(3):328–336. doi: 10.1016/j.ijfoodmicro.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Lorkowski M, Felipe-Lopez A, Danzer CA, Hansmeier N, Hensel M. Salmonella enterica invasion of polarized epithelial cells is a highly cooperative effort. Infect Immun. 2014;82(6):2657–2667. doi: 10.1128/iai.00023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangoni ML, Papo N, Barra D, Simmaco M, Bozzi A, Di Giulio A, Rinaldi A. Effects of the antimicrobial peptide temporin L on cell morphology, membrane and viability of Escherichia coli. Biochem J. 2004;380:859–865. doi: 10.1042/bj20031975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova M, Molimard P, Courau S, Crociani J, Dufour C, Le Vacon F, Carton T. Beneficial effects of probiotics in upper respiratory tract infections and their mechanical actions to antagonize pathogens. J Appl Microbiol. 2012;113(6):1305–1318. doi: 10.1111/j.1365-2672.2012.05394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan K, Shi Z, Ricke SC. Current aspects of Salmonella contamination in the US poultry production chain and the potential application of risk strategies in understanding emerging hazards. Crit Rev Microbiol. 2017;43(3):370–392. doi: 10.1080/1040841x.2016.1223600. [DOI] [PubMed] [Google Scholar]
- Rohde A, Hammerl JA, Al Dahouk S. Detection of foodborne bacterial zoonoses by fluorescence in situ hybridization. Food Control. 2016;69:297–305. doi: 10.1016/j.foodcont.2016.05.008. [DOI] [Google Scholar]
- Robertson J, McGoverin C, Vanholsbeeck F, Swift S. Optimisation of the protocol for the LIVE/DEAD® bacLightTM bacterial viability kit for rapid determination of bacterial load. Front Microbiol. 2019;10:801. doi: 10.3389/fmicb.2019.00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Qi Z, Sheng T, Tu J, Shao Y, Qi K. Antagonistic trait of Lactobacillus reuteri S5 against Salmonella enteritidis and assessment of its potential probiotic characteristics. Microb Pathog. 2019;137:103773. doi: 10.1016/j.micpath.2019.103773. [DOI] [PubMed] [Google Scholar]
- Umaraw P, Prajapati A, Verma AK, Pathak V, Singh VP. Control of campylobacter in poultry industry from farm to poultry processing unit: a review. Crit Rev Food Sci Nutr. 2017;57(4):659–665. doi: 10.1080/10408398.2014.935847. [DOI] [PubMed] [Google Scholar]
- Upadhyaya I, Upadhyay A, Kollanoor-Johny A, Darre MJ, Venkitanarayanan K. Effect of plant derived antimicrobials on Salmonella enteritidis adhesion to and invasion of primary chicken oviduct epithelial cells in vitro and virulence gene expression. Int J Mol Sci. 2013;14(5):10608–10625. doi: 10.3390/ijms140510608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente GLC, Acurcio LB, Freitas LPV, Nicoli JR, Silva AM, Souza MR, Penna CFAM. Short communication: In vitro and in vivo probiotic potential of Lactobacillus plantarum B7 and Lactobacillus rhamnosus D1 isolated from Minas artisanal cheese. J Dairy Sci. 2019;102(7):5957–5961. doi: 10.3168/jds.2018-15938. [DOI] [PubMed] [Google Scholar]
- Wang C, Chang T, Yang H, Cui M. Antibacterial mechanism of lactic acid on physiological and morphological properties of Salmonella Enteritidis, Escherichia coli and Listeria monocytogenes. Food Control. 2015;47:231–236. doi: 10.1016/j.foodcont.2014.06.034. [DOI] [Google Scholar]
- Warnes SL, Caves V, Keevil CW. Mechanism of copper surface toxicity in Escherichia coli O157:H7 and Salmonella involves immediate membrane depolarization followed by slower rate of DNA destruction which differs from that observed for Gram-positive bacteria. Environ Microbiol. 2012;14(7):1730–1743. doi: 10.1111/j.1462-2920.2011.02677.x. [DOI] [PubMed] [Google Scholar]