Abstract
HIV-1 pol sequences from antiretroviral therapy (ART)-naive and ART-experienced people living with HIV-1 are fundamental to understanding the genetic correlates and epidemiology of HIV-1 drug resistance (HIVDR). To assess the public availability of HIV-1 pol sequences and ART histories of the individuals from whom sequenced viruses were obtained, we performed a systematic review of PubMed and GenBank for HIVDR studies published between 2010 and 2019 that reported HIV-1 pol sequences. 934 studies met inclusion criteria, including 461 studies of ART-naive adults, 407 of ART-experienced adults, and 66 of ART-naive and ART-experienced children. Sequences were available for 317 (68·8%) studies of ART-naive individuals, 190 (46·7%) of ART-experienced individuals, and 45 (68·2%) of children. Among ART-experienced individuals, sequences plus linked ART histories were available for 82 (20·1%) studies. Sequences were available for 21 (29·2%) of 72 clinical trials. Among journals publishing more than ten studies, the proportion with available sequences ranged from 8·3% to 86·9%. Strengthened implementation of data sharing policies is required to increase the number of studies with available HIVDR data to support the enterprise of global ART in the face of emerging HIVDR.
Editorial note:
the Lancet Group takes a neutral position with respect to territorial claims in published maps and institutional affiliations.
Introduction
The ability to successfully scale up antiretroviral therapy (ART) to 25 million people living with HIV-1 worldwide is a remarkable achievement. However, the HIV-1 pandemic continues to expand, with nearly 2 million new infections occurring annually.1 Moreover, virological failure and the emergence of HIV-1 drug resistance (HIVDR) remains an ever-present threat to the successful control of the pandemic. Perhaps as many as 5 million people living with HIV-1 have already not responded to one or more ART regimens.2–5
HIV-1 pol sequences encoding reverse transcriptase, protease, and integrase obtained from ART-naive and ART-experienced individuals are fundamental to understanding the genetic correlates and epidemiology of HIVDR. In high-income countries, the results of genotypic resistance testing guide initial and subsequent choice of ART regimens for individual patients. In low-income and middle-income countries, knowledge of the most likely patterns of drug-resistance mutations associated with transmitted and acquired HIVDR are instead used to inform ART guidelines for public health programmes.
Sharing published biomedical data with the research community accelerates knowledge discovery and promotes individual and public health.6,7 Considering the importance of HIV-1 pol sequence data from ART-naive and ART-experienced individuals in defining the genetic correlates of HIVDR and the epidemiology of transmitted and acquired HIVDR, we sought to determine how often such data are being made publicly available.
We performed a systematic review of published HIVDR studies reporting HIV-1 reverse transcriptase, protease, and integrase sequences and determined the public availability of virus sequences and ART histories of the individuals from whom these virus sequences were obtained according to ART history, geographical region, journal, and whether the study was a clinical trial.
Methods
Search strategy and selection criteria
We searched PubMed and GenBank for studies of HIVDR or reporting HIV-1 pol sequences, published between Jan 1, 2010, and Dec 31, 2019. PubMed search terms are listed in the appendix (p 1). The GenBank search entailed a BLAST search using the consensus HIV-1 group M pol amino acid sequence comprising reverse transcriptase, protease, and integrase (see appendix p 1 for full search strategy).8 Retrieved sequences with the same GenBank “Author” and “Title” fields were grouped into submission sets. PubMed studies linked to a GenBank submission set were also included for analysis.
During an initial review of titles and abstracts, we excluded studies of non-group M HIV-1 pol sequences; review articles and meta-analyses not containing primary data; and studies of in-vitro susceptibility, biochemistry, mathematical modelling, and sequencing methods that lacked a description of the population from which virus sequences were obtained. A small number of studies devoted to HIV-1 quasispecies but not to HIVDR or published in the National Center for Biotechnology Information Sequence Read Archive but not in GenBank were also excluded.
For studies passing the title and abstract review, we reviewed the full text and extracted the geographical locations and ART histories of study participants, median sample year, and HIV-1 pol sequence availability. We also recorded which studies described clinical trials registered at ClinicalTrials.gov. We then excluded the following types of studies from further analysis: (1) studies for which more than half of samples were obtained before 2007; (2) studies providing no information on drug classes comprising the ART regimens received by study participants or that did not report the proportion of participants who were ART-naive or ART-experienced; and (3) studies reporting sequences from fewer than 25 individuals, unless they included previously protease inhibitor-naive individuals receiving atazanavir–ritonavir or darunavir–ritonavir, previously non-nucleoside reverse transcriptase inhibitor (NNRTI)-naive individuals receiving rilpivirine or doravirine, or previously integrase strand transfer inhibitor (INSTI)-naive individuals receiving elvitegravir, dolutegravir, or bictegravir. S-YR performed the searches of PubMed and GenBank. S-YR extracted data from all studies. SGK, MRJ, DK, and VK each extracted data from approximately a quarter of studies. Discrepancies were resolved jointly with RWS.
Data analysis
Studies meeting inclusion criteria were classified into the following categories: (1) ART-naive individuals with reverse transcriptase or protease sequences (or both) or INSTI-naive individuals with integrase sequences; (2) ART-experienced individuals with reverse transcriptase or protease sequences (or both) or INSTI-experienced individuals with integrase sequences; (3) infants and children, henceforth referred to as children, were included in a separate category because most were infected perinatally and might have been exposed to maternal ART—therefore, it would be difficult to know if their HIVDR was transmitted or acquired. Studies were also classified according to whether they reported the results of a clinical trial.
ART-experienced individuals were assigned to one or more of the following based on the regimen of ART that they were receiving: (1) WHO-recommended first-line NNRTI (nevirapine or efavirenz)-containing regimen; (2) pharmacologically boosted protease inhibitor-containing regimen (ie, boosted with ritonavir); (3) second-generation NNRTI-containing regimen; (4) INSTI-containing regimen; and (5) uncertain regimen.
Individuals receiving a WHO first-line NNRTI-containing regimen were assigned to one of five categories according to the nucleoside reverse transcriptase inhibitor (NRTI) used in combination with an NNRTI and lamivudine or emtricitabine: (1) individuals receiving a thymidine analogue (zidovudine or stavudine)-based regimen only; (2) individuals receiving a tenofovir-based regimen only; (3) individuals receiving an abacavir-based regimen only; (4) individuals receiving two or more regimens belonging to the preceding categories; and (5) individuals in studies for which the WHO first-line NNRTI regimen(s) received were not described.
Individuals receiving a protease inhibitor-containing regimen were assigned to one of five categories: (1) previously protease inhibitor-naive individuals receiving lopinavir–ritonavir; (2) previously protease inhibitor-naive individuals receiving atazanavir or atazanavir–ritonavir; (3) previously protease inhibitor-naive individuals receiving darunavir–ritonavir; (4) individuals receiving two or more protease inhibitors; and (5) individuals in studies for which the specific protease inhibitor(s) received were not described.
Individuals receiving a second-generation NNRTI-containing regimen were assigned to one of four categories: (1) previously NNRTI-naive individuals receiving rilpivirine; (2) previously NNRTI-naive individuals receiving doravirine; (3) individuals receiving a first-generation and second-generation NNRTI sequentially; and (4) individuals in studies for which a second-generation NNRTI was received, but for which previous NNRTI use was uncertain.
Individuals receiving an INSTI-containing regimen were assigned to one of six categories: (1) previously INSTI-naive individuals receiving raltegravir; (2) previously INSTI-naive individuals receiving elvitegravir; (3) previously INSTI-naive individuals receiving dolutegravir; (4) previously INSTI-naive individuals receiving bictegravir; (5) individuals receiving two or more INSTIs; and (6) individuals in studies for which INSTI history was uncertain.
Sequence availability was defined as having been submitted to GenBank or the Stanford HIV Drug Resistance Database.9 For studies with available sequences, we further evaluated whether the ART history category as described above could be linked to the individuals whose viruses were sequenced. For studies containing both ART-naive and ART-experienced individuals or containing individuals receiving different ART regimens, we reviewed the paper and Stanford HIV Drug Resistance Database to determine which sequences could be linked to ART history.
Retrieved studies were managed using a Zotero library shared among all authors. Statistical analyses were conducted with R version 3.4. This systematic review was reported in accordance with PRISMA guidelines.
Results
Search results
Searches yielded 2764 studies; 2139 were identified in PubMed and another 625 in GenBank (figure 1). 934 studies met inclusion criteria, including 461 studies of individuals who were ART-naive (or INSTI-naive and had integrase sequences), 407 of individuals who were ART-experienced (including 128 with both ART-naive and ART-experienced individuals) and 66 that included children only. 72 (7·7%) studies were registered clinical trials. The median sample year was 2010 (IQR 2008–2013). The complete list of 934 included studies is provided as a supplementary table (appendix pp 2–21).
Figure 1: Study selection and primary findings.
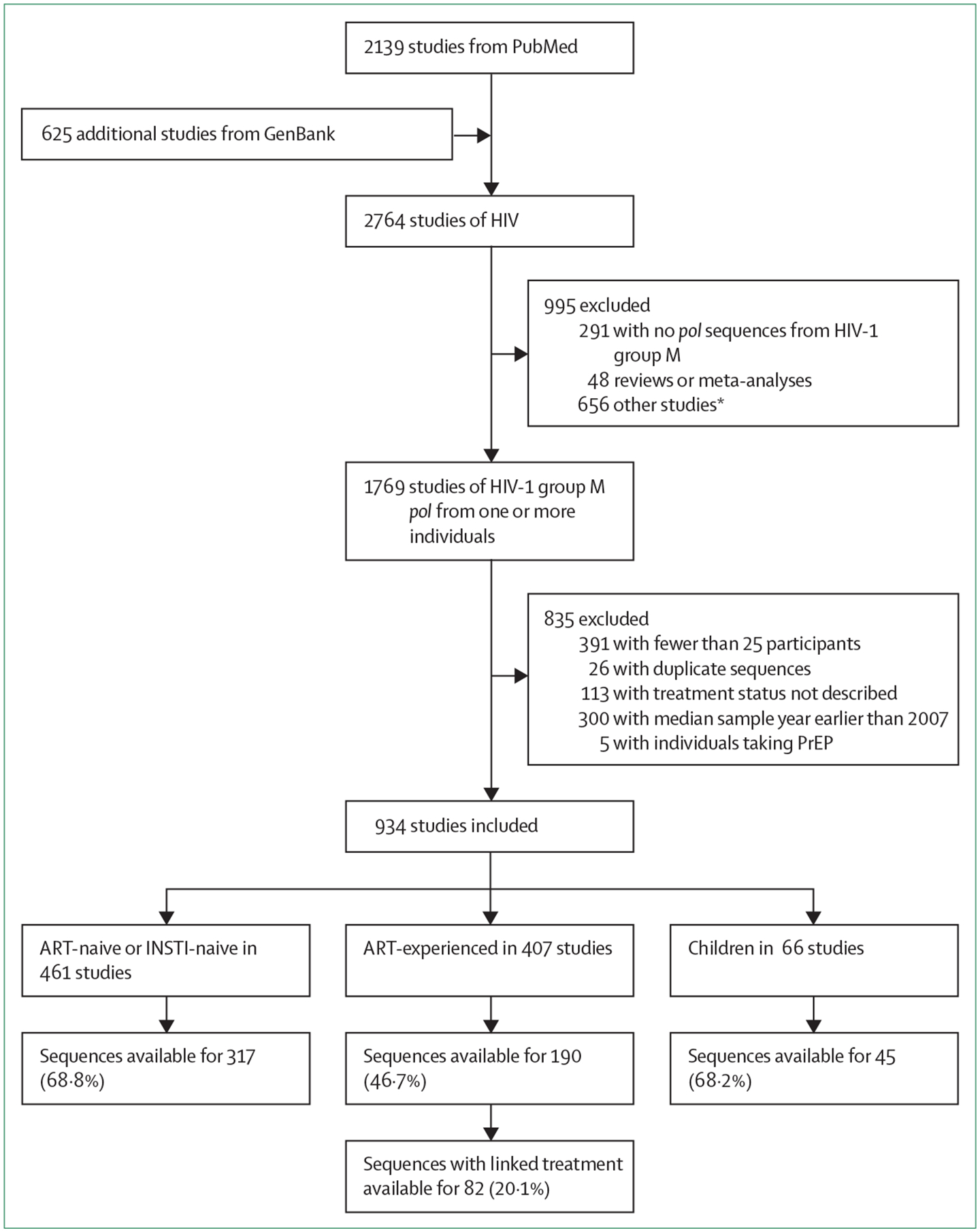
ART=antiretroviral therapy. INSTI=integrase strand transfer inhibitor. PrEP=pre-exposure prophylaxis.
*656 excluded studies comprised: studies of in-vitro susceptibility, biochemistry, mathematical modelling, and sequencing methods that lacked a description of the population from which virus sequences were obtained; studies devoted to HIV-1 quasispecies but not to HIV-1 drug resistance; and studies published in the National Center for Biotechnology Information Sequence Read Archive but not in GenBank.
Studies of ART-naive individuals
The 461 studies of ART-naive individuals included 385 studies of ART-naive individuals who underwent protease or reverse transcriptase sequencing, 40 studies of ART-naive individuals who underwent protease or reverse transcriptase sequencing and integrase sequencing, and 36 studies of INSTI-naive (but ART-experienced) individuals who underwent integrase sequencing. The median number of participants per study was 131 (IQR 61–337); the total number of individuals was 203 326. Sequences were publicly available for 317 (68·8%) studies, including 89 838 reverse transcriptase, 88 647 protease, and 8111 integrase sequences. There was no change in the proportion of studies with available sequences over time (odds ratio [OR] 0·98; 95% CI 0·92–1·06; p=0·6).
Studies of ART-experienced individuals
The 407 studies of ART-experienced individuals included (1) 126 studies of individuals receiving a WHO first-line NNRTI (nevirapine or efavirenz)-containing regimen; (2) 51 of individuals receiving a protease inhibitor-containing regimen; (3) eight of individuals receiving a second-generation NNRTI; (4) 48 of individuals receiving an INSTI-containing regimen; (5) 49 of individuals receiving more than one of the preceding types of ART; and (6) 125 of individuals receiving multiple ART regimens or having uncertain ART histories. The median number of individuals per study was 70 (IQR 34–178); the total number of individuals with sequences was 147 005. Sequences were publicly available for 190 (46·7%) studies, including 37 403 reverse transcriptase, 29 178 protease, and 2298 integrase sequences. Sequences plus linked ART histories were available for 82 (20·1%) studies, including 13 386 reverse transcriptase, 7238 protease, and 2256 integrase sequences. There was no change in the proportion of studies with available sequences over time (OR 1·00; 95% CI 0·93–1·08; p=0·9).
There were 160 studies reporting 26 792 individuals receiving a WHO first-line NNRTI regimen, including 126 in which all individuals received such a regimen and 34 containing individuals who also received other regimens: 104 included individuals receiving a thymidine analogue-containing regimen, 54 included individuals receiving a tenofovir-containing regimen, 11 included individuals receiving an abacavir-containing regimen, eight included individuals receiving more than one type of WHO first-line NNRTI regimen, and 42 included individuals with an uncertain NRTI history (table 1). Reverse transcriptase sequences were available from 101 (63·1%) of 160 studies and 17 721 (66·1%) of the 26 792 individuals. Linked sequences and treatment were available from 56 (35·0%) studies and 11 472 (42·8%) individuals.
Table 1:
Description of studies of ART-experienced individuals living with HIV-1—ART histories and HIV-1 pol sequence availability
| Number of individuals | Number of studies | Sequence and treatment availability | ||
|---|---|---|---|---|
| Number of individuals with available sequences (%) | Number of individuals with available linked sequences and treatment (%) | |||
| WHO-recommended first-line NNRTI-containing regimens (n=160 studies) * | ||||
| Zidovudine or stavudine | 12567 | 104 | 6110 (48·6%) | 4393 (35·0%) |
| Tenofovir | 7995 | 54 | 7360 (92·1%) | 6587 (82·4%) |
| Abacavir | 181 | 11 | 160 (88·4%) | 76 (42·0%) |
| ≥2 NRTIs | 416 | 8 | 416 (100%) | 416 (100%) |
| Uncertain† | 5633 | 42 | 3675 (65·2%) | 0 (0%) |
| Total | 26 792 | ·· | 17721 (66·1%) | 11472 (42·8%) |
| PI-containing regimens in previously PI-naive individuals (n=91 studies) * | ||||
| Lopinavir-ritonavir | 2182 | 43 | 1459 (66·9%) | 1170 (53·6%) |
| Atazanavir-ritonavir | 744 | 17 | 375 (50·4%) | 229 (30·8%) |
| Darunavir-ritonavir | 424 | 15 | 111 (26·2%) | 111 (26·2%) |
| ≥2 PIs or older PIs | 110 | 9 | 78 (70·9%) | 4 (3·6%) |
| Uncertain† | 4027 | 33 | 1803 (44·8%) | 0 (0%) |
| Total | 7487 | ·· | 3826 (51·1%) | 1514 (20·2%) |
| Second-generation NNRTI-containing regimens in previously NNRTI-naive individuals (n=14 studies) | ||||
| Rilpivirine | 110 | 2 | 0 (0%) | 0 (0%) |
| Etravirine | 0 | 0 | 0 (0%) | 0 (0%) |
| Doravirine | 7 | 1 | 0 (0%) | 0 (0%) |
| ≥2 NNRTIs | 1053 | 3 | 14 (1·3%) | 14 (1·3%) |
| Uncertain† | 250 | 8 | 0 (0%) | 0 (0%) |
| Total | 1420 | ·· | 14 (1·0%) | 14 (1·0%) |
| INSTI-containing regimens in previously INSTI-naive individuals (n=62 studies) * | ||||
| Raltegravir | 2818 | 34 | 1223 (43·4%) | 1199 (42·5%) |
| Elvitegravir | 334 | 11 | 224 (67·1%) | 224 (67·1%) |
| Dolutegravir | 154 | 15 | 113 (73·4%) | 113 (73·4%) |
| Bictegravir | 8 | 3 | 0 (0%) | 0 (0%) |
| ≥2 INSTIs | 209 | 14 | 158 (75·6%) | 158 (75·6%) |
| Uncertain† | 579 | 4 | 9 (1·6%) | 0 (0%) |
| Total | 4102 | ·· | 1727 (42·1%) | 1694 (41·3%) |
| Uncertain or unspecific regimens (n=125 studies) | ||||
| Total | 100143 | 125 | 15811 (15·8%) | 276 (0·3%) |
ART=antiretroviral therapy. INSTI=integrase strand transfer inhibitor. NRTI=nucleoside reverse transcriptase inhibitor. NNRTI=non-nucleoside reverse transcriptase inhibitor. PI=protease inhibitor.
The sum of the number of studies for each regimen is greater than n because some studies included individuals receiving different regimens.
Number of individuals in studies for which the number of individuals for each regimen was not described or the number of individuals with previous exposure to the drug class was not available.
There were 91 studies reporting 7487 individuals receiving a protease inhibitor-containing regimen, including 51 in which all individuals received such a regimen and 40 containing individuals who also received other regimens. Nine of the 91 studies included individuals receiving protease inhibitor monotherapy as simplification (n=7) or second-line regimens (n=2). Among the 91 studies, 43 included previously protease inhibitor-naive individuals receiving a lopinavir–ritonavir-containing regimen, 17 included previously protease inhibitor-naive individuals receiving an atazanavir-containing or atazanavir–ritonavir-containing regimen, 15 included previously protease inhibitor-naive individuals receiving a darunavir–ritonavir-containing regimen, nine included individuals receiving more than one protease inhibitor, and 33 included individuals with an uncertain protease inhibitor history (table 1). Protease sequences were available from 37 (40·7%) of 91 studies and 51·1% of the study population. Linked sequences and treatment were available from 17 (18·7%) studies and from 1514 (20·2%) individuals.
There were 14 studies reporting 1420 individuals receiving a second-generation NNRTI-containing regimen, including eight in which all individuals received such a regimen and six containing individuals who also received other regimens. Two studies included NNRTI-naive individuals receiving a rilpivirine-containing regimen, one included NNRTI-naive individuals receiving a doravirine-containing regimen, three included individuals receiving more than one NNRTI, and eight included individuals with an uncertain NNRTI history (table 1). Reverse transcriptase sequences were available from one (7·1%) study containing 14 individuals for which linked treatment was also available.
There were 62 studies reporting 4102 individuals receiving an INSTI-containing regimen, including 48 in which all individuals received such a regimen and 14 containing individuals who also received other regimens. 34 studies included previously INSTI-naive individuals receiving raltegravir, 11 included previously INSTI-naive individuals receiving elvitegravir, 15 included previously INSTI-naive individuals receiving dolutegravir, three included previously INSTI-naive individuals receiving bictegravir, 14 included individuals receiving more than one INSTI, and four included individuals with an uncertain INSTI history (table 1). Integrase sequences were available from 17 (27·4%) studies and 1727 (42·1%) individuals. Linked sequences and treatment histories were available from 15 (24·2%) studies and 1694 (41·3%) individuals.
There were 125 studies reporting individuals receiving multiple or uncertain ART regimens. They contained a median of 96 individuals (IQR 40–461). Sequences were available from 61 (48·8%) studies; sequences and linked treatment was available for five (4·0%) studies (table 1).
Children
66 studies reported sequences only from children. The median number of children per study was 85 (IQR 47–125) and the total number was 9950. The median sample year was 2010 (IQR 2009–2012). 22 studies included ART-naive children, 25 included ART-experienced children, 12 included both ART-naive and ART-experienced children, and seven included children receiving ART for prevention of mother-to-child transmission. Of 37 studies of ART-experienced children, 14 included children receiving first-generation NNRTI-containing regimens, seven included children receiving a lopinavir–ritonavir-containing regimen, nine included children receiving a first-generation NNRTI-containing or lopinavir–ritonavir-containing regimen, one included children receiving an etravirine-containing regimen, and six included children receiving an uncertain ART regimen. Sequences were available for 45 (68·2%) studies and 6289 (63·2%) children, including 6289 reverse transcriptase and 5712 protease sequences. Linked sequences and treatment were available from 32 (48·5%) studies and 4853 (48·8%) of the children, including 4853 reverse transcriptase and 4426 protease sequences. There was no change in the proportion of studies with available sequences over time (OR 0·85, 95% CI 0·69–1·05; p=0·1).
Geographical region
The distribution of studies by region was: sub-Saharan Africa (n=255 studies, 27·3% of studies), Asia (n=222, 23·8%), Europe (n=207, 22·2%), Latin America and the Caribbean (n=102; 10·9%), North America (n=52, 5·6%), Middle East (n=34, 3·6%), and former Soviet Union (n=19, 2·0%). An additional 43 studies (4·6%) included individuals from more than one of these regions. For those regions reporting the most studies, the proportions of studies with available sequences were 71·8% for sub-Saharan Africa, 70·6% for Latin America and the Caribbean, 68·5% for Asia, 38·5% for North America, and 34·8% for Europe (figure 2). The proportions with available linked sequences and treatment histories in studies containing ART-experienced individuals were 37·3% for sub-Saharan Africa, 21·7% for North America, 18·3% for Asia, 8·2% for Europe, and 8·1% for Latin America and the Caribbean.
Figure 2: Availability of sequences by geographical region.
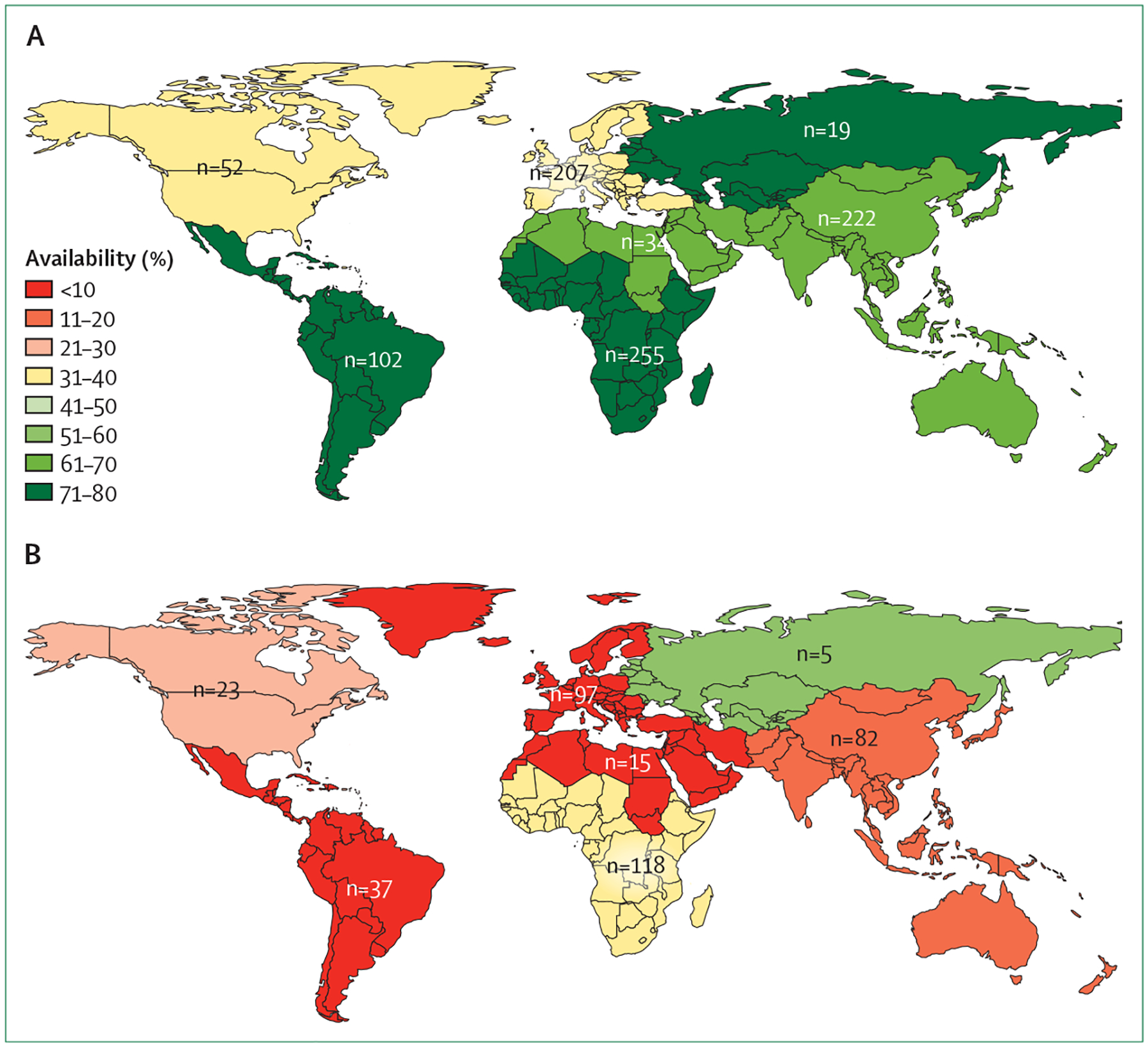
(A) Availability of sequences for all studies and (B) availability of sequences plus linked ART histories for studies of ART-experienced individuals, by geographical region. Numbers indicate total number of studies.
ART=antiretroviral therapy.
Clinical trials
72 studies described sequences from 16 574 individuals in 66 clinical trials. Sequences were available for 21 (29·2%) studies, including 15 (20·8%) with linked ART histories. These studies included 13·8% (n=56) of the 407 studies in ART-experienced individuals. 40 (56·0%) studies were sponsored by an academic institution or government agency, and 32 (44·4%) were sponsored by a pharmaceutical company (appendix pp 22–26).
Journals
The 934 studies were published in 138 journals. 17 journals published the largest number of studies (range 12–176 studies per journal) and accounted for 672 (71·9%) studies. The proportion of studies in these journals with available sequences ranged from 8·3% to 86·9% (table 2). The median proportion of studies with available sequence data in GenBank was 79·0% (range 32·3–86·9%) for nine journals that unequivocally stated that sequence submission was required and 32·8% (range 8·3–63·3%) for eight journals that made no mention of data sharing or that encouraged but did not appear to require sequence submission (p=0·003, Wilcoxon rank-sum test).
Table 2:
Proportion of studies with available sequences among journals publishing ten or more studies
| Number of studies | Number of studies with available sequences (%) | |
|---|---|---|
| AIDS Research and Human Retroviruses | 176 | 153 (86·9%) |
| PLoS One | 107 | 86 (80·4%) |
| Journal of Antimicrobial Chemotherapy | 81 | 23 (28·4%) |
| AIDS | 42 | 18 (42·9%) |
| Journal of Medical Virology | 37 | 31 (83·8%) |
| Journal of Acquired Immune Deficiency Syndromes | 33 | 20 (60·6%) |
| Journal of the International AIDS Society | 31 | 10 (32·3%) |
| Clinical Infectious Diseases | 30 | 19 (63·3%) |
| Antiviral Therapy | 23 | 6 (26·1%) |
| BMC Infectious Diseases | 18 | 14 (77·8%) |
| Archives of Virology | 15 | 12 (80%) |
| Current HIV Research | 15 | 8 (53·3%) |
| Infection, Genetics and Evolution | 14 | 11 (78.6%) |
| AIDS Research Therapy | 13 | 6 (46·2%) |
| Scientific Reports | 13 | 9 (69·2%) |
| HIV Medicine | 12 | 1 (8·3%) |
| Journal of Clinical Virology | 12 | 4 (33·3%) |
Discussion
Data sharing is a priority for all stakeholders in biomedical research, including regulatory and funding agencies, journal editors, individual researchers, and patients.6,7,10–12 In 2018, the International Committee of Medical Journal Editors announced the requirement of a data sharing statement from submissions reporting the results of clinical trials.12 In 2020, the National Institutes of Health published a new data management and sharing policy designed to foster more comprehensive data sharing (NOT-OD-21-013).13
The sharing of viral sequence data is crucial for the development of a public health response to epidemic and pandemic viral diseases, including the development of antiviral drugs and vaccines.14–17 This study is the first to benchmark data sharing in the field of HIVDR, and is probably one of the largest to benchmark data sharing in a single research area. Most previous studies evaluating data sharing empirically examined a subset of publications in a field or small number of journals.18–20
The analysis of HIV-1 pol sequences from newly diagnosed ART-naive individuals makes it possible to characterise naturally occurring genetic variability in the targets of ART in the absence of selective drug pressure and to estimate population-level prevalence of transmitted HIVDR.21 The analysis of HIV-1 pol sequences and linked treatment histories from ART-experienced individuals makes it possible to determine the genotypic correlates of HIVDR and to estimate the population-level prevalence of acquired HIVDR. The analysis of pol sequences from ART-experienced individuals is complicated because there are many different ART regimens consisting of various antiretroviral drug combinations, and because many individuals have received more than one ART regimen. In addition, the drug-resistance mutations that emerge during ART are influenced by the HIV-1 subtype.22–25
As the sampling framework of most studies is small and geographically limited, meta-analyses of sequence data from individuals with well characterised ART histories are required to identify the genotypic correlates of resistance and to evaluate emerging trends in global HIVDR. Establishing the genotypic correlates of resistance to an antiretroviral drug also requires a sufficient number of cases for which a single antiretroviral drug can be shown to select for the emergence of a drug-resistance mutation. For example, if an individual received the INSTIs raltegravir and dolutegravir, it is not possible to determine which INSTI-associated drug-resistance mutations were selected by raltegravir, which were selected by dolutegravir, and which might have arisen only as a result of exposure to both drugs.
In this systematic review of 934 studies published over a 10-year period, HIV-1 pol sequences were made publicly available for 69% of 461 studies of ART-naive individuals and 47% of 407 studies of ART-experienced individuals. Sequences plus linked ART histories were available for just 20% of studies of ART-experienced individuals. Sequence availability varied widely among the journals publishing the largest number of studies and was particularly low for clinical trials.
The proportion of studies with available sequences was biased upwards because a subset of studies was identified solely through a search of GenBank which, by definition, contains the sequences for a study. The proportion of studies with linked ART histories was also biased upwards because unless all of the individuals in a study received the same ART regimen, the ART histories were obtained from the Stanford HIV Drug Resistance Database, which recruited data through collaborative meta-analyses and individual author requests.26,27
There are major gaps in HIVDR knowledge that could be filled if the sequences and ART histories from a greater proportion of published studies were made available. For example, in a recent meta-analysis, we reported that sequences were submitted to GenBank for just two of the 63 viruses from dolutegravir-treated individuals with virological failure and INSTI-resistance mutations, making it likely that many dolutegravir-selected mutations were not reported.28 In addition, knowledge of the spectrum of mutations developing in individuals with virological failure while receiving atazanavir, darunavir, and the second-generation NNRTIs etravirine, rilpivirine, and doravirine is based on limited publicly available data.
This analysis provides several insights into how data sharing can be improved. First, the presence of unequivocal statements in the instructions to authors for a journal was associated with an increased likelihood that sequences would be made publicly available. Journal editors have substantial leverage over authors and have recently recognised the importance of data sharing policies to increase the transparency and reproducibility of published studies and to the advancement of science.29,30 However, there remains a wide gap between declared data sharing policies and their implementation.31 In addition, journal editors might not be aware of the unique data sharing requirements associated with different areas of research, such as the requirement for linked ART histories in studies of HIVDR.
Second, the low proportion of clinical trials with available data is disconcerting. HIV-1 sequences and linked ART history data in the setting of a clinical trial are particularly valuable because of the high reliability of ART histories in this setting. Moreover, most clinical trials have dedicated staff for data management, which would minimise the workload involved in making the sequence data and linked ART histories available. It has been speculated that the authors of some clinical trials might wish to publish additional findings, such as those pertaining to HIVDR, as part of a follow-up publication. However, we did not identify follow-up publications for the 72 clinical trials in this review. As a result, few of the sequences and ART histories from these clinical trials were ever made available.
Third, the lower levels of data sharing in the upper-income regions of North America and Europe compared with other geographical regions is also disconcerting. Studies from these regions are more likely than those from low-income and middle-income countries to report sequences from people receiving novel antiretroviral drugs, novel ART regimens, and multiple ART regimens.
In conclusion, published data on HIVDR are of paramount importance for making treatment decisions and guiding the selection of sequential HIV treatment regimens, especially in areas where individualised patient HIVDR testing is not feasible. This study demonstrates that despite the notion that submission of genetic sequence data is required for publication, sequences were not made publicly available for approximately half of the reviewed studies. In particular, studies reporting clinical trials and studies reported in certain journals were associated with low rates of sequence availability. Strengthened implementation of existing data sharing policies and eliminating barriers to data sharing would increase the proportion of studies with publicly available sequences and linked ART histories, resulting in improved interpretation of genotypic resistance tests and enhanced support for global ART delivery in the face of emerging HIVDR.
Supplementary Material
Acknowledgments
S-YR and RWS were supported in part by the National Institute of Allergy and Infectious Diseases of the US National Institutes of Health (award number AI136618). The funder had no role in study design, data collection, analysis, interpretation, or writing of the report. This work was previously presented, in part, at the Conference on Retroviruses and Opportunistic Infection, held virtually March 6–10, 2021.
Footnotes
Declaration of interests
We declare no competing interests.
See Online for appendix
References
- 1.Frank TD, Carter A, Jahagirdar D, et al. Global, regional, and national incidence, prevalence, and mortality of HIV, 1980–2017, and forecasts to 2030, for 195 countries and territories: a systematic analysis for the Global Burden of Diseases, Injuries, and Risk Factors Study 2017. Lancet HIV 2019; 6: e831–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boender TS, Sigaloff KCE, McMahon JH, et al. Long-term virological outcomes of first-line antiretroviral therapy for HIV-1 in low- and middle-income countries: a systematic review and meta-analysis. Clin Infect Dis 2015; 61: 1453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boender TS, Hamers RL, Ondoa P, et al. Protease inhibitor resistance in the first 3 years of second-line antiretroviral therapy for HIV-1 in sub-Saharan Africa. J Infect Dis 2016; 214: 873–83. [DOI] [PubMed] [Google Scholar]
- 4.Gupta A, Juneja S, Vitoria M, et al. Projected uptake of new antiretroviral (ARV) medicines in adults in low- and middle-income countries: a forecast analysis 2015–2025. PLoS One 2016; 11: e0164619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.UNAIDS. Global HIV & AIDS statistics—2020 fact sheet. https://www.unaids.org/en/resources/fact-sheet (accessed Jan 18, 2021).
- 6.Walport M, Brest P. Sharing research data to improve public health. Lancet 2011; 377: 537–39. [DOI] [PubMed] [Google Scholar]
- 7.Modjarrad K, Moorthy VS, Millett P, Gsell P-S, Roth C, Kieny M-P. Developing global norms for sharing data and results during public health emergencies. PLoS Med 2016; 13: e1001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Los Alamos National Laboratory. HIV sequence alignments. Los Alamos National Laboratory, 2019. https://www.hiv.lanl.gov/content/sequence/NEWALIGN/align.html (accessed Jan 18, 2021). [Google Scholar]
- 9.Rhee S-Y, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res 2003; 31: 298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilkinson MD, Dumontier M, Aalbersberg IJJ, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 2016; 3: 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Resnik DB, Morales M, Landrum R, et al. Effect of impact factor and discipline on journal data sharing policies. Account Res 2019; 26: 139–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taichman DB, Sahni P, Pinborg A, et al. Data sharing statements for clinical trials: a requirement of the International Committee of Medical Journal Editors. PLoS Med 2017; 14: e1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Institutes of Health. NOT-OD-21–013: Final NIH policy for data management and sharing. 2020; published online Oct 29. https://grants.nih.gov/grants/guide/notice-files/NOT-OD-21-013.html (accessed Jan 18, 2021).
- 14.Gostin LO, Phelan A, Stoto MA, Kraemer JD, Reddy KS. Virus sharing, genetic sequencing, and global health security. Science 2014; 345: 1295–96. [DOI] [PubMed] [Google Scholar]
- 15.Rambaut A, Holmes EC, O’Toole Á, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 2020; 5: 1403–07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill 2017; 22: 30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hadfield J, Megill C, Bell SM, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 2018; 34: 4121–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savage CJ, Vickers AJ. Empirical study of data sharing by authors publishing in PLoS journals. PLoS One 2009; 4: e7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alsheikh-Ali AA, Qureshi W, Al-Mallah MH, Ioannidis JPA. Public availability of published research data in high-impact journals. PLoS One 2011; 6: e24357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vines TH, Albert AYK, Andrew RL, et al. The availability of research data declines rapidly with article age. Curr Biol 2014; 24: 94–97. [DOI] [PubMed] [Google Scholar]
- 21.Bennett DE, Camacho RJ, Otelea D, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS One 2009; 4: e4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brenner BG, Oliveira M, Doualla-Bell F, et al. HIV-1 subtype C viruses rapidly develop K65R resistance to tenofovir in cell culture. AIDS 2006; 20: F9–13. [DOI] [PubMed] [Google Scholar]
- 23.Brenner B, Turner D, Oliveira M, et al. A V106M mutation in HIV-1 clade C viruses exposed to efavirenz confers cross-resistance to non-nucleoside reverse transcriptase inhibitors. AIDS 2003; 17: F1–5. [DOI] [PubMed] [Google Scholar]
- 24.Kolomeets AN, Varghese V, Lemey P, Bobkova MR, Shafer RW. A uniquely prevalent nonnucleoside reverse transcriptase inhibitor resistance mutation in Russian subtype A HIV-1 viruses. AIDS 2014; 28: F1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doyle T, Dunn DT, Ceccherini-Silberstein F, et al. Integrase inhibitor (INI) genotypic resistance in treatment-naive and raltegravir-experienced patients infected with diverse HIV-1 clades. J Antimicrob Chemother 2015; 70: 3080–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gregson J, Tang M, Ndembi N, et al. Global epidemiology of drug resistance after failure of WHO recommended first-line regimens for adult HIV-1 infection: a multicentre retrospective cohort study. Lancet Infect Dis 2016; 16: 565–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tzou PL, Rhee S-Y, Descamps D, et al. Integrase strand transfer inhibitor (INSTI)-resistance mutations for the surveillance of transmitted HIV-1 drug resistance. J Antimicrob Chemother 2020; 75: 170–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhee S-Y, Grant PM, Tzou PL, et al. A systematic review of the genetic mechanisms of dolutegravir resistance. J Antimicrob Chemother 2019; 74: 3135–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vasilevsky NA, Minnier J, Haendel MA, Champieux RE. Reproducible and reusable research: are journal data sharing policies meeting the mark? PeerJ 2017; 5: e3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Byrd JB, Greene AC, Prasad DV, Jiang X, Greene CS. Responsible, practical genomic data sharing that accelerates research. Nat Rev Genet 2020; 21: 615–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danchev V, Min Y, Borghi J, Baiocchi M, Ioannidis JPA. Evaluation of data sharing after implementation of the International Committee of Medical Journal Editors data sharing statement requirement. JAMA Netw Open 2021; 4: e2033972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


