Abstract
A bioluminescent reporter strain, Ralstonia eutropha ENV307(pUTK60), was constructed for the detection of polychlorinated biphenyls by inserting the biphenyl promoter upstream of the bioluminescence genes. In the presence of a nonionic surfactant, which enhances the solubility of chlorinated biphenyls, bioluminescence was induced three- to fourfold over background by biphenyl, monochlorinated biphenyls, and Aroclor 1242. The minimum detection limits for these compounds ranged from 0.15 mg/liter for 4-chlorobiphenyl to 1.5 mg/liter for Aroclor 1242.
Due to the long-term persistence of polychlorinated biphenyls (PCBs) in the environment and the continuing interest in microbial PCB biodegradation, the construction of a reporter strain for the detection of PCBs is warranted. Bioluminescent reporter strains have been used to study genetic regulation of metabolic operons (6, 15, 28) and also have potential environmental applications, such as for detecting specific organic and inorganic pollutants (2, 11, 32) and for monitoring organisms in the field (29, 35). Bioluminescent reporter strains have been developed by creating appropriate lux transcriptional fusions with promoters or operons regulated by specific chemical agents or their pathway intermediates.
The major aerobic pathway for PCB degradation is via the biphenyl operon which converts the biphenyl/chlorobiphenyl compound to benzoic acid or the corresponding chlorobenzoic acid (CBA) (10). The enzymes encoded by some biphenyl operons are also capable of cometabolizing many of the more highly chlorinated biphenyls found in PCB mixtures, such as Aroclor 1242. In this well-studied catabolic pathway, the regulatory unit may be an open reading frame (orf0) which is upstream of the bphA gene in the strains Burkholderia cepacia (16) (formerly Pseudomonas sp.) LB400 (7) and Pseudomonas pseudoalcaligenes KF707 (10, 14). In Rhodococcus sp. strain M5, the biphenyl operon is proposed to be under a two-component regulatory system (17) as is found in the closely related toluene dioxygenase pathway (21).
One of the limitations in the degradation of PCBs may be their low bioavailability due to their low aqueous solubility (34). In other applications, surfactants have been used to enhance the solubility of hydrophobic organic compounds including polycyclic aromatic hydrocarbons and PCBs (1, 23, 25, 36). In this study, nonionic surfactants were used to enhance the solubility of PCBs in order to minimize the effect of differences in aqueous solubilities of PCB congeners on the induction of the PCB operon.
Construction of bioluminescent strains.
The biphenyl/PCB-bioluminescent reporter strain was made by cloning a 2.8-kb EcoRI fragment containing the orf0-bphA1 genes from plasmid C14-15 (22) into a promoterless lux cassette (luxCDABE) from Vibrio fischeri carried on a broad-host-range vector pUCD615 (28) to create pUTK60. The orf0-bphA1 genes used in this construct were derived from the PCB-degrading strain Ralstonia eutropha ENV307 (19) (partial 16S rRNA gene sequence deposited as GenBank accession no. AF092087) (Envirogen, Inc., Lawrenceville, N.J.). The complete biphenyl operon from R. eutropha ENV307 has been cloned and expressed in other bacterial strains (19, 20). This operon is nearly identical to the biphenyl operon from B. cepacia LB400 based on restriction enzyme analysis (18, 19), the range of PCB congeners metabolized (18, 19, 20, 23), and partial sequencing of the orf0-bphA1 region (the DNA sequence is >94% similar [unpublished data]). Plasmid pUTK60 was transformed into competent Escherichia coli DH5α cells (Gibco BRL, Gaithersburg, Md.) and mated from E. coli into ENV307 by triparental mating by using plasmid pRK2073 (9) and a filter mating technique (2). Transconjugants were selected on phosphate-buffered minimal salts medium (PAS) agar plates (3, 19) containing 250 μg of kanamycin per ml and 500 μg of ampicillin per ml, with biphenyl supplied as a vapor as the sole carbon source. A control strain, ENV307(pUTK2), was created to measure potential toxic effects of the test compounds on ENV307 by mating E. coli (pUTK2) with strain ENV307 and selecting for tetracycline resistance and the ability to grow on biphenyl. Plasmid pUTK2 is an Inc Pβ plasmid with the bioluminescence genes inserted into a genetic region involved in plasmid replication and transfer (5). This plasmid produces continuous bioluminescence in several different host strains (13, 24) and has been used to measure acute chemical toxicity, which results in a reduction of bioluminescence (13, 24).
The experimental strain ENV307(pUTK60) and the control strain ENV307(pUTK2) were grown in 25 ml of PAS medium (3) with 40 mM pyruvate as the carbon source. Ampicillin (500 μg/ml) and kanamycin (250 μg/ml) were added to ENV307(pUTK60), and tetracycline (12.5 μg/ml) was added to ENV307(pUTK2). Both strains were grown in shake cultures at 26°C to an optical density at 600 nm of approximately 0.5 to 1.0. The cells were centrifuged at 12,000 × g for 10 min, washed twice with 20 ml of PAS medium, and resuspended in PAS medium to an optical density of 600 nm of approximately 1.5.
Solutions of biphenyl, monochlorinated biphenyls (CB), and the PCB mixture Aroclor 1242 were prepared as 10,000-mg/liter stock solutions in acetone. Appropriate amounts of the stock solutions were put in 200-ml bottles, the acetone was evaporated, and 100 ml of a 1% surfactant solution (polyoxyethylene 10 lauryl ether [Pol]) was added to make 800 mg of Aroclor 1242 per liter in 1% Pol and 500 mg of CB or biphenyl per liter in 1% Pol. The concentrations of CBs and Aroclor 1242 in these solutions were determined by gas chromatography-electron capture detection analysis as previously described (19). The biphenyl concentration was determined by gas chromatography-flame ionization detection analysis as previously described (20).
Solutions of 2-, 3-, and 4-CBA were prepared as 10,000-mg/liter stock solutions in dimethyl sulfoxide. Appropriate amount of the stock solutions were put into 200-ml bottles, the dimethyl sulfoxide was evaporated, and 500 ml of a 1% Pol solution prepared in PAS medium was added. The stock solutions were adjusted with 5 N NaOH to obtain a pH of 7. Sodium benzoate was prepared as a 10,000-mg/liter solution in water and then diluted to 500 mg/liter in 1% Pol. The benzoic acids were neutralized to prevent toxic responses associated with the acidic nature of these compounds (27).
A series of 1:2 dilutions in 1% Pol were made for each test compound, and 40-μl samples were added to the wells of a Microfluor B (Dynex Technologies, Chantilly, Va.) flat-bottom plate, along with a 160-μl solution of cells from either the experimental or control strain. Bioluminescence production in the sample wells was determined hourly for 6 h with a model 1450 Microbeta Plus Liquid Scintillation Counter (Wallac, Gaithersburg, Md.). Bioluminescence measurements were assayed as photon counts per minute in each sample. The level of induction at each time point was based on the comparison of bioluminescence of the bacteria in wells with test substrate versus the bioluminescence of the bacteria in wells with no test substrate. This was done to normalize the data for increases in bioluminescence due to cell growth. In each sample well, the final concentration of Pol was 0.2%. This concentration of surfactant was not toxic to strain ENV307.
Bioluminescent responses to biphenyl and CBs.
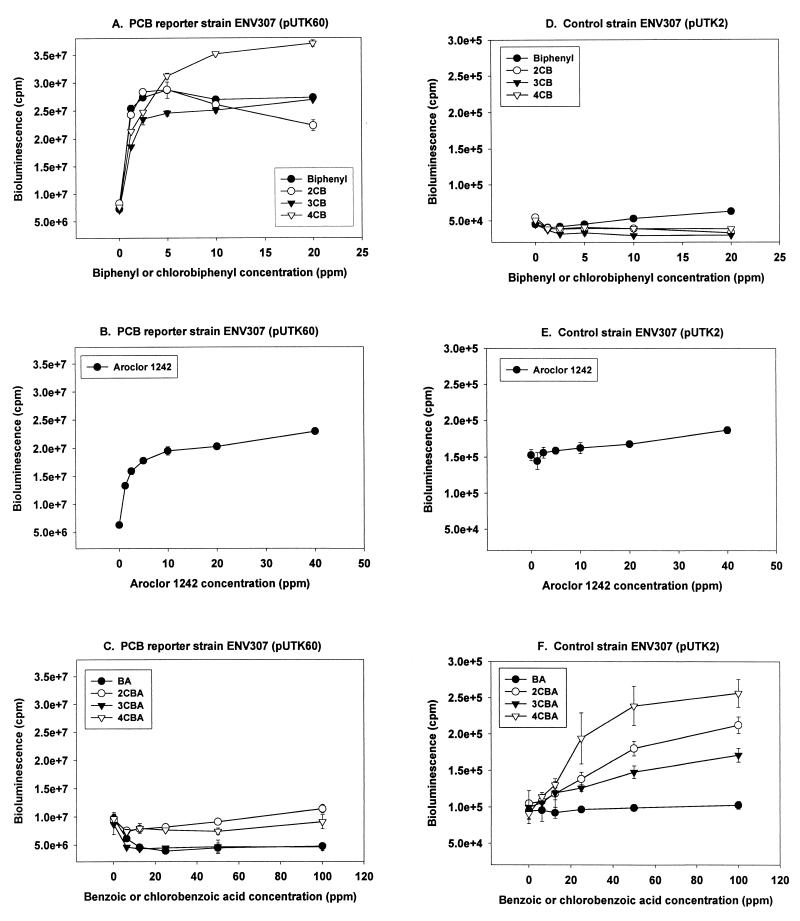
The bioluminescent responses of ENV307(pUTK60) to biphenyl and CBs were specific and sensitive (Fig. 1A; Table 1). The highest bioluminescent responses occurred when ENV307(pUTK60) was incubated with 4-CB followed by incubation with biphenyl. The high bioluminescent response to 4-CB generally began after 2 h of incubation and lasted for 2 to 3 h whereas the bioluminescent response to biphenyl generally began after 1 h and lasted for only 1 to 2 h. 4-CB and biphenyl were detected at concentrations lower than those for 2-CB, 3-CB, or Aroclor 1242, with a 1.5-fold induction in bioluminescence of ENV307(pUTK60) occurring at <0.3 mg/liter for the two compounds (Table 1). No significant bioluminescent responses were seen when control strain ENV307(pUTK2) was exposed to biphenyl and CBs (Fig. 1D).
FIG. 1.
Response of bioluminescent strain ENV307(pUTK60) and control strain ENV307(pUTK2) to biphenyl and CBs, Aroclor 1242, and benzoic acid and CBAs after 2 h of incubation. ppm, parts per million. cpm, counts per minute based on photon counting.
TABLE 1.
Maximum induction and minimum detection of bioluminescence for the PCB reporter strain ENV307(pUTK60)
| Compound | Maximum induction ± SDa | Minimum detection limit (mg/liter) ± SDb |
|---|---|---|
| Biphenyl | 4.3 ± 1.3c | 0.29 ± 0.15 |
| 2-CB | 3.6 ± 1.0c | 0.40 ± 0.3 |
| 3-CB | 3.3 ± 0.3c | 0.83 ± 0.3 |
| 4-CB | 4.8 ± 1.6d | 0.15 ± 0.1 |
| Aroclor 1242 | 3.3 ± 0.8d | 1.5 ± 0.7 |
Fold increase is the increase in bioluminescence at a substrate concentration relative to the control without substrate. These values are the averages for three or more experiments.
The minimum detection limit was determined from the linear regression of bioluminescence versus the substrate concentration from 0 to 5 mg/liter. The minimum detection value was calculated as the concentration resulting in a 1.5-fold increase. These values are the averages for three experiments.
Maximum induction was measured at 5 mg/liter.
Maximum induction was measured at 10 mg/liter.
The potential for the induction of the biphenyl operon by PCB mixtures was tested with Aroclor 1242 (Fig. 1B). ENV307(pUTK60) showed a greater-than-threefold induction of bioluminescence with exposure to 10 mg of Aroclor 1242 per liter (Table 1; Fig. 1B), whereas there was no bioluminescent response by control strain ENV307(pUTK2) in the same concentration range (Fig. 1E). This induction response is somewhat unexpected because PCB degradation is generally considered cometabolic and the growth substrates, biphenyl or CBs, were not included in the test mixture. The response of ENV307(pUTK60) to the PCB mixture Aroclor 1242 was enhanced by surfactant solubilization of the PCBs since the aqueous solubility of Aroclor 1242 is <1 mg/liter (26), and in these experiments bioluminescence increased with an Aroclor 1242 concentration of up to 10 mg/liter.
Bioluminescent responses to benzoic acid and CBAs.
The effect of benzoic acid and CBAs on the biphenyl promoter was tested because benzoic acids inhibit PCB degradation (4, 8, 12, 31, 33) (Fig. 1C). In strain ENV307(pUTK60), benzoic acid and 3-CBA reduced bioluminescence 2.5-fold at concentrations greater than 12.5 mg/liter, whereas 2-CBA and 4-CBA had no effect on the biphenyl operon (Fig. 1C). These bioluminescent responses to benzoic acid and 3-CBA were not likely to be acute toxicity responses because decreases in bioluminescence were not seen for the control strain ENV307(pUTK2) (Fig. 1F).
Summary.
The results of these experiments with the biphenyl-bioluminescent reporter strain are consistent with other findings regarding the effect of biphenyl on aerobic PCB-degrading bacteria in that a specific induction response was seen with biphenyl. Benzoic acid, the product of the biphenyl degradation pathway, was shown to repress bioluminescence in the reporter strain. This is consistent with the findings of studies which demonstrate the negative effect of benzoic acid on PCB degradation (8, 12, 31). Compared to benzoic acid and 3-CBA, 2-CBA and 4-CBA did not repress bioluminescence in the reporter strain. This result may explain why bioluminescence induction by 4-CB extended over a longer period than bioluminescence induction by biphenyl.
The bioluminescent responses of the ENV307(pUTK60) reporter strain supports the hypothesis that the promoter region and a putative regulator gene (orf0) are upstream of the bphA1 gene in the B. cepacia LB400 and P. pseudoalcaligenes KF707 biphenyl operons as proposed by other investigators (7, 14). However, the role of orf0 in the regulation of the biphenyl operon is unclear because the translated orf0 regions from these two organisms are not closely related to other known regulatory proteins and show only 80% homology to each other (14). The creation of additional bioluminescent reporter strains with smaller portions of the promoter region of the biphenyl operon may provide additional information on the regulation of the biphenyl operon. Sensor modules, such as those being created for other bioluminescent reporter strains, may eventually allow online detection of PCB contaminants (30).
Acknowledgments
This work was supported by WMERI Institute at the University of Tennessee and the Department of Energy grant no. DE-FG02-97ER62350.
We would like to thank Envirogen, Inc., Lawrenceville, N.J., for permission to use strain ENV307.
REFERENCES
- 1.Abdul A S, Gibson T L, Ang C C, Smith J C, Sobczynski R E. In situ surfactant washing of polychlorinated biphenyls and oil from a contaminated site. Groundwater. 1992;30:219–231. [Google Scholar]
- 2.Applegate B, Kelly C, Lackey L, McPherson J, Kehrmeyer S, Menn F-M, Bienkowski P, Sayler G. Pseudomonas putida B2: a tod-lux bioluminescent reporter for toluene and trichloroethylene co-metabolism. J Ind Microbiol Biotechnol. 1997;18:4–9. doi: 10.1038/sj.jim.2900334. [DOI] [PubMed] [Google Scholar]
- 3.Bopp L H. Degradation of highly chlorinated PCBs by a Pseudomonas strain LB400. J Ind Microbiol. 1986;1:23–29. [Google Scholar]
- 4.Brenner V, Arensdorf J J, Focht D D. Genetic construction of PCB degraders. Biodegradation. 1994;5:359–377. doi: 10.1007/BF00696470. [DOI] [PubMed] [Google Scholar]
- 5.Burlage R S, Bemis L A, Layton A C, Sayler G S, Larimer F. Comparative genetic organization of incompatibility group P degradative plasmids. J Bacteriol. 1990;172:6818–6825. doi: 10.1128/jb.172.12.6818-6825.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burlage R S, Sayler G S, Larimer F. Monitoring of naphthalene catabolism by bioluminescence with nah-lux transcriptional fursions. J Bacteriol. 1990;172:4749–4757. doi: 10.1128/jb.172.9.4749-4757.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erickson B D, Mondello F J. Nucleotide sequencing and transcriptional mapping of the genes encoding biphenyl dioxygenase, a multicomponent polychlorinated-biphenyl-degrading enzyme in Pseudomonas strain LB400. J Bacteriol. 1992;174:2903–2912. doi: 10.1128/jb.174.9.2903-2912.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fava F, Di Gioia D, Marchetti L, Quattroni G, Marraffa V. Aerobic mineralization of chlorobenzoates by a natural polychlorinated biphenyl-degrading mixed bacterial culture. Appl Microbiol Biotechnol. 1993;40:541–548. [Google Scholar]
- 9.Figurski D H, Helsinki D R. Replication of an origin-containing derivative of plasmid RK2 is dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furukawa K. Molecular genetics and evolutionary relationship of PCB-degrading bacteria. Biodegradation. 1994;5:289–300. doi: 10.1007/BF00696466. [DOI] [PubMed] [Google Scholar]
- 11.Heitzer A, Applegate B, Kehymeyer S, Pinkart H, Webb O F, Phelps T J, Sayler G S, White D C. Physiological considerations of environmental applications of lux reporter fusions. J Microbiol Methods. 1998;33:45–47. [Google Scholar]
- 12.Hickey W J, Searles D B, Focht D D. Enhanced mineralization of polychlorinated biphenyls in soil inoculated with chlorobenzoate-degrading bacteria. Appl Environ Microbiol. 1993;59:1194–1200. doi: 10.1128/aem.59.4.1194-1200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelly, C. J., C. A. Lajoie, A. C. Layton, and G. S. Sayler. Bioluminescent reporter bacterium for toxicity monitoring in biological wastewater treatment systems. Water Environment Res., in press.
- 14.Kimura N, Nishi A, Goto M, Furukawa K. Functional analyses of a variety of chimeric dioxygeneases constructed from two biphenyl dioxygeneases that are similar structurally but different functionally. J Bacteriol. 1997;179:3936–3943. doi: 10.1128/jb.179.12.3936-3943.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King J M H, DiGrazia P M, Applegate B, Burlage R, Sanseverino J, Dunbar P, Larimer F, Sayler G S. Rapid, sensitive bioluminescent reporter technology for naphthalene exposure and biodegradation. Science. 1990;249:778–781. doi: 10.1126/science.249.4970.778. [DOI] [PubMed] [Google Scholar]
- 16.Kumamaru T, Suenaga H, Mitsouka M, Watanabe T, Furukawa K. Enhanced degradation of polychlorinated biphenyls by directed evolution of biphenyl dioxygenease. Nat Biotechnol. 1998;16:663–666. doi: 10.1038/nbt0798-663. [DOI] [PubMed] [Google Scholar]
- 17.Labbe D, Garnon J, Lau P C K. Characterization of the genes encoding a receptor-like histidine kinase and a cognate response regulator from a biphenyl/polychlorobiphenyl-degrading bacterium Rhodococcus sp. strain M5. J Bacteriol. 1997;179:2772–2776. doi: 10.1128/jb.179.8.2772-2776.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lajoie C A, Zylstra G J, DeFlaun M F, Strom P F. Development of field application vectors for bioremediation of soils contaminated with polychlorinated biphenyls. Appl Environ Microbiol. 1993;59:1735–1741. doi: 10.1128/aem.59.6.1735-1741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lajoie C A, Layton A C, Sayler G S. Cometabolic oxidation of polychlorinated biphenyls in soil with a surfactant-based field application vector. Appl Environ Microbiol. 1994;60:2826–2833. doi: 10.1128/aem.60.8.2826-2833.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lajoie C A, Layton A C, Easter J P, Menn F-M, Sayler G S. Degradation of nonionic surfactants and polychlorinated biphenyls by recombinant field application vectors. J Ind Microbiol Biotechnol. 1997;19:252–262. doi: 10.1038/sj.jim.2900454. [DOI] [PubMed] [Google Scholar]
- 21.Lau P C K, Wang Y, Patel A, Labbe D, Bergeron H, Brousseau R, Konishi Y, Rawlings M. A bacterial basic region leucine zipper histidine kinase regulating toluene degradation. Proc Natl Acad Sci USA. 1997;94:1453–1458. doi: 10.1073/pnas.94.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Layton A C, Lajoie C A, Easter J P, Jernigan R, Sanseverino J, Sayler G S. Molecular diagnostics and chemical analysis for assessing biodegradation of polychlorinated biphenyls in contaminated soils. J Ind Microbiol. 1994;13:392–401. doi: 10.1007/BF01577225. [DOI] [PubMed] [Google Scholar]
- 23.Layton A C, Lajoie C A, Easter J P, Muccini M, Sayler G S. An integrated surfactant solubilization and PCB bioremediation process for soils. Bioremediation J. 1998;2:43–56. [Google Scholar]
- 24.Layton, A. C., B. Gregory, T. W. Schultz, and G. S. Sayler. The validation of genetically engineered bioluminescent surfactant resistant bacteria as toxicity assessment tool. Submitted for publication. [DOI] [PubMed]
- 25.Liu Z, Jacobson A M, Luthy R G. Biodegradation of naphthalene in aqueous nonionic surfactant systems. Appl Environ Microbiol. 1995;61:145–151. doi: 10.1128/aem.61.1.145-151.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacKay D, Shiu W Y, Billington J, Huang G L. Physical chemical properties of polychlorinated biphenyls. In: Mackay D, Paterson S, Eisenreich S J, Simmons M S, editors. Physical behavior of PCBs in the Great Lakes. Ann Arbor, Mich: Ann Arbor Science Publishers; 1983. pp. 59–69. [Google Scholar]
- 27.Muccini, M., A. C. Layton, G. S. Sayler, and T. W. Schultz. Ecotoxicity of metabolites of PCBs: halogenated benzoic acids to Tetrahymena. Submitted for publication.
- 28.Rogowsky P M, Close T J, Chimera J A, Shaw J J, Kado C I. Regulation of the vir genes of Agrobacterium tumefaciens plasmid pTiC58. J Bacteriol. 1987;169:5101–5112. doi: 10.1128/jb.169.11.5101-5112.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sayler G, Cox C D, Burlage R, Ripp S, Nivens D, Werner C, Ahn Y, Matrubutham U. Field application of a genetically engineered microorganism for polycyclic aromatic hydrocarbon bioremediation process monitoring and control. In: Fass R, Flashner Y, Reuveny S, editors. Novel approaches for bioremediation of organic pollution. Proceedings of the 42nd OHOLO Conference. New York, N.Y: Plenum Publishing Corp.; 1998. [Google Scholar]
- 30.Simpson M L, Sayler G S, Ripp S, Nivens D E, Applegate B M, Paulus M J, Jellison G E., Jr Bioluminescent bioreporter integrated circuits: novel whole-cell biosensors. Trends Biotechnol. 1998;16:332–338. [Google Scholar]
- 31.Sondossi M, Sylvestre M, Ahmad D. Effects of chlorobenzoate transformation on the Pseudomonas tesosteroni biphenyl and chlorobiphenyl degradation pathway. Appl Environ Microbiol. 1992;58:485–495. doi: 10.1128/aem.58.2.485-495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steinberg S M, Poziomek E J, Englemann W H, Rogers K R. A review of environmental applications of bioluminescence measurements. Chemosphere. 1995;30:2155–2197. [Google Scholar]
- 33.Stratford J, Wright M A, Reineke W, Mokross H, Havel J, Knowles C J, Robinson G K. Influence of chlorobenzoates on the utilization of chlorobiphenyls and chlorobenzoate mixtures by chlorobiphenyl/chlorobenzoate-mineralizing hybrid bacterial strains. Arch Microbiol. 1996;165:213–218. doi: 10.1007/BF01692864. [DOI] [PubMed] [Google Scholar]
- 34.Unterman R. A history of PCB biodegradation. In: Crawford R L, Crawford D L, editors. Bioremediation: principles and applications. New York, N.Y: Cambridge University Press; 1996. pp. 209–253. [Google Scholar]
- 35.Van Dyke M I, Lee H, Trevors J T. Survival of luxAB-marked Alcaligenes eutrophus H850 in PCB-contaminated soil and sediment. J Chem Technol Biotechnol. 1996;65:115–22. doi: 10.1002/(SICI)1097-4660(199602)65:2<115::AID-JCTB391>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 36.Yeom I T, Ghosh M M, Cox C C. Kinetic aspects of surfactant solubilization of soil-bound polycyclic aromatic hydrocarbons. Environ Sci Technol. 1996;30:1589–1595. [Google Scholar]