Abstract
Glioblastoma is the most frequent, as well as aggressive kind of high-grade malignant glioma. Chemoresistance is posing a significant clinical barrier to the efficacy of temozolomide-based glioblastoma treatment. By suppressing xeroderma pigmentosum group A (XPA), a pivotal DNA damage recognition protein implicated in nucleotide excision repair (NER), we devised a novel method to enhance glioblastoma therapy and alleviate temozolomide resistance. On the basis of preliminary assessment, we found that XPA dramatically increased in glioblastoma compared with normal cells and contributed to temozolomide resistance. By constructing XPA stably knockdown cells, we illustrate that XPA protects glioma cells from temozolomide-triggered reproductive cell death, apoptosis, as well as DNA repair. Besides, XPA silencing remarkably enhances temozolomide efficacy in vivo. This study revealed a crucial function of XPA-dependent NER in the resistance of glioma cells to temozolomide.
Keywords: glioblastoma, temozolomide, XPA, nucleotide excision repair
Introduction
Glioblastoma (GBM) is a very well-known and fatal kind of primary malignant brain tumor in adults, with a 1-year median survival period. Even in the best of situations, the majority of patients die within 2 years1,2. Surgical excision along with temozolomide (TMZ) chemotherapy as well as radiation are the conventional therapies for GBM, and they are more successful relative to radiation alone 3 . TMZ causes apoptosis by methylating guanine and causing DNA damage, which increases the median survival time from 12 to 15 months 4 . Most patients, however, suffer refractory illness along with tumor relapse as a result of GBM cells’ innate or acquired chemoresistance 5 . As a result, understanding the molecular mechanisms accounting for GBM cell chemoresistance is pivotal for developing more effective therapeutic approaches.
The chemotherapeutic drug TMZ is an SN1 methylating agent that can cross the blood-brain barrier 6 . It spontaneously hydrolyzes at physiological pH to make 3-methyl-(triazen-1-yl) imidazole-4-carboxamide (MTIC), which then produces 4-amino-5-imidazole carboxamide and the methyl diazonium (MDI) ion. Although MDI methylates DNA at several locations, O6-methylguanine (O6MeG) is the most important DNA lesion for treatment7,8. In gliomas, O6MeG does not immediately cause cell death; instead, it needs DNA replication coupled with mismatch repair to enable the creation of DNA double-strand breaks (DSBs)9,10. The ultimate deadly lesions are assumed to be these DSBs. TMZ resistance may be mediated by a number of mechanisms 11 , consisting of DNA methyltransferase (MGMT) along with DNA repair, and speeding up the repair of DSBs can improve GBM cells’ TMZ chemical resistance11,12. It has been reported that NHEJ (non-homologous end-joining) 13 , HR (homologous recombination) 14 , and BER (base excision repair) 15 are all involved in the formation of TMZ resistance. However, whether NER (nucleotide excision repair) is involved in TMZ resistance is still unclear.
NER is a flexible and ubiquitous repair process that can remove an extensive range of DNA helix-distorting lesions, including bulky DNA adducts16,17. More than 30 proteins make up the NER cascade, which is responsible for DNA damage detection, verification, incision, excision, gap filling, as well as ligation 18 . NER’s core protein, xeroderma pigmentosum group A (XPA), is involved in DNA damage verification as well as the mobilization of other NER proteins. DNA docking is the only biological action attributed to XPA. Damaged along with undamaged DNA strands are bound by XPA 19 . XPA works with replication protein A (RPA) to provide a scaffold for the construction and stability of the NER pre-incision complex, which organizes damaged DNA and this complex to guarantee lesions are excised properly 20 . XPA has been found to cross talk with proteins that are involved in every step of the NER process, from damage recognition through DNA synthesis 21 . XPA also cross talks with proteins that are not involved in the repair process22–24. In germ cell cancers, elevated XPA contents may be the cause of cisplatin resistance 25 . Via the activation of PARP1, XPA increases autophagy in melanoma cells to aid cisplatin resistance 26 .
Given its dual role in detecting and mobilizing other DNA repair proteins to the damaged template for NER, we speculate that XPA modulation may be pivotal in determining sensitivity to TMZ. According to analysis of publicly available database, the contents of XPA may harbor a role in TMZ resistance. We show that silencing XPA makes glioma cells more susceptible to TMZ-triggered cell death, apoptosis, and DSB repair. We established that XPA elevated glioma cell resistance to TMZ emphasizing the role of NER in TMZ-triggered DNA damage tolerance and suggests an approach for targeting this repair mechanism during TMZ-based glioma treatment.
Materials and Methods
Cell Culture and Reagents
U-87 MG, U-251, HEK-293T, the primary HUVECs (human umbilical cord endothelial cells), and the smooth muscle cells (HUASMCs) were commercially provided by the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. U-87 MG and HEK-293T, along with U-251 cells, were inoculated in the Dulbecco’s modified eagle medium (DMEM) medium enriched with 10% fetal bovine serum (FBS) along with 1% pen/strep for growth. HUVECs were inoculated in HUVEC media (R&D Systems, Minneapolis, MN, USA, CCM027) for growth. Human aortic smooth muscle cells (HASMCs) were inoculated in HASMC complete medium (Procell, Wuhan, China, Cat# CM-H081).TMZ-resistant clones (U-87 MG-R, U-251-R) were originated from the TMZ-sensitive U-87 MG or U-251 cells by culturing them with increasing doses of TMZ, as documented previously 27 . All cell lines were incubated at 37°C along with 5% CO2 settings. Cells were assessed for contamination with mycoplasma every 2 months, and we only used the mycoplasma-negative cells. TMZ, T4N5, and UCN-01 were provided by Sigma-Aldrich, USA and dispersed in dimethyl sulfoxide (DMSO), then diluted using DMEM to its final level.
Lentiviral Systems for XPA Silencing
Addgene provided the pLKO.1 purobased lentiviral vectors (containing a distinct shRNA coding sequence, packaging plasmid pCMVR8.91, as well as pMD). Packaging recombinant lentiviruses was done as documented by the manufacturer. Using LipofectamineTM 2000 Transfection Reagent, lentivirus was created by transfecting HEK-293T cells with the lentiviral vector (4 g) along with the packaging plasmids, pCMVR8.91 (4 μg), as well as pMD (0.4 μg) (Thermo Fisher Scientific, USA). The lentiviral plasmids targeting XPA were TRCN0000083194 (shXPA #1: GCATTAGAAGAAGCAAAGGAA), TRCN0000083196 (shXPA #2: CATGAGTATGGACCAGAAGAA), and pLKO.1 (scrambled shCon). U‑87 MG along with U-87 MG-R cells were inoculated with lentiviral supernatants harboring 8 μg/ml polybrene for 24 h. Afterward, we replaced the medium, followed by another 48 h of incubation. To establish the stable cell lines, puromycin (5μg/ml) was introduced 48 h post transfection. Collection of stable cells was done for western blotting to assess the efficiency of silencing, and a CCK-8 assay was adopted to assess the influence on TMZ sensitivity.
Western Blot
Lysing of cells or tumor tissues was done with the radioimmunoprecipitation assay (RIPA) buffer and span at 13,000 × g for 20 min. Fractionation of the proteins was done on the sodium dodecyl sulfate polyacrylamide gel electrophores (SDS/PAGE) gels, and subsequently blotted onto a polyvinylidene fluoride (PVDF) membrane, and inoculated with probed overnight with antibodies against mouse anti-GAPDH (1:5000; Santa Cruz, USA, sc-32233), mouse anti-XPA (1:1000, Invitrogen, USA, MA1-21460), mouse anti-XPB (1:500, Santa Cruz, USA, sc-271500), mouse anti-XPC (1:1000, Novus Biologicals, England, NB100-477), mouse anti-XPD (1:1000, Abcam, Cambridge, England, ab54676), mouse anti-XPF (1:1000, Invitrogen, USA, MA5-12054), mouse anti-XPG (1:1000, Santa Cruz, USA, sc-13563), mouse anti-ERCC1 (1:500, Santa Cruz, USA, sc-17809), rabbit anti-DDB1 (1:1000, Abcam, Cambridge, England, ab109027), mouse anti-DDB2 (1:1000, Abcam, Cambridge, England, ab51017), and rabbit anti-γH2AX (1:1000, Abcam, Cambridge, England, ab11174) at 4°C. Afterward, we inoculated the stripped membranes with a secondary antibody of goat anti-mouse or anti-rabbit IgG (1:5000, Thermo Fisher, USA) and then visualization was done with enhanced chemiluminescence.
Cell-Proliferation Assays
Cells (3 × 103) were inoculated for 12 h in the DMEM medium enriched with 10% FBS. We rinsed the cells twice in phosphate buffered saline (PBS), and then inoculated them in DMEM medium enriched with 10% FBS along with indicated inhibitors or agonists and cultured for additional 48 h. The growth rates were detected by the CCK-8 assay as documented by the manufacturer. Each experiment was performed in triplicate.
Apoptosis Determination by Flow Cytometry
Cells were inoculated with specified levels of TMZ for 48 h, and then they were prepared for assessment. Annexin V/fluorescein isothiocyanate (FITC) was employed to label the unfixed cells, followed by staining in 1 μg/ml propidium iodide (PI) prior and subsequent cytometry analysis was done. Apoptotic cells were categorized as Annexin V positive, while necrotic/late-apoptotic cells were categorized as Annexin V and PI double-positive cells. A fluorescence-activated cell sorting (FACS) flow cytometer was utilized for the flow cytometric analyses (Miltenyi, Germany). FlowJo software was adopted to analyze the data.
Reverse Transcription–Quantitative Polymerase Chain Reaction Assays
Isolation of total RNA was done with the RNeasy kit (Qiagen, Hilden, Germany, #74104). After that, cDNA was generated from the RNA with the PrimeScript RT reagent kit (TaKaRa, Shiga, Japan, #RR037A) as documented by the manufacturer. Next, we used the SYBR Green PCR Master Mix (Thermo Fisher, #4368706) to quantify the mRNA contents via quantitative polymerase chain reaction (qPCR), with GAPDH serving as the normalization control.
Immunofluorescence Staining
We fixed the cells with 4% PFA. After that, rinsing of the cells with PBS harboring 0.1% (v/v) Triton-X-100 solution was done. PBS harboring 2% BSA was employed to block the cells. After that, cells were inoculated with rabbit anti-H2AX antibody (1:200, Abcam, ab11174). Subsequently, we inoculated the cells with a secondary antibody linked to Alexa 488 (Molecular Probes, Life Technologies, Japan). Fluorescence microscopy was adopted to capture digital pictures after counterstaining the cells with 4′,6-diamidino-2-phenylindole (DAPI; DP72, Olympus, USA).
Immunohistochemistry
In brief, 5 µm slices were fixed at room temperature in paraformaldehyde (PFA) (4%) for 30 min. Thereafter, 2% bovine serum albumin (BSA) was employed to block the cells for 1 h. Next, we overnight inoculated the slides with rabbit anti-γH2AX (1:200, Abcam, ab11174), mouse anti-XPA (1:100, Invitrogen, MA1-21460), and rabbit anti-Ki67 (1:1000, Abcam, ab15580) at 4°C. Afterward, we inoculated the cells with biotinylated secondary antibody for 1 h, and we used diaminobenzidine as the chromogen substrate.
Tumor Growth Assays
Female BALB/c nude mice (8 weeks old, about 22 g) were provided food along with water ad libitum and housed at pathogen-free conditions at 20°C with a humidity of 60%, alternating between light and dark for 12 h. Each group consisted of five mice, all provided with appropriate food along with water and did not die normally. To generate tumors, U-87 MG shCon, U-87 MG shXPA, U-87 MG-R shCon, and U-87 MG-R shXPA cells were subcutaneously inoculated into nude mice, and then we treated subcutaneously inoculated mice with TMZ at a dosage of 1 mM per day for 2 weeks. After that, we excised the tumors and determined the volume along with weight of each tumor. Approval of this study was granted by the Ethics Committee of Wenzhou Medical University.
Statistics
All analyses were done in the GraphPad 6.0 software. All data herein were representative of at least three independent experiments. Data are given as mean ± standard error of the mean. * designates P < 0.05, ** designates P < 0.01, and *** designates P < 0.001.
Results
NER Participates in TMZ Resistance
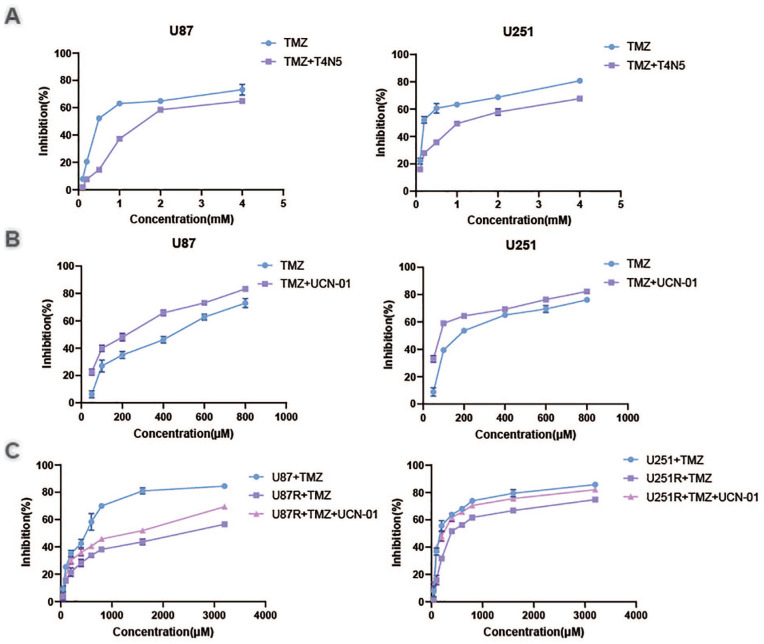
To investigate whether NER regulates TMZ sensitivity in glioblastomas, we assessed the effect of NER activator T4N5 and NER inhibitor UCN-01 on U87 and U251 cells combined with TMZ. CCK8 assays showed that T4N5 inhibited TMZ-triggered cell death (Fig. 1A), while UCN-01 sensitized U87 and U251 cells to TMZ-triggered cell death (Fig. 1B). These results suggested that NER may participate in TMZ resistance. We created TMZ-resistant U87 cell lines (U87-R cells) and U251 cell lines to better understand cellular TMZ resistance mechanisms (U251-R cells). When compared with their parent cells, these cells had a 5.0-fold and 2.4-fold increase in TMZ resistance, respectively, according to CCK8 tests. In U87-R along with U251-R cells, UCN-01 reversed TMZ resistance. Taken together, enhanced NER triggered TMZ resistance in GBM cells.
Figure 1.
Nucleotide excision repair participates in temozolomide resistance. U87 cells and U251 cells were treated with temozolomide and T4N5 (A) or UCN-01 (B). The cell viability was analyzed by CCK8 assay. (C) U87 cells and U87-R cells, U251 cells and U251-R cells were treated with temozolomide and UCN-01. The cell viability was analyzed by CCK8 assay.
TMZ: temozolomide.
TMZ-Resistant GBM Cells Exhibit a High Expression Level of XPA
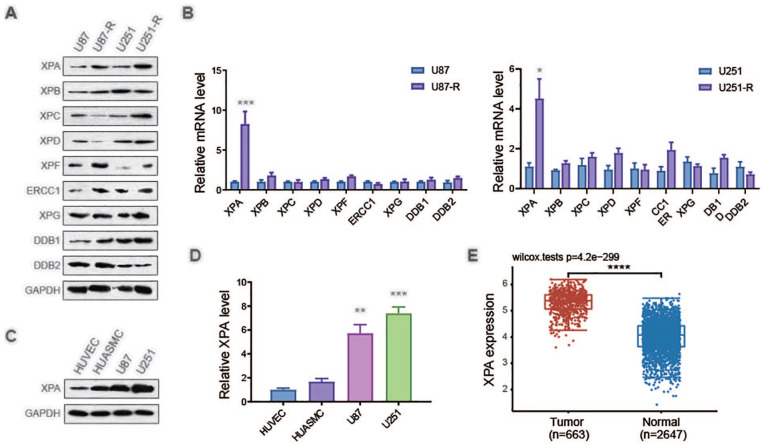
To determine the mechanism of how NER regulates TMZ resistance, the protein levels and the mRNA levels of NER-related genes in U87-R, U251-R cells, and their parent cells were analyzed. The expression of XPA was remarkably increased in both mRNA level and protein level (Fig. 2A, B). We also assess the expression of XPA in tumor cells and untransformed cells. Primary HUVECs and HUASMCs both had elevated contents of XPA in contrast with U87 along with U251 glioblastoma cells (Fig. 2C, D). Consistent with our findings, The Cancer Genome Atlas (TCGA) data analysis showed that the expression of XPA exhibited the strongest, as well as most remarkable increase in contrast with the normal tissue (Fig. 2E). These results indicated a high expression level of XPA in glioblastoma cells and TMZ-resistant cells.
Figure 2.
Temozolomide-resistant GBM cells exhibits a high expression level of XPA. (A) Representative western blot analysis of NER-related proteins and GAPDH in U87 cells, U87-R cells, U251 cells and U251-R cells. (B) qRT-PCR analysis of NER-related genes and GAPDH in U87 cells, U87-R cells, U251 cells and U251-R cells. ***P ≤ 0.001, compared with U87 group. (C) Representative western blot analysis of XPA and GAPDH in HUVEC, HUASMC, U87 cells and U251 cells. *P ≤ 0.05, compared with U251 group. (D) qRT-PCR analysis of XPA and GAPDH in HUVEC, HUASMC, U87 cells and U251 cells. **P ≤ 0.01, ***P ≤ 0.001, compared with HUVEC group. (E) Expression pattern of XPA in glioblastoma tumor tissue and brain tissue based on datasets in TCGA. ***P ≤ 0.001, compared with Tumor group.
Silencing XPA Attenuates TMZ Resistance
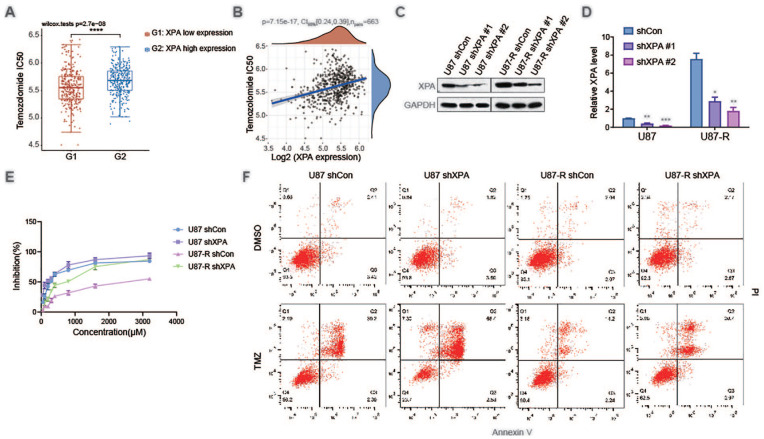
After establishing that glioblastoma cells over-express XPA in situ, we investigated whether XPA shields glioblastoma cells against the chemotherapy TMZ. To this end, we predicted the chemotherapeutic response and the spearman correlation analysis of IC50 score and XPA gene expression on the basis of the largest pharmacogenomics data resource [the Genomics of Drug Sensitivity in Cancer (GDSC) data resource, https://www.cancerrxgene.org/]. High expression of XPA exhibited a high IC50 of TMZ, and the IC50 of TMZ showed a correlation with XPA gene expression (Fig. 3A, B). To further examine this correlation, XPA was stably silenced in the glioblastoma cell line U87 cells along with U87-R cells (Fig. 3C, D). Knockdown of XPA remarkably sensitized U87 cells to TMZ-triggered cell death and reversed TMZ resistance of U87-R cells (Fig. 3E). Apoptosis has been proven to be responsible for TMZ-triggered cell death in glioblastoma cells 28 . As a result, whether the enhanced cell death in XPA silenced cells in response to TMZ inoculation is attributable to an increase in apoptosis was assessed. U87 shCon cells, U87 shXPA cells, U87-R shCon cells, and U87-R shXPA cells were inoculated with TMZ and quantification of apoptosis, as well as necrosis response were performed via annexin V/PI labeling along with flow cytometry. XPA silencing remarkably sensitized glioma cells to TMZ-triggered apoptosis along with necrosis (Fig. 3F). These data illustrated that XPA makes glioblastoma cells resistant to the methylating agent TMZ’s lethal action.
Figure 3.
Silencing XPA attenuates temozolomide resistance. (A) The distribution of IC50 scores in different groups, where the horizontal axis represents samples of different groups, and the vertical axis represents the distribution of IC50 scores, ***P ≤ 0.001, compared with XPA low expression group. (B) Spearman correlation analysis of IC50 score and XPA gene expression. (C) Representative western blot analysis of XPA and GAPDH in U87 cells and U87-R cells stably transfected with shCon or 2 independent shRNAs targeting XPA (shXPA #1 or shXPA #2). (D) qRT-PCR analysis of XPA and GAPDH in U87 cells and U87-R cells stably transfected with shCon or 2 independent shRNAs targeting XPA (shXPA #1 or shXPA #2). *P ≤ 0.05, ***P ≤ 0.01, ****P ≤ 0.001, compared with shCon group. (E) U87 cells and U87-R cells stably transfected with shCon or shXPA were treated with temozolomide. The cell viability was analyzed by CCK8 assay. (F) U87 cells and U87-R cells stably transfected with shCon or shXPA were treated with temozolomide. Apoptotic cells were detected by Annexin V and PI staining.
CI: confidence interval; TMZ: temozolomide; DMSO: dimethyl sulfoxide.
XPA Triggers the Repair of TMZ-Triggered DNA DSBs
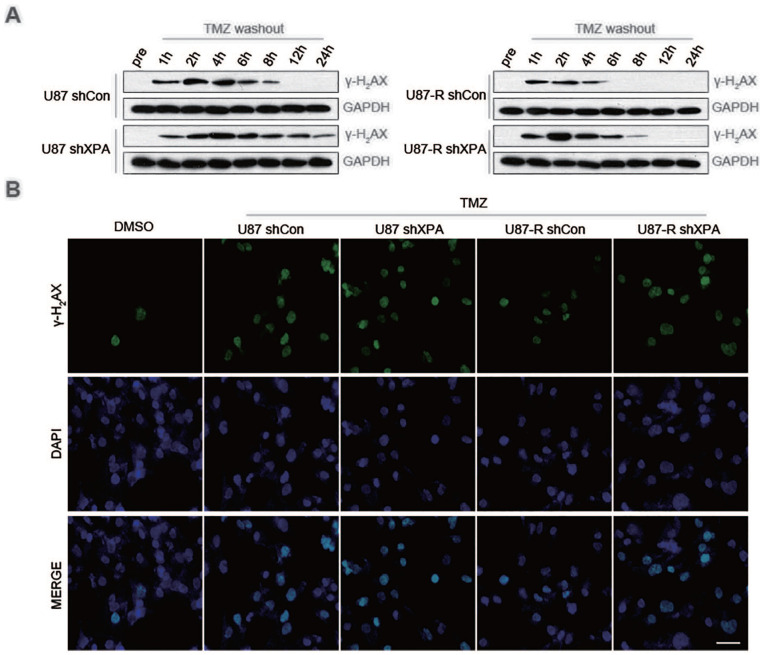
Furthermore, we assessed the status of DNA damage of U87 and U87-R cells stably transfected with shCon or shXPA by analyzing the phosphorylation of H2AX (γ-H2AX). Depletion of XPA in both U87 and U87-R cells resulted in persistently high contents of γ-H2AX up to 24 h after TMZ inoculation, in contrast with the control cells, where γ-H2AX could not be identified beyond 12 h after inoculation with TMZ (Fig. 4A). Immunofluorescence investigation validated these findings, exhibiting that TMZ treatment remarkably enhanced the active foci of γ-H2AX in XPA-depleted U87 along with U87-R cells (Fig. 4B). In conclusion, our data illustrate that XPA deficiency causes enhanced DNA damage coupled with TMZ sensitivity in glioblastoma cells.
Figure 4.
XPA stimulates the repair of temozolomide-induced DNA double-strand breaks. (A) Representative western blot analysis of γ-H2AX and GAPDH in U87 cells and U87-R cells stably transfected with shCon or shXPA treated with TMZ. (B) Representative immunofluorescence assay of γ-H2AX in U87 cells and U87-R cells stably transfected wih shCon or shXPA treated with TMZ.
TMZ: temozolomide; DMSO: dimethyl sulfoxide; DAPI: 4′,6-diamidino-2-phenylindole.
XPA Silencing Increases TMZ Sensitivity in Vivo
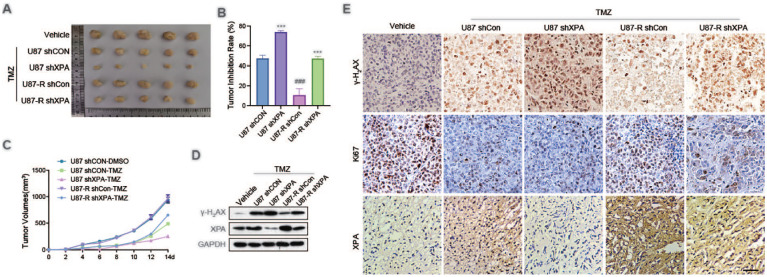
To elucidate the functional importance of XPA-triggered sensitization of TMZ in glioblastoma tumor, a xenografted model of tumor derived from U87 and U87-R cells transfected stably with shCon or shXPA was used. The data exhibited that sequential administration of TMZ tremendously reduced tumor growth compared with vehicle group except for U87-R shCon group (Fig. 5A–C). XPA silencing remarkably promoted the TMZ cytotoxicity in U87 shXPA group compared with U87 shCon group. Similar data were obtained between U87-R shCon group and U87-R shXPA group. Western blot analysis and immunohistochemistry assays showed that XPA knockdown obviously upregulated the expression of γ-H2AX (Fig. 5D, E). Moreover, low expression of XPA reduced cell proliferation (Ki67 expression) (Fig. 5E).
Figure 5.
XPA silencing increases temozolomide sensitivity in vivo. (A) U87 and U87-R cells stably transfected with shCon or shXPA, and subsequently injected into nude mice. Tumors were excised and volume was calculated every week. Representative images of isolated tumors were displayed. (B) The tumor inhibition rates according to the weight of explanted tumors in each experimental group. ***P ≤ 0.001, compared with shCon group. ###P ≤ 0.001, compared with U87 group. (C) Analysis of tumor growth in each experimental group. (D) Representative immunoblot analysis of XPA, γ-H2AX and GAPDH of xenografted tumors. (E) Representative immunohistochemical analysis of Ki67 and γ-H2AX protein levels (brown) in each experimental group.
TMZ: temozolomide.
Altogether, these data illustrate that silencing XPA improves the efficacy of TMZ and reverses the TMZ resistance in vitro along with in vivo.
Discussion
GBM constitutes a very aggressive brain tumor that is certainly a common fatal malignant tumor with a dismal prognosis 29 . TMZ is currently the only chemotherapeutic medication that has been illustrated to remarkably improve overall survival in individuals with GBM 28 . However, because glioblastoma develops resistance to TMZ fast, its efficacy is usually limited to a short time span 30 . Herein, we uncovered a potential role of NER in TMZ resistance. Combined with NER inhibitor UCN-01, TMZ exhibited a higher inhibition rate of cell proliferation, suggesting that NER inhibitor UCN-01 can be used in combination as a sensitizer for TMZ in clinical practice.
By comparing the expression data from non-malignant tissue and brain tumors, we found that gliomas showed a high expression level of XPA. Tumor radiation therapy can also cause DNA damage in tumor cells. Some studies have found that XPA is highly expressed in glioma radiation-resistance 31 . However, its mechanism remains unclear. In our current research, we also confirmed this phenomenon by further analysis through western blotting along with reverse transcription–quantitative polymerase chain reaction (RT-qPCR) assay. The over-expression of XPA seen in gliomas might make this tumor resistant to TMZ-centered treatments. To prove this hypothesis, we established TMZ-resistant U87 cells and U251 cells, and then stably transfected with interference RNA to downregulate the expression of XPA. XPA is overexpressed in TMZ-resistant U87 and U251 cells. Upon XPA knockdown, U87 cells exhibited a remarkable increase in cell killing after inoculation with TMZ, as well as U87-R cells. These findings illustrate that XPA plays an indispensable role in glioma cell resistance to TMZ-triggered cell death, as well as suggests a prospective mechanism by which this is accomplished.
By serving as a DNA-binding factor component, XPA plays a key role in NER 21 . In germ cell cancers, elevated contents of XPA may be the cause of cisplatin resistance 25 . In subtypes of head and neck squamous cell carcinoma, XPA might be a candidate for overcoming chemotherapy resistance 32 . In a replication and mismatch repair-dependent approach, TMZ methylates DNA at position 6 of guanine, and the methylation product, O6MeG, leads to the creation of DSBs 33 . Some constituents of the HR cascade34,35, the NHEJ cascade36,37, ligase IV 38 , and DNA-PKcs 39 participate in the tolerance of DSBs resulting from a response to TMZ. Our data illustrate that silencing XPA in GBM cells can exacerbate TMZ-triggered DNA damage, indicating that the XPA and NER cascades are implicated in TMZ-triggered DNA damage repair. More recently, molecular biomarkers have gained importance in providing both ancillary and defining diagnostic information 40 . Our study suggests that XPA can be used as a molecular biomarker of TMZ resistance, which can be treated in groups.
Ultimately, we established a pivotal mechanism that contributes to TMZ resistance in GBM cells. Upregulation of XPA improves the repair of TMZ-triggered DNA damage, dampens TMZ-triggered cell death and apoptosis, and leads to TMZ resistance. Our data revealed a prospective therapeutic target for treating individuals with GBM harboring TMZ resistance.
Footnotes
Ethical Approval: This study was approved by our institutional review board.
Statement of Human and Animal Rights: This article does not contain any studies with human or animal subjects.
Statement of Informed Consent: There are no human subjects in this article and informed consent is not applicable.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Science and Technology Project Foundation of Quzhou city (2020K46).
ORCID iD: An Wu  https://orcid.org/0000-0001-9571-3019
https://orcid.org/0000-0001-9571-3019
References
- 1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. [DOI] [PubMed] [Google Scholar]
- 2. Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. [DOI] [PubMed] [Google Scholar]
- 3. Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005; 352(10):987–96. [DOI] [PubMed] [Google Scholar]
- 4. Zhang J, Stevens MF, Bradshaw TD. Temozolomide: mechanisms of action, repair and resistance. Curr Mol Pharmacol. 2012;5(1):102–14. [DOI] [PubMed] [Google Scholar]
- 5. Happold C, Weller M. New insights into acquired temozolomide resistance in glioblastoma? Brain. 2015;138(Pt 12):3468–70. [DOI] [PubMed] [Google Scholar]
- 6. Nie E, Jin X, Miao F, Yu T, Zhi T, Shi Z, Wang Y, Zhang J, Xie M, You Y. TGF-beta1 modulates temozolomide resistance in glioblastoma via altered microRNA processing and elevated MGMT. Neuro Oncol. 2021;23(3):435–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Christmann M, Nagel G, Horn S, Krahn U, Wiewrodt D, Sommer C, Kaina B. MGMT activity, promoter methylation and immunohistochemistry of pretreatment and recurrent malignant gliomas: a comparative study on astrocytoma and glioblastoma. Int J Cancer. 2010;127(9):2106–18. [DOI] [PubMed] [Google Scholar]
- 8. Baer JC, Freeman AA, Newlands ES, Watson AJ, Rafferty JA, Margison GP. Depletion of O6-alkylguanine-DNA alkyltransferase correlates with potentiation of temozolomide and CCNU toxicity in human tumour cells. Br J Cancer. 1993;67(6):1299–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Roos W, Baumgartner M, Kaina B. Apoptosis triggered by DNA damage O6-methylguanine in human lymphocytes requires DNA replication and is mediated by p53 and Fas/CD95/Apo-1. Oncogene. 2004;23(2):359–67. [DOI] [PubMed] [Google Scholar]
- 10. Happold C, Roth P, Wick W, Schmidt N, Florea AM, Silginer M, Reifenberger G, Weller M. Distinct molecular mechanisms of acquired resistance to temozolomide in glioblastoma cells. J Neurochem. 2012;122(2):444–55. [DOI] [PubMed] [Google Scholar]
- 11. Yang B, Fu X, Hao J, Sun J, Li Z, Li H, Xu H. PAXX participates in base excision repair via interacting with pol beta and contributes to TMZ resistance in glioma cells. J Mol Neurosci. 2018;66(2):214–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Roos WP, Frohnapfel L, Quiros S, Ringel F, Kaina B. XRCC3 contributes to temozolomide resistance of glioblastoma cells by promoting DNA double-strand break repair. Cancer Lett. 2018;424:119–26. [DOI] [PubMed] [Google Scholar]
- 13. Sharma AB, Erasimus H, Pinto L, Caron MC, Gopaul D, Peterlini T, Neumann K, Nazarov PV, Fritah S, Klink B, Herold-Mende CC, et al. XAB2 promotes Ku eviction from single-ended DNA double-strand breaks independently of the ATM kinase. Nucleic Acids Res. 2021;49(17):9906–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gil Del Alcazar CR, Todorova PK, Habib AA, Mukherjee B, Burma S. Augmented HR repair mediates acquired temozolomide resistance in glioblastoma. Mol Cancer Res. 2016; 14(10):928–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qin L, Huiwen M, Wang J, Wang Y, Khan SA, Zhang Y, Qiu H, Jiang L, He L, Zhang Y, Jia S. A novel polymerase beta inhibitor from phage displayed peptide library augments the anti-tumour effects of temozolomide on colorectal cancer. J Chemother. Epub 2021 Dec. doi: 10.1080/1120009X.2021.2009987. [DOI] [PubMed] [Google Scholar]
- 16. Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15(7):465–81. [DOI] [PubMed] [Google Scholar]
- 17. Liu Y, Wang X, Li W, Xu Y, Zhuo Y, Li M, He Y, Wang X, Guo Q, Zhao L, Qiang L. Oroxylin A reverses hypoxia-induced cisplatin resistance through inhibiting HIF-1alpha mediated XPC transcription. Oncogene. 2020;39(45):6893–905. [DOI] [PubMed] [Google Scholar]
- 18. Spivak G. Nucleotide excision repair in humans. DNA Repair (Amst). 2015;36:13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sugitani N, Shell SM, Soss SE, Chazin WJ. Redefining the DNA-binding domain of human XPA. J Am Chem Soc. 2014;136(31):10830–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fadda E. Role of the XPA protein in the NER pathway: a perspective on the function of structural disorder in macromolecular assembly. Comput Struct Biotechnol J. 2016;14:78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Borszekova Pulzova L, Ward TA, Chovanec M. XPA: DNA repair protein of significant clinical importance. Int J Mol Sci. 2020;21(6):2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu X, Shell SM, Yang Z, Zou Y. Phosphorylation of nucleotide excision repair factor xeroderma pigmentosum group A by ataxia telangiectasia mutated and Rad3-related-dependent checkpoint pathway promotes cell survival in response to UV irradiation. Cancer Res. 2006;66(6):2997–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Park JM, Choi JY, Yi JM, Chung JW, Leem SH, Koh SS, Kang TH. NDR1 modulates the UV-induced DNA-damage checkpoint and nucleotide excision repair. Biochem Biophys Res Commun. 2015;461(3):543–48. [DOI] [PubMed] [Google Scholar]
- 24. Fang EF, Scheibye-Knudsen M, Brace LE, Kassahun H, SenGupta T, Nilsen H, Mitchell JR, Croteau DL, Bohr VA. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell. 2014;157(4):882–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cierna Z, Miskovska V, Roska J, Jurkovicova D, Pulzova LB, Sestakova Z, Hurbanova L, Machalekova K, Chovanec M, Rejlekova K, Svetlovska D, et al. Increased levels of XPA might be the basis of cisplatin resistance in germ cell tumours. BMC Cancer. 2020;20(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ge R, Liu L, Dai W, Zhang W, Yang Y, Wang H, Shi Q, Guo S, Yi X, Wang G, Gao T, et al. Xeroderma pigmentosum group A promotes autophagy to facilitate cisplatin resistance in melanoma cells through the activation of PARP1. J Invest Dermatol. 2016;136(6):1219–28. [DOI] [PubMed] [Google Scholar]
- 27. Hayashi T, Adachi K, Ohba S, Hirose Y. The Cdk inhibitor flavopiridol enhances temozolomide-induced cytotoxicity in human glioma cells. J Neurooncol. 2013;115(2):169–78. [DOI] [PubMed] [Google Scholar]
- 28. Xu S, Tang L, Li X, Fan F, Liu Z. Immunotherapy for glioma: current management and future application. Cancer Lett. 2020;476:1–12. [DOI] [PubMed] [Google Scholar]
- 29. Jimenez-Pascual A, Siebzehnrubl FA. Fibroblast growth factor receptor functions in glioblastoma. Cells. 2019;8(7):715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee SY. Temozolomide resistance in glioblastoma multiforme. Genes Dis. 2016;3(3):198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sun C, Wang Z, Song W, Chen B, Zhang J, Dai X, Wang L, Wu J, Lan Q, Huang Q, Dong J. Alteration of DNA damage signaling pathway profile in radiation-treated glioblastoma stem-like cells. Oncol Lett. 2015;10(3):1769–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Prochnow S, Wilczak W, Bosch V, Clauditz TS, Muenscher A. ERCC1, XPF and XPA-locoregional differences and prognostic value of DNA repair protein expression in patients with head and neck squamous cell carcinoma. Clin Oral Investig. 2019;23(8):3319–29. [DOI] [PubMed] [Google Scholar]
- 33. Karachi A, Dastmalchi F, Mitchell DA, Rahman M. Temozolomide for immunomodulation in the treatment of glioblastoma. Neuro Oncol. 2018;20(12):1566–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rajesh P, Rajesh C, Wyatt MD, Pittman DL. RAD51D protects against MLH1-dependent cytotoxic responses to O(6)-methylguanine. DNA Repair (Amst). 2010;9(4):458–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oza J, Doshi SD, Hao L, Musi E, Schwartz GK, Ingham M. Homologous recombination repair deficiency as a therapeutic target in sarcoma. Semin Oncol. 2020;47(6):380–89. [DOI] [PubMed] [Google Scholar]
- 36. Dong W, Li L, Teng X, Yang X, Si S, Chai J. End processing factor APLF promotes NHEJ efficiency and contributes to TMZ- and ionizing radiation-resistance in glioblastoma cells. Onco Targets Ther. 2020;13:10593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang B, Han N, Sun J, Jiang H, Xu HY. CtIP contributes to non-homologous end joining formation through interacting with ligase IV and promotion of TMZ resistance in glioma cells. Eur Rev Med Pharmacol Sci. 2019;23(5):2092–102. [DOI] [PubMed] [Google Scholar]
- 38. Kondo N, Takahashi A, Mori E, Ohnishi K, McKinnon PJ, Sakaki T, Nakase H, Ohnishi T. DNA ligase IV as a new molecular target for temozolomide. Biochem Biophys Res Commun. 2009;387(4):656–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roos WP, Batista LF, Naumann SC, Wick W, Weller M, Menck CF, Kaina B. Apoptosis in malignant glioma cells triggered by the temozolomide-induced DNA lesion O6-methylguanine. Oncogene. 2007;26(2):186–97. [DOI] [PubMed] [Google Scholar]
- 40. Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM, Reifenberger G, Soffietti R, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23(8): 1231–51. [DOI] [PMC free article] [PubMed] [Google Scholar]