Abstract
Denaturing gradient gel electrophoresis revealed changes in the bacterial species obtained from enrichment cultures with different inoculum dilutions. This inoculum dilution enrichment approach may facilitate the detection and isolation of a greater number of bacterial species than traditional enrichment techniques.
Traditional enrichment techniques underestimate the diversity of bacteria within natural environments (1, 7, 13). However, few studies have varied the enrichment approach in order to detect or isolate a greater number of species. One possible variation would be to alter the concentration of the initial inoculum. By diluting the sample, one should select for those organisms that are numerically abundant, not just those that show superior growth in a given medium. An inoculum dilution enrichment series should, therefore, show shifts in the bacteria cultured at each dilution level. At low dilutions, cultures will be dominated by species that show competitively superior growth in that medium. At higher dilutions, these superior growers should be diluted out of the inoculum to be replaced by populations that were numerically dominant in the original sample but whose growth has been inhibited at smaller dilutions (2). In order to detect the presence of different populations in such cultures, one could subculture each dilution and characterize isolates or use a molecular approach. Santegoeds et al. used denaturing gradient gel electrophoresis (DGGE) of PCR-amplified 16S rRNA genes to examine serial dilution cultures from a hot-spring microbial mat sample and found changes in the bacterial populations that were cultured at different dilutions (9).
We used DGGE to monitor changes in the bacterial populations present in serial dilution enrichments from a wetland biofilm sample. The biofilm sample was obtained from water lily (Nymphaea odorata) leaves collected from a wetland in Hale County, Ala. The leaves were shaken to dislodge attached cells, the sample was serially diluted (10-fold), and culture tubes were inoculated with sample suspension to yield enrichments with inoculum concentrations of from 10−1 to 10−8. The remaining sample suspension was retained for analysis of the original community.
An aerobic complex medium (15 g of tryptic soy broth [TSB], 1 liter of H2O) was used for enrichment with three replicate cultures at each dilution level. Tubes were shaken (150 rpm, 20°C) until visible growth was observed (5 days). Growth occurred down to the 10−6 dilution, giving a most probable number estimate for the original biofilm of 937,500 cells ml−1 (confidence factor, 4.67) or 16,175 cells cm−2. Following growth, a subsample of each culture was plated onto solid medium (as described above, with 20 g of agar) to obtain isolates, which were subsequently transferred to liquid culture to obtain sufficient biomass for DNA extraction. The remaining mixed cultures were centrifuged (5,000 × g), and DNA was extracted from each pellet by using three cycles of freeze-thaw (11), followed by extended heating according to the method described by Zhou et al. (14).
Following precipitation in alcohol, samples were resuspended in 100 μl of Tris-EDTA (TE) buffer (8), and the suspension was incubated with 10 μl of RNase (20 mg/ml) at 37°C for 2 h. Following phenol-chloroform (1:1) extraction and alcohol precipitation, samples were resuspended in TE buffer. DNA was similarly extracted from isolates and from the original biofilm. Because of contamination with humic substances, DNA from the biofilm sample was purified by using Sepharose 4B columns (3).
The region of 16S ribosomal DNA (rDNA) corresponding to helices 15 to 20 (12) in each sample was amplified with the 341 forward and 534 reverse primers described by Muyzer et al. (5) by using reactant concentrations and procedures described previously (3). PCR products from each sample were analyzed by DGGE along a urea-formamide gradient (25 to 60%) in 6% acrylamide gels at 130 V and 60°C for 4 h under conditions described by Muyzer et al. (6). Samples were initially run through the gel at 20 V for 10 min to improve resolution. The gels were stained in ethidium bromide and photographed with UV transillumination. DGGE bands were excised from some gels, and DNA was eluted and reamplified (6). Amplification products were purified with Wizard systems (Promega, Madison, Wis.) and sequenced.
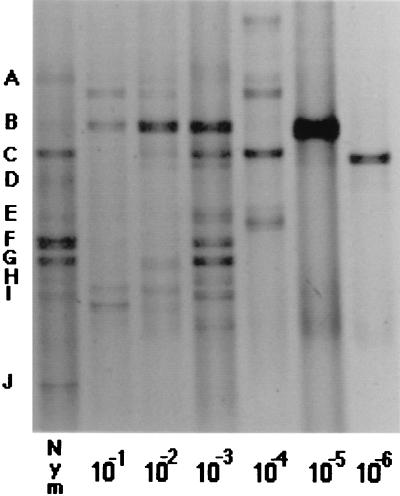
DGGE revealed various bacterial genotypes, as shown by the different bands representing 16S rDNA fragments (Fig. 1). DGGE analyses were repeatable. Enrichment selected for some bands that appeared to be present in the original sample (e.g., Fig. 1, bands B and C), as well as for others that were not detectable before enrichment (e.g., Fig. 1 [10−6 dilution]). This result is not unexpected, given that enrichment methods are unlikely to detect many bacteria in environmental samples (1, 7, 13). The bands observed in the most dilute enrichments were not visible in the original sample, confirming that enrichment may not detect the most abundant bacteria. Conversely, while DGGE is useful in examining abundant populations, it may not detect minor populations (6) which might require enrichment prior to their detection. A biphasic approach utilizing enrichment and molecular techniques may be necessary to detect some organisms (4).
FIG. 1.
DGGE analysis of 16S rRNA sequences obtained from 10−2 to 10−6 dilutions of biofilms from N. odorata leaf homogenate in TSB enrichment cultures sampled after 5 days. Nym, the original biofilm sample before enrichment; single letters, dominant 16S rRNA sequences within this sample. The photograph was scanned through Adobe PhotoDeluxe 1.0.1 with a PowerPC Macintosh.
Dilution of the inoculum affected the outcome of enrichment cultures (Fig. 1). This result can be attributed to the tradeoff between numerical abundance in the original sample and competitive ability in the growth medium (9). Changes in the species composition of enrichment cultures occurred throughout the dilution series, and these changes could be detected by DGGE. Santegoeds et al. observed the same phenomenon (9), and we have observed similar changes with other samples (data not shown).
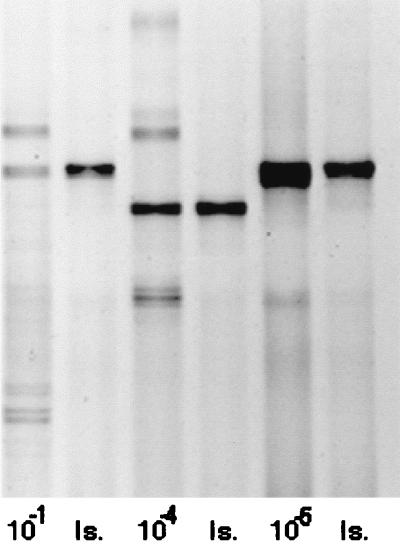
Few isolates could be obtained on solid media, with typically just one to three isolates obtained from each mixed culture. There was strong evidence for the presence of these isolates within the original enrichments, as shown by gels comparing the isolate to the enrichment from which it was obtained (e.g., Fig. 2). However, bands in mixed cultures could not always be matched to individual isolates.
FIG. 2.
DGGE analysis of 16S rRNA sequences obtained from three dilutions (10−1, 10−4, and 10−5) of biofilms from N. odorata leaf homogenate in TSB enrichment cultures sampled after 5 days and from isolates subsequently obtained from such cultures. Is, the DGGE profile of a bacterial isolate obtained from the enrichment culture shown in the preceding lane. The isolates were designated TSB013 (from the 10−1 dilution), TSB026 (from the 10−4 dilution), and TSB027 (from the 10−5 dilution). The photograph was scanned by using Adobe PhotoDeluxe 1.0.1 with a PowerPC Macintosh.
An assumption of this inoculum dilution approach is that an organism should be cultured only at consecutive dilution levels. From DGGE profiles, it appears that this was not so. A band corresponding to band B in the original inoculum appeared at the 10−1 to 10−3 dilutions but was absent at 10−4 and was present again in the 10−5 dilution (Fig. 1). We were able to isolate representatives from the dilutions involved in this discrepancy and verify that they did show similar migrations on DGGE gels (isolates TSB013 from the 10−1 dilution and TSB027 from the 10−5 dilution [Fig. 2]). Sequencing of these bands revealed that their sequences differed by 5%, suggesting that while they migrated to virtually identical locations, they were in fact different sequences. Although the sequences differed, they shared the same GC content (50%) and melting temperature (83°C), which presumably accounts for their similar migration patterns.
Combining variations in enrichment procedures with molecular methodologies may reveal that more bacteria can be cultured than we currently believe. By varying the inoculum dilution, we were able to detect 15 to 20 bands across all dilution levels, compared to an average of 6 bands in any individual culture or 10 in the original sample. The actual number of genomes present may be higher if different sequences can show similar migration patterns. The distinction between culturable and unculturable is relevant only within defined conditions (10). By altering these conditions and combining traditional and molecular approaches, we may gain a greater understanding of microbial diversity in environmental samples.
Nucleotide sequence accession numbers.
The two sequences reported here have been submitted to GenBank under accession numbers AF061035 (isolate TSB013) and AF061036 (TSB027).
REFERENCES
- 1.Colwell R R, Clayton R A, Ortiz-Conde B A, Jacobs D, Russek-Cohen E. The microbial species concept and biodiversity. In: Allsopp D, Colwell R R, Hawksworth D L, editors. Microbial diversity and ecosystem function. Wallingford, United Kingdom: CAB International; 1995. pp. 3–15. [Google Scholar]
- 2.Großkopf R, Janssen P H, Liesack W. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl Environ Microbiol. 1998;64:960–969. doi: 10.1128/aem.64.3.960-969.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jackson C R, Harper J P, Willoughby D, Roden E E, Churchill P F. A simple, efficient method for the separation of humic substances and DNA from environmental samples. Appl Environ Microbiol. 1997;63:4993–4995. doi: 10.1128/aem.63.12.4993-4995.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liesack L, Janssen P H, Rainey F A, Ward-Rainey N L, Stackebrandt E. Microbial diversity in soil: the need for a combined approach using molecular and cultivation techniques. In: van Elsas J D, Trevors J T, Wellington E M H, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker, Inc.; 1997. pp. 375–439. [Google Scholar]
- 5.Muyzer G, De Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muyzer G, Hottenträger S, Teske A, Wawer C. Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA—a new molecular approach to analyse the genetic diversity of mixed microbial communities. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology, manual 3.4.4. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 1–23. [Google Scholar]
- 7.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 8.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 9.Santegoeds C M, Nold S C, Ward D M. Denaturing gradient gel electrophoresis used to monitor the enrichment culture of aerobic chemoorganotrophic bacteria from a hot spring cyanobacterial mat. Appl Environ Microbiol. 1996;62:3922–3928. doi: 10.1128/aem.62.11.3922-3928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki M T, Rappé M S, Haimberger Z W, Winfield H, Adair N, Ströbel J, Giovannoni S J. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai Y-L, Olson B H. Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol. 1991;57:1070–1074. doi: 10.1128/aem.57.4.1070-1074.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van de Peer Y, Nicolaï S, De Rijk P, De Wachter R. Database on the structure of small ribosomal subunit RNA. Nucleic Acids Res. 1996;24:86–91. doi: 10.1093/nar/24.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ward N, Rainey F A, Goebel B, Stackebrandt E. Identifying and culturing the ‘unculturables’: a challenge for microbiologists. In: Allsopp D, Colwell R R, Hawksworth D L, editors. Microbial diversity and ecosystem function. Wallingford, United Kingdom: CAB International; 1995. pp. 89–110. [Google Scholar]
- 14.Zhou J, Bruns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]