Abstract
Bioluminescence, mRNA levels, and toluene degradation rates in Pseudomonas putida TVA8 were measured as a function of various concentrations of toluene and trichloroethylene (TCE). TVA8 showed an increasing bioluminescence response to increasing TCE and toluene concentrations. Compared to uninduced TVA8 cultures, todC1 mRNA levels increased 11-fold for TCE-treated cultures and 13-fold for toluene-treated cultures. Compared to uninduced P. putida F1 cultures, todC1 mRNA levels increased 4.4-fold for TCE-induced cultures and 4.9-fold for toluene-induced cultures. Initial toluene degradation rates were linearly correlated with specific bioluminescence in TVA8 cultures.
Trichloroethylene (TCE) has been extensively used as an industrial extraction solvent, a dry cleaning fluid, a degreaser (20), and a heat transfer fluid (8). The widespread use and improper disposal of TCE eventually led to its classification as a groundwater priority pollutant with potential health hazards (16, 21). Consequently, the bioremediation potential of TCE has received significant attention. Much of the recent TCE bioremediation research has focused on the oxygenase enzymes due to the relatively benign nature of their by-products in comparison to by-products of anaerobic degradation, primarily carcinogenic vinyl chloride (5). Examples of oxygenase enzymes are soluble methane monooxygenase from Methylosinus trichosporium OB3b (15, 22), toluene 2-monooxygenase from Pseudomonas cepacia G4 (6), toluene 4-monooxygenase from Pseudomonas mendocina (25), and toluene dioxygenase from P. putida F1 (7, 24).
The degradation of TCE by P. putida F1 and its dioxygenase encoded by the tod operon has been studied extensively (5, 23, 24, 26). However, TCE-mediated induction of the tod operon is one area of research that has yet to be fully elucidated. In 1994, Heald and Jenkins (9) reported the first evidence of the induction of toluene degradation by TCE in a wild-type P. putida strain. However, in 1995, McClay et al. (14) reported that TCE did not induce toluene oxidation activity in P. putida F1. In 1996, Leahy et al. (13) presented evidence that suggested that TCE partially induces toluene-degradative activity in F1. The experiments of Leahy et al. were conducted with resting cells that were previously grown in the presence of a noncompetitive growth substrate, lactate, and 2.35 mM TCE. In the experiments of McClay et al., activity was measured with resting cells that had no prior exposure to TCE in the presence of a growth substrate. When P. putida B2, a tod-lux bioluminescent reporter strain, was encapsulated in alginate beads and loaded into a differential-volume reactor, Applegate et al. (2) saw no increase in bioluminescence or TCE degradation in the absence of toluene. Neither Applegate nor McClay detected toluene dioxygenase induction by TCE in resting P. putida strains. However, the studies of Heald and Leahy show strong evidence for induction of toluene dioxygenase by TCE in P. putida strains. The objective of the present study was to determine, by using both biochemical and molecular techniques, whether TCE can induce the tod operon in the bioluminescent reporter P. putida TVA8, a P. putida F1 derivative containing a modified mini-Tn5 chromosomal insertion of a tod-lux fusion (1).
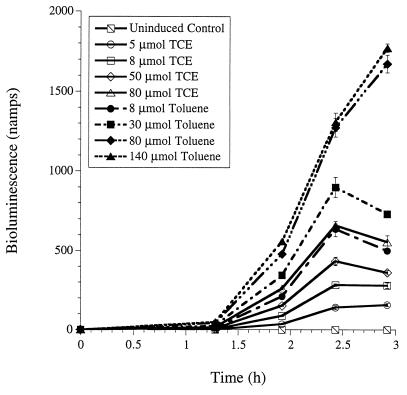
Bioluminescence response of P. putida TVA8 to toluene and TCE.
The bioluminescence response over time of TVA8 to various concentrations of TCE and toluene was measured by using the growing-cell assay adapted from Heitzer et al. (10, 11). Cultures were prepared from a frozen stock of TVA8 by inoculating 1.0 ml of the stock solution into 100 ml of yeast extract-peptone-glucose medium (18) amended with 10 ml of a 50 mM phosphate buffer. From a fresh overnight culture, a subculture was grown to an optical density at 546 nm of 0.35 at 30°C, and 2.0-ml aliquots of the culture were added to 20-ml scintillation vials containing 2.0 ml of a mineral salts medium (MSM) (19). TCE and toluene were added by supplementing the 4 ml of culture with known volumes of TCE-saturated MSM and toluene-saturated MSM. The volume of TCE or toluene-saturated MSM required for a given liquid-phase concentration was calculated on the basis of mass balance. Gas-liquid equilibrium was predicted by using Henry’s Law coefficients at 20°C (HTCE = 0.36; Htoluene = 0.27) (3). The vials were placed in a constant-temperature room at 21°C and shaken at 200 rpm for 3 h. Bioluminescence was measured with an Oriel (Stratford, Conn.) detection system (model 7070) as described by Heitzer et al. (10). After the final bioluminescence measurement, 0.5 ml of culture was removed and the final optical density at 546 nm was measured, converted to milligrams of protein based on a standard curve, and used to calculate the specific bioluminescence (nanoamperes per milligram of protein) by dividing the sample bioluminescence by total protein. The sample bioluminescence response of cells versus time for various concentrations of TCE and toluene was plotted (Fig. 1). For toluene concentrations of 8 to 140 μM, the specific bioluminescence response was 5,400- to 20,000-fold, respectively, over the response for the uninduced culture. Previous studies correlated toluene concentration with specific bioluminescence, and the same is true for this study (1, 2). For TCE concentrations of 5 to 80 μM, the bioluminescence response was 1,700- to 6,000-fold over that of the uninduced culture, respectively, and was linearly correlated with TCE concentration (r2 = 0.9626).
FIG. 1.
Bioluminescence response of TVA8 versus time in growing-cell assays with various TCE and toluene concentrations. μmol, micromolar.
mRNA expression of todC1 and luxA.
Because toluene dioxygenase induction by TCE in strain F1 had not been conclusively reported in previous literature, induction of the tod operon in TVA8 and F1 was validated by using mRNA slot blot analysis. After the 3-h time point in the growing-cell assay described above, 3.0 ml of culture was removed for mRNA analysis. Total RNA was isolated with an RNeasy Total RNA Kit (Qiagen, Chatsworth, Calif.) in accordance with the manufacturer’s protocol. RNA slot blots were prepared as outlined by Sambrook et al. (17). Five micrograms of total RNA from the various treatments was loaded onto a Biotrans nylon membrane (ICN, Irvine, Calif.) in triplicate along with luxA and todC1 DNA standards. The blots were prehybridized at 55°C for 4 h in a hybridization solution as previously described (4). The blots were then hybridized overnight with either a luxA (295 bases) or todC1 (900 bases) PCR-generated antisense 32P-DNA probe as previously described (10). The blots were washed with a 2× SSC–0.1% sodium dodecyl sulfate solution three times at 55°C (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The mRNA levels were quantified with a Storm 840 PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) and determined by using standard curves generated from the appropriate DNA standards. The data were normalized to the uninduced values and are expressed as relative todC1 and luxA mRNA levels (Table 1). The results show that TCE induces transcription of todC1 in both TVA8 and F1. Compared to the todC1 mRNA levels in the uninduced TVA8 cultures, the todC1 mRNA levels were 11-fold higher for the TCE-treated cultures and 13-fold higher for the toluene-treated cultures. In the TVA8 cultures, the TCE-induced treatments showed a statistically higher luxA mRNA level (α = 0.05) than the uninduced cultures, but the toluene-induced treatments showed a statistically higher todC1 mRNA level (α = 0.05). Compared to the todC1 mRNA levels in uninduced F1 cultures, the todC1 levels were 4.4-fold higher for the 80 μM TCE treatment and 4.9-fold higher for the 110 μM toluene treatment. In the F1 cultures, the difference between the todC1 levels in the TCE-induced and toluene-induced treatments was not significant (α = 0.05).
TABLE 1.
Specific bioluminescence and relative todC1 and luxA mRNA transcript levels in TVA8 and relative todC1 mRNA transcript levels in F1 for various inducer concentrations after 3 h in the growing-cell assay
| Inducing compound | Concn (μM) | Relative mRNA transcript levela (mean ± SD)
|
||
|---|---|---|---|---|
| TVA8 luxA | TVA8 todC1 | F1 todC1 | ||
| None (uninduced) | 0 | 1.0 ± 2 | 1.0 ± 0.3 | 1.0 ± 0.5 |
| TCE | 5 | 21 ± 4 | 9.2 ± 4 | NDb |
| 8 | 22 ± 11 | 11 ± 3 | 1.4 ± 0.4 | |
| 50 | 25 ± 4 | 12 ± 2 | ND | |
| 80 | 16 ± 6 | 10 ± 2 | 4.4 ± 1 | |
| Toluene | 8 | 12 ± 8 | 13 ± 2 | ND |
| 10 | ND | ND | 2.1 ± 0.9 | |
| 30 | 10 ± 2 | 12 ± 3 | ND | |
| 80 | 15 ± 3 | 13 ± 2 | ND | |
| 110 | ND | ND | 4.9 ± 0.8 | |
| 140 | 11 ± 8 | 13 ± 3 | ND | |
A 32P-labeled universal 16S oligonucleotide probe for all eubacteria was used to verify total RNA normalization (12).
ND, not determined.
Correlation between initial toluene degradation rates and bioluminescence.
Toluene degradation rates were used as an indicator of toluene dioxygenase activity in TVA8 cultures exposed to various concentrations of inducer. To determine the degradation rates, 3.5 ml of culture from a growing-cell assay was centrifuged, washed once with 4.0 ml of MSM, resuspended in 4.0 ml of MSM, and added to 25-ml glass vials with Teflon-lined caps. Known volumes of toluene-saturated MSM were added to give an initial concentration of 70 μM toluene (liquid phase). Headspace toluene concentration was measured over time by removing 100-μl samples from the vials and injecting the samples into a Hewlett-Packard 5890 gas chromatograph equipped with an electron capture detector and a flame ionization detector. The toluene degradation rate was determined by the change in headspace toluene concentration after 50 min, normalized to values for total protein. The toluene degradation rates (Table 2) show that TCE- and toluene-induced cells do have significantly more activity than uninduced cells. As the toluene concentration was increased from 1 to 330 μM toluene, specific bioluminescence increased approximately fivefold and the toluene degradation rate increased over fourfold. There is a linear correlation (r2 = 0.9690) between specific bioluminescence and toluene degradation rate for toluene treatments. The 1 μM TCE treatment showed no induction effect, as indicated by both the specific bioluminescence and toluene degradation rate (Table 2); however, TCE concentrations between 8 and 230 μM produced an increased bioluminescence response and an average toluene degradation rate of 84 nmol/min/mg of protein (Table 2).
TABLE 2.
Specific bioluminescence and toluene oxidation activity of P. putida TVA8 as a function of TCE and toluene concentrations after 3 h in the growing-cell assay
| Inducing compound | Concn (μM) | Specific bioluminescence (nA/mg of protein)a | Toluene degradation rate (nmol of toluene/min/mg of protein)a |
|---|---|---|---|
| None (uninduced) | 0 | 0.16 ± 0.01 | 0 |
| TCE | 1 | 0.15 ± 0.01 | 0 |
| 8 | 616 ± 27 | 87 ± 40 | |
| 80 | 1,027 ± 42 | 72 ± 11 | |
| 230 | 2,690 ± 53 | 92 ± 8 | |
| Toluene | 1 | 818 ± 320 | 123 ± 7 |
| 10 | 1,390 ± 32 | 134 ± 33 | |
| 110 | 2,190 ± 170 | 198 ± 10 | |
| 330 | 3,811 ± 30 | 423 ± 16 |
Values are means ± standard deviations.
Conclusions.
Based on molecular and biochemical techniques, this study shows that TCE induces the tod operon in the bioluminescent reporter TVA8 and its parent strain F1. Specific bioluminescence was linearly correlated with TCE concentration in the growing-cell assay, and the TCE detection limit with this system is apparently between 1 and 5 μM TCE. The mRNA studies showed increased levels of luxA and todC1 mRNA in TCE-induced samples relative to the control levels. While toluene and TCE concentrations are linearly correlated with bioluminescence after 3 h, TVA8 mRNA levels are constant for both increasing toluene and TCE concentrations after 3 h. The constant mRNA levels may be explained by the combination of a maximum transcription level and a relatively short half-life of mRNA. Because mRNA transcription has apparently reached a maximum level and because mRNA levels may not reflect any previous differential expression due to the relatively short half-life, the mRNA levels are relatively constant at 3 h. If the mRNA samples are taken at an earlier time, the mRNA levels may show a dependence on inducer concentration. While the quantitative differences between expression levels are difficult to ascertain, the qualitative difference between the induced and uninduced samples is definite.
Based on the relatively constant enzyme activity for the TCE-induced samples shown in Table 2, it is concluded that the Tod enzyme activity is relatively constant for increasing TCE concentrations in the growing-cell assay. The constancy in enzyme activity may be explained by two scenarios. The first is TCE-associated toxicity to the Tod enzyme, documented in previous studies (13, 23), which can deactivate the enzyme system. More enzyme is produced at the higher TCE concentrations, which is supported by the bioluminescence data, but more TCE is then present to deactivate the enzymes. If the amount of enzyme deactivated increases with TCE concentration, then the relatively constant enzyme activity may be explained by a balance between the production and toxicity of Tod enzyme. For a specific bioluminescence of 2,690 nA/mg of protein, enzyme activity for TCE-induced samples decreases threefold relative to that of the toluene-induced samples. The decrease may be attributed to the enzyme toxicity associated with TCE.
Alternately, the interaction of TCE with the regulatory proteins, such as TodS and TodT, is expected to be different than the interaction of toluene with these regulatory proteins. If the regulatory proteins do play a significant role in the level of enzyme activity or enzyme concentration present in the cell, then certainly the regulatory system’s effect on enzyme activity would be different in the presence of TCE as the sole inducer. Because the level of enzyme activity is constant for increasing TCE concentrations, the data suggest that the Tod regulatory system is insensitive to TCE concentration at 3 h.
This is the first report of a correlation between specific bioluminescence and initial toluene degradation rate, which is a measure of toluene dioxygenase enzyme activity. Based on these types of correlations as standard curves, specific bioluminescence from the growing-cell assays may be used to measure enzyme activity in toluene-induced bioreactor and groundwater samples.
Acknowledgments
This work was supported by the Waste Management Research and Education Institute of the University of Tennessee and in part by U.S. Department of Energy Office of Biological and Environmental Research grant DE-FG05-94ER61870 and by a SPHERE award from the Dow Foundation.
We thank Steve Ripp, David Nivens, Jim Fleming, Claudia Werner, and Rebecca Eisele for reviewing the manuscript. Hae-Jin Woo and Nathan Bright provided excellent technical assistance.
REFERENCES
- 1.Applegate B M, Kehrmeyer S R, Sayler G S. A chromosomally based tod-luxCDABE whole-cell reporter for benzene, toluene, ethylbenzene, and xylene (BTEX) sensing. Appl Environ Microbiol. 1998;64:2730–2735. doi: 10.1128/aem.64.7.2730-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Applegate B M, Kelly C, Lackey L, McPherson J, Kehrmeyer S, Menn F-M, Bienkowski P, Sayler G S. Pseudomonas putida B2: a tod-lux bioluminescent reporter for toluene and trichloroethylene co-metabolism. J Ind Microbiol Biotechnol. 1997;18:4–9. doi: 10.1038/sj.jim.2900334. [DOI] [PubMed] [Google Scholar]
- 3.Chang H-L, Alvarez-Cohen L. Transformation capacities of chlorinated organics by mixed cultures enriched on methane, propane, toluene, or phenol. Biotechnol Bioeng. 1995;45:440–449. doi: 10.1002/bit.260450509. [DOI] [PubMed] [Google Scholar]
- 4.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ensley B D. Biochemical diversity of trichloroethylene metabolism. Annu Rev Microbiol. 1991;45:283–299. doi: 10.1146/annurev.mi.45.100191.001435. [DOI] [PubMed] [Google Scholar]
- 6.Folsom B R, Chapman P J, Pritchard P H. Phenol and trichloroethylene degradation by Pseudomonas cepacia G4: kinetics and interaction between substrates. Appl Environ Microbiol. 1990;56:1279–1285. doi: 10.1128/aem.56.5.1279-1285.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson D T, Yeh W, Liu T, Subramanian V. Toluene dioxygenase: a multicomponent enzyme system from Pseudomonas putida. In: Nozaki M, et al., editors. Oxygenases and oxygen metabolisms. New York, N.Y: Academic Press, Inc.; 1982. pp. 51–61. [Google Scholar]
- 8.Green D W, editor. Perry’s chemical engineers’ handbook. 6th ed. New York, N.Y: McGraw-Hill, Inc.; 1984. pp. 12–45. [Google Scholar]
- 9.Heald S, Jenkins R O. Trichloroethylene removal and oxidation toxicity mediated by toluene dioxygenase of Pseudomonas putida. Appl Environ Microbiol. 1994;60:4634–4637. doi: 10.1128/aem.60.12.4634-4637.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heitzer A, Applegate B M, Kehrmeyer S, Pinkart H, Webb O F, Phelps T J, White D C, Sayler G S. Physiological considerations of environmental applications of lux reporter fusions. J Microbiol Methods. 1998;33:45–57. [Google Scholar]
- 11.Heitzer A, Webb O F, Thonnard J E, Sayler G S. Specific and quantitative assessment of naphthalene and salicylate bioavailability by using a bioluminescent catabolic reporter bacterium. Appl Environ Microbiol. 1992;58:1839–1846. doi: 10.1128/aem.58.6.1839-1846.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lane D J, Pace J, Olsen G J, Stahl D A, Sogin M L, Pace N R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leahy J G, Byrne A M, Olsen R H. Comparison of factors influencing trichloroethylene degradation by toluene-oxidizing bacteria. Appl Environ Microbiol. 1996;62:825–833. doi: 10.1128/aem.62.3.825-833.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClay K, Streger S H, Steffan R J. Induction of toluene oxidation activity in Pseudomonas mendocina KR1 and Pseudomonas sp. strain ENVPC5 by chlorinated solvents and alkanes. Appl Environ Microbiol. 1995;61:3479–3481. doi: 10.1128/aem.61.9.3479-3481.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oldenhuis R, Vink R L, Vink J M, Janssen D B, Witholt B. Degradation of chlorinated aliphatic hydrocarbons by Methylosinus trichosporium OB3b expressing soluble methane monooxygenase. Appl Environ Microbiol. 1989;55:2819–2826. doi: 10.1128/aem.55.11.2819-2826.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petura L C. Trichloroethylene and methylchloroform in ground water: a problem assessment. J Am Water Works Assoc. 1981;73:200–205. [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 1. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 7.53–7.55. [Google Scholar]
- 18.Sayler G S, Lund L C, Shiaris M P, Sherrill T W, Perkins R E. Comparative effects of AroClor 1254 (polychlorinated biphenyls) and phenanthrene on glucose uptake by freshwater microbial populations. Appl Environ Microbiol. 1979;37:878–885. doi: 10.1128/aem.37.5.878-885.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stanier R Y, Palleroni N J, Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966;41:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 20.Stecher P G. The Merck index. 8th ed. Rahway, N.J: Merck and Co., Inc.; 1968. p. 1069. [Google Scholar]
- 21.Storck W. Chlorinated solvents use hurt by federal rules. Chem Eng News. 1987;65:11. [Google Scholar]
- 22.Tsien H-C, Brusseau G A, Hanson R S, Wackett L P. Biodegradation of trichloroethylene by Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1989;55:3155–3161. doi: 10.1128/aem.55.12.3155-3161.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wackett L P. Bacterial co-metabolism of halogenated compounds. In: Young L Y, Cerniglia C E, editors. Microbial transformation and degradation of toxic chemicals. New York, N.Y: Wiley-Liss, Inc.; 1995. pp. 217–241. [Google Scholar]
- 24.Wackett L P, Gibson D T. Degradation of trichloroethylene by toluene dioxygenase in whole cells by Pseudomonas putida F1. Appl Environ Microbiol. 1988;54:1703–1708. doi: 10.1128/aem.54.7.1703-1708.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winter R B, Yen K-M, Ensley B D. Efficient degradation of trichloroethylene by a recombinant Escherichia coli. Biotechnology. 1989;7:282–285. [Google Scholar]
- 26.Zylstra G J, Wackett L, Gibson D T. Trichloroethylene degradation by Escherichia coli containing the cloned Pseudomonas putida F1 toluene dioxygenase genes. Appl Environ Microbiol. 1989;55:3162–3166. doi: 10.1128/aem.55.12.3162-3166.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]