Abstract
CA-074 is a selective inhibitor of cathepsin B, a lysosomal cysteine protease. CA-074 has been utilized in numerous studies to demonstrate the role of this protease in cellular and physiological functions. Cathepsin B in numerous human disease mechanisms involves its translocation from acidic lysosomes of pH 4.6 to neutral pH 7.2 of cellular locations including the cytosol and extracellular environment. To gain in-depth knowledge of CA-074 inhibition at these different pH conditions, this study evaluated the molecular features, potency, and selectivity of CA-074 for cathepsin B inhibition at acidic and neutral pH conditions. This study demonstrated that CA-074 is most effective at inhibiting cathepsin B at the acidic pH of 4.6 with nM potency, which was more than 100-fold more potent than its inhibition at the neutral pH of 7.2. The pH-dependent inhibition of CA-074 was abolished by methylation of its C-terminal proline, indicating the requirement for the free C-terminal carboxyl group for pH-dependent inhibition. At these acidic and neutral pH conditions, CA-074 maintained its specificity for cathepsin B over other cysteine cathepsins, displayed irreversible inhibition, and inhibited diverse cleavages of peptide substrates of cathepsin B assessed by profiling mass spectrometry. Molecular docking suggested that pH-dependent ionic interactions of the C-terminal carboxylate of CA-074 occur with His110 and His111 residues in the S2' subsite of the enzyme at pH 4.6, but these interactions differ at pH 7.2. While high levels of CA-074 or CA-074Me (converted by cellular esterases to CA-074) are used in biological studies to inhibit cathepsin B at both acidic and neutral pH locations, it is possible that adjusted levels of CA-074 or CA-074Me may be explored to differentially affect cathepsin B activity at these different pHs. Overall, results of this study demonstrates the molecular, kinetic, and protease specificity features of CA-074 pH-dependent inhibition of cathepsin B.
Graphical Abstract
Introduction
Cathepsin B is a lysosomal cysteine protease that participates in protein degradation for cellular protein homeostasis (1-3). Significantly, cathepsin B generates biologically active protein fragments that regulate functions in numerous human diseases including neurological disorders such as Alzheimer’s disease and traumatic brain injury (TBI) (3-7), cancer (8-10), autoinflammatory diseases conditions such as rheumatoid arthritis (11-13), atherosclerosis (14, 15), and others (16-19). Use of the selective cathepsin B inhibitor, CA-074, has facilitated studies of cathepsin B inhibition that ameliorates dysfunctional phenotypes in cell and animal models of human disease conditions (3). For example, CA-074 improves memory deficits and neuropathology in an Alzheimer’s disease (AD) mouse model (5), and CA-074 reduces the severity of motor dysfunction and accelerates recovery in the control cortical impact (CCI) model of traumatic brain injury (TBI) (6). These animal studies administered the pro-inhibitor CA-074Me that is converted by esterases in vivo to the potent CA-074 inhibitor (5, 6).
CA-074 is an epoxysuccinyl peptide known as N-(L-3-trans-propylcarbamoyloxirane-2-carbonyl)-L-isoleucyl-L-proline that was designed as a derivative of E-64 (L-trans-epoxysuccinyl-leucylamido(4-guanidino)butane (20,21), a natural product molecule originally isolated from Aspergillus japonicus (22). CA-074 has been reported to potently inhibit cathepsin B with nanomolar potency (20, 21) and selectivity for inhibition of cathepsin B compared to the related cysteine cathepsins L, H, and S (20-22). These inhibitory properties of CA-074 used purified rat cathepsin B activity monitored at pH 5.5 with the fluorogenic substrate Z-Arg-Arg-AMC (21).
While cathepsin B normally functions at the lysosomal pH of 4.6 (23, 24), evidence shows that in numerous human disease conditions, lysosomal leakage results in movement of cathepsin B to the cytosol of neutral pH 7.2 (25, 26). Cathepsin B in the cytosol is proteolytically active and initiates inflammation and cell death in numerous neurological disorders (3). Cell death lysis results in extracellular cathepsin B that causes damage through proteolysis at neutral pH (27, 28). Cathepsin B also translocates to the nucleus of neutral pH (29) where it results in telomere-related chromosome segregation defects (30) and degradation of sirtuins that promote aging (31). Furthermore, cathepsin B functions in secretory vesicles at a moderately acidic pH of 5.5 for production of peptide neurotransmitters (32). These results indicate that cathepsin B functions at neutral pH conditions which differs from its function in acidic lysosomes.
To gain more in-depth knowledge of CA-074 inhibition of cathepsin B at neutral pH compared to acidic pH conditions, this study examined the potency, molecular properties, and protease specificity of CA-074 inhibition at pH 4.6, 5.5, and 7.2. At pH 4.6, CA-074 was 7-fold and 120-fold more potent than at pH 5.5 and pH 7.2, respectively, for inhibiting cathepsin B. CA-074 retained specificity for inhibition of cathepsin B compared to other cysteine cathepsin family members at the acidic and neutral pH conditions. Notably, the methylated form of CA-074, CA-074Me (33), did not display pH-dependent inhibition and was much less potent than CA-074. These findings reveal the pH-dependent inhibitory properties of CA-074 for specific inhibition of cathepsin B.
Results
CA-074 displays activity at acidic to neutral pH conditions that correspond to subcellular locations of cathepsin B functions.
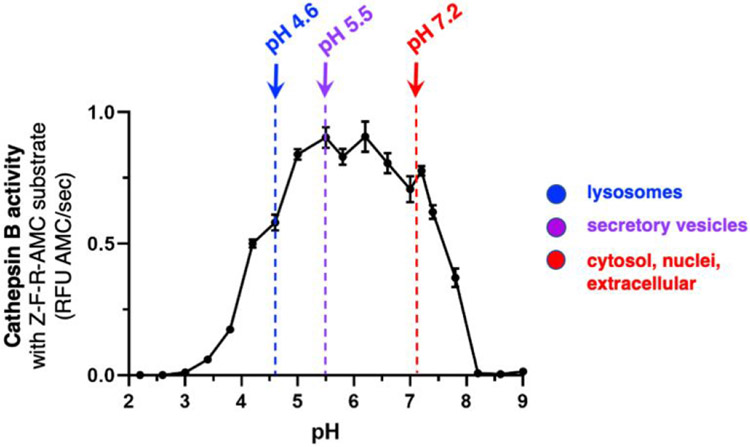
We assessed cathepsin B activity at acidic to neutral pH conditions using the fluorogenic substrate, Z-Phe-Arg-7-amino-4-methylcoumarin (Z-Phe-Arg-AMC). Z-Phe-Arg-AMC monitors cathepsin B activity from acidic to neutral pH conditions (Figure 1). Data shows that robust cathepsin B activity occurs between pH 4.6 to pH 7.2 (Figure 1). These results showed that cathepsin B is active at pH 4.6 of acidic lysosomes (23, 24), pH 5.5 of mildly acidic secretory vesicles (29), and neutral pH 7.2 of cytosol (25, 26), nuclei (29), and extracellular locations (29). Thus, cathepsin B is active over a wide pH range that correlated with the environment of multiple subcellular and extracellular locations.
Figure 1. Cathepsin B activity at acidic to neutral pH conditions representing the varying pH environments of cellular organelles from lysosomes to cytosol and extracellular locations.
Cathepsin B activity was monitored at pH 2.2 to pH 9.0 in increments of 0.4 pH units with addition of pH 7.2. The substrate Z-Phe-Arg-AMC (Z-F-R-AMC) was utilized in the cathepsin B assays. The biological pH conditions of lysosomes at pH 4.6 (23, 24), secretory vesicles at pH 5.5 (29, 32), and neutral cellular compartments of the cytosol, nuclei, and extracellular locations (28-30) are indicated.
Differential potencies of CA-074 inhibition of cathepsin B at acidic to neutral pHs.
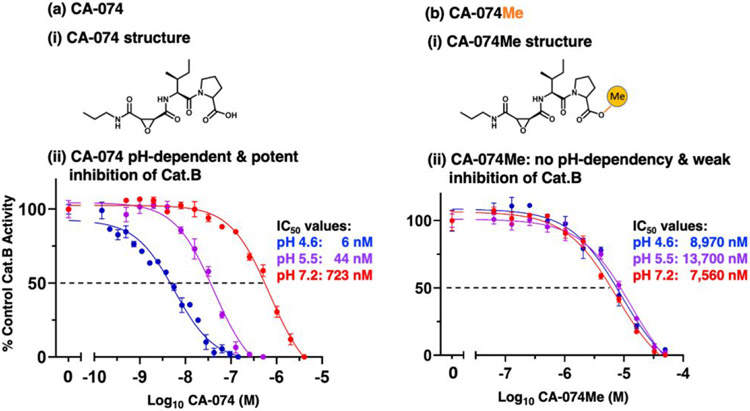
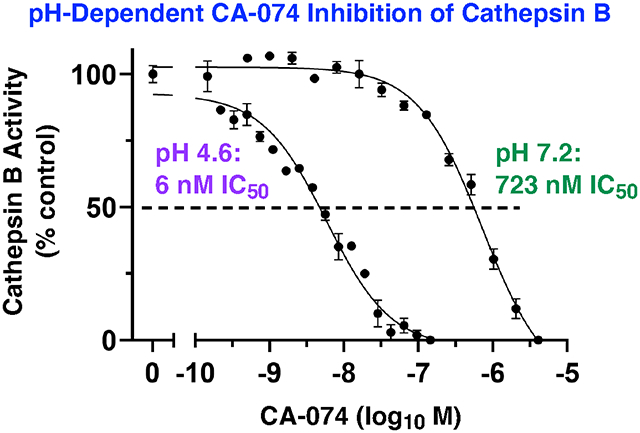
The potency of CA-074 inhibition of cathepsin B was assessed at pH 4.6, 5.5, and 7.2 (Figure 2a). At the lysosomal pH of 4.6, CA-074 was an effective inhibitor with an IC50 value of 6 nM (IC50, concentration of inhibitor for 50% inhibition), but at neutral pH 7.2 it was 120-fold less potent with an IC50 value of 723 nM (Figure 2a.ii). Inhibition at pH 5.5 was also assessed since cathepsin B is present in secretory vesicles (32). The pH of 5.5 was used in the original studies of CA-074 (21) and is also the pH condition that is routinely used in the field (34, 35). At pH 5.5, CA-074 inhibited cathepsin B with an IC50 value of 44 nM (Figure 2a.ii). These data show that CA-074 at pH 4.6 is 7-fold and 120-fold more potent than at pH 5.5 and pH 7.2, respectively, demonstrating the pH-dependent properties of CA-074 for cathepsin B inhibition.
Figure 2. CA-074 pH-dependent inhibition of cathepsin B at acidic to neutral pH conditions requires unmodified C-terminal proline.
(a) Effective pH-dependent CA-074 inhibition of cathepsin B at pH 4.6, 5.5, and 7.2.
(i) CA-074 structure. The chemical structure of CA-074 [N-3-trans-propylcarbamoyl-exirane-2-carbonyl)-L-isoleucyl-L-proline] is illustrated. CA-074 is an epoxysuccinyl inhibitor of cathepsin B (20, 21).
(ii) Potent, pH-dependent CA-074 inhibition of cathepsin B at acidic to neutral pH conditions. Potencies of CA-074 inhibition of cathepsin B were assessed at pH 4.6, pH 5.5, and pH 7.2 over a range of concentrations of CA-074. Inhibitory potencies were assessed by IC50 values, representing inhibitor concentrations that reduced cathepsin B activity by 50%.
(b) Poor CA-074Me inhibition of cathepsin B at pH 4.6, 5.5, and 7.2.
(i) CA-074Me structure. The chemical structure of CA-074Me is illustrated, showing methylation of the carboxylate group of the C-terminal proline of CA-074 (33).
(ii) Poor CA-074Me inhibition of cathepsin B at acidic to neutral pH conditions. Potencies of CA-074 inhibition of cathepsin B were assessed at pH 4.6, pH 5.5, and pH 7.2 over a range of concentrations of CA-074Me. Inhibitory potencies were evaluated by IC50 values, representing inhibitor concentrations that reduced cathepsin B by 50%.
Methylation of CA-074 abolishes its pH-dependent inhibition and reduces potency.
Importantly, methylation of the C-terminal proline of CA-074, generating CA-074Me, resulted in no pH-dependent inhibition of cathepsin B (Figure 2b). These data indicate that the non-methylated C-terminal carboxylate of the proline residue of CA-074 is necessary for its potency and pH-dependent inhibition. Compared to CA-074, CA-074Me was a weak inhibitor of cathepsin B at acidic and neutral pH conditions, shown by its high IC50 values of 8.9 μM, 13.7 μM, and 7.6 μM at pH 4.6, 5.5, and 7.2, respectively (Figure 2b.ii). Thus, CA-074Me was less potent than CA-074 by 1495-fold, 311-fold, and 10-fold at pH 4.6, pH 5.5, and pH 7.2, respectively.
Irreversible mechanism of CA-074 inhibition at acidic and neutral pH conditions.
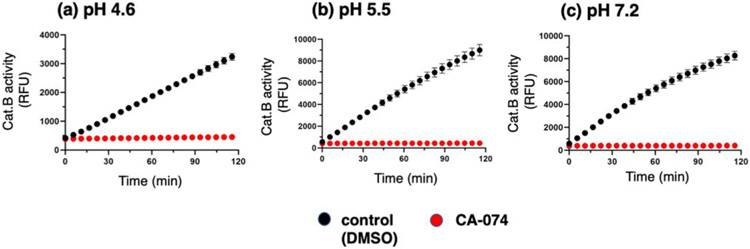
CA-074 has been proposed as an irreversible inhibitor based on structural binding analyses of the inhibitor to the cathepsin B protein structure (36, 37). However, direct evaluation of the irreversible nature of CA-074 inhibition has not yet been assessed. Therefore, the irreversible or reversible nature of protease inhibitors was assessed for CA-074. CA-074 was incubated with cathepsin B for 30 minutes at pH 4.6, pH 5.5, and pH 7.2 at 10-times the IC50 concentrations, then diluted 100-fold followed by addition of Z-F-R-AMC substrate (Figure 3). The lack of cathepsin B activity after dilution demonstrates the irreversible inhibitory mechanism of CA-074 at all three pH conditions. As a control, cathepsin B was preincubated without inhibitor and displayed a linear formation of product in the assay (Figure 3). These data show that inhibition of cathepsin B with CA-074 is irreversible at acidic and neutral pH conditions.
Figure 3. Irreversible CA-074 inhibition of cathepsin B at pH 4.6, pH 5.5, and pH 7.2.
CA-074 at pH 4.6 (panel a), pH 5.5 (panel b), and pH 7.2 (panel c) was evaluated for irreversible or reversible inhibition of cathepsin B by dilution experiments. Cathepsin B was pre-incubated with inhibitor at 10 times the IC50 concentration (corresponding to 58 nM at pH 4.6, 440 nM at pH 5.5, and 7230 nM at pH 7.2), followed by dilution to 1/10 the IC50 concentration, addition of substrate (Z-F-R-AMC), and measurement of activity in time-course assays. Inhibition of cathepsin B following dilution indicates the irreversible inhibitory mechanism of CA-074 at acidic and neutral pH conditions.
In addition, CA-074Me also displays irreversible inhibition of cathepsin B at pH 4.6, 5.5, and 7.2 (supplemental Figure S1). These results illustrate the irreversible mechanism of the weak inhibitor CA-074Me.
Kinetic evaluation of CA-074 KI and kinact values for inhibition of cathepsin B at acidic and neutral pHs.
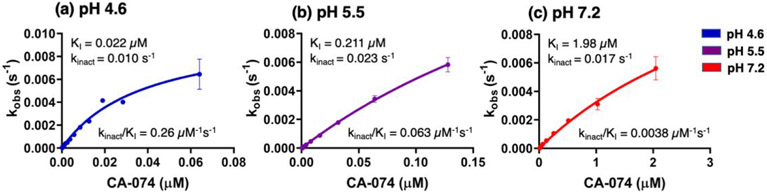
Kinetic characterization of CA-074 irreversible inhibition was conducted to assess KI and kinact values based on plots of inhibitor concentration and kobs values. The kobs values were assessed from plots of cathepsin B activity at different inhibitor concentrations (supplemental Figure S2). Data showed that CA-074 was most potent at pH 4.6 with a KI of 22 nM (Figure 4), and was a less effective inhibitor at pH 7.2 with a KI of 1.98 μM (Figure 4). Inhibition at pH 5.5 was observed with a moderate KI value of 211 nM. These KI values showed that the potency of inhibition at pH 4.6 was about 10-fold and 90-fold greater compared to that at pH 5.5 and pH 7.2, respectively (Table 1). Thus, KI and IC50 values both indicated the greater potency of CA-074 at acidic compared to neutral pH conditions (Table 1). Evaluation of kinact/KI values showed more effective inhibition kinetics at pH 4.6 than at pH 5.5 or pH 7.2. At pH 4.6, the kinact /KI value of 4.5 x 105 M−1s−1 was 4-fold and 52-fold greater than such values at pH 5.5 or pH 7.2 with values of 1.1 x 105 and 8.6 x 103 M−1s−1, respectively (Table 1). These data illustrate the kinetic properties of pH-dependent inhibition by CA-074.
Figure 4. Kinetics of CA-074 inhibition of cathepsin B at acidic and neutral pH conditions.
Kinetic analyses of CA-074 inhibition of cathepsin B was conducted at pH 4.6 (panel a), pH 5.5 (panel b), and pH 7.2 (panel c) to determine the kinetic values of KI, kinact, and kinact /KI, conducted as described in the methods.
Table 1.
Kinetic Values for CA-074 and CA-074Me Inhibition of Cathepsin B at pH 4.6, pH 5.5, and pH 7.2
| Kinetic constant |
Cathepsin B CA-074 |
Cathepsin B CA-074Me |
||||
|---|---|---|---|---|---|---|
| pH 4.6 | pH 5.5 | pH 7.2 | pH 4.6 | pH 5.5 | pH 7.2 | |
| IC50 (nM) | 6 | 44 | 723 | 8,970 | 13,700 | 7,560 |
| KI (nM) | 22 | 211 | 1,980 | 52,000 | 56,000 | 29,000 |
| kinact (s−1) | 0.010 | 0.023 | 0.017 | 0.016 | 0.014 | 0.014 |
| kinact/KI(M−1s−1 ) | 4.5 x 105 | 1.1 x 105 | 8.6 x 103 | 3.1 x 102 | 2.5 x 102 | 4.8 x 102 |
IC50 values were calculated for cathepsin B based on assays with Z-Phe-Arg-AMC substrate in the presence of a range of inhibitor concentrations (from Figure 2). The KI, kinact, and kinact/KI values were calculated as described in the methods to measure kobs values at different inhibitor concentrations (supplemental Figure S2 and Figure S3) for determination of KI, kinact, and kinact/KI values shown in this Table. KI calculation utilized Km values for Z-Phe-Arg-AMC substrate at pH 4.6, 5.5, and 7.2 (supplemental Figure S6).
The kinetic properties of CA-074Me were assessed, indicating its poor inhibition of cathepsin B (Table 1, and supplemental Figure S3). CA-074Me inhibition of cathepsin B at pH 4.6, pH 5.5, and pH 7.2 occurred with KI values of 52 μM, 56 μM, and 29 μM, respectively, which represented lower potencies by 1/2,363, 1/265, and 1/15 at these respective pHs compared to CA-074. The kinact/KI values of CA-074Me also indicated substantially weaker inhibition compared to CA-074 (Table 1).
Specificity of CA-074 inhibition of cathepsin B compared to other cysteine cathepsins at acidic and neutral pHs.
Cathepsin B is one among 11 lysosomal cysteine cathepsin proteases consisting of cathepsins B, C, F, H, K, L, O, S, V, W, and X (also known as cathepsin Z) (1, 38). While previous studies of CA-074 assessed its selectivity for cathepsin B compared to cathepsins L and H at pH 5.5 (20, 21), selectivity for the full spectrum of cysteine cathepsins was not assessed at both acidic and neutral pH conditions. Therefore, we evaluated the selectivity of CA-074 among the cysteine cathepsins at pH 4.6, pH 5.5, and pH 7.2 (Table 2). At pH 4.6, 16 μM CA-074 had no effect on cathepsins C, H, and L; CA-074 at 16 μM displayed minor effects on cathepsins K, S, V, and X of 5-20% inhibition. At pH 5.5, 16 μM CA-074 μM had no effect on these cysteine cathepsins C, H, L, K, V, and X; but cathepsin S was inhibited with an IC50 of 4.8 μM, showing 109-fold less potency than inhibition of cathepsin B. At pH 7.2, CA-074 at 16 μM had no effect on cathepsin C, H, K, V, and partial inhibition of cathepsin S by 29%; cathepsins L and X were inactive at pH 7.2. These findings illustrate the high specificity of CA-074 for cathepsin B over other cysteine cathepsin enzymes at acidic and neutral pH conditions.
Table 2.
Specificity of CA-074 and CA-074Me for inhibition of cathepsin B compared to other cysteine cathepsins.
| Protease | CA-074 IC50 (nM) (% inhibition) |
CA-074Me IC50 (nM) (% inhibition) |
||||
|---|---|---|---|---|---|---|
| pH 4.6 | pH 5.5 | pH 7.2 | pH 4.6 | pH 5.5 | pH 7.2 | |
| Cathepsin B | 6 | 44 | 723 | 8,970 | 13,700 | 7,560 |
| Cathepsin C | >16,000 0% |
>16,000 0% |
>16,000 0% |
>16,000 5% |
>16,000 0% |
>16,000 4% |
| Cathepsin H | >16,000 0% |
>16,000 0% |
>16,000 0% |
>16,000 0% |
>16,000 0% |
>16,000 19% |
| Cathepsin L | >16,000 0% |
>16,000 0% |
NA | >16,000 0% |
>16,000 0% |
NA |
| Cathepsin K | >16,000 8% |
>16,000 0% |
>16,000 0% |
>16,000 0% |
>16,000 0% |
>16,000 16% |
| Cathepsin S | >16,000 20% |
4,800 | >16,000 29% |
>16,000 22% |
5,500 | 3,800 |
| Cathepsin V | >16,000 10% |
>16,000 0% |
>16,000 0% |
12,000 33% |
>16,000 22% |
>16,000 0% |
| Cathepsin X | >16,000 5% |
>16,000 0% |
NA | >16,000 0% |
>16,000 0% |
NA |
Inhibitors were evaluated for protease selectivity among members of the cysteine cathepsin family. The activity of each cathepsin was assessed in the presence of CA-074 or CA-074Me at 0.5 nM to 16 μM (with the exception of 50 μM CA-074Me for cathepsin B). IC50 values were determined for CA-074 and CA-074Me for each protease. IC50 values are indicated as >16,000 nM when partial inhibition or no inhibition was observed. NA indicates that the enzyme had no activity at the indicated pH.
With respect to CA-074Me, it displayed nearly no inhibition of the cysteine cathepsins assessed at up to 16 μM CA-074Me (Table 2). Cathepsin S, however, was inhibited by high concentrations illustrated by the micromolar IC50 values of 5.5 μM and 3.8 μM CA-074Me at pH 5.5 and pH 7.2, respectively.
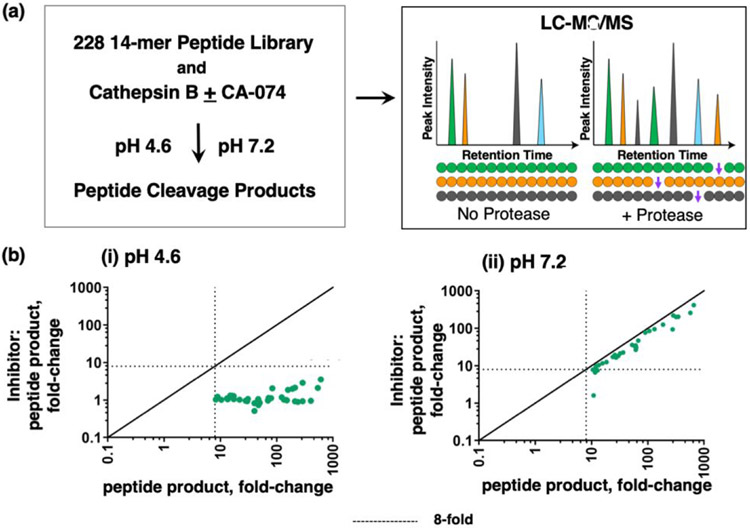
CA-074 inhibition of peptide cleavages of cathepsin B assessed by multiplex substrate profiling by mass spectrometry (MSP-MS).
The inhibitory properties of CA-074 have largely been evaluated with dipeptide-AMC fluorogenic substrates such as Z-Phe-Arg-AMC and Z-Arg-Arg-AMC (20, 21), but information about CA-074 inhibition of diverse cathepsin B cleavages is needed to characterize the inhibitor effects. To gain knowledge of cathepsin B peptide cleavages that can be inhibited by CA-074 at pH 4.6 and pH 7.2 for broader substrates, we evaluated the ability of CA-074 to inhibit cathepsin B cleavages by a global, unbiased cleavage profiling approach known as multiplex substrate profiling mass spectrometry (MSP-MS) (39, 40). The MSP-MS approach uses a substrate library consisting of 228 peptides (14 residues in length) containing 2,964 distinct cleavage sites. In the absence and presence of CA-074, cathepsin B cleavage products of the library were identified and quantified by nano-liquid chromatography tandem mass spectrometry (nano-LC-MS/MS). In the presence of 10 nM CA-074, cleavages of peptide substrates of the library were blocked by CA-074 (Figure 5). At pH 7.2, 10 nM CA-074 had no effect on cathepsin B cleavage the Z-Phe-Arg-AMC substrate (Figure 2) and, similarly, had no effect on cleaving peptide substrates in the MSP-MS assay (Figure 5). These findings confirm that CA-074 has selectivity for inhibiting cathepsin B peptide cleavages at pH 4.6 over pH 7.2 for broader substrates.
Figure 5. CA-074 inhibition of peptide cleavages by cathepsin B analyzed by multiplex substrate profiling by mass spectrometry (MSP-MS).
(a) MSP-MS approach for cleavage profiling of cathepsin B in the presence or absence of CA-074. Cathepsin B was preincubated with CA-074 (10 nM) at pH 4.6 and pH 7.2 for 30 min at RT, and without inhibitor as control. These cathepsin B conditions were then subjected to MSP-MS assays by addition of the peptide library and incubation (at RT) for 60 min, followed by LC-MS/MS identification and quantification of peptide cleavage products.
(b) Cleaved peptide products of cathepsin B in MSP-MS at pH 4.6 (panel i) and pH 7.2 (panel ii). The quantities of each cleaved peptide product generated in the absence of inhibitor or in the presence of inhibitor were plotted as the fold-change of each cleaved peptide product relative to no enzyme activity control. Cleaved peptides were defined as those with intensity scores of 8-fold or more (----) above the quenched inactive cathepsin B, evaluated using the ratio of log2(Cat.B/inactivated enzyme) for each peptide product, with p < 0.05 by 2-tailed homoscedastic t-test.
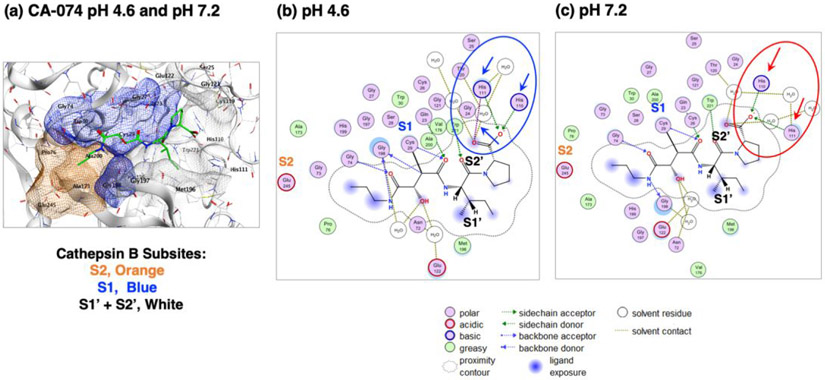
Molecular docking of CA-074 interactions with cathepsin B at acidic pH 4.6 and neutral pH 7.2.
Modeling of CA-074 binding interactions to cathepsin B at acidic pH 4.6 and neutral pH 7.2 was evaluated by the Molecular Operating Environment (MOE) docking software (41, 42). MOE modeled CA-074 binding to bovine cathepsin B (PDB: 1QDQ) (36) at pH 4.6 and pH 7.2, showing similar orientation at the active site (Figure 6a). Mature human and bovine cathepsin B share 88.3% protein sequence identity and 100% sequence identity for the amino acids (His 110 and His111) that directly interact with CA-074 (37). The docking assessment shows that CA-074 interacts with the S2, S1, S1' and S2' subsites of the enzyme according to the Schechter-Berger nomenclature (43), where the subsites interact with the corresponding amino acids adjacent to the cleavage site indicated as P2-P1-↓P1'-P2'. At pH 4.6, the P2' residue of CA-074 at the C-terminus (proline residue) shows a strong ionic interaction His110 and His111 in the S2' subsite of the enzyme (Figure 6b), reflected by total binding energy of −58.3 kcal/mol (Table 3, and supplemental Figure S4). However, at pH 7.2, the P2' residue of CA-074 at the C-terminus partially loses ionic interaction with His111 in the S2' subsite of the enzyme (Figure 6c), reflected by the altered binding energy of −43 kcal/mol (Table 3, and supplemental Figure S5), when compared to the binding energy at pH 4.6. These MOE docking analyses showing that CA-074 binds more favorably to cathepsin B at pH 4.6 than at pH 7.2, which supports our experimental data illustrating the greater inhibition by CA-074 at acidic compared to neutral pH conditions.
Figure 6. Molecular docking of CA-074 to cathepsin B at pH 4.6 and pH 7.2.
(a) Model of CA-074 docking to cathepsin B at pH 4.6 and pH 7.2. Analyses of CA-074 inhibitor binding to enzyme was conducted by MOE at pH 4.6 and pH 7.2 to illustrate the predicted docking orientation of the inhibitor to the cathepsin B structure of PDB 1QDQ as template for analyses (36, 37). CA-074 interacts with S2 to S2’ subsites of the enzyme substrate binding region (Schechter-Berger nomenclature (41)), containing the active site Cys29 residue. Cathepsin B enzyme subsites are shown as the S2 subsite in orange, the S1 subsite in blue, and the S1 and S2’ subsites in gray-white.
(b) 2-Dimensional illustration of CA-074 and cathepsin B binding interactions at pH 4.6. At pH 4.6, the P2’ proline residue of CA-074 at its C-terminus shows strong ionic interactions with His110 and His111 in the S2’ subsite of the occluding loop region of the enzyme (panel b, indicated by blue arrows)), indicated by the total binding energy of −58.3 kcal/mol (Table 3, and supplemental Figure S4).
(b) 2-Dimensional illustration of CA-074 and cathepsin B binding interactions at pH 7.2. At pH 7.2, the P2’ proline residue of CA-074 at the C-terminus partially loses ionic interaction with His111 in the S2’ subsite of the enzyme (panel c, indicated by red arrows), reflected by the altered binding energy of −43 kcal/mol (Table 3, and supplemental Figure S5), when compared to the binding energy at pH 4.6. These MOE docking analyses predict the differential binding features of CA-074 to the occluding loop domain of cathepsin B at pH 4.6 compared to pH 7.2.
Table 3.
Binding energies of CA-074 to cathepsin B at pH 4.6 and pH 7.2
| pH condition |
Binding energy |
|---|---|
| pH 4.6 | − 58.3 kca/mol |
| pH 7.2 | − 43.0 kcal/mol |
The more negative binding energy calculated for pH 4.6 compared to pH 7.2 predict the more favorable interaction of CA-074 to cathepsin B at pH 4.6.
Discussion
This study demonstrates the potency, molecular features, and protease specificity of CA-074 inhibition of cathepsin B, showing that CA-074 displays preference for inhibition at the acidic lysosomal pH of 4.6 compared to the mildly acidic pH 5.5 of secretory vesicles and the neutral pH of 7.2 of cytosol and extracellular locations. KI values indicate that CA-074 is 90-fold more potent for inhibition of cathepsin B at acidic pH 4.6 compared to neutral pH 7.2. CA-074 at pH 5.5 was about 10-fold less effective than at pH 4.6 (based on KI), and was 9-fold more potent than at pH 7.2. These results demonstrate that the potency of CA-074 increases as the pH decreases from pH 7.2 to 4.6. At these acidic and neutral pH conditions, CA-074 maintains its specificity for cathepsin B over other cysteine cathepsins, displays irreversible inhibition, and inhibits diverse peptide cleavages of cathepsin B assessed by MSP-MS profiling. Molecular docking suggested pH-dependent ionic interactions of the carboxylate group of the C-terminal proline of CA-074 with His110 and His111 residues at the S2 subsite of the enzyme. Notably, CA-074Me showed no pH dependent inhibition, indicating that methylation of the C-terminal proline of CA-074 abolishes its pH dependent inhibition of cathepsin B. These results demonstrate the pH-dependent structural and kinetic properties of CA-074 for specific inhibition of cathepsin B.
The distinct features of cathepsin B that result in its preferential inhibition by CA-074 at acidic pH suggest that this enzyme possesses different properties at acidic compared to neutral pH environments. Indeed, cathepsin B has been shown to possess different cleavage specificities at pH 4.6 compared to pH 7.2 (44), observed during unbiased cleavage profiling analyses by MSP-MS (44). Based on the peptide substrate preferences of cathepsin B at the two different pH conditions, a neutral pH selective substrate was modified with the AOMK warhead to generate Z-Arg-Lys-AOMK that displayed neutral pH selective inhibition of cathepsin B with high potency (42). These new findings distinguish cathepsin B properties in its normal acidic environment of lysosomes, versus its abnormal location in the cytosol and extracellularly in numerous human diseases. Together, these data support the hypothesis that neutral pH cathepsin B may represent a pathogenic form of the enzyme involved in disease mechanisms.
Our molecular docking studies by MOE demonstrated the more favorable interaction of the C-terminal region of CA-074 with the occluding loop of cathepsin B at pH 4.6 rather than at pH 7.2. Our molecular docking assessment by MOE at pH 4.6 and pH 7.2 using X-ray crystallography data for cathepsin B (36) shows the importance of the enzyme occluding loop interaction with the C-terminal carboxylate of CA-074. Binding energies of docking at pH 4.6 compared to pH 7.2 support the hypothesis that CA-074 at pH 4.6 displays preferable interaction with the occluding loop through its C-terminal proline carboxylate interaction with His110 and His111 of the enzyme. His110 and His111 have been shown to be key residues for cathepsin B exopeptidase activity. Mutagenesis of His110 to Gln, or His111 to Ala, results in reduction of cathepsin B activity to less than 1% of the wild-type cathepsin B activity when using substrates containing P2' residues with a C-terminal carboxylic acid group (45). Notably, methylation at the carboxylate residue of CA-074, forming CA-074Me, abolished the pH-dependent property of CA-074 with substantially decreased potency at acidic and neutral pHs. These findings indicate that the C-terminus of CA-074 participates in its pH-dependent inhibition and potency. These findings advance previous knowledge (20, 21, 36, 46, 47) concerning the importance of His110 and His111 of the occluding loop for interacting with the C-terminus of CA-074 in a pH-dependent manner.
In cellular and animal studies, the prodrug CA-074Me is widely used for CA-074 inhibition of cathepsin B (3). Treatment of cells with CA-074Me has been shown to more effectively inhibit intracellular cathepsin B compared to incubating cells with CA-074 (33). It has been hypothesized that CA-074Me is more cell permeable, and is converted by intracellular esterases to CA-074 (33). In numerous cellular studies, the concentration of CA-074Me used to inhibit intracellular cathepsin B generally ranges from 10 μM to 50 μM (48-51). With the assumption that most of the pro-inhibitor is converted to CA-074, the high concentration of CA-074 would be sufficient to inhibit cathepsin B located in different subcellular pH conditions of acidic lysosomes and neutral pH locations including cytosol. In animal studies, CA-074Me concentrations in the approximate range of 4-10 mg/kg and higher have been administered (52-56), which may correspond to micromolar and greater levels of the inhibitor that would inhibit cathepsin B at acidic to neutral pH cellular locations. Measurements of CA-074 and CA-074Me in vivo levels and clearance in animal studies will be important in future studies to assess in vivo inhibitor concentrations.
The new findings of this study suggest that adjustment of CA-074 concentrations may allow assessment of cathepsin B functions in acidic lysosomes compared to neutral pH locations such as the cytosol, nuclei, and extracellular environment (3, 57-62). We previously discovered that Z-Arg-Lys-AOMK is >100-fold selective at inhibiting cathepsin B at pH 7.4 over pH 4.6 (44), while in this study CA-074 has the opposite property for preferring inhibition at acidic pH 4.6. Use of these two inhibitors in cellular studies, at concentrations where they maintain pH selectivity, may allow evaluation of the functional roles of lysosomal compared to non-lysosomal cathepsin B.
In summary, this study has demonstrated the pH-dependent inhibitory properties of CA-074 inhibition of cathepsin B with respect to its potency, molecular features, and specificity. At appropriate concentrations, CA-074 may potentially serve as a selective acidic pH inhibitor of cathepsin B, with specificity for cathepsin B over other cysteine cathepsins. These findings proof-of-concept that it may be possible to develop other pH selective inhibitors. Novel pH selective inhibitors may allow studies in the future to specifically probe the role of cathepsin B in distinct pH cellular compartments such as the cytosol that have implicated to be involved in brain disorders and other human diseases.
Methods and Materials
Inhibitors, enzymes, peptides, and reagents.
CA-074 inhibitor was purchased from Calbiochem (Millipore Sigma #205530, St. Louis, MO). CA-074Me was purchased from Sigma (#205531). Recombinant human cathepsin B (accession NP_001899.1) and cysteine cathepsin proteases were from R & D Systems (Minneapolis, MN) or Abcam (Cambridge, MA) consisting of cathepsin B (R & D #953-CY-010), cathepsin L (R & D #952-CY-010), cathepsin V (R & D #1080-CY-010), cathepsin S (R & D #1183-CY-010), cathepsin K (Abcam #ab157067), cathepsin C (R & D #1071-CY-010), cathepsin H (R & D #75116-CY-010), and cathepsin X (R & D #934-CY)-010). Fluorogenic substrates consisted of Z-Phe-Arg-AMC ((#AS-24096, Anaspec, Fremont, CA), Gly-Arg-AMC and Arg-AMC from Bachem ((#4002196 and (#I-1050, respectively, Torrance, CA), and MCA-Arg-Pro-Pro-Gly-Phe-Ser-Ala-Phe-Lys(Dnp)OH ((#AMYD-111A, CPC Scientific, San Jose, CA). MSP-MS (multiplex substrate profiling mass spectrometry) assays utilized a library of 228 peptides of 14 amino acids in length, designed and synthesized to contain all neighbor and near-neighbor diversity in residues, as described previously (37, 38). These assays utilized buffer components of citric acid monohydrate (Merck #1.00244.0500, Burlington, MA), sodium phosphate dibasic anhydrous (EMD #SX-072305, Burlington, MA), sodium acetate (Fisher Scientific #BP-333-500, Fair Lawn, NJ), EDTA (Calbiochem #324503, Burlington, MA), sodium chloride (Fisher Chemical #S271-1, Pittsburgh, PA), dithiothreitol (DTT) (Promega #V351, Madison, WI), and urea (Teknova #U2222, Hollister, CA). Peptide extraction reagents consisted of C18 LTS Tips (Rainin #PT-LC18-960, Oakland, CA) and C18 for SPE stage-tips (3M company #2215-C18, Maplewood, MN), and (3) nano-LC-MS/MS reagents consisting of BEH C18 packing material (Waters Corporation #186004661, Milford, MA), acetonitrile (Fisher Chemical #A955-4, Pittsburgh, PA), formic acid (FA) (Fisher Chemical #A117-50, Pittsburgh, PA), trifluoroacetic acid (TFA) (Fisher Chemical #A116-50, Pittsburgh, PA), and HPLC-grade water (Fisher Chemical #W6-4).
Cathepsin B activity assay.
Pro-cathepsin B (recombinant) was activated to mature cathepsin B by incubation at 37°C for 30 minutes in 20 mM Na-acetate pH 5.5, 1 mM EDTA, 5 mM DTT, 100 mM NaCl. Assay of cathepsin B activity utilized final buffer conditions of 0.04 ng/μL cathepsin B, 40 μM Z-Phe-Arg-AMC, 40 mM citrate phosphate (pH 4.6, pH 5.5, or pH 7.2), 1 mM EDTA, 100 mM NaCl, 5 mM DTT, and 0.01% Tween-20. Assays were conducted at room temperature (25°C) in quadruplicate, and relative fluorescence readings (RFU) (excitation 360 nm, emission 460 nm) were recorded every 46 secs over a period of 30 min (for inhibitor assays) in a Biotek HTX microplate plate reader. For other assays, RFU readings were recorded every 101 secs, 30 secs, and 70 secs for studies of pH dependence (Figure 1), irreversibility (Figure 3), and Km values (SI Figure S6), respectively. Enzyme velocity (RFU/sec) calculations used the highest slope recorded for 10 consecutive fluorescent readings. RFU/s were converted to specific activity of pmol/min/μg using AMC standards. Specific activity was defined as pmol/min/μg enzyme. Data analysis was conducted using Prism GraphPad software.
For the pH curve experiment (Figure 1), cathepsin B activity was also monitored over the entire pH range of 2.2 to 9.0 in 40 mM citrate phosphate (pH 2.2 to pH 7.4) or 40 mM Tris-HCl (pH 7.8 to 9.0), 1 mM EDTA, 5 mM DTT, 100 mM NaCl, and 0.01% Tween-20, with preincubation in each pH buffer for 10 min prior to initiating the assay by adding substrate. We noted that pre-incubation at pH values of 3.0 or 8.0 resulted in reduced activity when compared with our previous studies (44) that had no pre-incubation step.
Inhibition of cathepsin B by CA-074 and CA-074Me in kinetic studies.
Kinetic analyses of CA-074 and CA-074Me inhibition of cathepsin B was conducted at pH 4.6, pH 5.5 and pH 7.2 to determine IC50, KI, kobs, and kinact/KI. The CA-074 inhibitor concentrations ranged from 4096 to 0.5 nM (2-fold serial dilution) or 144 to 0.5 nM (1.5-fold serial dilution). CA-074Me inhibitor concentrations ranged from 65,536 to 64 nM (2-fold serial dilution) or 50,000 to 64 nM (1.5-fold dilution). Inhibitors were added to the enzyme at the start of the incubation period (at RT), thus, there was no preincubation. A vehicle control assay without inhibitor contained enzyme, substrate and 2% (v/v) DMSO. Cathepsin B proteolytic assays with CA-074 were conducted as described above. IC50 values were calculated as the inhibitor concentration that reduced cathepsin B activity by 50%.
For determination of KI and kinact/KI kinetic inhibition constants, kobs constants were determined for each inhibitor concentration by plots of cathepsin B activity in time courses with various inhibitor concentrations by curve fitting slope data of RFU versus time into Y=Y0*e(−kobs*X), where Y0 is the activity for the control with no inhibitor, Y is the activity in the presence of inhibitor, and X is time. KI and kinact values were determined by plots of kobs and inhibitor concentration, combined with curve fitting the kobs values with the equation kobs=kinact*[I]/(KI+[I]), where [I] is inhibitor concentration, kinact is the maximum rate of inactivation at saturating inhibitor concentrations, and KI,app is the x-axis inhibitor concentration where y = kinact/2 (63, 64). KI, values were determined from KI,app and Km values by the equation KI,app= KI *(1+[S]0/Km. Determination of Km values is described in the next paragraph. These analyses are for irreversible inhibitor kinetic characterization.
Km values were determined for Cathepsin B (Z-Phe-Arg-AMC substrate), Cathepsin C (Gly-Arg-AMC), Cathepsin H (Arg-AMC), Cathepsin L (Z-Phe-Arg-AMC), Cathepsin K (Z-Phe-Arg-AMC), Cathepsin S (Z-Phe-Arg-AMC), Cathepsin V (Z-Phe-Arg-AMC), Cathepsin X (MCA-Arg-Pro-Pro-Gly-Phe-Ser-Ala-Phe-Lys(Dnp)-OH) at pH 4.6, pH 5.5 and pH 7.2 (supplemental Figure S6). Enzymes were assayed at different substrate concentrations ranging from 225 to 4 μM (1.5-fold serial dilution) and the slope of RFU per unit time was determined for each substrate concentration; plots of substrate concentration and velocity were assessed by Prism 9 by curve fitting using the equation v0=Vmax*[S]/(Km+[S]) to determine Km values. These kinetic methods are for irreversible inhibitor kinetic characterization.
Irreversible inhibition mechanism.
The irreversible mechanisms of CA-074 and CA-074Me inhibition of cathepsin B were conducted at pH 4.6, pH 5.5, and pH 7.2. Cathepsin B was pre-incubated with each inhibitor at 10 times the IC50 concentration, followed by dilution to 1/10 the IC50 concentration, addition of substrate (Z-F-R-AMC), and monitoring activity in time-course assays. Inhibition of cathepsin B following dilution indicates the irreversible inhibitory mechanism of CA-074 and CA-074Me at acidic and neutral pH conditions.
Selectivity of CA-074 for cathepsin B and cysteine cathepsin proteases.
The selectivity of CA-074 and CA-074Me was assessed by comparing its inhibitory potencies for cathepsin B compared to other cysteine cathepsin proteases consisting of cathepsin V, L, K, S, X, H and C. IC50 values were determined for pH 4.6 and pH 7.2 conditions, consisting of 40 mM citrate phosphate, 1 mM EDTA, 5 mM DTT, and 100 mM NaCl. The inhibitor concentrations ranged from 16.38 μM to 256 nM in 2-fold serial dilutions. When activity (RFU/s) in the presence of 16.38 μM inhibitor was reduced by <50%, the IC50 value was indicated as >16 μM. Cathepsin L (0.03 ng/μL), cathepsin K (0.10 ng/μL), cathepsin S (0.20 ng/μL), and cathepsin V (0.04 ng/μL), were assayed with 40 μM Z-Phe-Arg-AMC. Cathepsin C (0.51 ng/uL), cathepsin H (0.1 ng/μL), and cathepsin X (0.20 ng/uL) were assayed with 40 μM of MCA-Arg-Pro-Pro-Gly-Phe-Ser-Ala-Phe-Lys(Dnp)OH, Gly-Arg-AMC, and Arg-AMC, respectively. Activation of pro-cathepsin H to cathepsin H was achieved by incubation of cathepsin H (4.4 ng/μL) with cathepsin L (1.1 ng/μL) at RT for 2 hrs in 20 mM citrate phosphate pH 6.0, 5 mM DTT, and 100 mM NaCl. Cathepsin C (13.78 ng/μL) was activated by incubation with cathepsin L (3.4 ng/μL) at RT for one hr in 20 mM citrate phosphate, pH 6.0, 100 mM NaCl, 5 mM DTT. Cathepsin L did not cleave the cathepsin C and cathepsin H substrates Gly-Arg-AMC and Arg-AMC, respectively. These fluorogenic assays used the fluorescent microplate reader as described for cathepsin B. For assay of cathepsin X, the reader was set to excitation 320 nm and emission 400 nm. To convert RFU/s to picomol/min, 10 μM to 0.005 μM (in 2-fold dilutions) of MCA-Arg-Pro-Pro-Gly-Phe-Ser-Ala-Phe-Lys(Dnp)OH was fully hydrolyzed with excess Cathepsin X and a standard curve was generated using the fluorescence values measured at each concentration.
CA-074 inhibition of cathepsin B peptide cleavages assessed by MSP-MS (multiplex substrate profiling by mass spectrometry).
CA-074 inhibition of peptide cleavages was evaluated by MSP-MS assays of cathepsin B. Cathepsin B was pre-incubated at 25° C for 30 min with 10 nM of CA-074 in pH 4.6 buffer or pH 7.2 buffer. As a control, cathepsin B was pre-incubated with 2.5 % DMSO in each assay buffer. In a total volume of 10 μL, cathepsin B (0.1 ng/μL) pre-incubated with CA-074 was incubated with a mixture of 228 14-mer peptides (0.5 μM for each peptide) in assay buffer composed of 40 mM citrate phosphate at pH 4.6 or pH 7.2, 1 mM EDTA, 100 mM NaCl and 5 mM DTT for 60 minutes at 25°C. After 60 min incubation, the 10 μL aliquot was combined with 60 μL of 8 M urea. A control assay used activated cathepsin B in each assay buffer mixed with 8 M urea for 60 minutes at 25°C for inactivation, prior to addition of the peptide library. Assays were conducted in quadruplicate. Samples were acidified by addition of 40 μL of 2% TFA, enriched and desalted using C18 LTS Tips (Rainin), evaporated to dryness in a vacuum centrifuge, and placed at −70°C. Samples were resuspended in 40 μL of 0.1% FA (solvent A) and 4 μL was used for LC-MS/MS.
LC-MS/MS was performed on a Q-Exactive Mass Spectrometer (Thermo) equipped with an Ultimate 3000 HPLC (Thermo Fisher). Peptides were separated by reverse phase chromatography on a C18 column (1.7 μm bead size, 75 μm x 20 cm, 65°C) heated to 65°C at a flow rate of 400 nL/min using solvent A and 0.1% FA in acetonitrile (solvent B). LC separation used a 50-minute linear gradient of 5% to 30% solvent B with a subsequent 15-minute linear gradient of 30% to 75% solvent B. Survey scans were recorded over a 200–2000 m/z range (70,000 resolutions at 200 m/z, AGC target 1×106, 75 ms maximum). MS/MS was performed in data-dependent acquisition mode with HCD fragmentation (30 normalized collision energy) on the 10 most intense precursor ions (17,500 resolutions at 200 m/z, AGC target 5×104, 120 ms maximum, dynamic exclusion 15 s).
MS/MS data analysis was performed using PEAKS (v 8.5) software (Bioinformatics Solutions Inc.). MS2 data were searched against the 228-member tetradecapeptide library sequences and a decoy search was conducted with sequences in reverse order. A precursor tolerance of 20 ppm and 0.01 Da for MS2 fragments was defined. No protease digestion was specified. Data were filtered to 1% peptide and protein level false discovery rates with the target-decoy strategy. Peptides were quantified with label free quantification and data normalized by Loess-G algorithm and filtered by 0.5 peptide quality. Using R scripts, outliers from replicates were removed by Dixon’s Q testing. Missing and zero values are imputed with random normally distributed numbers in the range of the average of smallest 5% of the data ± SD. ANOVA testing was performed for peptide data of control and 60 min incubation conditions; those with p<0.05 were considered for further analysis. Criteria for cleaved peptide products were those with intensity scores of 8-fold or more above the quenched inactive cathepsin B, evaluated by log2(active/inactivated enzyme) ratios for each peptide product; with p < 0.05 by 2-tailed homoscedastic t-test
MOE modeling of CA-074 interactions with cathepsin B at acidic and neutral pH conditions.
The Molecular Operating Environment (MOE) molecular modeling tool was used to model CA-074 binding to cathepsin B using the crystal structure of bovine cathepsin B (PDB 1QDQ), co-crystal template with the inhibitor CA-074 as default binding ligand. The builder function of MOE was used to examine binding poses that considered polar contacts and hydrogen bonds between ligand and the active site pocket of 1QDQ at pH 4.6 and pH 7.2. Docking simulations used energy-minimized structures to assess ligand flexibility and poses.
Supplementary Material
Acknowledgments
This research was supported by NIH grant R01NS109075 awarded to VH). M. Yoon was supported by NIH T32GM067550 (awarded to W. Gerwick). V. Hook and G. Hook (spouse) have equity positions at American Life Science Pharmaceuticals (ALSP) and are founders of ALSP. V. Hook is an advisor to ALSP. G. Hook at ALSP is vice president of research, corporate counsel, and member of the board of directors. V. Hook's conflict has been disclosed and is managed by her employer, the University of California, San Diego. The other authors have no conflicts of interest.
Footnotes
Supporting Information
Supplemental Figure S1 illustrates the irreversible mechanism of CA-074Me inhibition of cathepsin B. Figure S2 and Figure S3 display kobs inhibition kinetics of CA-074 and CA-074Me, respectively. Figures S4 and S5 display calculated binding energies of CA-074 interactions to cathepsin B at pH 4.6 and 7.2, respectively.
Data Availability
LC-MS/MS files for the MSP-MS experiments can be accessed at www.proteomexchange.org under the dataset identifier numbers PXD022494 and PXD022493. Alternatively, the data files can be obtained through www.massive.ucsd.edu under the dataset identifier numbers MSV000086449 and MSV000086447.
References
- 1.Turk V, Stoka V, Vasiljeva O, Renko M, Sun T, Turk B, and Turk D (2012) Cysteine cathepsins: from structure, function and regulation to new frontiers, Biochim. Biophys. Acta 1824, 68–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reiser J, Adair B, and Reinheckel T (2010) Specialized roles for cysteine cathepsins in health and disease, J. Clin. Invest 120. 3421–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hook V, Yoon M, Mosier C, Ito G, Podvin S, Head BP, Rissman R, O'Donoghue AJ, and Hook G (2020) Cathepsin B in neurodegeneration of Alzheimer's disease, traumatic brain injury, and related brain disorders, Biochim. Biophys. Acta. Proteins Proteom 1868, 140428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Z, Ni J, Liu Y, Teeling JL, Takayama F, Collcutt A, Ibbett P, Nakanishi H (2017) Cathepsin B plays a critical role in inducing Alzheimer's disease-like phenotypes following chronic systemic exposure to lipopolysaccharide from Porphyromonas gingivalis in mice. Brain Behav Immun. 65, 350–361. [DOI] [PubMed] [Google Scholar]
- 5.Kindy MS, Yu J, Zhu H, El-Amouri SS, Hook V, and Hook GR (2012) Deletion of the cathepsin B gene improves memory deficits in a transgenic Alzheimer's disease mouse model expressing AβPP containing the wild-type β-secretase site sequence, J. Alzheimers Dis 29, 827–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hook GR, Yu J, Sipes N, Pierschbacher MD, Hook V, and Kindy MS (2014) The cysteine protease cathepsin B is a key drug target and cysteine protease inhibitors are potential therapeutics for traumatic brain injury, J. Neurotrauma 31, 515–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boutté AM, Hook V, Thangavelu B, Sarkis GA, Abbatiello BN, Hook G, Jacobsen JS, Robertson CS, Gilsdorf J, Yang Z, Wang KKW, and Shear DA (2020) Penetrating Traumatic Brain Injury Triggers Dysregulation of Cathepsin B Protein Levels Independent of Cysteine Protease Activity in Brain and Cerebral Spinal Fluid, J. Neurotrauma 37, 1574–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aggarwal N and Sloane BF (2014) Cathepsin B: multiple roles in cancer, Proteomics Clin. Appl 8, 427–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Victor BC, Anbalagan A, Mohamed MM, Sloane BF, and Cavallo-Medved D (2011) Inhibition of cathepsin B activity attenuates extracellular matrix degradation and inflammatory breast cancer invasion, Breast Cancer Res. 13, R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bian B, Mongrain S, Cagnol S, Langlois MJ, Boulanger J, Bernatchez G, Carrier JC, Boudreau F, and Rivard N (2016) Cathepsin B promotes colorectal tumorigenesis, cell invasion, and metastasis, Mol. Card nog 55, 671–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mort JS, Recklies AD, and Poole AR (1984) Extracellular presence of the lysosomal proteinase cathepsin B in rheumatoid synovium and its activity at neutral pH, Arthritis Rheum. 27, 509–15. [DOI] [PubMed] [Google Scholar]
- 12.Fujisawa A, Kambe N, Saito M, Nishikomori R, Tanizaki H, Kanazawa N, Adachi S, Heike T, Sagara J, Suda T, Nakahata T, and Miyachi Y (2007) Disease-associated mutations in CIAS1 induce cathepsin B-dependent rapid cell death of human THP-1 monocytic cells, Blood. 109, 2903–11. [DOI] [PubMed] [Google Scholar]
- 13.Toomey CB, Cauvi DM, Hamel JC, Ramirez AE, Pollard KM (2014) Cathepsin B regulates the appearance and severity of mercury-induced inflammation and autoimmunity. Toxicol Sci. 142, 339–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajamäki K, Lappalainen J, Oörni K, Välimäki E, Matikainen S, Kovanen PT, and Eklund KK (2010) Cholesterol crystals activate the NLRP3 inflammasome in human macrophages: a novel link between cholesterol metabolism and inflammation, PLoS. One 5, e11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez EA, Martins GR, Tavares AMV, Viegas M, Poletto E, Giugliani R, Matte U, and Baldo G (2018) Cathepsin B inhibition attenuates cardiovascular pathology in mucopolysaccharidosis I mice, Life Sci. 196, 102–109. [DOI] [PubMed] [Google Scholar]
- 16.Van Acker GJ, Saluja AK, Bhagat L, Singh VP, Song AM, Steer ML (2002) Cathepsin B inhibition prevents trypsinogen activation and reduces pancreatitis severity. Am J Physiol Gastrointest Liver Physiol. 283, G794–800. [DOI] [PubMed] [Google Scholar]
- 17.Cantres-Rosario YM, Ortiz-Rodríguez SC, Santos-Figueroa AG, Plaud M, Negron K, Cotto B, Langford D, Melendez LM (2019) HIV Infection Induces Extracellular Cathepsin B Uptake and Damage to Neurons. Sci Rep. 9, 8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amaral EP, Riteau N, Moayeri M, Maier N, Mayer-Barber KD, Pereira RM, Lage SL, Kubler A, Bishai WR, D'Império-Lima MR, Sher A, and Andrade BB (2018) Lysosomal Cathepsin Release Is Required for NLRP3-Inflammasome Activation by Mycobacterium tuberculosis in Infected Macrophages, Front Immunol. 9, 1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu M, Yang L, Rong JG, Ni Y, Gu WW, Luo Y, Ishidoh K, Katunuma N, Li ZS, Zhang HL (2014) Inhibition of cysteine cathepsin B and L activation in astrocytes contributes to neuroprotection against cerebral ischemia via blocking the tBid-mitochondrial apoptotic signaling pathway. Glia 62, 855–80. [DOI] [PubMed] [Google Scholar]
- 20.Towatari T, Nikawa T, Murata M, Yokoo C, Tamai M, K. Hanada, and Katunuma N (1991) Novel epoxysuccinyl peptides. A selective inhibitor of cathepsin B, in vivo, FEBS. Lett 280, 311–5. [DOI] [PubMed] [Google Scholar]
- 21.Murata M, Miyashita S, Yokoo C, Tamai M, Hanada K, Hatayama K, Towatari T, Nikawa T, and Katunuma N (1991) Novel epoxysuccinyl peptides. Selective inhibitors of cathepsin B, in vitro, FEBS. Lett 280, 307–10. [DOI] [PubMed] [Google Scholar]
- 22.Barrett AJ, Kembhavi AA, Brown MA, Kirschke H, Knight CG, Tamai M, and Hanada K (1982) L-trans-Epoxysuccinyl-leucylamido(4-guanidino)butane (E-64) and its analogues as inhibitors of cysteine proteinases including cathepsins B, H and L, Biochem. J 201, 189–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mindell JA (2012) Lysosomal acidification mechanisms, Annu. Rev. Physiol 74, 69–86. [DOI] [PubMed] [Google Scholar]
- 24.Ishida Y, Nayak S, Mindell JA, and Grabe M (2013) A model of lysosomal pH regulation, J. Gen. Physil 141, 705–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bright GR, Fisher GW, Rogowska J, and Taylor DL (1987) Fluorescence ratio imaging microscopy: temporal and spatial measurements of cytoplasmic pH, J. Cell Biol 104, 1019–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madshus IH (1988) Regulation of intracellular pH in eukaryotic cells, Biochem. J 250, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang F, Gómez-Sintes R, and Boya P (2018) Lysosomal membrane permeabilization and cell death, Traffic. 19, 918–931. [DOI] [PubMed] [Google Scholar]
- 28.Porter K, Lin Y, and Liton PB (2013) Cathepsin B is up-regulated and mediates extracellular matrix degradation in trabecular meshwork cells following phagocytic challenge, PLoS. One 8, e68668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casey JR, Grinstein S, Orlowski J (2010) Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol. 11, 50–61. [DOI] [PubMed] [Google Scholar]
- 30.Hämälistö S, Stahl JL, Favaro E, Yang Q, Liu B, Christoffersen L, Loos B, Guasch Boldú C, Joyce JA, Reinheckel T, Barisic M, Jäättelä M. (2020) Spatially and temporally defined lysosomal leakage facilitates mitotic chromosome segregation. Nat Commun. 11, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meng J, Liu YC, Xie Z, Qing H, Lei P, Ni J. (2020) Nucleus distribution of cathepsin B in senescent microglia promotes brain aging through degradation of sirtuins. Neurobiol. Aging 96, 255–266. [DOI] [PubMed] [Google Scholar]
- 32.Jiang Z, Lietz CB, Podvin S, Yoon MC, Toneff T, Hook V, and O'Donoghue AJ (2021) Differential Neuropeptidomes of Dense Core Secretory Vesicles (DCSV) Produced at Intravesicular and Extracellular pH Conditions by Proteolytic Processing, ACS. Chem. Neurosci 12, 2385–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buttle DJ, Murata M, Knight CG, and Barrett AJ (1992) CA074 methyl ester: a proinhibitor for intracellular cathepsin B, Arch Biochem. Biophys 299, 377–80. [DOI] [PubMed] [Google Scholar]
- 34.Choe Y, Leonetti F, Greenbaum DC, Lecaille F, Bogyo M, Brömme D, Ellman JA, and Craik CS (2006) Substrate profiling of cysteine proteases using a combinatorial peptide library identifies functionally unique specificities, J. Biol. Chem 281, 12824–32. [DOI] [PubMed] [Google Scholar]
- 35.Poreba M, Groborz K, Vizovisek M, Maruggi M, Turk D, Turk B, Powis G, Drag M, and Salvesen GS (2019) Fluorescent probes towards selective cathepsin B detection and visualization in cancer cells and patient samples, Chem. Sci 10, 8461–8477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamamoto A, Tomoo K, Hara T, Murata M, Kitamura K, and Ishida T (2000) Substrate specificity of bovine cathepsin B and its inhibition by CA074, based on crystal structure refinement of the complex, J. Biochem 127, 635–43. [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto A, Hara T, Tomoo K, Ishida T, Fuji T, Hata Y, Murata M, and Kitamura K (1997) Binding mode of CA074, a specific irreversible inhibitor, to bovine cathepsin B as determined by X-ray crystal analysis of the complex, J. Biochem 121, 974–7. [DOI] [PubMed] [Google Scholar]
- 38.Hsu A, Podvin S, and Hook V (2018) Lysosomal Cathepsin Protease Gene Expression Profiles in the Human Brain During Normal Development, J. Mol. Neurosci 65, 420–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Donoghue AJ, Eroy-Reveles AA, Knudsen GM, Ingram J, Zhou M, Statnekov JB, Greninger AL, Hostetter DR, Qu G, Maltby DA, Anderson MO, Derisi JL, McKerrow JH, Burlingame AL, and Craik CS (2012) Global identification of peptidase specificity by multiplex substrate profiling, Nat. Methods 9, 1095–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lape k J. D. Jr., Jiang Z, Wozniak JM, Arutyunova E, Wang SC, Lemieux MJ, Gonzalez DJ, and O'Donoghue AJ (2019) Quantitative Multiplex Substrate Profiling of Peptidases by Mass Spectrometry, Mol. Cell Proteomics 18, 968–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vilar S, Cozza G, and Moro S (2008) Medicinal chemistry and the molecular operating environment (MOE): application of QSAR and molecular docking to drug discovery, Curr Top Med Chem. 8, 1555–72. [DOI] [PubMed] [Google Scholar]
- 42.Roy U and Luck LA (2007) Molecular modeling of estrogen receptor using molecular operating environment, Biochem. Mol. Biol. Educ 35, 238–43. [DOI] [PubMed] [Google Scholar]
- 43.Schechter I (2005) Mapping of the active site of proteases in the 1960s and rational design of inhibitors/drugs in the 1990s, Curr. Protein Pept. Sci 6, 501–12. [DOI] [PubMed] [Google Scholar]
- 44.Yoon MC, Solania A, Jiang Z, Christy MP, Podvin S, Mosier C, Lietz CB, Ito G, Gerwick WH, Wolan DW, Hook G, O'Donoghue AJ, and Hook V (2021) Selective Neutral pH Inhibitor of Cathepsin B Designed Based on Cleavage Preferences at Cytosolic and Lysosomal pH Conditions, ACS. Chem. Biol 16, 1626–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krupa JC, Hasnain S, Nägler DK, Ménard R, Mort JS (2002) S2' substrate specificity and the role of His110 and His111 in the exopeptidase activity of human cathepsin B, Biochem J. 361, 613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turk D, Podobnik M, Popovic T, Katunuma N, Bode W, Huber R, and Turk V (1995) Crystal structure of cathepsin B inhibited with CA030 at 2.0-A resolution: A basis for the design of specific epoxysuccinyl inhibitors, Biochemistry. 34, 4791–7. [DOI] [PubMed] [Google Scholar]
- 47.Katunuma N (2011) Structure-based development of specific inhibitors for individual cathepsins and their medical applications, Proc. Jpn. Acad Ser. B Phys. Biol. Sci 87, 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang M, Meng J, Zeng F, Qing H, Hook G, Hook V, Wu Z, and Ni J (2020) Cathepsin B inhibition blocks neurite outgrowth in cultured neurons by regulating lysosomal trafficking and remodeling, J. Neurochem 155, 300–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matarrese P, Ascione B, Ciarlo L, Vona R, Leonetti C, Scarsella M, Mileo AM, Catricalà C, Paggi MG, and Malorni W (2010) Cathepsin B inhibition interferes with metastatic potential of human melanoma: an in vitro and in vivo study, Mol. Cancer 9, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Asai M, Yagishita S, Iwata N, Saido TC, Ishiura S, and Maruyama K (2011) An alternative metabolic pathway of amyloid precursor protein C-terminal fragments via cathepsin B in a human neuroglioma model, FASEB. J 25, 3720–30. [DOI] [PubMed] [Google Scholar]
- 51.Wu Z, Ni J, Liu Y, Teeling JL, Takayama F, Collcutt A, Ibbett P, and Nakanishi H (2017) Cathepsin B plays a critical role in inducing Alzheimer's disease-like phenotypes following chronic systemic exposure to lipopolysaccharide from Porphyromonas gingivalis in mice, Brain Behav. Immun 65, 350–361. [DOI] [PubMed] [Google Scholar]
- 52.Yamashima T, Kohda Y, Tsuchiya K, Ueno T, Yamashita J, Yoshioka T, and Kominami E (1998) Inhibition of ischaemic hippocampal neuronal death in primates with cathepsin B inhibitor CA-074: a novel strategy for neuroprotection based on 'calpain-cathepsin hypothesis', Eur. J. Neurosci 10, 1723–33. [DOI] [PubMed] [Google Scholar]
- 53.Yoshida M, Yamashima T, Zhao L, Tsuchiya K, Kohda Y, Tonchev AB, Matsuda M, and Kominami E (2002) Primate neurons show different vulnerability to transient ischemia and response to cathepsin inhibition, Acta. Neuropathol 104, 267–72. [DOI] [PubMed] [Google Scholar]
- 54.Zhang L, Fu XH, Yu Y, Shui RH, Li C, Zeng HY, Qiao YL, Ni LY, and Wang Q (2015) Treatment with CA-074Me, a Cathepsin B inhibitor, reduces lung interstitial inflammation and fibrosis in a rat model of polymyositis, Lab Invest. 95, 65–77. [DOI] [PubMed] [Google Scholar]
- 55.Van Acker GJ, Weiss E, Steer ML, and Perides G (2007) Cause-effect relationships between zymogen activation and other early events in secretagogue-induced acute pancreatitis, Am. J. Physiol. Gastrointest. Liver Physiol 292, G1738–46. [DOI] [PubMed] [Google Scholar]
- 56.Hook VY, Kindy M, and Hook G (2008) Inhibitors of cathepsin B improve memory and reduce beta-amyloid in transgenic Alzheimer disease mice expressing the wild-type, but not the Swedish mutant, beta-secretase site of the amyloid precursor protein, J. Biol. Chem 283, 7745–53. [DOI] [PubMed] [Google Scholar]
- 57.Stoka V, Turk V, and Turk B (2016) Lysosomal cathepsins and their regulation in aging and neurodegeneration, Ageing Res. Rev 32, 22–37. [DOI] [PubMed] [Google Scholar]
- 58.Wang F, Gómez-Sintes R, and Boya P (2018) Lysosomal membrane permeabilization and cell death, Traffic. 19, 918–931. [DOI] [PubMed] [Google Scholar]
- 59.Tedelind S, Poliakova K, Valeta A, Hunegnaw R, Yemanaberhan EL, Heldin NE, Kurebayashi J, Weber E, Kopitar-Jerala N, Turk B, Bogyo M, and Brix K (2010) Nuclear cysteine cathepsin variants in thyroid carcinoma cells, Biol. Chem 391, 923–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hämälistö S, Stahl JL, Favaro E, Yang Q, Liu B, Christoffersen L, Loos B, Guasch, Boldú C, J. A. Joyce, Reinheckel T, Barisic M, and Jäättelä M (2020) Spatially and temporally defined lysosomal leakage facilitates mitotic chromosome segregation, Nat. Commun 11, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Victor BC, Anbalagan A, Mohamed MM, Sloane BF, and Cavallo-Medved D (2011) Inhibition of cathepsin B activity attenuates extracellular matrix degradation and inflammatory breast cancer invasion, Breast Cancer Res. 13, R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sloane BF, Yan S, Podgorski I, Linebaugh BE, Cher ML, Mai J, Cavallo-Medved D, Sameni M, Dosescu J, and Moin K (2005) Cathepsin B and tumor proteolysis: contribution of the tumor microenvironment, Semin. Cancer Biol 15, 149–57. [DOI] [PubMed] [Google Scholar]
- 63.Strelow JM (2017) A Perspective on the Kinetics of Covalent and Irreversible Inhibition, SLAS Discov. 22, 3–20. [DOI] [PubMed] [Google Scholar]
- 64.Tonge PJ (2019) Quantifying the Interactions between Biomolecules: Guidelines for Assay Design and Data Analysis, ACS Infect. Dis 5, 796–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
LC-MS/MS files for the MSP-MS experiments can be accessed at www.proteomexchange.org under the dataset identifier numbers PXD022494 and PXD022493. Alternatively, the data files can be obtained through www.massive.ucsd.edu under the dataset identifier numbers MSV000086449 and MSV000086447.