Abstract
The strategies for nucleic acid sensing based on nucleic acid hybridization between the target sequence and the capture probe sequence are considered to be largely successful as far as detection of a specific target of known sequence is concerned. However, when compared with other complementary methods, like direct sequencing, a number of results are still found to be either “false positives” or “false negatives”. This suggests that modifications in these strategies are necessary to make them more accurate. In this minireview, we propose that one way toward improvement could be replacement of the DNA capture probes with the xeno nucleic acid or XNA capture probes. This is because the XNAs, especially the locked nucleic acid, the peptide nucleic acid, and the morpholino, have shown better single nucleobase mismatch discrimination capacity than the DNA capture probes, indicating their capacity for more precise detection of nucleic acid sequences, which is beneficial for detection of gene stretches having point mutations. Keeping the current trend in mind, this minireview will include the recent developments in nanoscale, fluorescent label-free applications, and present the cases where the XNA probes show clear advantages over the DNA probes.
Introduction
The modifications in nucleic acid sequences, for example, single nucleobase mutations, can be the basis of a number of human diseases of genetic origin, for example, sickle-cell anemia, cystic fibrosis, β thalassemia, Huntington disease and color-blindness. In fact, the number of disorders that are identified to be genetic in origin is increasing every year.1 The single nucleobase mutations may also create susceptibility toward critical diseases like cancer, tuberculosis and neurological disorders like Alzheimer’s disease.1 Therefore, sequence-specific and sensitive detection of nucleic acids is vital for early and reliable diagnosis of a genetic disease. The identification of certain nucleic acid sequences both in vitro and in vivo is important also for the discovery of hitherto unknown genetic diseases, diagnosis of pathogen infection and monitoring of disease treatment.2,3 The nucleic acid biosensors (NABs) or the genosensors, where detection is based on nucleic acid hybridization, have shown considerable potential, especially for clinical analysis,4 for example, in the cases of metabolic disorders like diabetes, cancer and cardiovascular disease; contagious diseases like tuberculosis, dengue and hepatitis; and food borne diseases like diarrhea, salmonellosis, cholera etc.2,3
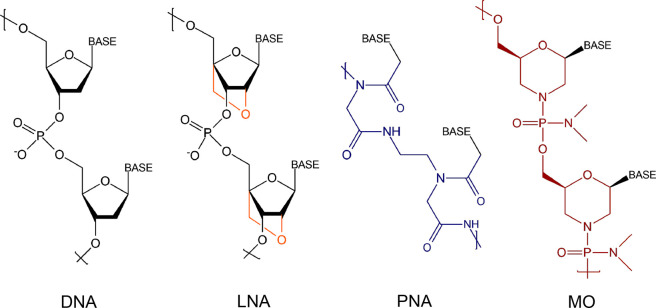
Though the DNA capture probes have been widely used for nucleic acid sensing (see Table 1),5 there are certain limitations of the DNA probes, for example, reduced bioactivity, nonspecific signals, lack of reproducibility and susceptibility to degradation by the nuclease enzyme.6,7 Therefore, the necessity of an alternative probe arises for developing a robust assay with high target specificity and detection sensitivity. A number of synthetic alternative nucleic acid analogs or xeno nucleic acids (XNAs), where “xeno” stands for “alien”, having non-natural backbones, such as arabinonucleic acid (ANA), cyclohexene nucleic acid (CeNA), fluoro-arabinonucleic acid (F-ANA), glycol nucleic acid (GNA), hexitol nucleic acid (HNA), peptide nucleic acid (PNA), morpholino (MO), threose nucleic acid (TNA), click nucleic acid (CNA) and locked nucleic acid (LNA), have been developed over the last three decades. Recently, the capabilities of different XNAs as capture probes have been explored, including in testing liquid biopsy samples (blood, saliva, urine, ascites and pleural effusion) for the presence of circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), cell-free DNA (cfDNA) or exosomes and micro-RNA (miRNA). Among all the alternative nucleic acids, LNA, PNA and MO (Figure 1) have emerged as the most popularly used capture probes.6
Table 1. Overview of Various DNA Capture Probe-Based Nucleic Acid Sensing Methods and Their Applicationsa.
| Capture probe | Target (limit of detection) | Method of signal transduction | Application |
|---|---|---|---|
| ssDNA | DNA (1.38 fM) | Surface plasmon resonance spectroscopy (optical) | Detection of p53 gene mutation5a |
| DNA molecular beacon | DNA (0.17 nM) | Fluorescence spectroscopy (optical) | Detection of nucleic acid sequences5b |
| Pyrene-excimer labeled DNA | DNA (256 fM) | Hybridization chain reaction-induced fluorescence emission spectroscopy (optical) | Detection of nucleobase mismatches, deletion and insertion mutations in complex biological fluid5c |
| Single walled carbon nanotubes (SWNT) associated ssDNA | DNA (4 nM) | Fluorescence spectroscopy (optical) | Detection of single nucleobase mismatch5d |
| Quantum dot (QD)-linked DNA | DNA (4.8 fM) | Fluorescence resonance energy transfer-based spectroscopy (optical) | Detection of Kras point mutation5e |
| BaGdF5: Yb/Er upconversion nanoparticles conjugated DNA probe | AuNP-linked virus oligonucleotide (300 fM) | Luminescence resonance energy transfer-based spectroscopy (optical) | Ebola virus detection5f |
| ssDNA | HIV-1 DNA (0.24 pg/mL) | Surface-enhanced Raman spectroscopy (optical) | Detection of viral DNA5g |
| DNA hairpin | miRNA (10 fM at 37 °C and 1 aM at 4 °C) | Quadratic isothermal amplification strategy-based fluorescence spectroscopy (optical) | Cancer-specific miRNA detection5h |
| Stem loop DNA | Genomic DNA (10 fM) | Amperometry (electrochemical) | Sequence-specific DNA detection5i |
| Tetrahedral DNA probe | miRNA (1 fM) | Cyclic voltammetry and amperometry (electrochemical) | Detection of cancer related biomarker-miRNA 1415j |
| ssDNA | DNA (1 pM DNA and genomic DNA from 2.7 × 102 CFU/mL bacterial culture) | Quartz crystal microbalance (mechanical) | Pathogenic E. coli detection5k |
| dsDNA scaffold | DNA (1 pM) | Gel electrophoresis (chemical, via hybridization-induced conformational change) | SNP detection5l |
The relevant reference is shown in superscript for each application.
Figure 1.
Chemical structures of DNA, LNA, PNA and MO. The structural differences in LNA, PNA and MO, compared to DNA, are highlighted by using different colors.
The LNA is a conformationally restricted DNA analogue where the sugar moiety is a bicyclic ribose derivative with a bridging methylene group between the 4′-carbon and the 2′-oxygen8 (Figure 1). LNA is nuclease-resistant in nature as being unnatural to such an extent that nuclease cannot recognize it as a substrate.9 The PNA10 contains a “peptide-like” backbone where the negatively charged sugar–phosphate backbone is substituted with nonionic, repeating N-(2-aminoethyl) glycine units, which are linked by amide bonds (Figure 1). Such a nonionic, achiral structure of PNA is not susceptible to degradation by nuclease or protease enzymes, which makes PNA a highly stable biological molecule.11 Another nonionic XNA is MO, where modifications are made to both the sugar and the phosphodiester linkage.12 It is composed of morpholino rings, nucleobases and the nonionic phosphorodiamidates (Figure 1). MO possesses a nonionic backbone similar to PNA, but it exhibits better solubility in water12 in comparison to PNA because the morpholine ring is hydrophilic in nature. In addition, MO is resistant to a number of nucleases.12 MO is less expensive than most of the other XNAs as it can be synthesized by starting with less expensive ribonucleosides. The advantageous properties of MO such as solubility in aqueous medium, nuclease-resistance, highly target-specific hybridization and activity inside cells, have made MO an ideal probe for developing NABs.
While DNA has remained a popular capture probe due to its relatively easy accessibility, low cost, and the already available extensive literature, difficulties associated with DNA-based sensing as mentioned earlier persist. Some difficulties in nucleic acid sensing have also been associated with the use of a fluorescent label. The fluorescence signals may not always be commensurate with the target concentration and/or false positive/false negative signals may arise from nonspecific origins for the following reasons. First, the organic fluorescent dyes used as fluorescent labels may exhibit poor photostability and pH sensitivity, thereby having altered or compromised functions in different experimental conditions. Second, the characteristics of the linker, i.e. its nature and length as well as the attachment position of the fluorophore (end or internal modification), may play a crucial role in signal development as dye–nucleotide interaction can influence the sensitivity of the fluorescence intensity to environmental conditions. Third, nonspecific association of the labeled target or the presence of unreacted dye may give rise to a false positive signal. Fourth, a poor degree of labeling of the target may give rise to weak or no signal even when sequence-specific binding occurs resulting in false negative signals. Fifth, though the low photodegradation rate of quantum dots (QDs) makes the QDs a superior alternative to the traditional organic fluorophores, they too have drawbacks like poor water solubility, and easy aggregation in an aqueous medium that reduces their fluorescence quantum yields. Sixth, in fluorescence quenching approaches (such as in molecular beacons), the quencher molecule that is covalently attached to the fluorescent oligonucleotide may suffer from low quenching efficiency when the distance between the fluorophore and the quencher is relatively large. For all these reasons, it is useful to explore fluorescent label-free approaches while testing the applicability of XNA probes in nucleic acid sensing. In our laboratory, notable progress has been made in the recent years toward developing a fluorescent label-free approach for detection of nucleic acid sequences using the XNA capture probes.6 This approach is an obvious shift from the conventionally applied ensemble methods, since here detection is performed in molecule-by-molecule manner using the single molecule force spectroscopy (SMFS) technique.
While most of the approaches for identification of point mutations are based on polymerase chain reaction (PCR)-based methods, the PCR-dependent approaches can suffer from amplification-related errors that are caused by mispriming, insufficient accuracy in discriminating single nucleotides, an inadequate multiplexing capability, contamination-related false positive signals, and false negative signals due to the presence of a very low amount of the sequence of interest. Therefore, there is a need for identification of the specific and characteristic point mutations with alternative approaches, such as the biosensor-based techniques that do not require PCR-based amplification of nucleic acid sequences. In a biosensor-based strategy, the binding of an analyte to a specific bioreceptor, which is the recognition element, is converted into a measurable electronic signal by the transducer. The importance of biosensor technologies, especially the miniaturized ones that are capable of rapid and point-of-care testing, is increasingly being felt in the healthcare management nowadays. Development of accurate and sensitive biosensors means that early detection of disease is possible and preventive measures can be taken at the right time. At present, the reverse transcription polymerase chain reaction (RT-PCR) is routinely used as the most standard approach for nucleic acid detection, for example, in the diagnosis of COVID-19 disease. However, false-positive cases (due to amplification errors, contamination) and false-negative cases (particularly in the initial stages of the viral infection, when the viral load is the minimum, and/or when a novel mutant virus causes the infection) are also reported. To surmount the limitations of RT-PCR, different biosensors techniques have been developed with promising capacities. For example, a plasmonic biosensor having dual functions has been developed, where the plasmonic photothermal (PPT) effect has been combined with localized surface plasmon resonance (LSPR)-based transduction. Here, the two-dimensional gold nanoislands (AuNIs) with surface-anchored complementary DNA receptors have been used for sequence-specific and sensitive detection of SARS-CoV-2-derived sequences.13 The SMFS-based approach that has been developed in our laboratory is a PCR-amplification-free approach since very low amount of sample is needed for the molecularly resolved detection by SMFS, for example, in the case of identification of the most common mutations of the multidrug-resistant Mycobacterium tuberculosis bacteria, where a 15-mer LNA capure probe sequence was used for the detection of a 45-mer long target DNA with overhang regions.14
Typically an efficient biosensor should exhibit a combination of the following characteristics: high sensitivity, selectivity, reproducibility, and reusability, where reusability is often not considered to be essential, especially in clinical diagnostics, where cross-contamination from another patient’s sample should be avoided. In order to improve sensitivity in nucleic acid sensing, considerable development in the use of nanoscale sensing surfaces has been made over the last decade.6 Because of the high surface-to-volume ratio of the nanomaterials, the sensing surface area effectively increases, thereby increasing the total amount of immobilized capture probes. The resulting increase in the capture probe density improves the prospect of enhancement in nucleic acid recognition signal. Also, because of the confinement of a nanoscale region, a low concentration of analytes (sub-nanomolar concentration) that are present within that region can be detected, which makes the limit of detection better. The most significant clinical applications of the DNA-based nanobiosensors available at this time are in the areas of diagnosis of infectious diseases like Mycobacterium tuberculosis, genetic diseases like cystic fibrosis, cancer diagnosis and biomarker discovery and immunodeficiency related diseases.15 Keeping this advantage of applying nanoscale detection approaches in mind, this review focuses on the recent developments, where nanoscale strategies, especially those that are fluorescent label-independent and potentially PCR-amplification-free, have been followed.
The Basic Design of a Nanoscale XNA-Based Nucleic Acid Sensor
In nucleic acid detection, the target analyte is usually a section of a nucleic acid sequence (e.g., ssDNA or ssRNA or dsDNA), or a nucleic acid pattern (e.g., the RNA G-quadruplex). The capture probes are usually fully or partly complementary single-stranded nucleic acid sequences that are used to detect target sequences via sequence-specific hybridization. These probes can be of short sequences (10–30 mer) and long sequences (40–90 mer). The capture probes can also be nucleic acid aptamers (mostly 10–100 mer in size) with specific 3-D structures. It has been observed that the nanoscale detection approaches based on hybridization/dehybridization processes occurring on surface can be generally faster, more sensitive and sequence-specific than the traditional approaches, for example, those that involve detection of electrophoresis-separated DNA/RNA fragments by Southern/Northern blotting methods. In addition, they can be made fluorescent label-free, when fluorescence-independent transduction methods are combined.
The basic steps in the nanoscale hybridization assays are (a) capture probe immobilization on the surface, (b) target hybridization and duplex formation and (c) monitoring hybridization/dehybridization event via signal transduction (Figure 2A). The optimization of the capture probe density is essential so that a high probe density with favorable probe-to-probe spacing can be achieved that ensures minimization of nonspecific interactions with the substrate and improvement in sensitivity of target-specific detection.6 Significant advantage of the LNA probe has been observed in this respect.6 Careful consideration is necessary also in following proper immobilization chemistry to ensure stability and functionality of the surface-confined capture probe molecules. Various immobilization methods have been developed that are based on the following three major approaches: (a) simple adsorption or physisorption, (b) immobilization via covalent bond formation or chemisorption, and (c) avidin (or streptavidin)–biotin interaction. Among these, the simplest immobilization method is based on physisorption, for example, via ionic interactions between the negatively charged DNA capture probe and positively charged surface like amino-silanized glass surface/cationic polymer-coated glassy electrode. However, because desorption of the physisorbed capture probes can happen under environmental changes such as in pH, temperature and ionic strength, even when the probes are adsorbed in a controlled manner under the action of electric potential, the covalent coupling of the capture probes with the surface is generally considered to be more useful. Electrostatics-driven physisorption of the capture probes also does not ensure oriented attachment of the probes on surface, because the capture probe with a charged backbone can interact with the oppositely charged surface through the total stretch of the probe and not a specific point on it. Biosensing approaches have been developed using thiol-containing DNA probes6,16 and XNA probes6 because the thiol groups (-SH) exhibit a high affinity toward the gold surface allowing covalent bond formation between sulfur and gold atoms of a variety of gold-coated surfaces including those of gold-interdigitated ultra-micro-electrode arrays or gold micropads (Figure 2B). Other covalent coupling chemistries involve the use of 5′ amino-modified ssDNA to attach onto functionalized (carboxyl, aldehyde, sulfonic, epoxy, isothiocyanate etc.) surfaces (Figure 2B).6,16 Another strategy for capture probe immobilization has been developed using avidin (streptavidin)–biotin interactions where biotin, which is a small molecule, binds with very high affinity to the tetrameric proteins like avidin/streptavidin. This has been done by modifying the 3′ or the 5′ end of DNA probe with a biotin molecule and then exposing it to the avidin/streptavidin-modified surface. A range of methods for the development of an avidin/streptavidin-functionalized electrode surface have been established for attachment of the biotinylated DNA probes, for example, using N-hydroxysuccinimide (NHS) and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) via NHS-EDC coupling between activated carboxyl on the surface and the amine on avidin/streptavidin (Figure 2B).6,16
Figure 2.
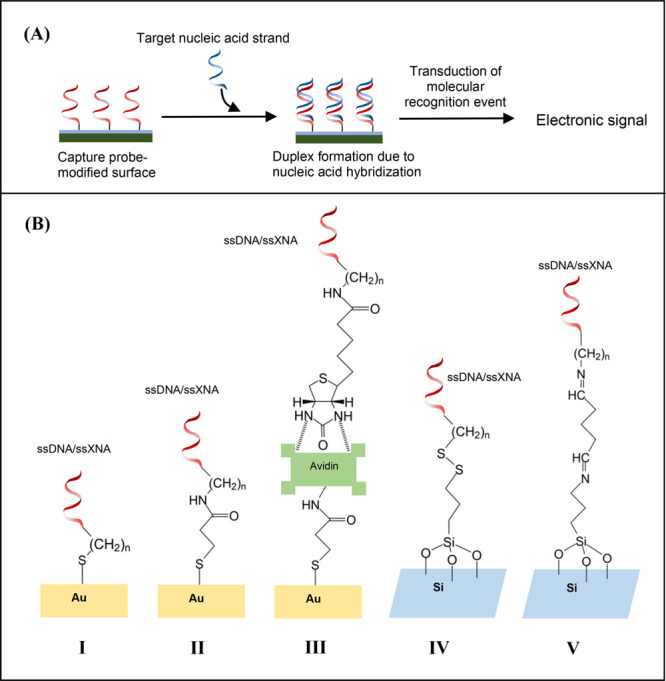
(A) Basic experimental design for nucleic acid sensing. (B) Different strategies for immobilization of ssDNA/ssXNA on surface via (I) gold–thiol linkage formation, (II) NHS-EDC activated immobilization, (III) affinity binding of avidin and biotinylated nucleic acid strand, (IV) dithiol linkage formation on 3-MPTMS-modified silicon surface, and (V) formation of secondary imine using glutaraldehyde cross-linker on 3-APTES-modified silicon surface.
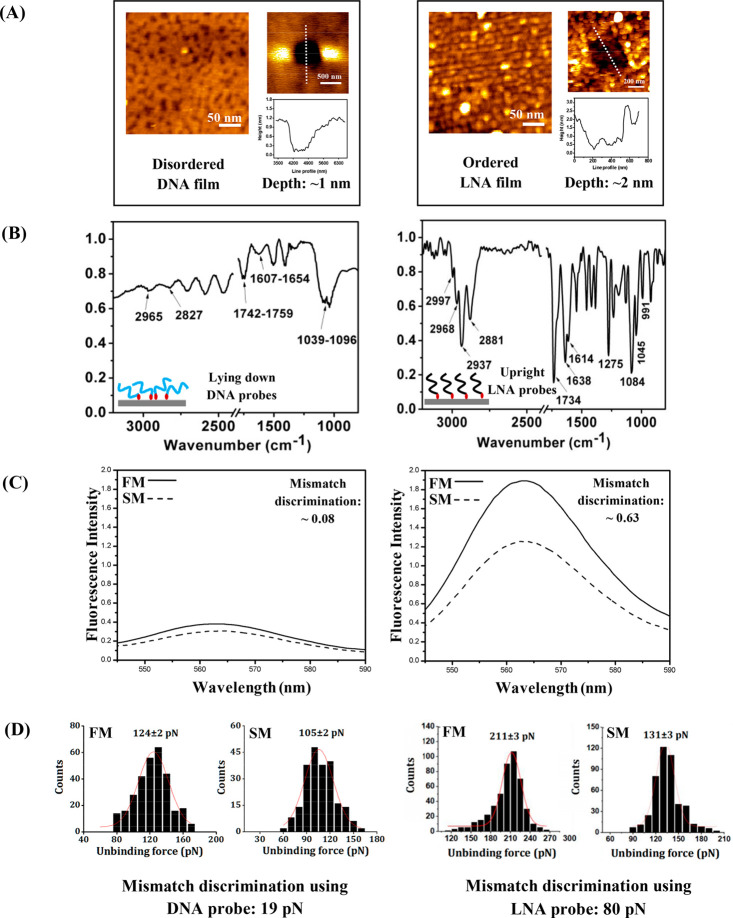
For effective target hybridization and duplex formation, an efficient design of the sensing surface is necessary, for example, construction of an ordered and self-assembled monolayer (SAM) of the capture probes via controlled preparation of the sensing film where the capture probes are oriented upright can be crucial. It has been observed that DNA films are mostly disordered (Figure 3A), where the ssDNA probes undergo back bending on the surface, as evident from the cyclic voltammetry (CV) and ellipsometry studies.17 This disposition of the ssDNA backbone means that nonspecific interactions between DNA and the underlying surface may occur through the relatively exposed nucleobases, resulting in reduced biosensor activity of the film, and thereby weakening the target-specific signal strength.7 Poorly ordered probes may even lead to false positive and reproducible signals that originate from these nonspecific interactions.7 These issues have been resolved to a significant extent by replacing DNA with XNA probes. For example, more ordered SAMs (Figure 3A) with more upright backbone orientation of the surface-anchored LNA probes18 has been observed compared to the DNA probes, as demonstrated by the reflection absorption infrared spectroscopy (RAIRS) experiments19 (Figure 3B). The more upright orientation of the capture probes not only makes the target’s passage to the total length of the capture probes more direct and accessible to the target sequences, but it also helps in reducing the nonspecific interactions of the capture probes with the underlying substrate. Another excellent hybridization probe is molecular beacons (MBs) for target-specific detection of nucleic acids, but DNA-based MBs suffer from limitations such as poor stability with poor sensitivity during surface-based sensing. The MB–surface interactions that partially disrupt the loop and destabilize the stem structure can result in a considerable reduction of the fluorescence enhancement exhibited by the surface-immobilized DNA-MBs.20 As a result, inefficient quenching can take place, which is reflected in background fluorescence signal, thus affecting the overall detection sensitivity of the MBs.20 Such a limitation has been overcome using the LNA-based MBs,20 which increase the sensitivity of target-specific detection events, because nonspecific interactions with the surface can largely be avoided due to the rigid backbone of LNA. The greater capacity of single nucleobase mismatch discrimination of the surface-anchored LNA probes over the corresponding surface-anchored DNA capture probes was observed both in an ensemble experiment (Figure 3C) and a molecularly resolved SMFS experiment (Figure 3D), meaning that the LNA-designed sensing surface retained the solution-phase capacity of the LNA probes, as a result of optimized surface preparation.14,18,21
Figure 3.
(A) The AFM topographic images of DNA and LNA films, along with the respective images of scratched areas generated by the nanoshaving method and their corresponding cross-sectional profiles that indicate monolayer formation. (B) The reflection absorption infra-red (RAIR) spectra of the DNA (left) and LNA (right) layers, that indicate a more upright orientation in case of the LNA probes compared to the DNA probes. (C) The fluorescence spectra for the full match (FM: solid line) and single base mismatch (SM: dotted line) cases for the DNA probe (left) and the LNA probe (right). (D) The single base mismatch discrimination ability of the DNA probe (see two force distributions at the left) and the LNA probe (see two force distributions at the right) using single molecule force spectroscopy (SMFS) technique. The data presented here are reproduced/adapted with permission from ref (19) (Copyright 2016, The Royal Society of Chemistry) for RAIRS data, ref (18) (Copyright 2012, American Chemical Society) for fluorescence data, and ref (26b) (Copyright 2022, The Royal Society of Chemistry) and ref (14) (Copyright 2019, American Chemical Society) for SMFS data. The AFM topographic images, DNA scratching image and LNA scratching image with corresponding cross sectional profile are reproduced/adapted with permission from ref (18) (Copyright 2012, American Chemical Society), ref (26b) (Copyright 2022, Royal Society of Chemistry) and ref (23) (Copyright 2016, Oxford University Press), respectively.
The nucleic acid-based sensors can be divided into two categories, namely, label-dependent and label-independent sensors. In label-dependent sensors, the label can be an enzyme, a redox indicator (e.g., ferrocene or Fc), radioisotopes, cationic metal complexes or intercalating organic molecules (e.g., methylene blue), fluorophores, UV-absorbing molecules like nanoparticles, etc. Here, the sensor signal is proportional to the amount of labels. As a result, an estimate of the number of bound target molecules can be obtained. The label-dependent methods are often not very effective as far as labor, cost and time are concerned. In addition, labeling of biomolecules may obstruct the active binding sites and alter the nature of binding. All together, this may unfavorably affect the affinity-based interaction between the recognition partner and the target partner. In contrast, the label-independent technologies do not require the labels to assist the measurements. Instead, they use intrinsic physical nature of the analytes based on their size, molecular weight, dielectric permittivity, charge, electrical impedance, and refractive index, or chemical properties like their capacity of chemical interaction with the capture probe, for detection. Label-independent biosensing methods have made considerable progress in the recent years due to their capacity of fast and cost-effective tests. Moreover, they can be integrated in the lab-on-a-chip type platforms that allow real-time monitoring of changes in target analyte concentration. A variety of label-free nucleic acid detection techniques based on sequence-selective DNA hybridization/dehybridization have been developed, where the transduction methods can be optical, electrochemical, mechanical, magnetic, thermal, electronic, chemical and acoustic in nature (Table 1). Among all these, the microcantilever movement that is associated with a DNA–DNA/XNA–DNA duplex dehybridization event, where nanomechanical transduction is employed for nucleic acid sensing using an atomic force spectroscopy (AFS)-based SMFS strategy, is a recent development.14 In this approach, the target sequence is detected by acquiring the unbinding force value from the force-induced unbinding (or forced dehybridization) event that occurs during the cantilever retraction step. The SMFS-based mechanical transduction offers an important advantage over the other transduction methods, i.e., molecule-by-molecule quantification of the interaction force and eliciting the associated free energy of the interaction from therein.14 Because biological interactions are generally weak in nature, involving forces of pN order, SMFS is an ideal approach for studying biological interactions. Apart from information on the interaction force, structural information can also be obtained because SMFS is highly sensitive to the nanoscale changes in the molecular conformation/assembly structure. A key advantage of the SMFS-based sensing is that nucleic acid amplification is not needed because the method allows molecularly resolved detection and requires only ∼10 nM to as low as ∼45 zM22 of target concentration. Among the other advantages, each duplex unbinding event for detection can be recorded very fast because every hybridization/dehybridization event takes place within a small time period of 100 ms.23
The XNAs as troubleshooter
As mentioned earlier, the XNA capture probes are chemically modified for providing greater stability including resistance to restriction enzymes, and improved target-specificity and detection sensitivity, compared to the DNA capture probes. Consequently, applications of the XNAs as nucleic acid hybridization probes have been realized in a number of applications (see Table 2).14,18,22,24−26 Because LNA, PNA and MO are the most popularly used XNAs with modified backbones, their applications as hybridization probes in the construction of advanced NABs will be presented here.
Table 2. Overview of XNA Capture Probe-Based Nucleic Acid Sensing Methods and Their Applicationsa.
| Capture probe | Target (limit of detection) | Method of signal transduction | Application |
|---|---|---|---|
| LNA | miRNA (10 fM) | AuNP-amplified surface plasmon resonance imaging (optical) | Quantitative measurement of miRNA24a |
| LNA | DNA amplicon (104 copies of plasmid/10 μL PCR reaction) | Absorbance measured by ELISA reader (optical) | Detection of factor V leiden mutation24b |
| LNA | DNA (50 nM) | Fluorescence polarization (optical) | SNP detection24c |
| LNA | DNA (1 μM, nucleobase mismatch discrimination ratio w.r.t. DNA probe ∼2) | Fluorescence spectroscopy after on-surface melting-induced dehybridization (optical) | Single nucleobase mismatch discrimination18 |
| Hairpin LNA | DNA (83 fM) | Amperometry (electrochemical) | Detection of promyelocytic leukemia/retinoic acid receptor alpha (PML/RARα) fusion gene in acute promyelocytic leukemia24d |
| Hairpin LNA | DNA (6 pM) | Chronoamperometry (electrochemical) | Single nucleobase mismatch discrimination24e |
| LNA | DNA (50 pM) | Single molecule force spectroscopy (mechanical) | Detection of point mutations associated with multiple drug-resistant tuberculosis14 |
| LNA | DNA (45 zM) | Single molecule force spectroscopy (mechanical) | Quantitative detection of BCR-ABL fusion gene22 |
| PNA | DNA (1 μM) | On-surface melting temperature analysis by measuring fluorescence intensity (optical) | Single nucleobase mismatch discrimination25a |
| PNA-functionalized silicon nanowires (SiNWs) | miRNA (1 fM) | Potentiometry (electrochemical) | Single nucleobase mismatch discrimination in miRNA and detection of miRNA from total RNA extracted from HeLa cell line25b |
| PNA | DNA (500 fM) | Impedance spectroscopy (electrochemical) | Detection of genomic DNA sequence from methicillin drug-resistant Staphylococcus aureus(25c) |
| PNA | DNA (100 fM) | Field effect transistor-based potentiometry (electrochemical) | Single nucleobase mismatch detection25d |
| PNA | DNA (10 nM) | Cantilever array-based sensing (mechanical) | Single nucleobase mismatch discrimination25e |
| MO | miRNA (2 fM) | Impedance spectroscopy (electrochemical) | miRNA expression profiling, detection of circulating miRNA in blood and cultured cells26a |
| MO | DNA (10 nM) | Single molecule force spectroscopy (mechanical) | Single nucleobase mismatch discrimination26b |
The relevant reference is shown in superscript for each application.
1. Application of LNA Probes
It has been found that the more rigid LNA backbone can support an upright backbone orientation,18,19 which is beneficial for preventing nonspecific interactions of the surface-anchored LNA capture probes with the underlying solid substrate. The standing orientation of the LNA probes also assists in making complete access of the capture probe by the target sequence possible. Consequently, recognition of the target sequences is achieved with improved sequence specificity, higher affinity toward target sequence and enhanced thermal stability of the LNA-containing duplexes.6 The nuclease-resistant nature, water solubility and low toxicity of LNA allow useful applications as probes for diagnostics (both in vivo and in vitro). A 16-mer LNA sequence has been used in microarray-based experiments for the detection of a 22-mer gene expression regulatory miRNA.24a A DNA (15-mer)–LNA (8–10 mer) chimera was used for detection of 150-mer PCR amplicons in solid phase hybridization experiments for the identification of the factor V leiden mutation responsible for venous thromboembolism (blood clotting).24b A novel 31-mer LNA-based MB has been designed that exhibits very high melting temperature, sensing with a 25-fold enhancement in the limit of detection reaching the nanomolar concentration range, enhanced single nucleobase mismatch discrimination capacity and resistance against digestion by nuclease.20 Traditional DNA-based MBs generally exhibit poor stability, sometimes false positive results, and weak enhancement of the signal once immobilized onto a solid surface.20 High affinity toward target nucleic acids with a mismatch discrimination ability makes the LNA probes suitable for the development of hybridization-based single nucleotide polymorphism (SNP) genotyping. Simeonov et al. have used short LNA (6–8 mer) probes labeled with Rhodamine or hexachlorofluorescein (HEX) dye for detection of a 107 bp long DNA target from a PCR-amplified genomic DNA sample by a fluorescence polarization (FP) measurement.24c An electrochemical sensor for the detection of the promyelocytic leukemia/retinoic acid receptor alpha fusion gene in severe promyelocytic leukemia has been reported that employed 31-mer LNA-modified hairpin probes (labeled with biotin and carboxyfluorescein).24d This assay showed a high degree of specificity for single nucleobase mismatches and an ability to detect 18-mer target DNA (detection limit 83 fM) in the presence of human serum. Another strategy based on electrochemical discrimination between target DNA sequences having full match, full mismatch and single mismatch combinations using LNA-modified hairpin capture probes with the 32-mer capture region has been presented, where the electrode is modified with gold nanoparticles and horseradish peroxidase in order to enhance the electrochemical readout.24e The 24-mer LNA-integrated probe has been utilized also for detection of SNPs using the toehold-mediated strand displacement reaction.27 The proposed detection platform is based on ON–OFF switching mechanism and it allows for target-specific, rapid detection of SNPs with high selectivity and specificity toward single nucleobase mismatches in detection of a part of codon 273 in the p53 tumor suppressor gene. The LNA nucleotides have been incorporated as sensing elements in DNA/RNA aptamers to overcome limitations of the conventional nucleic acid aptamers which are not only prone to nuclease-mediated degradation but also to damage by proteases. However, Darfeuille et al. studied LNA nucleotides as nuclease-resistant aptamers that are targeted against the HIV-1 trans-activation response RNA, which indicates that LNA units alternated with DNA could increase their stability and impart the capacity of resistance to nuclease action without reducing their affinity for the target.28
The reports on nucleic acid sensing using XNA probes at the nanoscale, especially at the single or few molecules level, are limited in number. In our laboratory, we have developed simple methods of generating self-assembled and ordered LNA monolayers18 that are capable of producing a stronger DNA detection signal and improved single nucleobase mismatch recognition, compared to the case of DNA monolayers, as detected by the molecularly resolved SMFS measurements14,19,23 (Table 3). The molecule-by-molecule detection by SMFS is largely dependent on the architecture of the capture probe film where the capture probe backbone orientation should be as upright as possible. We proposed that this is one of the primary reasons behind the improved performance of LNA as a capture probe, compared to the DNA capture probe, because a more standing backbone means fewer nonspecific interactions between the capture probe and the substrate and more ease of access of the capture probe strands to the target sequences. Another important reason behind LNA’s improved performance for molecularly resolved nucleic acid recognition, in comparison to the DNA probe, is a favorable probe density in the case of the LNA probe21 due to its effective molecular arrangement at the solid–liquid interface. An optimal probe density means that the space available per capture probe is sufficiently high to allow full target entry inside the capture probe film so that complete hybridization can take place between the target sequence and the capture probe strand, but not too high that the target can nonspecifically interact with the bare substrate surface. Consequently, the optimal LNA probe density assists in maximizing single nucleobase mismatch discrimination capacity as detected by the SMFS approach.23 The favorable LNA probe density also contributed in acquiring a high proportion (∼80%) of the SMFS-based molecule-by-molecule recognition events, whereas the success rate was lower in the case of DNA-based sensing.23
Table 3. SMFS-Derived Unbinding Force Values and Single Base Mismatch Discrimination Capacities as Demonstrated by the DNA and the XNA Probes.
| Mean
unbinding force (pN) |
|||
|---|---|---|---|
| Nucleic acid duplexes | Full match | Single base mismatch | Mismatch discrimination (pN) |
| DNA–DNA | 124 ± 3 | 105 ± 2 | ∼19 |
| PNA–DNA | 95 ± 2 | 64 ± 1 | ∼31 |
| MO–DNA | 157 ± 2 | 108 ± 1 | ∼49 |
| LNA–DNA | 203 ± 2 | 123 ± 2 | ∼80 |
In the SMFS-based approach, it was found that the surface-anchored LNA probes can discriminate between the different types of nucleobase mismatches.19 This is an important achievement because identifying different mismatch types are essential for development of a diagnostic approach for genetic screening. It is observed that any variation from the most usually formed pairs of nucleobases A-T and G-C can be identified and discriminated from each other.14,19 The LNA-based assay has been further employed for identification of mutant gene sequences as in the case of multiple drug-resistant tuberculosis (MDR-TB) bacteria.14 The most prevalent codon mutations of Mycobacterium tuberculosis leading to multiple drug resistance, as in the cases of first-line drugs, i.e., rifampicin and isoniazid, were considered, and the unbinding force data was translated into the corresponding free energy estimates using Jarzinsky’s equality treatment.14 In another SMFS-based study for the detection of the BCR-ABL fusion gene that is responsible for chronic myeloid leukemia, a 12-mer fully LNA-modified capture probe and a 21-mer LNA–DNA chimera probe were used to detect 154- to 160-mer target DNA sequences at the level of the best limit of detection so far, which is 45 zM.22 Ideally, the SMFS assay using LNA probes should be able to detect any sequence, provided the correct LNA capture probe sequence is applied and that the target sequence is not too long (a sequence of the maximum length of 80-mer has been tested so far). This approach could be a general one, which is potentially suitable for early detection of gene-related illnesses and for the prediction of vulnerability to such diseases.
Very recently, a direct SMFS-based approach to identify the KRAS G12D mutation in the circulating tumor DNA (ctDNA) samples that are extracted from patients’ plasma with very low mutant allele frequencies (0.006–0.2%) at high sensitivity and specificity (near 100%) has been reported.4 In this work, a LNA/DNA chimeric blocker has been added into the ctDNA sample before the denaturation step to make the desired target sequence available to the capture probe. Applicability of this assay for EGFR mutated DNA in cell-free DNA (cfDNA) samples was also assessed. This seminal work affirms that the XNA-based detection platform will open up new perspectives for early diagnosis of cancer and patient follow-ups based on liquid biopsy sample testing.
2. Application of PNA Probes
The PNA, which is a nonionic XNA, opens up important applications that are not feasible with conventional oligonucleotides. This is because it is resistant to the degrading actions of nuclease and protease enzymes, and is capable of target-specific binding with complementary DNA/RNA with high thermal stability and capacity of single nucleobase mismatch discrimination. Although earlier PNA was regarded to be useful primarily as a drug molecule for gene therapy, its other applications as a tool in molecular biology, biotechnology and diagnostic purposes (e.g., in testing liquid biopsy samples) where short PNA sequences can be applied as capture probes have been proposed in recent times. PNA has been utilized as a biorecognition element in a number of nucleic acid sensor technologies,6,29 yet PNA-based nanoscale, fluorescence label-free sensing studies are limited in number.6,29 Application of a surface-tethered 12-mer PNA capture probe in improving single nucleobase mismatch identification, in comparison to the DNA probe, has been exemplified by fluorescence measurement.25a Label-free direct recognition of single nucleobase mismatches in 22-mer miRNA by PNA (22-mer)-modified silicon nanowires (SiNWs) has been demonstrated at the level of 1 fM miRNA concentration.25b Importantly, the label-free SiNW sensor allows miRNA recognition in total RNA obtained from HeLa cells which shows potential application in early cancer diagnostics via detection of the miRNA biomarker.25b The development of a sensitive assay employing 23-mer PNA for point-of-care testing of methicillin-resistant Staphylococcus aureus (MRSA) using electrochemical impedance spectroscopy (EIS) has enabled the detection of fragmented MRSA gDNA in a label-independent manner in the femtomolar concentration range.25c A field-effect transistor (FET) having PNA (22-mer)-functionalized reduced graphene oxide (R-GO) has been reported for highly sensitive and label-independent detection of 22-mer DNA sequences having single nucleobase mismatches.25d In this work, detection at the level of 100 fM has been achieved using a PNA capture probe. In the case of the corresponding DNA probe, the detection limit is found to be 1 order of magnitude higher, i.e., 1 pM.25d A thermally controlled PNA-based nanoplasmonic refractometric biosensor for recognition of the E542K and E545K tumor-specific mutations and methylation of ctDNA (related to cancers in brain, lung, breast, colon, liver and stomach) of the PIK3CA gene has been introduced where PNA is used as a capture probe and enrich the 69-bp PIK3CA ctDNA.30 The application of PNA-modified AuNP to ctDNA generates shift in the LSPR-peak, matching to the primary response. For detection of the epigenetic changes, the plasmon coupling-based enhancement by using immunogold colloid was exploited that led to four times improvement in detection sensitivity (until 50 fM). An electrochemical approach has been developed using nanostructured microelectrodes functionalized with PNA probes that are specific for a definite mutant DNA sequence, and a number of PNA clamps were used to achieve high sequence-specificity in successful detection of mutated ctDNA in lung cancer (KRAS mutations) and melanoma (BRAF mutations) samples.31 When the complementary mutant target sequences were bound to the PNA probes, the signal generated from the discrete sensors was measured using an electrocatalytic reporter composed of Ru(NH3)63+ and Fe(CN)63– with differential pulse voltammetry. This assay demonstrates excellent specificity and sensitivity in the detection of mutated ctDNA as it detects 1 fg/μL of a mutant sequence in the background of 100 pg/μL of the wild-type DNA. Another gold nanorod-based plasmonic detection of ctDNA point mutation (G12V mutation in KRAS gene related to pancreatic ductal adenocarcinoma) has been demonstrated where the PNA probe specific to that point mutation was bound to the gold nanorods, and the LSPR measurement was performed after exposure to ctDNA.32 Using this method, the mutant type could be clearly discriminated from the wild type ctDNA, where the limit of detection was found to be 2 pg/μL.
In our laboratory, from comparative investigations using gold nanoparticles (AuNPs),33 nanomechanical cantilever arrays,25e and the SMFS approach,26b we found that the surface-tethered ssPNA probes exhibited better single nucleobase mismatch discrimination capacity compared to the corresponding DNA capture probes. In the AuNP-based approach, the AuNPs were surface-affixed that assists in overcoming the problem of AuNP aggregation.33 It was observed that the mismatch discrimination ability of the PNA capture probes that were immobilized onto the surface-affixed AuNPs was improved on the NP-modified surface, compared to the bare surface. The NP-facilitated improvement in the mismatch differentiation by the PNA probes was the highest when the smallest NP (10 nm) was used.33 In the piezoresistive cantilever array-based assay, which is a fluorescent label-free approach for nanomechanical sensing, the recognition event led to bending of the cantilever at nanometer length scale.25e Here, short ssPNA (9-mer to 12-mer) strands were used that are capable of direct detection of 12-mer target oligonucleotides in a sequence-specific manner including single nucleobase mismatch discrimination. The sensitivity of the PNA-based assay was found to improve about twenty times compared to that of the corresponding DNA-based assay.25e From the molecularly resolved studies using the fluorescent label-free SMFS approach, it was found that the surface-confined ssPNA strands could recognize and discriminate fully matched, singly mismatched and fully mismatched target DNA sequences (10–20 nM).26b Importantly, the mismatch discrimination ability of PNA was found to be greater in comparison to that of the DNA probe (Table 3). Therefore, in overall, we propose that use of the PNA capture probe can be beneficial for nanoscale nucleic acid sensing.
3. Application of MO Probes
Morpholino, or MO, is another nonionic DNA analogue,12 but unlike PNA, its backbone is conformationally rigid. The nuclease resistance of MO, along with its backbone rigidity, and single nucleobase mismatch discrimination capacity, has encouraged assessment of the nucleic acid sensing capacity of the MO-containing capture probes. A simple and ultrasensitive EIS-based detection of miRNA (22-mer) by the MO (22-mer) capture probe has been reported.26a Under optimized conditions, miRNA expression profiling (the recognition of circulating miRNAs in blood and miRNAs in total RNA extracted from cultured cells) was performed at the concentration level of 2 fM.26a A simple, ultrasensitive electrochemical (voltammetric- and amperometric detection) DNA sensor that employed a 24-mer MO probe and a cationic redox-active polymer as a signal generator reported better mismatch discrimination capacity than the case of the corresponding DNA capture probes with a detection limit of 1 pM.34 Zhang et al. have employed a 22-mer MO-functionalized SiNW for target-specific fluorescence label-free detection of 22-mer DNA with high recognition specificity.35 Fully matched versus mismatched DNA sequences could be identified by the MO-modified SiNW by time-dependent conductance measurements.35 The target DNA sensing performance of MO was found to be better than DNA as target concentration as low as 100 fM could be detected with the MO-functionalized SiNW whereas 100 pM could be detected by the DNA-modified SiNW surface.35 We have very recently employed the SMFS approach, which is capable of molecularly resolved detection of DNA sequences using duplex unbinding force values, and tested the ability of the MO probes for sequence-specific recognition of target DNA sequences.26b It was found that the surface-tethered MO probes are capable of differentiating between the fully matched, singly mismatched and the fully mismatched sequences. Importantly, the single nucleobase mismatch discrimination was significantly improved by using the MO probes compared to the DNA probes26b (Table 3).
4. Application of the XNAs other than LNA, PNA and MO
Although a number of other XNAs, for example, GNA, CeNA, HNA, CNA, TNA, ANA and F-ANA have been reported, there is very little information available on their applications in nucleic acid sensing, especially where a distinct advantage over the DNA probes has been reported. In the case of GNA, though the thermal stability of the GNA–GNA duplex (TM = 63 °C) surpasses the stability of the corresponding duplex of DNA (TM = 40.5 °C) and RNA (TM = 42.5 °C), GNA cannot form stable antiparallel cross-pairs with DNA.36 In the case of CeNA, inclusion of cylcohexenyl nucleosides in the DNA chain can enhance the stability of a DNA/RNA hybrid. CeNA is a new type of oligonucleotide capture probe combining the benefit of duplex stabilization and stability in serum with the potential to activate RNaseH, which means that these oligomers are important candidates for evaluation as antisense molecules for application in a cellular environment.37 The HNA is stable toward nuclease degradation with the ability to hybridize with DNA/RNA sequences selectively. The order of duplex stability was found to be HNA/HNA > HNA/RNA > HNA/DNA.38 This makes HNA an appropriate probe for nucleic acids, especially RNA. In order to improve the hybridization capacity, d-altritol nucleic acid that differs from HNA by the presence of an additional hydroxyl group in the 3′-α-position, has been designed. The high-affinity array-based detection has been performed applying HNA and altritol NA probes to detect DNA and RNA. Both the signal intensity and single nucleobase mismatch discrimination increased up to 5-fold and 3–3.5-fold for DNA and RNA targets, respectively, by applying HNA or altritol NA arrays (altritol NA > HNA > DNA).38 Altritol NA arrays could be beneficial for the growing field of RNA detection. Therefore, chemically and enzymatically stable HNA and altritol NA could be suitable for long storage and chip reuse, if and when needed. Recently, CNAs, an inexpensive XNA, has been synthesized. CNA has the ability to hybridize with DNA via Watson–Crick base pairing with high sequence specificity even at the single nucleobase mismatch discrimination level.39 However, the CNA–DNA stability is independent of salt concentration over a range of NaCl concentration (1.25–30 mM) unlike the DNA–DNA duplexes, which are largely unstable at low salt concentrations. The CNA also exhibits limited water solubility.39 Another interesting XNA is TNA, which is completely resistant to digestion by nuclease enzyme. TNA can participate in antiparallel duplex formation and cross-pairing with DNA and RNA in a sequence-specific manner following the Watson–Crick base pairing rule.40 The characteristics like stability in the physiological environment, high specificity and affinity toward DNA/RNA sequences, nuclease resistance, and effective cellular uptake may make TNA an important candidate for the development of robust XNA-based biosensors in the near future.40 The appeal of the arabinose-modified oligonucleotides, i.e., ANA and F-ANA, is based on their resistance to nuclease enzymes and their capacity to bind to target RNA. Their binding affinities for RNA are drastically different as ANA binds to RNA with comparatively low affinity, whereas F-ANA forms thermally stable duplexes with RNA.41 However, ANA and F-ANA can generate extensive RNaseH cleavage during hybrid formation with RNA.41
Conclusions
Improved performance in terms of single nucleobase mismatch discrimination has been observed in case of the XNA-based sensing in comparison to the DNA-based sensing. Applicability of the XNAs in developing nanoscale sensing strategies for detection of a variety of samples including the liquid biopsy samples from patients has been exemplified. The lowest target concentration detected so far is found to be 45 zM, as observed in an SMFS study.22 A number of these nanoscale approaches are fluorescent label-free, adding an extra advantage to the sensing strategy. However, it is yet to be seen if these approaches can be made fully PCR-independent because that will potentially lead to a method that is faster, simpler, more cost-effective and free of false positive/negative signals. We found in our studies that cost can further be lowered by preparing the XNA films using a small amount of XNA (0.1 μM–1 nM). Because the natural nucleic acids are susceptible to enzymatic degradation, whereas XNA probes (here, LNA, PNA and MO) are resistant to degradation by nuclease and/or protease enzymes, the XNA-modified sensing surfaces should be ideal for long storage and application in clinical settings. It is plausible that developing a fully PCR-independent, fluorescent label-free, nanoscale, XNA-based sensing strategy would lead to a useful tool, for example, for clinically relevant detection of the low-abundance SNPs. Indeed, because the XNAs other than LNA, PNA and MO are also capable of pairing with complementary nucleic acid sequences, and therefore, could be potential candidates as nucleic acid hybridization probes, effective applications of these XNAs too need to be explored. The application of the XNAs in point-of-care testing (POCT) using paper-based strategies is another direction that is worth to be explored. So far, for the POCTs only DNA has been used as the capture probe. Therefore, it would be interesting to use XNA capture probes and see if the target specificity and sensitivity can be improved. The XNA probe like PNA has already been integrated in paper-based assays, for example, in a multiplexed colorimetric assay for recognition of MERS-CoV, MTB and HPV-specific oligonucleotides,42 where the limit of detection is found to be about 1 nM. The paper-based POCT using XNA capture probes is an attractive option for nucleic acid sensing. However, more studies are necessary to develop it to a clinically applicable stage.
Acknowledgments
Rupa Mukhopadhyay acknowledges financial and infrastructural supports, and the fellowships of Tanushree Mana and Budhaditya Bhattacharya from Indian Association for the Cultivation of Science (IACS), Kolkata.
Biographies
Tanushree Mana received a B.Sc. (2012) and an M.Sc. (2014) in Chemistry from Vidyasagar University, Midnapore, West Bengal, India. She joined the School of Biological Sciences at the Indian Association for the Cultivation of Science for reading Ph.D. in the research group of Professor Rupa Mukhopadhyay. Her present research interest is exploring the application of xeno nucleic acid probes, especially LNA, PNA and MO for nanoscale detection of target nucleic acid sequences using AFM-based force spectroscopy.
Budhaditya Bhattacharya received a B.Sc. (2017) in Zoology from the University of Calcutta, Kolkata, India and an M.Sc. (2019) in Biotechnology from Kalinga Institute of Industrial Technology (Deemed to be University), Bhubaneswar, India. He joined the School of Biological Sciences at the Indian Association for the Cultivation of Science in 2020 for reading Ph.D. in the research group of Professor Rupa Mukhopadhyay. His research interest is utilization of the XNA probes for detection of genetic variations in clinically relevant disease models at nanoscale using a PCR-free, fluorescent label-independent approach using single molecule force spectroscopy.
Hiya Lahiri received a B.Sc. (2010) in Genetics and an M.Sc. (2012) in Molecular Biology from West Bengal University of Technology, Kolkata, India. She joined the School of Biological Sciences at the Indian Association for the Cultivation of Science for reading Ph.D. as a DST-INSPIRE fellow in the research group of Professor Rupa Mukhopadhyay in 2014 and obtained a Ph.D. in 2021. Currently, she is a postdoctoral fellow in Hebrew University of Jerusalem, Israel in the group of Professor Isaiah T Arkin.
Rupa Mukhopadhyay received a B.Sc. in Chemistry from Presidency College (University of Calcutta) with special paper in biochemistry and an M.Sc. in Chemistry from IIT Kanpur in India. She obtained a D.Phil. in Biophysical and Bioinorganic Chemistry under the supervision of Professor Allen Hill, FRS, from University of Oxford, United Kingdom. She performed postdoctoral research in nanobiotechnology under the mentorship of Professor Flemming Besenbacher at the Interdisciplinary Nanoscience Centre (iNANO), Denmark. She joined at the Indian Association for the Cultivation of Science (IACS), India, as an assistant professor in 2007 and became a professor in 2016. Her present research interests are the combined use of biomolecular/biomaterial assemblies- and nanostructures for precision surface engineering meant for nanoscale nucleic acid sensing, bioelectronics, and single molecule structural biology. This involves explorations with xeno nucleic acids as biosensing probes, protein modifications and assemblies, and single-molecule level force-based microscopy/spectroscopy.
Author Present Address
§ Hebrew University of Jerusalem, Israel
The authors declare no competing financial interest.
References
- Jackson M.; Marks L.; May G. H. W.; Wilson J. B. The Genetic Basis of Disease. Essays Biochem. 2018, 62, 643–723. 10.1042/EBC20170053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin M. L. Y.; Mach K. E.; Wong P. K.; Liao J. C. Advances and Challenges in Biosensor-based Diagnosis of Infectious diseases. Expert Rev. Mol. Diagn. 2014, 14, 225–244. 10.1586/14737159.2014.888313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrabi N.; Valizadeh A.; Farkhani S. M.; Akbarzadeh A. Basics of DNA Biosensors and Cancer Diagnosis. Artif. Cells, Nanomed., Biotechnol. 2016, 44, 654–663. 10.3109/21691401.2014.976707. [DOI] [PubMed] [Google Scholar]
- Mishra S.; Jeon J.; Kang J.-K.; Song S.-H.; Kim T.-Y.; Ban C.; Choi H.; Kim Y.; Kim M.; Park J. W. Direct Detection of Low Abundance Genes of Single Point Mutation. Nano Lett. 2021, 21, 9061–9068. 10.1021/acs.nanolett.1c02728. [DOI] [PubMed] [Google Scholar]
- a Yao X.; Li X.; Toledo F.; Zurita-Lopez C.; Gutova M.; Momand J.; Zhou F. Sub-attomole Oligonucleotide and p53 cDNA Determinations via a High-resolution Surface Plasmon Resonance Combined with Oligonucleotide-capped Gold Nanoparticle Signal Amplification. Anal. Biochem. 2006, 354, 220–228. 10.1016/j.ab.2006.04.011. [DOI] [PubMed] [Google Scholar]; b Zhang P.; Beck T.; Tan W. H. Design of a Molecular Beacon DNA Probe with Two Fluorophores. Angew. Chem. 2001, 40, 416–419. . [DOI] [PubMed] [Google Scholar]; c Huang J.; Wu Y. R.; Chen Y.; Zhu Z.; Yang X. H.; Yang C. J.; Wang K. M.; Tan W. H. Pyrene-Excimer Probes Based on the Hybridization Chain Reaction for the Detection of Nucleic Acids in Complex Biological Fluids. Angew. Chem., Int. Ed. 2011, 50, 401–404. 10.1002/anie.201005375. [DOI] [PubMed] [Google Scholar]; d Yang R. H.; Jin J. Y.; Chen Y.; Shao N.; Kang H. Z.; Xiao Z.; Tang Z. W.; Wu Y. R.; Zhu Z.; Tan W. H. Carbon Nanotube Quenched Fluorescent Oligonucleotides: Probes That Fluoresce upon Hybridization. J. Am. Chem. Soc. 2008, 130, 8351–8358. 10.1021/ja800604z. [DOI] [PubMed] [Google Scholar]; e Zhang C. Y.; Yeh H. C.; Kuroki M. T.; Wang T. H. Single Quantum-Dot-Based DNA Nanosensor. Nat. Mater. 2005, 4, 826–831. 10.1038/nmat1508. [DOI] [PubMed] [Google Scholar]; f Tsang M. K.; Ye W. W.; Wang G. J.; Li J. M.; Yang M.; Hao J. H. Ultrasensitive Detection of Ebola Virus Oligonucleotide Based on Upconversion Nanoprobe/Nanoporous Membrane System. ACS Nano 2016, 10, 598–605. 10.1021/acsnano.5b05622. [DOI] [PubMed] [Google Scholar]; g Fu X.; Cheng Z.; Yu J.; Choo P.; Chen L.; Choo J. A SERS Based Lateral Flow Assay Biosensor for Highly Sensitive Detection of HIV-1 DNA. Biosens. Bioelectron. 2016, 78, 530–537. 10.1016/j.bios.2015.11.099. [DOI] [PubMed] [Google Scholar]; h Duan R. X.; Zuo X. L.; Wang S. T.; Quan X. Y.; Chen D. L.; Chen Z. F.; Jiang L.; Fan C. H.; Xia F. Lab in a Tube: Ultrasensitive Detection of microRNAs at the Single-Cell Level and in Breast Cancer Patients Using Quadratic Isothermal Amplification. J. Am. Chem. Soc. 2013, 135, 4604–4607. 10.1021/ja311313b. [DOI] [PubMed] [Google Scholar]; i Liu G.; Wan Y.; Gau V.; Zhang J.; Wang L. H.; Song S. P.; Fan C. H. An Enzyme-Based E-DNA Sensor for Sequence-Specific Detection of Femtomolar DNA Targets. J. Am. Chem. Soc. 2008, 130, 6820–6825. 10.1021/ja800554t. [DOI] [PubMed] [Google Scholar]; j Lin M.; Wen Y.; Li L.; Pei H.; Liu G.; Song H.; Zuo X.; Fan C.; Huang Q. Target-Responsive, DNA Nanostructure-Based E-DNA Sensor for microRNA Analysis. Anal. Chem. 2014, 86, 2285–2288. 10.1021/ac500251t. [DOI] [PubMed] [Google Scholar]; k Mao X. L.; Yang L. J.; Su X. L.; Li Y. B. A Nanoparticle Amplification based Quartz Crystal Microbalance DNA Sensor for Detection of Escherichia coli O157:H7. Biosens. Bioelectron. 2006, 21, 1178–1185. 10.1016/j.bios.2005.04.021. [DOI] [PubMed] [Google Scholar]; l Chandrasekaran A. R.; Zavala J.; Halvorsen K. Programmable DNA Nanoswitches for Detection of Nucleic Acid Sequences. ACS Sens. 2016, 1, 120–123. 10.1021/acssensors.5b00178. [DOI] [Google Scholar]
- Lahiri H.; Mishra S.; Mukhopadhyay R. Nanoscale Nucleic Acid Recognition at the Solid–Liquid Interface Using Xeno Nucleic Acid Probes. Langmuir 2019, 35, 8875–8888. 10.1021/acs.langmuir.8b02770. [DOI] [PubMed] [Google Scholar]
- Dauphin-Ducharme P.; Arroyo-Currás N.; Plaxco K. W. High-Precision Electrochemical Measurements of the Guanine-, Mismatch-, and Length-Dependence of Electron Transfer from Electrode-Bound DNA are Consistent with a Contact-Mediated Mechanism. J. Am. Chem. Soc. 2019, 141, 1304–1311. 10.1021/jacs.8b11341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshkin A. A.; Nielsen P.; Meldgaard M.; Rajwanshi V. K.; Singh S. K.; Wengel J. LNA (Locked Nucleic Acid): An RNA Mimic Forming Exceedingly Stable LNA: LNA Duplexes. J. Am. Chem. Soc. 1998, 120, 13252–13253. 10.1021/ja9822862. [DOI] [Google Scholar]
- Vester B.; Wengel J. LNA (Locked Nucleic Acid): High-Affinity Targeting of Complementary RNA and DNA. Biochemistry 2004, 43, 13233–13241. 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- Nielsen P. E.; Egholm M.; Berg R. H.; Buchardt O. Sequence-Selective Recognition of DNA by Strand Displacement with a Thymine-Substituted Polyamide. Science 1991, 254, 1497–1500. 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- Demidov V. V.; Potaman V. N.; Frank-Kamenetskil M.D.; Egholm M.; Buchard O.; Sonnichsen S. H.; Nlelsen P. E. Stability of Peptide Nucleic Acid in Human Serum and Cellular Extracts. Biochem. Pharmacol. 1994, 48, 1310–1313. 10.1016/0006-2952(94)90171-6. [DOI] [PubMed] [Google Scholar]
- Summerton J. E. Morpholinos and PNAs compared. Lett. Pept. Sci. 2003, 10, 215–236. 10.1007/s10989-004-4913-y. [DOI] [Google Scholar]
- Qiu G.; Gai Z.; Tao Y.; Schmitt J.; Kullak-Ublick G. A.; Wang J. Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano 2020, 14, 5268–5277. 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- Lahiri H.; Banerjee S.; Mukhopadhyay R. Free-energy-based Gene Mutation Detection using LNA probes. ACS Sens. 2019, 4, 2688–2696. 10.1021/acssensors.9b01115. [DOI] [PubMed] [Google Scholar]
- Abu-Salah K. M.; Zourob M. M.; Mouffouk F.; Alrokayan S. A.; Alaamery M. A.; Ansari A. A. DNA-Based Nanobiosensors as an Emerging Platform for Detection of Disease. Sensors 2015, 15, 14539–14568. 10.3390/s150614539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassolas A.; Leca-Bouvier B. D.; Blum L. J. DNA Biosensors and Microarrays. Chem. Rev. 2008, 108, 109–139. 10.1021/cr0684467. [DOI] [PubMed] [Google Scholar]
- Gupta N. K.; Wilkinson E. A.; Karuppannan S. K.; Bailey L.; Vilan A.; Zhang Z.; Qi D.-C.; Tadich A.; Tuite E. M.; Pike A. R.; Tucker J. H. R.; Nijhuis C. A. Role of Order in the Mechanism of Charge Transport across Single-Stranded and Double-Stranded DNA Monolayers in Tunnel Junctions. J. Am. Chem. Soc. 2021, 143, 20309–20319. 10.1021/jacs.1c09549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S.; Ghosh S.; Mukhopadhyay R. Ordered Self Assembled Locked Nucleic Acid (LNA) Structures on Gold(111) Surface with Enhanced Single Base Mismatch Recognition Capability. Langmuir 2012, 28, 4325–4333. 10.1021/la204026j. [DOI] [PubMed] [Google Scholar]
- Lahiri H.; Mishra S.; Mana T.; Mukhopadhyay R. Discriminating unalike Single Nucleobase Mismatches using a Molecularly Resolved, Label-free, Interfacial LNA-based Assay. Analyst 2016, 141, 4035–4043. 10.1039/C6AN00484A. [DOI] [PubMed] [Google Scholar]
- Martinez K.; Estevez M. C.; Wu Y. R.; Phillips J. A.; Medley C. D.; Tan W. H. Locked Nucleic Acid Based Beacons for Surface Interaction Studies and Biosensor Development. Anal. Chem. 2009, 81, 3448–3454. 10.1021/ac8027239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S.; Ghosh S.; Mukhopadhyay R. Regulating On-surface LNA Probe Density for the Highest Target Recognition Efficiency. Langmuir 2014, 30, 10389–10397. 10.1021/la502860g. [DOI] [PubMed] [Google Scholar]
- Mishra S.; Lee Y.; Park J. W. Direct Quantification of Trace Amounts of a Chronic Myeloid Leukemia Biomarker Using Locked Nucleic Acid Capture Probes. Anal. Chem. 2018, 90, 12824–12831. 10.1021/acs.analchem.8b03350. [DOI] [PubMed] [Google Scholar]
- Mishra S.; Lahiri H.; Banerjee S.; Mukhopadhyay R. Molecularly Resolved Label-free Sensing of Single Nucleobase Mismatches by Interfacial LNA Probes. Nucleic Acids Res. 2016, 44, 3739–3749. 10.1093/nar/gkw197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Fang S.; Lee H. J.; Wark A. W.; Corn R. M. Attomole Microarray Detection of microRNAs by Nanoparticle-amplified SPR Imaging Measurements of Surface Polyadenylation Reactions. J. Am. Chem. Soc. 2006, 128, 14044–14046. 10.1021/ja065223p. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Orum H.; Jakobsen M. H.; Koch T.; Vuust J.; Borre M. B. Detection of the Factor V Leiden Mutation by Direct Allele Specific Hybridization of PCR Amplicons to Photoimmobilized Locked Nucleic Acids. Clin. Chem. 1999, 45, 1898–1905. [PubMed] [Google Scholar]; c Simeonov A.; Nikiforov T. T. Single Nucleotide Polymorphism Genotyping Using Short, Fluorescently Labeled Locked Nucleic Acid (LNA) Probes and Fluorescence Polarization Detection. Nucleic Acids Res. 2002, 30, e91. 10.1093/nar/gnf090. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Lin L.; Liu Q.; Wang L.; Liu A.; Weng S.; Lei Y.; Chen W.; Lin X.; Chen Y. Enzyme-amplified Electrochemical Biosensor for Detection of PML-RARα Fusion Gene Based on Hairpin LNA Probe. Biosens. Bioelectron. 2011, 28, 277–283. 10.1016/j.bios.2011.07.032. [DOI] [PubMed] [Google Scholar]; e Meng X. M.; Xu M. R.; Zhu J. Y.; Yin H. S.; Ai S. Y. Fabrication of DNA Electrochemical Biosensor Based on Gold Nanoparticles, Locked Nucleic Acid Modified Hairpin DNA and Enzymatic Signal Amplification. Electrochim. Acta 2012, 71, 233–238. 10.1016/j.electacta.2012.03.143. [DOI] [Google Scholar]
- a Ghosh S.; Mishra S.; Banerjee T.; Mukhopadhyay R. Facilitating Mismatch Discrimination by Surface-Affixed PNA Probes via Ionic Regulation. Langmuir 2013, 29, 3370–3379. 10.1021/la400125x. [DOI] [PubMed] [Google Scholar]; b Zhang G. J. Silicon Nanowire Biosensor for Ultrasensitive and Label-free Direct Detection of miRNAs. Methods Mol. Biol. 2011, 676, 111–121. 10.1007/978-1-60761-863-8_9. [DOI] [PubMed] [Google Scholar]; c Corrigan D. K.; Schulze H.; Henihan G.; Hardie A.; Ciani I.; Giraud G.; Terry J. G.; Walton A. J.; Pethig R.; Ghazal P.; Crain J.; Campbell C. J.; Templeton K. E.; Mount A. R.; Bachmann T. T. Development of a PCR-free Electrochemical Point of Care test for Clinical Detection of Methicillin Resistant Staphylococcus aureus (MRSA). Analyst 2013, 138, 6997–7005. 10.1039/c3an01319g. [DOI] [PubMed] [Google Scholar]; d Cai B.; Wang S.; Huang L.; Ning Y.; Zhang Z.; Zhang G. J. Ultrasensitive Label-Free Detection of PNA-DNA Hybridization by Reduced Graphene Oxide Field-Effect Transistor Biosensor. ACS Nano 2014, 8, 2632–2638. 10.1021/nn4063424. [DOI] [PubMed] [Google Scholar]; e Ghosh S.; Mishra S.; Mukhopadhyay R. Enhancing Sensitivity in a Piezoresistive Cantilever Based Label-free DNA Detection Assay Using ssPNA Sensor Probes. J. Mater. Chem. B 2014, 2, 960–970. 10.1039/c3tb21392g. [DOI] [PubMed] [Google Scholar]
- a Gao Z.; Deng H.; Shen W.; Ren Y. A Label-free Biosensor for Electrochemical Detection of Femtomolar microRNAs. Anal. Chem. 2013, 85, 1624–1630. 10.1021/ac302883c. [DOI] [PubMed] [Google Scholar]; b Mana T.; Kundu J.; Lahiri H.; Bera S.; Kolay J.; Sinha S.; Mukhopadhyay R. Molecularly Resolved, Label-free Nucleic Acid Sensing at Solid-liquid Interface using Non-ionic DNA Analogues. RSC Adv. 2022, 12, 9263–9274. 10.1039/D2RA00386D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z. F.; Ling Y.; Lu L.; Chen N. Y.; Luo H. Q.; Li N. B. Detection of Single-Nucleotide Polymorphisms Using an ON–OFF Switching of Regenerated Biosensor Based on a Locked Nucleic Acid Integrated and Toehold-Mediated Strand Displacement Reaction. Anal. Chem. 2014, 86, 2543–2548. 10.1021/ac500362z. [DOI] [PubMed] [Google Scholar]
- Darfeuille F.; Hansen J. B.; Orum H.; Primo C. D.; Toulme J. J. LNA/DNA Chimeric Oligomers Mimic RNA Aptamers Targeted to the TAR RNA Element of HIV-1. Nucleic Acids Res. 2004, 32, 3101–3107. 10.1093/nar/gkh636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadati A.; Hassanpour S.; Guardia M.; Mosafer J.; Hashemzaei M.; Mokhtarzadeh A.; Baradaran B. Recent Advances on Application of Peptide Nucleic Acids as a Bioreceptor in Biosensors Development. Trends Anal. Chem. 2019, 114, 56–68. 10.1016/j.trac.2019.02.030. [DOI] [Google Scholar]
- Nguyen A. H.; Sim S. J. Nanoplasmonic Biosensor: Detection and Amplification of Dual Bio-signatures of Circulating Tumor DNA. Biosens. Bioelectron. 2015, 67, 443–449. 10.1016/j.bios.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Das J.; Ivanov I.; Sargent E. H.; Kelley S. O. DNA Clutch Probes for Circulating Tumor DNA Analysis. J. Am. Chem. Soc. 2016, 138, 11009–11016. 10.1021/jacs.6b05679. [DOI] [PubMed] [Google Scholar]
- Tadimety A.; Zhang Y.; Kready K. M.; Palinski T. J.; Tsongalis G. J.; Zhang J. X. Design of Peptide Nucleic Acid Probes on Plasmonic Gold Nanorods for Detection of Circulating Tumor DNA Point Mutations. Biosens. Bioelectron. 2019, 130, 236–244. 10.1016/j.bios.2019.01.045. [DOI] [PubMed] [Google Scholar]
- Ghosh S.; Mishra S.; Mukhopadhyay R. Enhancing On-Surface Mismatch Discrimination Capability of PNA Probes by AuNP Modification of Gold(111) Surface. Langmuir 2013, 29, 11982–11990. 10.1021/la4019579. [DOI] [PubMed] [Google Scholar]
- Gao Z.; Ting B. P. A DNA Biosensor based on a Morpholino Oligomer Coated Indium-tin Oxide Electrode and a Cationic Redox Polymer. Analyst 2009, 134, 952–957. 10.1039/b816123b. [DOI] [PubMed] [Google Scholar]
- Zhang G. J.; Luo Z. H. H.; Huang M. J.; Tay G. K. I.; Lim E. J. A. Morpholino-functionalized Silicon Nanowire Biosensor for Sequence-specific Label-free Detection of DNA. Biosens. Bioelectron. 2010, 25, 2447–2453. 10.1016/j.bios.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Zhang L.; Peritz A.; Meggers E. A Simple Glycol Nucleic Acid. J. Am. Chem. Soc. 2005, 127, 4174–4175. 10.1021/ja042564z. [DOI] [PubMed] [Google Scholar]
- Wang J.; Verbeure B.; Luyten I.; Lescrinier E.; Froeyen M.; Hendrix C.; Rosemeyer H.; Seela F.; Van Aerschot A.; Herdewijn P. Cyclohexene Nucleic Acids (CeNA): Serum Stable Oligonucleotides that Activate RNase H and Increase Duplex Stability with Complementary RNA. J. Am. Chem. Soc. 2000, 122, 8595–8602. 10.1021/ja000018+. [DOI] [Google Scholar]
- Abramov M.; Schepers G.; Van Aerschot A.; Van Hummelen P.; Herdewijn P. HNA and ANA High-affinity Arrays for Detections of DNA and RNA Single-base Mismatches. Biosens. Bioelectron. 2008, 23, 1728–1732. 10.1016/j.bios.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Culver H. R.; Sinha J.; Prieto T. R.; Calo C. J.; Fairbanks B. D.; Bowman C. N. Click Nucleic Acid–DNA Binding Behavior: Dependence on Length, Sequence, and Ionic Strength. Biomacromolecules 2020, 21, 4205–4211. 10.1021/acs.biomac.0c00996. [DOI] [PubMed] [Google Scholar]
- Wang F.; Liu L. S.; Li P.; Leung H. M.; Tam D. Y.; Lo P. K. Biologically Stable Threose Nucleic Acid-Based Probes for Real-Time microRNA Detection and Imaging in Living Cells. Mol. Ther.-Nucleic Acids 2022, 27, 787–796. 10.1016/j.omtn.2021.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damha M. J.; Wilds C. J.; Noronha A.; Brukner I.; Borkow G.; Arion D.; Parniak M. A. Hybrids of RNA and Arabinonucleic Acids (ANA and 2′ F-ANA) are Substrates of Ribonuclease H. J. Am. Chem. Soc. 1998, 120, 12976–12977. 10.1021/ja982325+. [DOI] [Google Scholar]
- Teengam P.; Siangproh W.; Tuantranont A.; Vilaivan T.; Chailapakul O.; Henry C. S. Multiplex Paper-based Colorimetric DNA Sensor using Pyrrolidinyl Peptide Nucleic Acid-induced AgNPs Aggregation for Detecting MERS-CoV, MTB, and HPV Oligonucleotides. Anal. Chem. 2017, 89, 5428–5435. 10.1021/acs.analchem.7b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]