Abstract
Verticillium wilt, caused by the soil-borne fungus Verticillium dahliae, is one of the most devastating diseases in cotton (Gossypium spp.). Lignin in the cell wall forms a physical barrier to inhibit pathogen invasion, and defense-induced lignification reinforces secondary cell wall to prevent pathogens from further spreading. Cinnamyl alcohol dehydrogenases (CADs) catalyze the production of three main monolignols, p-coumaryl- (H), coniferyl- (G), and sinapyl-alcohols (S), which are the fundamental blocks of lignin. Here, we identified CAD genes in G. hirsutum, analyzed their expression profiles in cotton leaf, stem, and root from different developmental stages, and selected GhCAD35, GhCAD45, and GhCAD43, which were consistently induced by V. dahliae inoculation in G. hirsutum cultivars resistant or susceptible to V. dahliae. On the basis of confirmation of the in vitro enzymatic activity of the three proteins in generation of the three monolignols, we used virus-induced gene silencing (VIGS) to investigate the effects of silencing of GhCAD35, GhCAD45, or GhCAD43 on resistance to V. dahliae as well as on deposition and the composition of lignin. Silencing each of the three CADs impaired the defense-induced lignification and salicylic acid biosynthesis in stem, and compromised resistance to V. dahliae. Moreover, our study showed that silencing the three GhCADs severely affected the biosynthesis of S-lignin, leading to a decrease of the syringyl/guaiacyl (S/G) ratio. Heterogeneous overexpression of GhCAD35, GhCAD45, or GhCAD43 in Arabidopsis enhanced disease resistance. Taken together, our study demonstrates a role of the three GhCADs in defense-induced lignin biosynthesis and resistance to V. dahliae in G. hirsutum.
Keywords: lignin biosynthesis, CAD, enzymatic assay, monolignols, Gossypium, Verticillium wilt
Introduction
Cotton (Gossypium spp.) is the most important fiber crop worldwide. Upland cotton (G. hirsutum L.) and Sea Island cotton (G. barbadense L.) are the two domesticated allotetraploid species used for fiber production (Wendel, 1989). Upland cotton constitutes approximately 90% of world’s cotton production. However, most Upland cotton cultivars are susceptible to Verticillium dahliae, a soil-borne fungus pathogen causing the disease of Verticillium wilt that leads to plant vascular disease and significant economic loss worldwide. V. dahliae infects over 200 dicotyledon plant species such as cotton, tobacco, tomatoes, Arabidopsis (Song et al., 2020). Among tomato diseases, Verticillium wilt is a major biotic stressor of tomato production (Acharya et al., 2020). In cotton, the average yield loss caused by Verticillium wilt is approximately 10–35% annually; therefore it is urgent to understand the interaction mechanism between plants, especially crops, and V. dahliae. Most crops infected by V. dahliae show typical disease symptoms of leaf chlorosis, wilting, and defoliation, as well as vascular browning and even final plant death (Sink and Grey, 1999; Song et al., 2020). Because the dormant microsclerotia of V. dahliae can survive in soil for many years, it is very difficult to control Verticillium wilt in field once it occurred (Klosterman et al., 2009). Therefore, breeding of disease-resistant cultivars is considered to be one of the most effective and efficient approaches to control the threat of the pathogen (Cianchetta and Davis, 2015; Sanogo and Zhang, 2016).
Plants possess complex defense systems to counteract pathogens. When perceiving pathogen signals, plants will timely react and transduce the signal through a serial of signaling systems, which eventually lead to the induction of the expression of plant defense related genes and the production of phytoalexin, such as terpenoids and phenylpropanoids (Cui et al., 2000; Tan et al., 2000; McFadden et al., 2001). Phenylpropanoids could act as preformed and inducible antimicrobial compounds during plant defense against pathogens (Dixon et al., 2002; Koeduka et al., 2014; Tohge et al., 2017). The phenylpropanoid pathway synthesizes monolignols, the fundamental blocks of lignin biosynthesis, and phenolic compounds, which are essential for plant disease resistance (Mottiar et al., 2016; Dong and Lin, 2021). Lignin forms a physical barrier by enhancing plant mechanical strength and thickening cell walls to inhibit pathogen invasion and colonization (Xie et al., 2018; Cesarino, 2019); moreover, defense-induced lignification is important for plant innate immunity, which reinforces the secondary cell wall and contributes to pathogen- and insect-resistance (Bhuiyan et al., 2009; Eynck et al., 2012; Chezem et al., 2017). V. dahliae induced lignification has been reported to confer resistance to the pathogen in many plant species. For instance, a tomato line LA3038 (Ve/Ve) resistant to V. dahliae exhibited induction of lignin synthesis in V. dahliae inoculated roots (Gayoso et al., 2010). Application of elicitor from V. dahliae increases the synthesis and deposition of lignin and lignin-like phenolic polymers in cotton hypocotyl tissues (Smit and Dubery, 1997). Transcriptomic study in cotton has also found V. dahliae induced up-regulation of lignin biosynthesis genes (Xu et al., 2011). Most recently, GhMYB4 and Gh4CL30 were reported to regulate Verticillium wilt resistance in cotton by interfering lignification (Xiao et al., 2021; Xiong et al., 2021). Salicylic acid (SA) and jasmonic acid (JA) are necessary for the activation and mediation of plant defense signaling and responses to various biotic stresses (Loake and Grant, 2007; Miao et al., 2019; Zheng et al., 2019; Song et al., 2020). Several studies have shown that lignin associated resistance to Verticillium wilt is related to the SA mediated plant defense responses by activation of reactive oxygen species (ROS) signaling (Li et al., 2019; Tang et al., 2019).
Lignin consists of a polymeric structure which forms from the oxidative coupling of different monolignols. p- Coumaryl-, coniferyl-, and sinapyl-alcohols are the most prevalent monolignols, giving rise to hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units, respectively, in lignin polymer (Boudet et al., 1995). The relative proportion of these three units in lignin composition is usually up to the plant species, tissue types, and developmental stages (Campbell and Sederoff, 1996). The biosynthesis pathway of monolignols in plants has been well characterized (Dong and Lin, 2021). Cinnamyl alcohol dehydrogenase (CAD) is a class of NADPH-dependent enzymes involved in the last and key step of monolignol biosynthesis (Kim et al., 2004, 2007; Dong and Lin, 2021). Elevated expression of CAD gene(s) leads to increased lignin deposition. CAD typically presents as a multiple-gene family in plant species. Numerous CAD homologues have been described in different plant species with distinct or redundant functions in lignin biosynthesis. The Arabidopsis genome contains nine CADs, and several of them have been shown to be involved in lignin biosynthesis (Kim et al., 2004, 2007). AtCAD4 and AtCAD5 function predominantly and redundantly in lignin synthesis in floral stems, while AtCAD1 seems only having a minor effect on the lignin content in elongating stems (Sibout et al., 2005). Of the 15 CADs identified in Populus, PoptrCAD4 and PoptrCAD10 are preferentially expressed in xylem, the major tissue of lignin deposition (Barakat et al., 2010; Shi et al., 2010). Cotton GhCAD7 has a major role in biosynthesis of S-lignin and consequently changes the ratio of syringyl/guaiacyl (S/G) lignin (Zhang S. et al., 2020). Several studies in different species have reported roles of CADs in regulation of stress- and pathogen-induced lignification. For instance, three melon CADs, i.e., CmCAD1, CmCAD2, and CmCAD3, were found to be induced by drought stress to enhance lignin biosynthesis (Jin et al., 2014; Liu et al., 2020). In Arabidopsis and wheat, suppression of the expression of CADs enhances susceptibility to pathogens (Tronchet et al., 2010; Rong et al., 2016). Several GhCADs have been shown to be induced upon V. dahliae infection (Xu et al., 2011); however, we still know little of the cotton CAD genes, their function in lignin biosynthesis, and Verticillium wilt resistance.
In this study, we carried out a genome-wide identification of CAD genes in G. hirsutum and investigated their expression levels in different developmental stage of cotton plants. Three GhCADs that are consistently induced by V. dahliae infection in G. hirsutum cultivars resistant or susceptible to V. dahliae were selected for functional characterization. We showed that, based on in vitro enzymatic assays, all the three GhCADs possess catalyzing activity. Knocking down the expression level by virus-induced gene silencing (VIGS) compromised V. dahliae induced lignin biosynthesis and consequently resistance to V. dahliae infection. Therefore, our study demonstrates a clear role of three GhCADs in defense-induced lignin biosynthesis and resistance to V. dahliae infection in G. hirsutum.
Results
Genome-Wide Identification of CAD Genes in G. hirsutum
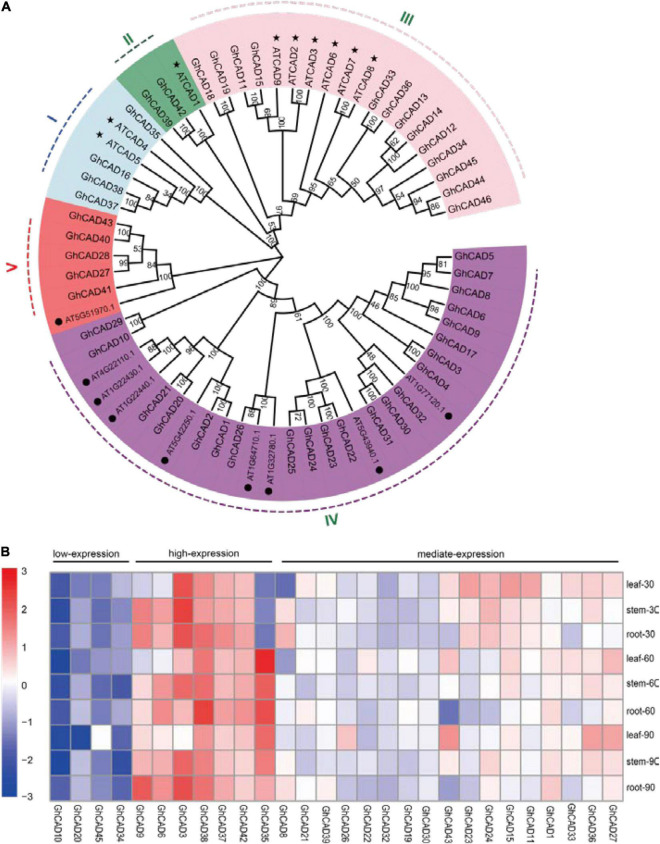
Using the nine Arabidopsis CAD protein sequences as queries, we searched the TAIR database1 and identified another nine CAD-like proteins in Arabidopsis based on domain analysis (Figure 1A). The nine CAD-like proteins were originally annotated as aldehyde dehydrogenases. We then used these Arabidopsis CAD and CAD-like protein sequences to search against the proteins annotated in the Upland cotton genome [Gossypium hirsutum (AD1) “TM-1” genome NAU-NBI_v1.1_a1.1] (Zhang et al., 2015). All hits were further validated based on the presence of the two key domains (PF08240, alcohol dehydrogenase GroES-like domain; PF00107, zinc-binding dehydrogenase) responsible for the CAD catalytic activity. Finally, we identified 46 cotton CAD and CAD-like proteins which were classified into five groups (group I to V) according to the phylogenetic tree and the conserved motifs analysis of the amino acids of this gene family (Figure 1A and Supplementary Figure 1). Groups I, II, and III contain the nine Arabidopsis CADs previously reported and 19 GhCADs. Groups IV and V contain the nine Arabidopsis CAD-like proteins identified in this study and 27 GhCAD-like proteins (Figure 1A).
FIGURE 1.
The phylogenetic analysis and tempo-spatial expression pattern of the CAD genes in G. hirsutum. (A) Phylogenetic tree of CAD proteins from Arabidopsis and G. hirsutum. The phylogenetic tree was generated by the Neighbor-Joining (NJ) method with 1,000 bootstrap replicates. CAD proteins were divided into I-V groups distinguished by different colors. The nine bona fide CADs of Arabidopsis were marked by “⬤”; the nine candidate CADs of Arabidopsis identified in this study were marked by “★.” (B) The tempo-spatial expression pattern of 28 GhCAD genes. The expression of different GhCADs in the leaf, stem, and root tissues of 30-, 60-, 90-day old plants, respectively. Three biological replicates were performed for each treatment. The expression of GhCADs was relative to that of GhUBQ7.
We investigated the tempo-spatial expression pattern of the 46 candidate GhCADs in root, stem, and leaf of 30-, 60-, and 90-day old cotton plants (cv. TM-1) by qRT-PCR. Of the 46 genes, 28 were found to be expressed in at least one of the samples tested (Figure 1B). Generally, the 28 GhCADs could be classified as low- (4), mediate- (17), and high-expression (7) three groups based on their overall expression levels in the tissues analyzed (Figure 1B). A specific expression pattern was observed for some GhCADs, for example, GhCAD27 and GhCAD43 seemed to be leaf specific, and GhCAD6 seemed to be mainly expressed in stem and root, while GhCAD35 showed an obvious developmental stage specific expression pattern (Figure 1B).
Identification of GhCADs Responding to V. dahliae Infection
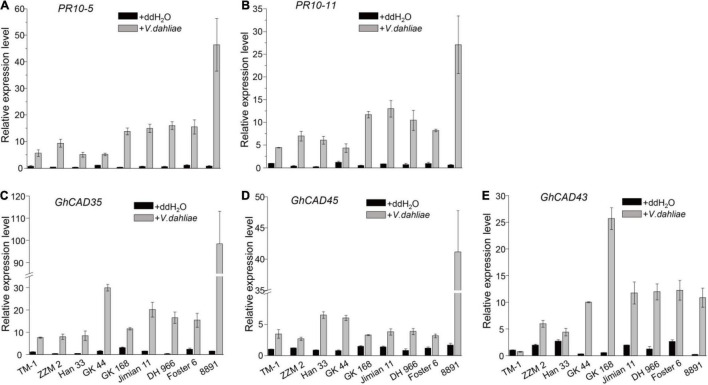
Lignin biosynthesis and deposition in plants induced by V. dahliae infection contribute to resistance to the pathogen (Zhang et al., 2019). To identify GhCADs responsible for V. dahliae induced lignin biosynthesis, we firstly identified four resistant cultivars, including Zhongzhimian 2 (ZZM 2), Han 333, GK 44, and GK 164, and four susceptible cultivars, including Jimian 11, 8891, Foster 6, and DH 966 from a collection of G. hirsutum cultivars (Supplementary Figure 2). ZZM 2 and Jiman11 have been previously reported to be resistant and susceptible to V. dahliae, respectively (Zhang et al., 2014; Zhang X.Y. et al., 2020). To exaggerate the expression of GhCADs after the induction of V. dahliae infection, we next compared the expression of the 28 GhCADs upon V. dahliae infection in the four resistant and the four susceptible cultivars, as well as TM-1, using plants at 18 days-post-infection (dpi) by qRT-PCR. PR10-5 and PR10-11 (Dowd et al., 2004), two pathogen defense-related genes, were used as positive controls. As expected, the expression level of PR10-5 and PR10-11 was highly up-regulated in all tested cultivars infected by V. dahliae (Figures 2A,B). Of the 28 GhCADs, three, including GhCAD35, GhCAD45, and GhCAD43, were consistently highly induced in almost all tested cultivars (Figures 2C–E), while the other GhCADs were either repressed or no consistent responsive trend in different cultivars after V. dahliae inoculation (Supplementary Figures 3, 4).
FIGURE 2.
V. dahliae inoculation induces the expression of several GhCADs in different G. hirsutum cultivars. (A,B) The transcriptional levels of two pathogen defense-related genes PR10-5 and PR10-11 under control (ddH2O) and V. dahliae treatment conditions in different G. hirsutum cultivars. (C–E) The transcriptional levels of GhCAD35, GhCAD45, GhCAD43, and GhCAD40 under control (ddH2O) and V. dahliae treatment conditions in different G. hirsutum cultivars. Four Verticillium wilt resistant cultivars (ZZM 2, Han 333, GK 44, and GK168), four Verticillium wilt susceptive cultivars (Jimian 11, DH 966, Foster 6, and 8891) and TM-1 were used in the V. dahliae inoculation experiment; two-weeks-old plants 18 days after V. dahliae inoculation were used for qRT-PCR analysis. Three biological replicates were performed for each treatment. The gene expression level of each gene in TM-1 under the control (ddH2O) treatment was normalized as 1. GhUBQ7 was used as reference gene.
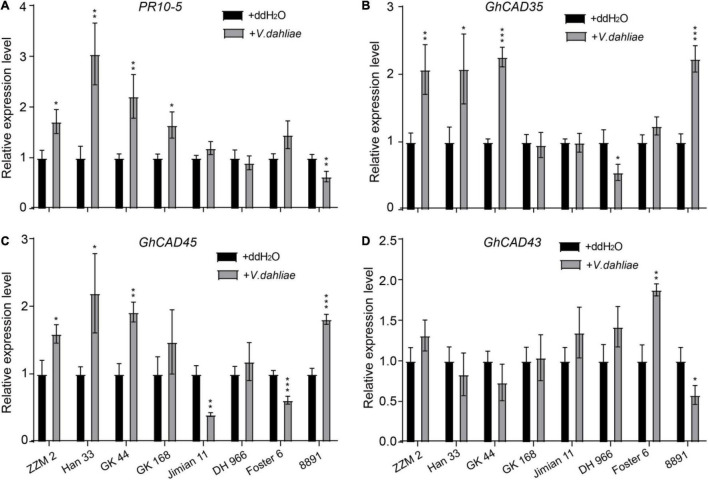
We further investigated the expression profiles of GhCAD35, GhCAD45, and GhCAD43, as well as PR10-5, 24 h after V. dahliae infection in the four resistant and the four susceptible cultivars. Interestingly, PR10-5 was significantly induced in all the four resistant but not in the four susceptible cultivars (Figure 3A). GhCAD35 and GhCAD45 were significantly induced in three (ZZM 2, Han 33, and GK 44) of the four resistant cultivars and one (8891) of the susceptible cultivars although they were repressed or not affected in different susceptible cultivars (Figures 3B,C); while the expression of GhCAD43 was only significantly induced in one (Foster6 24) of the cultivars (Figure 3D). Together, these results suggest a potential role of these three genes, particularly GhCAD35 and GhCAD45, in response to V. dahliae infection at the early infection stage.
FIGURE 3.
Different expression patterns of GhCAD35 and GhCAD45 in G. hirsutum resistant- and susceptible-cultivars 24 h after V. dahliae inoculation. (A–D) The transcriptional levels of PR10-5, GhCAD35, GhCAD45, and GhCAD43 under the control (ddH2O) and V. dahliae treatment conditions in different G. hirsutum cultivars. Four Verticillium wilt resistant cultivars (ZZM 2, Han 333, GK 44, and GK 168) and four Verticillium wilt susceptive cultivars (Jimian 11, DH 966, Foster 6, and 8891) were used in the V. dahliae inoculation experiment; two-weeks-old plants 24 h after V. dahliae inoculation were used in qRT-PCR analysis. Three biological replicates were performed for each treatment. The gene expression level in each cultivar under the control (ddH2O) conditions was normalized as 1. GhUBQ7 was used as reference gene. The data represent the mean ± SD (n = 3 biological replicates), *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t-test).
GhCAD35, GhCAD45, and GhCAD43 Possess Cinnamyl Alcohol Dehydrogenase Activities
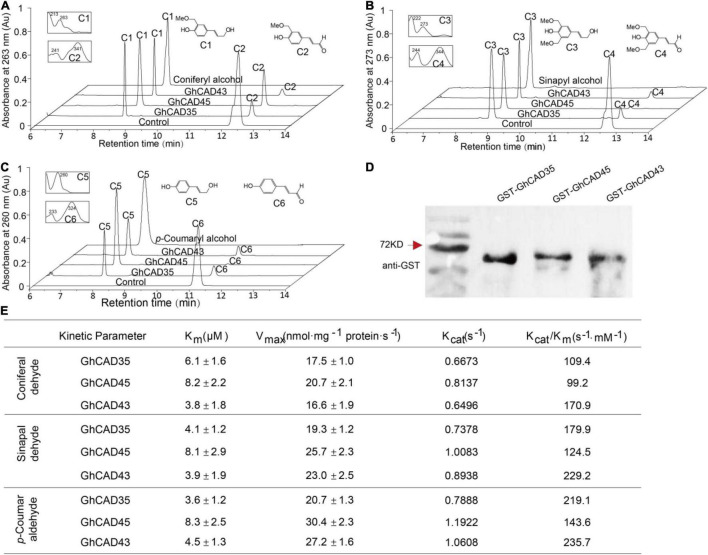
In plants, CADs catalyze the conversion of p-coumaraldehyde, coniferaldehyde, and sinapaldehyde into p- coumaryl-, coniferyl-, and sinapyl-alcohol, respectively, in the lignin biosynthesis pathway (Sibout et al., 2005; Li et al., 2012; Jun et al., 2017). To test whether GhCAD35, GhCAD45, and GhCAD43 have CAD catalytic activities, we generated recombinant GhCAD35, GhCAD45, and GhCAD43 fusion proteins with a GST tag (Figure 4D), incubated individual recombinant GhCAD35, GhCAD45, or GhCAD43 protein with each of the three substrates, p-coumaraldehyde, coniferaldehyde, and sinapaldehyde in vitro, and analyzed the final reaction products using HPLC. As shown in Figures 4A–C, GhCAD35, GhCAD45, and GhCAD43 all clearly showed CAD catalytic activities to efficiently catalyze p-coumaraldehyde, coniferaldehyde, and sinapaldehyde into their corresponding alcohol products, p- coumaryl-, coniferyl-, and sinapyl-alcohol, respectively.
FIGURE 4.
Characterization of the cinnamyl alcohol dehydrogenase (CAD) catalytic activities of GhCAD35, GhCAD45, and GhCAD43. (A–C) HPLC analyses of in vitro incubation of GhCAD35, GhCAD45, and GhCAD43 with coniferaldehyde, sinapaldehyde and p-coumaraldehyde, to yield corresponding monolignols, coniferyl alcohol, sinapyl alcohol, and p-coumaryl alcohol, respectively. C1-C6 indicate coniferyl alcohol, coniferaldehyde, sinapyl alcohol, sinapaldehyde, p-coumaryl alcohol, and p-coumaraldehyde, respectively. Controls in A-C were incubated with GST protein. (D) Western blot showing the levels of recombinant GhCAD35, GhCAD45, or GhCAD43 protein fused with a GST tag. GST antibody was used. (E) The catalytic parameters of GhCAD35, GhCAD45, and GhCAD43 toward the substrates p-coumaraldehyde, coniferaldehyde, and sinapaldehyde, respectively.
To further address the question of the substrate preference and catalytic efficiency, we determined the Km and Vmax values of GhCAD35, GhCAD45, and GhCAD43 for the three substrates p-coumaraldehyde, coniferaldehyde, and sinapaldehyde, respectively. The three enzymes displayed slight differences in their kinetic parameters (Figure 4E). GhCAD45 generally showed a less affinity toward all the three substrates compared with GhCAD35 and GhCAD43. GhCAD35 showed a little higher affinity toward p-coumaraldehyde and sinapaldehyde than coniferaldehyde. GhCAD43 showed a little higher affinity toward coniferaldehyde and sinapaldehyde than p-coumaraldehyde; while GhCAD45 showed no clear affinity preference toward the three substrates (Figure 4E). The three enzymes showed no significant differences in their Vmax values, although GhCAD45 generally showed higher catalytic rates toward all the three substrates compared with GhCAD35 and GhCAD43 (Figure 4E).
Together, these results demonstrate that GhCAD35, GhCAD45, and GhCAD43 have significant CAD catalytic activities to generate monolignol products.
Silencing GhCAD35, GhCAD45, or GhCAD43 Enhances Susceptibility to V. dahliae
To investigate whether GhCAD35, GhCAD45, and GhCAD43 are involved in Verticillium wilt resistance, we used the tobacco rattle virus (TRV)-based VIGS to knock down the expression of individual gene in the Verticillium wilt resistant G. hirsutum cultivar ZZM 2. The TRV:CLA construct was used as a positive control to validate the gene silencing system. As expected, TRV:CLA plants displayed a clear bleaching phenotype from the first pair of true leaves (Supplementary Figure 5A), indicating the VIGS system used here was effective and reliable. Next, we compared the expression level of GhCAD35, GhCAD45, and GhCAD43 in their corresponding VIGS plants with those in TRV:00 plants 2 weeks after Agrobacterium infiltration by qRT-PCR. The average expression level of GhCAD35, GhCAD45, and GhCAD43 in their corresponding VIGS plants was 21.3, 17.4, and 13.0% relative to that in TRV:00, respectively, indicating a successful silencing of the target genes (Supplementary Figures 5B,C). In addition, to check the specificity of the gene silencing in the VIGS plants, we also compared the expression of another three most homologous GhCADs of GhCAD35, GhCAD45, and GhCAD43, respectively, in the corresponding TRV:CAD35, TRV:CAD45 and TRV:CAD43 VIGS plants relative to those of TRV:00. As shown in Supplementary Figure 6, the expression of detected GhCADs were not altered in all the VIGS plants, suggesting TRV:GhCAD35, TRV:GhCAD45, and TRV:GhCAD43 plants specifically silenced the expression of the target gene, respectively.
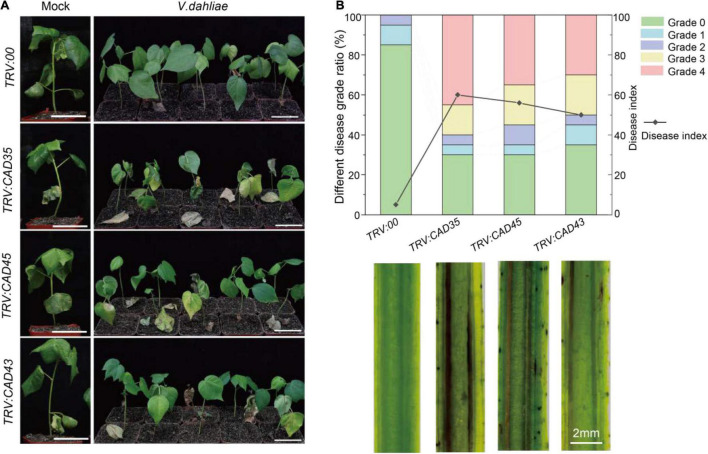
To investigate the responses of gene silencing to V. dahliae infection, V. dahliae was inoculated in the TRV:00, TRV:GhCAD35, TRV:GhCAD45, and TRV:GhCAD43 plants 2 weeks after Agrobacterium infiltration. The disease symptoms were examined at 18 dpi. As shown in Figure 5A, very weak disease symptoms were observed in TRV:00 plants; while obvious and typical disease symptoms of Verticillium wilt, including leaf chlorosis, wilting, and defoliation were observed in TRV:GhCAD35, TRV:GhCAD45, and TRV:GhCAD43 plants. We further quantified the ratio of different disease-grades and disease index (DI) to evaluate the degree of Verticillium wilt. Half or more of the TRV:GhCAD35 (60%), TRV:GhCAD45 (55%), and TRV:GhCAD43 (50%) plants displayed the severe grades 3 and 4, while no TRV:00 plant of these grades were observed and only 15% of TRV:00 plants displayed the weak grades 1 and 2 (Figure 5B). In addition, TRV:GhCAD35 plants (DI: 60.0) displayed slightly more severe disease symptoms compared with TRV:GhCAD45 (DI: 56.3) and TRV:GhCAD43 (DI: 50.0) plants (Figure 5B). Furthermore, we compared vascular browning of TRV:00, TRV:CAD35, TRV:CAD45, and TRV:CAD43 plants using longitudinal sections of stems. The vascular tissues of TRV:GhCAD45 and TRV:GhCAD43 plants, especially TRV:GhCAD35, displayed obviously dark-brown, typical symptom of V. dahliae infection, while no such symptom was observed in TRV:00 plants (Figure 5B). These results indicate that silencing GhCAD35, GhCAD45, or GhCAD43 compromised the resistance to V. dahliae infection in ZZM 2.
FIGURE 5.
Silencing of GhCAD35, GhCAD45, and GhCAD43 in Zhongzhimian 2 (Resistant to V. dahliae) resulted in susceptible to V. dahliae. (A) Phenotypes of the TRV:00, TRV:CAD35, TRV:CAD45, and TRV:CAD43 VIGS plants at 18 days post V. dahliae infection (dpi). (B) Ratio of different diseased grades and disease index for TRV:00, TRV:CAD35, TRV:CAD45, and TRV:CAD43 VIGS plants at 18 dpi (upper panel); the images showing the longitudinal stem sections of TRV:00, TRV:CAD35, TRV:CAD45, and TRV:CAD43 VIGS plants 18 dpi (lower panel). Two-weeks-old VIGS-plants were inoculated with V. dahliae at an inoculum of 1 × 107 spores/mL using the root-dipping method. n = 20.
In addition, we chose GhCAD8 and GhCAD1, which were repressed by V. dahliae infection in most cultivars 18 dpi (Supplementary Figure 4), and analyzed their roles in resistance to V. dahliae infection by examining their VIGS plants. Compared to TRV:00 plants, TRV:GhCAD8 plants displayed slight susceptibility to V. dahliae infection, and TRV:GhCAD1 plants showed no Verticillium wilt disease symptoms as indicated by plant growth, quantification of disease grades and DI, and observation of vascular tissues 18 dpi (Supplementary Figure 7). Together, these results suggest that GhCAD35, GhCAD45, and GhCAD43 are positively involved in Verticillium wilt resistance in G. hirsutum.
Silencing GhCAD35, GhCAD45, or GhCAD43 Impairs Defense-Induced Lignification in Stems
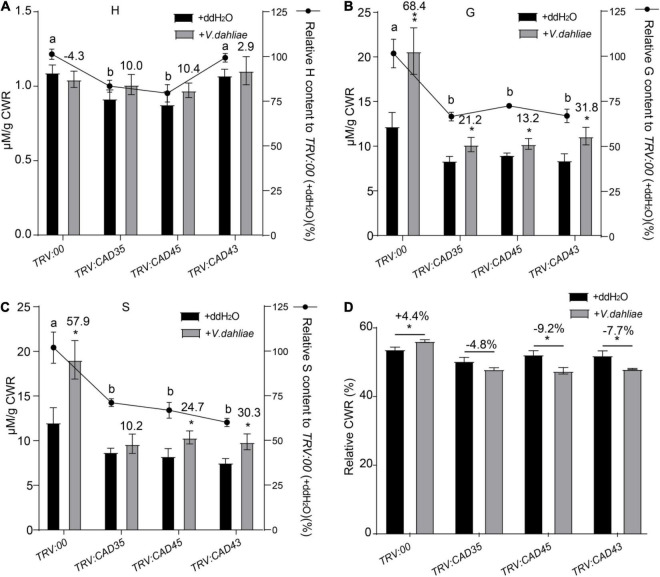
Verticillium dahliae induced lignification has been reported to confer resistance to Verticillium wilt in plants (Smit and Dubery, 1997; Gayoso et al., 2010). To evaluate the involvement of GhCAD35, GhCAD45, and GhCAD43 in lignin biosynthesis, we quantitatively measured the composition of the three lignin monomers, i.e., H-, G-, and S-unit, assayed by thioacidolysis and analyzed by gas chromatography mass spectrometry (GC-MS) using stems of TRV:00, TRV:GhCAD35, TRV:GhCAD45, and TRV:GhCAD43 plants with or without infection by V. dahliae (48 h after inoculation). In TRV:00 plants, V. dahliae infection significantly induced lignin biosynthesis and deposition (+60.0%), mainly the G- and S-unit lignin (+68.4% and +57.9%, respectively) (Figures 6B,C). Compared to TRV:00 plants, TRV:GhCAD35, TRV:GhCAD45, and TRV:GhCAD43 plants showed a decrease of the total lignin content, mainly due to decreased G- and S-unit, especially under V. dahliae infected condition (Supplementary Table 1). In addition, the levels of the G-unit and S-unit were increased in V. dahliae infected TRV:GhCAD35, TRV:GhCAD45, and TRV:GhCAD43 plants compared to their corresponding control plants, despite not as significant as that in TRV:00 plants (Figures 6B,C). No significant difference was observed for the effect of V. dahliae infection on the H-unit lignin between VIGS plants and controls (Figure 6A). Cell wall residue (CWR) mainly indicates the ratio of lignin in the whole tissue. Notably, CWR showed a slight but significant increase in TRV:00 plants after V. dahlia inoculation; in contrast, a decrease trend of CWR was observed in the V. dahliae infected TRV:GhCAD35, TRV:GhCAD45, and TRV:GhCAD43 plants compared to their corresponding controls (Figure 6D).
FIGURE 6.
Quantitative analyses of H-, G-, and S-units of lignin in the stem tissues from TRV:00, TRV:CAD35, TRV:CAD45, and TRV:CAD43 VIGS plants under the control (ddH2O) and V. dahliae treatment conditions. (A–C) Quantitative analyses of H-, G-, and S-units of lignin in TRV:00, TRV:CAD35, TRV:CAD45, and TRV:CAD43 VIGS plants, respectively. The y axel on the left indicates the contents of individual unit; the y axel on the right indicates the relative content of individual unit, TRV:CAD35, TRV:CAD45, and TRV:CAD43 to that of TRV:00. The number on top of the gray bars indicates the increase percentage of each unit under V. dahliae infection relative to the control (ddH2O); *P < 0.05, **P < 0.01 (Student’s t-test). The different letters above the black bars indicate significant differences; P < 0.05 (Student’s t-test). (D) The relative cell wall residue (CWR) in TRV:00, TRV:CAD35, TRV:CAD45, and TRV:CAD43 VIGS plants under the control (ddH2O) and V. dahliae treatment conditions. The number on top of the Ebars indicates the increase percentage of each unit under V. dahliae infection relative to the control (ddH2O); *P < 0.05 (Student’s t-test); the data represent the mean ± SD (n = 3 biological replicates). Two-weeks-old VIGS-plants were inoculated with V. dahliae at an inoculum of 1 × 107 spores/mL using the root-dipping method. The samples were collected 48 h after the inoculation.
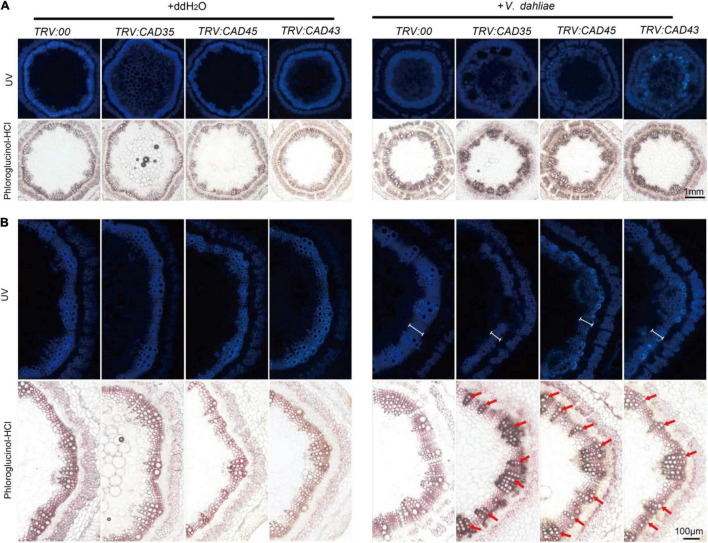
To further examine the effects of GhCAD35, GhCAD45, and GhCAD43 on lignin deposition upon V. dahliae infection, we sectioned the stems of the TRV:00, TRV:GhCAD35, TRV:GhCAD45, and TRV:GhCAD43 plants at 18 dpi, and checked deposition of lignin in xylem based on the autofluoresence under UV light and by phloroglucinol-HCl staining. Under the control (ddH2O) conditions, no difference in lignin deposition was observed in stems among TRV:00, TRV:GhCAD35, TRV:GhCAD45, and TRV:GhCAD43 plants (Figure 7); however, under the V. dahliae infection conditions, the xylem width of the stems from the TRV:GhCAD35, TRV:GhCAD45 and TRV:GhCAD43 plants was much thinner compared to that of TRV:00 indicated by autofluorescence under UV illumination and the color stained by phloroglucinol-HCl (Figure 7). Moreover, the V. dahlia infection could be observed surrounding the vessels in the V. dahlia infected VIGS plants, indicating spreading of the V. dahlia infection from the root to the shoot through vessels (Figure 7). These results were further confirmed by the quantification of the lignin composition of H-, G-, and S-unit. As shown in Table 1, under control (ddH2O) conditions, the contents of the three units in TRV:GhCAD35, TRV:GhCAD45, and TRV:GhCAD43 plants were slightly lower than that of TRV:00; however, upon V. dahliae infection, all the three units significantly reduced at 18 dpi in TRV:GhCAD35, TRV:GhCAD45, and TRV:GhCAD43 compared with TRV:00 plants. Moreover, the reduction of the S unit was more severe than the H- and G-units, leading to a significantly reduced S/G ratio in TRV:GhCAD35, TRV:GhCAD45, and TRV:GhCAD43 plants but not in TRV:00 plants (Table 1). In addition, to check whether the silencing of GhCAD genes disrupts the expression of phenylpropanoid metabolic and lignin biosynthesis genes, we checked the expression of several genes in phenylpropanoid metabolic pathway, such as GhPAL1, Gh4CL2, GhHCT, GhCOMT1, GhCCR1, etc. However, no clear induction or repression was observed for the investigated genes (Supplementary Figure 8).
FIGURE 7.
Histochemical analyses of lignin in the stem cross-sections from TRV:00, TRV:CAD35, TRV:CAD45, and TRV:CAD43 VIGS plants at 18 dpi. (A) The handcut stems were illuminated by UV or stained with phloroglucinol-HCl to detect lignin. (B) Close-up of UV autofluorescence and phloroglucinol-HCl stain of stems corresponding to images in A. Two-weeks-old VIGS plants were inoculated with V. dahliae at an inoculum of 1 × 107 spores/mL using the root-dipping method. Scale bars in A and B represent 1 mm and 100 μM, respectively. The white scales in V. dahliae inoculated stems under UV indicate the width of xylem; the red arrows indicate the vessels.
TABLE 1.
Quantification of H-, G-, and S-unit of lignin deposition in stems of TRV:00, TRV:CAD35, TRV:CAD45, and TRV:CAD43 VIGS plants at 18 dpi by V. dahliae.
| Sample name | Monomer yields |
Total yields | Molar ratios | |||
| H | G | S | S/G | |||
| +ddH2O | TRV:00 | 5.5 ± 0.9 | 67.8 ± 9.5 | 66.7 ± 8.3 | 140 | 0.98 |
| TRV:CAD35 | 4.6 ± 0.5 | 59.5 ± 7.2 | 61.5 ± 8.8 | 125.6 | 1.03 | |
| TRV:CAD45 | 6.1 ± 0.3 | 55.6 ± 5.0 | 57.3 ± 5.2 | 119 | 1.03 | |
| TRV:CAD43 | 4.7 ± 0.2 | 61.8 ± 6.0 | 58.5 ± 9.7 | 125 | 0.95 | |
| +V. dahliae | TRV:00 | 5.1 ± 0.2 | 62.1 ± 4.9 | 46.9 ± 5.2 | 114.1 | 0.76 |
| TRV:CAD35 | 5.0 ± 0.2 | 52.0 ± 8.8 | 25.0 ± 4.5 | 75 | 0.48 | |
| TRV:CAD45 | 5.5 ± 0.7 | 51.9 ± 4.7 | 31.2 ± 3.6 | 88.6 | 0.6 | |
| TRV:CAD43 | 5.9 ± 0.6 | 55.5 ± 7.0 | 39.2 ± 5.3 | 100.6 | 0.71 | |
Yields (μmol/g of Klason Lignin) of Lignin-Derived Thioacidolysis Monomers from Various Lignocellulosic Materials, Measured by GC–MS.
H, p-hydroxyphenyl monomer; G, guaiacyl monomer; S, syringyl monomer.
Taken together, these results demonstrate GhCAD35, GhCAD45, and GhCAD43 positively regulate V. dahliae induced lignification in the stems of Upland cotton.
Silencing GhCAD35, GhCAD45, or GhCAD43 Alters V. dahliae Induced Salicylic Acid Responses
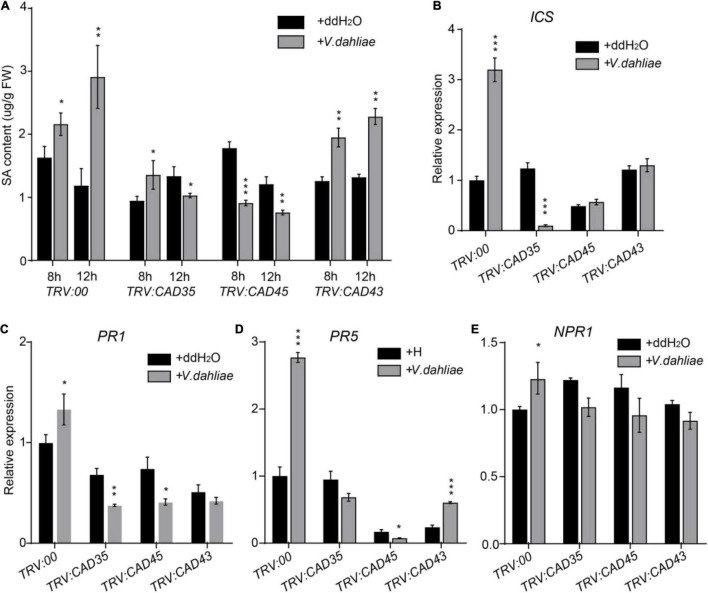
Salicylic acid and JA play key roles in plant defense against pathogen infection (Shaban et al., 2018; Song et al., 2020). Previous studies have shown that disturb of lignin metabolism in the cell wall of root and stem tissues in G. hirsutum results in the change of SA synthesis and signaling upon V. dahliae infection. Therefore, we investigated the effects of silencing GhCAD35, GhCAD45, or GhCAD43 on the SA biosynthesis and signaling. As shown in Figure 8A, SA content was significantly increased in TRV:00 plants 8-h- and 12-h-post inoculation of V. dahliae (Figure 8A); however, SA contents were lower or less induced in the TRV:GhCAD35, TRV:GhCAD45, and TRV:GhCAD43 plants compared to that of TRV:00 plants after V. dahliae infection. We further investigated the expression of SA biosynthesis gene ICS1, and SA signaling genes PR1, PR5, and NPR1 12-h-post V. dahliae infection by qRT-PCR. Consistently, the expression levels of ICS1, PR1, PR5, and NPR1 were all induced by V. dahliae infection in TRV:00 plants; while in TRV:GhCAD35 and TRV:GhCAD45, the expression levels of ICS1, PR1, and PR5 were inhibited or no significant change after V. dahliae infection. In TRV:GhCAD43, a less induction or no significant change were observed for the expression of ICS, PR1, and PR5 after V. dahliae infection compared to those of TRV:00 plants (Figures 8B–D). We also investigated the effects of silencing GhCAD35, GhCAD45, or GhCAD43 on the gene expression of JA signaling components, such as GhJAZ3, GhJAZ10, GhMYB2-2, and GhMYB2-3. As shown in Supplementary Figure 9, GhJAZ10 was slightly induced after V. dahliae infection in TRV:GhCAD35 and TRV:GhCAD45, but the other genes did not show consistent changes in the GhCAD silencing plants after V. dahliae infection compared to that of control. These results indicate that silencing GhCAD35, GhCAD45, or GhCAD43 might affect the defense induced SA responses at the early infection stage.
FIGURE 8.
Silencing of GhCAD35, GhCAD45, and GhCAD43 impaired V. dahliae induced SA responses. (A) Quantification of SA contents in TRV:00, TRV:CAD35, TRV:CAD45, and TRV:CAD43 VIGS plants 8- and 12-h post-infection by V. dahliae. (B–E) The expression level of SA biosynthesis gene ICS and SA responsive genes PR1, PR5, and NPR1 in TRV:00, TRV:CAD35, TRV:CAD45, and TRV:CAD43 VIGS plants 12-h post-infection by V. dahliae. The data represent the mean ± SD (n = 3 biological replicates), *P < 0.05, **P < 0.01, ***P < 0.001 (Student’s t-test).
Heterogeneous Overexpression of GhCAD35, GhCAD45, or GhCAD43 in Arabidopsis Enhances Resistance to V. dahliae
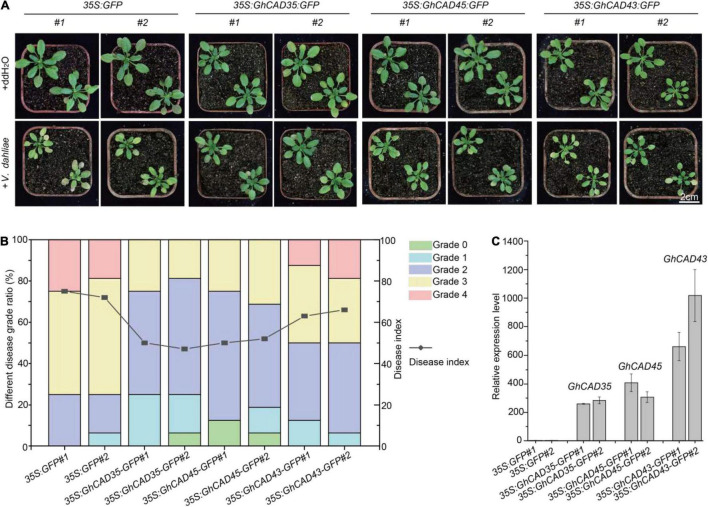
The above results indicate that GhCAD35, GhCAD45, and GhCAD43 are positive regulators of resistance to V. dahliae by affecting lignin biosynthesis and deposition. To further confirm this conclusion, we heterogeneously overexpressed GhCAD35, GhCAD45, or GhCAD43 in Arabidopsis to examine their effects on V. dahliae infection. We analyzed two independent Arabidopsis transgenic lines overexpressing GhCAD35, GhCAD45, or GhCAD43, and challenged the transgenics with V. dahliae. As shown in Figures 9A–C, compared to the control 35S:GFP plants, all Arabidopsis transgenic lines displayed overexpressed expression of corresponding GhCADs and enhanced resistance to V. dahliae infection, as indicated by plant growth, disease infection grades, and disease index statistics, although the resistance of the transgenics overexpressing GhCAD43 was not as significant as those of the transgenes overexpressing GhCAD35 or GhCAD45. Furthermore, we chose one line of each GhCADs overexpressing transgenic Arabidopsis plants and measured the H-, G-, and S-units of lignin contents in their stems. As shown in Supplementary Figure 10, S-unit of lignin was significantly elevated in the GhCADs overexpressing transgenic Arabidopsis plants compared with that of control, suggesting overexpression of GhCAD35, GhCAD45 or GhCAD43 enhanced the lignin biosynthesis in Arabidopsis.
FIGURE 9.
Heterogeneously expressing of GhCAD35, GhCAD45, and GhCAD43 in Arabidopsis enhanced resistance to V. dahliae infection. (A) Phenotypes of the transgenic Arabidopsis plants overexpressing GhCAD35, GhCAD45, or GhCAD43 at 14 days post-infection (dpi) by V. dahliae. (B) Ratio of different diseased grades and disease index for GhCAD35, GhCAD45, and GhCAD43 overexpressing transgenic Arabidopsis plants at 14 dpi by V. dahliae. Two independent Arabidopsis transgenic lines of 35S:GhCAD35-GFP, 35S:GhCAD45-GFP, and 35S:GhCAD43-GFP were used for V. dahliae infection. 35S:GFP Arabidopsis transgenic plants were used as control. Two-weeks-old Arabidopsis transgenic plants were inoculated with V. dahliae via root-dipping method; 16 plants for each line were used. (C) The expression level of GhCAD35, GhCAD45, and GhCAD43 in the corresponding Arabidopsis transgenic plants determined by qRT-PCR.
Discussion
As one key component of plant cell wall, lignin is important for plant immunity and defense responses (Xie et al., 2018; Cesarino, 2019; Dong and Lin, 2021). The lignin deposition in the cell wall provides the first barrier for pathogen invasion (Dong and Lin, 2021). In addition, the induced lignification by pathogen invasion further inhibits the growth, movement and infection ability of the pathogens (Xie et al., 2018; Li et al., 2020). In cotton, lignin biosynthesis is significantly induced upon V. dahliae infection and has a significant role in resistance to V. dahliae (Zhu et al., 2018; Li et al., 2020; Xiao et al., 2021; Xiong et al., 2021). Therefore, it is important to identify the genes responsible for the V. dahliae induced lignification. The CAD gene family encode a class of NADPH-dependent enzymes involved in the last step of monolignol biosynthesis, therefore important for lignin deposition (Kim et al., 2007; Dong and Lin, 2021). Previously study has identified 20 GhCADs in G. hirsutum, which are divided into three groups based on homology analysis with the nine bona fide Arabidopsis CADs (Zhang S. et al., 2020). In the present study, we reexamined the CAD gene family in G. hirsutum and identified 46 GhCAD or GhCAD-like genes that were classified into five groups (Figure 1A). By comparing the response of GhCADs to V. dahliae infection in cultivars resistant or susceptible to V. dahliae, GhCAD35, GhCAD45, and GhCAD43 were selected for further functional characterization.
The in vitro enzymatic assay showed that GhCAD35, GhCAD45, and GhCAD43 are active enzymes for catalyzing p-coumaraldehyde, coniferaldehyde, and sinapaldehyde into p- coumaryl-, coniferyl-, and sinapyl-alcohol, respectively (Figure 4), with no obvious enzyme kinetic difference. Notably, GhCAD43 is a member of the group V CAD-like protein (Figure 1A), suggesting a role of CAD-like genes in biosynthesis of lignin in planta.
Verticillium dahliae induced lignin biosynthesis has been shown in various plant species, such as pepper, tomato, and cotton (Pomar et al., 2004; Gayoso et al., 2010; Xiong et al., 2021). Consistently, we observed significantly elevated G-unit and S-unit in V. dahliae infected Upland cotton cultivar (ZZM 2) resistant to V. dahliae, but the V. dahliae induced lignin deposition was compromised in VIGS plants of GhCAD35, GhCAD45, or GhCAD43 (Figure 6 and Supplementary Table 1). Consequently, the TRV:GhCAD35, TRV:GhCAD45, and TRV:GhCAD43 plants were more susceptible to V. dahliae compared to TRV:00 (Figure 5). At the latter infection stage (18 dpi), lignin deposition was decreased in all V. dahliae infected plants compared to those of uninfected, which could attribute to the breakdown of lignin upon Verticillium wilt disease spreading (Figure 7). These results suggest GhCAD35, GhCAD45, and GhCAD43 play a pivotal role in V. dahliae induced lignification and resistance to the pathogen.
The biosynthesis of lignin mainly results from the oxidative coupling of three monolignols, giving rise to G-, H-, and S-unit, respectively. The proportions of these three units in the cell wall are significantly diverse depending on plant species, developmental stages, and tissue types (Bhuiyan et al., 2009). Plants have adopted an effective mechanism to restrict the spread of pathogen, by restructuring the monomer composition of lignin in the vascular tissues (Pomar et al., 2004; Gayoso et al., 2010). Generally, the lignin in cotton stems contains much higher proportions of G- and S-unit than H-unit (Xiong et al., 2021). Consistently, our study showed that G- and S-unit were predominant compared to H-unit in cotton stems (Table 1 and Supplementary Table 1). In addition, it was reported that G-rich lignin has a more pronounced impact on cotton defense responses (Xu et al., 2011; Zhang et al., 2019). However, several studies also indicate a similar role of S-rich lignin in plant immunity (Wang et al., 2018). Our study showed that the contents of both G- and S-unit were significantly induced 48 h after V. dahliae infection, with a slight higher induction of G- than S-unit (+68.4% vs. +57.9%), indicating both G- and S-unit play pivotal roles in the early defense responses (Figure 6 and Supplementary Table 1). However, at the latter infection stage, the S/G ratio greatly decreased in the V. dahliae infected plants compared to that of control (Table 1), suggesting that G-rich lignin might play a more pronounced role for resistance at the latter infection stage.
The Upland cotton genome contains 46 GhCAD genes, how do they coordinate with each other to regulate lignin biosynthesis is an interesting question. Here, we showed that silencing a single GhCAD (GhCAD35, GhCAD45, or GhCAD43) seems to be sufficient to repress V. dahliae induced lignin biosynthesis (Figure 5), suggesting they likely act synergistically rather than redundantly in lignin biosynthesis. The tempo-spatial expression of genes provides key and important insights for their biological functions. Most GhCADs are expressed in the root, leaf, and stem tissues at different developmental stages (Figure 1B). Interestingly, GhCAD35 and GhCAD45 belong to the mediate- and low-expression subgroup, respectively; while GhCAD43 displayed an obvious developmental stage dependent manner with low expression at seedling stage but high expression at the latter developmental stage (Figure 1B). Therefore, the induction of their expression by V. dahliae infection indicates GhCAD35, GhCAD45, and GhCAD43 are tightly regulated by internal and external factors. Transcriptional factors (TFs) usually act as master switches by controlling gene expression under different developmental or environmental conditions. Numerous TFs have been illustrated to be involved in plant lignin biosynthesis, including many members of the MYB family (Zhong et al., 2008; Zhou et al., 2009; Dong and Lin, 2021). TFs from the WRKY- and NAC-family are also involved in the regulation of lignin biosynthesis (Wang et al., 2020; Dong and Lin, 2021; Hu et al., 2021). Therefore, it is important to study how the expression of GhCADs is regulated by these TFs in the future.
Phytohormones such as abscisic acid, jasmonate acid, ethylene, and SA, play critical roles in the complex defense responses (Bari and Jones, 2009). Previous studies have shown that the alteration of lignin metabolism in the cell wall of root and stem tissues in G. hirsutum results in the change of SA synthesis and signaling upon V. dahliae infection (Li et al., 2019; Tang et al., 2019). Here we showed that the expression level of the SA biosynthesis gene ICS1, and the SA signaling genes PR1 PR5, and NPR1 were altered in TRV:GhCAD35, TRV:GhCAD45, and TRV:GhCAD43 plants compared to TRV:00 (Figure 8). How does silencing GhCADs affect SA content upon V. dahliae infection is still an open question. The cross-talk between SA and lignin metabolism on fine-tuning the defense response of cotton toward V. dahliae infection remains to be determined. We speculate that silencing CADs results in impaired defense responses, which might indirectly affected the SA levels upon V. dahliae infection. On the other hand, GhCAD35, GhCAD45, and GhCAD43 catalyze the generation of various monolignols, which likely affects the turnover of intermediates in lignin biosynthesis pathway that might act as phytoalexins or signal molecules in the plant defense responses (Dixon et al., 2002). One of the main goals of studying defense-induced lignification is to utilize the generated knowledge to develop crops with improved tolerance to V. dahliae. Future work by modulating the expression of GhCADs involved in lignin biosynthesis through bioengineering or molecular breeding strategies might be able to develop cotton cultivars resistant to Verticillium wilt.
Experimental Procedures
Gossypium hirsutum and Arabidopsis Growth Conditions
Nine Upland cotton (G. hirsutum L.) accessions, including four (Zhongzhimian 2, Han 333, GK 44, and GK 164) resistant to V. dahliae and five (Jimian 11, DH 966, Foster 6, 8891, and TM-1) susceptible to V. dahliae, were used in disease assay. Plants were grown at 25°C in a growth room with 80–100 μmol⋅m–2⋅s–1 light and a 16-hr light/8-hr dark photoperiod. Two-weeks-old cotton plants were used in V. dahliae infection with the strain of V991.
The V. dahliae resistant cultivar Zhongzhimian 2 (ZZM 2) was used in the VIGS experiment and disease assay. The experiment was performed in a growth room with a temperature of 23–24°C, light intensity of 100 μmol⋅m–2⋅s–1, and a photoperiod of 16-hr light/8-hr dark.
The Arabidopsis (Col-0 background) plants were grown at 22°C in a growth chamber under 100 μmol⋅m–2⋅s–1 light and a 16-hr light/8-hr dark photoperiod. Arabidopsis transformation was performed using the flower dipping method as described previously (Clough and Bent, 1998).
Identification of the CAD Genes in G. hirsutum
Two approaches were adopted to identify cotton CAD genes in G. hirsutum. First, nine bona fide Arabidopsis CAD proteins were used to search Arabidopsis TAIR database (see text footnote 1) and identified another nine CAD-like proteins in Arabidopsis based on domain analysis. Arabidopsis CAD and CAD-like protein sequences were used as queries to search against the annotated proteins of G. hirsutum (Gossypium hirsutum (AD1) ‘TM-1’ genome NAU-NBI_v1.1_a1.1) (Zhang et al., 2015) downloaded from the CottonGen database2 by local blast tool with the default settings (E-value < 1e–10). All hits were considered as potential candidate CAD genes. Second, the hidden Markov model (HMM) profile of the two conserved domains (PF08240, alcohol dehydrogenase GroES-like domain; PF00107, zinc-binding dehydrogenase)3,4 (Potter et al., 2018) was used to search the G. hirsutum (Gossypium hirsutum (AD1) ‘TM-1’ genome NAU-NBI_v1.1_a1.1) (Zhang et al., 2015) using the HMMER 3.0 software. All candidate sequences derived from the two searches were subjected to domain analysis using the InterProScan5 and SMART6 tools with the default parameters. Only protein sequences with both domains were considered to be CADs. The gene ID and corresponding gene name for all GhCADs are listed in Supplementary Table 2.
Verticillium dahliae Inoculation
The V. dahliae strain V991 was cultivated on potato dextrose agar (PDA) plate at 25°C for 4–5 days, and was then transferred to potato dextrose broth (PDB) liquid media and incubated on a shaker (120 rpm, 25°C) for 3 days. The conidia of V. dahliae were collected and adjusted to a concentration of ∼1 × 107 spores/mL. Cotton seedlings were inoculated by root dipping with the inoculum. Three replicates were set up for each genotype with at least 20 plants in each replicate.
Disease Index Calculation
Disease index was calculated according to the method descripted previously (Miao et al., 2019). Disease symptom severity was scored as level 0 to level 4 at 18 dpi according to the percentage of wilted true leaves and leaf chlorosis or defoliation. A score of zero means no visible wilting or chlorosis symptoms; a score of 1–4 indicates 0–25% (inclusive), 25–50% (inclusive), 50–75% (inclusive), and 75–100% of wilted true leaves and leaf chlorosis or defoliation, respectively. DI was calculated according to the following formula:
n represents disease levels ranging from 0 to 4.
RNA Isolation and Quantitative Real-Time PCR Analysis
Total RNA was extracted from cotton stems using a RNAprep Pure Plant Plus Kit (Polysaccharides and Polyphenolics-rich) (Tiangen, Beijing, China). Then 1 μg total RNA was taken for reverse transcription to synthesize cDNA using HiScript II Q Select RT SuperMix (Vazyme, Nanjing, China). The quantitative real-time PCR (qRT-PCR) experiment was conducted using ChamQ SYBR Color qPCR Master Mix (Vazyme, Nanjing, China) and analyzed using the Roche LightCycler 480 (Beijing, China). The thermal profile for qRT-PCR was 95°C for 2 min, followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. Primers used for qRT-PCR are listed in Supplementary Table 3.
Gene Cloning, Protein Expression and Purification
Full-length GhCADs were amplified with a KOD-Plus kit (Toyobo) using the cDNA generated from TM-1. The gene specific primers were designed according to the TM-1 genome sequence of NAU-NBI_v1.1. GhCADs were expressed in E. coli (BL21) by cloning individual GhCAD into a glutathione (GST)-tagged expression construct pGEX4T-1. The recombinant GST-GhCADs were purified using a column containing GST coupled with agarose medium. Western blot was used to detect the recombinant GST-GhCADs using anti-GST antibody on an SDS-PAGE Gel.
Enzyme Assay and Kinetics
The reaction (200 μL) used in assay of GhCAD activity contains: 50 mM Tris HCl (pH = 7.5), 0.5 mM NADPH, 0.1 mM substrate (coniferyl aldehyde, p-coumaryl aldehyde, or sinapyl aldehyde), 2.0 mM dithiotreitol (DTT), and 1.3 μg recombinant GST-GhCAD protein. The reaction was incubated at 30°C for 30 min and terminated by adding 200 μL methanol. The reaction mixture was filtrated through a 0.22 μm organic diameter filter. A volume of 100 μL filtrated reaction mixture was then subjected to high-performance liquid chromatograph (HPLC) analysis with an XBridge BEH C18 column (2.5 μm, 3.0 × 150 mm XP). The experimental procedure established to separate the products (three alcohols) from the substrates (three aldehydes) was as follows: two chromatographic grade solvents, water (containing 1% formic acid) and acetonitrile, were used as the mobile phase A and B, respectively; the proportion of A:B was set to change from 95:5 to 5:95 from 0 to 30 min gradually; the flow rate was set to be 0.5 mL/min; the section of full wavelengths was set to be from 210 nm to 400 nm.
In order to analyze the GhCAD catalytic kinetics, nine concentrations (2, 6, 8, 17, 25, 33, 42, 50, and 100 μM) of each substrate were chosen to conduct the catalytic experiment as mentioned above, and the enzyme kinetic curves were fitted using the Origin 2018 software (Pan et al., 2014; Ma et al., 2018).
Virus-Induced Gene Silencing
A gene-specific fragment encoding 100–200 amino acids was amplified from GhCAD35, GhCAD43, or GhCAD45, and cloned into the pTRV2 plasmid between the EcoRI and KpnI restriction sites. Primers used are listed in Supplementary Table 4. After confirmation by sequencing, the recombinant vector was introduced into Agrobacterium tumefaciens GV3101 by thermal activation. For VIGS, the Agrobacterium culture was resuspended in the VIGS buffer (10 mM MgCl2, 10 mM MES and 200 μM acetosyringone) and mixed with Agrobacterium tumefaciens transformed with pTRV1 in equal proportion. After 3 h of incubation at room temperature, various Agrobacterium combinations were injected into cotyledons of 1 week old seedlings of ZZM 2. Two weeks later, the treated cotton seedlings were used in confirmation of silencing of the target gene (based on qRT-PCR) and then used in disease assay by inoculation of V. dahliae inoculum (as mentioned above).
Phloroglucinol Staining
Stems collected from the same position of different cotton plants were cross sectioned by hand with a section thickness of 100–120 μM. Sections were immersed in 1% phloroglucinol (0.1 g phloroglucinol in 10 mL 95%EtOH) for 3–5 min; afterward, sections were gently rinsed with water and acidified with 20% HCl solution. Finally, the sections were examined by inverted microscope.
Cell Wall Preparation
Freeze-dried stem materials were fully ground to fine powder and extracted in 70% EtOH (5 × volume) at 65°C for 3 h. After repeated twice the extraction, the final extraction was carried out with chloroform/methanol (1:1 v/v) to get rid of water and pigments. The products were washed once by acetone (2–3 × volume) and dried for 3 days in an oven in a hood.
GC-MS Measurement of Lignin G-, S-, and H-Units
Lignin G-, S-, and H-units were depolymerized and separated with a streamlined thioacidolysis, and analyzed by gas chromatography–mass spectrometry (GC–MS) as previously described (Robinson and Mansfield, 2009). For each sample, 10 ± 1 mg of ground, oven-dried wood powder was filled into a 2-mL HPLC glass vial. The vials were blanked with nitrogen gas, added with 1 mL of freshly made reaction buffer [2.5% boron trifluoride etherate and 10% ethanethiol, in recently distilled dioxane (v/v)], and tightly capped. The vials were then placed in a dry heating block (98°C) for 4 h, with periodic manual agitation. The reaction was stopped by placing on ice for 15 min and adjusted to have a pH value between 3 and 4 by adding appropriate volume of 0.4 M sodium bicarbonate (∼0.3 mL, as determined by pH indicator paper). The solution was transferred to a new 10 mL vial. Internal standard (methyl heptadecanoate, 1 mg) dissolved in 1 mL methylene chloride was then added to each vial. To extract the reaction products from the aqueous mixture, the vials were recapped, vortexed and kept still for half an hour, phase-separating the upper (aqueous) and lower (organic, containing lignin breakdown products) phases. An aliquot (1.5 mL) of the organic phase was removed and simultaneously cleared of residual water and filtered by passing through a Pasteur pipette packed with a small tissue-paper plug and an inch (∼50 mg) of granular anhydrous sodium sulfate, and transferred directly into a 2-mL polypropylene microfuge tube. Samples were then collectively evaporated to dryness in a low pressure oven (50°C) overnight and resuspended in 0.5 mL of methylene chloride. A volume of 50 μL samples was transferred to new microcentrifuge tube and dried in the same condition as above; the dried samples were then combined with 50 μL of pyridine and 50 μL of N-methyl-N-(trimethylsilyl) trifluoroacetamide. After incubation for at least 5 h at room temperature, 1 μL of the reaction product was analyzed by GC-MS.
GC-MS was conducted on gas chromatography tandem mass spectrometry Xevo TQ-GC (Waters, United States). The analyses used were as previously described (Rolando et al., 1992). In brief, the GC-MS was equipped with a DB-5 column (30 m × 0.25 mm × 0.25 μm, Agilent Technologies). The oven initial temperature on the DB-5 column was maintained at 130°C for 3 min and then increased to 250°C at 3°C/min, and finally maintained 250°C for 5 min. The injector temperature was maintained at 250°C with a constant flow rate of 1.0 mL/min of helium. The inlet was operated in pulsed splitless injection mode. The GC–MS electron impact source was operated in scan mode with the MS source temperature at 280°C and MS Quad at 150°C.
Salicylic Acid Measurement
Endogenous SA was extracted from the stems of cotton plants as previously described (Hu et al., 2018; Miao et al., 2019), and measured by LC-MS. In brief, three replicates, each with 100 mg (fresh weight) of stem samples, were harvested at 8 and 12 h after V. dahliae inoculation. The samples were ground to a fine powder in liquid nitrogen, mixed with 750 μL of ice-cold 80% methanol containing 1% acetic acid, and placed on a shaker for 16 h at 4°C in darkness. The samples were then centrifuged at 12,000 rpm (4°C) for 15 min, and the supernatant was collected and transferred into a new tube and was then evaporated to dryness, and the residue was reconstituted in 200 μL of 80% methanol. Finally, the extracts were analyzed using a Q Exactive Plus HPLC-MS system (Thermo, United States).
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
YM, K-PJ, and HL conceived the study. SZ performed bioinformatic analysis. HL, XlZ, WX, YG, and YW performed the other experiments. YM, K-PJ, HL, SZ, and YZ analyzed the data. K-PJ, HL, and SZ wrote the manuscript. YM, YZ, Q-HZ, XeZ, KL, and JG revised the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
A patent application about the application of GhCAD35, GhCAD45 and GhCAD43 on Upland cotton breeding has been filed. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31770300 and 31900245), the key research and promotion projects (202102110002), and 111 project of China and Natural Science Foundation of Henan Province (202300410012).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.840397/full#supplementary-material
References
- Acharya B., Ingram T. W., Oh Y., Adhikari T. B., Dean R. A., Louws F. J. (2020). Opportunities and challenges in studies of host-pathogen interactions and management of Verticillium dahliae in tomatoes. Plants 9:1622. 10.3390/plants9111622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barakat A., Bagniewska-Zadworna A., Frost C. J., Carlson J. E. (2010). Phylogeny and expression profiling of CAD and CAD-like genes in hybrid Populus (P. deltoides × P. nigra): evidence from herbivore damage for subfunctionalization and functional divergence. BMC Plant Biol. 10:100. 10.1186/1471-2229-10-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R., Jones J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69 473–488. 10.1007/s11103-008-9435-0 [DOI] [PubMed] [Google Scholar]
- Bhuiyan N. H., Selvaraj G., Wei Y., King J. (2009). Gene expression profiling and silencing reveal that monolignol biosynthesis plays a critical role in penetration defence in wheat against powdery mildew invasion. J. Exp. Bot. 60 509–521. 10.1093/jxb/ern290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudet A. M., Lapierre C., Grima-Pettenati J. (1995). Biochemistry and molecular biology of lignification. New Phytol. 129 203–236. 10.1111/j.1469-8137.1995.tb04292.x [DOI] [PubMed] [Google Scholar]
- Campbell M. M., Sederoff R. R. (1996). Variation in lignin content and composition (mechanisms of control and implications for the genetic improvement of plants). Plant Physiol. 110 3–13. 10.1104/pp.110.1.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarino I. (2019). Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr. Opin. Biotechnol. 56 209–214. 10.1016/j.copbio.2018.12.012 [DOI] [PubMed] [Google Scholar]
- Chezem W. R., Memon A., Li F. S., Weng J. K., Clay N. K. (2017). SG2-Type R2R3-MYB transcription factor MYB15 controls defense-induced lignification and basal immunity in Arabidopsis. Plant Cell 29 1907–1926. 10.1105/tpc.16.00954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianchetta A. N., Davis R. M. (2015). Fusarium wilt of cotton: management strategies. Crop Protect. 73 40–44. 10.1016/j.cropro.2015.01.014 [DOI] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Cui Y., Bell A. A., Joost O., Magill C. (2000). Expression of potential defense response genes in cotton. Physiol. Mol. Plant Pathol. 56 25–31. 10.1006/pmpp.1999.0245 [DOI] [Google Scholar]
- Dixon R. A., Achnine L., Kota P., Liu C. J., Reddy M. S., Wang L. (2002). The phenylpropanoid pathway and plant defence-a genomics perspective. Mol. Plant Pathol. 3 371–390. 10.1046/j.1364-3703.2002.00131.x [DOI] [PubMed] [Google Scholar]
- Dong N. Q., Lin H. X. (2021). Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Integr. Plant Biol. 63 180–209. 10.1111/jipb.13054 [DOI] [PubMed] [Google Scholar]
- Dowd C., Wilson L. W., Mcfadden H. (2004). Gene expression profile changes in cotton root and hypocotyl tissues in response to infection with Fusarium oxysporum f. sp vasinfectum. Mol. Plant Microbe Interact. 17 654–667. 10.1094/MPMI.2004.17.6.654 [DOI] [PubMed] [Google Scholar]
- Eynck C., Séguin-Swartz G., Clarke W. E., Parkin I. A. (2012). Monolignol biosynthesis is associated with resistance to Sclerotinia sclerotiorum in Camelina sativa. Mol. Plant Pathol. 13 887–899. 10.1111/j.1364-3703.2012.00798.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayoso C., Pomar F., Novo-Uzal E., Merino F., De Ilárduya O. M. (2010). The Ve-mediated resistance response of the tomato to Verticillium dahliae involves H2O2, peroxidase and lignins and drives PAL gene expression. BMC Plant Biol. 10:232. 10.1186/1471-2229-10-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q., Min L., Yang X., Jin S., Zhang L., Li Y., et al. (2018). Laccase GhLac1 modulates broad-spectrum biotic stress tolerance via manipulating phenylpropanoid pathway and Jasmonic acid synthesis. Plant Physiol. 176 1808–1823. 10.1104/pp.17.01628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q., Xiao S., Wang X., Ao C., Zhang X., Zhu L. (2021). GhWRKY1-like enhances cotton resistance to Verticillium dahliae via an increase in defense-induced lignification and S monolignol content. Plant Sci. 305:110833. [DOI] [PubMed] [Google Scholar]
- Jin Y., Zhang C., Liu W., Qi H., Chen H., Cao S. (2014). The cinnamyl alcohol dehydrogenase gene family in melon (Cucumis melo L.): bioinformatic analysis and expression patterns. PLoS One 9:e101730. 10.1371/journal.pone.0101730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S. Y., Walker A. M., Kim H., Ralph J., Vermerris W., Sattler S. E., et al. (2017). The enzyme activity and substrate specificity of two major cinnamyl alcohol dehydrogenases in Sorghum (Sorghum bicolor), SbCAD2 and SbCAD4. Plant Physiol. 174 2128–2145. 10.1104/pp.17.00576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. J., Kim K. W., Cho M. H., Franceschi V. R., Davin L. B., Lewis N. G. (2007). Expression of cinnamyl alcohol dehydrogenases and their putative homologues during Arabidopsis thaliana growth and development: lessons for database annotations? Phytochemistry 68 1957–1974. 10.1016/j.phytochem.2007.02.032 [DOI] [PubMed] [Google Scholar]
- Kim S. J., Kim M. R., Bedgar D. L., Moinuddin S. G., Cardenas C. L., Davin L. B., et al. (2004). Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101 1455–1460. 10.1073/pnas.0307987100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosterman S. J., Atallah Z. K., Vallad G. E., Subbarao K. V. (2009). Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 47 39–62. 10.1146/annurev-phyto-080508-081748 [DOI] [PubMed] [Google Scholar]
- Koeduka T., Sugimoto K., Watanabe B., Someya N., Kawanishi D., Gotoh T., et al. (2014). Bioactivity of natural O-prenylated phenylpropenes from Illicium anisatum leaves and their derivatives against spider mites and fungal pathogens. Plant Biol. 16 451–456. 10.1111/plb.12054 [DOI] [PubMed] [Google Scholar]
- Li C., He Q., Zhang F., Yu J., Zhao T., Zhang Y., et al. (2019). Melatonin enhances cotton immunity to Verticillium wilt via manipulating lignin and gossypol biosynthesis. Plant J. 100 784–800. 10.1111/tpj.14477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Wang K., Chern M., Liu Y., Zhu Z., Liu J., et al. (2020). Sclerenchyma cell thickening through enhanced lignification induced by OsMYB30 prevents fungal penetration of rice leaves. New Phytol. 226 1850–1863. 10.1111/nph.16505 [DOI] [PubMed] [Google Scholar]
- Li X., Ma D., Chen J., Pu G., Ji Y., Lei C., et al. (2012). Biochemical characterization and identification of a cinnamyl alcohol dehydrogenase from Artemisia annua. Plant Sci. 194 85–95. 10.1016/j.plantsci.2012.05.011 [DOI] [PubMed] [Google Scholar]
- Liu W., Jiang Y., Wang C., Zhao L., Jin Y., Xing Q., et al. (2020). Lignin synthesized by CmCAD2 and CmCAD3 in oriental melon (Cucumis melo L.) seedlings contributes to drought tolerance. Plant Mol. Biol. 103 689–704. 10.1007/s11103-020-01018-7 [DOI] [PubMed] [Google Scholar]
- Loake G., Grant M. (2007). Salicylic acid in plant defence–the players and protagonists. Curr. Opin. Plant Biol. 10 466–472. 10.1016/j.pbi.2007.08.008 [DOI] [PubMed] [Google Scholar]
- Ma D., Xu C., Alejos-Gonzalez F., Wang H., Yang J., Judd R., et al. (2018). Overexpression of Artemisia annua cinnamyl alcohol dehydrogenase increases lignin and coumarin and reduces Artemisinin and other sesquiterpenes. Front. Plant Sci. 9:828. 10.3389/fpls.2018.00828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden H. G., Chapple R., de Feyter R., Dennis E. (2001). Expression of pathogenesis-related genes in cotton stems in response to infection by Verticillium dahliae. Physiol. Mol. Plant Pathol. 58 119–131. 10.1006/pmpp.2001.0320 [DOI] [Google Scholar]
- Miao Y., Xu L., He X., Zhang L., Shaban M., Zhang X., et al. (2019). Suppression of tryptophan synthase activates cotton immunity by triggering cell death via promoting SA synthesis. Plant J. 98 329–345. 10.1111/tpj.14222 [DOI] [PubMed] [Google Scholar]
- Mottiar Y., Vanholme R., Boerjan W., Ralph J., Mansfield S. D. (2016). Designer lignins: harnessing the plasticity of lignification. Curr. Opin. Biotechnol. 37 190–200. 10.1016/j.copbio.2015.10.009 [DOI] [PubMed] [Google Scholar]
- Pan H., Zhou R., Louie G. V., Mühlemann J. K., Bomati E. K., Bowman M. E., et al. (2014). Structural studies of cinnamoyl-CoA reductase and cinnamyl-alcohol dehydrogenase, key enzymes of monolignol biosynthesis. Plant Cell 26 3709–3727. 10.1105/tpc.114.127399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomar F., Novo M., Bernal M. A., Merino F., Barceló A. R. (2004). Changes in stem lignins (monomer composition and crosslinking) and peroxidase are related with the maintenance of leaf photosynthetic integrity during Verticillium wilt in Capsicum annuum. New Phytol. 163 111–123. 10.1111/j.1469-8137.2004.01092.x [DOI] [PubMed] [Google Scholar]
- Potter S. C., Luciani A., Eddy S. R., Park Y., Lopez R., Finn R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204. 10.1093/nar/gky448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A. R., Mansfield S. D. (2009). Rapid analysis of poplar lignin monomer composition by a streamlined thioacidolysis procedure and near-infrared reflectance-based prediction modeling. Plant J. 58 706–714. 10.1111/j.1365-313X.2009.03808.x [DOI] [PubMed] [Google Scholar]
- Rolando C., Monties B., Lapierre C. (1992). “Thioacidolysis,” in Methods in Lignin Chemistry, eds Lin S. Y., Dence C. W. (Berlin: Springer Berlin Heidelberg; ), 334–349. 10.1007/978-3-642-74065-7_23 [DOI] [Google Scholar]
- Rong W., Luo M., Shan T., Wei X., Du L., Xu H., et al. (2016). A wheat cinnamyl alcohol dehydrogenase TaCAD12 contributes to host resistance to the sharp eyespot disease. Front. Plant Sci. 7:1723. 10.3389/fpls.2016.01723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanogo S., Zhang J. (2016). Resistance sources, resistance screening techniques and disease management for Fusarium wilt in cotton. Euphytica 207 255–271. 10.1007/s10681-015-1532-y [DOI] [Google Scholar]
- Shaban M., Miao Y., Ullah A., Khan A. Q., Menghwar H., Khan A. H., et al. (2018). Physiological and molecular mechanism of defense in cotton against Verticillium dahliae. Plant Physiol. Biochem. 125, 193–204. 10.1016/j.plaphy.2018.02.011 [DOI] [PubMed] [Google Scholar]
- Shi R., Sun Y. H., Li Q., Heber S., Sederoff R., Chiang V. L. (2010). Towards a systems approach for lignin biosynthesis in Populus trichocarpa: transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol. 51 144–163. 10.1093/pcp/pcp175 [DOI] [PubMed] [Google Scholar]
- Sibout R., Eudes A., Mouille G., Pollet B., Lapierre C., Jouanin L., et al. (2005). CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17 2059–2076. 10.1105/tpc.105.030767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sink K. C., Grey W. E. (1999). A root-injection method to assess Verticillium wilt resistance of peppermint (Mentha × piperita L.) and its use in identifying resistant somaclones of cv. Black Mitcham. Euphytica 106 223–230. 10.1023/A:1003591908308 [DOI] [Google Scholar]
- Smit F., Dubery I. A. (1997). Cell wall reinforcement in cotton hypocotyls in response to a Verticillium dahliae elicitor. Phytochemistry 44 811–815. 10.1016/S0031-9422(96)00595-X [DOI] [Google Scholar]
- Song R., Li J., Xie C., Jian W., Yang X. (2020). An overview of the molecular genetics of plant resistance to the Verticillium wilt pathogen Verticillium dahliae. Int. J. Mol. Sci. 21:1120. 10.3390/ijms21031120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X. P., Liang W. Q., Liu C. J., Luo P., Heinstein P., Chen X. Y. (2000). Expression pattern of (+)-delta-cadinene synthase genes and biosynthesis of sesquiterpene aldehydes in plants of Gossypium arboreum L. Planta 210 644–651. 10.1007/s004250050055 [DOI] [PubMed] [Google Scholar]
- Tang Y., Zhang Z., Lei Y., Hu G., Liu J., Hao M., et al. (2019). Cotton WATs modulate SA biosynthesis and local lignin deposition participating in plant resistance against Verticillium dahliae. Front. Plant Sci. 10:526. 10.3389/fpls.2019.00526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T., De Souza L. P., Fernie A. R. (2017). Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 68 4013–4028. 10.1093/jxb/erx177 [DOI] [PubMed] [Google Scholar]
- Tronchet M., Balagué C., Kroj T., Jouanin L., Roby D. (2010). Cinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol. Plant Pathol. 11 83–92. 10.1111/j.1364-3703.2009.00578.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Zhu X., Wang K., Lu C., Luo M., Shan T., et al. (2018). A wheat caffeic acid 3-O-methyltransferase TaCOMT-3D positively contributes to both resistance to sharp eyespot disease and stem mechanical strength. Sci. Rep. 8:6543. 10.1038/s41598-018-24884-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Mao Y., Guo Y., Gao J., Liu X., Li S., et al. (2020). MYB transcription factor161 mediates feedback regulation of secondary wall-associated NAC-domain1 family genes for wood formation. Plant Physiol. 184 1389–1406. 10.1104/pp.20.01033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J. F. (1989). New World tetraploid cottons contain Old World cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 86 4132–4136. 10.1073/pnas.86.11.4132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Hu Q., Shen J., Liu S., Yang Z., Chen K., et al. (2021). GhMYB4 downregulates lignin biosynthesis and enhances cotton resistance to Verticillium dahliae. Plant Cell Rep. 40 735–751. 10.1007/s00299-021-02672-x [DOI] [PubMed] [Google Scholar]
- Xie M., Zhang J., Tschaplinski T. J., Tuskan G. A., Chen J. G., Muchero W. (2018). Regulation of lignin biosynthesis and its role in growth-defense tradeoffs. Front. Plant Sci. 9:1427. 10.3389/fpls.2018.01427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X. P., Sun S. C., Zhu Q. H., Zhang X. Y., Li Y. J., Liu F., et al. (2021). The cotton lignin biosynthetic gene Gh4CL30 regulates lignification and phenolic content and contributes to Verticillium wilt resistance. Mol. Plant Microbe Interact. 34 240–254. [DOI] [PubMed] [Google Scholar]
- Xu L., Zhu L., Tu L., Liu L., Yuan D., Jin L., et al. (2011). Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J. Exp. Bot. 62 5607–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Jia T., Zhang Z., Zou X., Fan S., Lei K., et al. (2020). Insight into the relationship between S-lignin and fiber quality based on multiple research methods. Plant Physiol. Biochem. 147 251–261. [DOI] [PubMed] [Google Scholar]
- Zhang T., Hu Y., Jiang W., Fang L., Guan X., Chen J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33 531–537. [DOI] [PubMed] [Google Scholar]
- Zhang X. J., Yuan Y. C., Wei Z., Guo X., Guo Y. P., Zhang S. Q., et al. (2014). Molecular mapping and validation of a major QTL conferring resistance to a defoliating isolate of Verticillium wilt in cotton (Gossypium hirsutum L.). PLoS One 9:e96226. 10.1371/journal.pone.0096226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Y., Cheng W. H., Feng Z. D., Zhu Q. H., Sun Y. Q., Li Y. J., et al. (2020). Transcriptomic analysis of gene expression of Verticillium dahliae upon treatment of the cotton root exudates. BMC Genomics 21:155. 10.1186/s12864-020-6448-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wu L., Wang X., Chen B., Zhao J., Cui J., et al. (2019). The cotton laccase gene GhLAC15 enhances Verticillium wilt resistance via an increase in defence-induced lignification and lignin components in the cell walls of plants. Mol. Plant Pathol. 20 309–322. 10.1111/mpp.12755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Koopmann B., Von Tiedemann A. (2019). Role of salicylic acid and components of the phenylpropanoid pathway in basal and cultivar-related resistance of oilseed rape (Brassica napus) to Verticillium longisporum. Plants 8:491. 10.3390/plants8110491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Lee C., Zhou J., Mccarthy R. L., Ye Z. H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20 2763–2782. 10.1105/tpc.108.061325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Lee C., Zhong R., Ye Z. H. (2009). MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21 248–266. 10.1105/tpc.108.063321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Gao E., Shaban M., Wang Y., Wang H., Nie X., et al. (2018). GhUMC1, a blue copper-binding protein, regulates lignin synthesis and cotton immune response. Biochem. Biophys. Res. Commun. 504 75–81. 10.1016/j.bbrc.2018.08.128 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.