Abstract
A 15-mer peptide fragment derived from pediocin PA-1 (from residue 20 to residue 34) specifically inhibited the bactericidal activity of pediocin PA-1. The fragment did not inhibit the pediocin-like bacteriocins sakacin P, leucocin A, and curvacin A to nearly the same extent as it inhibited pediocin PA-1. Enterocin A, however, was also significantly inhibited by this fragment, although not as greatly as pediocin PA-1. This is consistent with the fact that enterocin A contains the longest continuous sequence identical to that of pediocin PA-1 in the region spanned by the fragment. The fragment inhibited pediocin PA-1 to a much greater extent than did the other 29 possible 15-mer fragments that span pediocin PA-1. The results suggest that the fragment—by interacting with the target cells and/or pediocin PA-1—interferes specifically with pediocin-target cell interaction.
Bacteria produce ribosomally synthesized antimicrobial polypeptides, termed bacteriocins. Bacteriocins produced by gram-positive bacteria are often membrane-permeabilizing cationic peptides with less than 50 amino acid residues (1, 18, 21, 23, 25, 31). These peptide bacteriocins may roughly be classified into two main groups. Group I consists of bacteriocins, often termed lantibiotics, that contain lanthionine and or lanthionine-related residues, whereas group II consists of bacteriocins that lack modified residues. The pediocin-like bacteriocins constitute a large subgroup within group II (23): they are all unmodified, they have similar primary structures, and they exert their bactericidal activity by permeabilizing the target cell membrane (6, 7).
The first pediocin-like bacteriocins to be characterized were pediocin PA-1 (14, 19, 22), sakacin P (27, 29), leucocin A (11), curvacin A (2, 15, 27, 28), and mesentericin Y105 (13), all produced by lactic acid bacteria. More recently identified pediocin-like bacteriocins are carnobacteriocin BM1 and B2 (26), enterocin A (3) and P (8), bavaricin MN (17), piscicolin 126 (16), piscicocin V1a (4), and bacteriocin 31 (30). All of these bacteriocins exhibit 40 to 60% sequence similarity. The similarity is especially pronounced in the hydrophilic N-terminal half of the peptides. In contrast to the N-terminal half, the C-terminal half is hydrophobic and/or amphiphilic (9, 10). Thus, it is the C-terminal half of the pediocin-like bacteriocins which may interact with the hydrophobic part of the target cell membrane, thereby causing membrane leakage. The recent three-dimensional nuclear magnetic resonance structural analysis of the pediocin-like bacteriocin leucocin A shows that upon exposure to dodecylphosphocholine micelles, the hydrophilic N-terminal half forms a three-stranded antiparallel β-sheet and the C-terminal half forms an amphiphilic α-helix (10).
Despite similar primary structures, the pediocin-like bacteriocins differ in their target cell specificity (i.e., they differ in their antimicrobial spectra) (9). This difference in target cell specificity, combined with the extensive similarity in amino acid sequence, makes the pediocin-like bacteriocins well suited for analyzing the relationship between target cell specificity and primary structure. Such an analysis may eventually enable the identification of peptide-cell interactions that are general and of prime importance for determining whether or not a cell is sensitive to an antimicrobial peptide.
By determining the target cell specificity of hybrid bacteriocins containing N- and C-terminal regions from different pediocin-like bacteriocins, it has been shown that the C-terminal half of these bacteriocins is an important determinant of target cell specificity (9). Thus, the C-terminal half must interact in a specific manner with an entity on the target cell membrane, an entity which might perhaps also be recognized by peptide fragments derived from the C-terminal half. In this study, we have identified a 15-mer peptide fragment derived from the C-terminal half of the pediocin-like bacteriocin, pediocin PA-1, which inhibits the bactericidal activity of pediocin PA-1, but not the activity of other closely related pediocin-like bacteriocins. The results indicate that this fragment spans a region in the pediocin-like bacteriocins which is important for target cell specificity. Thus, a target cell specificity-determining region has been more closely localized within the C-terminal half of these bacteriocins.
Peptide fragments, bacteriocins, and bacteriocin assay.
Thirty peptide fragments derived from the sequence of pediocin PA-1 (Fig. 1) were synthesized by standard methods of solid-phase multiple peptide synthesis with an F-moc strategy. The peptides were 15 amino acids in length, with an overlap of 14 residues. Thus, the first peptide (fragment 1) spans amino acid residues 1 to 15 of pediocin PA-1, the second (fragment 2) spans residues 2 to 16, and so on to fragment 30, which starts with residue 30 and ends with residue 44—the last residue in pediocin PA-1. All fragments were synthesized as acetylated peptide-amides to avoid the effects of terminal functional groups, and an acetylating capping step was employed after each synthesis cycle in order to reduce the chances of “failure sequences” containing internal deletions. Cysteine residues in pediocin PA-1 were substituted for with α-aminobutyric acid in order to avoid the formation of disulfide linkages between fragments (and between fragments and bacteriocins) that contain cysteine residues. All peptides were characterized by electrospray ionization mass spectrometry (Perkin-Elmer Sciez API III) and high-performance liquid chromatography; the purities of the final products were >70%. The synthesized peptides were solubilized to a concentration of 1 to 10 mg/ml in 0.1% (vol/vol) trifluoroacetic acid and 10 to 30% (vol/vol) 2-propanol.
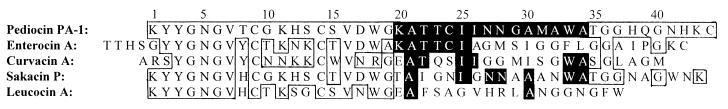
FIG. 1.
Amino acid sequences of pediocin PA-1 (19), enterocin A (3), curvacin A (2, 28), sakacin P (29), and leucocin A (11). The regions where the sequences are identical to the sequence in pediocin PA-1 are boxed. Regions where the sequences are identical to the sequence of fragment 20 are in black.
Pediocin PA-1, sakacin P, curvacin A, and enterocin A were purified to homogeneity from 0.5-liter cultures of the producer strain by ammonium sulfate precipitation and cation exchange, hydrophobic interaction, and reverse-phase chromatography, essentially as previously described (3, 22). Enterocin A was produced by Enterococcus faecium CTC492 (3), whereas the producing strains for the other bacteriocins were as described in references 22 and 27. Leucocin A was synthesized and purified to 80 to 90% purity as described in reference 9.
Bacteriocin activity was measured with a microtiter plate assay system (24). A 200-μl volume of culture medium, bacteriocin fractions at twofold dilutions, various amounts of peptide fragments derived from pediocin PA-1, and the indicator strain (Lactobacillus sake NCDO 2714 [type strain] at an optical density at 610 nm of 0.01) were added to each well of a microtiter plate. The microtiter plate cultures were incubated for 10 to 15 h at 30°C, after which growth of the indicator strain was measured spectrophotometrically at 610 nm with a microtiterplate reader. When this assay system is standardized with respect to the amount of indicator cells used and the incubation time and temperature, determination of bacteriocin activity is reproducible to within a twofold dilution. The MIC was defined as the bacteriocin concentration that inhibited the growth of the indicator strain by 50% (50% of the turbidity of the control culture without bacteriocin). The indicator strain and the bacteriocin-producing strains were all grown at 30°C in MRS broth (Oxoid).
A fragment that spans pediocin PA-1 from residue 20 to residue 34 specifically inhibits bacteriocin activity.
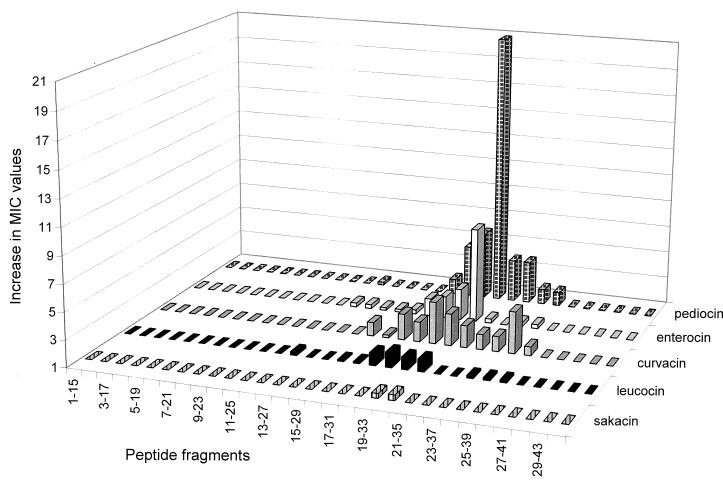
The peptide fragments derived from pediocin PA-1 were all assayed for bacteriocin activity and for their ability to inhibit the activity of pediocin-like bacteriocins. None of the fragments showed any bacteriocin activity when tested at concentrations of 100 μM (data not shown). However, when tested for their ability to inhibit bacteriocin activity, the fragment which spans residues 20 to 34 of pediocin PA-1 (fragment 20) clearly inhibited the bacteriocin activity of pediocin PA-1 (Fig. 2). Adjacent fragments (fragments 18, 19, 21, and 22) also inhibited the bacteriocin activity of pediocin PA-1, but to a much lesser extent than fragment 20. The other fragments did not significantly inhibit the bacteriocin activity (Fig. 2).
FIG. 2.
Inhibition of the bactericidal activities of pediocin-like bacteriocins due to the presence of 5 μM peptide fragments derived from pediocin PA-1. Inhibition of the bactericidal activities is quantitated as the increase in MIC due to the presence of a fragment. The increase in MIC is defined as the MIC obtained in the presence of a fragment divided by the MIC obtained in the absence of a fragment. The MICs were between 0.5 and 0.05 nM for all of the bacteriocins in the absence of a fragment, with sakacin P having the highest MIC, enterocin A the lowest, and curvacin A, pediocin PA-1, and leucocin A having MICs between those of sakacin P and enterocin A.
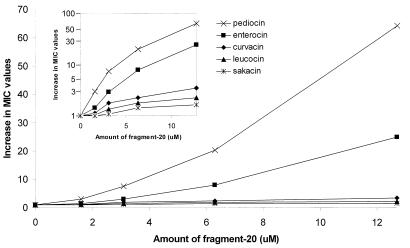
Fragment 20, and its adjacent fragments, inhibited the bacteriocin activity of pediocin PA-1 in a specific manner, in the sense that these fragments inhibited sakacin P, leucocin A, and curvacin A to a much lesser extent than pediocin PA-1 (Fig. 2 and 3). Enterocin A, however, was also significantly inhibited by fragment 20, although not as greatly as pediocin PA-1 (Fig. 3). Interestingly, in the region spanned by fragment 20, enterocin A contains the longest continuous sequence that is identical to that of pediocin PA-1 (Fig. 1). Moreover, there is a cysteine residue in this region in both pediocin PA-1 and enterocin A, but not in the other three bacteriocins, and this enables the formation of a C-terminally-located disulfide bond unique to pediocin PA-1 and enterocin A (Fig. 1). The fact that fragment 20 inhibited pediocin PA-1 to a much greater extent than other bacteriocins suggests that inhibition by fragment 20 is not merely due to nonspecific hydrophobic interactions between fragment 20 and pediocin-like bacteriocins in general. It rather suggests that fragment 20 (by interacting with the target cells and or pediocin PA-1) somehow interferes specifically with pediocin-target cell interaction.
FIG. 3.
Inhibition (measured as increase in MIC [see the legend to Fig. 2]) of the bactericidal activities of pediocin-like bacteriocins as a function of increasing amounts of fragment 20. The y axis in the inset has a logarithmic scale, whereas the y axis in the main figure is linear.
The primary and three-dimensional structures of these bacteriocins suggest that their polypeptide chains may be divided into two functional domains: the relatively well-conserved hydrophilic N-terminal β-sheet domain and the somewhat more diverse hydrophobic or amphiphilic C-terminal α-helical domain (9, 10). The well-conserved N-terminal β-sheet domain must have an important function common to the pediocin-like bacteriocin, perhaps to mediate the initial unspecific binding of the bacteriocins to target cells through electrostatic interactions (5). The C-terminal domain, due to its hydrophobic or amphiphilic character, must be the domain which interacts with the hydrophobic part of the membrane. This is consistent with the observation that chimeric pediocin PA-1, which has maltose-binding protein fused to its N terminus, displayed bactericidal activity, suggesting that the N-terminal part of the bacteriocin does not enter the target cell membrane (20). Earlier studies using hybrid bacteriocins indicate that the C-terminal domain is an important determinant of target cell specificity (9). The present results suggest that residues in pediocin that are important for determining target cell specificity are present in the region spanned by fragment 20.
A specificity-determining region must interact in a specific manner with an entity on the target cell membrane. This entity might simply be a cell surface binding site, with increased binding to the cell surface resulting in higher concentrations of bacteriocins in the vicinity of the membrane, which in turn leads to increased membrane permeabilization and cytotoxicity. Alternatively, the entity might be a membrane component which interacts with residues in the specificity-determining region and thereby makes the membrane more susceptible to permeabilization. The interaction may be nonchiral, for instance, between membrane lipids and side chains of bacteriocin residues, since strict target cell specificity does not per se prove that the antagonistic activity depends on a stereo-specific interaction. Short membrane-permeabilizing amphiphilic α-helical bacteriocin-like peptides display strain-specific antagonistic activities, which clearly do not depend on stereo-specific interactions (12). Work which is now in progress to identify residues that are particularly important for specificity indicates that residues within the region spanned by fragment 20 are of importance for the target cell specificity of pediocin PA-1 and sakacin P (9a).
Acknowledgments
This work was supported by a Norwegian Research Council grant and the Deutsche Forschungsgemeinschaft (DFG), Sonderforschungsbereich (SFB) 323.
We thank Linda Bross for expert technical assistance.
REFERENCES
- 1.Abee T. Pore-forming bacteriocins of gram-positive bacteria and self-protection mechanisms of producer organisms. FEMS Microbiol Lett. 1995;129:1–10. doi: 10.1016/0378-1097(95)00137-T. [DOI] [PubMed] [Google Scholar]
- 2.Axelsson L, Holck A, Birkeland S-E, Aukrust T, Blom H. Cloning and nucleotide sequence of a gene from Lactobacillus sake Lb706 necessary for sakacin A production and immunity. Appl Environ Microbiol. 1993;59:2868–2875. doi: 10.1128/aem.59.9.2868-2875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aymerich T, Holo H, HÅvarstein L S, Hugas M, Garriga M, Nes I F. Biochemical and genetic characterization of enterocin A from Enterococcus faecium, a new antilisterial bacteriocin in the pediocin family of bacteriocins. Appl Environ Microbiol. 1996;62:1676–1682. doi: 10.1128/aem.62.5.1676-1682.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhugaloo-Vial P, Dousset X, Metivier A, Sorokine O, Anglade P, Boyaval P, Marion D. Purification and amino acid sequences of piscicocins V1a and V1b, two class IIa bacteriocins secreted by Carnobacterium piscicola V1 that display significantly different levels of specific inhibitory activity. Appl Environ Microbiol. 1996;62:4410–4416. doi: 10.1128/aem.62.12.4410-4416.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, Ludescher R D, Montville T J. Electrostatic interactions, but not the YGNGV consensus motif, govern the binding of pediocin PA-1 and its fragments to phosphoslipid vesicles. Appl Environ Microbiol. 1997;63:4770–4777. doi: 10.1128/aem.63.12.4770-4777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Y, Shapira R, Eisenstein M, Montville T J. Functional characterization of pediocin PA-1 binding to liposomes in the absence of a protein receptor and its relationship to a predicted tertiary structure. Appl Environ Microbiol. 1997;63:524–531. doi: 10.1128/aem.63.2.524-531.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chikindas M L, García-Garcerá M J, Driessen A J M, Ledeboer A M, Nissen-Meyer J, Nes I F, Abee T, Konings W N, Venema G. Pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0, forms hydrophilc pores in the cytoplasmic membrane of target cells. Appl Environ Microbiol. 1993;59:3577–3584. doi: 10.1128/aem.59.11.3577-3584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cintas L M, Casaus P, Håvarstein L S, Hernández P E, Nes I F. Biochemical and genetic characterization of enterocin P, a novel sec-dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl Environ Microbiol. 1997;63:4321–4330. doi: 10.1128/aem.63.11.4321-4330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fimland G, Blingsmo O R, Sletten K, Jung G, Nes I F, Nissen-Meyer J. News biologically active hybrid bacteriocins constructed by combining regions from various pediocin-like bacteriocins: the C-terminal region is important for determining specificity. Appl Environ Microbiol. 1996;62:3313–3318. doi: 10.1128/aem.62.9.3313-3318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Fimland, G., et al. Unpublished results.
- 10.Fregeau Gallagher N L, Sailer M, Niemczura W P, Nakashima T T, Stiles M E, Vederas J C. Three-dimensional structure of leucocin A in trifluoroethanol and dodecylphosphocholine micelles: spatial location of residues critical for biological activity in type IIa bacteriocins from lactic acid bacteria. Biochemistry. 1997;36:15062–15072. doi: 10.1021/bi971263h. [DOI] [PubMed] [Google Scholar]
- 11.Hastings J W, Sailer M, Johnson K, Roy K L, Vederas J C, Stiles M E. Characterization of leucocin A-UAL 187 and cloning of the bacteriocin gene from Leuconostoc gelidum. J Bacteriol. 1991;173:7491–7500. doi: 10.1128/jb.173.23.7491-7500.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hauge, H. H., D. Mantzilas, G. Moll, W. N. Konings, A. J. M. Driessen, V. G. H. Eijsink, and J. Nissen-Meyer. Plantaricin A is an amphiphilic α-helical bacteriocin-like pheromone which exerts antimicrobial and pheromone activities through different mechanisms. Biochemistry, in press. [DOI] [PubMed]
- 13.Héchard Y, Dérijard B, Letellier F, Cenatiempo Y. Characterization and purification of mesentericin Y105, an anti-Listeria bacteriocin from Leuconostoc mesenteroides. J Gen Microbiol. 1992;138:2725–2731. doi: 10.1099/00221287-138-12-2725. [DOI] [PubMed] [Google Scholar]
- 14.Henderson J R, Chopko A L, van Wassenaar D. Purification and primary structure of pediocin PA-1 produced by Pediococcus acidilactici PAC-1.0. Arch Biochem Biophys. 1992;295:5–12. doi: 10.1016/0003-9861(92)90480-k. [DOI] [PubMed] [Google Scholar]
- 15.Holck A, Axelsson L, Birkeland S-E, Aukrust T, Blom H. Purification and amino acid sequence of sakacin A, a bacteriocin from Lactobacillus sake Lb706. J Gen Microbiol. 1992;138:2715–2720. doi: 10.1099/00221287-138-12-2715. [DOI] [PubMed] [Google Scholar]
- 16.Jack R W, Wan J, Gordon J, Harmark K, Davidson B E, Hillier A J, Wettenhall R E H, Hickey M W, Coventry M J. Characterization of the chemical and antimicrobial properties of piscicolin126, a bacteriocin produced by Carnobacterium piscicola JG126. Appl Environ Microbiol. 1996;62:2897–2903. doi: 10.1128/aem.62.8.2897-2903.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaiser A L, Montville T J. Purification of the bacteriocin bavaricin MN and characterization of its mode of action against Listeria monocytogenes Scott A cells and lipid vesicles. Appl Environ Microbiol. 1996;62:4529–4535. doi: 10.1128/aem.62.12.4529-4535.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klaenhammer T R. Genetics of bacteriocins produced by lactic acid bacterial FEMS Microbiol. Rev. 1993;12:39–86. doi: 10.1111/j.1574-6976.1993.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 19.Marugg J D, Gonzalez C F, Kunka B S, Ledeboer A M, Pucci M J, Toonen M Y, Walker S A, Zoetmulder L C M, Vandenbergh P A. Cloning, expression, and nucleotide sequence of genes involved in production of pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0. Appl Environ Microbiol. 1992;58:2360–2367. doi: 10.1128/aem.58.8.2360-2367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller K W, Schamber R, Chen Y, Ray B. Production of active chimeric pediocin AcH in Escherichia coli in the absence of processing and secretion genes from the Pediococcus pap operon. Appl Environ Microbiol. 1998;64:14–20. doi: 10.1128/aem.64.1.14-20.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nes I F, Diep D B, Håverstein L S, Brurberg M B, Eijsink V, Holo H. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:113–128. doi: 10.1007/BF00395929. [DOI] [PubMed] [Google Scholar]
- 22.Nieto Lozano J C, Nissen-Meyer J, Slettten K, Peláz C, Nes I F. Purification and amino acid sequence of a bacteriocin produced by Pediococcus acidilactici. J Gen Microbiol . 1992;138:1985–1990. doi: 10.1099/00221287-138-9-1985. [DOI] [PubMed] [Google Scholar]
- 23.Nissen-Meyer J, Hauge H H, Fimland G, Eijsink V G H, Nes I F. Ribosomally synthesized antimicrobial peptides produced by lactic acid bacteria: their function, structure, biogenesis, and their mechanism of action. Recent Res Dev Microbiol. 1998;1:141–154. [Google Scholar]
- 24.Nissen-Meyer J, Holo H, Håvarstein L S, Sletten K, Nes I F. A novel lactococcal bacteriocin whose activity depends on the complementary action of two peptides. J Bacteriol. 1992;174:5686–5692. doi: 10.1128/jb.174.17.5686-5692.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nissen-Meyer J, Nes I F. Ribosomally synthesized antimicrobial peptides: their function, structure, biogenesis, and mechanism of action. Arch Microbiol. 1997;167:67–77. [PubMed] [Google Scholar]
- 26.Quadri L E N, Sailer M, Roy K L, Vederas J C, Stiles M E. Chemical and genetic characterization of bacteriocins produced by Carnobacterium piscicola LV17B. J Biol Chem. 1994;269:12204–12211. [PubMed] [Google Scholar]
- 27.Tichaczek P S, Nissen-Meyer J, Nes I F, Vogel R F, Hammes W P. Characterization of the bacteriocins curvacin A from Lactobacillus curvatus LTH1174 and sakacin P from L. sake LTH673. Syst Appl Microbiol. 1992;15:460–468. [Google Scholar]
- 28.Tichaczek P S, Vogel R F, Hammes W P. Cloning and sequencing curA encoding curvacin A, the bacteriocin produced by Lactobacillus curvatus LTH1174. Arch Microbiol. 1993;160:279–283. doi: 10.1007/BF00292077. [DOI] [PubMed] [Google Scholar]
- 29.Tichaczek P S, Vogel R F, Hammes W P. Cloning and sequencing of sakA encoding sakacin P, the bacteriocin produced by Lactobacillus sake LTH673. Microbiology. 1994;140:361–367. doi: 10.1099/13500872-140-2-361. [DOI] [PubMed] [Google Scholar]
- 30.Tomita J, Fujimoto S, Tanimoto K, Ike Y. Cloning and genetic organization of the bacteriocin 31 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pYI17. J Bacteriol. 1996;178:3585–3593. doi: 10.1128/jb.178.12.3585-3593.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venema K, Venema G, Kok J. Lactococcal bacteriocins: mode of action and immunity. Trends Microbiol. 1995;3:299–304. doi: 10.1016/s0966-842x(00)88958-1. [DOI] [PubMed] [Google Scholar]