Abstract
Genetic studies of DNA have been unable to explain a significant portion of the variance of the estimated heritability of blood pressure (BP). Epigenetic mechanisms, particularly DNA methylation, have helped explain additional biological processes linked to BP phenotypes and diseases. Candidate gene methylation studies and genome-wide methylation studies of BP have highlighted impactful cytosine-phosphate-guanine (CpG) markers across different ethnicities. Furthermore, many of these BP-related CpG sites are also linked to metabolism-related phenotypes. Integrating epigenome-wide association study data with other layers of molecular data such as genotype data (from single nucleotide polymorphism arrays or sequencing), other epigenetic data, and/or transcriptome data can provide additional information about the significance and complexity of these relationships. Recent data suggest that epigenetic changes can be consequences rather than causes of BP variation. Finally, these data can give insight into downstream effects of long-standing high BP (due to target organ damage (TOD)). The current review provides a literature overview of epigenetic modifications in BP and TOD. Recent studies strongly support the importance of epigenetic modifications, such as DNA methylation, in BP and TOD for relevant biological insights, reliable biomarkers, and possible future therapeutics.
Keywords: blood pressure, genomics, hypertension, methylation
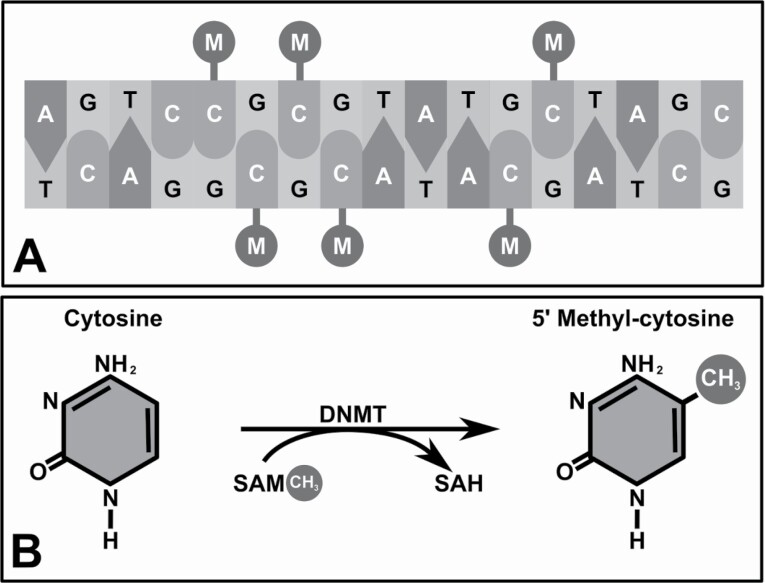
High blood pressure (BP) is a common condition faced by over a billion people worldwide.1 It is a serious risk factor for multiple cardiovascular and renal disorders.2 A 20 mm Hg increase in the systolic BP (SBP) in patients within the 40–49 year age group doubles the risk of mortality from stroke and other vascular diseases.3 Likewise, achieving a 10 mm Hg lower SBP (through lifestyle changes and/or antihypertensive treatment) within the same age group has been associated with a 41% decreased risk of stroke or other vascular disease.4 Effective antihypertensive treatments exist, but unfortunately only about ~50% of those treated for high BP achieve treatment goals.5 High BP has no clear cause but is thought to be linked to genomics, poor diet, lack of exercise, and obesity. While lifestyle factors affecting high BP are well understood, the genomic factors linked to high BP are still under intense study.6 Importantly, it is becoming clear that strictly studying DNA sequence variation does not fully account for the complex mechanisms of regulation of gene expression.7 Epigenetic mechanisms, including DNA methylation, histone modification, and various RNA-mediated processes, are major contributors to gene expression and respond to both genetic (underlying DNA variants) and environmental effects (e.g., smoking, alcohol, stress). The most commonly studied epigenetic modification is the addition of a methyl group to the cytosine of a cytosine-phosphate-guanine (CpG) dinucleotide pair (see Figure 1). DNA methylation is a component of the 1-carbon metabolism pathway and is dependent upon several enzymes (S-adenosyl methionine is converted to S-adenosyl homocysteine by DNA methyltransferases) and dietary micronutrient cofactors, including folate, choline, and betaine. Functionally, hypermethylation of cytosine at CpG sites, particularly in the gene promoter region of genes, most often leads to gene silencing by altering DNA structure and/or by inhibiting transcription factor binding. Thus, epigenetic markers play a crucial role in defining the genetically active or inactive regions of the genome.8,9 As DNA methylation is a stable, covalently bound mark, it is consistently measurable in fresh and archived tissue and biofluid samples with various polymerase chain reaction, array, and sequencing methods.10 High-density regions of CpG dinucleotides are found in the regulatory regions of about 60% of known genes.11 Over the last decade, the availability of DNA methylation (at CpG sites) arrays has led to an explosion of studies characterizing new markers for cardiovascular diseases (CVDs, including high BP).12–15 Notably, not all mammalian cell types show the same degree of methylation, which can result in the expression of diverse phenotypes (which is especially important during development). In comparison to genetic epidemiology studies, epigenetic epidemiology studies are complicated by tissue type and the population distribution of epigenetic differences (especially among age, sex, and ethnic groups16–18). Despite this heterogeneity, statistically significant findings have been generally easier to replicate in genetic epidemiology studies of CVDs (even across ethnic groups).19,20 This may be because many epigenetic variations associated with CVD traits are suspected to be environmentally driven as determined by Mendelian randomization studies (which report methylation variations at CVD trait associated CpG sites are the consequence of the trait rather than the cause).21,22 Additionally, CpGs associated with continuous CVD traits at the cross-sectional level have been associated with incident disease (e.g., methylation sites associated with blood lipids are associated with future coronary heart disease; see Figure 2).23 Given the strong findings in this field, methylation is a recognized target for the continuation of advancements in individualized risk assessments and even therapeutic targets.24 Here, we describe published studies of methylation and BP traits, including a section on target end organ damage (TOD), which have focused on candidate genes as well as hypothesis free (i.e., genome-wide) investigations.
Figure 1.
DNA methylation. (Panel a) Hypermethylation of cytosine (C) at CpG sites often leads to gene silencing by altering DNA structure and/or by inhibiting transcription factor binding. Abbreviations: A, adenine; G, guanine; M, methyl group; T, thymine. (Panel b) S-Adenosyl methionine (SAM) is converted to S-adenosyl homocysteine (SAH) by DNA methyltransferases (DNMT), thereby donating the methyl group (CH3) to the fifth-position carbon of cytosine.
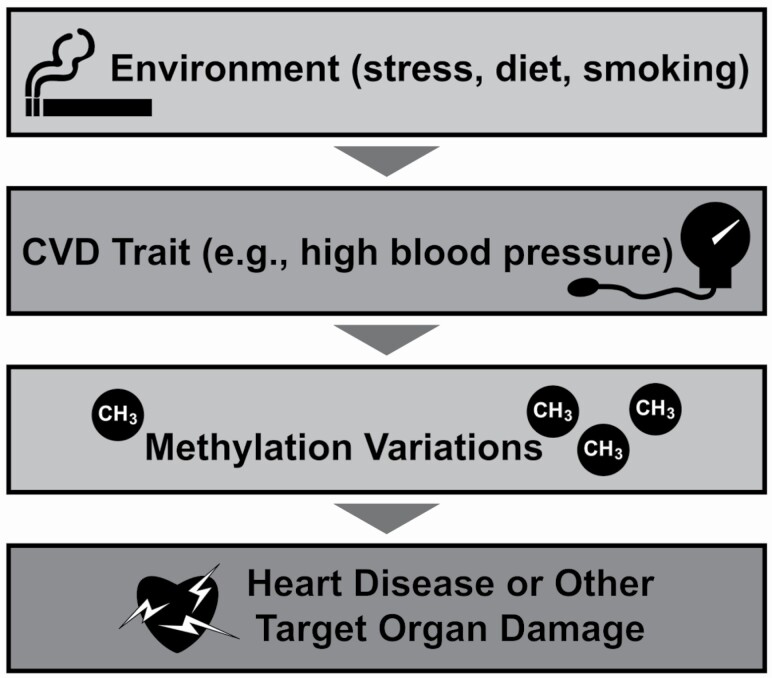
Figure 2.
Epigenetic factors can contribute to cardiovascular diseases (CVDs) in dynamic ways. One possible pathway is that environmental factors contribute to CVD, and DNA methylation changes are a consequence of elevated CVD traits. Those methylation variations could potentially be used as risk markers or therapeutic targets to prevent CVD-related organ damage.
CANDIDATE GENE ASSOCIATION STUDIES
Many candidate gene studies of epigenetic determinants of BP have examined genes associated with the renin–angiotensin–aldosterone system (RAAS). Key RAAS genes associated with methylation alterations in humans or human cell models have included AGT (angiotensinogen), AGTR1 (angiotensin II type 1 receptor), ACE (angiotensin-converting enzyme), NOS3 (for eNOS, endothelial nitric oxide synthase), and SCNN1A and SCNN1B (epithelial sodium channel (ENaC) alpha and beta subunits). In one study, 3 different stimulatory signals (interleukin 6 (IL6), excess circulating aldosterone, and high salt intake) led to changes in DNA methylation of AGT around a transcription factor binding site and a transcription start site (in 3 different tissue-based models).25 Specifically, in cultured human H295R cells, IL6 stimulation caused DNA demethylation around a CCAAT/Enhancer Binding Protein (CEBP) binding site accompanied by increased CEBP-β recruitment and chromatin accessibility of the AGT promoter. In a second model in human visceral adipose tissue, excess circulating aldosterone upregulated AGT expression and was accompanied by DNA hypomethylation around a CEBP binding site. Finally, high salt intake led to upregulation of AGT expression, DNA hypomethylation around the transcription start site, and decreased DNA methylation activity in rat visceral adipose tissue.25 Similarly, studies from rat (lungs and liver) and in vitro models (such as human liver (HepG2), colon (HT29), microvascular endothelial (HMEC-1), and lung (SUT) cell lines) demonstrated that hypermethylation of the ACE gene promoter resulted in transcriptional repression, indicating potential epigenetic regulation of ACE-mediated hypertension.26
Several small case–control studies of essential hypertension (EH, the most common form of high BP) have also shown methylation differences in RAAS-related genes. In a study with ~200 EH cases vs. normotensive controls, Fan et al. found that 2 of 5 CpGs in the promoter of ACE2 (angiotensin-converting enzyme 2, chrx:15579156–15620192) were associated with EH after accounting for confounding (P < 0.05).27 ACE2 plays a key protective role in high BP. In a related study from the same author group, a CpG island in the promoter region of AGTR1 (human GRCh37/hg19 assembly: chr3:148,415,443–148,415,932) was assayed. The methylation level in 1 of 5 successfully measured CpGs was significantly lower in the cases (cases vs. controls: 6.74 ± 4.32% vs. 9.66 ± 5.45%, adjusted P = 0.007).28 Moreover, in a case–control study that compared both incident and prevalent EH cases to non-EH controls, the mean methylation level of SCNN1A (in the gene body chr12: 6473058–6473092) was highest among incident cases compared with the non-EH controls (16.15 ± 4.51 vs. 13.66 ± 4.08, P = 0.041) and prevalent EH cases (16.15 ± 4.51 vs. 13.77 ± 3.90, P = 0.002). Logistic regression analysis showed that higher SCNN1A methylation was a risk factor for EH compared with non-EH (odds ratio = 1.157, P = 0.01), as well as for incident EH compared with prevalent EH (odds ratio = 1.149, P = 0.013). The authors suggested that the differences in methylation of SCNN1A between the prevalent and incident hypertension cases may be due to antihypertensive treatment but suggested more research was needed as information on the class and duration of treatment was not collected in that study.29 In a similar study that included non-EH controls and prevalent and incident EH cases, Zhong et al. observed significant elevated methylation at 2 of 6 CpGs in the promoter region of SCNN1B (chr16: 23313436–23313460)—the beta subunit of ENaC.30 Among those 6 sites, a significant difference in CpG1 and CpG2 methylation levels was detected between controls and incident cases (CpG1: β-standardized = 0.17, adjusted P = 0.015; CpG2: β-standardized = 0.41, adjusted P = 0.001). Additionally, a significant difference was detected in CpG1 methylation levels between incident and prevalent cases (β-standardized = 0.25, adjusted P = 3.77 × 10−4).30
Epigenetic processes have been demonstrated to play important roles in the cell-specific expression of eNOS in the vasculature.31 In one study, chromatin immunoprecipitation demonstrated the presence of transcription factors Sp1, Sp3, and Ets1 at the native NOS3 promoter in endothelial cells but not in vascular smooth muscle cells.32 Finally, robust expression of eNOS mRNA was induced in nonendothelial cell types following inhibition of DNA methyltransferase activity with 5-azacytidine, demonstrating the importance of DNA methylation-mediated repression.32 Overall, these studies suggest methylation at genes involved in the RAAS may contribute to the genomic determinants of BP and EH risk. However, these results also highlight the complexity of these relationships at the molecular level, especially the potential for variability across tissues and duration of disease. While the RAAS has the major role in BP regulation, studies are still discovering new pathways that influence BP. Genome-wide analyses of CpGs are advancing knowledge and continue to highlight other mechanisms involved in BP regulation and relevant disease processes.
EPIGENOME-WIDE ASSOCIATION STUDIES
Large, epigenome-wide studies have revealed several genes that contribute to the variation in both SBP and diastolic BP (DBP). For example, in genome-wide DNA methylation meta-analysis from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) for SBP and DBP—including individuals of European (EA), African (AA), and Hispanic ancestries—(N ~ 17,000) DNA methylation was measured in peripheral blood samples using the Illumina Methyl450 array in all cohorts (targeting across gene regions with sites in the promoter region, 5′ untranslated region (UTR), first exon, gene body, and 3′UTR).12 In each cohort, race-stratified linear mixed effect models were used to estimate CpG associations adjusting for age, sex, estimated blood cell counts, body mass index, smoking, and genetic ancestry, as well as fixed and/or random effects for technical covariates to control for batch effects. Effect estimates from all cohorts were combined using inverse variance fixed effects meta-analysis. Heterogeneity of effect estimates between ethnic strata, sexes, and methylation tissue source among discovery cohorts was evaluated (using a 1 degree of freedom chi-square test for effect differences between strata) and no heterogeneous effects were observed. In the 2-stage discovery (N ~ 10,000) and replication (N ~ 7,000) meta-analyses, 13 of 31 identified CpGs replicated for association with SBP or DBP, and these CpGs were located in 8 intragenic regions and 3 intergenic regions. Further, an overall meta-analysis of the discovery and replication cohorts identified 126 CpG sites associated with BP. A methylation profile score based on the replicated CpG sites explained an additional 1.4% and 2.0% of the interindividual variation in SBP and DBP, respectively, beyond age, sex, and BMI among additional samples (N = 1,516, Third Generation Cohort) not included in the discovery or replication set. Expanding the DNA methylation risk score to include the 126 CpGs that were significant in the overall meta-analysis did not explain additional phenotypic variance in any ancestry group. The top 2 CpGs for both SBP and DBP were at the PHGDH (phosphoglycerate dehydrogenase) locus, cg14476101 (SBP: 0.03% decrease in DNA methylation per 1 mm Hg increase in BP, P = 2.7 × 10−34; DBP: 0.04% decrease in DNA methylation per 1 mm Hg increase in BP, P = 2.1 × 10−21), and the SLC7A11 (solute carrier family 7 member 11) locus, cg06690548 (SBP: 0.02% decrease in DNA methylation per 1 mm Hg increase in BP, P = 1.6 × 10−32; DBP: 0.03% decrease in DNA methylation per 1 mm Hg increase in BP, P = 7.9 × 10−26). Cg14476101 is located on chromosome 1 in the first intron of PHGDH, which encodes a phosphoglycerate dehydrogenase that catalyzes the rate-limiting step of serine biosynthesis.33 A contribution of PHGDH to cancer cell metabolism has been well studied, but more recently a role for PHGDH in lipid homeostasis in normal tissues has been identified.33 Located on chromosome 4, cg06690548 is in the first intron of SLC7A11, which encodes a sodium-independent cysteine/glutamate antiporter that aids in protection from oxidative stress and ferroptotic cell death.34 In whole-blood gene expression analyses, 4 of the 13 replicated CpG sites were found to have 1 or more cis-located genes (TSPAN2 (tetraspanin 2), SLC7A11, UNC93B1 (unc-93 homolog B1, TLR signaling regulator), CPT1A (carnitine palmitoyltransferase 1A), PTMS (parathymosin), and LPCAT3 (lysophosphatidylcholine acyltransferase 3)) where transcription levels were associated with both CpG methylation and SBP, DBP, or hypertension. The direction of effects for all 6 gene transcripts was consistent with the negative associations of BP with DNA methylation at each CpG. Among all transcripts tested, expression of TSPAN2 showed the strongest associations with both CpG methylation (cg23999170) and BP. TSPAN2 is involved in signal transduction, and it is not only highly expressed in vascular tissues but also implicated in the contractile ability and differentiation of vascular smooth muscle cells.35 Sequence variation mapped to TSPAN2 has previously been associated with large artery atherosclerosis-related stroke,36 migraine,37,38 and anti-inflammatory pathways in the central nervous system.39 Other notable top hits from the CHARGE meta-analyses were linked to metabolic traits, particularly lipids and adiposity (CPT1A, cg00574958; PHGDH, cg16246545/cg14476101; TXNIP (thioredoxin interacting protein), cg19693031; SLC7A11, cg06690548), in previous epigenome-wide associations studies (EWAS). An effect of triglycerides on methylation at cg00574958 has been identified,21,40–43 which supports the authors’ hypothesis that an underlying cardiometabolic disease process related to BP and lipids alters DNA methylation at CPT1A.
In a follow-up to the CHARGE analysis, Huang et al. conducted a meta-EWAS in 4,820 EA and AA individuals across 14 cohorts (including 2 youth cohorts).44 The EWAS meta-analysis identified 39 BP-related CpGs with P < 1 × 10−5. In silico replication in the CHARGE consortium of ~17,000 individuals validated 16 of the CpGs, of which 13 showed novel association with BP. Conversely, of the 126 CpGs identified as being associated (P < 1 × 10−7) with BP in the CHARGE consortium, 21 were replicated in the Huang et al. study. Methylation levels of 34 CpGs (13 from the Huang et al. study and 21 cross-validated from CHARGE) were heritable and 6 showed association with gene expression. Additionally, 9 CpGs also had an association with BP with P < 0.05 and were consistent in the direction of the effect in a meta-analysis of the Finnish Twin Cohort and the Netherlands Twin Register. Interestingly, bivariate quantitative genetic modeling of the twin data demonstrated that the phenotypic correlations between methylation levels of the CpGs and BP could be explained by shared, unique environmental characteristics rather than genetic factors.44
In a third EWAS of BP,45 samples from 364 EA and 348 South Asian men (first generation migrants to the United Kingdom) from the Southall And Brent REvisited cohort were assayed using the Illumina Methyl450 array. In a trans-ancestry analysis, DBP was associated with methylation at 1 CpG site (cg07598370 near OR5AP2 (olfactory receptor family 5 subfamily AP member 2)) below the Bonferroni-corrected threshold. A genetic variant near the OR5AP2 has been previously reported to be associated with hematological phenotypes46; in addition, olfactory receptors are known to regulate BP via their renal expression.47 The SBP and hypertension analyses did not uncover statistically significant CpGs after adjustment for confounders (estimated cell counts, and ancestry principal components). The authors also examined the 126 associations reported earlier in the CHARGE analysis.12 The evidence of association from these sites was generally weaker, potentially due to the smaller sample size, except for cg19693031 (near TXNIP) and cg18120259 (in gene body LOC100132354). These CpG sites were consistent in direction of effect with the previous analysis with a slightly larger magnitude of association in the Southall And Brent REvisited cohort study.
TARGET END ORGAN DAMAGE STUDIES
TOD is the resultant injury (often vascular) to organ systems associated with long-standing hypertension including left ventricular hypertrophy, renal disease, retinopathy, and vascular dementia.48 Damage to these organs typically manifests as coronary heart disease, heart failure, stroke, other CVDs, or end-stage kidney failure. Importantly, disease progression and severity may be influenced by DNA methylation alterations that are associated with high BP itself. Alternatively, the vascular and metabolic dysfunction due to high BP could alter DNA methylation patterns in affected organ systems.49 Overall, human cohort studies are limited to access high BP-affected tissue samples (e.g., cardiomyocytes) from subjects at risk, making peripheral blood the only accessible tissue. However, Kato et al. reported on blood and cross-tissue DNA methylation, suggesting that blood and other tissues (liver, muscle, subcutaneous and visceral fat) often exhibit comparable patterns.50 These results suggest DNA methylation in peripheral blood can capture information about disease processes and gene expression changes underlying TOD. In one study, the associations between epigenome-wide DNA methylation and 5 measures of TOD traits (estimated glomerular filtration rate (eGFR), urinary albumin–creatinine ratio (UACR), left ventricular mass index (LVMI), relative wall thickness (RWT), and white matter hyperintensity (WMH)) were assessed in 961 AAs from the Genetic Epidemiology Network of Arteriopathy (GENOA) on the Illumina Infinium MethylationEPIC BeadChip (which assays >850,000 CpGs in genes and regulatory regions). A multivariate model of eGFR, UACR, LVMI, and RWT identified 7 CpGs near 4 genes associated with at least 1 of the traits (cg21134922, cg04816311 near C7orf50; cg09155024, cg10254690 near OAT (ornithine aminotransferase); cg07660512, cg12661888 near IFT43 (intraflagellar transport 43); and cg02264946 near CATSPERD (cation channel sperm associated auxiliary subunit delta)) with false discovery rate q < 0.1. Adjusting for BP, body mass index, and type 2 diabetes attenuated the association for 4 CpGs (cg21134922, cg04816311 near C7orf50; cg09155024, cg10254690 near OAT). When analyzing individual CpG sites, 4 of the 7 (cg04816311, cg09155024, cg07660512, cg12661888) were significantly associated with 2 TOD traits (P < 0.007), while the other 3 were associated with only LVMI. None of the 7 CpGs were associated with WMH, which was only measured in a subset of the GENOA participants. Both cg04816311 and cg12661888 had a consistent effect in which increased methylation was associated with worse TOD outcomes (higher UACR, lower eGFR, and/or higher LVMI). DNA methylation was associated with cis-gene expression for all 7 of the CpGs, but no significant mediation by gene expression was detected. Mendelian randomization analyses suggested causality of 3 CpGs on eGFR (cg04816311, cg10254690, and cg07660512). Replication was attempted in 614 AAs in the Hypertension Genetic Epidemiology Network (HyperGEN) study, whose blood was assayed with the Illumina Infinium Methyl450K array. Out of 3 CpGs available for replication (cg09155024, cg10254690, cg04816311), cg04816311 near C7orf50 was significantly associated with eGFR (P = 0.0003), LVMI (P = 0.0003), and RWT (P = 0.002). Cg04816311 near OAT was associated with eGFR (P = 2 × 10−4) in HyperGEN in a minimally adjusted model (age, sex, smoking, genetic ancestry principal components, and family relatedness), but not after further adjustment for SBP, DBP, and antihypertensive treatment. Among the top findings, OAT was the most notable with respect to biological plausibility for the traits under study. OAT codes ornithine aminotransferase, a key mitochondrial enzyme that converts arginine and ornithine into glutamate and GABA (found in the liver, intestine, brain, and kidney).51,52 Ornithine is involved in the urea cycle and synthesis of nitric oxide.53 Additionally, genetic variants in OAT have been associated with DBP.54IFT43, near cg12661888, encodes a subunit of the intraflagellar transport complex A, a multiprotein complex involved in cilia assembly and maintenance. This subunit is essential in regulating the Sonic Hedgehog signaling pathway, which is involved in regulating the growth, differentiation, and patterning of cells, especially during embryonic development.55CATSPERD, near cg02264946, encodes an auxiliary subunit of sperm calcium channel pore-forming proteins required for the motility of spermatozoa and male fertility.56 The study of TOD in AAs was a hypothesis-free genome-wide study; thus, the authors did not further validate previously published associations for SBP, DBP, or hypertension as part of the analysis. However, a follow-up study of the HyperGEN population (N = 614 AAs) did not show association of any of the 13 replicated CpGs from the CHARGE BP EWAS with echocardiographic traits (LV mass, midwall shortening, RWT, left atrial systolic dimension, LV internal diastolic dimension, ejection fraction). Still, some of the CpGs trended toward significance (P < 0.1, data not published), and further validation is needed to determine if BP-related CpGs are further associated with TOD.
CONCLUSION AND FUTURE DIRECTIONS
The study of epigenetic modifications remains a promising area of research in vascular-related diseases. Current scientific knowledge does not completely explain the molecular mechanisms dictating high BP and related diseases. Epigenetic modifications, such as DNA methylation, might form an additional path to understanding the relevant disease processes. Despite notable challenges in conducting epigenetic research in human populations (potential confounding effects and tissue of study), important findings have been replicated.
Overall, the published work related to methylation variations associated with high BP is promising. Still current research has mostly considered single CpG sites and largely omitted studies of differentially methylated regions, which could provide additional insight into groups of nearby CpGs (e.g., CpG islands) and how they jointly are associated with relevant traits. Additionally, studies have not integrated other epigenetic layers (histone modifications and silencing RNAs) that can provide information on the interplay between other molecular mechanisms dictating gene expression. Furthermore, published studies have included multiple ancestry groups; however, studies of methylation changes across the life course—in the same individuals—are needed to give additional insight into when disease processes start and organ damage related to these conditions occurs. Finally, functional follow-up studies of confirmed loci will help unravel the precise molecular mechanisms at specific CpG sites, including the identification of methylation-specific binding proteins and characterization of their mode of action. Only then can these biological insights be translated to clinical benefits, including reliable biomarkers and effective strategies for disease prevention.
DISCLOSURE
The authors declared no conflict of interest.
REFERENCES
- 1. World Health Organization. Hypertension Fact Sheet. World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- 2. Kearney PM, Whelton M, Reynolds K, Muntner P, Whelton PK, He J. Global burden of hypertension: analysis of worldwide data. Lancet 2005; 365:217–223. [DOI] [PubMed] [Google Scholar]
- 3. Lewington S, Clarke R, Qizilbash N, Peto R, Collins R; Prospective Studies Collaboration . Age-specific relevance of usual blood pressure to vascular mortality: a meta-analysis of individual data for one million adults in 61 prospective studies. Lancet 2002; 360:1903–1913. [DOI] [PubMed] [Google Scholar]
- 4. Pezzini A, Grassi M, Lodigiani C, Patella R, Gandolfo C, Zini A, Delodovici ML, Paciaroni M, Del Sette M, Toriello A, Musolino R, Calabrò RS, Bovi P, Adami A, Silvestrelli G, Sessa M, Cavallini A, Marcheselli S, Bonifati DM, Checcarelli N, Tancredi L, Chiti A, Del Zotto E, Spalloni A, Giossi A, Volonghi I, Costa P, Giacalone G, Ferrazzi P, Poli L, Morotti A, Rasura M, Simone AM, Gamba M, Cerrato P, Micieli G, Melis M, Massucco D, De Giuli V, Iacoviello L, Padovani A; Italian Project on Stroke in Young Adults (IPSYS) Investigators . Predictors of long-term recurrent vascular events after ischemic stroke at young age: the Italian Project on Stroke in Young Adults. Circulation 2014; 129:1668–1676. [DOI] [PubMed] [Google Scholar]
- 5. Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Makuc DM, Marcus GM, Marelli A, Matchar DB, Moy CS, Mozaffarian D, Mussolino ME, Nichol G, Paynter NP, Soliman EZ, Sorlie PD, Sotoodehnia N, Turan TN, Virani SS, Wong ND, Woo D, Turner MB; American Heart Association Statistics Committee and Stroke Statistics Subcommittee . Heart disease and stroke statistics—2012 update: a report from the American Heart Association. Circulation 2012; 125:e2–e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cabrera CP, Ng FL, Nicholls HL, Gupta A, Barnes MR, Munroe PB, Caulfield MJ. Over 1000 genetic loci influencing blood pressure with multiple systems and tissues implicated. Hum Mol Genet 2019; 28:R151–R161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Husquin LT, Rotival M, Fagny M, Quach H, Zidane N, McEwen LM, MacIsaac JL, Kobor MS, Aschard H, Patin E, Quintana-Murci L. Exploring the genetic basis of human population differences in DNA methylation and their causal impact on immune gene regulation. Genome Biol 2018; 19:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ballestar E, Wolffe AP. Methyl-CpG-binding proteins. Targeting specific gene repression. Eur J Biochem 2001; 268:1–6. [DOI] [PubMed] [Google Scholar]
- 9. Ballestar E, Paz MF, Valle L, Wei S, Fraga MF, Espada J, Cigudosa JC, Huang TH, Esteller M. Methyl-CpG binding proteins identify novel sites of epigenetic inactivation in human cancer. EMBO J 2003; 22:6335–6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA 2008; 105:17046–17049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 2002; 16:6–21. [DOI] [PubMed] [Google Scholar]
- 12. Richard MA, Huan T, Ligthart S, Gondalia R, Jhun MA, Brody JA, Irvin MR, Marioni R, Shen J, Tsai PC, Montasser ME, Jia Y, Syme C, Salfati EL, Boerwinkle E, Guan W, Mosley TH Jr, Bressler J, Morrison AC, Liu C, Mendelson MM, Uitterlinden AG, van Meurs JB, Franco OH, Zhang G, Li Y, Stewart JD, Bis JC, Psaty BM, Chen YI, Kardia SLR, Zhao W, Turner ST, Absher D, Aslibekyan S, Starr JM, McRae AF, Hou L, Just AC, Schwartz JD, Vokonas PS, Menni C, Spector TD, Shuldiner A, Damcott CM, Rotter JI, Palmas W, Liu Y, Paus T, Horvath S, O’Connell JR, Guo X, Pausova Z, Assimes TL, Sotoodehnia N, Smith JA, Arnett DK, Deary IJ, Baccarelli AA, Bell JT, Whitsel E, Dehghan A, Levy D, Fornage M; BIOS Consortium . DNA methylation analysis identifies loci for blood pressure regulation. Am J Hum Genet 2017; 101:888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liang M. Epigenetic mechanisms and hypertension. Hypertension 2018; 72:1244–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mittelstraß K, Waldenberger M. DNA methylation in human lipid metabolism and related diseases. Curr Opin Lipidol 2018; 29:116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chu AY, Tin A, Schlosser P, Ko YA, Qiu C, Yao C, Joehanes R, Grams ME, Liang L, Gluck CA, Liu C, Coresh J, Hwang SJ, Levy D, Boerwinkle E, Pankow JS, Yang Q, Fornage M, Fox CS, Susztak K, Köttgen A. Epigenome-wide association studies identify DNA methylation associated with kidney function. Nat Commun 2017; 8:1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suñer D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA 2005; 102:10604–10609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, Berdasco M, Fraga MF, O’Hanlon TP, Rider LG, Jacinto FV, Lopez-Longo FJ, Dopazo J, Forn M, Peinado MA, Carreño L, Sawalha AH, Harley JB, Siebert R, Esteller M, Miller FW, Ballestar E. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res 2010; 20:170–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rozek LS, Dolinoy DC, Sartor MA, Omenn GS. Epigenetics: relevance and implications for public health. Annu Rev Public Health 2014; 35:105–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chambers JC, Loh M, Lehne B, Drong A, Kriebel J, Motta V, Wahl S, Elliott HR, Rota F, Scott WR, Zhang W, Tan ST, Campanella G, Chadeau-Hyam M, Yengo L, Richmond RC, Adamowicz-Brice M, Afzal U, Bozaoglu K, Mok ZY, Ng HK, Pattou F, Prokisch H, Rozario MA, Tarantini L, Abbott J, Ala-Korpela M, Albetti B, Ammerpohl O, Bertazzi PA, Blancher C, Caiazzo R, Danesh J, Gaunt TR, de Lusignan S, Gieger C, Illig T, Jha S, Jones S, Jowett J, Kangas AJ, Kasturiratne A, Kato N, Kotea N, Kowlessur S, Pitkäniemi J, Punjabi P, Saleheen D, Schafmayer C, Soininen P, Tai ES, Thorand B, Tuomilehto J, Wickremasinghe AR, Kyrtopoulos SA, Aitman TJ, Herder C, Hampe J, Cauchi S, Relton CL, Froguel P, Soong R, Vineis P, Jarvelin MR, Scott J, Grallert H, Bollati V, Elliott P, McCarthy MI, Kooner JS. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol 2015; 3:526–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Demerath EW, Guan W, Grove ML, Aslibekyan S, Mendelson M, Zhou YH, Hedman ÅK, Sandling JK, Li LA, Irvin MR, Zhi D, Deloukas P, Liang L, Liu C, Bressler J, Spector TD, North K, Li Y, Absher DM, Levy D, Arnett DK, Fornage M, Pankow JS, Boerwinkle E. Epigenome-wide association study (EWAS) of BMI, BMI change and waist circumference in African American adults identifies multiple replicated loci. Hum Mol Genet 2015; 24:4464–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dekkers KF, van Iterson M, Slieker RC, Moed MH, Bonder MJ, van Galen M, Mei H, Zhernakova DV, van den Berg LH, Deelen J, van Dongen J, van Heemst D, Hofman A, Hottenga JJ, van der Kallen CJ, Schalkwijk CG, Stehouwer CD, Tigchelaar EF, Uitterlinden AG, Willemsen G, Zhernakova A, Franke L, ‘t Hoen PA, Jansen R, van Meurs J, Boomsma DI, van Duijn CM, van Greevenbroek MM, Veldink JH, Wijmenga C, van Zwet EW, Slagboom PE, Jukema JW, Heijmans BT; BIOS Consortium . Blood lipids influence DNA methylation in circulating cells. Genome Biol 2016; 17:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dicorpo DA, Lent S, Guan W, Hivert MF, Pankow JS; CHARGE Epigenetics Working Group , CHARGE Diabetes Working Group. Mendelian randomization suggests causal influence of glycemic traits on DNA methylation. Diabetes 2018; 67(Suppl 1).:1707-P [Google Scholar]
- 23. Pfeiffer L, Wahl S, Pilling LC, Reischl E, Sandling JK, Kunze S, Holdt LM, Kretschmer A, Schramm K, Adamski J, Klopp N, Illig T, Hedman ÅK, Roden M, Hernandez DG, Singleton AB, Thasler WE, Grallert H, Gieger C, Herder C, Teupser D, Meisinger C, Spector TD, Kronenberg F, Prokisch H, Melzer D, Peters A, Deloukas P, Ferrucci L, Waldenberger M. DNA methylation of lipid-related genes affects blood lipid levels. Circ Cardiovasc Genet 2015; 8:334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van der Harst P, de Windt LJ, Chambers JC. Translational perspective on epigenetics in cardiovascular disease. J Am Coll Cardiol 2017; 70:590–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang F, Demura M, Cheng Y, Zhu A, Karashima S, Yoneda T, Demura Y, Maeda Y, Namiki M, Ono K, Nakamura Y, Sasano H, Akagi T, Yamagishi M, Saijoh K, Takeda Y. Dynamic CCAAT/enhancer binding protein-associated changes of DNA methylation in the angiotensinogen gene. Hypertension 2014; 63:281–288. [DOI] [PubMed] [Google Scholar]
- 26. Rivière G, Lienhard D, Andrieu T, Vieau D, Frey BM, Frey FJ. Epigenetic regulation of somatic angiotensin-converting enzyme by DNA methylation and histone acetylation. Epigenetics 2011; 6:478–489. [DOI] [PubMed] [Google Scholar]
- 27. Fan R, Mao SQ, Gu TL, Zhong FD, Gong ML, Hao LM, Yin FY, Dong CZ, Zhang LN. Preliminary analysis of the association between methylation of the ACE2 promoter and essential hypertension. Mol Med Rep 2017; 15:3905–3911. [DOI] [PubMed] [Google Scholar]
- 28. Fan R, Mao S, Zhong F, Gong M, Yin F, Hao L, Zhang L. Association of AGTR1 promoter methylation levels with essential hypertension risk: a matched case-control study. Cytogenet Genome Res 2015; 147:95–102. [DOI] [PubMed] [Google Scholar]
- 29. Mao S, Fan R, Gu T, Zhong Q, Gong M, Dong C, Hao L, Zhang L. Hypermethylation of SCNN1A gene-body increases the risk of essential hypertension. Int J Clin Exp Med 2016; 9:8047–8056. [Google Scholar]
- 30. Zhong Q, Liu C, Fan R, Duan S, Xu X, Zhao J, Mao S, Zhu W, Hao L, Yin F, Zhang L. Association of SCNN1B promoter methylation with essential hypertension. Mol Med Rep 2016; 14:5422–5428. [DOI] [PubMed] [Google Scholar]
- 31. Fish JE, Marsden PA. Endothelial nitric oxide synthase: insight into cell-specific gene regulation in the vascular endothelium. Cell Mol Life Sci 2006; 63:144–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chan Y, Fish JE, D’Abreo C, Lin S, Robb GB, Teichert AM, Karantzoulis-Fegaras F, Keightley A, Steer BM, Marsden PA. The cell-specific expression of endothelial nitric-oxide synthase: a role for DNA methylation. J Biol Chem 2004; 279:35087–35100. [DOI] [PubMed] [Google Scholar]
- 33. Kang YP, Falzone A, Liu M, González-Sánchez P, Choi BH, Coloff JL, Saller JJ, Karreth FA, DeNicola GM. PHGDH supports liver ceramide synthesis and sustains lipid homeostasis. Cancer Metab 2020; 8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lewerenz J, Hewett SJ, Huang Y, Lambros M, Gout PW, Kalivas PW, Massie A, Smolders I, Methner A, Pergande M, Smith SB, Ganapathy V, Maher P. The cystine/glutamate antiporter system x(c)(−) in health and disease: from molecular mechanisms to novel therapeutic opportunities. Antioxid Redox Signal 2013; 18:522–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhao J, Wu W, Zhang W, Lu YW, Tou E, Ye J, Gao P, Jourd’heuil D, Singer HA, Wu M, Long X. Selective expression of TSPAN2 in vascular smooth muscle is independently regulated by TGF-β1/SMAD and myocardin/serum response factor. FASEB J 2017; 31:2576–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.NINDS Stroke GeneticsNetwork (SiGN), International Stroke Genetics Consortium (ISGC). Loci associated with ischaemic stroke and its subtypes (SiGN): a genome-wide association study. Lancet Neurol 2016; 15:174– 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Esserlind AL, Christensen AF, Le H, Kirchmann M, Hauge AW, Toyserkani NM, Hansen T, Grarup N, Werge T, Steinberg S, Bettella F, Stefansson H, Olesen J. Replication and meta-analysis of common variants identifies a genome-wide significant locus in migraine. Eur J Neurol 2013; 20:765–772. [DOI] [PubMed] [Google Scholar]
- 38. Anttila V, Winsvold BS, Gormley P, Kurth T, Bettella F, McMahon G, Kallela M, Malik R, de Vries B, Terwindt G, Medland SE, Todt U, McArdle WL, Quaye L, Koiranen M, Ikram MA, Lehtimäki T, Stam AH, Ligthart L, Wedenoja J, Dunham I, Neale BM, Palta P, Hamalainen E, Schürks M, Rose LM, Buring JE, Ridker PM, Steinberg S, Stefansson H, Jakobsson F, Lawlor DA, Evans DM, Ring SM, Färkkilä M, Artto V, Kaunisto MA, Freilinger T, Schoenen J, Frants RR, Pelzer N, Weller CM, Zielman R, Heath AC, Madden PAF, Montgomery GW, Martin NG, Borck G, Göbel H, Heinze A, Heinze-Kuhn K, Williams FMK, Hartikainen AL, Pouta A, van den Ende J, Uitterlinden AG, Hofman A, Amin N, Hottenga JJ, Vink JM, Heikkilä K, Alexander M, Muller-Myhsok B, Schreiber S, Meitinger T, Wichmann HE, Aromaa A, Eriksson JG, Traynor B, Trabzuni D, Rossin E, Lage K, Jacobs SBR, Gibbs JR, Birney E, Kaprio J, Penninx BW, Boomsma DI, van Duijn C, Raitakari O, Jarvelin MR, Zwart JA, Cherkas L, Strachan DP, Kubisch C, Ferrari MD, van den Maagdenberg AMJM, Dichgans M, Wessman M, Smith GD, Stefansson K, Daly MJ, Nyholt DR, Chasman D, Palotie A; North American Brain Expression Consortium; UK Brain Expression Consortium . Genome-wide meta-analysis identifies new susceptibility loci for migraine. Nat Genet 2013; 45:912–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zeis T, Enz L, Schaeren-Wiemers N. The immunomodulatory oligodendrocyte. Brain Res 2016; 1641:139–148. [DOI] [PubMed] [Google Scholar]
- 40. Aslibekyan S, Demerath EW, Mendelson M, Zhi D, Guan W, Liang L, Sha J, Pankow JS, Liu C, Irvin MR, Fornage M, Hidalgo B, Lin LA, Thibeault KS, Bressler J, Tsai MY, Grove ML, Hopkins PN, Boerwinkle E, Borecki IB, Ordovas JM, Levy D, Tiwari HK, Absher DM, Arnett DK. Epigenome-wide study identifies novel methylation loci associated with body mass index and waist circumference. Obesity (Silver Spring) 2015; 23:1493–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Irvin MR, Zhi D, Joehanes R, Mendelson M, Aslibekyan S, Claas SA, Thibeault KS, Patel N, Day K, Jones LW, Liang L, Chen BH, Yao C, Tiwari HK, Ordovas JM, Levy D, Absher D, Arnett DK. Epigenome-wide association study of fasting blood lipids in the Genetics of Lipid-lowering Drugs and Diet Network study. Circulation 2014; 130:565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Frazier-Wood AC, Aslibekyan S, Absher DM, Hopkins PN, Sha J, Tsai MY, Tiwari HK, Waite LL, Zhi D, Arnett DK. Methylation at CPT1A locus is associated with lipoprotein subfraction profiles. J Lipid Res 2014; 55:1324–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Das M, Sha J, Hidalgo B, Aslibekyan S, Do AN, Zhi D, Sun D, Zhang T, Li S, Chen W, Srinivasan SR, Tiwari HK, Absher D, Ordovas JM, Berenson GS, Arnett DK, Irvin MR. Association of DNA methylation at CPT1A locus with metabolic syndrome in the Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) Study. PLoS One 2016; 11:e0145789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huang Y, Ollikainen M, Muniandy M, Zhang T, van Dongen J, Hao G, van der Most PJ, Pan Y, Pervjakova N, Sun YV, Hui Q, Lahti J, Fraszczyk E, Lu X, Sun D, Richard MA, Willemsen G, Heikkila K, Mateo Leach I, Mononen N, Kähönen M, Hurme MA, Raitakari OT, Drake AJ, Perola M, Nuotio ML, Huang Y, Khulan B, Räikkönen K, Wolffenbuttel BHR, Zhernakova A, Fu J, Zhu H, Dong Y, van Vliet-Ostaptchouk JV, Franke L, Eriksson JG, Fornage M, Milani L, Lehtimäki T, Vaccarino V, Boomsma DI, van der Harst P, de Geus EJC, Salomaa V, Li S, Chen W, Su S, Wilson J, Snieder H, Kaprio J, Wang X. Identification, heritability, and relation with gene expression of novel DNA methylation loci for blood pressure. Hypertension 2020; 76:195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kazmi N, Elliott HR, Burrows K, Tillin T, Hughes AD, Chaturvedi N, Gaunt TR, Relton CL. Associations between high blood pressure and DNA methylation. PLoS One 2020; 15:e0227728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang Q, Kathiresan S, Lin JP, Tofler GH, O’Donnell CJ. Genome-wide association and linkage analyses of hemostatic factors and hematological phenotypes in the Framingham Heart Study. BMC Med Genet 2007; 8(Suppl 1):S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen Z, Zhao H, Fu N, Chen L. The diversified function and potential therapy of ectopic olfactory receptors in non-olfactory tissues. J Cell Physiol 2018; 233:2104–2115. [DOI] [PubMed] [Google Scholar]
- 48. Ammous F, Zhao W, Ratliff SM, Kho M, Shang L, Jones AC, Chaudhary NS, Tiwari HK, Irvin MR, Arnett DK, Mosley TH, Bielak LF, Kardia SLR, Zhou X, Smith JA. Epigenome-wide association study identifies DNA methylation sites associated with target organ damage in older African Americans. Epigenetics 2020:1– 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM. DNA methylation in white blood cells: association with risk factors in epidemiologic studies. Epigenetics 2011; 6:828–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kato N, Loh M, Takeuchi F, Verweij N, Wang X, Zhang W, Kelly TN, Saleheen D, Lehne B, Leach IM, Drong AW, Abbott J, Wahl S, Tan ST, Scott WR, Campanella G, Chadeau-Hyam M, Afzal U, Ahluwalia TS, Bonder MJ, Chen P, Dehghan A, Edwards TL, Esko T, Go MJ, Harris SE, Hartiala J, Kasela S, Kasturiratne A, Khor CC, Kleber ME, Li H, Yu Mok Z, Nakatochi M, Sapari NS, Saxena R, Stewart AFR, Stolk L, Tabara Y, Teh AL, Wu Y, Wu JY, Zhang Y, Aits I, Da Silva Couto Alves A, Das S, Dorajoo R, Hopewell JC, Kim YK, Koivula RW, Luan J, Lyytikäinen LP, Nguyen QN, Pereira MA, Postmus I, Raitakari OT, Scannell Bryan M, Scott RA, Sorice R, Tragante V, Traglia M, White J, Yamamoto K, Zhang Y, Adair LS, Ahmed A, Akiyama K, Asif R, Aung T, Barroso I, Bjonnes A, Braun TR, Cai H, Chang LC, Chen CH, Cheng CY, Chong YS, Collins R, Courtney R, Davies G, Delgado G, Do LD, Doevendans PA, Gansevoort RT, Gao YT, Grammer TB, Grarup N, Grewal J, Gu D, Wander GS, Hartikainen AL, Hazen SL, He J, Heng CK, Hixson JE, Hofman A, Hsu C, Huang W, Husemoen LLN, Hwang JY, Ichihara S, Igase M, Isono M, Justesen JM, Katsuya T, Kibriya MG, Kim YJ, Kishimoto M, Koh WP, Kohara K, Kumari M, Kwek K, Lee NR, Lee J, Liao J, Lieb W, Liewald DCM, Matsubara T, Matsushita Y, Meitinger T, Mihailov E, Milani L, Mills R, Mononen N, Müller-Nurasyid M, Nabika T, Nakashima E, Ng HK, Nikus K, Nutile T, Ohkubo T, Ohnaka K, Parish S, Paternoster L, Peng H, Peters A, Pham ST, Pinidiyapathirage MJ, Rahman M, Rakugi H, Rolandsson O, Ann Rozario M, Ruggiero D, Sala CF, Sarju R, Shimokawa K, Snieder H, Sparsø T, Spiering W, Starr JM, Stott DJ, Stram DO, Sugiyama T, Szymczak S, Tang WHW, Tong L, Trompet S, Turjanmaa V, Ueshima H, Uitterlinden AG, Umemura S, Vaarasmaki M, van Dam RM, van Gilst WH, van Veldhuisen DJ, Viikari JS, Waldenberger M, Wang Y, Wang A, Wilson R, Wong TY, Xiang YB, Yamaguchi S, Ye X, Young RD, Young TL, Yuan JM, Zhou X, Asselbergs FW, Ciullo M, Clarke R, Deloukas P, Franke A, Franks PW, Franks S, Friedlander Y, Gross MD, Guo Z, Hansen T, Jarvelin MR, Jørgensen T, Jukema JW, Kähönen M, Kajio H, Kivimaki M, Lee JY, Lehtimäki T, Linneberg A, Miki T, Pedersen O, Samani NJ, Sørensen TIA, Takayanagi R, Toniolo D, Ahsan H, Allayee H, Chen YT, Danesh J, Deary IJ, Franco OH, Franke L, Heijman BT, Holbrook JD, Isaacs A, Kim BJ, Lin X, Liu J, März W, Metspalu A, Mohlke KL, Sanghera DK, Shu XO, van Meurs JBJ, Vithana E, Wickremasinghe AR, Wijmenga C, Wolffenbuttel BHW, Yokota M, Zheng W, Zhu D, Vineis P, Kyrtopoulos SA, Kleinjans JCS, McCarthy MI, Soong R, Gieger C, Scott J, Teo YY, He J, Elliott P, Tai ES, van der Harst P, Kooner JS, Chambers JC; BIOS-consortium; CARDIo GRAMplusCD; LifeLines Cohort Study; InterAct Consortium . Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat Genet 2015; 47:1282–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ginguay A, Cynober L, Curis E, Nicolis I. Ornithine aminotransferase, an important glutamate-metabolizing enzyme at the crossroads of multiple metabolic pathways. Biology 2017; 6: 1–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huynh NN, Chin-Dusting J. Amino acids, arginase and nitric oxide in vascular health. Clin Exp Pharmacol Physiol 2006; 33:1–8. [DOI] [PubMed] [Google Scholar]
- 53. Mori M, Gotoh T, Nagasaki A, Takiguchi M, Sonoki T. Regulation of the urea cycle enzyme genes in nitric oxide synthesis. J Inherit Metab Dis 1998; 21(Suppl 1):59–71. [DOI] [PubMed] [Google Scholar]
- 54. Parmar PG, Taal HR, Timpson NJ, Thiering E, Lehtimäki T, Marinelli M, Lind PA, Howe LD, Verwoert G, Aalto V, Uitterlinden AG, Briollais L, Evans DM, Wright MJ, Newnham JP, Whitfield JB, Lyytikäinen LP, Rivadeneira F, Boomsma DI, Viikari J, Gillman MW, St Pourcain B, Hottenga JJ, Montgomery GW, Hofman A, Kähönen M, Martin NG, Tobin MD, Raitakari O, Vioque J, Jaddoe VWV, Jarvelin MR, Beilin LJ, Heinrich J, van Duijn CM, Pennell CE, Lawlor DA, Palmer LJ; Early Genetics and Lifecourse Epidemiology Consortium . International genome-wide association study consortium identifies novel loci associated with blood pressure in children and adolescents. Circ Cardiovasc Genet 2016; 9:266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liem KF Jr, Ashe A, He M, Satir P, Moran J, Beier D, Wicking C, Anderson KV. The IFT-A complex regulates Shh signaling through cilia structure and membrane protein trafficking. J Cell Biol 2012; 197:789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chung JJ, Navarro B, Krapivinsky G, Krapivinsky L, Clapham DE. A novel gene required for male fertility and functional CATSPER channel formation in spermatozoa. Nat Commun 2011; 2:153. [DOI] [PMC free article] [PubMed] [Google Scholar]