Abstract
The auxin-inducible degradation system in C. elegans allows for spatial and temporal control of protein degradation via heterologous expression of a single Arabidopsis thaliana F-box protein, transport inhibitor response 1 (AtTIR1). In this system, exogenous auxin (Indole-3-acetic acid; IAA) enhances the ability of AtTIR1 to function as a substrate recognition component that adapts engineered degron-tagged proteins to the endogenous C. elegans E3 ubiquitin ligases complex [SKR-1/2-CUL-1-F-box (SCF)], targeting them for degradation by the proteosome. While this system has been employed to dissect the developmental functions of many C. elegans proteins, we have found that several auxin-inducible degron (AID)-tagged proteins are constitutively degraded by AtTIR1 in the absence of auxin, leading to undesired loss-of-function phenotypes. In this manuscript, we adapt an orthogonal auxin derivative/mutant AtTIR1 pair [C. elegans AID version 2 (C.e.AIDv2)] that transforms the specificity of allosteric regulation of TIR1 from IAA to one that is dependent on an auxin derivative harboring a bulky aryl group (5-Ph-IAA). We find that a mutant AtTIR1(F79G) allele that alters the ligand-binding interface of TIR1 dramatically reduces ligand-independent degradation of multiple AID*-tagged proteins. In addition to solving the ectopic degradation problem for some AID-targets, the addition of 5-Ph-IAA to culture media of animals expressing AtTIR1(F79G) leads to more penetrant loss-of-function phenotypes for AID*-tagged proteins than those elicited by the AtTIR1-IAA pairing at similar auxin analog concentrations. The improved specificity and efficacy afforded by the mutant AtTIR1(F79G) allele expand the utility of the AID system and broaden the number of proteins that can be effectively targeted with it.
Keywords: C. elegans, AID system, CRISPR/Cas9, targeted degradation, heterochronic, RNA pol II inhibition, auxin
Introduction
Detailed analyses of developmental and other dynamic biological events, processes, and mechanisms have been facilitated in part by advancements in techniques to precisely control the products of gene expression. In Caenorhabditis elegans, a variety of tools have been developed to examine the stage- and tissue-specific function of genes, including FLP-mediated recombination to control gene activation (Davis et al. 2008; Voutev and Hubbard 2008) and RNAi to degrade the RNA gene product (Qadota et al. 2007). While these techniques are indirect, limited by the stability of the target protein, and are prone to off-target effects, more modern approaches have leveraged the power of CRISPR genome editing to engineer gene products to harbor epitopes that enable proteins to be directly targeted for degradation.
The auxin-inducible degradation system allows for the rapid and conditional degradation of target proteins in yeast, vertebrate cells, and C. elegans (Nishimura et al. 2009; Holland et al. 2012; Zhang et al. 2015). This two-component system relies on the heterologous expression of a plant-specific F-box protein, TIR1, that binds the plant hormone auxin (Ruegger et al. 1998; Gray et al. 1999) and an encoded 44-amino acid minimal degron sequence [auxin-inducible degron (AID*)] derived from the Arabidopsis thaliana IAA17 protein (Morawska and Ulrich 2013). The tractability and specificity of this system to provide exquisite spatiotemporal control of target protein levels rely on two key features of the TIR1/AID*-tagged protein interaction. First, stable association of TIR1 with proteins harboring an AID* sequence is allosterically regulated by auxin (indole-3-acetic acid, or IAA; Tan et al. 2007). Second, A. thaliana TIR1 can interact with endogenous members of the Skp1 and Cullin family of proteins to form a functional, ligand-gated ubiquitin E3 ligase that targets AID*-tagged proteins to the 26S proteosome (Nishimura et al. 2009; Kanke et al. 2011; Holland et al. 2012; Zhang et al. 2015).
The AID system has proven to be extremely powerful in the C. elegans model where targeted inactivation of proteins can complement the already robust genetics of this system. The utility in this model comes from the application of many tissue-specific drivers that have been employed to drive TIR1 expression in various cell types(Ashley et al. 2021). Three issues have limited the applicability of this system for general use. First, the required dose of natural auxin (IAA) typically required for efficient degradation of target proteins is relatively high (1 mM) and can elicit defined biological responses in C. elegans independent of AtTIR1 expression (Bhoi et al. 2021; Loose and Ghazi 2021). The current system also exhibits a substantial amount of activity against a simple, heterologous AID*::GFP reporter even in the absence of added IAA (Martinez et al. 2020). Whether ectopic AtTIR1-mediated degradation is a general issue of the AID system or a reflection of latent properties of individual AID*-tagged target proteins in C. elegans is unknown. Alternatively, endogenously or environmentally derived partially activating ligands may be present in C. elegans and these metabolites may trigger aberrant AID-target degradation in the presence of AtTIR1. Finally, some AID-targets are inefficiently degraded and fail to generate strong loss-of-function phenotypes (Patel and Hobert 2017; Serrano-Saiz et al. 2018; Duong et al. 2020).
In this manuscript, we aimed to improve the C. elegans AID system by altering the ligand-binding specificity of TIR1. Using a previously established TIR1 variant that can target degradation in A. thaliana using a synthetic, modified auxin/IAA ortholog (Uchida et al. 2018), we demonstrate that the AtTIR1(F79G) variant combined with a 5-Ph-IAA auxin analog also functions in the C. elegans model to degrade AID*-tagged proteins. Importantly, the substitution of this single amino acid in AtTIR1 alleviates the ligand-independent activity of this protein for multiple, biologically relevant AID*-tagged fusion proteins. Finally, we demonstrate that the AtTIR1(F79G)/5-Ph-IAA (C.e.AIDv2 System) also exhibits elevated activity, enabling strong loss-of-function phenotypes to be elicited with low levels of exogenously added ligand. Because the AIDv2 system efficiently targets existing AID*-tagged target genes, we suggest that this modification functions as an improved experimental platform to dissect spatial and temporal gene functions.
Materials and methods
Caenorhabditis elegans maintenance and genetics
Caenorhabditis elegans strains were maintained on standard media at 20°C and fed Escherichia coli OP50 (Brenner 1974). Some strains were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). A complete list of strains outlined in this manuscript can be found in Supplementary Table S1. Four hundred millimolar IAA (Sigma; Product #I3750) and 100 mM 5-Ph-IAA (Bioacademia; Product #30-003-10) stocks were made in 95% ethanol diluted into NGM media at the indicated concentration.
CRISPR editing
cshIs140[TIR1(F79G)]
The single copy rps-28pro::AtTIR1::T2A::mCherry::his-11 (cshIs128) was integrated using standard CRISPR-mediated genomic editing to the ttTi5605 landing site following standard protocols (Dickinson et al. 2013). CRISPR editing of the cshIs128 allele to generate the AtTIR1(F79G) variant was accomplished using standard procedures (Paix et al. 2017). Briefly, recombinant nlsCas9 obtained from the University of California, Berkley MacroLab was used in conjunction with a recombinant sgRNA (Synthego, Menlo Park, CA, USA) with the following guide sequence (PAM sequence underlined): TTCCCTTGAGCTCGACGGAACGG (Oligo #1; see Table S2). A single-stranded, HPLC-purified repair template (Oligo #2; see Table S2 was used to edit the TIR1 coding sequence by homologous repair. The allele was verified by standard Sanger sequencing.
lin-28::AID*(ae157)
The AID*-tagged lin-28 allele was constructed in a similar manner as above using a separate commercially available Cas9 protein (EnGen® Spy Cas9 NLS, M0646T) with gRNA targeting a genomic sequence in the 3′ coding sequence of lin-28 (5′-ATATCATCGTCAGATGTAGT-3′). The sgRNA was synthesized using the Invitrogen™, MEGAshortscript™ T7 Transcription Kit (AM1354) and a single-stranded transcription oligo #1 template (oligo #3; see Table S2). The repair templates were generated using two template oligos #4 and #5 (see Table S2). Repair templates #2 and #3 were mixed together, heated to 96°C for 5 min, then placed on ice. The purpose was to create a “hybrid template” that is more efficient in the homology-directed repair (HDR) (Dokshin et al. 2018).
ama-1::AID*::GFP(ers49)
The 1752 bp repair template to generate the AID*::GFP C-terminally tagged ama-1 allele was made by amplifying the AID*::GFP sequences from pLZ29 (Zhang et al. 2015) using the oligos #6 and #7 (Table S2). Injection mixtures using recombinant Streptococcus pyogenes Cas9 3NLS (10 μg/μL, IDT), crRNA (GATGAATTTGGATCATAAGT, 2 nmol, IDT, oligo #8; see Table S2), tracrRNA (IDT, cat# 1072532), dsDNA donors, and pCFJ90 (coinjection marker pharynx mCherry was used at a final concentration of 3 ng/µL) were prepared as previously described (Dokshin et al. 2018).
hrpa-1::AID*(zen91)
To generate hrpa-1::linker::AID*::TEV::FLAG strain, N2 worms were injected with the CRISPR-Cas9 RNA-protein complex(Paix et al. 2017). The injection mix consisted of Alt-R Cas9 (IDT, cat# 1081058) along with hrpa-1 crRNA (oligo #9; see oligo table), dpy-10 crRNA (5′-GCUACCAUAGGCACCACGAG-3′; Arribere et al. 2014), tracer RNA (IDT, cat# 1072532), and hrpa-1::linker::AID*::TEV::FLAG donor (oligo #10; see Table S2). Four independent alleles of hrpa-1::linker::AID*::TEV::FLAG were obtained and the tagged endogenous loci were sequenced. Two lines were chosen, outcrossed twice, and assessed. One line was chosen for further experiments.
Image acquisition
Confocal microscopy
Images were acquired using a Hamamatsu Orca EM-CCD camera and a Borealis-modified Yokagawa CSU-10 spinning disk confocal microscope (Nobska Imaging, Inc.) with a Plan-APOCHROMAT x 100/1.4 or 40/1.4 oil DIC objective controlled by MetaMorph software (version: 7.8.12.0). Animals were anesthetized on 5% agarose pads containing 10 mM sodium azide and secured with a coverslip. Imaging on the microfluidic device was performed on a Zeiss AXIO Observer.Z7 inverted microscope using a 40× glycerol immersion objective and DIC and GFP filters controlled by ZEN software (version 2.5). Images were captured using a Hamamatsu C11440 digital camera. For scoring plate level phenotypes, images were acquired using a Moticam CMOS (Motic) camera attached to a Zeiss dissecting microscope.
Wide-field fluorescence microscopy
Images were acquired with a Zeiss Axio Observer microscope equipped with Nomarski and fluorescence optics as well as a Hamamatsu Orca Flash 4.0 FL Plus camera. An LED lamp emitting at 470 nm was used for fluorophore excitation. For single images, animals were immobilized on 2% agarose pads supplemented with 100 mM Levamisole (Sigma).
Image processing and analysis
All acquired images were processed using Fiji software (version: 2.0.0-rc-69/1.52p; Schindelin et al. 2012). To quantify VPC-specific AID*::GFP or AMA-1::AID*::GFP expression levels, images were captured at the P6.p 1-cell stage (early L3 stage). Images of AID*::GFP animals were obtained at time points 0, 30, 60, 90, and 120 min in the absence or presence of auxin, and images of AMA-1::AID*::GFP animals were obtained 2-h post-treatment. Expression levels were quantified by measuring the mean fluorescence intensity (MFI) of vulval precursor cells (VPCs) subtracted by the MFI of a background region in the image to account for camera noise. Cells were outlined using the freehand selection or wand (tracing) tool in Fiji. Kinetic data from AID*::GFP animals were normalized by dividing the MFI in treated or untreated animals at time points 30, 60, 90, and 120 min by the average MFI in untreated animals at 0 min.
Embryonic viability and brood size measurements
Brood size measurements were carried out at 20°C and 25°C for each indicated strain using standard protocols (Tissenbaum and Ruvkun 1998). Briefly, single, late L4-stage animals of each genotype were transferred to a new plate every 24 h for a total of 4 days. Plates were scored 24 and 48 h after each transfer for the number of eggs and number of viable, hatched worms. The embryonic viability from each animal was established by calculating the percentage of unhatched eggs after 48 h for each associated plate in the experiment. The total brood size was determined for individual worms by summing the number of viable offspring of each derived plate. In all cases, the stated brood size is the average of >15 worms for each genotype. For hrpa-1::linker::AID*::TEV::FLAG brood measurements, embryos of each genotype were plated onto control, IAA, or 5-ph-IAA plates and cultured until L4 stage. Single, late L4-stage animals of each genotype were transferred to a new plate. Larval arrest from each animal was established by calculating the percentage of arrested larvae that failed to grow after 24 h.
Motility assays
Motility assays are modified from a previously published protocol (Tsalik et al. 2003). Briefly, five animals (grown in the indicated conditions) were transferred to a standard NGM plate (± auxin analog) to a region in the center of a set of concentric rings (4, 6, 8, and 10 mm; see Template 1). Animals were allowed to freely move for the indicated time and the proportion of animals that left the first ring within the annotated time were recorded. Animals were scored positive even if they left the ring and returned within the boundary.
Western blotting
Forty to 50 worms were picked directly into the 2× Laemmli protein loading buffer and boiled at 95°C for 5 min. HRPA-1 was detected using the Monoclonal ANTI-FLAG M2-Peroxidase (HRP) antibody (A8592-2MG) from Sigma-Aldrich at a 1:500 dilution. Mouse anti-tubulin antibody (Sigma-Aldrich) was used at a 1:5000 dilution to detect tubulin as a loading control.
Graph plots and statistical analysis
Plots and diagrams were generated using GraphPad Prism v9 (GraphPad Software, San Diego, CA, USA). Statistical significance was determined using a two-tailed unpaired Student’s t-test. P < 0.05 was considered statistically significant. **** indicates P < 0.0001.
Results
AtTIR1(WT) triggers loss-of-function phenotypes of an LIN-28::AID* allele in the absence of exogenously added ligand
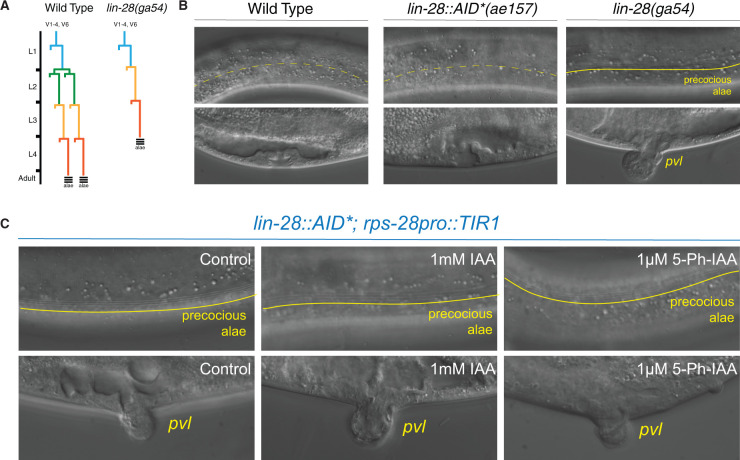
During postembryonic development, genes in the heterochronic pathway control the sequence of stage-specific cell divisions and cell fate specification (Rougvie and Moss 2013). A key feature of this pathway is that transitions from one stage-specific pattern of development to the next are controlled by the sharp temporal downregulation of protein-coding genes (Rougvie and Moss 2013). The lin-28 gene encodes a highly conserved RNA-binding protein that functions to modulate miRNA processing, turnover, and activity; lin-28 is acutely downregulated after the first larval stage (Moss et al. 1997; Lehrbach et al. 2009; Van Wynsberghe et al. 2011). lin-28 null mutants, lin-28(ga94), initiate postembryonic development normally but then skip L2-stage patterns of cell division and immediately execute L3 patterns of development after the L1 molt (Figure 1A; Moss et al. 1997). Consequently, lin-28(0) mutants exhibit vulval morphology defects (protruding vulva; pvl), fail to proliferate their lateral seam cells, and precociously deposit adult-specific alae structures one stage earlier than wild-type animals (Figure 1B and Table 1).
Figure 1.
AtTIR1(WT) induces a lin-28 loss-of-function phenotype in the absence of exogenously added auxin ligand. (A) The lateral seam cells of wild-type C. elegans larva exhibit a stereotyped cell division program that generates additional seam cell in the L2 stage. Mutations in the heterochronic gene lin-28 result in an altered seam cell division pattern due to the skipping of L2 stage-specific developmental programs. In addition, the lateral seam cells lin-28(0) mutants precociously exit the cell cycle and inappropriately deposit alae, adult-specific cuticle structures, during the L3–L4 molt. (B) In contrast to the normal skin and vulval developmental programs of wild-type animals and animals that harbor an AID-tagged lin-28 allele, lin-28(0) mutants exhibit precocious alae at the L4 stage and a protruding vulval phenotype (pvl). (C) Combining the AID-tagged lin-28 allele with a ubiquitously expressed AtTIR1(WT) allele results in strong heterochronic phenotypes in the absence of additional auxin. Dashed yellow lines indicate the absence of adult stage-specific alae structures whereas a solid line demarcates the presence of adult stage-specific alae structures (see Table 1 for details).
Table 1.
Measurement of TIR1 and TIR1(F79G) dependent heterochronic phenotypes in strains harboring an AID-tagged lin-28 allele
| Experimental |
% Animals |
% Animals |
Ave. seam |
Seam |
% pvl |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | Strain | Genotypea,b | Conditionc | L4 alaed | n | Adult alae | n | Numbere | Cell range | n | Animals | n |
| 1 | RG773 | Wild type | — | 0 | 20 | 100 | 20 | 15.8 | 15–17 | 22 | 0 | 100 |
| 2 | HML1023 | lin-28(ga54)I | — | 85 | 20 | 100 | 15 | 11.8 | 10–12 | 27 | 100 | 100 |
| 3 | ME462 | lin-28::AID(ae157) I | — | 0 | 20 | 100 | 20 | 16 | 15–17 | 26 | 0 | 100 |
| 4 | HML1030 | lin-28::AID(ae157) I; cshIs128[rps-28pro::TIR1_P2A_mCherryHis-11] II | — | 86 | 21 | 100 | 20 | 12.68 | 12–16 | 25 | 68 | 100 |
| 1 mM auxin | 90 | 20 | 100 | 20 | 11.7 | 10–12 | 26 | 85 | 100 | |||
| 1uM 5-Ph-IAA | 85 | 20 | 100 | 20 | 12.24 | 11–12 | 25 | 100 | 100 | |||
| 5 | HML1024 | lin-28::AID(ae157) I; cshIs140[rps-28pro::TIR1(F79G)_P2A_mCherryHis-11] II | — | 0 | 20 | 100 | 20 | 16 | 16–17 | 42 | 0 | 100 |
| 1 mM auxin | 0 | 20 | 100 | 20 | 15.8 | 15–16 | 31 | 0 | 100 | |||
| 1uM 5-Ph-IAA | 91 | 23 | 100 | 20 | 11.7 | 11–12 | 23 | 100 | 100 |
Animals contain wIS78 V [ajm-1::GFP and scm::GFP] which were used to visualize adherence junctions and lateral seam cells.
L-the ae157 allele of lin-28 harbors an AID tag fused in frame to the lin-28 coding sequence generating a carboxy-terminal tagged allele.
L-4-staged Po animals were plated onto indicated plates and F1 progeny of the indicated stage were scored for.
Presence and quality of cuticular alae structures were assayed by Normarski DIC optics. Only one side of each animals was scored.
Average seam cell numbers were scored by counting the number of SCM::GFP(+) cells on a single side of each animal.
To generate an lin-28 allele whose expression can be modulated by auxin, we used CRISPR genome editing to insert a DNA fragment encoding the AID* tag at the 3′ end of the lin-28 open reading frame. Examination of transgenic animals indicates that, in contrast to lin-28(ga54) animals, lin-28::AID*(ae127) animals do not exhibit heterochronic phenotypes, indicating that addition of the degron tag does not appreciably alter lin-28 activity (Figure 1B and Table 1). We next crossed the lin-28::AID*(ae127) allele into a strain harboring a ubiquitously expressed AtTIR1 (cshIs128 [rps-28pro::TIR1::T2A::mCherry::his-11]). Surprisingly, homozygous lin-28::AID*(ae127); cshIs128 animals exhibited highly penetrant heterochronic phenotypes (precocious vulval cell divisions, altered seam cell lineage (seam cell number and the precocious production of adult alae structures at the L4 stage)) on normal NGM plates indicating that that the TIR1 activity, in the absence of auxin, results in a reduction of lin-28 activity during development that phenocopies lin-28(lf) phenotypes.
Generation, validation, and characterization of an orthogonal auxin and AtTIR1(F79G) allele in C. elegans
Crystallographic studies indicate that IAA fills a hydrophobic cavity in between TIR1 and the AID/degron peptide sequence to form a stable trimeric complex (Dharmasiri et al. 2005; Kepinski and Leyser 2005; Tan et al. 2007). We hypothesized that the TIR1-dependent/auxin-independent degradation phenotypes of the lin-28::AID* allele could be caused either by a low-level interaction between the AID*-tag and TIR1 in the absence of auxin or by the inappropriate recognition of chemically related endogenously produced ligand that may bind to the pocket of AtTIR1. Uchida et al. (2018) have previously described a series of mutations of TIR1 that alter the auxin-binding pocket to enable ligand-dependent target degradation using IAA derivatives that have been modified by aryl groups on the fifth position of IAA. One of the engineered AtTIR1 mutants they identified, AtTIR1(F79G), failed to interact with AID*-tagged proteins in the presence of IAA but exhibited high-binding specificity for AID*-tagged substrates in the presence of 5-phenyl-indole-3-acetic acid (5-Ph-IAA; Uchida et al. 2018). We reasoned that the inappropriate activity of AtTIR1, mediated by either of the above mechanisms, may be alleviated by similar alterations in the AtTIR1-IAA binding pocket. A similar, orthogonal TIR1/auxin analog approach has been adapted for other systems using Oryza sativa TIR1 (OsTIR1) suggesting that this strategy is generally applicable (Yesbolatova et al. 2020).
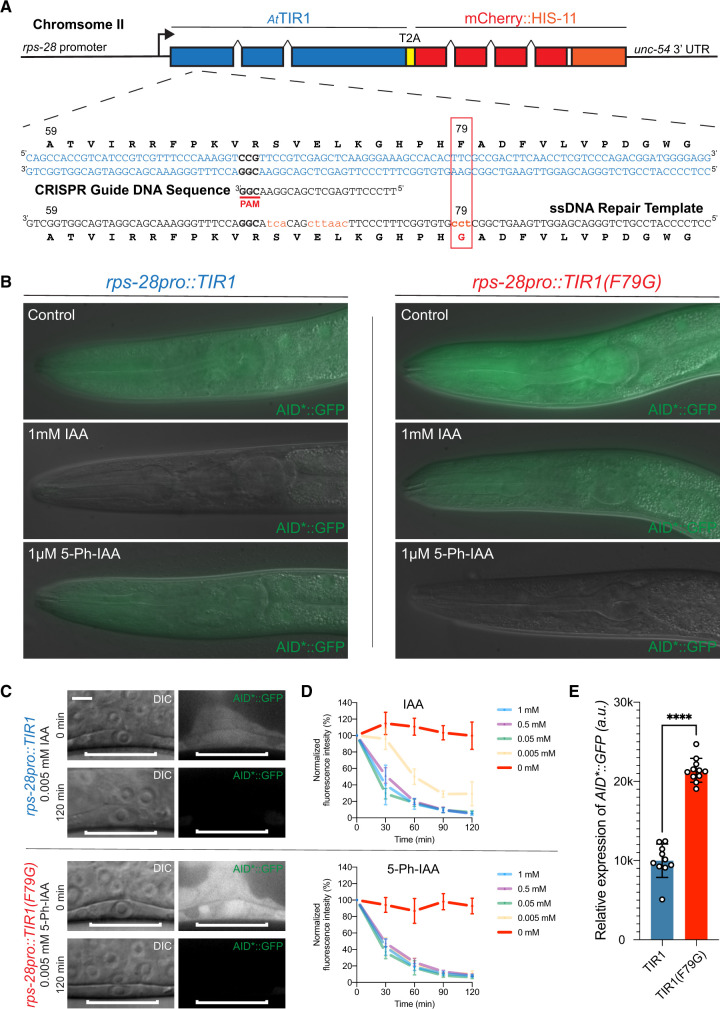
To test the activity of an AtTIR1(F79G) allele of TIR1 in C. elegans, we used CRISPR genome editing and HDR to edit an existing, single-copy TIR1 allele. Specifically, animals harboring a ubiquitously expressed rps-28pro::TIR1 allele (cshIs128) and an eft-3pro::AID*::GFP transgene (ieSi58) were injected with a recombinant nlsCas9/guide ribonucleoprotein complex that is predicted to induce dsDNA breaks approximately 30-bp upstream of the TIR1 F79 codon and an HRD repair oligo partially complementary to the TIR1 ORF overlapping with the TIR1 F79 codon (Figure 2). F1 progeny were then cloned onto NGM plates containing 1-mM IAA to identify GFP(+) F2 progeny that fails to efficiently degrade the AID*::GFP reporter. Candidate GFP(+) F2 animals were then plated on NGM plates containing 100 µM 5-Ph-IAA to identify clonal animals harboring a modified AtTIR1 allele that is capable of degrading the AID*::GFP reporter only in the presence of the 5-Ph-modified auxin (Figure 2B). A single recombinant AtTIR1(F79G) allele (cshIs140) was sequence-verified and used in further experiments.
Figure 2.
Mutation of phenylalanine 79 to glycine in the TIR1 protein switches the specificity of the auxin degradation system to one that is now responsive to 5-Ph-IAA. (A) Structure of cshIs128 that encodes both a AtTIR1(WT) protein and an autocatalytically cleaved nuclear-localized mCherry::HIS-11 reporter driven by a ubiquitously expressed ribosomal protein (rps-28) promoter. Below the gene structure is the coding sequence of the region of AtTIR1(WT) that was mutagenized via CRISPR and HDR using a coinjected ssDNA oligo. (B) Representative mid-larval staged animals expressing AID*::GFP and one of two indicated AtTIR1 variants. Animals were grown continuously on untreated NGM plates or NGM plates including the indicated auxin analog at the listed concentration. (C) Micrographs of early L3 staged animals expressing AID*::GFP and one of the two AtTIR1 variants before and 120 min after the addition of the auxin analog. (D) Rates of AID*::GFP degradation were determined by quantifying AID*::GFP in early L3 staged P6.p cells in animals coexpressing the indicated AtTIR1 variant following auxin analog treatment. Data presented as the mean and SD (n ≥ 10 animals examined for each time point). (E) Quantification of the relative expression levels of the AID*::GFP reporter in P6.p cells of early L3 staged animals that were grown on control plates. Data presented as the median with SD (n ≥ 10 animals examined for each TIR1 transgene, and ****P < 0.0001 by a Mann–Whitney U-test).
We next compared the relative expression levels and degradation kinetics of the AID*::GFP reporter in animals expressing the two AtTIR1 variants. Previous observations indicate that AtTIR1(WT) reduces AID*::GFP expression in the absence of IAA (Martinez et al. 2020). We then compared the expression levels of the AID*::GFP reporter in animals expressing the two different TIR1 alleles. As shown in Figure 2, C and E, AID*::GFP expression is significantly lower in the P6.p vulval precursor cells of animals expressing AtTIR1(WT) when compared to the levels in similarly staged AtTIR1(F79G)-expressing animals. Expression levels of AID*::GFP in AtTIR1(F79G) animals are similar to that observed in WT animals (Supplementary Figure S1). We next compared the efficiency of ligand-induced degradation mediated by these two AtTIR1 alleles by quantifying the relative changes in AID*::GFP expression in the presence of various IAA or 5-Ph-IAA concentrations over time. We monitored AID*::GFP expression in single vulval precursor cells of stage-matched (early mid-L3 stage) animals that have been exposed to different concentrations of auxin orthologs that were incorporated into standard C. elegans solid culture media (Figure 2, C and D). We found that elevated concentrations (≥0.05 mM) of both auxin analogs led to the efficient reduction of AID*::GFP with greater than 50% of the target protein depleted in P6.p cells within the first 30 min of the time course (Figure 2D). At lower concentrations of 5-Ph-IAA (0.05 mM), AtTIR1(F79G) maintained similar AID*::GFP depletion kinetics suggesting that this level of activating ligand is still saturating in this system and that overall kinetics are dependent on AtTIR1(F79G) expression levels. In contrast, the efficiency of the AtTIR1(WT)-mediated degradation when normal IAA was reduced (Figure 2D). These results suggest that the AtTIR1(F79G) (AIDv2) functions more efficiently with the 5-Ph-IAA ligand than the classical version of the AID system with unmodified IAA (Zhang et al. 2015).
The AIDv2/TIR1(F79G) version of the AID system efficiently regulates the expression of dosage-sensitive AID* targets
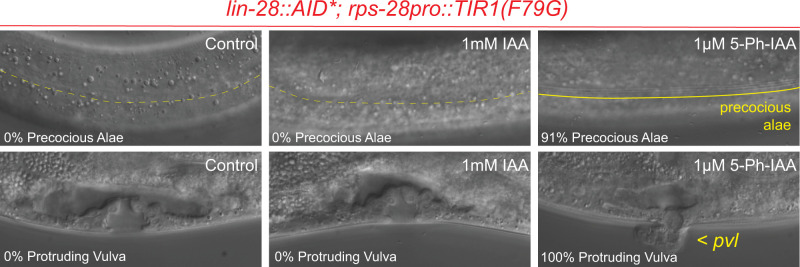
Given that the AtTIR1(F79G) allele of AtTIR1 does not lead to a significant reduction of AID*::GFP expression in the absence of ligand, we next tested whether this allele could be used to efficiently modulate the activity of AID*-tagged genes that exhibit dosage-sensitive phenotypes in a ligand-specific manner. First, we crossed the AtTIR1(F79G) allele into a strain harboring the lin-28::AID* degron-tagged allele. As opposed to the inappropriate lin-28(lf) phenotypes that are caused by AtTIR1(WT), lin-28::AID*; cshIs140 [AtTIR1(F79G)] animals exhibit completely wild-type seam cell lineage (seam cell number) and do not exhibit precocious alae formation or a protruding vulval phenotype (Figure 3 and Table 1). Importantly, treatment of lin-28::AID*; cshIs140 [AtTIR1(F79G)] parental animals with very low concentrations of 5-Ph-IAA (1 µM) lead to highly penetrant heterochronic phenotypes for each developmental feature (seam cell number, alae, and vulval development) in F1 progeny indicating that the AIDv2 system can elicit fully penetrant lin-28(lf) phenotypes. These phenotypes were not elicited when animals were grown on media containing 1000-fold higher concentration of normal auxin (IAA), demonstrating the sensitivity and specificity of the TIR1(F79G) allele.
Figure 3.
The AtTIR1(F79G) variant of TIR1 allows for ligand-dependent regulation of LIN-28::AID* activity by 5-Ph-IAA. Representative images depict the cuticular and vulval development at the L4-stage of F1 animals that have developed on NGM plates containing the auxin analog at the indicated concertation. Dashed yellow lines demarcate the absence of adult stage-specific alae structures whereas a solid line demarcates the presence of adult stage-specific alae structures. Pvl, protruding vulva (see Table 1 for details).
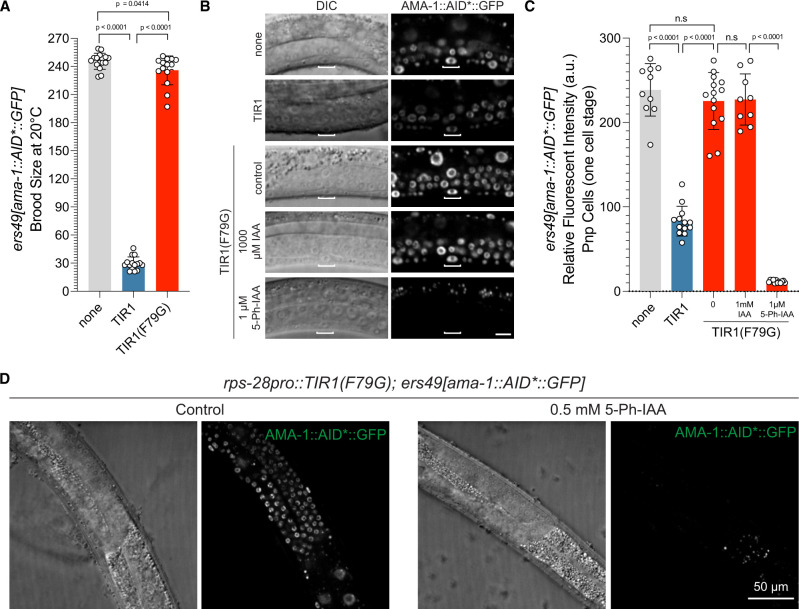
We have also previously tagged the large, catalytic subunit of the C. elegans pol II complex, ama-1, with the AID epitope with the idea that an auxin degradation system could be used to acutely inactivate transcription in a temporal and cell-type-specific manner. This would bypass the limitations of prior approaches to reduce transcriptional output via chemical inhibitors or by cell-type-specific RNAi (Rogalski and Riddle 1988; Rogalski et al. 1988; Firnhaber and Hammarlund 2013). Transgenic animals homozygous for a C-terminally tagged ama-1::AID*::GFP allele, ama-1(ers49), are indistinguishable from wild-type animals and exhibit normal development and brood sizes (Figure 4A). When the ama-1::AID*::GFP allele is combined with a ubiquitously expressed AtTIR1(WT), the pace of overall animal development is dramatically slowed and there is a significant reduction (greater than eightfold) in the fecundity of animals compared to isogenic strains lacking AtTIR1 expression (Figure 4A). Consistent with the assumption that these developmental phenotypes result from an inappropriate activity of AtTIR1(WT), AMA-1::AID*::GFP expression is reduced by approximately 2.9-fold in P6.p vulval precursor cells (1-cell stage) of animals expressing ubiquitous AtTIR1(WT) when compared to a strain only expressing the tagged pol II allele (Figure 4, B and C).
Figure 4.
AtTIR1(WT) allele of TIR1 generates ligand-independent loss-of-function phenotypes that are alleviated by the 5-Ph-IAA regulated AtTIR1(F79G) variant. (A) An AID-tagged, endogenous allele of ama-1, ama-1(ers49), exhibits a severe reduction in brood size when combined with AtTIR1(WT) even in the absence of added auxin. In contrast, the AtTIR1(F79G) variant does not alter the brood size of ama-1(ers49) animals (n = 15 for each genotype and significance was calculated using an unpaired t-test). (B) The expression of AMA-1::AID*::GFP is dramatically reduced in animals expressing AtTIR1(WT) and not in animals expressing AtTIR1(F79G). Shown are DIC and corresponding GFP images of vulval precursor cells (VPCs) (from early L3 stage animals at the P6.p 1-cell stage). The scale bar is 10 µ. (C) Quantification of AMA-1::AID*::GFP expression animals depicted in (B). Data presented as the mean ± SD (n ≥ 10 animals examined for each, and P < 0.0001 by a Student’s t-test). (D) AMA-1::AID*::GFP is degraded in the germline by 5-Ph-IAA. Representative DIC and GFP images of L4 staged animals that have been transferred to control NGM plates or NGM plates containing 0.5 mM 5-Ph-IAA for 2 h.
We next determined if the developmental phenotypes associated with the ectopic activity of AtTIR1(WT) would also be alleviated by mutating the binding pocket of the AtTIR1 ligand-binding domain. When we crossed the ama-1::AID*::GFP allele into a strain expressing AtTIR1(F79G), we found that animals exhibited only slightly reduced brood sizes, consistent with a dramatically reduction of ligand-independent activity for this TIR1 variant (Figure 4A). In addition, AMA-1::AID*::GFP expression levels in AtTIR1(F79G) expressing animals were similar to those observed in wild type (Figure 4, B and C). Importantly, AMA-1::AID*::GFP expression is extinguished when animals are incubated with 5-Ph-IAA, but not a 1000-fold higher concentration of regular IAA (Figure 4, B and C). AMA-1::AID*::GFP depletion results in a variety of pleiotropic, terminal phenotypes (sterility, slow growth, arrest, and lethality) that resemble phenotypes associated with ama-1 mutations or animals that have been treated with alpha-amanitin, a pol II-specific enzymatic inhibitor (Rogalski and Riddle 1988; Rogalski et al. 1988; Bird and Riddle 1989). These results suggest that the AtTIR1(F79G) allele of AtTIR1 lacks appreciable ligand-independent activity on AID*-tagged substrates while maintaining the ability to program target degradation for multiple substrates.
Exposure of C. elegans larva to high levels of IAA analogs elicits a transcriptional response
Previous experiments have demonstrated that indole (and likely indole derivatives) derived from commensal bacteria extends lifespan (Sonowal et al. 2017). Furthermore, recent reports utilizing the AID system have demonstrated that IAA at high physiological concentrations can also induce physiological responses in C. elegans that modulate a number of developmental and cellular activities. Specifically, continuous exposure of animals to auxin during development significantly extends the lifespan and can confer protection against endoplasmic reticulum (ER) stress (Bhoi et al. 2021; Loose and Ghazi 2021). Auxin antagonizes the negative effects of tunicamycin, a chemical inhibitor of N-linked glycosylation and a known inducer of ER stress, and this protective response requires the activity of the XBP-IRE-1 pathway of the Unfolded Protein Response (Bhoi et al. 2021). These results collectively suggest that high levels of exogenous auxin may activate latent genetic programs that are optimized to ensure that animals can normally survive in diverse environments.
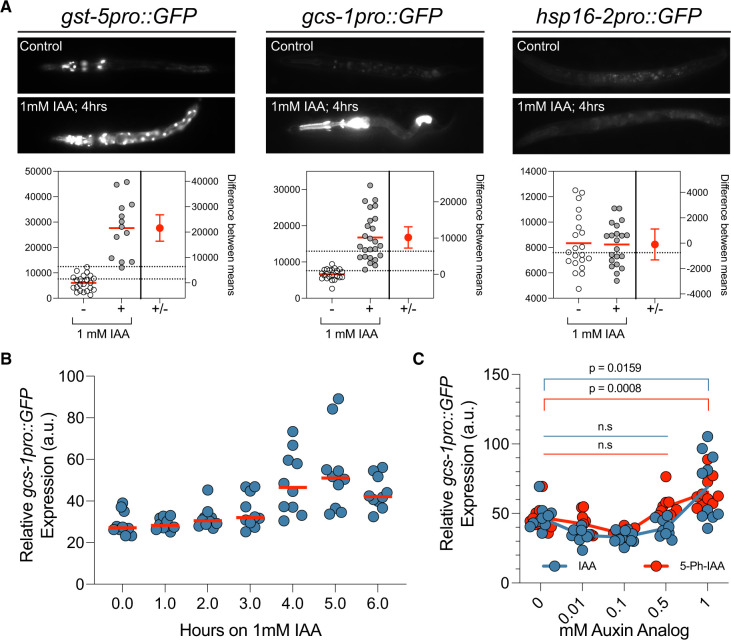
We discovered that auxin exposure induces the expression of multiple transcriptional reporters that are also induced in response to ER stress. These include transcriptional reporters for two glutathione S-transferases (gst-4 and gst-5) and gcs-1, encoding a gamma glutamyl-cysteine synthetase, that are upregulated by SKN-1/Nrf, a transcription factor that orchestrates both oxidative and xenobiotic stress responses in C. elegans after exposure to tunicamycin (Figure 5A; Papp et al. 2012; Glover-Cutter et al. 2013). The induction of this transcriptional response is not a generalized stress response as other transcriptional reporters of environmental stressors are not induced by auxin (e.g., hsp-16::GFP; Figure 5A). Induction of gcs-1pro::GFP is rapid and peaks after 4–5 h of 1-mM auxin exposure (Figure 5B). This rapid and quantifiable response enabled us to both determine if 5-Ph-IAA also induces this regulatory pathway and then to identify the minimal dose of each auxin analog required to activate this gcs-1pro::GFP expression We found that both IAA and 5-ph-IAA induced gcs-1pro::GFP expression, but this induction was not significant over background until the auxin analog reached a concentration of greater than 0.5 mM (Figure 5C). Therefore, an optimal AID system should work at exogenously added auxin analog concentrations that, at a minimum, do not trigger this transcriptional response.
Figure 5.
Elevated levels of auxin analogs induce stress-responsive gene expression. (A) Representative GFP images and estimation plots of the expression levels of each indicated stress-responsive reporter in animals that have been exposed to auxin/IAA for 4 h. For quantification of GFP expression, the relative fluorescence of whole, individual animals was measured (n > 13 for each reporter). (B) Transgenic gcs-1pro::GFP animals were exposed to 1 mM IAA for the indicated times and relative fluorescence of individual animals was calculated as in (A). (C) Quantification of gcs-1pro::GFP expression levels in animals that have been incubated on plates containing the indicated concentrations of IAA or 5-Ph-IAA for 4 h.
The AIDv2 degradation system generates more penetrant loss-of-function phenotypes at lower IAA analog concentrations when compared to the classical TIR1(WT)-IAA pairing
Given that the typical high levels of IAA or 5-Ph-IAA trigger the ectopic expression of gcs-1pro::GFP, we aimed to determine if AtTIR1(F79G) worked more efficiently than TIR1(WT) at lower auxin analog concentrations. Kinetic data comparing the two TIR1-auxin analog pairings in Figure 2C suggested that AtTIR1(F79G) more efficiently depletes the expression of AID*::GFP than AtTIR1(WT) in low auxin analog concentrations. To test this feature, we assayed both TIR1 variants for their ability to generate phenotypes on AID* targets that only display partially penetrant phenotypes with the AIDv1/TIR1(WT) system.
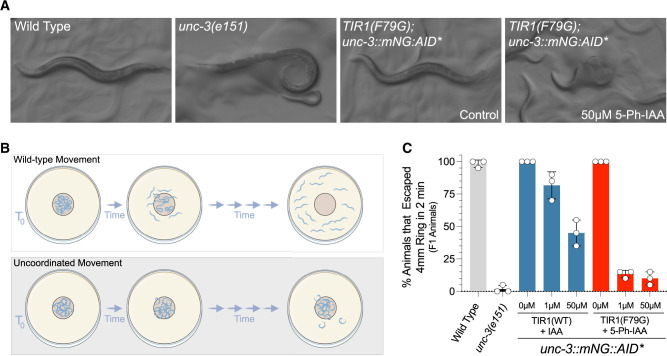
The unc-3 gene, encoding the sole C. elegans ortholog of the Collier/Olf/Ebf (COE) family of transcription factors, functions as a neuronal terminal selector gene that is required to both establish and maintain distinct cholinergic motor neuron cell fates in the ventral nerve chord (Kerk et al. 2017; Feng et al. 2020; Li et al. 2020). Animals harboring nonfunctional alleles of unc-3 display penetrant locomotion defects and exhibit a “coiler” phenotype where the posterior portions of the animal appear paralyzed (Figure 6A; Brenner 1974). To determine if AtTIR1(F79G) can elicit more penetrant phenotypes than AtTIR1(WT) at similar auxin analog concentrations, we scored uncoordinated phenotypes of AtTIR1(F79G) or AtTIR1(WT) parental animals exposed to varied auxin analog concentrations using a modified radial motility assay (Tsalik et al. 2003; Figure 6B). Specifically, late L4/adult F1 animals that had or had not been exposed to auxin analogs were transferred to a center point of a series of concentric circles of a marked NGM plate. Animals were then allowed to move freely for 2 min and the percentage of animals that moved greater than 2 mm from that center point were scored (Figure 6C). Using this assay, 100% of wild type and ∼2% of unc-3(e151) animals moved >2 mm within the recorded time course (Figure 6C). We found that treatment of AtTIR1(WT); unc-3::mNG::AID* animals with IAA induced an uncoordinated phenotype but only at elevated IAA concentrations [approximately 45% of animals were motile enough to travel outside of the assay area (Figure 6C)]. In contrast, 5-Ph-IAA treatment of AtTIR1(F79G) animals led to highly penetrant unc phenotypes in this assay (Figure 6C). Consistent with unc-3 activity being continuously required for normal motility, AtTIR1(F79G) animals treated after hatching also developed an unc phenotype by young adulthood, though the penetrance of these phenotypes was reduced when compared to continuous 5-Ph-IAA treatment (Supplementary Figure S1). We do note that the expressivity of the unc-3 phenotypes elicited by 5-Ph-IAA treatment in AtTIR1(F79G); unc-3::GFP::AID* animals, as measured by the proportion of animals that exhibiting a “coiler” phenotype, is lower than what is observed for strong unc-3 loss-of-function alleles and are consistent with residual unc-3 activity in 5-Ph-IAA treated animals.
Figure 6.
The C.eAIDv2 system generates more penetrant uncoordinated phenotypes than the AtTIR1(WT)/IAA pairing for an AID-tagged allele of UNC-3. (A) The plate phenotypes of wild type, unc-3(e151), and AtTIR1(F79G); unc-3::GFP::AID animals ± 5-Ph-IAA. Loss-of-functional alleles of unc-3 cause animals to exhibit the “coiler” phenotype in which the tails of animals are paralyzed and coiled. (B) Depiction of the modified motility assay in which animals are placed in the center of a defined region of a solid media plate and allowed to freely move for a defined period of time. Animals were scored as uncoordinated (unc) if they failed to move out of the prescribed circle. (C) Quantification of the animals in the modified mobility assay. Each dot represents the data from four separate experiments containing five animals per circle.
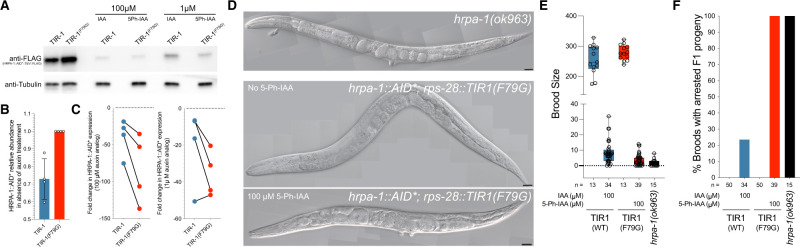
Genes encoding essential, ubiquitously expressed RNA-binding proteins are typically difficult to study using standard genetics or RNAi because maternal contribution and partial rescue of these highly expressed genes confounds analyses, and for RNAi, the half-life of these proteins is typically long. An example of this class of gene is the C. elegans ortholog of HNRNPA2, hrpa-1 (Ryan and Hart 2021; Ryan et al. 2021). Null mutations of hrpa-1, hrpa-1(ok963), can be propagated in balanced strains and homozygous hrpa-1(ok963) animals segregating from these animals exhibit a dramatic reduction in brood size and the resulting F1 progeny of these animals exhibit a fully penetrant larval lethal phenotype (Ryan and Hart 2021). Homozygous animals expressing a CRISPR-engineered, AID*-tagged version of hrpa-1, hrpa-1(zen91), exhibit a wild-type growth phenotype indicating that the AID* tag does not interfere with normal development. Consistent with previous AID*-tagged proteins, HRPA-1::AID*::TEV::FLAG animals expressing AtTIR1(WT) exhibit a reduction in expression of the AID*-tagged target in the absence of IAA, when compared to animals expressing the AtTIR1(F79G) allele (Figure 7, A and B). We next used the hrpa-1::AID*::TEV::FLAG allele to determine if the C.e.AIDv2 system was more efficient at degrading target proteins than the standard AtTIR1(WT) variant. To do this, we hatched transgenic animals on solid NGM media containing either IAA or 5-Ph-IAA (at the indicated concentration) and allowed these animals to grow to a young adult stage of development. Quantitative westerns (n = 4) indicate that both TIR1 variants strongly deplete HRPA-1::AID*::TEV::FLAG. We next calculated the fold reduction of HRPA-1::AID*::TEV::FLAG expression in each case and determined that the C.e.AIDv2 system was reproducibly more effective at depleting the AID*-target protein regardless of auxin analog concentration (Figure 7C). Treatment of AtTIR1(F79G); hrpa-1::AID*::TEV::FLAG with 5-Ph-IAA lead to phenotypes that superficially resemble phenotypes exhibited by homozygous hrpa-1(ok963) animals (Figure 7D). To compare the TIR1 variants at the phenotypic level, we quantified brood size and % of arrested F1 broods. Consistent with a strong depletion of hrpa-1::AID*::TEV::FLAG by both TIR1 alleles, the addition of auxin analogs to the growth media strongly affected brood sizes (Figure 7E). In contrast to the nonpenetrant F1 arrest phenotypes of AtTIR1(WT) animals on IAA-containing media, treatment of AtTIR1(F79G); HRPA-1::AID*::TEV::FLAG animals with 5-Ph-IAA lead to a fully penetrant F1 embryonic or larval arrest phenotype that is identical to those measured in homozygous hrpa-1(ok963) animals (Figure 7F). We conclude that the C.e.AIDv2 version of the auxin degradation system more efficiently depletes AID*-tagged target proteins.
Figure 7.
The C.eAIDv2 system phenocopies hrpa-1(0) phenotypes with an AID-tagged hrpa-1 allele. (A) Quantitative western blots depicting the levels of HRPA-1::AID::TEV::FLAG in transgenic animals exposed to the indicated auxin analogs. For drug treatments, indicated animals were incubated from hatching to young adulthood with the indicated concentration of auxin analogs. (B) Quantification of the relative levels of HRPA-1::AID::TEV::FLAG in animals expressing AtTIR1(WT) or AtTIR1(F79G) on normal solid NGM media (n = 4). (C) Calculation of the relative fold reduction in HRPA-1::AID::TEV::FLAG expression in AtTIR1(WT) or AtTIR1(F79G) expressing animals exposed to auxin or auxin analog. (D) Pictomicrographs of representative young adult hrpa-1(0) animal or AtTIR1(F79G); hrpa-1::AID::TEV::FLAG animals that have been grown on normal media or media containing 100 µM 5-Ph-IAA as described in (A) and (B). (E) Quantification of the brood size of wild type, hrp1-1(0) and animals expressing hrpa-1::AID::TEV::FLAG and one of the two TIR1 variants on or off the indicated auxin analogs. (F) Calculation of the percentage of broods with arrested F1 progeny.
Discussion
The ability to rapidly and conditionally deplete target proteins using the heterologous TIR1 degradation system has dramatically improved the utility of the C. elegans model for dissecting aspects of multicellular development (Zhang et al. 2015). This is in part due to the relatively modular structure of the system which requires a single genetic modification of a target gene to insert the AID* epitope and the heterologous expression of a single E3 adaptor protein, TIR1, that, in response to the addition of an auxin analog, rapidly targets the AID*-tagged protein for degradation. This approach has enabled many gene products that were refractory to genetic manipulation to be studied in detail. In the intervening 6 years from its first description in C. elegans, the number of AID-tagged genes that have already been generated is estimated to be in the hundreds with more than 20 currently available at the C. elegans Genome Stock Center (CGC). For a small number of target genes, two features of the original system still exist. First, the classical allele of TIR1 exhibits a level of activity in the absence of added auxin/IAA ligand that inappropriately reduces target gene expression. As outlined in this manuscript, this ligand-independent activity is substantial and can range from 20% (for AID*::GFP and HRPA-1::AID*) to 65% (in the case of AMA-1::AID*::GFP). Relatively mild reductions of AID*-tagged proteins may not be physiologically important for many AID* targets but for other important developmental genes, this off-target activity makes the classic AID system unusable. Second, we and others find that auxin/IAA concentrations typically used to inactivate AID* target genes in C. elegans induce a physiological response that may occlude or modulate phenotypic observations in as of yet, unanticipated ways (Bhoi et al. 2021; Loose and Ghazi 2021). Our analysis of gcs-1pro::GFP expression indicates that at least one of these responses can occur at levels above 0.5 mM IAA and are independent of TIR1 expression.
In this manuscript, we describe a modification to the TIR1 protein that solves both of these problems. Specifically, mutation of the phenylalanine 79 to a glycine residue prevents ectopic TIR1 activity while maintaining the ability to regulate AID*-tagged gene destruction through the addition of a synthetic IAA analog, 5-Ph-IAA. Further analysis of the activity of the AtTIR1(F79G) variant also indicates that it exhibits a fortuitous increase in relative activity that generates more penetrant loss-of-function phenotypes for several AID*-tagged genes than those elicited with the classical TIR1 (unc-3 and hrpa-1). This enables experiments to be performed at auxin analog concentrations that do not elicit undesired phenotypic consequences. While we were able to achieve near-complete degradation of some target proteins at micromolar 5-Ph-IAA concentrations, optimal 5-Ph-IAA concentrations for individual AID* targets should be determined empirically. The reduction in off-target effects and increase in activity can arise from a number of biochemical features that could be altered by modifying the ligand-binding pocket of TIR1. We hypothesize that the inappropriate degradation of AID*-targets that occurs without the addition of exogenously added auxin may be facilitated by auxin-related indoles, whose origin may be dependent on the E. coli bacterial food source. It is known that commensal bacteria provide indole-related compounds to developing larva and these indoles induce phenotypic changes (Sonowal et al. 2017). Furthermore, we favor this hypothesis because we have noted that several sensitive AID*-tagged genes exhibit variably penetrant phenotypes that track proportionally with the age of the bacterial lawn used to culture animals. By altering the binding pocket of TIR1, the TIR1(F79G) variant may no longer be able to bind these food-derived indoles. The TIR1(F79G) variant would then be activated exclusively by the engineered 5-Ph-IAA ligand leading to enhanced experimental efficacy and tractability.
An important feature of the C.e.AIDv2 system described here is that previously existing AID*-tagged genes remain targetable with this system. This feature can be exploited in two ways. Using the CRISPR guide and repair template we used to generate the initial F79G variant, any other single copy TIR1(WT) transgene derived from the original TIR1 sequences described in Zhang et al. can be easily engineered to express the 5-Ph-IAA inducible variant, including the recent array of tissue-specific drivers described in Ashley et al. (2021) (Figure 2A; Zhang et al. 2015; Ashley et al. 2021; Vo et al. 2021). Alternatively, we have engineered the ubiquitously expressed rps-28pro::TIR1(F79G) allele to contain unique, CRISPR targetable sites that flank the rps-28 promoter sequences that can be utilized for genomic engineering and promoter preplacement. In this case, alternative promoters can easily be exchanged by HDR (Supplementary Figure S3). This method has the added advantage in that the TIR1(F79G) allele expressed from the single copy cshIs140 allele also expresses a highly visible mCherry::HIS-11 reporter, whose ubiquitous expression will change to the expression pattern programed by any recombinant promoter sequences.
Finally, this engineered system is ripe for further enhancement at the molecular/genetic and chemical levels. In other heterologous systems, TIR1 has been modified to contain a nuclear localization sequence that targets TIR1 activity to the nucleus. This modification increases the ability of TIR1 to degrade several, high abundance nuclear proteins (Kanke et al. 2011). Fusion of an Skp1 ortholog, a component of the SCF complex, to TIR1 also improves degradation kinetics and loss-of-function phenotypes of AID*-tagged target genes that are refractory to TIR1 alone suggesting that these additions may also improve the TIR1(F79G) activity in C. elegans. Finally, 5-Ph-IAA does not appear to penetrate the eggshell. Previously described chemical modifications of IAA increase have been demonstrated to alleviate this limitation, enabling more sophisticated temporal studies of developmental processes to be addressed. It seems likely that similar modifications to 5-Ph-IAA may improve the activity of the C.e.AIDv2 system.
Data availability
Strains in Supplementary Table S1 are available through the Caenorhabditis Genetics Center. Other strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplementary material is available at GENETICS online.
Supplementary Material
Acknowledgments
We would like to thank members of the Hammell and Matus laboratories for the critical review of this manuscript. We received strains from Oliver Hobert and from the Caenorhabditis Genetics Center (CGC), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Funding
D.Q.M. is funded by the NIH NIGMS (R01GM121597) and the Damon Runyon Cancer Research Foundation (DRR-47–17). A.Y.Z. is supported by the NIH NIGMS (R35GM124828). M.A.Q.M. was supported by an NIH NIGMS Diversity Supplement (R01GM121597) and is currently funded by the NIH NCI (F30CA257383). T.N.M.-K. is supported by the NIH NICHD (F31HD100091). F.E.Q.M. is supported by an NIH NIGMS Diversity Supplement (R01GM121597). J.D.W. is supported by the NIH NIGMS (R01GM138701). S.E. and A.M. were supported by NIH NIGMS (R35 GM130311). Cold Spring Harbor Laboratory, the Rita Allen Foundation, The Maria Bainer Trust, and NIH NIGMS (R01GM117406) support C.M.H.
Conflicts of interest
The authors declare that there is no conflict of interest.
Literature cited
- Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, et al. 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 198:837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley GE, Duong T, Levenson MT, Martinez MAQ, Johnson LC, et al. 2021. An expanded auxin-inducible degron toolkit for Caenorhabditis elegans. Genetics. 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoi A, Palladino F, Fabrizio P.. 2021. Auxin confers protection against ER stress in Caenorhabditis elegans. Biol Open. 10:bio057992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird DM, Riddle DL.. 1989. Molecular cloning and sequencing of ama-1, the gene encoding the largest subunit of Caenorhabditis elegans RNA polymerase II. Mol Cell Biol. 9:4119–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MW, Morton JJ, Carroll D, Jorgensen EM.. 2008. Gene activation using FLP recombinase in C. elegans. PLoS Genet. 4:e1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M.. 2005. The F-box protein TIR1 is an auxin receptor. Nature. 435:441–445. [DOI] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, Goldstein B.. 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods. 10:1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokshin GA, Ghanta KS, Piscopo KM, Mello CC.. 2018. Robust genome editing with short single-stranded and long, partially single-stranded DNA donors in Caenorhabditis elegans. Genetics. 210:781–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong T, Rasmussen NR, Ballato E, Mote FS, Reiner DJ.. 2020. The Rheb-TORC1 signaling axis functions as a developmental checkpoint. Development. 147:dev181727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Li Y, Dao P, Aburas J, Islam P, et al. 2020. A terminal selector prevents a Hox transcriptional switch to safeguard motor neuron identity throughout life. Elife. 99:e50065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firnhaber C, Hammarlund M.. 2013. Neuron-specific feeding RNAi in C. elegans and its use in a screen for essential genes required for GABA neuron function. PLoS Genet. 9:e1003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter KM, Lin S, Blackwell TK.. 2013. Integration of the unfolded protein and oxidative stress responses through SKN-1/Nrf. PLoS Genet. 9:e1003701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, et al. 1999. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13:1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Fachinetti D, Han JS, Cleveland DW.. 2012. Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc Natl Acad Sci U S A. 109: E3350–E3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanke M, Nishimura K, Kanemaki M, Kakimoto T, Takahashi TS, et al. 2011. Auxin-inducible protein depletion system in fission yeast. BMC Cell Biol. 12:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O.. 2005. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 435:446–451. [DOI] [PubMed] [Google Scholar]
- Kerk SY, Kratsios P, Hart M, Mourao R, Hobert O.. 2017. Diversification of C. elegans motor neuron identity via selective effector gene repression. Neuron. 93:80–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrbach NJ, Armisen J, Lightfoot HL, Murfitt KJ, Bugaut A, et al. 2009. LIN-28 and the poly(U) polymerase PUP-2 regulate let-7 microRNA processing in Caenorhabditis elegans. Nat Struct Mol Biol. 16:1016–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Osuma A, Correa E, Okebalama MA, Dao P, et al. 2020. Establishment and maintenance of motor neuron identity via temporal modularity in terminal selector function. Elife. 9:e59464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loose JA, Ghazi A.. 2021. Auxin treatment increases lifespan in Caenorhabditis elegans. Biol Open. 10: 1242/bio.058703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez MAQ, Kinney BA, Medwig-Kinney TN, Ashley G, Ragle JM, et al. 2020. Rapid degradation of Caenorhabditis elegans proteins at single-cell resolution with a synthetic auxin. G3 (Bethesda). 10:267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska M, Ulrich HD.. 2013. An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast. 30:341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V.. 1997. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 88:637–646. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M.. 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 6:917–922. [DOI] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Seydoux G.. 2017. Precision genome editing using CRISPR-Cas9 and linear repair templates in C. elegans. Methods. 121–122:86–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp D, Csermely P, Sőti C.. 2012. A role for SKN-1/Nrf in pathogen resistance and immunosenescence in Caenorhabditis elegans. PLoS Pathog. 8:e1002673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel T, Hobert O.. 2017. Coordinated control of terminal differentiation and restriction of cellular plasticity. ELife. 6:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, Inoue M, Hikita T, Köppen M, Hardin JD, et al. 2007. Establishment of a tissue-specific RNAi system in C. elegans. Gene. 400:166–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski TM, Bullerjahn AM, Riddle DL.. 1988. Lethal and amanitin-resistance mutations in the Caenorhabditis elegans ama-1 and ama-2 genes. Genetics. 120:409–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski TM, Riddle DL.. 1988. A Caenorhabditis elegans RNA polymerase II gene, ama-1 IV, and nearby essential genes. Genetics. 118:61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Moss EG.. 2013. Developmental transitions in C. elegans larval stages. Curr Top Dev Biol. 105:153–180. [DOI] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, et al. 1998. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 12:198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan VH, Hart AC.. 2021. sym-2 loss-of-function causes glutamatergic neurodegeneration after oxidative stress. MicroPubl Biol. 2021. doi:10.17912/micropub.biology.000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan VH, Perdikari TM, Naik MT, Saueressig CF, Lins J, et al. 2021. Tyrosine phosphorylation regulates hnRNPA2 granule protein partitioning and reduces neurodegeneration. EMBO J. 40:e105001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods. 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E, Leyva-Diaz E, De La Cruz E, Hobert O.. 2018. BRN3-type POU homeobox genes maintain the identity of mature postmitotic neurons in nematodes and mice. Curr Biol. 28:2813–2823.e2. [DOI] [PubMed] [Google Scholar]
- Sonowal R, Swimm A, Sahoo A, Luo L, Matsunaga Y, et al. 2017. Indoles from commensal bacteria extend healthspan. Proc Natl Acad Sci U S A. 114: E7506–E7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, et al. 2007. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 446:640–645. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Ruvkun G.. 1998. An insulin-like signaling pathway affects both longevity and reproduction in Caenorhabditis elegans. Genetics. 148:703–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsalik EL, Niacaris T, Wenick AS, Pau K, Avery L, et al. 2003. LIM homeobox gene-dependent expression of biogenic amine receptors in restricted regions of the C. elegans nervous system. Dev Biol. 263:81–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Takahashi K, Iwasaki R, Yamada R, Yoshimura M, et al. 2018. Chemical hijacking of auxin signaling with an engineered auxin-TIR1 pair. Nat Chem Biol. 14:299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wynsberghe PM, Kai ZS, Massirer KB, Burton VH, Yeo GW, et al. 2011. LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat Struct Mol Biol. 18:302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo AA, Levenson MT, Ragle JM, Ward JD.. 2021. Efficient generation of a single-copy eft-3p::TIR1::F2A:: BFP::AID::NLS allele in the C. elegans ttTi5605 insertion site through recombination-mediated cassette exchange. MicroPubl Biol 2021. doi:10.17912/micropub.biology.000425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voutev R, Hubbard EJ.. 2008. A "FLP-Out" system for controlled gene expression in Caenorhabditis elegans. Genetics. 180:103–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesbolatova A, Saito Y, Kitamoto N, Makino-Itou H, Ajima R, et al. 2020. The auxin-inducible degron 2 technology provides sharp degradation control in yeast, mammalian cells, and mice. Nat Commun. 11:5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ward JD, Cheng Z, Dernburg AF.. 2015. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development. 142:4374–4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains in Supplementary Table S1 are available through the Caenorhabditis Genetics Center. Other strains and plasmids are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables.
Supplementary material is available at GENETICS online.