Abstract
L1CAMs are immunoglobulin cell adhesion molecules that function in nervous system development and function. Besides being associated with autism and schizophrenia spectrum disorders, impaired L1CAM function also underlies the X-linked L1 syndrome, which encompasses a group of neurological conditions, including spastic paraplegia and congenital hydrocephalus. Studies on vertebrate and invertebrate L1CAMs established conserved roles that include axon guidance, dendrite morphogenesis, synapse development, and maintenance of neural architecture. We previously identified a genetic interaction between the Caenorhabditis elegans L1CAM encoded by the sax-7 gene and RAB-3, a GTPase that functions in synaptic neurotransmission; rab-3; sax-7 mutant animals exhibit synthetic locomotion abnormalities and neuronal dysfunction. Here, we show that this synergism also occurs when loss of SAX-7 is combined with mutants of other genes encoding key players of the synaptic vesicle (SV) cycle. In contrast, sax-7 does not interact with genes that function in synaptogenesis. These findings suggest a postdevelopmental role for sax-7 in the regulation of synaptic activity. To assess this possibility, we conducted electrophysiological recordings and ultrastructural analyses at neuromuscular junctions; these analyses did not reveal obvious synaptic abnormalities. Lastly, based on a forward genetic screen for suppressors of the rab-3; sax-7 synthetic phenotypes, we determined that mutants in the ERK Mitogen-activated Protein Kinase (MAPK) pathway can suppress the rab-3; sax-7 locomotion defects. Moreover, we established that Erk signaling acts in a subset of cholinergic neurons in the head to promote coordinated locomotion. In combination, these results suggest a modulatory role for Erk MAPK in L1CAM-dependent locomotion in C. elegans.
Keywords: sax-7 L1CAM, MAPK, KSR-1, synaptic regulation, C. elegans
Introduction
The immunoglobulin superfamily of cell adhesion molecules (IgCAMs) is essential in the development and function of the nervous system, playing roles that range from axon guidance and fasciculation to synapse formation and regulation of synaptic activity. Single-pass transmembrane L1CAMs are a well-characterized subfamily of neuronal IgCAMs that are highly conserved in vertebrates, Drosophila, and Caenorhabditis elegans. Studies have established conserved L1CAM roles in axon guidance, myelination, and fasciculation, as well as dendrite morphogenesis and synaptogenesis (Chen and Zhou 2010; Hortsch et al. 2014; Sundararajan et al. 2019). Consistent with these neurodevelopmental roles, mutations in human L1CAM-encoding genes, L1, NrCAM, CHL1, and neurofascin, are strongly implicated in a variety of neurological disorders. Moreover, mutations in the L1 gene are directly linked to the congenital X-linked L1 syndrome that encompasses a range of neurological conditions, including hydrocephalus, spastic paraplegia, intellectual disability, and corpus callosum hypoplasia (Fransen et al. 1994, 1995; Vits et al. 1994; Van Camp et al. 1996). Interestingly, the severity or manifestation of these conditions can vary even among family members carrying the same L1 mutation, suggesting the presence of other genetic variants that can interact with L1 to modify disease expressivity or penetrance (Fryns et al. 1991; Jouet et al. 1995; Schrander-Stumpel et al. 1995). This variability, particularly with respect to hydrocephalus, was also observed in independently generated L1 knockout mice (Dahme et al. 1997; Fransen et al. 1998). Consistent with the notion that genetic interactions with L1 underlie disease severity, a subsequent study in mice uncovered genetic modifier loci that influenced L1-associated hydrocephalus (Tapanes-Castillo et al. 2010).
In addition to developmental disorders, both genome-wide association studies and individual patient analyses have also implicated L1CAMs in specific behavioral disorders. NrCAM is associated with autism spectrum disorder and susceptibility to addiction; CHL1 is linked to schizophrenia (Sakurai et al. 2002, 2006; Ishiguro et al. 2006; Tam et al. 2010; Ayalew et al. 2012; Shaltout et al. 2013; Zhong et al. 2015). Similarly, L1, NrCAM, and CHL1 knockout mice exhibit altered social and exploratory behaviors (Fransen et al. 1998; Law et al. 2003; Moy et al. 2009). As most genetic mutations implicated in behavioral disorders have mild effects on behavior individually, it has been challenging to determine how these genetic variants contribute to behavioral disorders. The prevailing hypothesis in the field posits it is the genetic interactions between two or more variants that lead to the manifestation of behavioral conditions (Owen et al. 2016; Wamsley and Geschwind 2020). Identifying such genetic interactions will assist in elucidating the function of the L1CAMs associated genetic networks in behavioral outputs.
We previously identified several distinct genetic interactions with the C. elegans canonical L1CAM encoded by the sax-7 gene (Opperman et al. 2015). Despite established roles in neural architecture maintenance, dendrite morphogenesis and organization, axon branching, and axon-dendrite fasciculation (Wang et al. 2005; Dong et al. 2013; Diaz-Balzac et al. 2015, 2016; Yip and Heiman 2018; Chen et al. 2019), sax-7 null animals do not have obvious locomotory abnormalities. However, in sensitized rab-3 or unc-13 genetic backgrounds, sax-7 null animals exhibit synthetic or synergistic uncoordinated (Unc) locomotion and neuronal dysfunction. Intriguingly, late-onset, transient SAX-7 expression can suppress these phenotypes but cannot rescue the neural architecture maintenance defects also exhibited in sax-7 mutants (Opperman et al. 2015). These findings are consistent with a postdevelopmental role for sax-7 in directing coordinated locomotion. Taken together with the known functions of rab-3 and unc-13 in exocytosis and a role for CHL1 in mammalian synaptic vesicle (SV) recycling (Nonet et al. 1997; Richmond et al. 1999; Leshchyns'ka et al. 2006), these findings suggest sax-7 may have a role in synaptic transmission. In this study, we use genetics, electrophysiology, and image analyses of C. elegans synapses to assess how sax-7 promotes coordinated locomotion. These results together with further genetic analyses of sax-7 function uncovered a role for the ERK Mitogen-Activated Protein Kinase (MAPK) pathway in central cholinergic neurons to promote coordinated locomotion.
Materials and methods
Strains
Caenorhabditis elegans strains, provided by the Caenorhabditis Genetics Center, were grown on nematode growth medium (NGM) plates at 21°C. N2 Bristol served as the wild-type strain (Brenner 1974). The alleles used in this study are listed by linkage groups as follows:
LGI: unc-13(n2813) (Brenner 1974)
LGII: rab-3(js49) (Nonet et al. 1997); syd-1(ju82) (Hallam et al. 2002)
LGIII: mpk-1(ga117) (Lackner et al. 1994)
LGIV: sax-7(eq1) (Wang et al. 2005)
LGV: rpm-1(ju41) (Zhen et al. 2000); snb-1(e1563) (Sandoval et al. 2006)
LGX: unc-10(md1117) (Koushika et al. 2001); ksr-1(ok786) (C. elegans Deletion Mutant Consortium 2012)
The strains generated in this study are as follows:
LH1038: sax-7(eq1); snb-1(e1563)
LH1055: rab-3(js49); sax-7(eq1); ksr-1(ok786)
LH1079: sax-7(eq1); unc-10(md1117)
LH1086: sax-7(eq22 [sax-7::mCherry + loxP::GFP:: loxP])
LH1087: sax-7(eq23 [sax-7::mCherry + loxP])
LH1098: rab-3(js49); sax-7(eq22); tmIs1087 [Pmyo-3::Cre]
LH1102: rab-3(js49); sax-7(eq23)
LH1118: syd-1(ju82); sax-7(eq1)
LH1119: sax-7(eq1); rpm-1(ju41)
LH1132: rab-3(js49); sax-7(eq22); tmIs1028 [Pdpy-7::Cre]
LH1145: rab-3(js49); sax-7(eq22); tmIs778 [Prgef-1::Cre]
LH1190: rab-3(js49); mpk-1(ga117)/hT2; sax-7(eq1)
LH1192: rab-3(js49); sax-7(eq22); eqIs4 [Prab-3::Cre]
LH1200: rab-3(js49); sax-7(eq1); ksr-1(eq7)
LH1217: rab-3(js49); sax-7(eq22)
LH1220: sax-7(eq27 [loxP::sax-7::mCherry + loxP])
LH1221: sax-7(eq28 [sax-7 + loxP])
LH1235: unc-13(n2813); sax-7(eq1); ksr-1(eq7)
LH1298: rab-3(js49); sax-7(eq1); ksr-1(ok786); eqSi1 [Punc-17H::ksr-1]
LH1299: rab-3(js49); sax-7(eq1); ksr-1(ok786); eqSi2 [Punc-17B::ksr-1]
LH1300: rab-3(js49); sax-7(eq1); ksr-1(ok786); eqSi3 [Punc-17::ksr-1]
LH1333: rab-3(js49); sax-7(eq27); ksr-1(ok786); eqEx606 [Punc-17H::Cre]
LH1337: rab-3(js49); sax-7(eq27); eqEx607 [Punc-17H::Cre]
LH1341: rab-3(js49); sax-7(eq1); eqSi13 [Prab-3::rab-3]
LH1342: rab-3(js49); sax-7(eq1); eqSi14 [Punc17H::rab-3]
Extrachromosomal arrays generated:
eqEx594–595: pLC776 (Punc-17::Cre, 50 ng/µl), Pmyo-2::tdTom (1.5 ng/µl)
eqEx606–608: pLC747 (Punc-17H::Cre, 50 ng/µl), Pmyo-2::tdTom (1.5 ng/µl)
Plasmids generated:
pLC739: sgRNA plasmid that was used to generate the conditional sax-7(eq22) knock-in strain via Crispr gene-editing. The sgRNA was generated by cloning DNA oligonucleotides LC1638/39 into pRB1017, a gift from Andrew Fire (Addgene plasmid number 59936; http://n2t.net/addgene:59936; RRID: Addgene_59936) (Arribere et al. 2014).
pLC740: repair plasmid carrying the homology arms that was used to generate the conditional sax-7(eq22) knock-in strain via Crispr gene-editing. 5′ and 3′ homology arms were amplified from genomic worm DNA via PCR using primers LC1634/35 and LC1636/37 and cloned by standard restriction digestion with SpeI and Not1 into the vector carrying mCherry_loxP_myo_neoR as described (Norris et al. 2015).
pLC743: sgRNA plasmid carrying primers LC1671 and LC1672 that is specific to β-lactamase.
pLC755: sgRNA plasmid carrying oligonucleotides LC1778 and LC1779 that is specific to mKate2.
pLC756: sgRNA plasmid carrying oligonucleotides LC1780 and LC1781 that is specific to mKate2.
pLC759: The Prab-3::Cre-recombinase repair plasmid was generated via Gibson assembly of the rab-3 promoter, CRE recombinase and the tbb-2 UTR into plasmid pCFJ150-pDESTttTi5605[R4-R3], a gift from Erik Jorgensen (Addgene plasmid number 19329; http://n2t.net/addgene:19329; RRID: Addgene_19329) (Frøkjaer-Jensen et al. 2008).
pLC763:sgRNA plasmid containing oligonucleotides LC1814 and LC1815 that was used to generate sax-7(eq27).
pLC776: For cholinergic neuron expression of Cre recombinase, Gibson assembly was used to replace the eft-3 promoter of plasmid pDD104 with the unc-17 promoter.
pLC777: For Cre recombinase expression in head cholinergic neurons, Gibson assembly was used to replace the eft-3 promoter of plasmid pDD104 with the unc-17H promoters.
pLC783: The rab-3 expression plasmids were generated via Gibson assembly of the rab-3 cDNA with the PUnc-17H.
pLC784: The rab-3 expression plasmids were generated via Gibson assembly of the rab-3 cDNA with the Prab-3 promoter.
Oligonucleotides used are listed 5′ to 3′:
LC1634 ggtgactagtatcgcaaaacgaaattctcc
LC1635 ggtgactagttcgcttttcatcatcatcgct
LC1636 ggtggcggccgcatccttaacgggctccaaagcc
LC1637 ggtggcggccgccgactcgggaagaaaggag
LC1638 tcttgatgaaaagcgatcattgac
LC1639 aaacgtcaatgatcgcttttcatc
LC1671 tcttgttaatagactggatggagg
LC1672 aaaccctccatccagtctattaac
LC1778 tcttgagtcaacttcccatccaa
LC1779 aaacttggatgggaagttgactc
LC1780 tcttgcacttcaagtgcacctccga
LC1781 aaactcggaggtgcacttgaagtgc
LC1814 tcttgtgaccatgaggaagcacgt
LC1815 aaacacgtgcttcctcatggtcac
LC1837 aatgaatcaaacaacagatttaatttcttattctcatcgttaatccaataacttcgtatagcatacattatacgaagttatacgtgcttcctcatggtcacttttcggtcagcggggtagattcccat
Generation of sax-7(eq22) and sax-7(eq23) alleles
mCherry coding sequence was added to the cytoplasmic tail of sax-7 using CRISPR/Cas9 (Norris et al. 2015). The sgRNA-containing plasmid (pLC739) along with the repair plasmid (pLC740) were injected into C. elegans gonads as described (Norris et al. 2015). Worms containing an integrated Pmyo-2::GFP cassette were identified, and individually picked until homozygous. This strain (eq22) was then injected with the Cre recombinase-expressing plasmid pDD104, a gift from Bob Goldstein (Addgene plasmid number 47551; http://n2t.net/addgene:47551; RRID: Addgene_47551) (Dickinson et al. 2013) to excise the gene-disrupting Pmyo-2::GFP cassette, generating the eq23 allele, which will result in SAX-7::mCherry expression.
Generation of sax-7(eq27) and sax-7(eq28) alleles
A single loxP site was inserted into the promoter region of sax-7(eq23) using dpy-10 co-CRISPR (Arribere et al. 2014) with guide plasmid pLC763 and repair primer LC1837 to generate sax-7(eq27). These animals were then injected with pDD104 for germline Cre-recombinase expression to excise the portion of sax-7 which lies between the loxP sites, resulting in the generation of sax-7(eq28). sax-7(eq27) animals were also crossed with tissue-specific Cre recombinase to conditionally delete sax-7 somatically.
Tissue-specific sax-7 knock-ins
To knock in somatic sax-7 expression in a tissue specific manner, we excised the gene-disrupting cassette in sax-7(eq22) using multicopy integrated arrays expressing Cre recombinase driven by promoters of interest. For neuronal, muscle, and hypodermal expression of sax-7, we used integrated multicopy arrays (tmIs778, tmIs1087, and tmIs1028) expressing Cre recombinase that is driven by the rgef-1, myo-3 or dpy-7 promoters, respectively (Kage-Nakadai et al. 2014). We also used another neuronally expressing Cre transgene we generated: the Prab-3::CRE. To generate this transgene, the pLC759 plasmid (50 ng/µl) was injected along with coinjection marker Psur-5::GFP (50 ng/µl) into eq22 animals to generate transgenic animals carrying extrachromosomal arrays; these arrays were then integrated into the genome using a variation of a published method (Yoshina et al. 2016). This adapted method is as follows: the extrachromosomal arrays are first crossed into CGC62, a strain carrying a single copy insertion of Pmyo-2::mKate2 on LGV. Animals homozygous for Pmyo-2::mKate2 also carrying the extrachromosomal array were then injected with a cocktail containing two sgRNA plasmids specific to mKate2 (pLC755 and pLC756, 50 ng/µl), an sgRNA plasmid specific to β-lactamase (plasmid pLC743, 50 ng/µl), Peft-3::Cas9 (50 ng/µl), and pCFJ90 (2.5 ng/µl). F2 animals with positive GFP expression from the array, but negative for mKate expression were individually picked and checked for Mendelian expression of Psur-5::GFP from the array, which indicates successful targeted transgene integration (eqIs4).
Site-specific single copy transgene insertions
Single-copy insertions of Punc-17::ksr-1::gfp, Punc-17H::ksr-1::gfp, Punc-17B::ksr-1::gfp, Prab-3::rab-3, and Punc-17H::rab-3 were generated using CRISPR/Cas9. Each transgene was inserted into the oxTi365 MoSCI site present in the EG8082 strain (Frøkjær-Jensen et al. 2014), which also contains a mutation in unc-119. To do so, we individually injected MosSCI plasmids with ksr-1 [pBC31, pBC32, and pBC33 (50 ng/µl)] (Coleman et al. 2018) or rab-3 [pLC783 and pLC784 (50 ng/µl)], each of which also encodes a rescuing copy of unc-119. Each of these plasmids was injected together with a cocktail containing 50 ng/µl each of sgRNA-containing vectors pXW7.01 and pXW7.02 (gift of Katya Voronina, University of Montana) to cleave the universal ttTi5605 MosSCI site and 2.5 ng/μl of selection marker pCFJ90 (Pmyo-2::mCherry). Injected animals were incubated at 25°C for approximately 7 days, after which unc-119-rescued, nonfluorescent F2 progeny were individually picked and screened for stable transmission of non-Unc, non-fluorescent progeny.
Locomotion assays
Crawling and radial locomotion assays were performed using L4-staged hermaphrodite animals transferred onto 60 mm NGM plates. After transfer onto plates, animals were given 2 min of recovery time before being recorded for 2 min using a Nikon AZ100 microscope with a 1× PlanApo objective and the Photometrics ES2 camera. The individual movement of worms in the videos was analyzed with NIS-Elements AR Analysis software. The coordinates of individual worms were tracked over a span of 1 min to calculate radial distance and speed. The radial graph was plotted using a custom MatLab script.
Swimming assays were performed using L4 stage hermaphrodites transferred into a depression slide containing 1 ml M9 buffer as described (Miller et al. 1996; Opperman et al. 2015). After 1 min of recovery time, the animals were recorded for 1 min usinga Nikon AZ100 microscope with a 1× PlanApo objective and Photometrics ES2 camera. The number of times an animal thrashed (bending at mid body) was counted manually by examining the video recording.
All locomotion assays were performed with the experimenter blind to the genotype of each strain. To prevent outliers from skewing data, animals immobile for more than 10 s were removed from the data pool. All strains were grown on NGM media with OP50 bacteria at 20°C incubator.
Isolation and mapping of ksr-1(eq7)
Screen for genetic suppressors of the rab-3; sax-7 uncoordinated locomotion. Synchronized L4-staged double mutant animals were incubated in 50 mM ethylmethylsulfonate for 4 h at 20°C as described (Brenner 1974). Gravid F1 animals were placed at one distal end of 100 mm petri dishes with OP50 bacteria seeded on the opposite end. After 24 h, we screened the F2 generation for animals that were able to traverse the plate to the bacterial lawn, indicating improved locomotion. From a screen of 10,000 mutagenized haploid genomes, we isolated four suppressors. We characterized the strongest suppressor,eq7, mapping it to the linkage group X.
Mapping of eq7: eq7 was further mapped using single nucleotide polymorphisms (SNPs) in the Hawaiian strain CB4856 as described (Davis et al. 2005). Cosegregation of SNPs with suppressed or nonsuppressed animals was used to map eq7 to a 250-kb interval on LGX bounded by WBVar00081116 and WBVar00054056. ksr-1 was identified as a candidate gene within this interval; sequencing of the exons of the gene verified eq7 as a nonsense mutation at codon 546. Additional confirmation was obtained via a complementation test with the ksr-1(ok786) deletion allele.
Quantitative analysis of fluorescence microscopy
GABA and cholinergic synapses in motor neurons were examined using the respective SNB-1::GFP transgenes, juIs1, wyIs92, and wdIs20 (Lickteig et al. 2001; Hallam et al. 2002; Klassen and Shen 2007). Young adult animals were mounted on 2% agarose pads and anesthetized using 10 mM levamisole in M9 buffer. Images were acquired with a Zeiss Axio Observer Z1 motorized microscope equipped with a C-ApoCHROMAT 63×/1.20 NA water immersion objective and a QuantEM512SC camera (Photometrics). The light source was an HXP-120 mercury halide lamp and the excitation and emission bands were 450–490 and 500–550 nm, respectively. Plane interval in Z acquisitions were 31 nm, for a voxel size of 254 × 254 × 31 nm. Acquisition software used was ZEN2 (Zeiss). To quantitate punctal fluorescence, images of the dorsal nerve cord were acquired by the posterior gonad bend. The images were then deconvolved with AutoQuantX software and sum slices projection was obtained using ImageJ. The average background intensity value was used to standardize the dorsal cord intensity. Punctal fluorescence values were scored over 100 μm using the Find Maxima Function of ImageJ, n > 10 animals for each strain. All images were taken under the same magnification, binning, bit depth, and exposure time.
Electrophysiology
Postsynaptic recordings were obtained from ventromedial body wall muscles anterior to the vulva from immobilized dissected worms, as previously described (Richmond and Jorgensen 1999). Briefly, spontaneous and electrically evoked synaptic responses were acquired from individual muscle cells, whole-cell voltage-clamped at a holding potential of −60 mV in the presence of a 1-mM Ca2+ extracellular solution, using a HEKA EPC-10 amplifier. Data were analyzed using Synaptosoft and Igor Pro.
Electron microscopy
Samples were prepared using the high-pressure freeze fixation and freeze substitution methods as previously described (Weimer 2006). Approximately 30 young adult hermaphrodites were placed in each specimen chamber containing Escherichia coli and rapidly frozen to −180°C under high pressure (Leica HPM 100). Frozen specimens then underwent freeze substitution (Leica Reichert AFS) during which samples were held at −90°C for 107 h in 0.1% tannic acid and 2% OsO4 in anhydrous acetone. The temperature was then raised at 5°C/h to −20°C, kept at −20°C for 14 h, and then raised by 10°C/h to 20°C. After fixation, samples were infiltrated with 50% Epon/acetone for 4 h, 90% Epon/acetone for 18 h, and 100% Epon for 5 h. Finally, samples were embedded in Epon and incubated for 48 h at 65°C. Ultrathin (40 nm) serial sections were cut using a Leica Ultracut 6 and collected on formvar-covered, carbon-coated copper grids (EMS, FCF2010-Cu). Poststaining was performed using 2.5% aqueous uranyl acetate for 4 min, followed by Reynolds lead citrate for 2 min. Images were acquired using a JEOL JEM-1220 transmission electron microscope operating at 80 kV using a Gatan digital camera at a magnification of 100k (1.8587 pixels/nm). Images were collected from the ventral nerve cord region anterior to the vulva for all genotypes. Serial micrographs were manually annotated using NIH ImageJ/Fiji software to quantify the number of plasma membrane docked SVs, and distance of docked SVs from the dense projection (DP) for each synaptic profile. Specimens were encrypted to ensure unbiased analysis. Cholinergic synapses were identified on the basis of their typical morphology (White et al. 1986). A synapse was defined as a series of sections (profiles) containing a DP as well as two flanking sections on both sides without DPs. SVs were identified as spherical, light gray structures with an average diameter of ∼30 nm. A docked SV was defined as an SV whose membrane was morphologically contacting the plasma membrane.
Results
sax-7 interacts with SV cycle genes, resulting in a coiling behavior
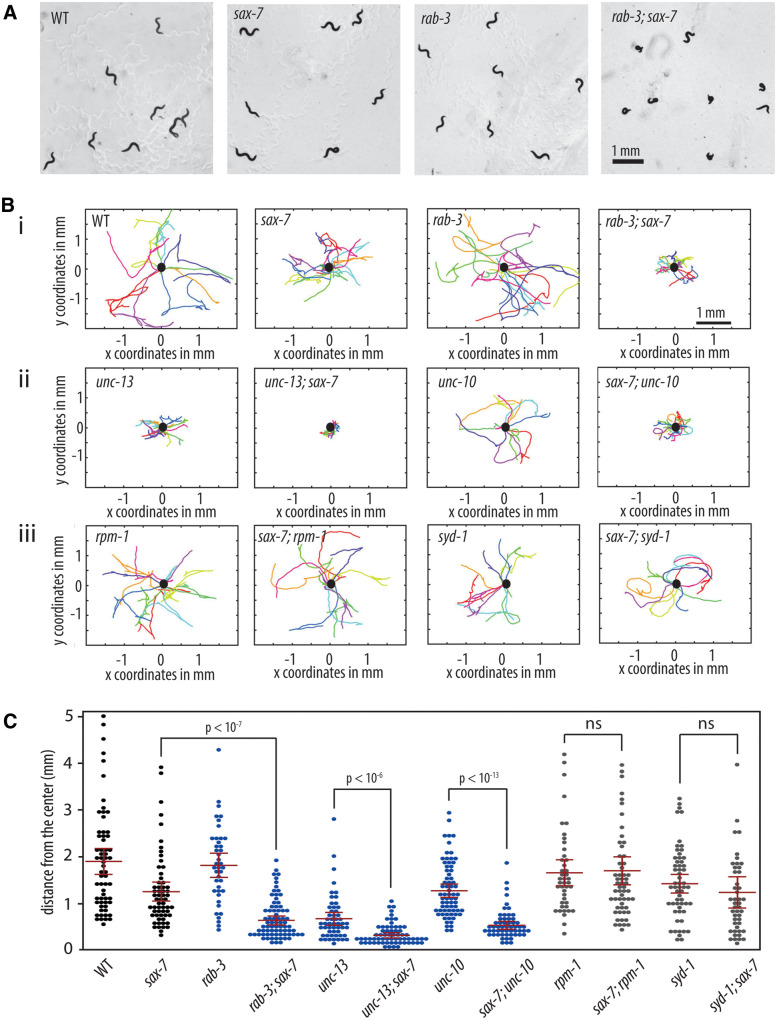
We previously identified a genetic interaction between sax-7 and rab-3, which encodes a monomeric G-protein that is associated with SVs in its GTP-bound state (Opperman et al. 2015). rab-3; sax-7 double mutant animals display synthetic uncoordinated (Unc), loopy locomotion with a tendency to coil that is not observed in either rab-3 or sax-7 single null animals (Figure 1A; Supplementary Video S1). Although not paralyzed, rab-3; sax-7 double mutants tend to meander locally, exhibiting apparent nonvectorial locomotion. As a result, rab-3; sax-7 mutant animals show severely limited radial displacement, in comparison to either rab-3 or sax-7 single mutant animals (Figure 1, Bi and C).
Figure 1.
sax-7 interacts with genes that function in the SV cycle but not in synaptogenesis. (A) rab-3; sax-7 mutant animals exhibit synthetic Unc locomotion with a tendency to coil that is not observed in either rab-3 or sax-7 single mutant animals. Here we used the sax-7(eq1) allele. (B) Graphs tracing the movements of 10 random animals per strain over a span of 1 min. Each colored line represents the tracks of an individual animal with the point of origin marked as a black circle in the center (0,0 coordinate). These graphs reveal that sax-7 interacts with SV cycle genes, (i) rab-3 as well as (ii) unc-13 and unc-10, resulting in a synergistic reduction in the ability to disperse. In contrast, sax-7 does not interact with genes that function in synaptogenesis, (iii) rpm-1 and syd-1. (C) The ability to disperse, quantified as the radial distance traveled by each animal over a span of 1 min, is illustrated in a scatter plot where each point is a data point for a single animal; the mean and the 95% confidence intervals are marked in red. The blue plots are data points for mutant strains of SV cycle genes while gray plots show data points for mutant strains of synaptogenesis genes. n = 50–75, P-values are calculated using one-way ANOVA with Bonferroni’s post hoc test and compared between the double mutant and the single mutant exhibiting the stronger phenotype; n.s., not significant.
The RAB-3 protein promotes neurotransmission, acting in the SV cycle with roles that include the trafficking and docking of SVs to the active zone at synapses (Nonet et al. 1997; Gracheva et al. 2008). The genetic interaction with rab-3 suggests that sax-7 may interact with additional genes that function in SV exocytosis. While the majority of SV cycle mutants exhibit severely Unc phenotypes that preclude enhancement, we were nonetheless able to test for an interaction with unc-10, which encodes for the RAB-3-interacting molecule that recruits SVs to the active zone via RAB-3::GTP interactions (Koushika et al. 2001; Gracheva et al. 2008). We also confirmed a previously identified interaction with unc-13, which functions in SV docking and priming at the active zone (Richmond et al. 1999; Weimer et al. 2006; Opperman et al. 2015). We observed synthetic coiling and poor dispersal behaviors in sax-7 animals homozygous for either unc-13 hypomorphic or unc-10 null alleles (Figure 1, Bii and C). These results are consistent with the notion that sax-7 interacts with SV cycle genes. In contrast, we did not observe an interaction between sax-7 and genes with roles in synaptogenesis, such as rpm-1 and syd-1 (Schaefer et al. 2000; Zhen et al. 2000; Hallam et al. 2002; Patel et al. 2006; Cherra and Jin 2015). Loss of either rpm-1 or syd-1 in sax-7 null animals does not produce the coiling and poor dispersal behavior exhibited by rab-3; sax-7 mutant animals (Figure 1, Biii and C). These results indicate that the sax-7 interaction with SV cycle genes is specific.
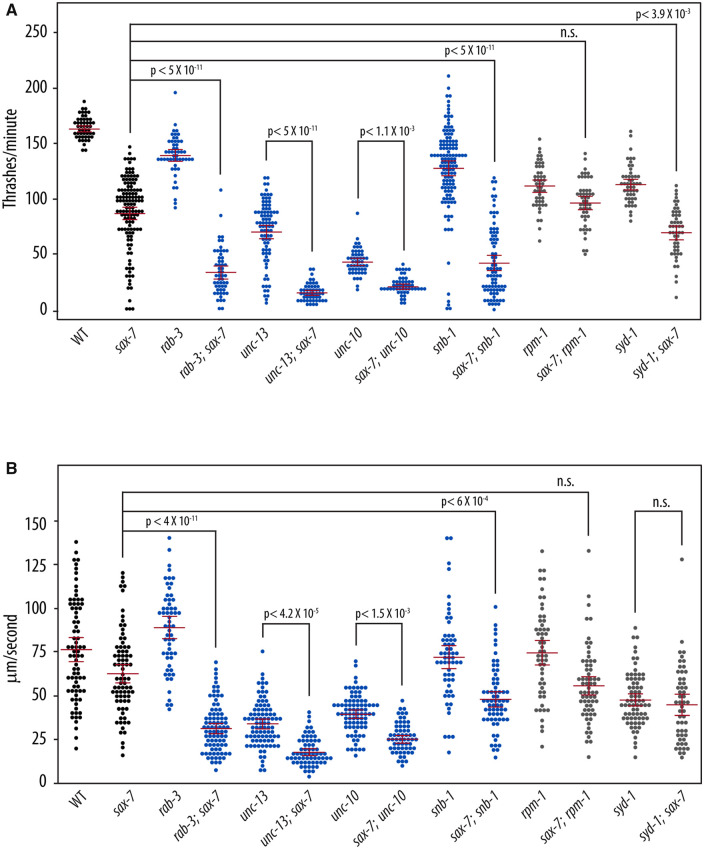
The abnormal locomotory behaviors in double mutants of sax-7 and SV cycle genes suggest underlying neuronal dysfunction. To test this notion, we measured the ability of rab-3; sax-7 mutant animals to swim in liquid and crawl on agar medium. The ability for animals to swim was quantified as the number of times an animal thrashes in liquid per minute. While wild-type animals have an average swim rate of 163 thrashes/min, the mean swim rate of rab-3; sax-7 mutant animals is 33 thrashes/min, which is only 20% of the wild-type rate (Figure 2A). By comparison, the average swim rates of sax-7 and rab-3 single mutants are 53% and 85% that of wild-type, respectively. rab-3; sax-7 double mutants also show a synergistic decrease in crawl rates (Figure 2B). At 35 μm/s, rab-3; sax-7 mutant animals crawl at an average rate that is 44% of the wild-type rate (80.5 μm/s) whereas the average crawl rates of sax-7 and rab-3 single mutants are 82% and 116% of the wild-type rate, respectively. Synergistic reductions in swim and crawl rates are also observed in sax-7 double mutant animals of unc-13, unc-10, and snb-1, which encodes synaptobrevin (Nonet et al. 1998). In contrast, sax-7 double mutants with synaptogenesis genes, rpm-1 and syd-1, exhibit modest, if any, synergism (Figure 2). These results are not only consistent with neuronal dysfunction contributing to the synthetic Unc and locomotion behaviors, but also underscore the specificity of the sax-7 genetic interaction with SV cycle genes.
Figure 2.
sax-7 double mutants with SV cycle genes display synergistic neuronal dysfunction. (A) sax-7 interacts with SV cycle but not synaptogenesis genes, showing synergistic reduction in swim rates (number of thrashes per minute) and (B) crawl rates (μm/s). Here we used the sax-7(eq1) allele. The blue plots are data points for mutant strains of SV cycle genes while gray plots show data points for mutant strains of synaptogenesis genes. The mean and the 95% confidence intervals are marked in red. n = 50 − 75, P-values are calculated using one-way ANOVA with Bonferroni’s post hoc test and compared between the double mutant and the single mutant exhibiting the stronger phenotype; n.s., not significant.
Sax-7 is required in the nervous system to promote coordinated locomotion
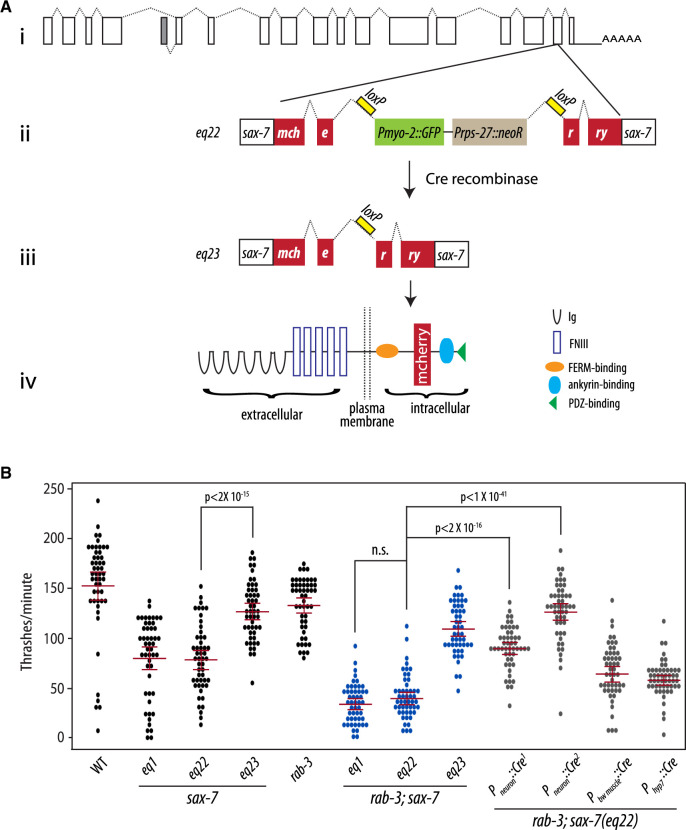
sax-7 is expressed both in the nervous system and non-neuronal tissues, including body-wall muscles and the hypodermis (Chen et al. 2001). Based on the locomotory deficiencies observed in sax-7 double mutants with SV cycle genes, we predicted that sax-7 expression in the nervous system would suppress the synthetic phenotypes exhibited by rab-3; sax-7 double mutant animals. However, non-neuronal sax-7 expression was shown previously to impact the maintenance of neural architecture, dendrite morphogenesis, and axon branching (Wang et al. 2005; Dong et al. 2013; Salzberg et al. 2013; Diaz-Balzac et al. 2016; Zhu et al. 2017). To determine where sax-7 is required for coordinated locomotion, we first established a conditional sax-7 knock-in system using the Crispr-Cas9-engineered sax-7(eq22) allele. eq22 is an in-frame DNA insertion in the penultimate exon of the sax-7 gene consisting of coding sequence for mCherry that is interrupted by a gene-disrupting cassette comprising Pmyo-3::GFP::unc-54 3′UTR and Prps-27::neoR::unc-54 3′UTR sequences (Figure 3A; Norris et al. 2015). As such, we expect disrupted sax-7 function in sax-7(eq22) animals, which can be identified as neomycin-resistant animals that express GFP in pharyngeal muscles. Indeed, sax-7(eq22) animals resemble sax-7(eq1) null animals, showing comparable reduced swim rates of 89.6 thrashes/min (Figure 3B). Importantly, eq22 also genetically interacts with rab-3, producing synthetic coiling and Unc locomotion as well as a synergistically low swim rate similar to that observed in rab-3; sax-7(eq1) animals (Figure 3B). These results are consistent with eq22 disrupting sax-7 function and further demonstrate that the sax-7 genetic interaction we observed with rab-3 is not allele-specific.
Figure 3.
Neuronal expression of sax-7 rescues rab-3; sax-7 uncoordinated locomotion and neuronal dysfunction. (A) A schematic of the conditional sax-7 knock-in allele, eq22. (i) Gene structure of the sax-7 locus, with boxes representing exons and dotted lines representing introns; the gray box represents an exon used in an alternatively spliced isoform of sax-7 (Chen et al. 2001). eq22 is an in-frame insertion into the penultimate sax-7 exon (ii). The insertion contains mCherry sequence and a gene-disrupting cassette (Pmyo-2::GFP and Prps-27::neoR) that interrupts the mCherry reading frame. This gene-disrupting cassette, which is flanked by loxP sites located in intronic sequences, can be excised out with tissue-specific Cre-recombinase expression (iii) to restore sax-7 expression as (iv) a full-length SAX-7::mCherry fusion protein. Cre-recombinase expression in the germline was used to generate the eq23 allele. (B) Quantitation of thrash rates reveal that eq22 resembles the eq1 null allele and similarly interacts with rab-3 for a synergistically reduced swim rate; in contrast, eq23 does not interact with rab-3. Conditional knock-in of sax-7 in neurons with pan neuronally expressed Cre-recombinase dramatically rescues the low rab-3; sax-7(eq22) thrash rate; in contrast, body-wall muscle or hypodermal knock-in results in modest rescue. Pan neuronally expressed Cre-recombinase, Pneuron::Cre1 and Pneuron::Cre2, was produced using the eqIs4 and tmIs778 transgenes, respectively. The red lines show the mean and the 95% confidence interval. n = 50–75, P-values are shown; n.s., not significant, one-way ANOVA with Bonferroni’s post hoc test.
In addition to the gene-disrupting cassette, the eq22 DNA insertion also contains loxP sites flanking the gene-disrupting cassette (Figure 3Aii). The loxP sites provide the ability to direct the excision of the gene-disrupting cassette with tissue-specific Cre-recombinase expression (Figure 3Aiii) for a targeted restoration of SAX-7 as a mCherry fusion protein (Figure 3Aiv). Using germline Cre-recombinase expression, we successfully excised the gene-disrupting cassette in the sax-7(eq22) germline to generate the eq23 allele. sax-7(eq23) animals are not neomycin-resistant and do not express GFP in the pharynx. Instead, they show SAX-7::mCherry expression and localization (Supplementary Figure S2A) similar to that of endogenous SAX-7 (Chen et al. 2001). Importantly, sax-7(eq22) phenotypes are suppressed in sax-7(eq23) animals (Figure 3B). For example, sax-7(eq23) animals display an increased average swim rate of 144.75 thrashes/min when compared with the mean sax-7(eq22) swim rate of 89.6 thrashes/min. As expected, rab-3; sax-7(eq23) animals do not display Unc locomotion or the synergistically low swim rate that are exhibited in both rab-3; sax-7(eq1) and rab-3; sax-7(eq22) animals. These results indicate that the SAX-7::mCherry protein expressed in sax-7(eq23) animals is functional.
To assess whether SAX-7 is required in the nervous system for proper locomotion in a rab-3 background, we introduced the eqIs4 transgene, which directs neuronal Cre-recombinase expression with the rab-3 promoter, into rab-3; sax-7(eq22) animals. In rab-3; sax-7(eq22); eqIs4 animals, neuronal sax-7 expression is recovered with mCherry fluorescence present only in the nervous system (Supplementary Figure S2B). Importantly, eqIs4 suppresses the rab-3; sax-7(eq22) Unc behavior and low swim rates, as does another neuronal-expressing Cre transgene, tmIs778, which directs Cre-recombinase expression with the rgef-1 promoter (Figure 3B; Kage-Nakadai et al. 2014). The rab-3; sax-7(eq22) average swim rate of 39.5 thrashes/min is restored to a mean rate of 89.7 thrashes/min by eqIs4 and 126 thrashes/min by tmIs778. In contrast, the low rab-3; sax-7(eq22) thrash rate is not rescued with the recovery of SAX-7 expression in either body-wall muscles (Supplementary Figure S2C) or hypodermis (not shown) using the tmIs1087 and tmIs1028 transgenes, respectively (Figure 3; Kage-Nakadai et al. 2014). These results demonstrate a requirement for SAX-7 in the nervous system for coordinated locomotion in a rab-3 null background.
A role for Erk signaling in locomotory behavior and function
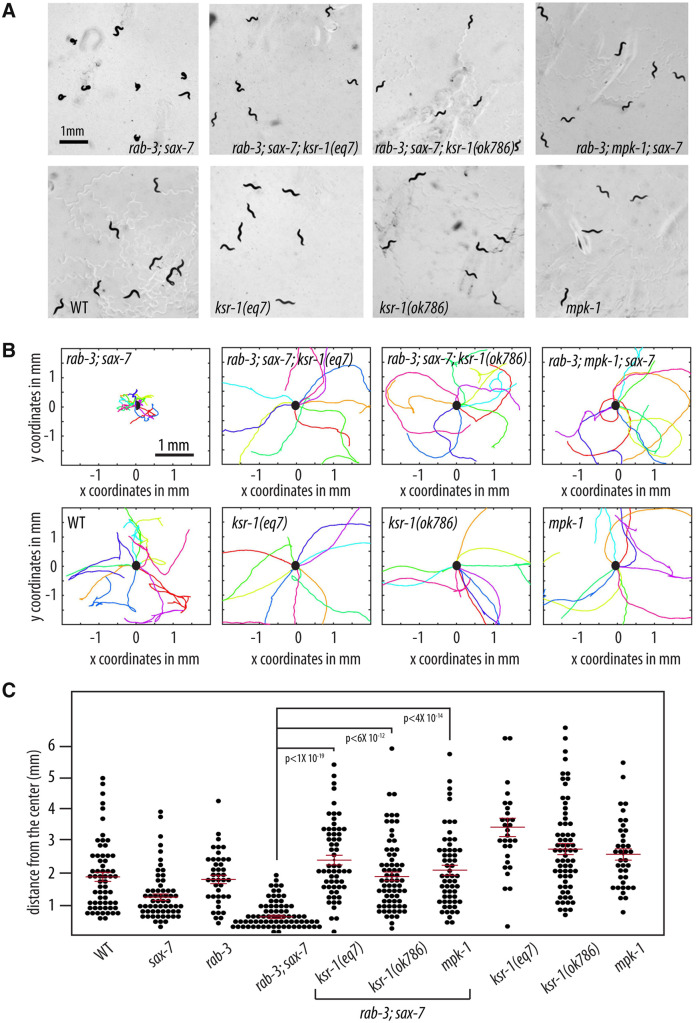
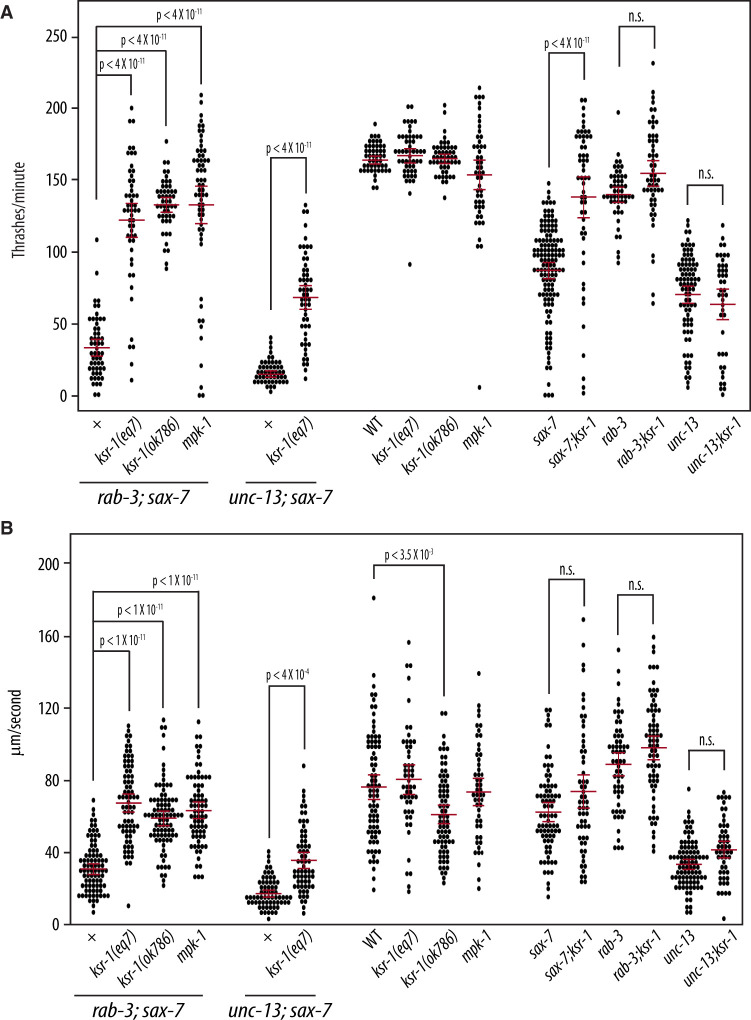
We performed a forward genetic screen for mutations that could suppress the Unc and coiling behavior of rab-3; sax-7 mutant animals as an approach to determine the basis of their abnormal locomotion and neuronal function. We isolated a strong suppressor, eq7, a C-to-T transition that converts codon 546 of the ksr-1 gene into a nonsense mutation (see Materials and methods). rab-3(js49); sax-7(eq1); ksr-1(eq7) triple mutant animals display similar locomotory and dispersal behaviors as wild-type animals (Figure 4; Supplementary Video S3A). ksr-1(eq7) also rescues the abnormal swim rates of rab-3; sax-7 mutant animals from 33 to 121 thrashes/min as well as crawl rates from 35 to 67 μm/s (Figure 5). The ksr-1 deletion allele, ok786 (Moerman and Barstead 2008; C. elegans Deletion Mutant Consortium 2012), similarly suppresses rab-3(js49); sax-7(eq1) phenotypes (Figures 4 and 5). These results are consistent with the loss of ksr-1 function suppressing the abnormal locomotion and neuronal defects exhibited by rab-3; sax-7 mutant animals. ksr-1 single mutants do not have apparent locomotion deficiencies, displaying thrash and crawl rates that are similar or modestly reduced when compared with wild-type animals.
Figure 4.
Reducing ERK MAPK signaling suppresses rab-3; sax-7 uncoordinated locomotion. (A) Null alleles of ksr-1 or mpk-1 suppress rab-3; sax-7 uncoordinated locomotion; yet ksr-1 or mpk-1 null animals themselves are not Unc. Here, we used the sax-7(eq1) allele. (B) Graphs tracing the movements of 10 random animals per strain over a span of 1 min. Each colored line represents the tracks of an individual animal with the point of origin marked as a black circle in the center (0,0 coordinate). These graphs reveal that loss of ksr-1 or mpk-1 effectively suppress rab-3; sax-7 reduced ability to disperse. (C) The ability to disperse, quantified as the radial distance traveled by each animal over a span of 1 min, is illustrated in a scatter plot where each point is a data point for a single animal; the mean with the 95% confidence interval is marked in red. n = 50–75, P-values are shown; n.s., not significant, one-way ANOVA with Bonferroni’s post hoc test.
Figure 5.
Reduced MAPK signaling suppresses sax-7-related neuronal dysfunction. Null alleles of ksr-1 and mpk-1 can suppress both rab-3; sax-7 and unc-13; sax-7 synergistically reduced (A) swim and (B) crawl rates; yet ksr-1 or mpk-1 null animals have similar swim and crawl rates as wild-type animals. Loss of ksr-1 can also suppress the reduced swim rate exhibited by sax-7 null animals, but not unc-13 mutants, suggesting that the suppression of rab-3; sax-7 and unc-13; sax-7 neuronal dysfunction is due to ksr-1 impinging on the sax-7 pathway. Here, we used the sax-7(eq1) allele. The mean with the 95% confidence interval is marked in red. n = 50–75; n.s., not significant, one-way ANOVA with Bonferroni’s post hoc test.
ksr-1(eq7) also suppresses the synergistic defects exhibited by unc-13; sax-7 mutant animals, including the low swim and crawl rates (Figure 5). This suppression in the double mutants raises the possibility that ksr-1 may act on the sax-7 pathway, the SV cycle, or function in a third as-yet-unidentified pathway promoting coordinated locomotion and neuronal function. Interestingly, the rescued swim and crawl rates of rab-3; sax-7; ksr-1 and unc-13; sax-7; ksr-1 mutant animals are more similar to those of rab-3 and unc-13 single mutants, respectively (Figure 5), suggesting that the loss of ksr-1 may be impinging on the sax-7 pathway rather than the SV cycle. If this hypothesis is correct, loss of ksr-1 should suppress sax-7 mutant phenotypes but not affect rab-3 or unc-13 mutant phenotypes. This hypothesis was best tested by examining the swim rates rather than crawl rates since the sax-7 phenotype is more robust in liquid than on solid media (Figures 2 and 5). In liquid, the swim rate of 86 thrashes/min observed in sax-7 mutant animals is 51% of wild-type while the swim rate of 154 thrashes/min observed in sax-7; ksr-1 double mutant animals is 94% of the wild-type rate, showing almost complete suppression by loss of KSR-1. By comparison, loss of KSR-1 has a negligible effect on the swim rate exhibited by unc-13 and rab-3 single mutant animals (Figure 5A). This genetic interaction observed between ksr-1 and sax-7 but not unc-13 or rab-3 is consistent with the notion that KSR-1 likely impinges on SAX-7 function.
KSR-1 is a molecule that interacts with core components of the MAPK cascade to facilitate the activation of the MAPK signaling pathway (Sundaram and Han 1995; Sundaram 2013). As both putative loss-of-function ksr-1 alleles suppressed rab-3; sax-7 phenotypes, we predicted that reducing the function of the MAPK, Erk, would similarly suppress the rab-3; sax-7 mutant phenotypes. To test this prediction, we crossed a null allele of Erk, encoded by the mpk-1 gene (Lackner et al. 1994), into rab-3; sax-7 mutant animals. Consistent with our hypothesis, rab-3; mpk-1; sax-7 triple mutant animals display locomotory behavior and ability to swim that are similar to that of wild-type animals (Figures 4 and 5; Supplementary Video S3B), revealing a role for MAPK signaling in regulating locomotion and neuronal function. Interestingly, both ksr-1 and mpk-1 single mutant animals do not display apparent locomotory deficiencies, suggesting that the role the MAPK signaling pathway plays in coordinating locomotion is likely modulatory.
SAX-7 and KSR-1 do not affect SV release at neuromuscular junctions
A delayed response to the paralytic effects of the cholinesterase inhibitor, aldicarb, is often observed in animals with impaired SV exocytosis due to a slower build-up of acetylcholine in the synaptic clefts of neuromuscular junctions, when compared with animals with wild-type SV exocytosis (Miller et al. 1996). Consistent with the role of rab-3 in the trafficking and recruitment of SVs to the active zone, rab-3 mutant animals are mildly resistant to inhibitors of cholinesterase (RIC) (Nonet et al. 1997; Gracheva et al. 2008). Although sax-7 mutant animals are not RIC, loss of SAX-7 synergistically enhances the RIC phenotype in rab-3 animals (Opperman et al. 2015). The rab-3; sax-7 RIC phenotype, taken together with the accompanying Unc behavior and reduced swim and crawl rates, suggests that rab-3; sax-7 mutant animals may have synergistic SV exocytosis defects.
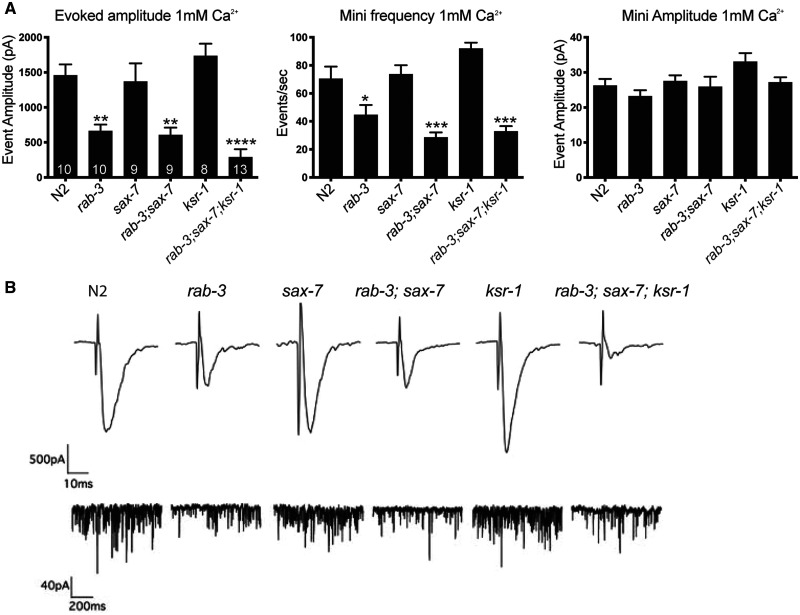
To resolve potential synergistic SV release abnormalities underlying the rab-3; sax-7 phenotypes, we performed electrophysiological analyses of neuromuscular synapses to assess functional consequences in rab-3; sax-7 double when compared with rab-3, sax-7, and ksr-1 single mutant animals as well as the rab-3; sax-7; ksr-1 triple mutant animals. Whole-cell voltage-clamped body recordings from medial ventral body wall muscles were directly assayed before and after electrical stimulation of the ventral nerve cord. First, the average endogenous miniature event amplitude observed in each genotype is similar to that of wild-type, suggesting that postsynaptic muscle receptor function does not account for the phenotypes observed in rab-3; sax-7 animals (Figure 6). This finding is consistent with the inability of SAX-7 expression in muscle to rescue rab-3; sax-7 phenotypes (Figure 3B). Second, we examined the evoked responses in each strain. As expected, rab-3 null animals exhibit reduced evoked responses, consistent with previous studies (Nonet et al. 1997; Gracheva et al. 2008), while sax-7 and ksr-1 mutant animals showed wild-type evoked responses. Surprisingly and contrary to our prediction, the evoked responses in rab-3; sax-7 double mutants were not significantly lower than that of rab-3 single mutants (Figure 6). In fact, there was no significant difference among the evoked responses of rab-3 single, rab-3; sax-7 double, and rab-3; sax-7; ksr-1 triple mutant animals. Similarly, analysis of the endogenous mini frequencies at the NMJ, showed no significant differences between rab-3 single mutants and either the rab-3; sax-7 double or the rab-3, sax-7: ksr-1 triple mutants.
Figure 6.
SAX-7 and KSR-1 do not affect neurotransmitter release in ventral cord cholinergic motor neurons. (A) Whole-cell voltage-clamped body recordings from medial ventral body wall muscles (held at −60 mV in 1 mM Ca2+) of dissected worms. The graphs show the average evoked amplitude, mini frequency, and mini-amplitude at the NMJ of each strain. The number of animals analyzed per genotype is shown within each bar. Data shown as mean ± SEM. (B) Representative evoked traces (top panel) and miniature traces (bottom panel) for genotypes labeled on the top. Here, we used the sax-7(eq1) and ksr-1(ok786) alleles. P-values are calculated using one-way ANOVA with Tukey’s multiple comparison test comparing the different strains to wild-type; *P < 0.05, **P < 0.01, ***P < 0.0001.
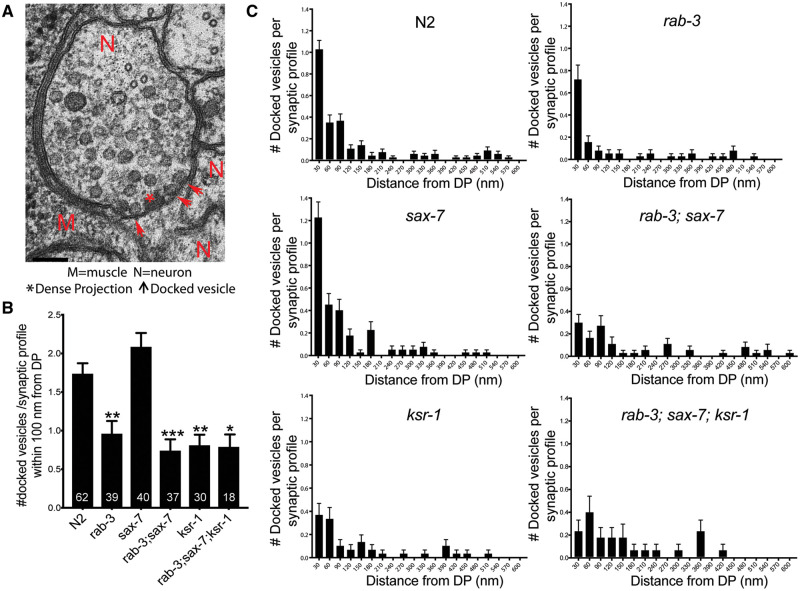
We also examined synapses in rab-3; sax-7 mutant animals at the ultrastructural level using high pressure freeze electron microscopy (Figure 7). As a control, we first compared cholinergic presynaptic terminals between rab-3 mutant and wild-type animals. Consistent with published studies, we observed reduced numbers of docked SVs within 100 nm of the presynaptic density in rab-3 synapses (Gracheva et al. 2008). Consistent with the electrophysiological analyses, the number of docked SVs within the same region were not significantly different in rab-3; sax-7 mutants, while synapses in sax-7 single mutants showed wild-type SV numbers (Figure 7). Moreover, the docking deficit of the rab-3; sax-7; ksr-1 triple mutant animals was not significantly different from rab-3 single and rab-3; sax-7 double mutant animals. Surprisingly, the synapses in ksr-1 mutants also presented a deficit in vesicle docking that is not consistent with the suppression of rab-3; sax-7 mutant phenotypes. These results are consistent with our findings obtained by examining the synapses using light microscopy. Using GFP-tagged synaptobrevin (SNB-1::GFP) as a fluorescence marker for SVs, we determined there was no apparent difference in SNB-1::GFP localization, number, or punctal intensity in GABA and cholinergic synapses in rab-3; sax-7 mutant animals when compared with wild-type animals (Supplementary Figure S4). In contrast, the punctal fluorescence intensity of SNB-1::GFP is increased in the synapses of unc-18 mutant animals, a result reflective of defective SV release; unc-18 encodes for a syntaxin-binding protein necessary for SV exocytosis (Richmond 2005; Ch'ng et al. 2008). Taken together, these data indicate that altered presynaptic function at the ventral cord cholinergic neuromuscular junctions is unlikely a contributing factor for the abnormal locomotory behaviors and neuronal dysfunction observed in rab-3; sax-7 animals, and cannot explain the ability of ksr-1 mutants to suppress these defects. This finding suggests that ksr-1 suppression of the rab-3; sax-7 phenotypes occur elsewhere in the nervous system.
Figure 7.
SAX-7 and KSR-1 do not affect number or distribution of docked SVs in ventral cord cholinergic motor neurons. (A) A representative electron micrograph of a wild type (N2) cholinergic synaptic profile. The presynaptic density is labeled with an asterisk, arrowheads indicate membrane docked SVs, N indicates neurons, and M body wall muscle. (B) Average number of docked SVs per synaptic profile within 100 nm distance from DP or presynaptic density. Number of synaptic profiles analyzed per genotype is mentioned within the bars. (C) Distribution of docked SVs with respect to the DP. Here, we used the sax-7(eq1) and ksr-1(ok786) alleles. Data shown as mean ± SEM, P-values are calculated using one-way ANOVA with Tukey’s multiple comparison test comparing the different strains to wild-type; *P < 0.05, **P < 0.01, ***P < 0.0001
ksr-1 functions in a subset of cholinergic neurons in the head for coordinated locomotion
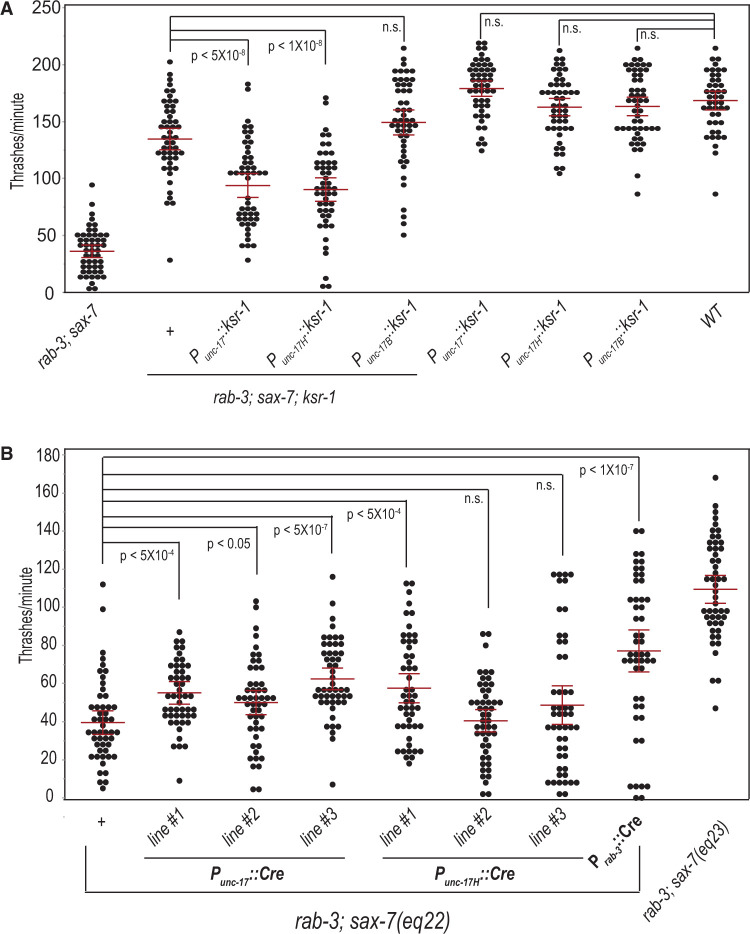
A previous study uncovered a novel neuronal role for ksr-1 (Coleman et al. 2018). Specifically, loss-of-function ksr-1 alleles were isolated in a genetic screen for suppressors of the hyperactive and loopy locomotion exhibited by activated Gq/EGL-30 animals. Moreover, ksr-1 expression restricted to a subset of cholinergic neurons in the head of egl-30; ksr-1 mutant animals was sufficient to reverse this suppression; in contrast, ksr-1 expression in ventral nerve cord cholinergic neurons in the body did not (Coleman et al. 2018).
Because of overlapping loopy locomotion shared by both egl-30 and rab-3; sax-7 animals, we hypothesized that ksr-1 might similarly act in cholinergic neurons to regulate locomotion in rab-3; sax-7 animals. If so, ksr-1 expression in cholinergic neurons should reverse the suppression observed in rab-3; sax-7; ksr-1 triple mutant animals. With the eqSi3 single-copy transgene, we drove KSR-1 expression in cholinergic neurons, using the promoter for the neuronal unc-17 acetylcholine transporter. As predicted, rab-3; sax-7; eqSi3; ksr-1 animals strongly resemble rab-3; sax-7 animals, exhibiting reduced swim rates as well as the Unc, loopy locomotion with a tendency to coil (Figure 8A). This reversal is also observed when ksr-1 expression is limited by the partial unc-17H promoter to cholinergic neurons just in the head, using the eqSi1 single-copy transgene; rab-3; sax-7; eqSi1; ksr-1 animals are Unc, displaying coiling locomotion and reduced swim rates (Figure 8A). In contrast, reversal of the suppression is not observed in rab-3; sax-7; ksr-1 animals with ksr-1 expression restricted by the unc-17B promoter to cholinergic neurons just in the body, using the eqSi2 single copy transgene. Importantly, ksr-1 expression by any of these single-copy transgenes does not impact locomotion or swim rates in wild-type animals (Figure 8A). These results indicate that ksr-1 expression in the head cholinergic neurons is sufficient to promote both coordinated locomotion and neuronal function, and likely accounts for the absence of neuronal dysfunction or abnormality in ventral nerve cord synapses assayed in our electrophysiological and ultrastructural analyses.
Figure 8.
ksr-1 functions in a subset of cholinergic neurons in the head to modulate rab-3; sax-7 neuronal function. (A) The loss of ksr-1 suppresses rab-3; sax-7(eq1) reduced swim rate. This suppression is reversed when ksr-1 is expressed in rab-3; sax-7(eq1); ksr-1(ok786) cholinergic neurons using unc-17 acetylcholine transporter promoter. This reversal is similarly observed when ksr-1 expression is restricted to cholinergic neurons in the head using the partial unc-17H promoter; in contrast, no reversal is observed when ksr-1 expression is confined to body cholinergic motor neurons using the partial unc-17B promoter. Wild-type animals carrying each transgene exhibit normal swim rates and show no apparent locomotion phenotype. (B) To determine whether sax-7 functions in the same neurons as ksr-1, we used the conditional sax-7(eq22) knock-in allele and Punc-17::Cre-recombinase to knock in sax-7 expression in cholinergic neurons. sax-7 expression in all or only head cholinergic neurons show modest or poor suppression of rab-3; sax-7 decreased swim rates. The mean with the 95% confidence interval is marked in red. n = 50–75. n.s., not significant, one-way ANOVA with Bonferroni’s post hoc test.
We next assessed whether sax-7 functions in the same set of neurons as ksr-1 in coordinating locomotion. Using multicopy transgenes on extrachromosomal arrays, we directed Cre-recombinase expression with the unc-17 promoter to restore sax-7 expression in cholinergic neurons of rab-3; sax-7(eq22) animals. In three independent transgenic lines, we did not observe suppression of the Unc locomotion, although there was a modest yet significant suppression of the mean swim rate that ranged from 50 to 62 thrashes/min when compared with the mean rate of 39.5 thrashes/min in rab-3; sax-7(eq22) animals (Figure 8B). Similarly, restoring sax-7 expression specifically in head cholinergic neurons resulted in little, if any, suppression of the rab-3; sax-7(eq22) swim rate. By comparison, Cre-recombinase expressed pan-neuronally resulted in a stronger suppression of the rab-3; sax-7(eq22) swim rate (Figure 8B; also Figure 3B). These results indicate that sax-7 expression in cholinergic neurons alone is not sufficient to rescue rab-3; sax-7 phenotypes, suggesting sax-7 function is also required in noncholinergic neurons.
In addition to knocking in sax-7, we assessed the site of sax-7 function by knocking out sax-7 in a tissue-specific manner in rab-3 animals. To do so, we used a conditional sax-7 knockout allele, eq27 (Supplementary Figure S5A). eq27 was generated by inserting a single loxP site 454 bp upstream of the start codon of sax-7(eq23), which already contains a loxP site in the 3′ end of the gene (Figure 3Aiii; Supplementary Figure S5A). sax-7(eq27) animals express SAX-7 as a mCherry fusion protein and behave similarly to sax-7(eq23) animals (Supplementary Figure S5B). Floxing out nearly the entire sax-7 gene with germline Cre-recombinase expression resulted in the eq28 allele (Supplementary Figure S5A). As expected, sax-7(eq28) animals no longer show mCherry expression, behaving similarly to sax-7(eq1) and sax-7(eq22) animals. Not surprisingly, rab-3; sax-7(eq28) animals show loopy, uncoordinated locomotion and severely reduced swim rate similar to rab-3; sax-7(eq1) and rab-3; sax-7(eq22) animals (Supplementary Figure S5B). Consistent with results of the knock-in sax-7 experiments, somatic knock out of sax-7 in the nervous system with pan-neuronal Cre-recombinase expression resulted in loopy, uncoordinated movement and severely reduced swim rates in rab-3; sax-7(eq27) animals, confirming a role for sax-7 in the nervous system. But somatic knockout of sax-7 function in cholinergic head neurons of rab-3; sax-7(eq27) animals resulted in a modest defect, indicating that loss of sax-7 in cholinergic head neurons is not sufficient to induce to the same level of uncoordinated locomotion and swim rate observed in rab-3; sax-7 double null animals.
We also assessed where rab-3 functions with respect to its interaction with sax-7. Not surprisingly, pan-neuronal rab-3 expression using the single-copy eqSi13 transgene completely rescues the synergistic rab-3; sax-7(eq1) phenotypes; however, rab-3 expression limited only to the cholinergic head neurons did not (Supplementary Figure S6). Taken together, these results point to both sax-7 and rab-3 acting in a larger set of neurons, in addition to the cholinergic head neurons, to promote coordinated locomotion and neuronal function.
Discussion
We previously uncovered a role for sax-7 in promoting coordinated locomotion, first revealed in sensitized rab-3 and unc-13 mutant backgrounds, in which sax-7 mutants produced a synergistic decrease in locomotion. Adult-onset, transient sax-7 expression could suppress the synergistic defects, consistent with a postdevelopmental role for the SAX-7/L1CAM protein. On the other hand, this transient sax-7 expression did not suppress the well-established loss-of-sax-7 defect in maintaining neuronal and axonal positioning, suggesting a novel role for sax-7 in promoting coordinated locomotion and neuronal function (Opperman et al. 2015). In this study, we examined these genetic interactions further and found them to be specific to genes that encode key players in the SV cycle necessary for neurotransmission. In contrast, genes that function in synapse formation do not interact with sax-7 (Figures 1 and 2). In addition to abnormal locomotion with a tendency to coil, sax-7 animals in sensitized backgrounds of SV cycle mutants showed synergistic neuronal dysfunction manifested as decreased crawl and swim rates (Figures 1 and 2). This sax-7 role in promoting coordinated locomotion and appropriate swim and crawl rates is dependent on sax-7 expression in the nervous system (Figure 3). We further demonstrate that this SAX-7 function in promoting coordinated locomotory behaviors is regulated by KSR-1 and MPK-1 (Figures 4 and 5).
In addition to the specific interaction with SV cycle genes, we previously showed rab-3; sax-7 animals also displayed strong resistance to the cholinesterase inhibitor, aldicarb, reflecting impaired cholinergic signaling (Opperman et al. 2015). Taken together, these findings suggested impaired synaptic activity as underlying these phenotypes and a potential role for sax-7 in synaptic modulation. However, the electrophysiological recordings and ultrastructural analyses of the neuromuscular junctions performed in this study did not reveal presynaptic abnormalities that could account for the observed behavioral phenotypes (Figures 6 and 7). Specifically, the cholinergic release defects associated with rab-3 mutants were not exacerbated by loss of sax-7, nor was the release defect of rab-3; sax-7 double mutants suppressed in the absence of ksr-1. Taken together, these findings suggest a potential role for SAX-7 and KSR-1 in synaptic modulation, but this does not appear to be at the level of the ventral cord neuromuscular junctions.
Our study also identified a role for the Erk MAPK pathway acting in head cholinergic neurons to modulate sax-7-dependent locomotion. Loss of either KSR-1 or MPK/Erk dramatically suppresses the abnormal locomotion, coiling tendency, and reduced swim and crawl rates of rab-3; sax-7 and unc-13; sax-7 mutant animals (Figures 4 and 5). Head cholinergic neurons comprise a subset of motor neurons that synapse on muscles in the head region as well as interneurons that synapse onto ventral nerve cord motor neurons to coordinate locomotion (White et al. 1986; Pereira et al. 2015). The Erk MAPK pathway was previously discovered to also modulate locomotion mediated by the heterotrimeric G protein Gq (Coleman et al. 2018). Hyperactive Gq signaling results in aberrant loopy locomotion that is also dependent on KSR-1 function in head cholinergic neurons. Furthermore, an activated form of the MAPK component, LIN-45/Raf, expressed in the same head neurons in wild-type animals not only phenocopies this loopy posture but also results in aldicarb hypersensitivity. The MAPK signaling pathway may thus promote coordinated locomotion by modulating central patterning of synaptic activity.
The overlapping loopy posture of rab-3; sax-7 and hyperactive Gq animals combined with the shared genetic interaction with the MAPK pathway suggest that SAX-7 may act to modulate the MAPK pathway specifically by dampening MAPK activity. In support of this notion, mammalian L1 is known to regulate MAPK signaling (Schmid et al. 2000; Cheng et al. 2005). It is conceivable that SAX-7 could influence MAPK activity either in the same cells or in adjacent cells with SAX-7 functioning in a noncell-autonomous fashion. Indeed, SAX-7 has been shown to influence the behavior of adjacent cells through physical interactions of its extracellular domain to diverse transmembrane proteins on adjacent cells (Sundararajan et al. 2019). In this study, we find that driving or knocking out sax-7 expression in head cholinergic neurons showed, at best, modest suppression or induction, respectively, of rab-3; sax-7 synergistic phenotypes (Figure 8; Supplementary Figure S5B), suggesting that sax-7 function is not limited to these head neurons in which ksr-1 functions. Alternatively, these modest effects may be due to technical issues. For example, a recent study revealed that the strength of the unc-17 promoter may be weak relative to other promoters for cholinergic interneuron expression (Bhardwaj et al. 2020). Additionally, somatic CRE activity may not be sufficient in our inducible knock-in and knockout system to affect SAX-7 levels accordingly in head cholinergic neurons for the respective rescue or induction of rab-3; sax-7 phenotypes.
How might sax-7 coordinate locomotion? Previously, the mammalian L1CAM encoded by the CHL1 gene was shown to function as a cochaperone, along with the 70 kDa heat shock protein (hsc70), the alpha cysteine string protein (αCSP), and the small glutamine-rich tetratricopeptide repeat-containing protein (αSGT), all chaperones of the exocytotic machinery that include the SNARE complex. CHL1 knockout mice exhibit reduced levels of assembled SNARE complex after stress or prolonged synaptic activity as well as impaired SV recycling, which became more apparent over time (Leshchyns'ka et al. 2006; Andreyeva et al. 2010). It is possible that sax-7 functions in synapses in a similar manner. Such a role may account for why neuronal dysfunction in sax-7 animals was apparent in the more rigorous swim assay and not with the crawl assay (Figure 2). Potential SV recycling defects in neuromuscular junctions of rab-3; sax-7 mutant animals would likely not have been detected in our ultrastructural analyses and electrophysiological recordings because these animals were not subjected to prolonged synaptic stress prior to examination. The presynaptic chaperones and exocytotic machinery also function in the regulated exocytosis of dense core vesicles, which contain neuropeptides and hormones as opposed to classical neurotransmitters (Johnson et al. 2010; Burgoyne and Morgan 2015). Indeed, CHL1 was demonstrated more recently also to influence the translocation and/or docking of insulin-containing DCV vesicles to the plasma membrane of pancreatic islet cells, with loss of CHL1 leading to impaired insulin secretion. Furthermore, CHL1 is implicated as a risk factor in type 2 diabetes (Taneera et al. 2012; Xin et al. 2016; Jiang et al. 2020). It is thus conceivable that SAX-7 could similarly influence exocytosis of neuropeptide-carrying DCVs. Neuropeptides play an important role in influencing C. elegans synaptic strength and behaviors (Li and Kim 2008). Intriguingly, loss of neuropeptides encoded by the flp-1 gene results in changes in synaptic activity and a similar loopy body posture exhibited by rab-3; sax-7 animals as well as by animals with elevated Gq signaling (Nelson et al. 1998; Stawicki et al. 2013; Buntschuh et al. 2018; Oranth et al. 2018).
To conclude, this study uncovered a role for the MAPK signaling pathway in SAX-7-dependent locomotion. While many questions remain regarding the mechanisms by which MAPK and SAX-7 coordinate locomotion and control body posture, the well-endowed genetic toolkit, mapped neural connectome, and well-characterized behaviors of C. elegans provides an excellent platform to further investigate the processes by which SAX-7 intersects the MAPK pathway.
Data availability
Strains and plasmids are available upon request. The authors affirm that all data necessary to support the conclusions of this study are included within the manuscript and figures.
Supplementary material is available at GENETICS online.
Supplementary Material
Acknowledgments
We thank Michael Ailion for the DNA constructs used to drive ksr-1 expression in acetylcholine neurons and Guillermo Marques at the University of Minnesota Imaging Center for assistance with imaging and use of the NIS-Elements AR Analysis software used to quantify crawl rates. Some strains were provided by the C. elegans Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). EN sample processing was partly performed at the BioCryo facility of Northwestern University’s NUANCE Center, which is supported by NSF (NSF ECCS-1542205 and NSF DMR-1720139) and the International Institute for Nanotechnology (IIN). EM images were acquired using instruments in the Electron Microscopy Core of the University of Illinois at Chicago’s Research Resources Center.
Funding
This work was supported by a grant from the National Institute of Neurological Disorders and Stroke (R01NS045873) to L.C.
Conflicts of interest
The authors declare that there is no conflict of interest.
Literature cited
- Andreyeva A, Leshchyns'ka I, Knepper M, Betzel C, Redecke L, et al. 2010. CHL1 is a selective organizer of the presynaptic machinery chaperoning the SNARE complex. PLoS One. 5:e12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere JA, Bell RT, Fu BX, Artiles KL, Hartman PS, et al. 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics. 198:837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, et al. 2012. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol Psychiatry. 17:887–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj A, Pandey P, Babu K.. 2020. Control of locomotory behavior of Caenorhabditis elegans by the immunoglobulin superfamily protein RIG-3. Genetics. 214:135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buntschuh I, Raps DA, Joseph I, Reid C, Chait A, et al. 2018. FLP-1 neuropeptides modulate sensory and motor circuits in the nematode Caenorhabditis elegans. PLoS One. 13:e0189320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne RD, Morgan A.. 2015. Cysteine string protein (CSP) and its role in preventing neurodegeneration. Semin Cell Dev Biol. 40:153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Deletion Mutant Consortium. 2012. Large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3 (Bethesda). 2:1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ch'ng Q, Sieburth D, Kaplan JM.. 2008. Profiling synaptic proteins identifies regulators of insulin secretion and lifespan. PLoS Genet. 4:e1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Hsu HW, Chang YH, Pan CL.. 2019. Adhesive L1CAM-robo signaling aligns growth cone F-actin dynamics to promote axon-dendrite fasciculation in C. elegans. Dev Cell. 48:215–228.e5. [DOI] [PubMed] [Google Scholar]
- Chen L, Ong B, Bennett V.. 2001. LAD-1, the Caenorhabditis elegans L1CAM homologue, participates in embryonic and gonadal morphogenesis and is a substrate for fibroblast growth factor receptor pathway-dependent phosphotyrosine-based signaling. J Cell Biol. 154:841–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhou S.. 2010. “CRASH”ing with the worm: insights into L1CAM functions and mechanisms. Dev Dyn. 239:1490–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Lemmon S, Lemmon V.. 2005. RanBPM is an L1-interacting protein that regulates L1-mediated mitogen-activated protein kinase activation. J Neurochem. 94:1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherra SJ III, Jin Y.. 2015. Advances in synapse formation: forging connections in the worm. Wiley Interdiscip Rev Dev Biol. 4:85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman B, Topalidou I, Ailion M.. 2018. Modulation of Gq-Rho signaling by the ERK MAPK pathway controls locomotion in Caenorhabditis elegans. Genetics. 209:523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahme M, Bartsch U, Martini R, Anliker B, Schachner M, et al. 1997. Disruption of the mouse L1 gene leads to malformations of the nervous system. Nat Genet. 17:346–349. [DOI] [PubMed] [Google Scholar]
- Davis MW, Hammarlund M, Harrach T, Hullett P, Olsen S, et al. 2005. Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics. 6:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Balzac CA, Lazaro-Pena MI, Ramos-Ortiz GA, Bulow HE.. 2015. The adhesion molecule KAL-1/anosmin-1 regulates neurite branching through a SAX-7/L1CAM-EGL-15/FGFR receptor complex. Cell Rep. 11:1377–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Balzac CA, Rahman M, Lazaro-Pena MI, Martin Hernandez LA, Salzberg Y, et al. 2016. Muscle- and skin-derived cues jointly orchestrate patterning of somatosensory dendrites. Curr Biol. 26:2379–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, Goldstein B.. 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods. 10:1028–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Liu OW, Howell AS, Shen K.. 2013. An extracellular adhesion molecule complex patterns dendritic branching and morphogenesis. Cell. 155:296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen E, D'Hooge R, Van Camp G, Verhoye M, Sijbers J, et al. 1998. L1 knockout mice show dilated ventricles, vermis hypoplasia and impaired exploration patterns. Hum Mol Genet. 7:999–1009. [DOI] [PubMed] [Google Scholar]
- Fransen E, Lemmon V, Van Camp G, Vits L, Coucke P, et al. 1995. CRASH syndrome: clinical spectrum of corpus callosum hypoplasia, retardation, adducted thumbs, spastic paraparesis and hydrocephalus due to mutations in one single gene, L1. Eur J Hum Genet. 3:273–284. [DOI] [PubMed] [Google Scholar]
- Fransen E, Schrander-Stumpel C, Vits L, Coucke P, Van Camp G, et al. 1994. X-linked hydrocephalus and MASA syndrome present in one family are due to a single missense mutation in exon 28 of the L1CAM gene. Hum Mol Genet. 3:2255–2256. [DOI] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, et al. 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 40:1375–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C, Davis MW, Sarov M, Taylor J, Flibotte S, et al. 2014. Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat Methods. 11:529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryns JP, Spaepen A, Cassiman JJ, van den Berghe H.. 1991. X linked complicated spastic paraplegia, MASA syndrome, and X linked hydrocephalus owing to congenital stenosis of the aqueduct of Sylvius: variable expression of the same mutation at Xq28. J Med Genet. 28:429–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracheva EO, Hadwiger G, Nonet ML, Richmond JE.. 2008. Direct interactions between C. elegans RAB-3 and Rim provide a mechanism to target vesicles to the presynaptic density. Neurosci Lett. 444:137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam SJ, Goncharov A, McEwen J, Baran R, Jin Y.. 2002. SYD-1, a presynaptic protein with PDZ, C2 and rhoGAP-like domains, specifies axon identity in C. elegans. Nat Neurosci. 5:1137–1146. [DOI] [PubMed] [Google Scholar]
- Hortsch M, Nagaraj K, Mualla R.. 2014. The L1 family of cell adhesion molecules: a sickening number of mutations and protein functions. Adv Neurobiol. 8:195–229. [DOI] [PubMed] [Google Scholar]
- Ishiguro H, Liu QR, Gong JP, Hall FS, Ujike H, et al. 2006. NrCAM in addiction vulnerability: positional cloning, drug-regulation, haplotype-specific expression, and altered drug reward in knockout mice. Neuropsychopharmacology. 31:572–584. [DOI] [PubMed] [Google Scholar]
- Jiang H, Liu Y, Qian Y, Shen Z, He Y, et al. 2020. CHL1 promotes insulin secretion and negatively regulates the proliferation of pancreatic β cells. Biochem Biophys Res Commun. 525:1095–1102. [DOI] [PubMed] [Google Scholar]
- Johnson JN, Ahrendt E, Braun JE.. 2010. CSPalpha: the neuroprotective J protein. Biochem Cell Biol. 88:157–165. [DOI] [PubMed] [Google Scholar]
- Jouet M, Moncla A, Paterson J, McKeown C, Fryer A, et al. 1995. New domains of neural cell-adhesion molecule L1 implicated in X-linked hydrocephalus and MASA syndrome. Am J Hum Genet. 56:1304–1314. [PMC free article] [PubMed] [Google Scholar]
- Kage-Nakadai E, Imae R, Suehiro Y, Yoshina S, Hori S, et al. 2014. A conditional knockout toolkit for Caenorhabditis elegans based on the Cre/loxP recombination. PLoS One. 9:e114680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen MP, Shen K.. 2007. Wnt signaling positions neuromuscular connectivity by inhibiting synapse formation in C. elegans. Cell. 130:704–716. [DOI] [PubMed] [Google Scholar]
- Koushika SP, Richmond JE, Hadwiger G, Weimer RM, Jorgensen EM, et al. 2001. A post-docking role for active zone protein Rim. Nat Neurosci. 4:997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner MR, Kornfeld K, Miller LM, Horvitz HR, Kim SK.. 1994. A MAP kinase homolog, mpk-1, is involved in ras-mediated induction of vulval cell fates in Caenorhabditis elegans. Genes Dev. 8:160–173. [DOI] [PubMed] [Google Scholar]
- Law JW, Lee AY, Sun M, Nikonenko AG, Chung SK, et al. 2003. Decreased anxiety, altered place learning, and increased CA1 basal excitatory synaptic transmission in mice with conditional ablation of the neural cell adhesion molecule L1. J Neurosci. 23:10419–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshchyns'ka I, Sytnyk V, Richter M, Andreyeva A, Puchkov D, et al. 2006. The adhesion molecule CHL1 regulates uncoating of clathrin-coated synaptic vesicles. Neuron. 52:1011–1025. [DOI] [PubMed] [Google Scholar]
- Li C, Kim K.. 2008. Neuropeptides. In: WormBook, editor. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.142.1, http://www.wormbook.org. 1–36. [Google Scholar]
- Lickteig KM, Duerr JS, Frisby DL, Hall DH, Rand JB, et al. 2001. Regulation of neurotransmitter vesicles by the homeodomain protein UNC-4 and its transcriptional corepressor UNC-37/groucho in Caenorhabditis elegans cholinergic motor neurons. J Neurosci. 21:2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KG, Alfonso A, Nguyen M, Crowell JA, Johnson CD, et al. 1996. A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc Natl Acad Sci U S A. 93:12593–12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerman DG, Barstead RJ.. 2008. Towards a mutation in every gene in Caenorhabditis elegans. Brief Funct Genomic Proteomic. 7:195–204. [DOI] [PubMed] [Google Scholar]
- Moy SS, Nonneman RJ, Young NB, Demyanenko GP, Maness PF.. 2009. Impaired sociability and cognitive function in Nrcam-null mice. Behav Brain Res. 205:123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson LS, Rosoff ML, Li C.. 1998. Disruption of a neuropeptide gene, flp-1, causes multiple behavioral defects in Caenorhabditis elegans. Science. 281:1686–1690. [DOI] [PubMed] [Google Scholar]
- Nonet ML, Saifee O, Zhao H, Rand JB, Wei L.. 1998. Synaptic transmission deficits in Caenorhabditis elegans synaptobrevin mutants. J Neurosci. 18:70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet ML, Staunton JE, Kilgard MP, Fergestad T, Hartwieg E, et al. 1997. Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J Neurosci. 17:8061–8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris AD, Kim HM, Colaiacovo MP, Calarco JA.. 2015. Efficient genome editing in Caenorhabditis elegans with a toolkit of dual-marker selection cassettes. Genetics. 201:449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman K, Moseley-Alldredge M, Yochem J, Bell L, Kanayinkal T, et al. 2015. A novel nondevelopmental role of the sax-7/L1CAM cell adhesion molecule in synaptic regulation in Caenorhabditis elegans. Genetics. 199:497–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oranth A, Schultheis C, Tolstenkov O, Erbguth K, Nagpal J, et al. 2018. Food sensation modulates locomotion by dopamine and neuropeptide signaling in a distributed neuronal network. Neuron. 100:1414–1428.e10. [DOI] [PubMed] [Google Scholar]
- Owen MJ, Sawa A, Mortensen PB.. 2016. Schizophrenia. Lancet. 388:86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MR, Lehrman EK, Poon VY, Crump JG, Zhen M, et al. 2006. Hierarchical assembly of presynaptic components in defined C. elegans synapses. Nat Neurosci. 9:1488–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, Kratsios P, Serrano-Saiz E, Sheftel H, Mayo AE, et al. 2015. A cellular and regulatory map of the cholinergic nervous system of C. elegans. Elife. 4:e12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond J. 2005. Synaptic function. In: WormBook, editor. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.69.1, http://www.wormbook.org. 1–14. [Google Scholar]
- Richmond JE, Davis WS, Jorgensen EM.. 1999. UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat Neurosci. 2:959–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond JE, Jorgensen EM.. 1999. One GABA and two acetylcholine receptors function at the C. elegans neuromuscular junction. Nat Neurosci. 2:791–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K, Migita O, Toru M, Arinami T.. 2002. An association between a missense polymorphism in the close homologue of L1 (CHL1, CALL) gene and schizophrenia. Mol Psychiatry. 7:412–415. [DOI] [PubMed] [Google Scholar]
- Sakurai T, Ramoz N, Reichert JG, Corwin TE, Kryzak L, et al. 2006. Association analysis of the NrCAM gene in autism and in subsets of families with severe obsessive-compulsive or self-stimulatory behaviors. Psychiatr Genet. 16:251–257. [DOI] [PubMed] [Google Scholar]
- Salzberg Y, Diaz-Balzac CA, Ramirez-Suarez NJ, Attreed M, Tecle E, et al. 2013. Skin-derived cues control arborization of sensory dendrites in Caenorhabditis elegans. Cell. 155:308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval GM, Duerr JS, Hodgkin J, Rand JB, Ruvkun G.. 2006. A genetic interaction between the vesicular acetylcholine transporter VAChT/UNC-17 and synaptobrevin/SNB-1 in C. elegans. Nat Neurosci. 9:599–601. [DOI] [PubMed] [Google Scholar]
- Schaefer AM, Hadwiger GD, Nonet ML.. 2000. rpm-1, a conserved neuronal gene that regulates targeting and synaptogenesis in C. elegans. Neuron. 26:345–356. [DOI] [PubMed] [Google Scholar]
- Schmid RS, Pruitt WM, Maness PF.. 2000. A MAP kinase-signaling pathway mediates neurite outgrowth on L1 and requires Src-dependent endocytosis. J Neurosci. 20:4177–4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrander-Stumpel C, Howeler C, Jones M, Sommer A, Stevens C, et al. 1995. Spectrum of X-linked hydrocephalus (HSAS), MASA syndrome, and complicated spastic paraplegia (SPG1): clinical review with six additional families. Am J Med Genet. 57:107–116. [DOI] [PubMed] [Google Scholar]
- Shaltout TE, Alali KA, Bushra S, Alkaseri AM, Jose ED, et al. 2013. Significant association of close homologue of L1 gene polymorphism rs2272522 with schizophrenia in Qatar. Asia Pac Psychiatry. 5:17–23. [DOI] [PubMed] [Google Scholar]
- Stawicki TM, Takayanagi-Kiya S, Zhou K, Jin Y.. 2013. Neuropeptides function in a homeostatic manner to modulate excitation-inhibition imbalance in C. elegans. PLoS Genet. 9:e1003472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram M, Han M.. 1995. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell. 83:889–901. [DOI] [PubMed] [Google Scholar]
- Sundaram MV. 2013. Canonical RTK-Ras-ERK signaling and related alternative pathways. In: WormBook, editor. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.80.2, http://www.wormbook.org. 1–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundararajan L, Stern J, Miller DM III. 2019. Mechanisms that regulate morphogenesis of a highly branched neuron in C. elegans. Dev Biol. 451:53–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam GW, van de Lagemaat LN, Redon R, Strathdee KE, Croning MD, et al. 2010. Confirmed rare copy number variants implicate novel genes in schizophrenia. Biochem Soc Trans. 38:445–451. [DOI] [PubMed] [Google Scholar]
- Taneera J, Lang S, Sharma A, Fadista J, Zhou Y, et al. 2012. A systems genetics approach identifies genes and pathways for type 2 diabetes in human islets. Cell Metab. 16:122–134. [DOI] [PubMed] [Google Scholar]
- Tapanes-Castillo A, Weaver EJ, Smith RP, Kamei Y, Caspary T, et al. 2010. A modifier locus on chromosome 5 contributes to L1 cell adhesion molecule X-linked hydrocephalus in mice. Neurogenetics. 11:53–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Camp G, Fransen E, Vits L, Raes G, Willems PJ.. 1996. A locus-specific mutation database for the neural cell adhesion molecule L1CAM (Xq28). Hum Mutat. 8:391. [DOI] [PubMed] [Google Scholar]
- Vits L, Van Camp G, Coucke P, Fransen E, Boulle KD, et al. 1994. MASA syndrome is due to mutations in the neural cell adhesion gene L1CAM. Nat Genet. 7:408–413. [DOI] [PubMed] [Google Scholar]
- Wamsley B, Geschwind DH.. 2020. Functional genomics links genetic origins to pathophysiology in neurodegenerative and neuropsychiatric disease. Curr Opin Genet Dev. 65:117–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kweon J, Larson S, Chen L.. 2005. A role for the C. elegans L1CAM homologue lad-1/sax-7 in maintaining tissue attachment. Dev Biol. 284:273–291. [DOI] [PubMed] [Google Scholar]
- Weimer RM, Gracheva EO, Meyrignac O, Miller KG, Richmond JE, et al. 2006. UNC-13 and UNC-10/rim localize synaptic vesicles to specific membrane domains. J Neurosci. 26:8040–8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S.. 1986. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 314:1–340. [DOI] [PubMed] [Google Scholar]
- Xin Y, Kim J, Okamoto H, Ni M, Wei Y, et al. 2016. RNA sequencing of single human islet cells reveals type 2 diabetes genes. Cell Metab. 24:608–615. [DOI] [PubMed] [Google Scholar]
- Yip ZC, Heiman MG.. 2018. Ordered arrangement of dendrites within a C. elegans sensory nerve bundle. Elife. 7:e35825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshina S, Suehiro Y, Kage-Nakadai E, Mitani S.. 2016. Locus-specific integration of extrachromosomal transgenes in C. elegans with the CRISPR/Cas9 system. Biochem Biophys Rep. 5:70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen M, Huang X, Bamber B, Jin Y.. 2000. Regulation of presynaptic terminal organization by C. elegans RPM-1, a putative guanine nucleotide exchanger with a RING-H2 finger domain. Neuron. 26:331–343. [DOI] [PubMed] [Google Scholar]
- Zhong X, Drgonova J, Li CY, Uhl GR.. 2015. Human cell adhesion molecules: annotated functional subtypes and overrepresentation of addiction-associated genes. Ann N Y Acad Sci. 1349:83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Liang X, Wang XM, Shen K.. 2017. Dynein and EFF-1 control dendrite morphology by regulating the localization pattern of SAX-7 in epidermal cells. J Cell Sci. 130:4063–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and plasmids are available upon request. The authors affirm that all data necessary to support the conclusions of this study are included within the manuscript and figures.
Supplementary material is available at GENETICS online.