Abstract
The formation of the digits is a tightly regulated process. During embryogenesis, disturbance of genetic pathways in limb development could result in syndactyly; a common congenital malformation consisting of webbing in adjacent digits. Currently, there is a paucity of knowledge regarding the exact developmental mechanism leading to this condition. The best studied canonical interactions of Wingless‐type–Bone Morphogenic Protein–Fibroblast Growth Factor (WNT–BMP–FGF8), plays a role in the interdigital cell death (ICD) which is thought to be repressed in human syndactyly. Animal studies have displayed other pathways such as the Notch signaling, metalloprotease and non-canonical WNT-Planar cell polarity (PCP), to also contribute to failure of ICD, although less prominence has been given. The current diagnosis is based on a clinical evaluation followed by radiography when indicated, and surgical release of digits at 6 months of age is recommended. This review discusses the interactions repressing ICD in syndactyly, and characterizes genes associated with non-syndromic and selected syndromes involving syndactyly, according to the best studied canonical WNT-BMP-FGF interactions in humans. Additionally, the controversies regarding the current syndactyly classification and the effect of non-coding elements are evaluated, which to our knowledge has not been previously highlighted. The aim of the review is to better understand the developmental process leading to this condition.
Keywords: Syndactyly, Limb, WNT-BMP-FGF, Synostosis, Syndactyly classification, Genetic screening
Background
Syndactyly, devised from the Greek word ‘syn’ meaning ‘together,’ and ‘dactyly’ meaning ‘digits’, is a congenital malformation, where there is failure of separation (aberrant webbing) between adjacent digits during embryological limb formation [1, 2]. It is one of the most common hereditary deformities, occurring in 1 in 2000 live births, and has twice the occurrence in males than in females. There is seen to be half the proportion of cases to have bilateral syndactyly, whilst 57% of cases involve the third web space [3]. The underlying aetiology for syndactyly is mostly genetic but causes such as maternal smoking, lower nutritional and economic status have also been elucidated [3].
This phenotype is present as soft tissue and/or bony fusions, or further characterized as complete fusions if adjacent digits are fused until the fingertips, else, as incomplete. It can be present in isolation or as a feature of more than 300 recognized syndromes. Non-syndromic syndactyly is currently classified into 9 main types and subtypes, some subtypes possessing loci with genes to be identified [1, 4, 5]. Syndactyly may also be a feature in syndromes including Acrocephalosyndactyly, Fraser or even associated with other digit abnormalities (Ex: polydactyly). A conglomeration of pathways for syndactyly pathogenesis have been stipulated, involving a plethora of genes [6–10].
The majority of them are diagnosed clinically, followed by radiographs when indicated to help determine if syndactyly is complete/complex [11]. Surgical release of joined digits is the current therapy, albeit recommended earlier release at 6 months to prevent angulatory rotational distortions of some digits. Prenatal diagnosis of syndactyly is difficult and hand syndactyly can be mistaken for clenched fists [12].
Through this article, we attempt to better understand the developmental process underlying this condition and the genes involved.
Digit separation interactions in normal vs. syndactyly phenotypes
Human limb structure regulation is controlled by two signaling centers; the zone of polarizing activity (ZPA controlling overall patterning in the anterior–posterior axis) and the apical ectodermal ridge (AER for limb growth) [13]. Different interactions including WNTs, Bone Morphogenetic Proteins (BMPs), Fibroblast Growth Factors (FGFs), and Retinoic acid metabolites (RA) have been elucidated to have a role in the ICD in the nascent autopod. Initial autopod formation is stipulated as the interactions arising from the FGF and WNT signaling.
The canonical WNT signaling in particular has been studied extensively. This is also controlled by the balance of Sonic hedgehog in the ZPA regions. Bone morphogenetic protein 2 (BMP2), 4, and 7 are expressed in the interdigit and they are associated with apoptosis in the interdigits [14, 15]. The BMPs produce retinoic acid metabolites, which may further downregulate FGF signaling. It is known that FGF signaling is a potential survival pathway with anti-apoptotic signals. They are represented mainly by FGF4 and FGF8 in the AER promoting growth of the underlying mesenchyme [16]. Retinoic acid, may induce ICD directly distally, but is thought to be induced by the BMP-FGF axis indirectly to induce ICD proximally [2, 17]. Together, the signaling interactions of WNT-BMP-FGF and retinoic acid are vital for autopod digit separation. To date, there is no specified axis of syndactyly pathogenesis. However, the absence of ICD during the 7th to 8th weeks of gestation in the interdigital mesenchyme involving the WNT-BMP-FGF axis seems to be a highly conjectured pathway for the pathogenesis of soft tissue syndactyly [17–19]. The first steps in cutaneous syndactyly are described as the overexpression of the WNT canonical pathway, or the suppression of BMPs in the interdigital mesenchyme. Lower levels of BMPs, which normally positively regulate interdigital apoptosis, now leads to an overexpression of FGF8 causing increased anti-apoptotic signals in the AER. This can indirectly inhibit retinoic acid and finally repress its function in the extracellular matrix (ECM), causing perturbed ICD inducing syndactyly phenotypes [7, 13, 17, 19].
Non canonical WNT-PCP pathway While FGF8 modulates only rapid disorganized movement of cells, WNT5A (3p14.3) expressed in the AER induces polarized orientation of cells via the WNT-PCP pathway and this was observed in cell culture and is known to be required for ICD [20, 21]. Most focus for the underlying pathology of complex syndactyly, is the WNT-BMP-FGF8 pathway. It is reported that ICD occurs at E12.5-E13.5 in mouse cell lines [22]. In a study by Zhu et al., WNTless; a key regulator of WNT trafficking was tested. The WNTless mutants (WIShh-Cre) show webbing of cartilage before E12.5, suggesting that abrogated ICD does not account for cartilage webbing in WNTless mutant mice. This deficiency of WNT5a could outride the Shh regulation (occurs half a day later), and could be a better suited cause for digit malformations in the WNTless mutant mice [22]. HOXD13 mutants displayed downregulated WNT5a signaling, which could mean WNT5a gradient in chondrocyte cell polarity could be an imperative aetiology [10, 22]. Hence a combined effect of both FGF and non-canonical WNT-PCP pathways is postulated for normal cell orienting, especially for synostosis syndactyly commonly seen in Malik–Percin type or complex syndactyly [20, 23].
Notch Signaling There is control of ICD by modulating the level of FGF8 expressing cells in the AER. This was exemplified in murine models deficient in the Notch ligands, Serrate or Jagged2, where syndactyly was observed as a result of expanded FGF8 domain and substantial decrease of BMPs in the interdigital tissue [6, 13].
A Disintegrin And Metalloproteinase with Thrombospondin Motifs (ADAMTS) Metalloproteases Proteolytic activity of the proteases ADAMTS5 (21q21.3), ADAMTS9 (3p14.1) and ADAMTS20 (12q12) have been postulated to be vital for completion of ICD. As ADAMTS 5/20 does not act upstream, this leaves the BMP/FGF axis unaffected. Thus, the ADAMTS Metalloproteases with cleaved Versican, prove as a promising alternative independent pathway to the BMP/FGF axis, and act concurrently to produce ICD in the developing limb bud [2, 8]. Finally, Hedgehog signaling critically influences the distribution of other signaling pathways. Interactions in Notch signaling and the ADAMTS pathway also seem to contribute to soft tissue syndactyly in humans but are less prioritized [8, 24]. This might be since the deletion of Notch1 is lethal before limb bud stages, its role in development remains elusive. Additionally, only animal models have been used to observe syndactyly pathogenesis in Notch and ADAMTS. Collectively, the perturbation of ICD may account for simple (cutaneous) syndactyly. However, it is questionable that complex (bony) syndactyly phenotypes arise solely from perturbed ICD, prompting further investigation [2].
The importance of analyzing genes in pathogenesis will help in understanding the genes involved in autopod formation, as well promote analyzing other gene–gene and protein–protein interactions which may contribute to syndactyly. To date, the canonical WNT pathway only justifies pathogenesis of cutaneous syndactyly in the developing limb bud, and does not account fully for synostosis syndactyly; such as the Malik Percin type. Since interdigital webbing occurs at day E12.5 before ICD in mice models, the WNT-PCP non-canonical pathway seems to be a promising mechanism for aberrant mesenchymal cell migration in synostosis syndactyly phenotypes [10, 22].
Syndromic syndactyly genetic determinants
The complexity of syndactyly classification is augmented since it can exist in 300 or more syndromic forms [1, 3]. Selected syndromic phenotypes having postulated mechanisms for pathogenesis, are described below and listed in Table 1.
Table 1.
Genetic determinants for selected syndromic syndactyly phenotypes
| Gene | Type of syndrome | OMIM | Common variant/s | Inheritance | Clinical presentation | Postulated contribution to pathogenesis | Key references |
|---|---|---|---|---|---|---|---|
| 1 (a) Genetic determinants in coding regions | |||||||
| ROR2 | Brachydactyly type B (BDB1) | 113000 | c.1324C.T; p.R441X | AD | Brachydactyly characterized by hypoplasia/aplasia of distal phalanges in combination with distal symphalangism, fusion of carpal/tarsal bones, and partial cutaneous syndactyly | Facilitate WNT Overexpression | [20, 40] |
| SOST | Sclerosteosis | 269500 | Nonsense mutations | AR | Presence of asymmetric cutaneous syndactyly of the index and middle fingers in many cases. The jaw has an unusually square appearance | [29, 41] | |
| LRP4 | Sclerosteosis, 2 | 614305 | c.3508C > T; p.R1170W and c.3557G > C; p.W1186S | AD, AR | Progressive skeletal overgrowth. Syndactyly is a variable manifestation | [26, 28, 42] | |
| GLI3 | Pallister–Hall syndrome | 146510 | Haploinsufficiency, c.1468_1469insG and c.1007_1008dupAC | AD | Hypothalamic hamartoma, pituitary dysfunction, central polydactyly, and variable degree of syndactyly | BMP suppression | [30, 43, 44] |
| Greig cephalopolysyndactyly syndrome | 175700 | c.2374C > T; p.Arg792* | AD | Frontal bossing, scaphocephaly, and hypertelorism associated with pre- and postaxial polydactyly and variable syndactyly | |||
| LMBR1 | Triphalangeal thumb-polysyndactyly syndrome | 174500 | Position 287 on ZRS enhancer | AD | Thumb in this malformation is usually opposable and possesses a normal metacarpal. Variable degree of syndactyly | [31, 32, 45] | |
| DHCR7 | Smith–Lemli–Opitz syndrome | 270400 | c.453G > A; p.W151X | AR | Affects multiple body systems with Syndactyly of toes 2 and 3 being a common finding | [33, 46] | |
| RAB23 | Carpenter syndrome | 201000 | Homozygous nonsense/frameshift pathogenic variants c.434 T > A; p.L145X | AR | Craniosynostosis, polysyndactyly, obesity, and cardiac defects | [39, 47–49] | |
| FGFR2 | Apert syndrome | 101200 | c.755C > G; p.S252W or c.758C > G; p.P253W | AD | Craniosynostosis, midface hypoplasia, and syndactyly of the hands and feet with a tendency to fusion of bony structures | FGF Overexpression | [7, 34, 50–52] |
| FGFR1/FGFR2 | Pfeiffer syndrome | 101600 | FGFR1 – p.P252R | AD | Type 1, the classic syndrome, is compatible with life and consists of craniosynostosis, midface deficiency, broad thumbs, broad great toes, brachydactyly, and variable syndactyly | ||
| TWIST1;FGFR3 | Saethre–Chotzen syndrome | 101400 | FGFR3 p.P250R, deletion of TWIST1 | AD | Craniosynostosis, facial dysmorphism, and hand and foot abnormalities. The degree of syndactyly is also variable | [35, 36, 53] | |
| HOXD13 | Brachydactyly-syndactyly syndrome | 610713 | Polyalanine constriction, c.950A > G; p.Q317K | n.r | Brachydactyly and syndactyly (partial cutaneous webbing) in association with oligodactyly | Retinoic acid suppression | [10, 37] |
| LRP4 | Sclerosteosis, 2 | 614305 | c.1151A > G; p.Tyr384Cys | AD, AR | Progressive skeletal overgrowth. Syndactyly is a variable manifestation | Repression of Notch signaling | [54] |
| Other loci/variants | Clinical presentation | Postulated contribution to pathogenesis | Key references |
|---|---|---|---|
| 1 (b) Genetic determinants in non-coding regions | |||
| Intron EMID2 | Holoprosencephaly spectrum disorder and severe upper limb syndactyly | Ectopic SHH expression | [55] |
| Intron 5 LMBR1 | Cutaneous syndactyly without polydactyly | Decreased ICD | [45] |
| Intron IRF6 | Van der Woude syndrome (MIM: 119,300) | Unclassified | [56] |
| Exonization of 22 intronic YY1AP1 | Grange syndrome (MIM: 602,531) with complete cutaneous syndactyly. Fingers: third, fourth, and fifth finger and third/fourth finger of the right hand. Toes: bilateral cutaneous syndactyly of her second/third toes | Unclassified | [57] |
| Intron 6 KATNB1 | Congenital microcephaly, lissencephaly, short stature, polysyndactyly, and dental abnormalities | Unclassified | [58] |
| Intron 8 FGFR2 | Apert syndrome (MIM 101,200) | Unclassified | [59] |
| IVS8-1G > C variant DHCR7 | Smith-Lemli-Opitz syndrome (MIM 270,400) | BMP suppression | [60] |
| Pre-ZRS region | TPS (MIM: 174,500) | Ectopic expression | [61] |
AR autosomal recessive, AD autosomal dominant, XLR X-Linked recessive, n.r not reported due to insufficient evidence, EMID2 EMI domain containing 2, LMBR1 Limb development membrane protein 1, IRF6 Interferon regulatory factor 6, YY1AP1 YY1 associated protein 1, KATNB1 Katanin,p80 subunit B1, FGFR2 Fibroblast growth factor receptor 2, TPS Thumb polysyndactyly syndrome
Variants in coding regions
Some of the syndromes are mentioned below with their documented involvement in each interaction for canonical pathogenesis, but one must note that many other syndromes are also present. The genetic variants for each syndrome are listed where found in Table 1.
Some of the main WNT overexpressing syndromes observed include Brachydactyly type B (BDB1) (MIM 113000), Sclerosteosis (MIM 269500) and Sclerosteosis, 2 (MIM 614305).
BDB1 is mapped to the ROR2 gene, (9q22.31) encoding a co-receptor to WNT ligands, such as WNT5a. It is postulated that ROR2 may implicate canonical WNT signaling since a striking resemblance of the CRD of ROR2 and the WNT binding domain of Frizzled receptors has been identified [25]. It is also seen to be involved in the non-canonical WNT-PCP interactions, since mice with mutant ROR2 produce similar phenotypes in comparison to WNT5a mutant mice [20]. Sclerosteosis 2 has been mapped to the LRP4 gene where there is direct interaction between sclerostin and LRP4, and LRP4 facilitates sclerostin mediated WNT inhibition; which is implicated in LRP4 mutants [26, 27]. Additionally, LRP4 interactions with the Notch pathway is known to b evolutionarily conserved and a homozygous missense variant in the highly conserved EGF-2 calcium binding domain of the LRP4 gene was also hypothesized to have abrogated Notch signaling [28].
Finally, in Sclerostosis, loss of function SOST gene variants may enable sclerostin to inhibit the LRP5/6 and frizzled receptor complex, thereby leading to phosphorylation and downregulation of β-catenin resulting in the inhibition of WNT signaling interactions [29].
The main syndromes seen to promote BMP suppression include Pallister-Hall syndrome (PHS) (MIM 146510) and Greig cephalopolysyndactyly syndrome (MIM 175700), Triphalangeal thumb-polysyndactyly syndrome (TPS) (MIM 174500), Smith-Lemli-Opitz syndrome (SLOS) (MIM 270400) and Carpenter Syndrome (MIM 201000). PHS and GCPS are both mapped to the GLI3 gene. However, Pallister-Hall syndrome is associated with central polydactyly while GCPS involves pre and post axial polydactyly. The GLI3 gene (7p14), encodes a transcription factor that is a bifunctional downstream modulator of SHH signaling; which is implicated in both syndromes. Most GLI3 gene variants are associated with haploinsufficiency of the GLI3 gene, which eventually cause the skewing of GLI3 Activator (GLIA) or Repressor (GLIR) formation [30]. Common genetic determinant for TPS includes duplications spanning the LMBR1 gene [7, 31, 32], and the RAB23 gene (6p12.1-q12), encodes a negative regulator of the signaling in sonic hedgehog (SHH), perturbed in Carpenters syndrome. As for SLOS, the gene encoding the enzyme 7-dehydrocholesterol (7-DHC) reductase (DHCR7: 11q13.4), is known to be deregulated. The DHCR7 gene is hypothesized to orchestrate cholesterol moiety and SHH signaling, which governs the severity of syndactyly phenotypes [33].
Syndromic dactylies shown to promote FGF8 overexpression mainly include Apert Syndrome (MIM 101200), Pfeiffer Syndrome (MIM 101600) and Saethre-Chotzen syndrome (MIM 101400) syndrome. The FGFR2 gene variants implicates the function of this receptor in both Apert and Pfeiffer syndromes. The P253W mutant may perturb ligand-binding specificity to FGF10 resulting in overstimulation of FGF8 via the FGF10-FGF8 loop in the ectoderm [7, 34]. As for Saethre-Chotzen syndrome, TWIST1 and FGFR1/3 genes have been mapped to this syndrome. TWIST1 is classified as an upstream regulator of FGFs, thus modulating FGF expression [35, 36].
Finally, the Brachydactyly-syndactyly syndrome (MIM 610713) associated with variants in the HOXD13 gene (polyalanine constrictions, and missense variants) [10, 37]. It is postulated that HOXD13 mutants, suppresses retinoic acid (RA) in the ECM.
Variants in non coding regions
The importance to screen the gene and its flanking regions remains imperative, since alternative splice variants in non-coding regions may lead to exon skipping, and epigenetic influences which underlie protein malformation [1, 5]. The presence of modifier genes has been postulated to influence phenotypes in gene variants of p63 [38] and RAB23 [39], which may underlie causes for intrafamilial variability. A number of intronic loci (Table 1), have been identified in syndactyly phenotypes, although most require classification of their pathway.
Non-syndromic syndactyly genetic determinants
Together, 9 types of non-syndromic syndactyly (I-IX), according to their phenotypical diversity have been classified (Table 2), with the respective gene/loci [1]. Some of these identified genes include HOXD13, FBLN1, GJA1, LMBR1, LRP4, GREM1 (BMP antagonist), FGF16 and recently discovered BHLHA9 [1, 62–65].
Table 2.
Genes/loci with postulated pathogenesis for non-syndromic syndactyly
| Type of non syndromic syndactyly | Locus/gene | OMIM | Common variant/s | Inheritance | Clinical presentation | Postulated contribution to pathogenesis | Key references | |
|---|---|---|---|---|---|---|---|---|
| I-a | ZD1; Zygodactyly; Weidenreich type | 3p21.31 | 609815 | n.r | AD |
Bilateral, symmetrical, Fingers: Normal Toes: 2/3 only |
n.r | [64] |
| I-b | SD1; Lueken type | 2q34-q36 | 185900 | n.r | AD |
Usually, bilateral Fingers: 3/4 Fingers, cutaneous/bony Toes: 2/3 Toes, cutaneous |
n.r | [68, 69] |
| I-c | Montagu type |
2q31-q32 HOXD13 |
n.r |
c.917G > A; p.R306Q, c.916C > G; p.R306G |
AD |
Typically, bilateral Fingers: 3/4 Fingers only, cutaneous/bony Toes: Normal |
Suppression of Retinoic Acid | [70] |
| I-d | Castilla type | n.r | n.r | n.r | AD |
Bilateral Fingers: Normal Toes: 4/5 Toes only, cutaneous |
n.r | [71] |
| II-a | SPD1; Vordingborg type |
2q31; HOXD13 |
186000 | Polyalanine repeat expansions, frameshift deletions, 2q31.1 microdeletion and G11A missense | AD |
Fingers: SPD, mesoaxial (3/4 fingers) Toes: SPD, postaxial (4/5 toes) |
Suppression of Retinoic Acid | [72] |
| II-b | SPD2; Debeer type |
22q13.3; FBLN1 |
608180 | t(12;22) (p11.2;q13.3) | AD |
Fingers: SPD is central and postaxial Toes: Postaxial syndactyly |
Increased expression of FGFR8 | [73] |
| II-c | SPD3; Malik type | 14q11.2-q13 | 610,234 | n.r | AD |
Fingers: SPD is central Toes: SPD postaxial |
n.r | [65] |
| III | SDTY3; Johnston-Kirby type |
6q21-q23; GJA1 |
186100 |
nt427G > A and c.T274C; p.Y92H |
AD |
Fingers: Complete, bilateral syndactyly of 4/5 Fingers; fifth finger short, middle digit missing or underdeveloped Toes: Normal |
Reduces downstream BMP2 expression, contributing to overexpression of the FGF4 and FGF8 | [74–76] |
| IV-a | SDTY4; Haas type | 7q36; ZRS (LMBR1) | 186200 | Heterozygous variants in the SHH regulatory element (ZRS), and duplications | AD |
Complete and Bilateral, often polydactyly associated Fingers: All fingers webbed; pre-/post-axial polydactyly, cup-shaped hand Toes: Normal |
Alter the ZRS control/limb-specific SHH expression | [77, 78] |
| IV-b | Andersen-Hansen type | n.r | n.r | n.r | n.r |
Fingers: All fingers webbed; pre-/post-axial polydactyly, cup-shaped hand Toes: Variable webbing of toes with polydactyly |
n.r | [79] |
| V | SDTY5; Dowd type |
2q31; HOXD13 |
186300 | c.950A > G; p.Q317R, and polyalanine expansions | AD |
Complete Fingers: 4/5 Fingers with metacarpals fusion; hypoplastic metacarpals 4/5 Toes: Mesoaxial webbing |
Suppression of Retinoic Acid | [37, 80] |
| VI | Mitten type | n.r | n.r | n.r | AD |
Unilateral Fingers: 2/5 Fingers Toes: 2/5 Toes |
n.r | [81] |
| VII-a | Cenani-Lenz type; spoon-hand type |
11p12–p11.2; LRP4 |
212780 | Missense variations are most common, c.1117C > T; p.R373W | AR |
Fingers: Total synostotic syndactyly with metacarpals fusion, spoon-head shape Toes: Total synostotic syndactyly with metatarsals fusion |
WNT overexpression /Postulated to cause repression of Notch signaling |
[82] |
| VII-b | Oligodactyly type |
15q13.3; GREM1-FMN1 |
n.r | 1.7 Mb duplication spanning both the GREM1 and FMN1 1 genes | AD |
Fingers: Few deformed digits Toes: Variable syndactyly of toes |
BMP antagonist GREM1(usually repressed by FGF); is active in mutant forms, suppressing BMP activity | [83] |
| VIII-a | Orel-Holmes type | Xq21.1; FGF16 | 309630 | Nonsense variants p.R179X and p.S157X in exon 3 | X-R |
Fingers: 4/5 Metacarpal fusion Toes: Normal |
Impair FGF16-FGFR1 interactions | [84, 85] |
| VIII-b | Lerch type | n.r | n.r | n.r | AD |
Fingers: 4/5 Metacarpal fusion Toes: Normal |
n.r | [1] |
| IX | MSSD; Malik-Percin type |
17p13.3/ BHLHA9 |
609432 | c.311T > C; p.Ile104Thr | AR |
Fingers: Mesoaxial synostotic syndactyly with phalangeal reduction Toes: Preaxial webbing; distal phalangeal hypoplasia |
Ectopic FGF8 expression restrict BMP suppression | [63, 86, 87] |
AR autosomal recessive, AD autosomal dominant, XLR X-Linked recessive, n.r not reported due to insufficient evidence, SPD synpolydactyly
However, some controversies regarding non-syndromic syndactyly classification have been identified. A report by Qattan et al. identified a case with cutaneous syndactyly occurring in digits two/five or three/five without polydactyly. This finding of isolated syndactyly without polydactyly is similar to type III syndactyly, caused by variants in the GJA1 gene and has been observed in digits four/five or three/five. However, although similar to type III, the authors observed a variant to be present in the ch7q36.3 (LMBR1 region) which is syntenic to the Hammertoe locus in mice of the similar phenotype [45]. This may highlight a case of genetic heterogeneity, and supports the notion that this type may be polygenic.
In another two reports regarding type IX syndactyly which is characterized by osseous synostosis of the third and fourth metacarpals, whereby slightly different phenotypes have been associated. Weinrich et al. highlighted a case of MSSD involving the second and third proximal phalanges which is different from Malik’s original classification and attributed it to being a novel non-syndromic type [66]. However, this notion was dismissed and re-classified by Malik as being a type of non-syndromic type IX associated with the BHLHA9 gene [63]. The second report, presented a case of MSSD with a triangular proximal phalanx of the second and third metacarpal similar to Weinrich’s case report, in addition with ulnar hemimelia (shortening), a novel finding [67]. Genetic testing was not performed in both MSSD cases, and conclusions on classification were based on hand radiography. If it maps to the same gene, this could be a form of phenotypic heterogeneity associated with the BHLHA9 gene. Hence it remains imperative to genetic testing to be able to further evaluate these inconsistencies.
Application to genetic screening
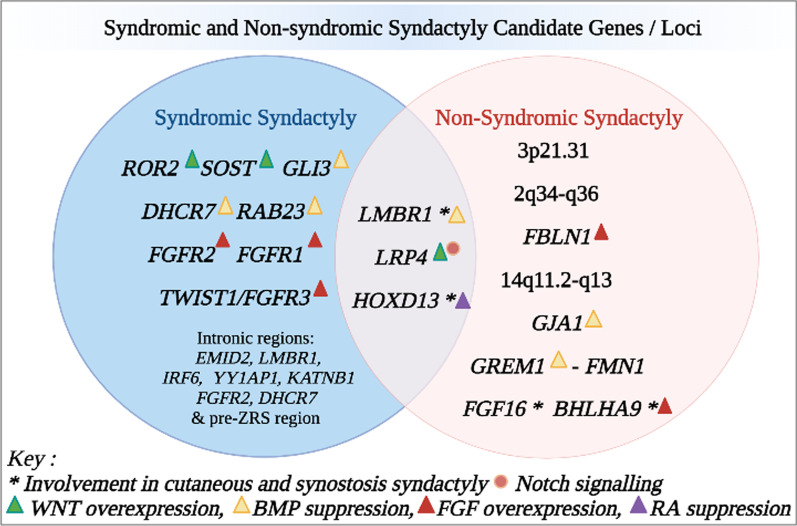
Emerging evidence into genes in syndactyly may provide an alternative gene screening panel as a diagnostic; especially prenatally when radiographic diagnosis is almost impossible. The GJA1, HOXD13 and FGF16 genes involved in non-syndromic syndactyly, involve the fourth webspace and can offer diagnostic choice in the pretext of prenatal screening; to prevent angulatory deformities later [3]. The LMBR1, LRP4 and HOX13 genes are seen to be deregulated in both syndromic and non-syndromic forms (Fig. 1). Additionally, HOXD13 may be able to modulate both cutaneous and osseous syndactyly pathways. Hence, inclusion of these genes in gene panels will expand diagnosis criteria. During diagnosis, if no conclusion is reached on gene testing, it can be important to screen flanking regions, and intronic regions. Not to forget the presence of syndactyly itself, in syndromes, can sometimes be a conclusive factor to categorize the syndrome (Ex: syndactyly of the second/third toes in SLOS) [46].
Fig. 1.
Representation of genes and loci in syndromic and non syndromic syndactyly analyzed in this review. These variants are further characterized according to involvement in documented syndactyly pathogenesis interactions. WNT Wingless‐type integration site family, BMP Bone Morphogenic Protein, FGF Fibroblast Growth Factor, RA Retinoic Acid
Further characterization of genetic determinants
Non syndromic syndactyly is thought to be well characterized, however there has been controversy over the associated MSSD phenotype, where both case reports did not include genetic testing for the BHLHA9, so the possibility of phenotypic heterogeneity cannot be excluded [63, 67]. Genetic Heterogeneity; although Cenani Lenz phenotype has been attributed to the gene LRP4, there have been reports with the same phenotype in APC gene [18]. Additionally, syndactyly Type III in Qattan’s study may prove another locus in addition to GLI3 [45]. Furthermore, since BMPs are secreted factors, gene expression patterns will not fully correlate with functional perspectives. Hence, protein distribution also needs to be considered. Moreover, understanding gene–gene interactions, non-coding variants and epigenetics further prompts better characterization of syndactyly phenotypes.
For syndactyly of the first and fourth webspaces, earlier release at approximately 6 months of age is advised [88]. Hence genes associated with non-syndromic Castilla type and Lerch type can be prioritized for further study, in addition to other known genes in syndromic syndactyly involving these webspaces. The complexity of syndactyly is furthered as one gene may involve more than one pathway. The LRP4 gene variants are known to be involved in overexpression of WNT, but also variants showing abrogated Notch signaling [28]. On the other hand, majority of syndromic syndactyly phenotypes remain unclassified in terms of likely involvement in pathways [73, 89–92].
Therapeutic targets from Mendelian traits and genome-wide association studies (GWASs) are more likely to be successful in clinical studies for multifactorial diseases [93]. Thus, efforts to analyze genes in this regard seem promising.
Conclusion
Autopod development is a tightly regulated process involving a plethora of genes. A classical belief is that loss of interdigital cell death by the WNT canonical pathway causes syndactyly. However, many genes with unclassified pathways still exist, with most other pathways being investigated in only animal studies. We propose that a conglomeration of genes in the canonical and non-canonical WNT signaling, Notch signaling, ADAMTS metalloprotease pathways and non-coding regions may work in concert to establish normal development of the limb bud. Candidate gene studies on HOXD13, LMBR1, FGF16, BHLHA9, to understand their contribution to both cutaneous and synostosis syndactyly phenotypes, could enhance screening. The genes GJA1, HOXD13 and FGF16 along with other genes involving either the first and fourth webspaces may also be prioritized as prenatal genetic testing here is urgently necessary. Collectively, the translation of these findings to human research, especially pathway mapping, could further resolve the genetic underpinnings associated with syndactyly. Furthermore, it will aid in the management of syndactyly and facilitate biomarker discovery.
Acknowledgements
All figures created with BioRender.com.
Abbreviations
- ADAMTS
A disintegrin and metalloproteinase with thrombospondin motifs
- AER
Apical ectodermal ridge
- BMP
Bone morphogenic protein
- CLSS
Cenani–Lenz syndactyly syndrome
- CRD
Cysteine-rich domain
- DHCR7
7-Dehydrocholesterol (7-DHC) reductase
- ECM
Extracellular matrix
- FGF
Fibroblast growth factor
- GLIA
GLI3 activator
- GLIR
GLI3 repressor
- GCPS
Greig cephalopolysyndactyly syndrome
- IHH
Indian hedgehog
- ICD
Interdigital cell death
- LRP4
Low-density lipoprotein receptor-related protein 4
- MDDS
Mesoaxial synostotic syndactyly syndrome
- MS4
Synostosis of the fourth and fifth metacarpals
- PCP
Planar cell polarity
- RA
Retinoic acid
- SCS
Saethre–Chotzen syndrome
- SHH
Sonic Hedgehog
- SLOS
Smith–Lemli–Opitz syndrome
- WNT
Wingless‐type integration site family
- ZPA
Zone of polarising activity
- ZRS
Zone of polarizing activity regulatory sequence
Author contributions
AC wrote the first draft of the manuscript and visualisations, with contributions from DH. VHWD critically revised the final manuscript for important intellectual content. All authors reviewed, modified and approved the final version of the manuscript.
Funding
This project did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors consent for publication.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Afraah Cassim, Email: c.afraah@gmail.com.
Dineshani Hettiarachchi, Email: dineshani@anat.cmb.ac.lk.
Vajira H. W. Dissanayake, Email: vajira@anat.cmb.ac.lk
References
- 1.Malik S. Syndactyly: phenotypes, genetics and current classification. Eur J Hum Genet. 2012;20:817–824. doi: 10.1038/ejhg.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hernández-Martínez R, Covarrubias L. Interdigital cell death function and regulation: new insights on an old programmed cell death model. Dev Growth Differ. 2011;53:245–258. doi: 10.1111/j.1440-169X.2010.01246.x. [DOI] [PubMed] [Google Scholar]
- 3.Jordan D, Hindocha S, Dhital M, Saleh M, Khan W. The epidemiology, genetics and future management of syndactyly. Open Orthop J. 2012;6:14–27. doi: 10.2174/1874325001206010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed H, Akbari H, Emami A, Akbari MR. Genetic overview of syndactyly and polydactyly. Plast Reconstr Surg. 2017;5:1–8. doi: 10.1097/GOX.0000000000001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Umair M, Ahmad F, Bilal M, Abbas S. Syndactyly genes and classification: a mini review. J Biochem Clin Genet. 2018;1:10–10. [Google Scholar]
- 6.Montero JA, Hurle JM. Sculpturing digit shape by cell death. Apoptosis. 2010;15:365–375. doi: 10.1007/s10495-009-0444-5. [DOI] [PubMed] [Google Scholar]
- 7.Al-Qattan MM. A review of the genetics and pathogenesis of syndactyly in humans and experimental animals: a 3-step pathway of pathogenesis. Biomed Res Int. 2019;2019:9652649. doi: 10.1155/2019/9652649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCulloch DR, Nelson CM, Dixon LJ, Silver DL, Wylie JD, Lindner V, et al. ADAMTS metalloproteases generate active versican fragments that regulate interdigital web regression. Dev Cell. 2009;17:687–698. doi: 10.1016/j.devcel.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bocci F, Onuchic JN, Jolly MK. Understanding the principles of pattern formation driven by Notch signaling by integrating experiments and theoretical models. Front Physiol. 2020;11:929. doi: 10.3389/fphys.2020.00929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuss P, Kraft K, Stumm J, Ibrahim D, Vallecillo-Garcia P, Mundlos S, et al. Regulation of cell polarity in the cartilage growth plate and perichondrium of metacarpal elements by HOXD13 and WNT5A. Dev Biol. 2014;385:83–93. doi: 10.1016/j.ydbio.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 11.Ermito S, Dinatale A, Carrara S, Cavaliere A, Imbruglia L, Recupero S. Prenatal diagnosis of limb abnormalities: role of fetal ultrasonography. J Prenat Med. 2009;3:18–22. [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Ma D, Sun Y, Meng L, Wang Y, Jiang T. Apert syndrome with FGFR2 758 C > G mutation: a Chinese case report. Front Genet. 2018;9:1–4. doi: 10.3389/fgene.2018.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaltcheva MM, Anderson MJ, Harfe BD, Lewandoski M. BMPs are direct triggers of interdigital programmed cell death. Dev Biol. 2016;411:266–276. doi: 10.1016/j.ydbio.2015.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pajni-Underwood S, Wilson CP, Elder C, Mishina Y, Lewandoski M. BMP signals control limb bud interdigital programmed cell death by regulating FGF signaling. Development. 2007;134:2359–2368. doi: 10.1242/dev.001677. [DOI] [PubMed] [Google Scholar]
- 15.Macias D, Gañan Y, Ros MA, Hurle JM. In vivo inhibition of programmed cell death by local administration of FGF-2 and FGF-4 in the interdigital areas of the embryonic chick leg bud. Anat Embryol. 1996;193:533–541. doi: 10.1007/BF00187925. [DOI] [PubMed] [Google Scholar]
- 16.Boulet AM, Moon AM, Arenkiel BR, Capecchi MR. The roles of Fgf4 and Fgf8 in limb bud initiation and outgrowth. Dev Biol. 2004;273:361–372. doi: 10.1016/j.ydbio.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 17.Hernández-Martínez R, Castro-Obregón S, Covarrubias L. Progressive interdigital cell death: regulation by the antagonistic interaction between fibroblast growth factor 8 and retinoic acid. Development. 2009;136:3669–3678. doi: 10.1242/dev.041954. [DOI] [PubMed] [Google Scholar]
- 18.Al-Qattan MM, Alkuraya FS. Cenani-Lenz syndrome and other related syndactyly disorders due to variants in LRP4, GREM1/FMN1, and APC: insight into the pathogenesis and the relationship to polyposis through the WNT and BMP antagonistic pathways. Am J Med Genet Part A. 2019;179:266–279. doi: 10.1002/ajmg.a.60694. [DOI] [PubMed] [Google Scholar]
- 19.Murgai A, Altmeyer S, Wiegand S, Tylzanowski P, Stricker S. Cooperation of BMP and IHH signaling in interdigital cell fate determination. PLoS ONE. 2018;13:e0197535. doi: 10.1371/journal.pone.0197535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gros J, Hu JKH, Vinegoni C, Feruglio PF, Weissleder R, Tabin CJ. WNT5A/JNK and FGF/MAPK pathways regulate the cellular events shaping the vertebrate limb bud. Curr Biol. 2010;20:1993–2002. doi: 10.1016/j.cub.2010.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montero JA, Hurle JM, Lorda-diez CI, Sanchez-fernandez C. Cell death in the developing vertebrate limb: a locally regulated mechanism contributing to musculoskeletal tissue morphogenesis and differentiation. 2020;1–12. [DOI] [PMC free article] [PubMed]
- 22.Zhu X-J, Fang Y, Xiong Y, Wang M, Yang X, Li Y, et al. Disruption of Wnt production in Shh lineage causes bone malformation in mice, mimicking human Malik–Percin-type syndactyly. FEBS Lett. 2018;592:356–368. doi: 10.1002/1873-3468.12963. [DOI] [PubMed] [Google Scholar]
- 23.Gao B, Song H, Bishop K, Elliot G, Garrett L, English MA, et al. Wnt signaling gradients establish planar cell polarity by inducing vangl2 phosphorylation through Ror2. Dev Cell. 2011;20:163–176. doi: 10.1016/j.devcel.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian J, Shao J, Liu C, Hou HY, Chou CW, Shboul M, et al. Deficiency of lrp4 in zebrafish and human LRP4 mutation induce aberrant activation of Jagged–Notch signaling in fin and limb development. Cell Mol Life Sci. 2019;76:163–178. doi: 10.1007/s00018-018-2928-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehawej C, Chouery E, Maalouf D, Baujat G, Le Merrer M, Cormier-Daire V, et al. Identification of a novel causative mutation in the ROR2 gene in a Lebanese family with a mild form of recessive Robinow syndrome. Eur J Med Genet. 2012;55:103–108. doi: 10.1016/j.ejmg.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 26.Leupin O, Piters E, Halleux C, Hu S, Kramer I, Morvan F, et al. Bone overgrowth-associated mutations in the LRP4 gene impair sclerostin facilitator function. J Biol Chem. 2011;286:19489–19500. doi: 10.1074/jbc.M110.190330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boudin E, Yorgan T, Fijalkowski I, Sonntag S, Steenackers E, Hendrickx G, et al. The Lrp4 R1170Q homozygous knock-in mouse recapitulates the bone phenotype of Sclerosteosis in humans. J Bone Miner Res. 2017;32:1739–1749. doi: 10.1002/jbmr.3160. [DOI] [PubMed] [Google Scholar]
- 28.Alrayes N, Aziz A, Ullah F, Ishfaq M, Jelani M, Wali A. Novel missense alteration in LRP4 gene underlies Cenani–Lenz syndactyly syndrome in a consanguineous family. J Gene Med. 2020;22:e3143. doi: 10.1002/jgm.3143. [DOI] [PubMed] [Google Scholar]
- 29.Sebastian A, Loots GG. Genetics of Sost/SOST in sclerosteosis and van Buchem disease animal models. Metabolism. 2018;80:38–47. doi: 10.1016/j.metabol.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Démurger F, Ichkou A, Mougou-Zerelli S, Le Merrer M, Goudefroye G, Delezoide AL, et al. New insights into genotype-phenotype correlation for GLI3 mutations. Eur J Hum Genet. 2015;23:92–102. doi: 10.1038/ejhg.2014.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi L, Huang H, Jiang Q, Huang R, Fu W, Mao L, et al. Sub-exome target sequencing in a family with syndactyly type IV due to a novel partial duplication of the LMBR1 gene: first case report in Fujian Province of China. Front Genet. 2020;11:130. doi: 10.3389/fgene.2020.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu J, Wu J, Teng X, Cai L, Yuan H, Chen X, et al. Large duplication in LMBR1 gene in a large Chinese pedigree with triphalangeal thumb polysyndactyly syndrome. Am J Med Genet Part A. 2020;182:2117–2123. doi: 10.1002/ajmg.a.61757. [DOI] [PubMed] [Google Scholar]
- 33.Koide T, Hayata T, Cho KWY. Negative regulation of Hedgehog signaling by the cholesterogenic enzyme 7-dehydrocholesterol reductase. Development. 2006;133:2395–2405. doi: 10.1242/dev.02393. [DOI] [PubMed] [Google Scholar]
- 34.Azoury SC, Reddy S, Shukla V, Deng C-X. Fibroblast growth factor receptor 2 (FGFR2) mutation related syndromic craniosynostosis. Int J Biol Sci. 2017;13:1479–1488. doi: 10.7150/ijbs.22373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Rourke MP, Soo K, Behringer RR, Hui CC, Tam PPL. Twist plays an essential role in FGF and SHH signal transduction during mouse limb development. Dev Biol. 2002;248:143–156. doi: 10.1006/dbio.2002.0730. [DOI] [PubMed] [Google Scholar]
- 36.Zuniga A, Quillet R, Perrin-Schmitt F, Zeller R. Mouse Twist is required for fibroblast growth factor-mediated epithelial-mesenchymal signalling and cell survival during limb morphogenesis. Mech Dev. 2002;114:51–59. doi: 10.1016/s0925-4773(02)00048-5. [DOI] [PubMed] [Google Scholar]
- 37.Zhao X, Sun M, Zhao J, Leyva JA, Zhu H, Yang W, et al. Mutations in HOXD13 underlie syndactyly type V and a novel brachydactyly-syndactyly syndrome. Am J Hum Genet. 2007;80:361–371. doi: 10.1086/511387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Avitan-Hersh E, Indelman M, Bergman R, Sprecher E. ADULT syndrome caused by a mutation previously associated with EEC syndrome. Pediatr Dermatol. 2010;27:643–645. doi: 10.1111/j.1525-1470.2010.01131.x. [DOI] [PubMed] [Google Scholar]
- 39.Alessandri J-L, Dagoneau N, Laville J-M, Baruteau J, Hébert J-C, Cormier-Daire V. RAB23 mutation in a large family from Comoros Islands with Carpenter syndrome. Am J Med Genet Part A. 2010;152A:982–986. doi: 10.1002/ajmg.a.33327. [DOI] [PubMed] [Google Scholar]
- 40.Schwarzer W, Witte F, Rajab A, Mundlos S, Stricker S. A gradient of ROR2 protein stability and membrane localization confers brachydactyly type B or Robinow syndrome phenotypes. Hum Mol Genet. 2009;18:4013–4021. doi: 10.1093/hmg/ddp345. [DOI] [PubMed] [Google Scholar]
- 41.van Lierop AH, Appelman-Dijkstra NM, Papapoulos SE. Sclerostin deficiency in humans. Bone. 2017;96:51–62. doi: 10.1016/j.bone.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 42.Boudin E, Yorgan ÃT, Fijalkowski ÃI, Sonntag S, Steenackers E, Hendrickx G, et al. The Lrp4 R1170Q homozygous knock-in mouse humans. J Bone Miner Res. 2017;32:1739–1749. doi: 10.1002/jbmr.3160. [DOI] [PubMed] [Google Scholar]
- 43.Sethi SK, Goyal D, Khalil S, Yadav DK. Two Indian families with Greig cephalopolysyndactyly with non-syndromic phenotype. Eur J Pediatr. 2013;172:1131–1135. doi: 10.1007/s00431-013-1938-2. [DOI] [PubMed] [Google Scholar]
- 44.Yousaf M, Azeem Z, Bilal M, Liaqat K, Hussain S, et al. Variants in GLI3 cause Greig Cephalopolysyndactyly syndrome. Genet Test Mol Biomark. 2019;23:744–750. doi: 10.1089/gtmb.2019.0071. [DOI] [PubMed] [Google Scholar]
- 45.Al-Qattan MM, Shamseldin HE, Al Mazyad M, Al Deghaither S, Alkuraya FS. Genetic heterogeneity in type III familial cutaneous syndactyly and linkage to chromosome 7q36. Am J Med Genet Part A. 2013;161:1579–1584. doi: 10.1002/ajmg.a.35956. [DOI] [PubMed] [Google Scholar]
- 46.Ko JS, Choi BS, Seo JK, Shin JY, Chae JH, Kang GH, et al. A novel DHCR7 mutation in a Smith–Lemli–Opitz syndrome infant presenting with neonatal cholestasis. J Korean Med Sci. 2010;25:159–162. doi: 10.3346/jkms.2010.25.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Twigg SRF, Lloyd D, Jenkins D, Elçioglu NE, Cooper CDO, Al-Sannaa N, et al. Mutations in multidomain protein MEGF8 identify a carpenter syndrome subtype associated with defective lateralization. Am J Hum Genet. 2012;91:897–905. doi: 10.1016/j.ajhg.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haye D, Collet C, Sembely-Taveau C, Haddad G, Denis C, Soulé N, et al. Prenatal findings in carpenter syndrome and a novel mutation in RAB23. Am J Med Genet Part A. 2014;164:2926–2930. doi: 10.1002/ajmg.a.36726. [DOI] [PubMed] [Google Scholar]
- 49.Jenkins D, Seelow D, Jehee FS, Perlyn CA, Alonso LG, Bueno DF, et al. RAB23 mutations in carpenter syndrome imply an unexpected role for Hedgehog signaling in cranial-suture development and obesity. Am J Hum Genet. 2007;80:1162–1170. doi: 10.1086/518047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh CB, Mishra B, Patel R, Kumar A, Ali A. Tripod-shaped syndactyly in Apert syndrome with FGFR2 p.P253R mutation. Indian J Plast Surg. 2021;54:370. doi: 10.1055/s-0041-1733808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki H, Suda N, Shiga M, Kobayashi Y, Nakamura M, Iseki S, et al. Apert syndrome mutant FGFR2 and its soluble form reciprocally alter osteogenesis of primary Calvarial osteoblasts. J Cell Physiol. 2012;227:3267–3277. doi: 10.1002/jcp.24021. [DOI] [PubMed] [Google Scholar]
- 52.Hackett A, Rowe L. FGFR1 Pfeiffer syndrome without craniosynostosis: an additional case report. Clin Dysmorphol. 2006;15:207–210. doi: 10.1097/01.mcd.0000220608.40155.d4. [DOI] [PubMed] [Google Scholar]
- 53.Chun K, Teebi AS, Jung JH, Kennedy S, Laframboise R, Meschino WS, et al. Genetic analysis of patients with the Saethre–Chotzen phenotype. Am J Med Genet. 2002;110:136–143. doi: 10.1002/ajmg.10400. [DOI] [PubMed] [Google Scholar]
- 54.Tian J, Shao J, Liu C, Hou HY, Chou CW, Shboul M, et al. Deficiency of lrp4 in zebrafish and human LRP4 mutation induce aberrant activation of Jagged–Notch signaling in fin and limb development. Cell Mol Life Sci. 2019;76:163–178. doi: 10.1007/s00018-018-2928-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lettice LA, Daniels S, Sweeney E, Venkataraman S, Devenney PS, Gautier P, et al. Enhancer-adoption as a mechanism of human developmental disease. Hum Mutat. 2011;32:1492–1499. doi: 10.1002/humu.21615. [DOI] [PubMed] [Google Scholar]
- 56.Charzewska A, Obersztyn E, Hoffman-Zacharska D, Lenart J, Poznański J, Bal J. Novel mutations in the IRF6 gene on the background of known polymorphisms in Polish patients with orofacial clefting. Cleft Palate-Craniofacial J. 2015;52:e161–e167. doi: 10.1597/14-030. [DOI] [PubMed] [Google Scholar]
- 57.Rath M, Spiegler S, Strom TM, Trenkler J, Kroisel PM, Felbor U. Identification of pathogenic YY1AP1 splice variants in siblings with Grange syndrome by whole exome sequencing. Am J Med Genet Part A. 2019;179:295–299. doi: 10.1002/ajmg.a.60700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yigit G, Wieczorek D, Bögershausen N, Beleggia F, Möller-Hartmann C, Altmüller J, et al. A syndrome of microcephaly, short stature, polysyndactyly, and dental anomalies caused by a homozygous KATNB1 mutation. Am J Med Genet Part A. 2016;170:728–733. doi: 10.1002/ajmg.a.37484. [DOI] [PubMed] [Google Scholar]
- 59.Torres L, Hernández G, Barrera A, Ospina S, Prada R. Molecular analysis of exons 8, 9 and 10 of the fibroblast growth factor receptor 2 (FGFR2) gene in two families with index cases of apert syndrome. Colomb Med. 2015;46:155–158. [PMC free article] [PubMed] [Google Scholar]
- 60.Waye JS, Nakamura LM, Eng B, Hunnisett L, Chitayat D, Costa T. Smith-Lemli-Opitz syndrome: carrier frequency and spectrum of DHCR7 mutations in Canada. J Med Genet. 2002;39:31. doi: 10.1136/jmg.39.6.e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Potuijt JWP, Baas M, Sukenik-Halevy R, Douben H, Nguyen P, Venter DJ, et al. A point mutation in the pre-ZRS disrupts sonic hedgehog expression in the limb bud and results in triphalangeal thumb–polysyndactyly syndrome. Genet Med. 2018;20:1405–1413. doi: 10.1038/gim.2018.18. [DOI] [PubMed] [Google Scholar]
- 62.Díaz-González F, Parrón-Pajares M, Barcia-Ramirez A, Heath KE. First case of compound heterozygous BHLHA9 variants in mesoaxial synostotic syndactyly with phalangeal reduction. Am J Med Genet Part A. 2020;182:628–631. doi: 10.1002/ajmg.a.61480. [DOI] [PubMed] [Google Scholar]
- 63.Malik S. Mesoaxial synostotic syndactyly with phalangeal reduction (MSSD): syndactyly type IX. Skelet Radiol. 2018;47:149–149. doi: 10.1007/s00256-017-2842-z. [DOI] [PubMed] [Google Scholar]
- 64.Malik S, Schott J, Ali SW, Oeffner F, Amin-Ud-Din M, Ahmad W, et al. Evidence for clinical and genetic heterogeneity of syndactyly type I: the phenotype of second and third toe syndactyly maps to chromosome 3p21.31. Eur J Hum Genet. 2005;13:1268–1274. doi: 10.1038/sj.ejhg.5201492. [DOI] [PubMed] [Google Scholar]
- 65.Malik S, Abbasi AA, Ansar M, Ahmad W, Koch MC, Grzeschik KH. Genetic heterogeneity of synpolydactyly: a novel locus SPD3 maps to chromosome 14q11.2–q12. Clin Genet. 2006;69:518–524. doi: 10.1111/j.1399-0004.2006.00620.x. [DOI] [PubMed] [Google Scholar]
- 66.Weinrich JM, Ajabnoor W, Bannas P. Case report of a novel nonsyndromic unilateral syndactyly of the hand. Skeletal Radiol. 2017;46:1741–1743. doi: 10.1007/s00256-017-2725-3. [DOI] [PubMed] [Google Scholar]
- 67.Özdemir M, Kavak P, Kaplanoğlu H. Case report an unusual association of ulnar Hemimelia with mesoaxial synostotic syndactyly. BJR Case Rep. 2020;6:20190073. doi: 10.1259/bjrcr.20190073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bosse K, Betz RC, Lee YA, Wienker TF, Reis A, Kleen H, et al. Localization of a gene for syndactyly type 1 to chromosome 2q34-q36. Am J Hum Genet. 2000;67:492–497. doi: 10.1086/303028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ghadami M, Majidzadeh-A K, Haerian BS, Damavandi E, Yamada K, Pasallar P, et al. Confirmation of genetic homogeneity of syndactyly type 1 in an Iranian family. Am J Med Genet. 2001;104:147–151. doi: 10.1002/ajmg.10061. [DOI] [PubMed] [Google Scholar]
- 70.Montagu MFA. A pedigree of Syndactylism of the middle and ring fingers. Hum Genet. 1953;5:70–72. [PMC free article] [PubMed] [Google Scholar]
- 71.Castilla EE, Paz JE, Orioli-Parreiras IM. Syndactyly: Frequency of specific types. Am J Med Genet. 1980;5:357–364. doi: 10.1002/ajmg.1320050406. [DOI] [PubMed] [Google Scholar]
- 72.Muragaki Y, Mundlos S, Upton J, Olsen BR. Altered growth and branching patterns in synpolydactyly caused by mutations in HOXD13. Science (80-). 1996;272:548–551. doi: 10.1126/science.272.5261.548. [DOI] [PubMed] [Google Scholar]
- 73.Debeer P, Schoenmakers EFPM, Twal WO, Argraves WS, De Smet L, Fryns JP, et al. The fibulin-1 gene (FBLN1)is disrupted in a t(12;22) associated with a complex type of synpolydactyly. J Med Genet. 2002;39:98–104. doi: 10.1136/jmg.39.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ngoc NT, Duong NT, Quynh DH, Ton ND, Duc HH, Huong LTM, et al. Identification of novel missense mutations associated with non-syndromic syndactyly in two vietnamese trios by whole exome sequencing. Clin Chim Acta. 2020;506:16–21. doi: 10.1016/j.cca.2020.03.017. [DOI] [PubMed] [Google Scholar]
- 75.Dobrowolski R, Sasse P, Schrickel JW, Watkins M, Kim JS, Rackauskas M, et al. The conditional connexin43G138R mouse mutant represents a new model of hereditary oculodentodigital dysplasia in humans. Hum Mol Genet. 2008;17:539–554. doi: 10.1093/hmg/ddm329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Johnston O, Kirby VV. Syndactyly of the ring and little finger. Am J Hum Genet. 1955;7:80–82. [PMC free article] [PubMed] [Google Scholar]
- 77.Haas SL. Bilateral complete syndactylism of all fingers. Am J Surg. 1940;50:363–366. [Google Scholar]
- 78.Wieczorek D, Pawlik B, Li Y, Akarsu NA, Caliebe A, May KJW, et al. A specific mutation in the distant sonic hedgehog (SHH) cis-regulator (ZRS) causes Werner Mesomelic Syndrome (WMS) while complete ZRS duplications underlie Haas type polysyndactyly and preaxial polydactyly (PPD) with or without triphalangeal thumb. Hum Mutat. 2010;31:81–89. doi: 10.1002/humu.21142. [DOI] [PubMed] [Google Scholar]
- 79.Wu L, Liang D, Niikawa N, Ma F, Sun M, Pan Q, et al. A ZRS duplication causes Syndactyly type IV with Tibial Hypoplasia. Am J Med Genet Part A. 2009;149:816–818. doi: 10.1002/ajmg.a.32740. [DOI] [PubMed] [Google Scholar]
- 80.Cn D. Cleft hand: a report of a case successfully treated by the use of periosteal flaps. Ann Surg. 1896;24:211–216. doi: 10.1097/00000658-189607000-00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Temtamy SA, McKusick VA. The genetics of hand malformations. 1978. [PubMed]
- 82.Li Y, Pawlik B, Elcioglu N, Aglan M, Kayserili H, Yigit G, et al. LRP4 mutations Alter Wnt/β-catenin signaling and cause limb and kidney malformations in Cenani–Lenz syndrome. Am J Hum Genet. 2010;86:696–706. doi: 10.1016/j.ajhg.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dimitrov BI, Voet T, De Smet L, Vermeesch JR, Devriendt K, Fryns JP, et al. Genomic rearrangements of the GREM1-FMN1 locus cause oligosyndactyly, radio-ulnar synostosis, hearing loss, renal defects syndrome and Cenanie–Lenz-like non-syndromic oligosyndactyly. J Med Genet. 2010;47:569–574. doi: 10.1136/jmg.2009.073833. [DOI] [PubMed] [Google Scholar]
- 84.Jamsheer A, Zemojtel T, Kolanczyk M, Stricker S, Hecht J, Krawitz P, et al. Whole exome sequencing identifies FGF16 nonsense mutations as the cause of X-linked recessive metacarpal 4/5 fusion. J Med Genet. 2013;50:579–584. doi: 10.1136/jmedgenet-2013-101659. [DOI] [PubMed] [Google Scholar]
- 85.Jones B, Byers H, Stewart Watson J, Newman WG. Identification of a novel familial FGF16 mutation in metacarpal 4–5 fusion. Clin Dysmorphol. 2014;23:95–97. doi: 10.1097/MCD.0000000000000043. [DOI] [PubMed] [Google Scholar]
- 86.Kantaputra PN, Carlson BM. Genetic regulatory pathways of split-hand/foot malformation. Clin Genet. 2019;95:132–139. doi: 10.1111/cge.13434. [DOI] [PubMed] [Google Scholar]
- 87.Khan A, Wang R, Han S, Ahmad W, Zhang X. A novel homozygous missense mutation in BHLHA9 causes mesoaxial synostotic syndactyly with phalangeal reduction in a Pakistani family. Hum Genome Var. 2017;4:1–4. doi: 10.1038/hgv.2017.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Adkinson JM, Chung KC. Release of Finger Syndactyly Using Dorsal Rectangular Flap. Oper Tech Hand Wrist Surg. 2018. p. 811–8.
- 89.Boczek NJ, Miller EM, Ye D, Nesterenko VV, Tester DJ, Antzelevitch C, et al. Novel Timothy syndrome mutation leading to increase in CACNA1C window current. Hear Rhythm. 2015;12:211–219. doi: 10.1016/j.hrthm.2014.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim H-G, Rosenfeld JA, Scott DA, Bénédicte G, Labonne JD, Brown J, et al. Disruption of PHF21A causes syndromic intellectual disability with craniofacial anomalies, epilepsy, hypotonia, and neurobehavioral problems including autism. Mol Autism. 2019;10:35. doi: 10.1186/s13229-019-0286-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Orge FH, Dar SA, Blackburn CN, Grimes-Hodges SJ, Mitchell AL. Ocular manifestations of X-linked dominant FAM58A mutation in toe syndactyly, telecanthus, anogenital, and renal malformations (‘STAR’) syndrome. Ophthalmic Genet. 2016;37:323–327. doi: 10.3109/13816810.2015.1071407. [DOI] [PubMed] [Google Scholar]
- 92.Vladimir Socolov R, Ioana Andreescu N, Maria Haliciu A, Vlad Gorduza E, Dumitrache F, Anca Balan R, et al. Intrapartum diagnostic of Roberts syndrome-case presentation. Rom J Morphol Embryol. 2015;56:585–588. [PubMed] [Google Scholar]
- 93.Struck TJ, Mannakee BK, Gutenkunst RN. The impact of genome-wide association studies on biomedical research publications. Hum Genom. 2018;12:1–9. doi: 10.1186/s40246-018-0172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.