SUMMARY
Microvilli are conserved actin-based surface protrusions that have been repurposed throughout evolution to fulfill diverse cell functions. In the case of transporting epithelia, microvilli are supported by a core of actin filaments bundled in parallel by villin, fimbrin, and espin. Remarkably, microvilli biogenesis persists in mice lacking all three of these factors, suggesting the existence of unknown bundlers. We identified Mitotic Spindle Positioning (MISP) as an actin-binding factor that localizes specifically to the rootlet end of the microvillus. MISP promotes rootlet elongation in cells, and purified MISP exhibits potent filament bundling activity in vitro. MISP-bundled filaments also recruit fimbrin, which further elongates and stabilizes bundles. MISP confinement to the rootlet is enforced by ezrin, which prevents decoration of the membrane-wrapped distal end of the core bundle. These discoveries reveal how epithelial cells optimize apical membrane surface area and offer insight on the remarkable robustness of microvilli biogenesis.
In brief
Morales et al. identify Mitotic Spindle Positioning (MISP) as an actin bundler in the rootlets of epithelial microvilli. MISP cooperates with other bundlers, and its rootlet-specific localization is enforced by membrane-actin linker ezrin. These findings illuminate mechanisms that drive the assembly and compartmentalization of actin bundle-supported protrusions.
Graphical Abstract
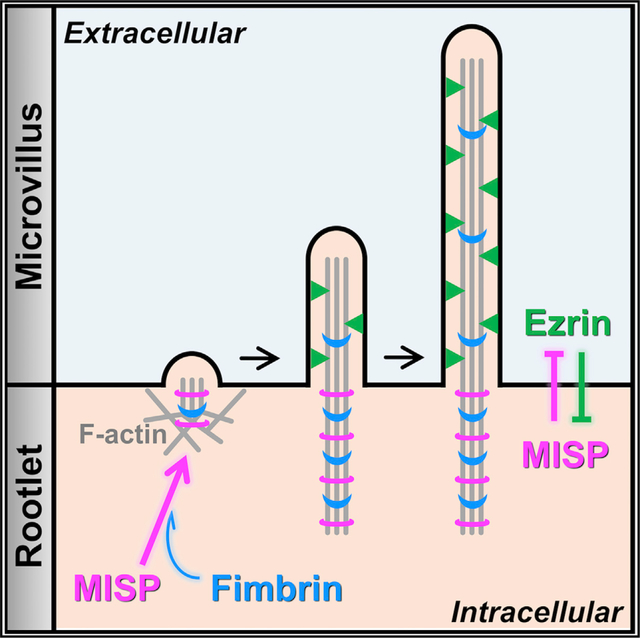
INTRODUCTION
Surface protrusions are essential features that enable cells to interact with the external environment in all domains of life. Choanoflagellates, among the closest living relatives of animals, are unicellular eukaryotes that developed one of the earliest known polarized feeding systems consisting of long-lived actin-based protrusions, which we now generally refer to as microvilli (Sebé-Pedrós et al., 2013). Multicellular eukaryotes eventually maximized solute transport by compartmentalizing large numbers of microvilli on the surface of specialized hollow organs (Peña et al., 2016). In animals, striking examples of such organization are found on the apical luminal surface of enterocytes in the small intestine, where densely packed arrays of thousands of microvilli extend from the surface of individual cells, collectively forming the brush border (BB) (Crawley et al., 2014). In other specialized cases, arrays of microvilli have been repurposed for diverse functions, including sperm recognition in oocytes, mechanosensation in inner ear hair cells, and light detection in photoreceptor cells, among others (Lange, 2011).
An individual microvillus extends from the cell surface as a finger-like membrane protrusion, supported by a core of 20–40 actin filaments bundled in parallel (Mooseker and Tilney, 1975; Ohta et al., 2012). Core actin bundles exhibit lengths on the micron scale and flexural rigidities high enough to deform the enveloping plasma membrane (Atilgan et al., 2006). Previous studies established that at least three bundlers assemble the microvillar core bundle: villin, fimbrin (also known as plastin-1), and espin (Bartles et al., 1998; Bretscher and Weber, 1979, 1980). Villin is the first bundler recruited apically during BB differentiation, followed by fimbrin and espin (Bartles et al., 1998; Ezzell et al., 1989). Single-villin or espin knockout (KO) mice exhibit near-normal microvillar morphology and organization (Ferrary et al., 1999; Pinson et al., 1998; Revenu et al., 2012). In contrast, fimbrin KO mice exhibit microvilli that are ~15% shorter (Grimm-Günter et al., 2009; Revenu et al., 2012). Remarkably, villin-espin-fimbrin triple-KO mice are viable, and their enterocytes still assemble apical BBs, although microvillar length is reduced by ~40% (Revenu et al., 2012). This latter finding underscores the remarkable robustness of microvillar growth and further suggests that BB assembly is driven by multiple factors operating in parallel, some of which may remain unidentified.
Within the microvillus, actin filaments that comprise the core bundle are oriented with their barbed ends toward the distal tip and pointed ends extending down into the subapical cytoplasm (Mooseker and Tilney, 1975). The barbed ends are the preferred site of new actin monomer incorporation, whereas the pointed ends are the favored site of disassembly (Pollard and Mooseker, 1981). Kinetic differences at the two ends create a system that allows subunits to flux retrograde through the core bundle in a process referred to as “treadmilling” (Kirschner, 1980). Indeed, recent studies with epithelial cell culture models revealed that treadmilling is crucial for microvilli assembly and motility (Gaeta et al., 2021; Meenderink et al., 2019).
Although a long segment of the core bundle protrudes from the cell surface enveloped in plasma membrane, a much shorter segment, the “rootlet,” extends down into the subapical cytoplasm. Core bundle rootlets are directly linked to a dense filamentous network called the “terminal web,” an organelle-free zone that presumably regulates trafficking to and from the apical plasma membrane (Mooseker et al., 1983). Ultrastructural studies first suggested that rootlets are interconnected with terminal web filaments at least in part by non-muscle myosin-2 and spectrin (Hirokawa et al., 1982). Deep in the terminal web, rootlets appear to be directly crosslinked with a meshwork of cytokeratins (Hirokawa et al., 1982, 1983). One possible crosslinking factor is the actin bundler fimbrin, which is found along the length of the core bundle with an apparent enrichment on the rootlet (Grimm-Günter et al., 2009). Based on the highly interconnected nature of filaments throughout the terminal web, this network likely serves as a physical platform that offers long-term stability and mechanical support for protruding BB microvilli. Although core bundle rootlets can interact with the terminal web filaments only if they remain free of membrane wrapping, factors that protect the proximal end of the bundle from membrane encapsulation during microvillar growth remain undefined.
Biophysical investigations have also established that the structural stability of microvilli is promoted by tethering core bundles to the surrounding plasma membrane (Nambiar et al., 2010). Core bundles are laterally bridged to their enveloping membrane by myosin-1a and myosin-6, as well as ezrin (Berryman et al., 1993; Hegan et al., 2012; Howe and Mooseker, 1983). Recent studies on the dynamics of growing microvilli revealed that ezrin accumulates in a nascent microvillus in parallel with core bundle elongation, and that loss of ezrin from the protrusion leads to microvillus collapse (Gaeta et al., 2021). Ezrin is a well-characterized membrane-cytoskeleton linker that adopts two states: an open “active” state when phosphorylated and a closed “inactive” state when dephosphorylated (Bretscher et al., 1997). Dynamic cycling between these two states allows ezrin to bridge treadmilling actin bundles to the enveloping plasma membrane (Viswanatha et al., 2012), while providing continuous mechanical support for the protrusion. However, mechanisms that constrain ezrin enrichment to the distal segment of the core bundle and control the extent of membrane wrapping remain poorly understood.
Here we report that Mitotic Spindle Positioning (MISP) is a BB component that targets specifically to the rootlets of microvillar core bundles. Previous studies revealed that MISP is an actin-binding and bundling protein that promotes mitotic spindle orientation and mitotic progression (Kumeta et al., 2014; Maier et al., 2013; Zhu et al., 2013), although a role in native tissues has yet to be reported. In intestinal epithelial cells, we found that MISP is enriched in the subapical region beneath the plasma membrane at the base of the BB, where it colocalizes with fimbrin along core bundle rootlets. Loss- and gain-of-function studies revealed that MISP elongates rootlets and limits the extent of membrane wrapping of core bundles. Consistent with these phenotypes, MISP bundles F-actin in vitro and in cells, creating structures that are primed for fimbrin recruitment. Overexpression of both factors leads to a striking overgrowth of hyper-stable rootlets from the subapical domain. Further, we found that MISP confinement to microvillar rootlets depends on the presence of active ezrin in the microvillus. Overall, our findings lead to a model for rootlet specification whereby ezrin and MISP exert mutual exclusivity to establish membrane-wrapped versus unwrapped segments of the core bundle. MISP confinement to rootlets, in turn, recruits fimbrin to further crosslink the proximal ends of core bundles in the terminal web. This work holds important implications for understanding the assembly and stabilization of actin-based protrusions in diverse epithelial systems and also provides a molecular rationale for the remarkable robustness of BB assembly alluded to in previous multi-gene loss-of-function mouse models (Delacour et al., 2016).
RESULTS
MISP localizes to the rootlets of BB microvilli
In a previous proteomic study, we identified peptides from MISP in BBs isolated from mouse small intestine (McConnell et al., 2011). To validate MISP as a bona fide BB resident and to examine its localization in native tissues at higher resolution, we immunostained paraffin sections of mouse small intestine. Confocal microscopy of stained sections revealed that MISP specifically localizes to the BB along the full length of the crypt-villus axis (Figure 1A). This was consistent with MISP localization to the apical surface of the intestinal epithelium in H&E staining from the Human Protein Atlas (Figure S1A) (Uhlén et al., 2015). Using an anti-villin antibody to label core actin bundles and a membrane marker to delineate the apical surface, we found that MISP is highly enriched in the terminal web and exhibits mutually exclusive labeling with the membrane-wrapped protruding microvilli (Figures 1A and 1B). As previously reported (Dudouet et al., 1987; Robine et al., 1985), we found that villin signal gradually increases along the crypt-villus axis, following the direction of enterocyte migration and differentiation (Figures 1A, 1C, and 1D, magenta labels). In contrast, MISP signal remains relatively constant along the crypt-villus axis (Figures 1A, 1C, and 1D, green labels), suggesting that this factor is apically targeted independent of differentiation state. We recapitulated this observation in cell culture using CACO-2BBE cells, which differentiate and take on an enterocyte-like phenotype after a prolonged period of confluent culture (Peterson and Mooseker, 1993; Peterson et al., 1993). In this system, MISP was also expressed and localized from early to late stages of microvilli assembly (Figures 1E–1G). These staining results are consistent with a previous study showing that MISP targets apically in 3D CACO-2 cyst cultures (Kschonsak and Hoffmann, 2018). Localization to the actin-rich apical domain is also broadly consistent with staining in non-polarized HeLa cells, where endogenous and overexpressed MISP localizes to actin-rich structures (Kumeta et al., 2014; Maier et al., 2013; Zhu et al., 2013). Together, our data indicate MISP is a BB component that is highly enriched in the subapical terminal web throughout the full course of enterocyte differentiation.
Figure 1. MISP localizes to microvillar rootlets.
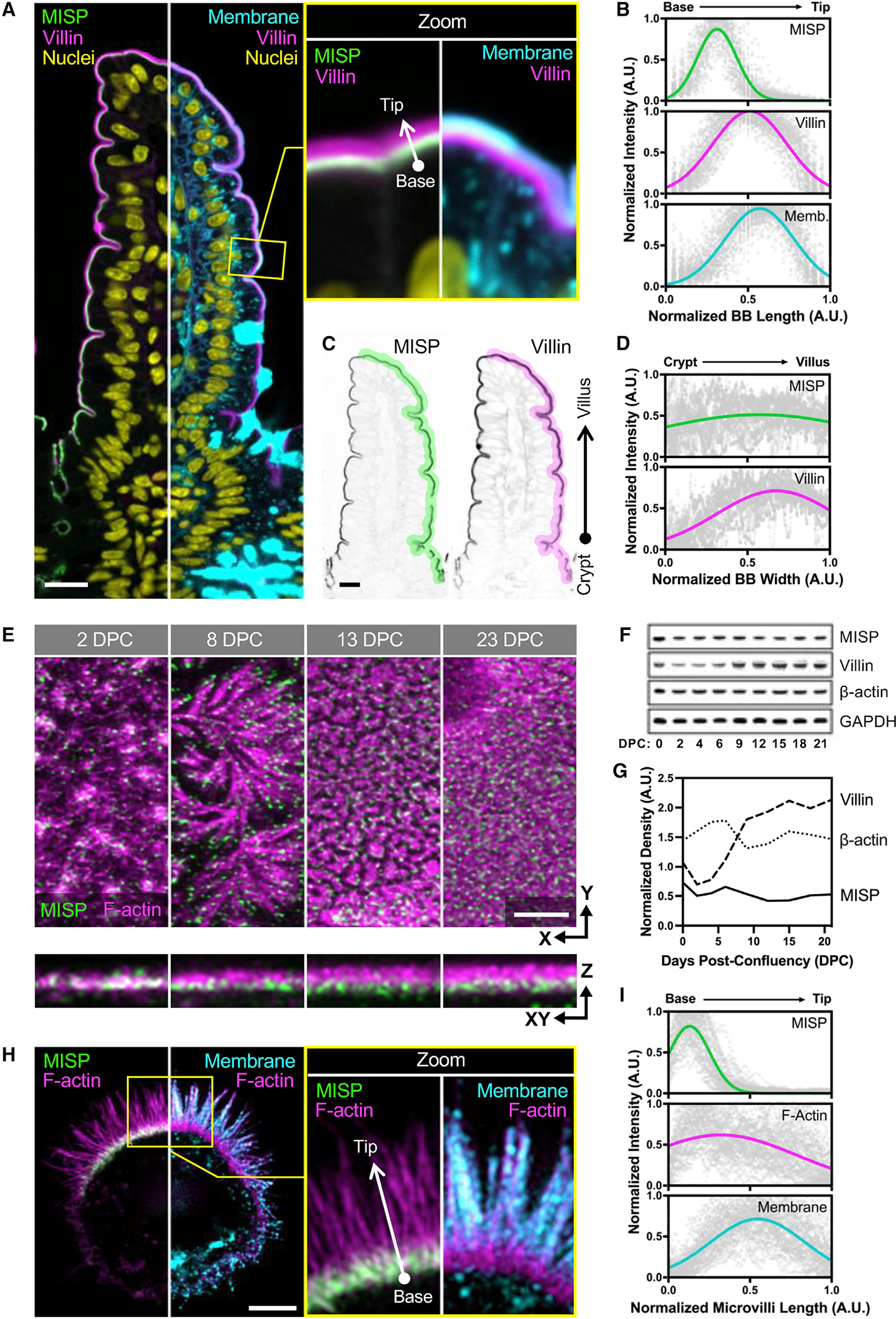
(A) Confocal images of small intestinal sections stained for MISP (green), villin (magenta), DNA with DRAQ5 (yellow), and membrane with WGA (cyan). Panel shows a split three-color merge. Scale bar: 15 μm.
(B) Fluorescence intensity distributions of MISP, villin, and apical membrane measured parallel to the microvillar axis as indicated by the white arrow in (A); n = 200 line scans measured on five villi.
(C) Inverted MISP and villin channels from (A). Scale bar: 15 μm.
(D) Fluorescence intensity distribution of MISP and villin measured relative to the crypt-villus axis as indicated by the highlighted area in (C); black arrow shows line scan orientation; n = 11 scans of six villi.
(E) Confocal maximum intensity projection (Max IP) of CACO-2BBE cells at different stages of differentiation stained for MISP (green) and F-actin with phalloidin (magenta). Upper panels show xy en face views at the indicated days post-confluency (DPC); bottom panels show resliced xz views. Scale bar: 3 μm.
(F) Western blot time series of CACO-2BBE cell lysates probed for MISP, villin, β-actin, and GAPDH at the indicated DPC.
(G) Density values of MISP, villin, and β-actin bands from F normalized to GAPDH and plotted as a function of DPC.
(H) SIM Max IP of a W4 cell stained for MISP (green), F-actin with phalloidin (magenta), and membrane with WGA (cyan). Panel shows a split two-color merge. Scale bar: 3 μm.
(I) Fluorescence intensity distributions of MISP, F-actin, and apical membrane from line scans measured parallel to the microvillar axis as indicated by the white arrow in (H); n = 58 line scans of single core bundles from 10 cells.
All plots in (B), (D), and (I) show Gaussian curve fits of the raw data for each channel.
Previous work established that MISP holds actin-binding and bundling potential (Kumeta et al., 2014). In light of these data and our imaging studies indicating highly specific terminal web enrichment, we next sought to determine whether MISP associates with core bundle rootlets at the base of microvilli. To examine this possibility, we turned to LS174T-W4 cells (herein referred to as W4 cells), a human intestinal epithelial cell line engineered to provide switch-like control over BB assembly (Baas et al., 2004). Using super-resolution structural illumination microscopy (SIM), we found that MISP was highly enriched on the rootlet segments of core bundles that extend immediately beneath the apical membrane (Figures 1H and 1I), which was consistent with its localization in mouse intestinal tissue. We also examined the localization of an EGFP-tagged variant of MISP expressed in LLC-PK1-CL4 cells, a pig kidney epithelial cell line that displays individual microvilli extending from the apical surface (Gaeta et al., 2021). In these cultures, we again noted a striking enrichment of MISP on microvillar rootlets, with a signal that was mutually exclusive with the membrane-wrapped protruding microvilli (Figures S1B and S1C). Together, these localization studies in native tissues and cell culture models uniformly indicate that MISP specifically targets to core bundle rootlets and is excluded from the membrane-wrapped segment of the core bundle.
MISP is required for maintaining rootlets at the base of microvilli
Core bundle rootlets are anchored in the terminal web, which likely provides mechanical support for BB assembly and long-term stability. To determine whether MISP is required for normal BB assembly and microvillar structure, we generated W4 cell lines with stable shRNA-mediated knockdown (KD) of MISP. Loss of MISP was confirmed by western blot analysis (Figures S2A and S2B). Using low-magnification confocal microscopy, we scored the fraction of cells that were BB positive as indicated by polarized F-actin staining. At a population level, the percentage of W4 cells forming a polarized BB decreased from 82% in the scramble control to 70% in MISP KD cells (Figures 2A and 2B). This modest phenotype was rescued when an EGFP-MISP construct refractory to KD was reintroduced (Figure 2B). However, the overall intensity of F-actin per cell decreased significantly in MISP KD cells (Figure 2C), suggesting a marked perturbation in F-actin network architecture even in cells that still exhibited polarized BB assembly. To further understand the impact of MISP loss of function, we look closer at individual MISP KD cells that still formed a polarized BB. Measurements of microvillar dimensions revealed that the overall length of core bundles did not change significantly between scramble control and MISP KD cells (Figure S2C). However, using a membrane marker to delineate the membrane-wrapped versus unwrapped segments of the core bundle, we found that the protruding microvillus increased in length (1.96 ± 0.37 μm in controls versus 2.22 ± 0.39 μm in KD) at the expense of rootlet length, which decreased significantly (0.43 ± 0.09 μm in controls versus 0.27 ± 0.07 μm in KD; Figures 2D and 2E). The microvillus/rootlet ratio calculated on a per-cell basis increased in MISP KD cells compared with control cells (Figure 2F), and this was consistent with the increased percent membrane coverage measured for MISP KD core bundles (Figure S2D). Thus, rootlet shortening in MISP KD cells is driven by membrane overwrapping of core bundles.
Figure 2. MISP is required for maintaining rootlets.
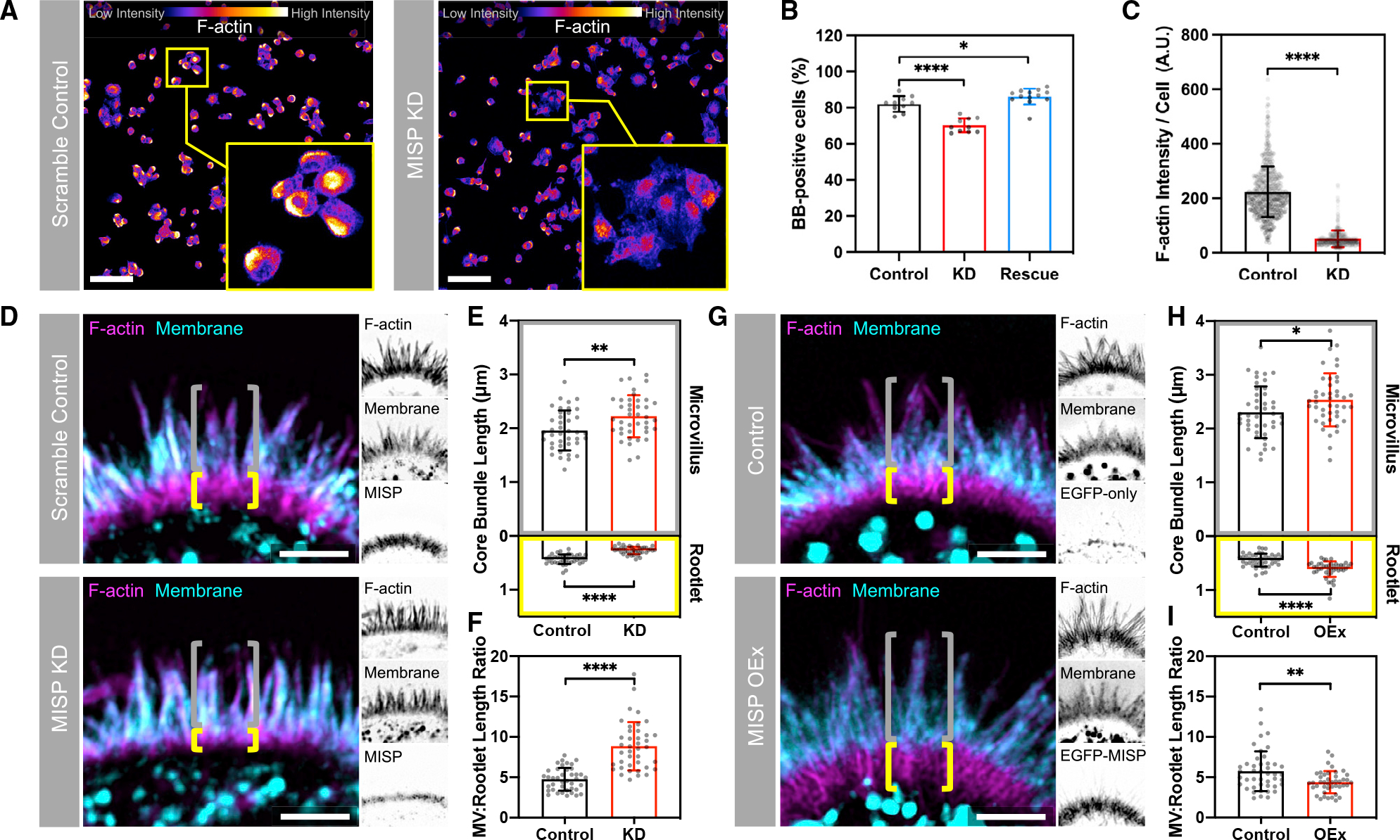
(A) Confocal Max IPs of scramble control and MISP knockdown (KD) W4 cells stained for F-actin; intensity color-coded so warmer colors denote higher intensities. Zooms show representative cells for each condition. Scale bar: 100 μm.
(B) Percentage of W4 cells forming BBs (BB-positive cells) from (A) comparing scramble control, MISP KD, and EGFP-MISP rescue conditions. Each dot represents the percentage of BB-positive cells in a single field of 620 μm2; n ≥ 10 fields per condition.
(C) F-actin intensity values of W4 cells from (A). Each dot represents the average F-actin intensity of a single cell; n > 600 cells per condition.
(D) SIM Max IPs of the BB of W4 cells in scramble control (top) and MISP KD (bottom) conditions stained for F-actin (magenta) and membrane (cyan); each panel shows the merge with their inverted single channels. Yellow brackets indicate the extension of actin rootlets; gray brackets show the extension of microvilli. Scale bars = 2 μm.
(E) Lengths of microvilli (gray, top plot) and rootlets (yellow, bottom plot) from scramble control and KD cells. Each dot represents the average of >10 length values per cell; n ≥ 40 cells per condition. All data represent three independent experiments.
(F) Microvillus/rootlet length ratios measured on a per-cell basis from (E).
(G) SIM Max IPs of the BB of W4 cells in control (top) and MISP-overexpressing (bottom) cells stained for F-actin (magenta) and membrane (cyan); each panel shows the merge with their inverted single channels. Yellow brackets indicate the extension of actin rootlets; gray brackets show the extension of microvilli. Scale bars = 2 μm.
(H) Lengths of microvilli (gray, top) and rootlets (yellow, bottom) from control and MISP OEx cells. Each dot represents the average of >10 length values per cell; n ≥ 40 cells per condition. All data represent three independent experiments.
(I) Microvillus/rootlet length ratios measured on a per-cell basis from (H).
All bar plots and error bars denote mean ± SD. p values were calculated using the unpaired t test (*p < 0.05; **p < 0.01; ****p < 0.0001).
Because loss of MISP shortened rootlets and increased membrane wrapping of core bundles, we sought to determine whether increasing MISP levels would elongate rootlets at the expense of membrane wrapping. To test this hypothesis, we stably overexpressed EGFP-MISP in W4 cells and examined microvillar structure using SIM. Similar to the localization studies described above, EGFP-MISP exhibited specific enrichment on microvillar rootlets. Relative to control cells, MISP overexpression (OEx) promoted a significant elongation of both the membrane-wrapped (2.30 ± 0.48 μm in controls versus 2.54 ± 0.49 μm in OEx) and rootlet (0.44 ± 0.12 μm in controls versus 0.61 ± 0.15 μm in OEx) segments of the core bundle (Figures 2G and 2H). These changes together drove a significant increase in the overall length of core bundles and a slight reduction in percent membrane coverage of total core bundles in MISP-overexpressing cells (Figures S2E and S2F). However, the marked elongation of rootlets decreased the microvillus/rootlet ratio under these conditions (Figure 2I). In combination, these findings show that MISP promotes microvillar rootlet elongation and protects this end of the core bundle from membrane wrapping.
Purified MISP assembles tightly packed linear actin bundles in vitro
Based on the terminal web localization of MISP and the impact of MISP perturbation on rootlet length, we sought to determine whether purified MISP is sufficient to drive the formation of linear F-actin bundles similar in structure to core bundle rootlets. Full-length human MISP was highly insoluble in previous purification attempts, and thus far only truncated fragments have been studied in vitro (Kumeta et al., 2014). Although MISP’s actin-binding potential might be distributed throughout the molecule, the C-terminal half of MISP was capable of linking F-actin together in mesh-like networks (Kumeta et al., 2014). To further develop our understanding of MISP’s actin-binding properties, we sought to express and characterize the activity of full-length human MISP. To aid with solubility, we tagged the N terminus of MISP and EGFP-MISP with maltose-binding protein (MBP) and purified these variants from Sf9 insect cells for further characterization (Figures S3A and S3B). We first confirmed that soluble MBP-MISP was sufficient to sediment F-actin (Figure S3C). Using a low-speed sedimentation assay, we found that MBP-tagged full-length MISP robustly bound to and bundled F-actin in a concentration-dependent manner (Figures 3A and 3B). To gain insight on MISP’s affinity for actin, we conducted a high-speed sedimentation assay and found that MISP binds to F-actin with a dissociation constant (KD) of 0.76 μM (Figures 3C and 3D), which is comparable with other bundlers expressed in epithelial cells (Bartles et al., 1998; Kitajiri et al., 2010). Interestingly, this KD is also comparable with that reported for the C-terminal half of MISP (amino acids [aa] 352–680), which suggests that this fragment might contribute to most of the binding affinity for F-actin in full-length MISP. To directly visualize the impact of MISP on F-actin organization and bundling, we mixed MBP-EGFP-MISP with phalloidin-stabilized F-actin and then examined the resulting structures using confocal microscopy. MISP/F-actin mixtures exhibited extensive bundling and crosslinking of filaments, particularly in regions that were heavily decorated with MBP-EGFP-MISP (Figure 3E, zoom 1). F-actin intensity in bundles was also significantly higher when MISP was present in solution (Figure 3F). Interestingly, MISP accumulated at the ends of some actin bundles where the phalloidin signal was lower (Figures 3E and 3G, red arrowheads in zoom 2). To examine the ultrastructural organization of these samples, we turned to transmission electron microscopy (TEM). We removed the MBP tag using TEV protease to reduce the possibility of functional interference from this moiety before TEM imaging (Figure S3D). In control samples (F-actin alone), TEM images revealed single-actin filaments that extended for many microns across the grid surface (Figure 3H, left panels). In contrast, MISP/F-actin mixtures exhibited extensive crosslinking of filaments and the formation of tightly packed linear actin bundles (Figure 3H, right panels). Although the tightly packed, 3D nature of these bundles precluded clear determination of filament polarity in our images, spacing measurements revealed that filaments in these bundles were separated by an average distance of 10.2 ± 2.5 nm (Figure 3I), which is shorter than the distances between filaments bundled by villin or espin (~12 nm), but comparable with the spacing produced by fimbrin (Bartles et al., 1998; Hampton et al., 2008; Matsudaira et al., 1983; Volkmann et al., 2001). Together, these findings demonstrate that MISP is sufficient to form tightly packed linear actin bundles with an inter-filament spacing similar to that of fimbrin.
Figure 3. Purified MISP assembles tightly packed linear actin bundles in vitro.
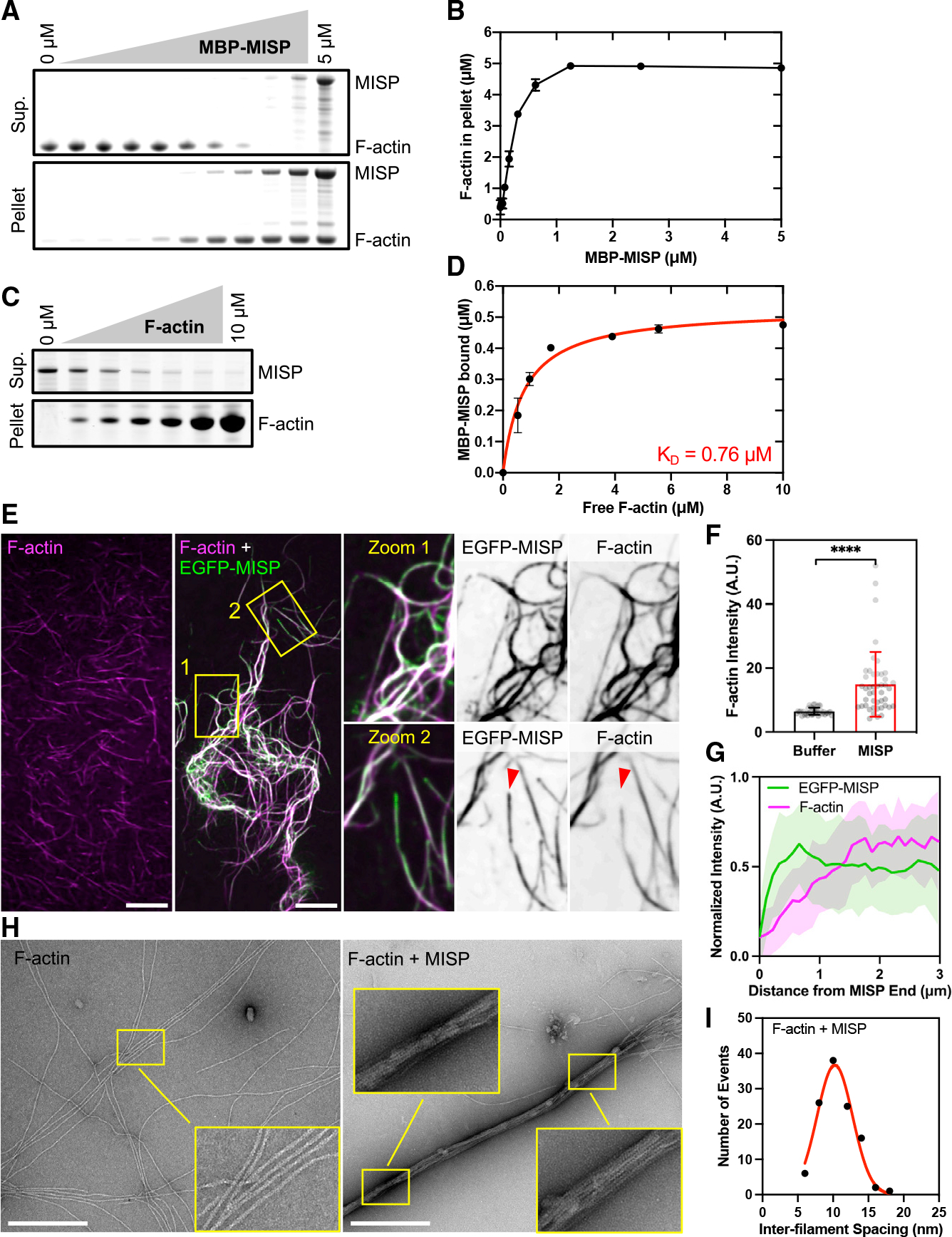
(A) Low-speed sedimentation assay of phalloidin-stabilized F-actin (5 μM) and increasing concentrations of MBP-MISP (0–5 μM). Coomassie-stained SDS-PAGE shows the supernatant and pellet fractions recovered after centrifugation.
(B) Density quantification of bands shown in (A). All data represent three independent experiments. Error bars denote mean ± SD.
(C) High-speed sedimentation assay of MBP-MISP (0.5 mM) and increasing concentrations of non-stabilized F-actin (0–10 μM). Coomassie-stained SDS-PAGE shows the supernatant and pellet fractions recovered after centrifugation.
(D) Density quantification of bands shown in (C). All data represent three independent experiments that were fit using a hyperbolic saturation binding model yielding a KD = 0.76 μM. Error bars denote mean ± SD.
(E) Confocal images of phalloidin-stabilized F-actin (0.5 μM; magenta) alone or pre-mixed with MBP-EGFP-MISP (0.1 μM; green). Zooms correspond to the yellow boxes shown in merge; single channels are shown as inverted images. Red arrowheads indicate the end of MISP-bundled F-actin. Scale bar: 10 μm.
(F) Fluorescence intensities of F-actin in buffer alone or with MISP from (E). Each dot represents the integrated intensity value of a 250-μm2 field; n ≥ 39. Bar plots and error bars denote mean ± SD. p values were calculated using the unpaired t test (****p < 0.0001).
(G) Line scan analysis of EGFP-MISP (green) and F-actin (magenta) intensities measured at bundles ends shown in (E). Data are shown as mean ± SD.
(H) Transmission electron microscopy images of negatively stained phalloidin-stabilized F-actin (0.2 μM) in buffer alone or pre-mixed with purified MISP (0.04 μM). Scale bar: 400 nm.
(I) Histogram of inter-filament spacing measurements from bundles shown in (H). Each dot represents the average of ≥ 110 values; bin size = 2. Average values were fit using a Gaussian curve.
MISP recruits fimbrin to actin bundles
Among the three previously characterized actin bundlers in the BB, fimbrin is the only one that appears to preferentially accumulate on core bundle rootlets, where it might mediate physical interactions with the terminal web cytokeratin network (Grimm-Günter et al., 2009). We therefore sought to determine whether MISP binding to F-actin depends on fimbrin, either cooperatively or competitively. To this end, we turned to HeLa cells, which do not typically form microvilli but can assemble a variety of other actin-based networks. Interestingly, mCherry-MISP expression alone promoted the formation of aberrant actin bundles throughout the cytoplasm (Figure 4A, top panel), whereas EGFP-fimbrin expression had little impact on existing actin networks (Figure 4A, middle panel). However, when MISP and fimbrin were co-expressed, fimbrin was robustly recruited to MISP-bundled F-actin (Figure 4A, bottom panel), showing a strong colocalization with MISP (Figure 4B). We conducted similar co-expression experiments using fluorescently tagged versions of villin or espin. Independent of the presence or absence of MISP, villin remained cytosolic (Figure S4A). In contrast, espin promoted the elongation of filopodia-like structures that were reminiscent of microvillar protrusions induced by espin overexpression in LLC-PK1-CL4 cells (Loomis et al., 2003). Interestingly, in MISP/espin-expressing HeLa cells, MISP enriched at the base of these filopodia-like protrusions, and the aberrant cytosolic bundles observed in MISP-only cells were absent (Figure S4B). Overall, these data indicate that MISP promotes the formation of actin bundles, which in turn recruit fimbrin, but not villin or espin, and further suggest a hierarchical functioning of these factors during microvillar assembly.
Figure 4. MISP recruits fimbrin to actin bundles.
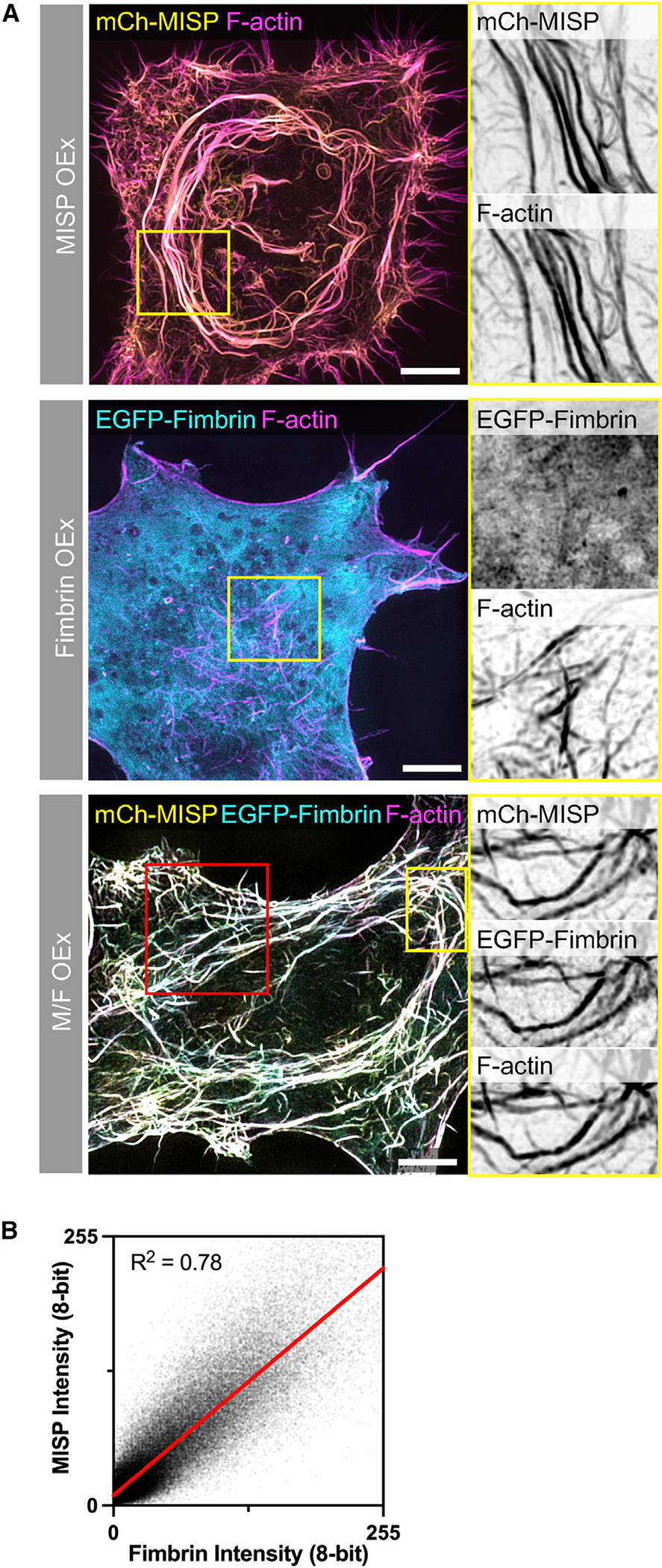
(A) SIM Max IPs of HeLa cells overexpressing mCherry-MISP (top panel), EGFP-fimbrin (middle panel), and mCherry-MISP and EGFP-fimbrin (bottom panel). All cells were stained for F-actin with phalloidin (magenta). Each panel shows the merge with their zooms as inverted single channels. Scale bars: 5 μm.
(B) Colocalization analysis of mCherry-MISP and EGFP-fimbrin intensities along actin bundles shown in the red box in (A); data were fit using linear regression.
MISP and fimbrin cooperate to elongate microvillar rootlets
We next sought to determine whether MISP and fimbrin cooperate to elongate rootlets in W4 cells. Indeed, HALO-MISP and EGFP-fimbrin co-expression resulted in a dramatic hyper-elongation of rootlets, which extended deep into the cell (Figure 5A). The tangled nature of these exaggerated rootlets prevented us from measuring the length of individual core bundles in these structures. Instead, we focused on measuring the length of protruding microvilli, as well as the maximum distance that rootlets reached into the cytoplasm using a membrane marker as a point of reference. Although the length of microvilli increased with moderate significance, the reach of rootlets strikingly increased by ~3-fold in cells co-expressing MISP and fimbrin compared with cells overexpressing either MISP or fimbrin alone, or untransfected control cells (3.01 ± 1.35 μm versus 0.91 ± 0.41 μm versus 1.18 ± 0.49 μm versus 0.70 ± 0.16 μm, respectively) (Figures 5A and 5B). We also observed that these exaggerated rootlet networks converged as they grew farther from the apical membrane (Figure S5A). Colocalization analysis showed a strong correlation between MISP and fimbrin signals throughout these structures (Figure S5B). To further define the properties of the exaggerated rootlets promoted by MISP and fimbrin co-expression, we conducted fluorescence recovery after photobleaching (FRAP) assays on W4 cells expressing HALO-β-actin alone or in combination with EGFP-fimbrin and mCherry-MISP. Photobleaching of HALO-β-actin allowed us to directly interrogate actin dynamics in distinct regions of interest (ROIs) in transfected cells. In the microvilli of control cells, β-actin turned over with a thalf of 126.7 s, which likely reflects the treadmilling rate of core bundles in this system (Figures 5C and 5E, green labels). However, in cells co-expressing MISP and fimbrin, we noted two distinct recovery rates: β-actin in protruding microvilli turned over at a rate that was 4-fold slower than controls (thalf = 529.1 s), whereas recovery in exaggerated rootlets was extremely slow to nonexistent (Figures 5D and 5E, magenta and cyan labels, respectively). Therefore, consistent with their actin bundling activities, MISP and fimbrin co-expression hyper-stabilized core bundles and reduced β-actin flux through both rootlets and protruding microvilli.
Figure 5. MISP and fimbrin cooperate to elongate rootlets.
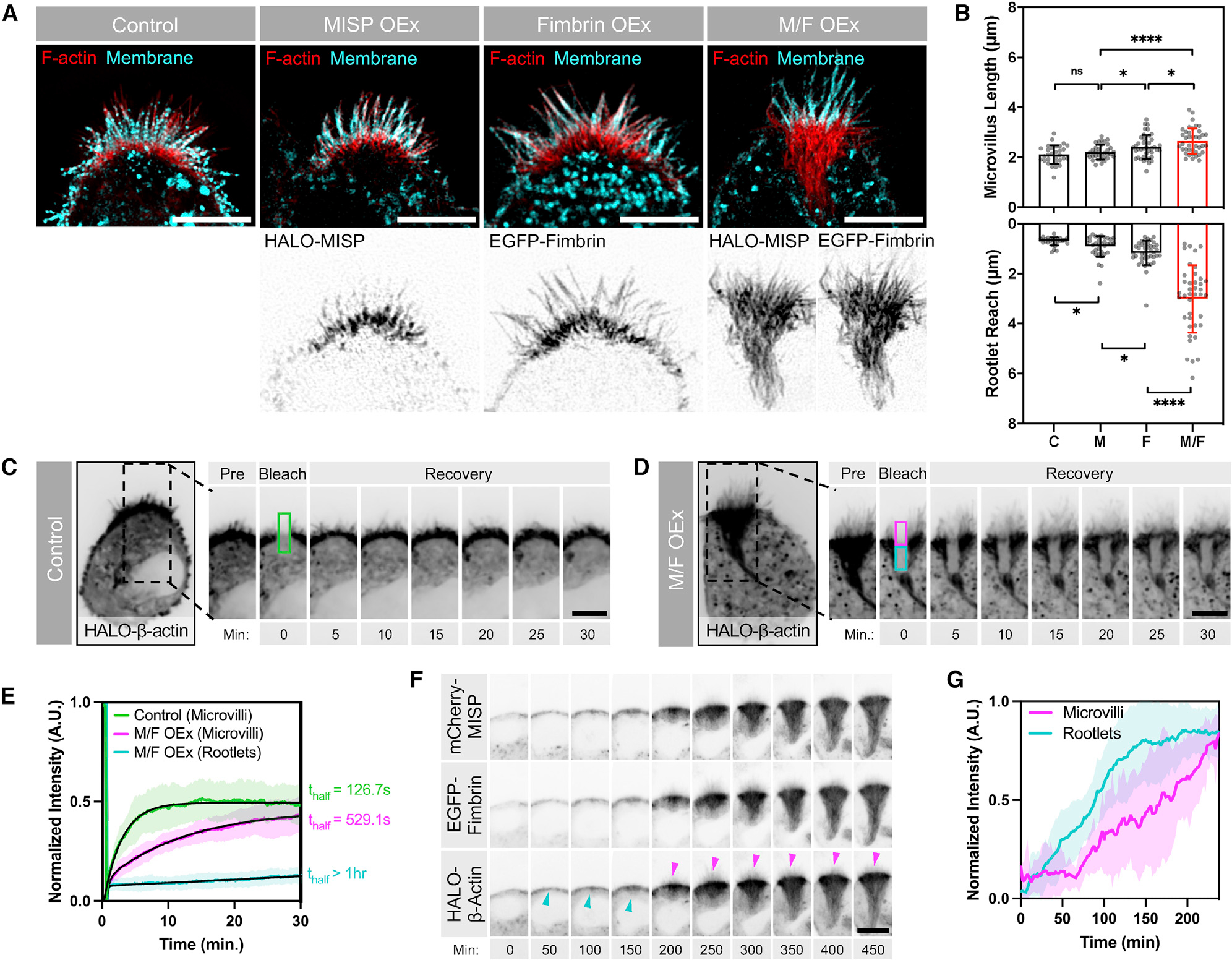
(A) SIM Max IPs of W4 cells non-transfected (control) and overexpressing HALO-MISP (MISP OEx), EGFP-fimbrin (Fimbrin OEx), or HALO-MISP and EGFP-fimbrin together (M/F OEx). All cells were stained for F-actin with phalloidin (red) and membrane with WGA (cyan). Each panel shows merges on top with inverted single channels along the bottom. Scale bars: 5 μm.
(B) Lengths of microvilli (“Microvillus Length,” top plot) versus distance that rootlets extend into the cytoplasm (“Rootlet Reach,” bottom plot) in W4 cells expressing the constructs described in (A). Each dot represents the average of >10 length values per cell; n ≥ 34 cells per condition. All data represent three independent experiments. Bar plots and error bars are mean ± SD. p values were calculated using the unpaired t test (*p < 0.05; ****p < 0.0001; ns, not significant.).
(C and D) Photobleaching analysis of W4 cells expressing HALO-β-actin alone (C) or HALO-β-actin with EGFP-fimbrin and mCherry-MISP (D). Although a single ROI was positioned on the BB and bleached in both conditions, the ROI in (D) was subdivided into two sub-ROIs to quantify differences in the recovery of the apical microvilli (magenta box) versus subapical rootlets (cyan box). Scale bar: 5 μm.
(E) Fluorescence intensity recovery of HALO-β-actin from the color-coded ROIs described in (C) and (D); n > 14 cells per condition. All intensity values for each condition are shown as mean ± SD. Average values for each condition were fit using two-phase association curves.
(F) Time-series montages of W4 cells expressing HALO-β-actin, EGFP-fimbrin, and mCherry-MISP after adding doxycycline to induce BB assembly. Cyan arrowheads denote initiation of terminal web actin network assembly. Magenta arrowheads denote microvilli assembly. Scale bar: 10 μm.
(G) Fluorescence intensity of HALO-β-actin during BB assembly from microvilli and terminal web actin network in (F); n = 10 cells. Data are shown as mean ± SD.
To further understand how these hyper-elongated and stable rootlets assemble relative to protruding microvilli, we used live imaging to visualize BB assembly in W4 cells expressing mCherry-MISP, EGFP-fimbrin, and HALO-β-actin (Video S1). During the first 2 h after the addition of doxycycline to promote BB assembly, we observed the assembly of a terminal web actin network immediately beneath the apical cap (Figures 5F and 5G, cyan labels). As this dense network accumulated sub-apically, microvilli began to emerge (Figures 5F and 5G, magenta labels). Consistent with our FRAP analysis, core bundle rootlets elongated from the subapical region below microvillar protrusions with no apparent actin turnover or disassembly from the basal ends (Figure 5F, β-actin channel). These results suggest that the assembly of a terminal web actin network precedes the assembly of microvilli, which is consistent with the proposed role of the terminal web in offering mechanical support for protrusion assembly (Tilney and Cardell, 1970).
MISP and ezrin exhibit mutually exclusive targeting along core actin bundles
Our localization studies in native tissues and cell culture models establish that MISP localizes specifically to the rootlet segment of the core bundle, which remains free of plasma membrane wrapping. Although such specific targeting for a BB component has not been described before, one possible explanation is that MISP is normally prevented from occupying the membrane-wrapped segment of the core bundle by other microvillar actin-binding factors. One potential competing factor is ezrin, a membrane-actin linker that provides structural stability to microvilli (Casaletto et al., 2011; Saotome et al., 2004). Interestingly, active ezrin was previously identified as a MISP binding partner, and these two proteins also demonstrate partial colocalization at the cortex in dividing HeLa cells and CACO-2 cysts (Kschonsak and Hoffmann, 2018). A close inspection of W4 cells using SIM revealed that endogenous ezrin localizes to the membrane-wrapped ends of microvillar core bundles as expected (Figure S6A) and shows a mutually exclusive signal relative to MISP rootlet labeling (Figure S6A). This is consistent with previous work indicating that KD or inactivation of ezrin increases MISP levels at the cell cortex (Kschonsak and Hoffmann, 2018). To determine whether MISP is confined to microvillar rootlets by ezrin, we expressed an ezrin-EGFP construct in W4 cells and monitored the localization of endogenous MISP. In control W4 cells, SIM images revealed that MISP signal was uniformly distributed at the base of the BB as expected (Figures 6A and 6B, left panels). However, in W4 cells expressing ezrin-EGFP, we noted that MISP signal was displaced toward the BB periphery; MISP was almost entirely excluded from the center of the apical domain where ezrin levels were highest (Figures 6A and 6B, right panels). 3D rendering of ezrin-overexpressing cells revealed that MISP signal appeared as a ring-like structure surrounding ezrin signal at the center of the BB (Video S2). When we stained ezrin-overexpressing cells with phalloidin and membrane marker to visualize F-actin and the plasma membrane, F-actin signal was reduced in regions lacking MISP signal (Figures 6A and 6B). This was also accompanied by drastic shortening of core bundle rootlets (0.43 ± 0.10 μm in controls versus 0.28 ± 0.06 μm in OEx) and a significant increase in length of protruding microvilli (2.16 ± 0.36 μm in controls versus 2.58 ± 0.40 μm in OEx; Figures 6A and 6C), which resulted in an increased microvillus/rootlet ratio in ezrin-overexpressing cells compared with control cells (Figure 6D). Interestingly, the length of membrane-wrapped versus unwrapped segments of core bundles and their ratio in ezrin-overexpressing cells are similar to what we observed in MISP KD cells (Figure 2). Using differentiated CACO-2BBE monolayers, we recapitulated these experiments and found that MISP levels were also reduced in ezrin-overexpressing conditions with no apparent redistribution to other cellular compartments (Figures S6B–S6D). Together, these findings indicate that normal levels of ezrin are required to maintain MISP targeting to core bundle rootlets, which in turn promotes their elongation.
Figure 6. MISP and ezrin exhibit mutually exclusive targeting at opposite ends of core actin bundles.
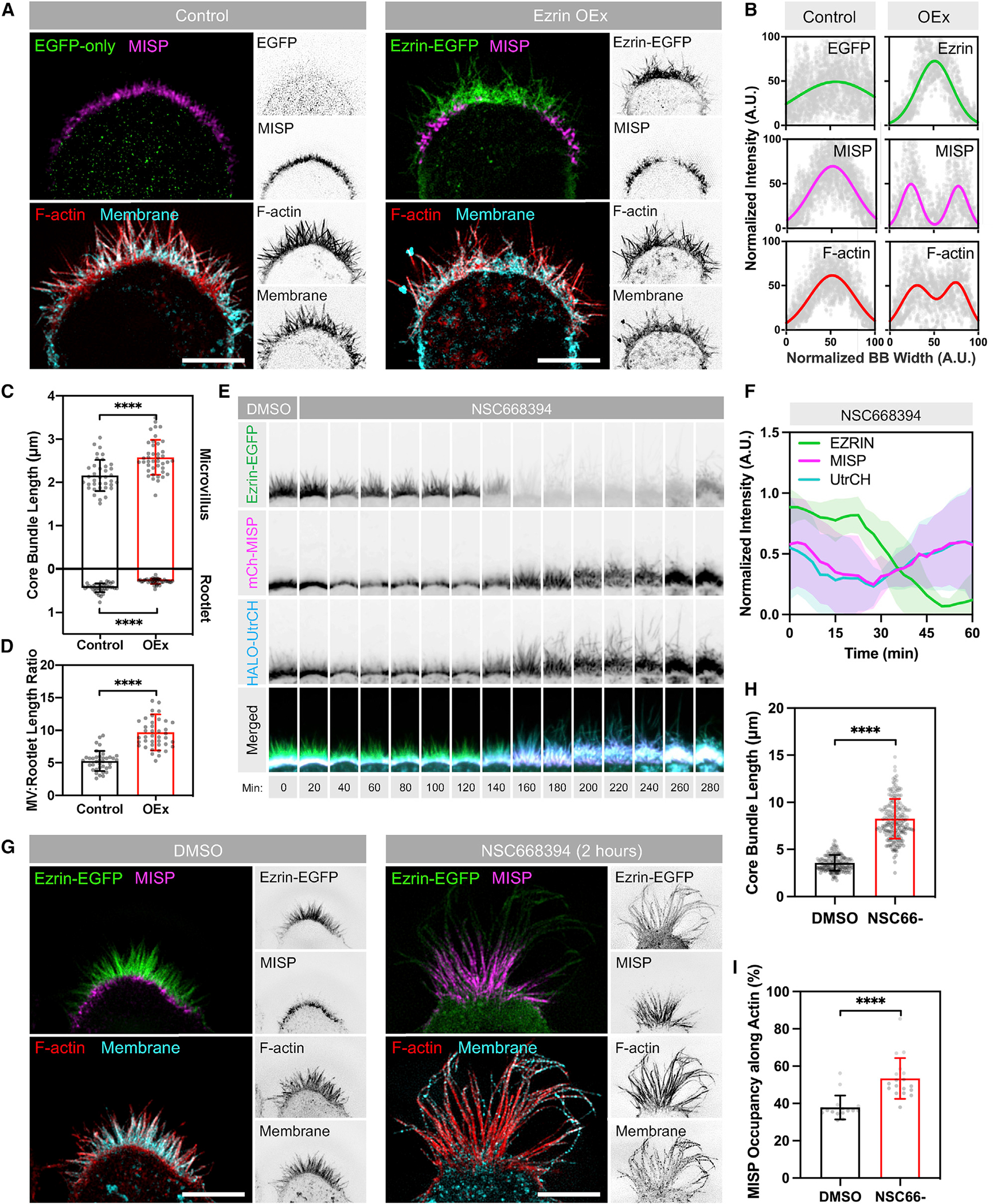
(A) SIM Max IPs of W4 cells overexpressing EGFP alone (green, left panel) or ezrin-EGFP (green, right panel) and stained for endogenous MISP (magenta), F-actin with phalloidin (red), and membrane with WGA (cyan). Each panel shows two-color merges with their inverted single channels. Scale bar: 5 μm.
(B) Intensity distributions across the BB from left to right, measured for each marker described in (A). Distributions were fit using single- or double-Gaussian curves. Number of cells per condition was ≥8.
(C) Lengths of microvilli (top plot) and rootlets (bottom plot) from W4 cells shown in (A). Each dot represents the average of >10 length values per cell; n ≥ 37. All data represent three independent experiments.
(D) Microvillus/rootlet length ratios measured on a per-cell basis from (C).
(E) Confocal Max IP time-series montages of a W4 cell expressing ezrin-EGFP (green), mCherry-MISP (magenta), and HALO-UtrCH (blue) before and after adding NSC668394 (ezrin inhibitor). The width of each box in the montage is 7 μm.
(F) Fluorescence intensity values of markers described in (E). Data are shown as mean ± SD.
(G) SIM Max IPs of W4 cells expressing ezrin-EGFP in DMSO (left panel) or NSC668394 (right panel) conditions after 2 h of exposure. Cells were stained for endogenous MISP (magenta), F-actin with phalloidin (red), and membrane with WGA (cyan). Each panel shows two-color merges with their inverted single channels. Scale bar: 5 μm.
(H) Lengths of core bundles from conditions described in (G). Each dot represents the length of a single core bundle; n > 190 length values.
(I) Percentages of MISP occupancy along core bundles from the conditions in (G). Each dot represents the percentage of the average MISP coverage along core bundles per cell; n > 16 cells per condition; length values per cell >10.
All bar plots and error bars denote mean ± SD. p values were calculated using the unpaired t test (****p < 0.0001).
Within microvillar protrusions, phosphorylated ezrin adopts an open state that bridges the plasma membrane to the underlying actin cytoskeleton (Bretscher et al., 1997). We hypothesized that the open/active state of ezrin within membrane protrusions restricts MISP to core bundle rootlets. To test this idea, we used an ezrin inhibitor (NSC668394), which disrupts its phosphorylation and actin-binding capacity (Bulut et al., 2012). We overexpressed ezrin-EGFP, mCherry-MISP, and HALO-UtrCH (an F-actin-binding probe based on the calponin homology domain of utrophin) in W4 cells and monitored their fluorescence intensity over time before and after the addition of 50 μM NSC668394. Using confocal microscopy, we observed that ezrin enrichment in the BB was lost within a 3-h window after exposure to NSC668394. Notably, in all these events, the loss of ezrin signal was followed by a striking increase of MISP and UtrCH signal throughout the BB (Figures 6E and 6F; Video S3). Moreover, overaccumulation of MISP and UtrCH in NSC668394-treated W4 cells also coincided with a dramatic increase in microvillar length (Figure 6E, UtrCH channel). However, these instances of elongation were temporary because protrusions eventually collapsed after 30–60 min of growth without impacting the accumulation of MISP and UtrCH at the base of the BB. To further define the impact of ezrin accumulation on MISP localization and microvillar structure, we used SIM to look closer at the BB of W4 cells fixed after 2 h of NSC668394 treatment. SIM images revealed a significant increase in the overall length of core bundles in NSC668394-treated cells compared with control cells (3.58 ± 0.83 μm versus 8.25 ± 2.11 μm) (Figures 6G and 6H). Interestingly, MISP occupancy along core bundles also increased from 38% in DMSO-treated cells to 53% in NSC668394-treated cells (Figure 6I). These findings indicate that ezrin and its associated membrane-actin linking activity confine MISP to the rootlets of microvilli.
DISCUSSION
The filament crosslinking activity of actin bundlers provides the microvillar core bundle with the flexural rigidity needed to overcome plasma membrane tension and protrude from the cell surface (Atilgan et al., 2006). Although villin, fimbrin, and espin are canonical actin bundlers that have been identified and characterized in the context of the epithelial BB, persistent microvillar growth in mice lacking all three of these factors suggested the existence of as-of-yet-unidentified bundlers (Revenu et al., 2012). Epidermal growth factor receptor pathway substrate 8 (EPS8) has been invoked to potentially compensate for crosslinking activity in the absence of other canonical bundlers (Revenu et al., 2012). However, its specific localization to the distal tips of microvilli is at odds with the need for canonical bundlers to be distributed along the length of the core bundle. Additionally, whereas certain cell types employ isoforms of fascin to drive robust parallel bundling of filaments in other related actin-based protrusions such as filopodia and stereocilia (Krey et al., 2016; Roy and Perrin, 2018; Svitkina et al., 2003), there is no evidence for fascin expression in transporting epithelia of the gut and kidney. Thus, the identity of other functional bundlers in the apical BB has remained an open question.
Here, we identify MISP as a BB resident that holds F-actin bundling potential. Previous studies on MISP focused on its role during mitotic progression (Kschonsak and Hoffmann, 2018; Maier et al., 2013; Zhu et al., 2013). In that context, cortically localized MISP contributes to anchoring the asters of spindle microtubules to cortical actin-rich structures during metaphase (Maier et al., 2013; Zhu et al., 2013). In native intestinal tissues and differentiating epithelial cell culture models, we found that MISP localizes to the base of core bundles that support microvilli. Moreover, confocal and super-resolution images revealed that MISP is restricted to the rootlet ends of core bundles, which are embedded in the subapical terminal web and thus are not wrapped by plasma membrane. These results also align well with MISP being enriched in the proximal region of neuronal growth cones, where the pointed ends of filopodial actin filaments coalesce (Kumeta et al., 2014). Thus, localization near pointed ends of actin filaments appears to be a conserved property of MISP. Interestingly, MISP labeling is observed not only in the terminal web of enterocytes along the villus but also in the subapical region of immature/differentiating enterocytes found in the crypt. Therefore, MISP is enriched at the cell apex during the window of differentiation when microvilli are actively growing. Having a rootlet-specific bundler present at early stages of differentiation is consistent with classic ultrastructural studies, which suggested that the growth of new microvilli is supported by a simultaneous maturation of the terminal web immediately beneath the apical membrane (Tilney and Cardell, 1970).
Our data indicate that MISP selectively stabilizes the rootlet ends of core bundles. Indeed, MISP KD in W4 cells led to significant shortening of rootlets when visualized with SIM, whereas overexpression promoted rootlet elongation. The bundle elongation induced by increasing MISP levels aligns well with previous studies on espin, which also drives microvillus elongation when overexpressed (Loomis et al., 2003). Previous MISP overexpression studies in HeLa cells reported a “thickening” of actin structures, which is generally consistent with our observations (Kumeta et al., 2014). However, that study also showed that depletion of MISP led to filopodial overgrowth, which seems to be at odds with our conclusion that MISP promotes the assembly of parallel actin bundle-based protrusions. These phenotypic distinctions are related to the prominent differences in actin network architecture found in unpolarized HeLa cells versus the polarized intestinal epithelial cells at the focus of our study.
We propose that the gain- and loss-of-function phenotypes observed in our experiments are explained by MISP’s F-actin bundling activity, which we reconstituted in vitro. F-actin bundles assembled with purified MISP demonstrate tight packing with an average inter-filament spacing of ~10.2 nm, which is close to that reported for fimbrin (~9–12 nm) (Matsudaira et al., 1983; Volkmann et al., 2001) but slightly shorter to the distance between filaments bundled by villin or espin (~12 nm) (Bartles et al., 1998; Hampton et al., 2008). This suggests that the arrangement of filaments in intact microvilli reflects the collective activity of multiple bundlers, each bringing their own characteristic spacing. Indeed, previous electron microscopy (EM) studies on filament packing and spacing in stereocilia core bundles, which are occupied by fascin-2, espin-1, fimbrin (Krey et al., 2016) and TRIOBP-4 and −5 (Kitajiri et al., 2010), are consistent with this general idea.
Intriguingly, co-expression of MISP and fimbrin cooperatively elongated core bundle rootlets deep into the cytoplasm of W4 cells. The exaggerated nature of these rootlets allowed us to capture the temporal details of their formation, which preceded microvilli assembly. Thus, the apical localization of bundlers early in enterocyte differentiation might provide mechanical stability to nascent, growing microvilli. Overexpression experiments in HeLa cells, which generally do not make microvilli, also revealed that MISP can drive the formation of aberrant actin bundles, and these structures in turn recruit fimbrin, but not villin or espin. How MISP binds to and bundles F-actin and recruits fimbrin remains unknown. In our analysis, we failed to identify recognizable actin-binding and bundling motifs in the MISP primary sequence, although previous studies point to actin-binding potential as being distributed throughout the molecule (Kumeta et al., 2014), which is consistent with the functional requirements of a bundler. Considering the cooperative effects of MISP and fimbrin on rootlet length and stability, it is tempting to speculate that these factors bind to different sites on F-actin. In contrast with MISP, the multiple actin-binding domains of fimbrin are well characterized (Klein et al., 2004), and their binding sites on F-actin in 2D arrays have been mapped using cryoelectron microscopy (cryo-EM) (Volkmann et al., 2001). Based on those structural studies, we speculate that MISP binds outside the canonical inter-monomer cleft that is targeted not only by fimbrin but also cofilin (Tanaka et al., 2018), myosin (Mentes et al., 2018), and even live imaging probes such as Lifeact (Belyy et al., 2020). Future cryo-EM studies aimed toward elucidating the structural details of the MISP binding site on F-actin will be needed to understand the nature of MISP/fimbrin cooperativity. Independent of a detailed actin-binding and bundling mechanism, the hierarchical targeting of MISP and fimbrin suggests an order of action for these two bundlers during microvillar assembly. We propose that MISP localization to rootlets leads to the arrival of fimbrin at the apical surface in differentiating enterocytes. Examining this possibility during microvilli biogenesis will require high-temporal-resolution live imaging as we recently described (Gaeta et al., 2021).
The highly restricted targeting of MISP to the rootlet is unique among epithelial actin bundlers, although previous studies revealed that fimbrin accumulates at higher levels at core bundle rootlets in the terminal web, relative to the distal end (Grimm Günter et al., 2009). In MISP KD cells, we noted that the membrane-wrapped segment of the core bundle elongated in parallel with the shortening of rootlets induced by loss of MISP. This finding suggested a previously unrecognized interplay between mechanisms that control the length of membrane-wrapped protruding microvilli and the activity of actin bundlers that dictate the length of rootlets. Thus, we hypothesized that factors that simultaneously bind to plasma membrane and F-actin would be well positioned to prevent MISP binding along the more distal membrane-wrapped segment of the core bundle. A common feature of actin-based protrusions is tethering of the cytoskeleton to the enveloping membrane by ERM (ezrin, radixin, moesin) proteins (Revenu et al., 2004). In the BB, ezrin is the most abundant ERM (McConnell et al., 2011). Interestingly, a previous study showed that ezrin holds the potential to limit MISP accumulation at the cell cortex (Kschonsak and Hoffmann, 2018). Paradoxically, that same work also reported that MISP binds directly to active ezrin, leaving open questions about the nature and mechanism of the functional interaction between these two proteins. Based on super-resolution imaging, our results unambiguously show that MISP and ezrin occupy spatially distinct domains along individual core bundles, with active and inactive ezrin (Hanono et al., 2006) occupying the membrane-wrapped domain and MISP residing on the rootlet, which is free of membrane wrapping. Thus, our data suggest that if MISP and active ezrin do physically interact, the resulting complex does not bind to the microvillar core bundle (Kschonsak and Hoffmann, 2018).
Remarkably, we found that inactivation of ezrin using a small molecule inhibitor led to the release of ezrin from the plasma membrane and immediate ectopic redistribution of MISP from the rootlets up to more distal regions of the core bundle. Ezrin and MISP redistribution also led to a drastic increase in microvillar length, which might reflect loss of the mechanical constraint that the membrane normally imposes on the distal barbed ends, the preferred site of actin monomer incorporation. Alternatively, MISP recruitment to more distal regions of the bundle might directly promote stabilization and slow the robust treadmilling and turnover that normally occur in this system (Meenderink et al., 2019; Tyska and Mooseker, 2002). Taken together, these data argue for a mutual exclusivity model where opposite ends of core bundles are decorated by either ezrin or MISP, and the balance between these populations ultimately dictates the extent of membrane coverage.
The fact that ezrin excludes MISP from binding along the membrane-wrapped segment of the microvillus may also offer additional insight on where MISP resides in a core bundle. Assuming that membrane-associated ezrin binds only to F-actin superficially exposed on the surface of the core bundle, MISP’s inability to occupy distal regions might suggest that this bundler also binds superficially. Although speculative, such superficial binding has been demonstrated for TRIOBP-4, a bundler that targets specifically to the rootlets of hair cell stereocilia (Kitajiri et al., 2010). MISP and TRIOBP do not share motifs or domain organization, but secondary structure analysis in Phyre2 predicted that MISP sequence is largely disordered, as has been reported for TRIOBP-4 (Bao et al., 2013). Thus, it remains possible that MISP bundles filaments using a similar mechanism. It is also worth noting that other well-characterized actin bundlers in microvilli, villin and espin, are uniformly found along the length of the core bundle, and they do not exhibit mutually exclusive localization with ezrin.
Collectively, the discoveries reported here point to an unconventional mechanism for bundling F-actin in the core bundles that support epithelial microvilli. These findings strengthen our molecular understanding of the biologically robust formation of evolutionary conserved microvillus-rich apical specializations. The emergence ofMISP as a linear actin bundler also offers amolecular explanation for the remarkable finding that triple-villin-espin-fimbrin KO mice are still capable of assembling BB microvilli (Revenu et al., 2012). Because MISP is also implicated in promoting mitotic progression (Maier et al., 2013; Zhu et al., 2013), future studies might focus on examining the role of MISP in coupling oriented cell division with differentiation in transporting epithelial cells.
Limitations of the study
The primary limitations of the current study are technical in nature. In the intestinal tract, microvillus-rich BBs assemble on the surface of nascent enterocytes during differentiation as these cells migrate out of stem cell-containing crypts and on to the villus. Recapitulating the biochemical and morphological transition that these cells undergo in a laboratory is a major challenge for researchers. In this paper, our conclusions are based largely on data derived from epithelial cell culture models, namely, the W4 (Baas et al., 2004) and CACO2-BBE cell lines (Peterson and Mooseker, 1993; Peterson et al., 1993), which are both limited in the extent to which they reflect the mechanistic details and nuances of the native process. Although not employed in the current study, intestinal organoid cultures promise the experimental accessibility afforded by cell culture models while more faithfully recapitulating the phenotypic details of differentiated intestinal epithelial cells in vivo (Date and Sato, 2015).
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Matthew J. Tyska (matthew.tyska@vanderbilt.edu).
Materials availability
Plasmids generated in this study will be made available from the lead contact on request.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
Ls174T-W4 cells (W4; human colon epithelial cancer cells), CACO-2BBE cells (human colorectal adenocarcinoma epithelial cell line), LLC-PK1-Cl4 cells (CL4; pig kidney epithelial cells), HeLa cells (human cervical cancer cell line), and HEK293T cells were cultured in Dulbecco’s modified Eagle’s (DMEM) medium with high glucose and 2 mM L-glutamine (Corning; 25–005-CI). Ls174T-W4 cells (a generous gift from Dr. Hans Clevers) were grown in media supplemented with 10% tetracyclin-free fetal bovine serum (Atlanta Biological, S10350), 1 mg/mL G418 (Gold Biotechnology; G-418), 10 μg/mL blasticidin (Gold Biotechnology; B-800), and 20 μg/mL phleomycin (InvivoGen; ant-ph-1). For CACO-2BBE cells, the media was supplemented with 20% fetal bovine serum. For LLC-PK1-Cl4, HeLa, and HEK293T cells, the media was supplemented with 10% fetal bovine serum. All cultured cells were grown at 37°C and 5% CO2.
METHOD DETAILS
Cloning and constructs
The full-length human MISP sequence harbored in a pCMV-SPORT plasmid (Harvard PlasmID Database; HsCD00326629) was subcloned by PCR and TOPO-cloned into a pCR™8 Gateway entry vector (Invitrogen; 46–0899). In-frame sequence insertion was confirmed by sequencing. MISP was then shuttled into Gateway-adapted plasmids: pEGFP-C1, pmCherry-C1, and pHALO-C1. Similarly, the human beta-actin and UtrCH sequences were cloned and shuttled into a Gateway adapted HALO-C1 plasmid. To create lentiviral expression vectors, the human MISP sequence was subcloned by PCR and inserted into a puromycin-resistant pLVX1-EGFP backbone by restriction enzyme digestion using XhoI and BamHI. The human fimbrin and villin sequences were cloned into a pEGFP-C1 plasmid (Clontech; 6084–1). The pEGFP-N1 construct harboring the human ezrin sequence was purchased from Addgene, plasmid# 20680. The pEGFP-C1-espin (rat small espin) was a generous gift from Dr. Jim Bartles. To create baculovirus expression vectors, the MISP and EGFP-MISP sequences were subcloned into modified pFastBac-6xHis-MBP LIC expression vector (Addgene; plasmid #30116). All constructs were confirmed by sequencing.
Transfection and lentivirus production
For overexpression experiments except in Figure 2G, cells were transfected using Lipofectamine2000 (Invitrogen; 11668019) according to the manufacturer’s instructions. For stable overexpression experiments in Figure 2G, cells were transduced with lentiviral particles expressing pLVX-EGFP or pLVX-EGFP-MISP. For KD experiments, cells were transduced with lentiviral particles expressing PLKO.1 scramble control and MISP-targeted shRNA plasmids (Sigma-Aldrich; TRCN0000422523, TRCN0000116527). For both stable overexpression and KD experiments, lentiviral particles were generated by transfecting HEK293T cells with 6 μg of the corresponding lentiviral expression vector alluded to above, 4 μg psPAX2 packing plasmid (Addgene, 12260), and 0.8 μg pMD2.G envelope plasmid (Addgene; #12259) using FuGENE 6 (Promega; E2691). Lentiviral particles were harvested and concentrated using a Lenti-X Concentrator (Clontech; 631231). Concentrated lentiviral particles were supplemented with polybrene (Sigma-Aldrich; H9268) and incubated with W4 cells at 80% confluency. After 24 h, the media was replaced with fresh media containing puromycin (Gold Biotechnology; P-600–100) for selection. Selection was applied for 14 days, replacing with fresh selection media every other day. For KD experiments, rescue assays were conducted using an EGFP-MISP construct designed to be refractory to shRNA KD.
Immunofluorescence
Cells grown on a glass coverlips were fixed with 4% paraformaldehyde (EMS; 15710) in 1X PBS for 15 min at 37°C. Fixed cells were washed with 1X PBS, and permeabilized with 0.1% Triton X-100 in 1X PBS for 15 min at room temperature. Cells were washed with 1X PBS and blocked with 5% Bovine Serum Albumin (BSA) in 1X PBS for 2 h at room temperature. Cells were washed and incubated with primary antibodies overnight at 4°C. Primary antibodies used were anti-MISP (Thermo Scientific; PA5–61995), anti-villin (Santa Cruz; sc-66022), anti-ezrin (CST; 3145). Cells were washed with 1X PBS four times for 5 min and incubate with secondary antibodies. Goat anti-rabbit Alexa Fluor 488 F(ab’)2 Fragment (Molecular Probes; A11070), goat anti-mouse Alexa Fluor 568 F(ab’)2 Fragment (Molecular Probes; A11019), Alexa Fluor 568-phalloidin (Invitrogen; A12380), Wheat Germ Agglutinin 405M (WGA) (Biotium; 29028–1), DRAQ5 (Thermo Scientific; 62251). Cells were washed again with 1X PBS and mounted on glass slides using ProLong Gold (Invitrogen; P36930).
Western blot analysis
Cell lysates were prepared using RIPA buffer (Sigma-Aldrich; R0278) supplemented with protease inhibitors (Roche; 04693124001). Samples were centrifuged at 20,000 × g for 15 min to remove cell debris. The resulting supernatant was boiled with Laemmli sample buffer for 5 min. Samples were then loaded on a 4%–12% NuPAGE gradient gel (Invitrogen; NP0322BOX). Gels were transferred onto a nitrocellulose membrane at 30V for 18 h. Membranes were blocked with 5% dry milk diluted in 1X PBS containing 0.1% Tween 20 (PBS-T) for 2 h at room temperature. The membranes were incubated with primary antibody diluted in 1X PBS-T containing 1% BSA overnight at 4C. Primary antibodies used were anti-MISP (Thermo Scientific; PA5–61995), anti-villin (Santa Cruz; sc-66022), anti-GAPDH (Cell Signaling; 2118), anti-β-actin (Sigma-Aldrich; A5316). Membranes were then washed with 1X PBS-T and incubated with secondary antibodies for 1 h at room temperature. Secondary antibodies used were IRdye 800 donkey anti-rabbit (LI-COR; 926–32213) or donkey anti-mouse (LI-COR; 926–32212). Membranes were washed with 1X PBS-T and imaged using the Odyssey CLx infrared scanner (LI-COR). Images were processed using the FIJI software (NIH). Protein expression levels were normalized to GAPDH.
Light microscopy and image processing
Laser scanning confocal imaging was conducted using Nikon A1 Microscope equipped with 405, 488, 561 and 645 nm LASERs, Apo TIRF 100×/1.45 NA, Plan Apo 60×/1.4 NA, Plan Fluor ELWD 40×/0.6 NA objectives. Live-cell imaging was conducted using a Nikon Ti2 Eclipse equipped with 488, 561 and 645 nm excitation LASERs, Apo TIRF 100×/1.49 NA and Plan Fluor 40×/1.3 NA objectives, a Hamamatsu X1 spinning disk, and Photometrics Prime 95B sCMOS or Hamamatsu Orca-Fusion BT sCMOS cameras. FRAP was also conducted using a Bruker mini-scanner module capable of producing ROI specific 405 nm photo-stimulation. Images were deconvolved and/or denoised using Nikon Elements software. Super-resolution imaging was performed using a Nikon Structured Illumination Microscope (N-SIM) equipped with 405, 488, 561 and 640 nm LASERs, an SR Apo TIRF 100×/1.49 NA objective, and an Andor iXon Ultra DU-897 EMCCD camera. Images were reconstructed using Nikon Elements software. For imaging in all microscope modalities, gain was matched between samples during image acquisition.
Protein purification
6xHis-MBP-MISP and 6xHis-EGFP-MBP-MISP constructs were expressed in Sf9 insect cells. Insect cell pellets were resuspended in lysis buffer (20 mM Tris HCl, 0.3 M KCl, 10 mM imidazole, 10% glycerol, 2 mM DTT, pH 7.5) supplemented with protease inhibitors (Roche, 5892953001). Resuspended samples were lysed using a Dounce homogenizer and passed through an 18-gauge needle to shear DNA. The resultant lysate was then centrifuged at 35,000 rpm in a Ti 50.2 rotor (Beckman) for 30 min at 4°C. Clarified lysates were then filtered using a 0.45 μm syringe filter. Samples were then loaded into a HisTrap column according to the manufacturer protocol and eluted with a 50–500 mM linear imidazole gradient (pH 7.5). Protein purity was assessed by SDS-PAGE. Eluted protein was concentrated using a centrifugal filter (Millipore; UFC803024). For in vitro EM experiments, the 6xHis-MBP tag was cleaved from 6xHis-MBP-MISP using a TEV protease (NEB; P8112) for 1 h at room temperature. The cleaved 6xHis-MBP tag was removed by incubating the solution with Ni-NTA magnetic beads (NEB; S1423) for 1 h at 4°C. The solution was then placed in a magnetic rack to separate the bead-bound 6xHis-MBP fraction from MISP. The purified full-length MISP was run in an SDS-PAGE gel to confirm successful cleavage.
Actin Co-Sedimentation assays
Rabbit skeletal G-actin (Cytoskeleton Inc., AKL99) was resuspended according to manufacturer instructions. Resuspended G-actin were centrifuged at 100,000 × g to remove aggregated monomers. G-actin was then polymerized according to the manufacturer instruction. For low-speed sedimentation assays, F-actin was stabilized with phalloidin (Sigma-Aldrich, A22287), and centrifuged at 20,000 × g to precipitate nonspecific aggregates. F-actin (5 μM) was incubated with increasing concentrations of 6xHis-MBP-MISP (0–5 μM) for 15 min at room temperature. Subsequently, all MISP/F-actin sample series were centrifuged at 10,000 × g for 20 min at room temperature. For high-speed sedimentation assays, MBP-MISP (0.5 μM) was incubated with increasing concentrations of non-stabilized F-actin (0–10 μM) for 2 h at 4°C. Subsequently, all MISP/F-actin sample series were centrifuge at 100,000 × g for 30 min at 4°C. In low- and high-sedimentation assays, the supernatant was carefully removed without disrupting the pellet. Both supernatant and pellet fractions were boiled with samples buffer and run into a 4%–12% NuPAGE gradient gel (Invitrogen, NP0322BOX). Gels were stained with Coomassie blue (Bio-Rad, 1610786) and imaged in a gel imaging system (Bio-Rad, Gel Doc™ EZ System).
Transmission electron microscopy
To prepare MISP/F-actin mixtures for electron microscopy (EM), F-actin was prepared as previously described. Phalloidin-stabilized F-actin was incubated with or without purified MISP at a 5:1 molar ratio overnight at 4°C. Carbon-coated copper grids (EMS; cat# CF300-Cu) were glow discharged and coated with 0.1% poly-lysine solution for 15 min and washed 2 times with ddH2O to remove free poly-lysine. Samples were incubated with the grids for 15 min, briefly washed with ddH2O, and negative stained with 2% uranyl acetate. Images were collected on a FEI Tecnai T-12 transmission electron microscope operating at 100 kV using an AMT CMOS camera.
QUANTIFICATION AND STATISTICAL ANALYSIS
All images were process and analyzed using Nikon Elements software or FIJI software package (https://fiji.sc). Time series volumes from live imaging experiments were registered using the StackReg plugin in FIJI as needed.
Analysis of signal intensity in intestinal tissue samples
To measure signal intensities along microvilli, a 1-pixel-wide line scans were drawn along the base-tip axis of BB. To measure signal intensities in the BB along the crypt-villus axis, an ROI containing the BB was drawn and straightened using the Straighten plugin in FIJI; average intensities were calculated across the resulting rectangle. All intensity values were normalized from 0 (base) to 1 (tip) and fit to a Gaussian curve using PRISM v. 9.0.
Analysis of BB assembly in W4 cells
To quantify the percentage of W4 cells capable of forming BBs, cells exhibiting a single polarized cap of F-actin (representing a BB) were counted manually. For rescue experiments, only W4 cells expressing an EGFP-MISP refractory construct were scored. To quantify the overall actin intensity in W4 cells, multiple ROIs containing single cells were generated using Nikon Elements software, and F-actin intensities measured in each ROI.
Measuring the lengths of microvilli and rootlets in W4 cells
For the purposes of quantification throughout the paper, we define a microvillus as the segment of a core bundle that is wrapped in plasma membrane, and ‘rootlet’ as the segment that is free of membrane wrapping. Microvilli and rootlet lengths were measured separately using a membrane marker to delineate the boundary of these regions. To calculate membrane coverage (i.e. fraction of the core bundle wrapped in membrane), we summed the average lengths of microvilli and rootlets per cell to obtain a total core bundle length. We then calculated membrane coverage as the ratio of average microvilli length to total core bundle length.
Inter-filament spacing
To quantify the spacing between MISP-bundled actin filaments, EM images were process using the FFT bandpass filter in FIJI. We used image filtering to remove small structures down to 10 pixels (5 nm) and large structures up to 100 pixels (50 nm). Line scans were then drawn perpendicular to tightly packed actin filaments, signal intensity was plotted, and the distance between peaks was measured. For control conditions, actin bundles with an inter-filament spacing of less than 20 nm were considered for quantification and processed as described above.
FRAP analysis
ROIs of similar area were drawn over the microvilli and rootlets of W4 cells, and bleached using a 405 nm LASER steered with a Bruker mini-scanner. Cells were imaged for 30 s before photobleaching, bleached over the course of 5 s, and then imaged every 10 s for 30 min to capture signal recovery dynamics. All intensity values for each condition were normalized from 0 to 1 and plotted together to facilitate comparison. Average values for each condition were fit using two-phase association curves.
Microvilli and rootlet assembly in W4 cells
To quantify the intensity of actin turnover in the microvilli and rootlets in W4 cells overexpressing mCherry-MISP, EGFP-fimbrin and HALO-β-actin, we drew ROIs delimiting these domains in the β-actin channel. As cells were not synchronized in their differentiation following doxycycline addition, in each cell we set ‘0’ as the time frame where the β-actin signal in the rootlet domain increased above background.
Ezrin inhibition in W4 cells
To quantify MISP enrichment to the BB upon ezrin inhibition in W4 cells overexpressing mCherry-MISP, ezrin-EGFP, and HALO-UtrCH, we drew ROI containing the BB in the β-actin channel. As the effect of NSC668394 on ezrin inhibition from the BB was not synchronized across cells, we arbitrarily set the time point ‘0’ as 9 frames (22.5 min) before ezrin signal dropped below background. All intensity values for each condition were normalized from 0 to 1 and plotted together to facilitate comparison.
Statistical analysis
Statistical significance was performed using the unpaired T test for pairwise comparison. Statistical correlation was conducted using the Pearson correlation coefficient for colocalization analysis. All statistical analysis was computed in PRISM v. 9.0. (GraphPad).
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Anti-MISP | Thermo Fisher Scientific | Cat# PA5-61995; RRID:AB_2638859 |
| Anti-villin | Santa Cruz Biotechnology | Cat# sc-66022; RRID:AB_2216247 |
| Anti-ezrin | Cell Signaling Technology | Cat# 3145; RRID:AB_2100309 |
| Anti-GAPDH | Cell Signaling Technology | Cat# 2118; RRID:AB_561053 |
| Anti-β-actin | Sigma-Aldrich | Cat# A5316; RRID:AB_476743 |
| F(ab’)2-Goat anti-rabbit Alexa Fluor 488 | Molecular Probes | Cat# A-11070; RRID:AB_142134 |
| F(ab’)2-Goat anti-mouse Alexa Fluor 568 | Molecular Probes | Cat# A-11019; RRID:AB_143162 |
| IRDye 800CW Donkey anti-rabbit IgG antibody | LI-COR Biosciences | Cat# 926-32213; RRID:AB_621848 |
| IRDye 800CW Donkey anti-mouse IgG antibody | LI-COR Biosciences | Cat# 926-32212; RRID:AB_621847 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| DH5-Alpha competent Escherichia coli cells | Molecular Cell Biology Resource Core, Vanderbilt University Medical Center | Cat# DH5-Alpha |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Blasticidin | Gold Biotechnology | Cat# B-800 |
| G418 Disulfate | Gold Biotechnology | Cat# G-418 |
| Phleomycin | InvivoGen | Cat# Ant-ph-1 |
| Doxycycline | RPI | Cat# D43020 |
| Puromycin | Gold Biotechnology | Cat# P-600-100 |
| NSC668394 | Sigma-Aldrich | Cat# 341216 |
| DMSO | Sigma-Aldrich | Cat# D8418 |
| TET-free FBS | Atlanta Biological | Cat# S10350 |
| FuGENE 6 | Promega | Cat# E2691 |
| Polybrene Infection Reagent | Sigma-Aldrich | Cat# H9268 |
| Lipofectamine 2000 | Thermo Fisher Scientific | Cat# 11668019 |
| Lenti-X Concentrator | Clontech | Cat# 631231 |
| Paraformaldehyde, 16% | Electron Microscopy Sciences | Cat# 15710 |
| ProLong Gold Antifade Mountant | Invitrogen | Cat# P36930 |
| cOmplete, Mini Protease Inhibitor Cocktail | Roche | Cat# 04693124001 |
| cOmplete ULTRA Tablets, Protease Inhibitor Cocktail | Roche | Cat# 5892953001 |
| Ni-NTA magnetic beads | NEB | Cat# S1423 |
| TEV protease | NEB | Cat# P8112 |
| Rabbit skeletal G-actin | Cytoskeleton Inc | Cat# AKL99 |
| Poly-L-lysine solution, 0.1% | Sigma-Aldrich | Cat# P8920 |
| Carbon-coated copper grids | Electron Microscopy Sciences | Cat# CF300-Cu |
| Alexa Fluor 568 Phalloidin | Invitrogen | Cat# A12380 |
| Alexa Fluor 647 Phalloidin | Invitrogen | Cat# A22287 |
| Wheat Germ Agglutinin 405M (WGA) | Biotium | Cat# 29028-1 |
| DRAQ5 Fluorescent Probe Solution | Thermo Fisher Scientific | Cat# 62251 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| LS174T-W4 | Gift from Dr. Hans Clevers (Utrecht University, Netherlands) | Baas et al. (2004); RRID CVCL J074 |
| CACO-2BBE | Gift from Dr. Mark Mooseker (Yale University) | N/A |
| LLC-PK1-CI4 | Gift from Dr. Carolyn Slayman (Yale University) | N/A |
| HEK293T | ATCC | CRL-11268 |
| HeLa | ATCC | CCL-2 |
| Sf9 | Thermo Fisher Scientific | Cat# 11496015 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mus musculus (C57BL/6J) | Jackson Laboratory | 000664 |
|
| ||
| Recombinant DNA | ||
|
| ||
| pCMV-SPORT-MISP | Harvard PlasmID Database | HsCD00326629 |
| pEGFP-C1-MISP | This paper | N/A |
| pmCherry-C1-MISP | This paper | N/A |
| pHALO-C1-MISP | This paper | N/A |
| pEGFP-C1-fimbrin | Tyska Laboratory | N/A |
| pEGFP-C1-villin | Tyska Laboratory | N/A |
| pEGFP-C1-espin | Gift from Dr. James Bartles (Northwestern University) | N/A |
| pHALO-C1-β-actin | Tyska Laboratory | N/A |
| pEGFP-N1-ezrin | Addgene | Cat# 20680 |
| PLKO.1 (scramble) | Dr. David Sabatini / Addgene | Cat# 1864 |
| PLKO.1 (MISP-KD) | Sigma-Aldrich | TRCN0000422523 |
| PLKO.1 (MISP-KD-2) | Sigma-Aldrich | TRCN0000116527 |
| psPAX2 packaging plasmid | Dr. Didier Trono / Addgene | Cat# 12260 |
| pDMD2.G | Dr. Didier Trono / Addgene | Cat# 12259 |
| pFastBac-6xHis-MBP | Addgene | Cat# 30116 |
| pFastBac-6xHis-MBP-MISP | This paper | N/A |
| pFastBac-6xHis-MBP-EGFP-MISP | This paper | N/A |
| pLVX-EGFP | This paper | N/A |
| pLVX-EGFP-MISP | This paper | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| FIJI | https://fiji.sc | N/A |
| NIS AR Elements Analysis | Nikon (https://bit.ly/3fFd8rz) | N/A |
| Prism 8.1.2 | GraphPad (https://graphpad.com) | N/A |
|
| ||
| Other | ||
|
| ||
| 35 mm glass bottom dishes | Invitro Scientific | Cat# D35-20-1.5-N |
Highlights.
MISP is an actin bundling protein found in the rootlets of epithelial microvilli
MISP expression levels tune the length of microvillar rootlets
MISP recruits fimbrin to cooperatively elongate and stabilize microvillar rootlets
Ezrin restricts MISP from the membrane-wrapped segment of core actin bundles
ACKNOWLEDGMENTS
The authors would like to thank all members of the Tyska laboratory, the Vanderbilt Microtubules and Motors Club, and the Vanderbilt Epithelial Biology Center for feedback and advice. Microscopy was performed in part using the Vanderbilt Cell Imaging Shared Resource. This work was supported by NIH grants R01-DK111949, R01-DK095811, and R01-DK125546 (to M.J.T.). M.Z. acknowledges the support of NIH grant R35-GM119552.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110692.
REFERENCES
- Atilgan E, Wirtz D, and Sun SX (2006). Mechanics and dynamics of actin-driven thin membrane protrusions. Biophys. J. 90, 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas AF, Kuipers J, van der Wel NN, Batlle E, Koerten HK, Peters PJ, and Clevers HC (2004). Complete polarization of single intestinal epithelial cells upon activation of LKB1 by STRAD. Cell 116, 457–466. [DOI] [PubMed] [Google Scholar]
- Bao J, Bielski E, Bachhawat A, Taha D, Gunther LK, Thirumurugan K, Kitajiri S, and Sakamoto T (2013). R1 motif is the major actin-binding domain of TRIOBP-4. Biochemistry 52, 5256–5264. [DOI] [PubMed] [Google Scholar]
- Bartles JR, Zheng L, Li A, Wierda A, and Chen B (1998). Small espin: a third actin-bundling protein and potential forked protein ortholog in brush border microvilli. J. Cell Biol. 143, 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyy A, Merino F, Sitsel O, and Raunser S (2020). Structure of the Lifeact–F-actin complex. PLoS Biol. 18, e3000925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berryman M, Franck Z, and Bretscher A (1993). Ezrin is concentrated in the apical microvilli of a wide variety of epithelial cells whereas moesin is found primarily in endothelial cells. J. Cell Sci. 105, 1025–1043. [DOI] [PubMed] [Google Scholar]
- Bretscher A, and Weber K (1979). Villin: the major microfilament-associated protein of the intestinal microvillus. Proc. Natl. Acad. Sci. U S A 76, 2321–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A, and Weber K (1980). Fimbrin, a new microfilament-associated protein present in microvilli and other cell surface structures. J. Cell Biol. 86, 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A, Reczek D, and Berryman M (1997). Ezrin: a protein requiring conformational activation to link microfilaments to the plasma membrane in the assembly of cell surface structures. J. Cell Sci. 110, 3011–3018. [DOI] [PubMed] [Google Scholar]
- Bulut G, Hong S-H, Chen K, Beauchamp EM, Rahim S, Kosturko GW, Glasgow E, Dakshanamurthy S, Lee H-S, Daar I, et al. (2012). Small molecule inhibitors of ezrin inhibit the invasive phenotype of osteosarcoma cells. Oncogene 31, 269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaletto JB, Saotome I, Curto M, and McClatchey AI (2011). Ezrin-mediated apical integrity is required for intestinal homeostasis. Proc. Natl. Acad. Sci. U S A 108, 11924–11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley SW, Mooseker MS, and Tyska MJ (2014). Shaping the intestinal brush border. J. Cell Biol. 207, 441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date S, and Sato T (2015). Mini-gut organoids: reconstitution of the stem cell niche. Annu. Rev. Cell Dev. Biol. 31, 269–289. [DOI] [PubMed] [Google Scholar]
- Delacour D, Salomon J, Robine S, and Louvard D (2016). Plasticity of the brush border — the yin and yang of intestinal homeostasis. Nat. Rev. Gastroenterol. Hepatol. 13, 161–174. [DOI] [PubMed] [Google Scholar]
- Dudouet B, Robine S, Huet C, Sahuquillo-Merino C, Blair L, Coudrier E, and Louvard D (1987). Changes in villin synthesis and subcellular distribution during intestinal differentiation of HT29–18 clones. J. Cell Biol. 105, 359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzell RM, Chafel MM, and Matsudaira PT (1989). Differential localization of villin and fimbrin during development of the mouse visceral endoderm and intestinal epithelium. Dev. Camb. Engl. 106, 407–419. [DOI] [PubMed] [Google Scholar]
- Ferrary E, Cohen-Tannoudji M, Pehau-Arnaudet G, Lapillonne A, Athman R, Ruiz T, Boulouha L, Marjou FE, Doye A, Fontaine J-J, et al. (1999). In vivo, villin is required for Ca2ϩ-dependent F-actin disruption in intestinal brush borders. J. Cell Biol. 146, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaeta IM, Meenderink LM, Postema MM, Cencer CS, and Tyska MJ (2021). Direct visualization of epithelial microvilli biogenesis. Curr. Biol. 31, 2561–2575.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm-Günter E-MS, Revenu C, Ramos S, Hurbain I, Smyth N, Ferrary E, Louvard D, Robine S, and Rivero F (2009). Plastin 1 binds to keratin and is required for terminal web assembly in the intestinal epithelium. Mol. Biol. Cell 20, 2549–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton CM, Liu J, Taylor DW, DeRosier DJ, and Taylor KA (2008). The 3D structure of villin as an unusual F-actin crosslinker. Structure 16, 1882–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanono A, Garbett D, Reczek D, Chambers DN, and Bretscher A (2006). EPI64 regulates microvillar subdomains and structure. J. Cell Biol. 175, 803–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegan PS, Giral H, Levi M, and Mooseker MS (2012). Myosin VI is required for maintenance of brush border structure, composition, and membrane trafficking functions in the intestinal epithelial cell. Cytoskeleton 69, 235–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Tilney LG, Fujiwara K, and Heuser JE (1982). Organization of actin, myosin, and intermediate filaments in the brush border of intestinal epithelial cells. J. Cell Biol. 94, 425–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Cheney RE, and Willard M (1983). Location of a protein of the fodrin-spectrin-TW260/240 family in the mouse intestinal brush border. Cell 32, 953–965. [DOI] [PubMed] [Google Scholar]
- Howe CL, and Mooseker MS (1983). Characterization of the 110-kdalton actin-calmodulin-, and membrane-binding protein from microvilli of intestinal epithelial cells. J. Cell Biol. 97, 974–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner MW (1980). Implications of treadmilling for the stability and polarity of actin and tubulin polymers in vivo. J. Cell Biol. 86, 330–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajiri S, Sakamoto T, Belyantseva IA, Goodyear RJ, Stepanyan R, Fujiwara I, Bird JE, Riazuddin S, Riazuddin S, Ahmed ZM, et al. (2010). Actin-bundling protein TRIOBP forms resilient rootlets of hair cell stereocilia essential for hearing. Cell 141, 786–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein MG, Shi W, Ramagopal U, Tseng Y, Wirtz D, Kovar DR, Staiger CJ, and Almo SC (2004). Structure of the actin crosslinking core of fimbrin. Structure 12, 999–1013. [DOI] [PubMed] [Google Scholar]
- Krey JF, Krystofiak ES, Dumont RA, Vijayakumar S, Choi D, Rivero F, Kachar B, Jones SM, and Barr-Gillespie PG (2016). Plastin 1 widens stereocilia by transforming actin filament packing from hexagonal to liquid. J. Cell Biol. 215, 467–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kschonsak YT, and Hoffmann I (2018). Activated Ezrin controls MISP levels to ensure correct NuMA polarization and spindle orientation. J. Cell Sci. 131, jcs.214544. [DOI] [PubMed] [Google Scholar]
- Kumeta M, Gilmore JL, Umeshima H, Ishikawa M, Kitajiri S, Horigome T, Kengaku M, and Takeyasu K (2014). Caprice/MISP is a novel F-actin bundling protein critical for actin-based cytoskeletal reorganizations. Genes Cells 19, 338–349. [DOI] [PubMed] [Google Scholar]
- Lange K (2011). Fundamental role of microvilli in the main functions of differentiated cells: outline of an universal regulating and signaling system at the cell periphery. J. Cell. Physiol. 226, 896–927. [DOI] [PubMed] [Google Scholar]
- Loomis PA, Zheng L, Sekerková G, Changyaleket B, Mugnaini E, and Bartles JR (2003). Espin cross-links cause the elongation of microvillus-type parallel actin bundles in vivo. J. Cell Biol. 163, 1045–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B, Kirsch M, Anderhub S, Zentgraf H, and Krämer A (2013). The novel actin/focal adhesion-associated protein MISP is involved in mitotic spindle positioning in human cells. Cell Cycle 12, 1457–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira P, Mandelkow E, Renner W, Hesterberg LK, and Weber K (1983). Role of fimbrin and villin in determining the interfilament distances of actin bundles. Nature 301, 209–214. [DOI] [PubMed] [Google Scholar]
- McConnell RE, Benesh AE, Mao S, Tabb DL, and Tyska MJ (2011). Proteomic analysis of the enterocyte brush border. Am. J. Physiol. Gastrointest. Liver Physiol 300, G914–G926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meenderink LM, Gaeta IM, Postema MM, Cencer CS, Chinowsky CR, Krystofiak ES, Millis BA, and Tyska MJ (2019). Actin dynamics drive microvillar motility and clustering during brush border assembly. Dev. Cell 50, 545–556.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentes A, Huehn A, Liu X, Zwolak A, Dominguez R, Shuman H, Ostap EM, and Sindelar CV (2018). High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing. Proc. Natl. Acad. Sci. U S A 115, 1292–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooseker MS, and Tilney LG (1975). Organization of an actin filament-membrane complex. Filament polarity and membrane attachment in the microvilli of intestinal epithelial cells. J. Cell Biol. 67, 725–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooseker MS, Keller TC 3rd, and Hirokawa N (1983). Regulation of cytoskeletal structure and contractility in the brush border. In Novartis Foundation Symposia, Porter R and Collins GM, eds. (John Wiley & Sons, Ltd.), pp. 195–215. [DOI] [PubMed] [Google Scholar]
- Nambiar R, McConnell RE, and Tyska MJ (2010). Myosin motor function: the ins and outs of actin-based membrane protrusions. Cell. Mol. Life Sci. 67, 1239–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta K, Higashi R, Sawaguchi A, and Nakamura K (2012). Helical arrangement of filaments in microvillar actin bundles. J. Struct. Biol. 177, 513–519. [DOI] [PubMed] [Google Scholar]
- Peña JF, Alié A, Richter DJ, Wang L, Funayama N, and Nichols SA (2016). Conserved expression of vertebrate microvillar gene homologs in choanocytes of freshwater sponges. EvoDevo 7, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson MD, and Mooseker MS (1993). An in vitro model for the analysis of intestinal brush border assembly. I. Ultrastructural analysis of cell contact-induced brush border assembly in Caco-2BBe cells. J. Cell Sci. 105, 445–460. [DOI] [PubMed] [Google Scholar]
- Peterson MD, Bement WM, and Mooseker MS (1993). An in vitro model for the analysis of intestinal brush border assembly. II. Changes in expression and localization of brush border proteins during cell contact-induced brush border assembly in Caco-2BBe cells. J. Cell Sci. 105, 461–472. [DOI] [PubMed] [Google Scholar]
- Pinson KI, Dunbar L, Samuelson L, and Gumucio DL (1998). Targeted disruption of the mouse villin gene does not impair the morphogenesis of microvilli. Dev. Dyn. 211, 109–121. [DOI] [PubMed] [Google Scholar]
- Pollard TD, and Mooseker MS (1981). Direct measurement of actin polymerization rate constants by electron microscopy of actin filaments nucleated by isolated microvillus cores. J. Cell Biol. 88, 654–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenu C, Athman R, Robine S, and Louvard D (2004). The co-workers of actin filaments: from cell structures to signals. Nat. Rev. Mol. Cell Biol. 5, 635–646. [DOI] [PubMed] [Google Scholar]
- Revenu C, Ubelmann F, Hurbain I, El-Marjou F, Dingli F, Loew D, Delacour D, Gilet J, Brot-Laroche E, Rivero F, et al. (2012). A new role for the architecture of microvillar actin bundles in apical retention of membrane proteins. Mol. Biol. Cell 23, 324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robine S, Huet C, Moll R, Sahuquillo-Merino C, Coudrier E, Zweibaum A, and Louvard D (1985). Can villin be used to identify malignant and undifferentiated normal digestive epithelial cells? Proc. Natl. Acad. Sci. U S A 82, 8488–8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy P, and Perrin BJ (2018). The stable actin core of mechanosensory stereocilia features continuous turnover of actin cross-linkers. Mol. Biol. Cell 29, 1856–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saotome I, Curto M, and McClatchey AI (2004). Ezrin is essential for epithelial organization and villus morphogenesis in the developing intestine. Dev. Cell 6, 855–864. [DOI] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Burkhardt P, Sánchez-Pons N, Fairclough SR, Lang BF, King N, and Ruiz-Trillo I (2013). Insights into the origin of metazoan filopodia and microvilli. Mol. Biol. Evol. 30, 2013–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, and Borisy GG (2003). Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 160, 409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Takeda S, Mitsuoka K, Oda T, Kimura-Sakiyama C, Maéda Y, and Narita A (2018). Structural basis for cofilin binding and actin filament disassembly. Nat. Commun. 9, 1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, and Cardell RR (1970). Factors controlling the reassembly of the microvillus border of the small intestine of the salamander. J. Cell Biol. 47, 408–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyska MJ, and Mooseker MS (2002). MYO1A (brush border myosin I) dynamics in the brush border of LLC-PK1-CL4 cells. Biophys. J. 82, 1869–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. (2015). Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- Viswanatha R, Ohouo PY, Smolka MB, and Bretscher A (2012). Local phosphocycling mediated by LOK/SLK restricts ezrin function to the apical aspect of epithelial cells. J. Cell Biol. 199, 969–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann N, DeRosier D, Matsudaira P, and Hanein D (2001). An atomic model of actin filaments cross-linked by fimbrin and its implications for bundle assembly and function. J. Cell Biol. 153, 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Settele F, Kotak S, Sanchez-Pulido L, Ehret L, Ponting CP, Gönczy P, and Hoffmann I (2013). MISP is a novel Plk1 substrate required for proper spindle orientation and mitotic progression. J. Cell Biol. 200, 773–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


