Summary
The accumulation of lipid droplets (LDs) in the liver is a hallmark of steatosis, which is often associated with lysosomal dysfunction. Nevertheless, the underlying mechanisms remain unclear. Here, using Huh7 cells loaded with oleate as a model to study LD metabolism, we show that cellular content and distribution of LDs are correlated with those of the lysosome and regulated by oleate and septin 9. High expression of septin 9 promotes perinuclear clustering of lysosomes which co-localized with Golgi and not with their surrounding LDs. On the other hand, knockdown of septin 9 disperses the two organelles which colocalize at the cell periphery. The Rab7 is present around these peripheral LDs. PtdIns5P which binds septin 9 and MTMR3 which converts PtdIns(3,5)P2 into PtdIns(5) recapitulates the effects of septin 9. By contrast, PtdIns(3,5)P2 promotes LD/lysosome co-localization. Overall, our data reveal a phosphoinositide/septin 9-dependent mechanism that regulates LD behavior through the control of their association with lysosomes.
Subject areas: Cell biology, Organizational aspects of cell biology, Functional aspects of cell biology
Graphical abstract
Highlights
-
•
Septin 9 is regulates oleate-induced lysosome perinuclear clustering
-
•
Septin 9 and MTs regulate oleate-induced lysosome co-localization with Golgi
-
•
LDs with high septin 9 have less interaction with Rab7 and LAMP1
-
•
PIs have specific effects on LD and lysosome
Cell biology; Organizational aspects of cell biology; Functional aspects of cell biology
Introduction
The liver plays a crucial role in the homeostasis of lipids (Bechmann et al., 2012) and thus alterations of the processes involved in their metabolism because of stressful conditions, such as exposure to a high-fat diet, alcohol, or infection by pathogens such as hepatitis C virus, may induce an accumulation of lipid in the liver and the subsequent development of diseases that include steatosis, nonalcoholic steatohepatitis (NASH) which can progress in cirrhosis and cancer (Benedict and Zhang, 2017; Michelotti et al., 2013). Most of the lipids within the liver are stored in the hepatocytes in the form of cytosolic lipid droplets (LDs). LDs perform diverse cellular functions, which include sequestering the toxic lipids and acting as dynamic lipid storage that enables the rapid mobilization of fatty acids for energy (Herms et al., 2015; Rambold et al., 2015), membrane biosynthesis (Chauhan et al., 2015; Kurat et al., 2009), and lipid signaling pathways (Haemmerle et al., 2011; Tang et al., 2013). LDs also associate with other organelles to maintain cell homeostasis (Barbosa et al., 2015; Barbosa and Siniossoglou, 2017; Schuldiner and Bohnert, 2017).
LDs consist of a hydrophobic core of neutral lipids (primarily composed of triacylglycerols and cholesterol esters) surrounded by a monolayer of phospholipid and cholesterol in which proteins are embedded. The lipid droplet proteins are dominated by enzymes involved in lipid metabolism and members of the perilipin family. However, proteins with other functions such as membrane trafficking, like small Rab-GTPases, and proteins for degradation are also well represented (Bersuker and Olzmann, 2017; Krahmer et al., 2013). There are about 70 human Rab GTPases, which are mostly involved in membrane trafficking and act as master regulators of organelle biogenesis and cellular homeostasis. The targeting of Rab to the membrane depends on the binding of their corresponding GEF (Guanosine Exchange factor) to specific phospholipids/phosphoinositides (PIs) (Blümer et al., 2013). Specific combinations of Rab GTPases and PIs in discrete membrane domains on the organelles and vesicles facilitate endosomal functions to ensure the maintaining and coordination of membrane trafficking (Jean and Kiger, 2012). Therefore, identifying the regulators of the mechanisms behind this complex regulation of Rabs and PIs combination and function on organelles such as LDs and their contribution to cellular homeostasis is a challenging issue.
Septins belong to a family of GTP-binding proteins with 13 members and are highly conserved in eukaryotes (Nishihama et al., 2011). Septins assemble in hetero-oligomeric complexes and form nonpolar filaments or rings that bind to actin and microtubules (MTs) (Kim et al., 2011; Kinoshita et al., 2002; Sellin et al., 2011). Septins are therefore essential for cytoskeleton-dependent cell processes and are considered as the fourth component of the cytoskeleton (Bai et al., 2013; Joo et al., 2007; Mostowy and Cossart, 2012; Smith et al., 2015). Septins associate with a variety of PIs at different intracellular membranes through a polybasic domain (PB1) (Akil et al., 2016; Dolat and Spiliotis, 2016; Pagliuso et al., 2016; Zhang et al., 1999). Recently, we reported that septin 9 has a second polybasic domain (PB2) conserved in the human septin family. This newly identified domain is critical to septin binding to PIs, filament formation, and the assembly and functionality of the Golgi apparatus (Omrane et al., 2019). Once attached to the membranes, septins can act as a scaffold that recruits cytosolic proteins and other cytoskeletal elements to control numerous cellular functions such as cytokinesis, ciliogenesis, vesicle trafficking, and cell polarity (Akil et al., 2016; Bridges et al., 2016; Fung et al., 2014; Gassama-Diagne et al., 2006; Mostowy and Cossart, 2012; Song et al., 2016; Tanaka-Takiguchi et al., 2009). Therefore, alterations of the expression of septins are associated with several diseases including infectious diseases and cancer (Angelis and Spiliotis, 2016; Henzi et al., 2021; Krokowski et al., 2018).
We reported a high expression of septin 9 in hepatitis C virus (HCV)-induced cirrhosis. We also showed that septin 9 is hijacked by HCV to induce the perinuclear accumulation of LDs by a phosphatidylinositol-5-phosphate and microtubule-dependent mechanism (Akil et al., 2016).
Excessive intracellular accumulation of LDs in liver cells generally results from increased uptake of hepatic free fatty acids (FFA), enhanced de novo lipogenesis, or impaired lipid catabolism (Bessone et al., 2019; Koo, 2013; Kwanten et al., 2014). The autophagy pathway plays an important role in lipid degradation and depends on the activities of the lysosome, whose dysfunction has been associated with steatosis in different studies (Inami et al., 2011; Miyagawa et al., 2016; Wang et al., 2018).
Lysosomes are highly dynamic organelles that are subject to bidirectional movements along microtubules between the center and periphery of cells and represent the main degradative compartments of eukaryotic cells (Cabukusta and Neefjes, 2018). The movements of lysosome are also modulated by contacts with other organelles such as the ER(Jongsma et al., 2016; Raiborg et al., 2015), Trans-Golgi network (TGN) (Wang and Hong, 2002), and peroxisomes (Chu et al., 2015).
Thus, according to the binding capacities of septin 9 to PI and its critical role in the microtubule-dependent assembly of Golgi and vesicle trafficking, we made a hypothesis that septin 9 could control LD perinuclear accumulation by regulating the lysosomes.
Results
Kinetic study of oleate-treated cells indicates that the size and perinuclear accumulation of LDs are related to lysosome localization and depend on oleate concentration
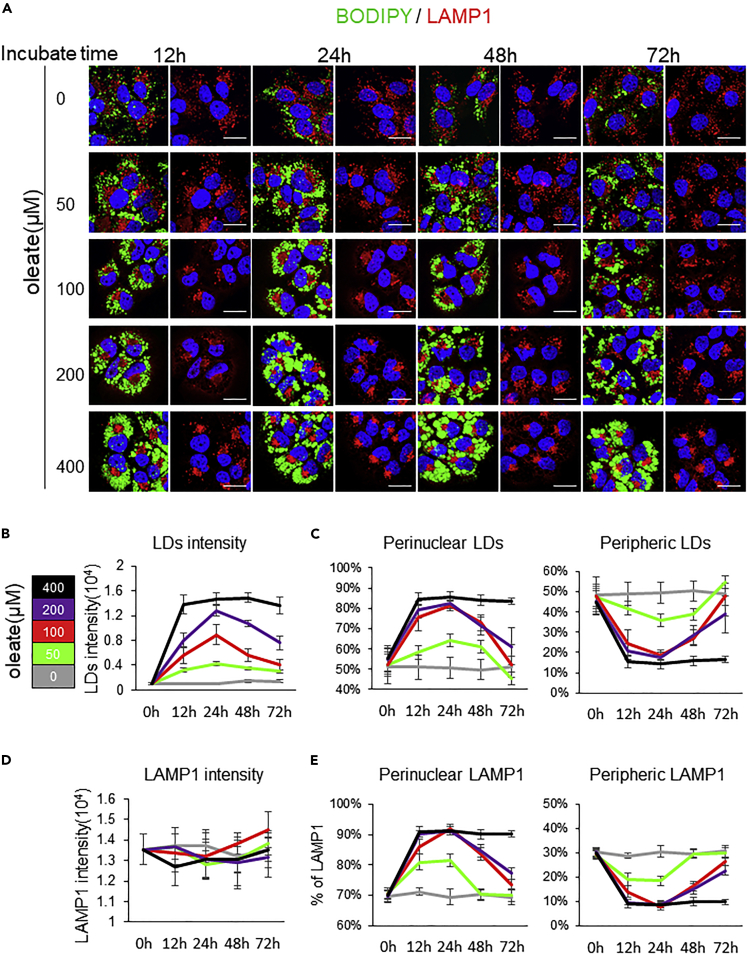
To understand how hepatocytes could respond to lipid overload and the mechanisms behind the cytoplasmic LD accumulation, we used hepatocellular carcinoma derived Huh7 cells. The cells were treated with sodium oleate at 50,100, 200, and 400μM and analyzed after 12h, 24, 48, and 72h of treatment. First, we used BODIPY to stain LDs and assess their total intensity and their time course distribution within cells. Immunofluorescence data revealed non-significant time-dependent changes of the LD behavior in non-treated (0μM) cells. However, at the different concentrations of oleate, the intensity of LDs significantly increased up to 24h after the treatment and then continued to decrease until 72h. Although the increase of LD intensity increased in an oleate dose-dependent manner to reach the maximum at 24h, the decrease was inversely correlated with the dose of oleate, and almost no change was observed in the kinetic for the dose of 400 μM of oleate (Figures 1A and 1B) (Table 1). Furthermore, we showed a correlation between the strong increase in the total intensity of LDs and their perinuclear localization (Figure 1C) (Table 2). Indeed, at 400μM of oleate, no significant change was observed over time for LDs distribution. In fact, nearly 83% of LDs are consistently distributed in the perinuclear area (Figure 1C).
Figure 1.
The kinetic of LD and lysosome intracellular distributions are correlated and dependent of oleate concentration
(A) Huh7 cells were grown overnight. Then culture medium was supplemented without or with 50, 100, 200, and 400 μM sodium oleate complex for different time (12, 24, 48, and 72 h), then stained for LDs (green) and LAMP1(red). Scale bar, 20μm.
(B) LDs intensity were analyzed in 25 cells from three experiments performed as described in (A).
(C) The percentage of LD intensity in perinuclear and peripheric are present in 25 cells from three experiments performed as described in (A).
(D) LAMP1 intensity of 25 cells from 3 independent experiments performed as described in (A).
(E) The percentage of LAMP1 intensity in perinuclear and peripheric are present in 25 cells from three experiments performed as described in (A). Data information: Linegraphs present Mean ± SEM. Student’s t-test was used. ∗p < 0.05, ∗∗p < 0.001, ∗∗∗p < 0.0001.
Table 1.
Analysis of BODIPY distribution
| LD |
Mean |
SEM |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Position | Dosage(μM) | 0h | 12h | 24h | 48h | 72h | 0h | 12h | 24h | 48h | 72h |
| Perinuclear | 0 | 0.5134 | 0.5102 | 0.5061 | 0.4975 | 0.5124 | 0.0865 | 0.0592 | 0.0522 | 0.0509 | 0.0534 |
| 50 | 0.5242 | 0.5822 | 0.6424 | 0.6112 | 0.4545 | 0.0507 | 0.0342 | 0.0302 | 0.0294 | 0.0308 | |
| 100 | 0.5234 | 0.7542 | 0.8143 | 0.7288 | 0.5215 | 0.0605 | 0.0734 | 0.0668 | 0.0402 | 0.0986 | |
| 200 | 0.5454 | 0.7945 | 0.8262 | 0.7126 | 0.6124 | 0.0623 | 0.0704 | 0.0572 | 0.0462 | 0.0598 | |
| 400 | 0.5545 | 0.8457 | 0.8571 | 0.8414 | 0.8349 | 0.0615 | 0.0318 | 0.0234 | 0.0254 | 0.0167 | |
| Peripheric | 0 | 0.4866 | 0.4898 | 0.4939 | 0.5025 | 0.4876 | 0.0865 | 0.0592 | 0.0522 | 0.0509 | 0.0534 |
| 50 | 0.4758 | 0.4178 | 0.3576 | 0.3888 | 0.5455 | 0.0507 | 0.0342 | 0.0302 | 0.0294 | 0.0308 | |
| 100 | 0.4766 | 0.2458 | 0.1857 | 0.2712 | 0.4785 | 0.0605 | 0.0734 | 0.0668 | 0.0402 | 0.0986 | |
| 200 | 0.4546 | 0.2055 | 0.1738 | 0.2874 | 0.3876 | 0.0623 | 0.0704 | 0.0572 | 0.0462 | 0.0598 | |
| 400 | 0.4455 | 0.1543 | 0.1429 | 0.1586 | 0.1651 | 0.0615 | 0.0318 | 0.0234 | 0.0254 | 0.0167 | |
Table 2.
Analysis of the correlation between LD and its perinuclear distribution
| Dosage(μM) | Correlation Total/perinuclear |
|---|---|
| 0 | −0.4629498 |
| 50 | 0.564126976 |
| 100 | 0.876392283 |
| 200 | 0.829894671 |
| 400 | 0.996684504 |
Lysosomes are broadly distributed throughout the cytoplasm, although a higher pool was found in the perinuclear region near the microtubule-organizing center (MTOC) and a very dynamic pool was at the periphery of the cells. These spatial different pools of lysosomes contribute to different biological roles (Cabukusta and Neefjes, 2018). Subsequently, we studied both the total intensity and the intracellular distribution of lysosomes by staining LAMP1 (Lysosomal-associated membrane protein 1). Surprisingly, no significant changes were observed on the total intensity of LAMP1 whatever the oleate concentration and the time after treatment (Figure 1D). Nevertheless, the increase of oleate concentration affected the intracellular distribution of LAMP1 which was mainly compact and formed a cluster in the perinuclear region. The maximum perinuclear distribution of LAMP1 was for the dose of 400 μM of oleate (Figure 1E) as observed for LDs (Figure 1C) (Table 3). Then we stained the cells using the red LysoTracker® probe which is a fluorescent probe for labeling and tracking acidic organelles in live cells such as lysosomes (Figure S1A). Data revealed a profile of LysoTracker® distribution in the cells similar to LAMP1 distribution. Thus, suggesting that the lysosomal components present in the cells and mainly those accumulated in the perinuclear area are still acidic.
Table 3.
Analysis of LAMP1 distribution
| LAMP1 |
Mean |
SEM |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Position | Dosage(μM) | 0h | 12h | 24h | 48h | 72h | 0h | 12h | 24h | 48h | 72h |
| Perinuclear | 0 | 0.6955 | 0.7119 | 0.6935 | 0.7022 | 0.6922 | 0.6955 | 0.7119 | 0.6935 | 0.7022 | 0.6922 |
| 50 | 0.7074 | 0.8096 | 0.8148 | 0.7022 | 0.7012 | 0.7074 | 0.8096 | 0.8148 | 0.7022 | 0.7012 | |
| 100 | 0.7046 | 0.8604 | 0.9201 | 0.8364 | 0.7361 | 0.7046 | 0.8604 | 0.9201 | 0.8364 | 0.7361 | |
| 200 | 0.6976 | 0.9024 | 0.9128 | 0.8476 | 0.7725 | 0.6976 | 0.9024 | 0.9128 | 0.8476 | 0.7725 | |
| 400 | 0.6992 | 0.9095 | 0.9117 | 0.9008 | 0.9018 | 0.6992 | 0.9095 | 0.9117 | 0.9008 | 0.9018 | |
| Peripheric | 0 | 0.3045 | 0.2881 | 0.3065 | 0.2978 | 0.3078 | 0.6955 | 0.7119 | 0.6935 | 0.7022 | 0.6922 |
| 50 | 0.2926 | 0.1904 | 0.1852 | 0.2978 | 0.2988 | 0.7074 | 0.8096 | 0.8148 | 0.7022 | 0.7012 | |
| 100 | 0.2954 | 0.1396 | 0.0799 | 0.1636 | 0.2639 | 0.7046 | 0.8604 | 0.9201 | 0.8364 | 0.7361 | |
| 200 | 0.3024 | 0.0976 | 0.0872 | 0.1524 | 0.2275 | 0.6976 | 0.9024 | 0.9128 | 0.8476 | 0.7725 | |
| 400 | 0.3008 | 0.0905 | 0.0883 | 0.0992 | 0.0982 | 0.6992 | 0.9095 | 0.9117 | 0.9008 | 0.9018 | |
Lysosomes accomplish their catabolic function through a wide range of enzymes including proteases, lipases, nucleases, and other hydrolytic enzymes that break down complex macromolecules (Perera and Zoncu, 2016; Settembre et al., 2013). Thus, to get further insights into the function of lysosomes in oleate treated cells, we analyzed by RT-PCR the mRNA expression of several lysosomal proteins at 24 and 72 h after the treatment of the cells with 0, 100, and 400 μM of oleate. Surprisingly, we observed an increase of the transcripts of those genes after treatment compared to non-treated cells (Figure S2A), indicating that the accumulation of LDs and lysosome in the perinuclear region is not associated with a decrease in the expression of lysosomal enzymes but probably to the organelle clustering which could not be associated with LDs for their degradation. Indeed, we observed that when LAMP1 were clustered in the perinuclear area, which was observed in the cells treated with 100,400μM oleate for 24h or 400μM for 72h, there is less interaction between LAMP1 and LD, compared to the control cells and 100μM oleate treated for 72h in which LAMP1 displayed disperse distribution (Figure S2B). This strongly suggests that the accumulation of LDs and lysosomes in the perinuclear region is not associated with a decrease in the expression of lysosomal enzymes but probably to the intracellular localization of the organelles.
Septin 9 regulates oleate-induced lysosome perinuclear clustering
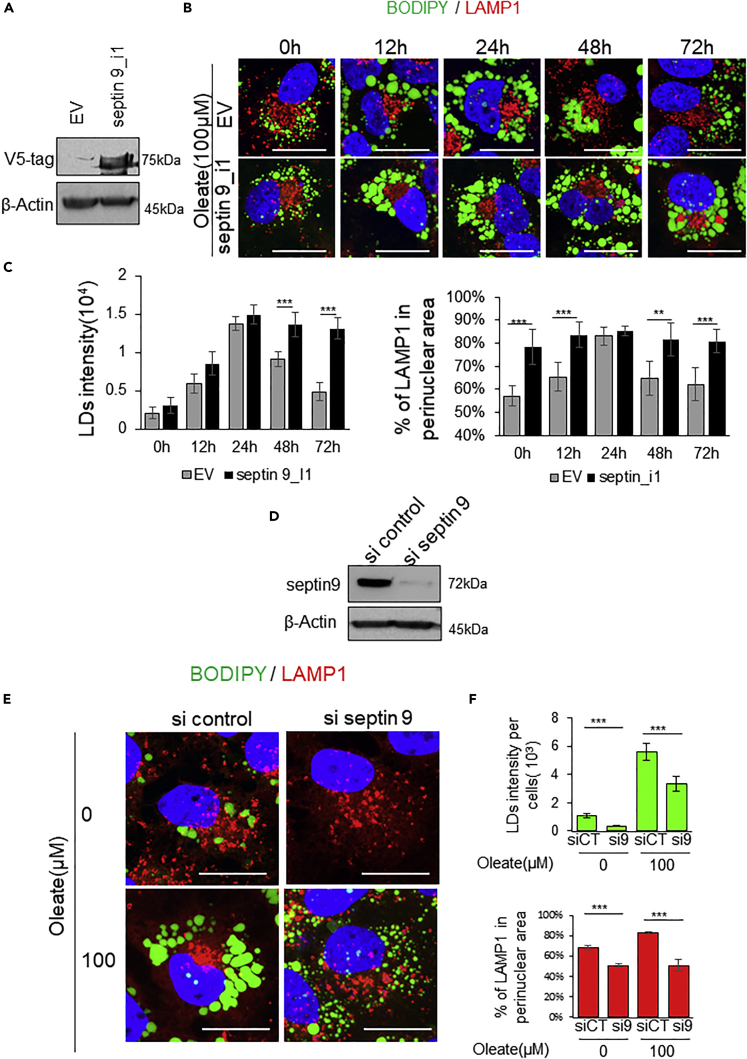
In line with our results described previously and given our previous report on the role of septin 9 in HCV infection or oleate treatment-induced LD perinuclear accumulation (Akil et al., 2016), we hypothesized that septin 9 might control the dynamic of LDs in our kinetic studies through changes in lysosome distribution. To validate that, Huh7 cells were transfected with cDNA of either the isoform 1 of septin 9 (septin 9_i1) or with the empty vector (EV) as a control and treated with 100μM of oleate. In EV transfected cells, the LD intensity and the perinuclear distribution increased until 24h and then decreased up to 72h after the treatment (Figures 2B and 2C) as shown in Figure 1A. However, the expression of septin 9_i1 in those cells blocked the decrease of LDs and accumulated them in the perinuclear region and sustains the perinuclear cluster of LAMP1 (Figures 2B and 2C). Subsequently, knockdown of septin 9 with septin 9 siRNA before treatment with 100μM oleate for 24h significantly decreased the intensity of cellular LDs and the clustering of lysosomes in the perinuclear region and the latter was dispersed throughout the cytoplasm (Figures 2D–2F). This effect of septin 9 knockdown was also confirmed in cells treated with 400μM oleate (Figure S3). Therefore, we concluded that septin 9 regulated oleate-induced lysosome clustering in the perinuclear area.
Figure 2.
Septin 9 regulates oleate-induced lysosome perinuclear clustering
(A) Huh7 cells were transfected with EV or septin 9_I1 for 48 h, then analyzed for the expression of septin 9_i1 by Immunoblot for V5-tag; β-Actin was used for loading control.
(B) Huh7 cells were transfected with EV or septin 9_I1 were grown for 24h. Then culture medium was supplemented with 100 μM sodium oleate complex for different time points (0, 12, 24, 48, and 72h). After treatment, cells were stained for LDs (green) and LAMP1(red). Scale bar, 20μm.
(C) Fluorescence intensity of LDs and the percentage of LAMP1 in perinuclear area were analyzed in 25 cells from three experiments performed as described in (B).
(D) Huh7 cells were transfected with or not septin 9-siRNA for 48h, and then the expression of septin 9 was analyzed by Immunoblot; β-Actin was used for loading control.
(E) Huh7 cells were transfected with or not septin 9-siRNA for 24h then culture with medium supplemented with or not 100 μM oleate for 24h, and then stained for BODIPY (green) and LAMP1 (red). Scale bar, 20μm.
(F) Fluorescence intensity of LDs and the percentage of LAMP1 in perinuclear area were analyzed in 25 cells from three experiments performed as described in (D). Data information: Bar graphs present Mean ± SEM. Student’s t test was used. ∗p < 0.05, ∗∗p < 0.001, ∗∗∗p < 0.0001.
Septin 9 and MTs regulate oleate-induced co-localization of perinuclear lysosomes with Golgi
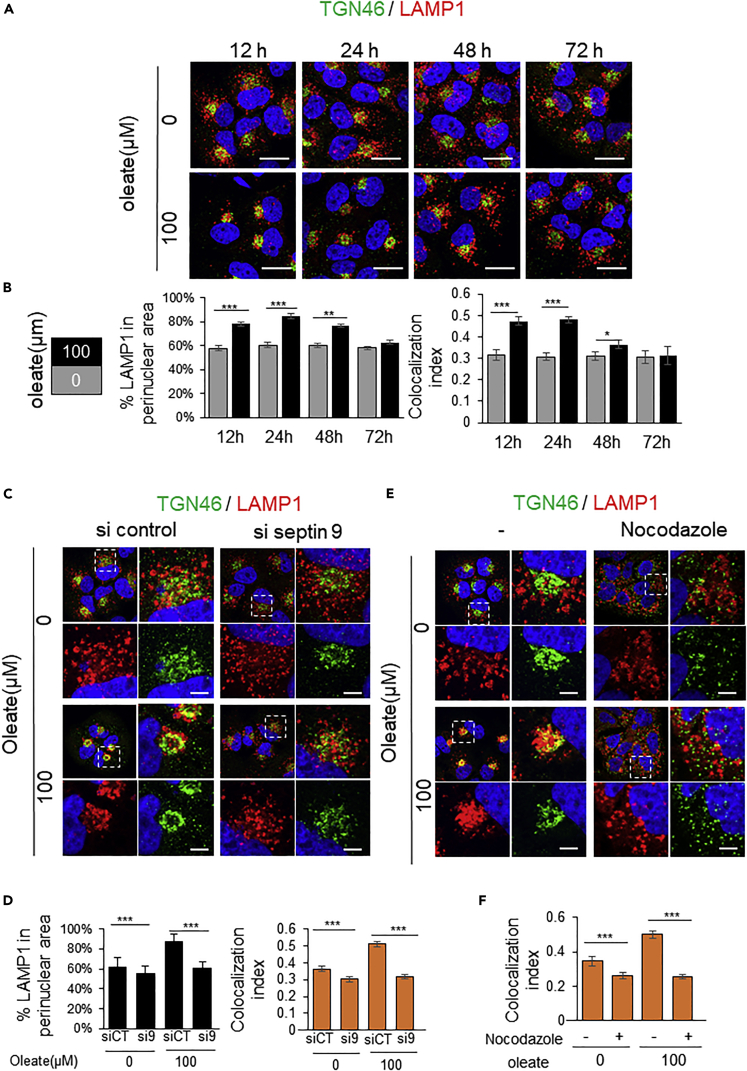
Several studies have shown that the cellular localization of lysosomes is regulated by the Golgi (Tapia et al., 2019; Wang and Hong, 2002). For example, the activation of KDELR, which has been described as the Golgi-localized G protein-coupled receptor (GPCR) that regulates Golgi homeostasis, induces lysosome redistribution in the perinuclear area (Tapia et al., 2019). Besides, we reported that septin 9 co-localized with Golgi, whereas depletion of septin 9 promoted Golgi fragmentation, thus impairing Golgi-dependent secretion (Omrane et al., 2019). Accordingly, we co-stained the cells treated with 100μM of oleate with LAMP1 and the trans-Golgi network marker (TGN46). As expected, after oleate treatment, the perinuclear lysosome co-localized with TGN46 at 12 and 24h, whereas the LAMP1 signal re-localized to the peripheral area of the cells from 48 to 72h, and its co-localization with Golgi decreased (Figures 3A and 3B). Interestingly, knockdown of septin 9 using siRNA markedly decreased LAMP1 perinuclear signal and its co-localization with Golgi, which was also dispersed in the cytoplasm as previously reported (Omrane et al., 2019) (Figures 3C and 3D). By contrast, overexpression of septin 9_i1 significantly increased the co-localization of LAMP1 with Golgi at 72h of treatment with oleate at 100μM (Figure S4). Taken together, these data revealed that oleate induced the septin 9-dependent perinuclear clustering of lysosomes which co-localized with the Golgi. Because lysosome trafficking is heavily reliant on MTs, nocodazole treatment was performed to assess whether lysosome to Golgi clustering at 24h posttreatment required MT dynamic. Interestingly, nocodazole treatment disrupted the perinuclear clustering and the co-localization of both lysosomes and Golgi (Figures 3E and 3F). Thus, indicating that oleate induced LAMP1 perinuclear clustering and its co-localization with the Golgi is also dependent on MTs.
Figure 3.
Septin 9-induced perinuclear cluster of lysosomes colocalizes with Golgi structure
(A) Huh7 cells were cultured overnight. Then culture medium was supplemented with or without 100 μM sodium oleate complex for different time points (12, 24, 48, and 72 h), after treatment, cells were stained for TGN46 (green) and LAMP1 (red). Scale bar, 20μm.
(B) Bar graphs show percentage of LAMP1 in the perinuclear area and Pearson’s correlation coefficient (Rr) for co-localization between LAMP1 and TGN46 of 30 cells from 3 independent experiments performed as described in (A).
(C) Huh7 cells were transfected with or not septin 9-siRNA for 24h then culture with medium supplemented with or not 100 μM oleate for 24 h and stained for TGN46 (green) and LAMP1 (red). The dot squares indicate the zoomed area shown right, TGN46 and LAMP1 are shown below. Scale bar, 5μm.
(D) Bar graphs show percentage of LAMP1 in perinuclear area and Pearson’s correlation coefficient (Rr) for co-localization between LAMP1 and TGN46 of 28 cells from 3 independent experiments performed as described in (C).
(E) Huh7 cells were culture medium supplemented with or without 100 μM oleate for 24h. 4 h before fixing, added 33 nM of nocodazole. After incubation cells were stained for LAMP1 (red) and TGN46 (green). The dot squares indicate the zoomed area shown right, TGN46 and LAMP1 were shown below. Scale bar, 5μm.
(F) Bar graphs show Pearson’s correlation coefficient (Rr) for co-localization between LAMP1 and TGN46 of 30 cells from 3 independent experiments performed as described in (C). Data information: Bar graphs present Mean ± SEM. Student’s t test was used. ∗p < 0.05, ∗∗p < 0.001, ∗∗∗p < 0.0001.
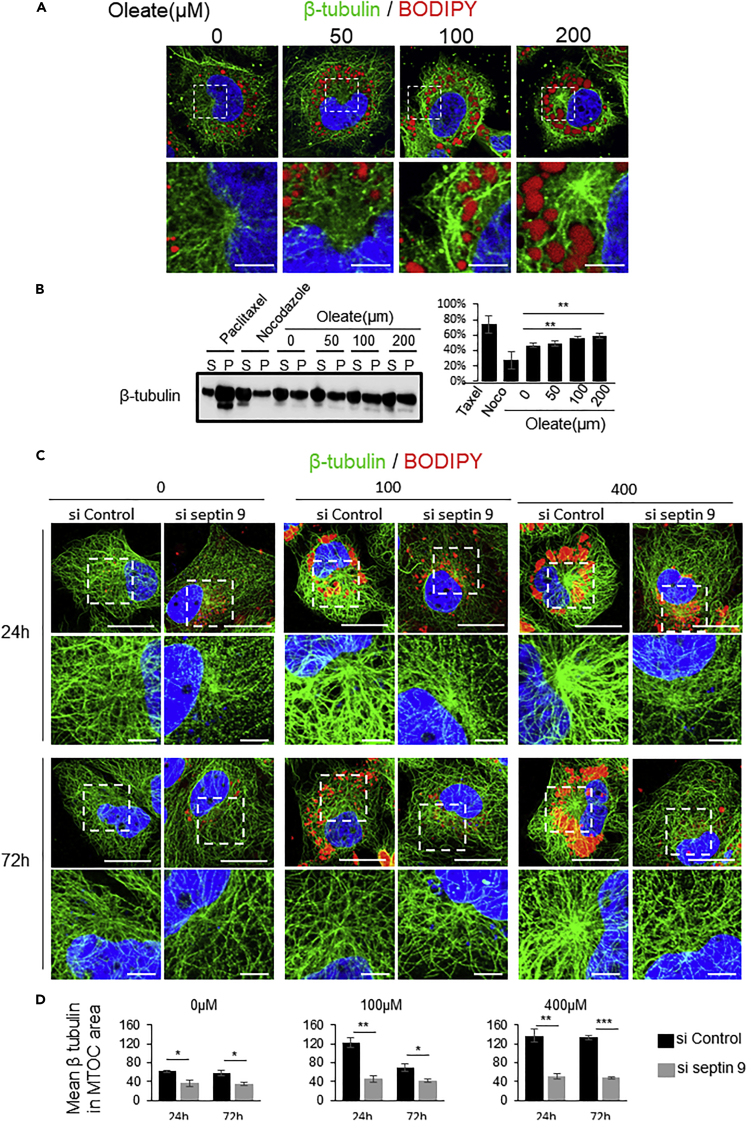
Oleate induces septin 9-dependent MT polymerization at MTOC
We sought to determine the potential effect of oleate treatment on MTs by staining the β-tubulin. The data indicated that oleate enhances the MT network around the MTOC (Figure 4A), suggesting that LD accumulation may have a positive effect on MT polymerization. We subsequently examined the effect of LDs on tubulin polymerization in vitro using paclitaxel and nocodazole treatments as positive and negative controls, respectively. Immunoblots for soluble (S) and polymerized (P) tubulin fractions showed that oleate treatment promoted a dose-dependent polymerization of MTs compared to control cells (Figure 4B), which were consistent with the observation of tubulin staining (Figure 4A). Therefore, we treated the cells with siRNA of septin 9 and then added oleate at 0, 100, and 400 μM for 24 and 72h, resulting in the decrease of the MTOC accumulation of MTs irrespective of the concentrations of oleate (Figures 4C and 4D). Thus, these results revealed that septin 9 regulated the MT stabilization at MTOC promoted by perinuclear accumulation of LDs. Furthermore, we co-stained endogenous septin 9 and β-tubulin in the same experimental conditions (Figure S5A). The results indicated that both the signal of MTs and septin 9 enhanced and co-localized at MTOC in cells treated with both 100 and 400μM oleate for 24h; however, after 72 h treatment, the MTs signal decreased in 100μM oleate treated cells but not in those treated with 400μM oleate (Figures S5A and S5B), remaining the data from LAMP1 staining in Figure 1. Interestingly we showed that the perinuclear clusters of LAMP1 were associated with the tubulin at MTOC (Figure S5C). Thus, suggesting that MT stabilization at MTOC is a determinant of the perinuclear localization of LDs, septin 9, and lysosomes.
Figure 4.
LD accumulation regulates microtubule polymerization
(A) Huh7 cells were cultured with medium supplemented with 0, 50, 100, or 200 μM oleate complex for 24h then stained for β tubulin (green) and LDs (red). White dot squares are presented in higher magnification. Scale bar, 5μm.
(B) Immunoblot analysis of soluble β-tubulin and polymerized β-tubulin in Huh7 cells treated with oleate. Taxol and nocodazole were used as polymerized positive and negative controls. Bar graph presents the densitometry analysis of the percentage of polymerized β-tubulin in total β-tubulin (P/(S + P)) from three independent experiments.
(C) Huh7 cells were transfected with or not septin 9-siRNA for 24h then culture with medium supplemented with 0.100 or 400 μM oleate complex for 24h or 72h then stained for β tubulin (green) and BODIPY (red). Scale bar, 20μm. White dot squares are presented in higher magnification. Scale bar, 5μm.
(D) Bar graphs show the mean intensity of β-tubulin in the MTOC area of 24 cells from 3 independent experiments performed as described in (C). Data information: Bar graphs present Mean ± SEM. Student’s t test was used. ∗p < 0.05, ∗∗p < 0.001, ∗∗∗p < 0.0001.
Septin 9 is associated with large perinuclear LDs
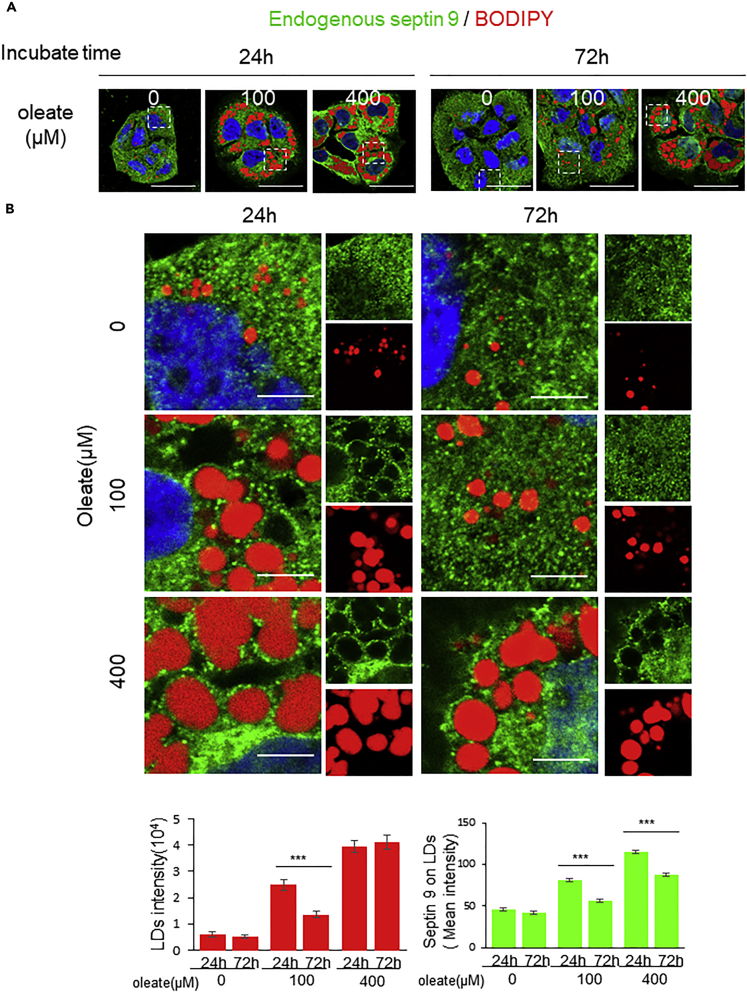
Subsequently, we stained endogenous septin 9 and LDs under conditions similar to that described to analyze MT in Figure 4 to assess their association. As shown in Figures 5A and 5B, in the cells treated with 100 or 400μM oleate, septin 9 was found around the LDs at 24h for both conditions. Interestingly, at 72h, the LD size decreased as well as their association with septin 9 in cells treated with 100μM oleate. However, septin 9 was still present around the large LDs in cells treated with 400μM oleate (Figure 5B). Therefore, we concluded that septin 9 is associated with the large clusters of LDs in the perinuclear region.
Figure 5.
Septin 9 is associated with large LDs in the perinuclear area
(A) Huh7 cells were grown overnight. Then culture medium was supplemented without or with 100 and 400 μM sodium oleate complex for different time (24 h, 72 h), then stained for LDs(red) and septin 9 (green). Scale bar, 20μm.
(B) Dot rectangles in (A) are presented in higher magnification. Scale bar, 5μm. Bar graphs show total intensity of LD and Mean intensity of Rab7 on LDs. Data information: Bar graphs present Mean ± SEM. Student’s t test was used. ∗p < 0.05, ∗∗p < 0.001, ∗∗∗p < 0.0001.
Septin 9 regulates Rab7 intracellular distribution and its association with LDs
Rab7 is an important regulator of lysosome biogenesis (Bucci et al., 2000). It has been reported that when placed under nutrient deprivation, Rab7 is markedly recruited to LDs and is required to promote direct physical interactions between lysosomes and LDs, which is essential for the subsequent utilization of LDs (Schroeder et al., 2015). Therefore, to further determine how septin 9 controlled the fate of LDs and lysosomes in oleate-treated cells, we studied Rab7. For that, Huh7 cells were transfected with the construct of GFP-Rab7 and treated with oleate. First, we validated the association of Rab7 with lysosome staining with LAMP1. In the control cells, both LAMP1 and GFP-Rab7 were co-distributed throughout the cytoplasm and treatment with oleate induced the co-localization and clustering of both proteins in the perinuclear area (Figure S6). Next, we treated the cells expressing GFP-Rab7 with 100 and 400μM oleate for 24 and 72h and we analyzed the GFP-Rab7 and LDs (Figures 6A and 6B). In control cells, the GFP-Rab7 was dispersed within the cytoplasm, and low levels of Rab7 surrounded the LDs surfaces. When treated with either 100μM or 400μM oleate for 24h, Rab7 was clustered in the perinuclear region and surrounded by large LDs. However, after 72h treatment, in cells treated with 100μM oleate, Rab7 signal was dispersed in the cytoplasm and appeared clearly around the LDs, whereas both LDs and Rab7 remained clustered in the perinuclear region when cells are treated with 400μM oleate (Figures 6A and 6B), consistent with data obtained with LAMP1 staining (Figure 1). Strikingly, knockdown of septin 9 in the cell treated with 400μM oleate at 72h re-localized Rab7 on LDs (Figure 6C). Those results were also validated for endogenous Rab7 (Figure S7). We also showed a co-localization of the perinuclear Rab7 clusters with Golgi and treatment with siRNA of septin 9 disrupted the colocalization (Figures S8A and S8B). This suggested that septin 9 also regulated oleate-induced co-localization of perinuclear Rab7 with Golgi.
Figure 6.
Septin 9 regulates the intracellular distribution of Rab7 and its interaction with LDs
(A) Huh7 cells transfected with GFP-Rab7 were grown for 24 h. Then culture medium was supplemented without or with 100 and 400 μM sodium oleate complex for different time (24 h, 72 h), then stained for LDs (red). Scale bar, 20μm.
(B) Dot rectangles in A are presented in higher magnification. Scale bar, 5μm. Bar graphs show total intensity of LD and Mean intensity of Rab7 on LDs of 24 cells from three independent experiments performed as described in (A).
(C) Huh7 cells were transfected with or not septin 9-siRNA for 24h then culture with medium supplemented with or not 400 μM oleate for 72h then stained for LD (red). The dot squares indicate the zoomed area shown right. Scale bar, 5μm. Bar graphs show total intensity of LD and Mean intensity of Rab7 on LDs of 15 cells from 3 independent experiments performed as described. Data information: Bar graphs present Mean ± SEM. Student’s t test was used. ∗p < 0.05, ∗∗p < 0.001, ∗∗∗p < 0.0001.
Together, these data strongly suggested that a high dose of oleate promoted the clustering of lysosomes, thus impairing the association of lysosomes with LDs. These data also highlighted a difference between Rab7 and septin 9 associations with LDs. Although septin 9 was found around the large and perinuclear LDs (Figure 5), Rab7 was found associated with dispersed and smaller LD (Figures 6 and S7), which strongly suggested that septin 9 inhibited Rab7-associated LD degradation by promoting Rab7 perinuclear cluster and subsequently its by binding to LDs.
Septin 9 controls the decrease of autophagy markers induced by oleate
Autophagy is a central mechanism by which hepatocytes catabolize LDs through lysosomal degradation, and Rab7 is a key regulatory component of this autophagy process (Schroeder et al., 2015). Thus, to reinforce the role of septin 9 in lysosome activity and LD catabolism, we assessed its potential contribution to autophagy. First, we knocked down septin 9 using siRNA ad cells and analyzed whether they were treated or not with 100 nM of bafilomycin A1, which is a V-ATPase inhibitor that blocks the autophagic flux acutely by inhibiting autolysosome acidification and autophagosome-lysosome fusion. Then we analyzed the microtubule-associated protein light chain 3 B (LC3B) involved in the formation of autophagosomes and autolysosomes. Knockdown of septin 9 decreased the signal of LC3B, which was particularly enhanced by treatment of cells with bafilomycin as expected (Figures S9A and S9B). Interestingly, treatment of cells with 100μM oleate also increased LC3B signal, which was further enhanced by bafilomycin. In these different conditions, knockdown of septin 9 significantly decreased the LC3B signal (Figures S9A and S9B). These results were confirmed by immunofluorescence studies (Figures S9C and S9D). Together, these data strongly suggested that septin 9 controlled the autophagy process probably through its effect on lysosomes.
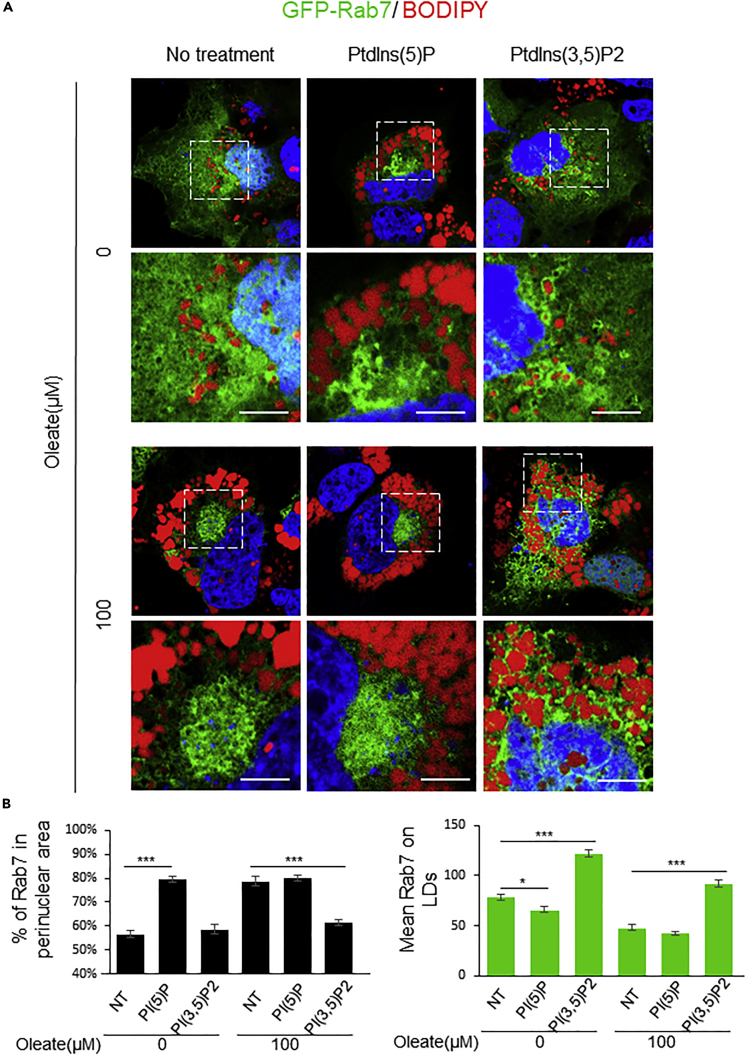
PtdIns(3,5)P2 and PtdIns(5)P regulate Rab7 and septin 9 association with LDs, respectively
We reported that different PIs have distinct effects on the LD size and their interaction with endogenous septin 9, and particularly PtdIns5P had the highest effects (Akil et al., 2016). To note, Rab7 is associated with lysosomes, whose membrane is enriched in PtdIns(3,5)P2. Therefore, we hypothesized that the presence of distinct PIs might regulate Rab7 distribution and its interaction with LDs. To explore this, we treated the cells which expressed GFP-Rab7 with PtdIns(5)P and PtdIns(3,5)P2 (Figures 7A and 7B). Consistent with our previous results (Akil et al., 2016), we observed a strong increase of LDs and a perinuclear clustering of Rab7 within the cells treated with PtdIns(5)P compared to non-treated (NT) cells. Interestingly, treatment of cells with PtdIns(3,5)P2 slightly decreased LD size and Rab7 was seen around the LDs (Figures 7A and 7B). The effects of PtdIns(5)P and PtdIns(3,5)P2 on Rab7/LDs interaction were also validated in the cells pretreated with oleate (Figures 7A and 7B). In addition, the effects of PtdIns(5)P and PtdIns(3,5)P2 on both endogenous septin 9 and septin 9_i1 and their association with LDs were also investigated (Figure S10). Here again, addition of PtdIns(5)P to the cells promoted LD clustering in the perinuclear area and promotes septin 9 filament formation surrounding LDs and particularly visible in septin 9_i1 overexpressing cells. However, addition of PtdIns(3,5)P2 dispersed LDs and decreased the septin 9 on their surfaces (Figure S10).
Figure 7.
PIs regulate LD behavior and intracellular distribution of lysosomal proteins
(A) Huh7 cells transfected with GFP-Rab7 were pretreated with or without 100μM oleate for 24h, then treated with cell-permeant PtdIns5P or PtdIns (3,5) P2 at 30 mM for 15 min before fixing and staining for LDs (red). The dot squares indicate the zoom area. Scale bar, 5μm.
(B) Bar graphs show the percentage of Rab7 in perinuclear area and Mean intensity of Rab7 on LDs of 24 cells from three independent experiments performed as described in (A).
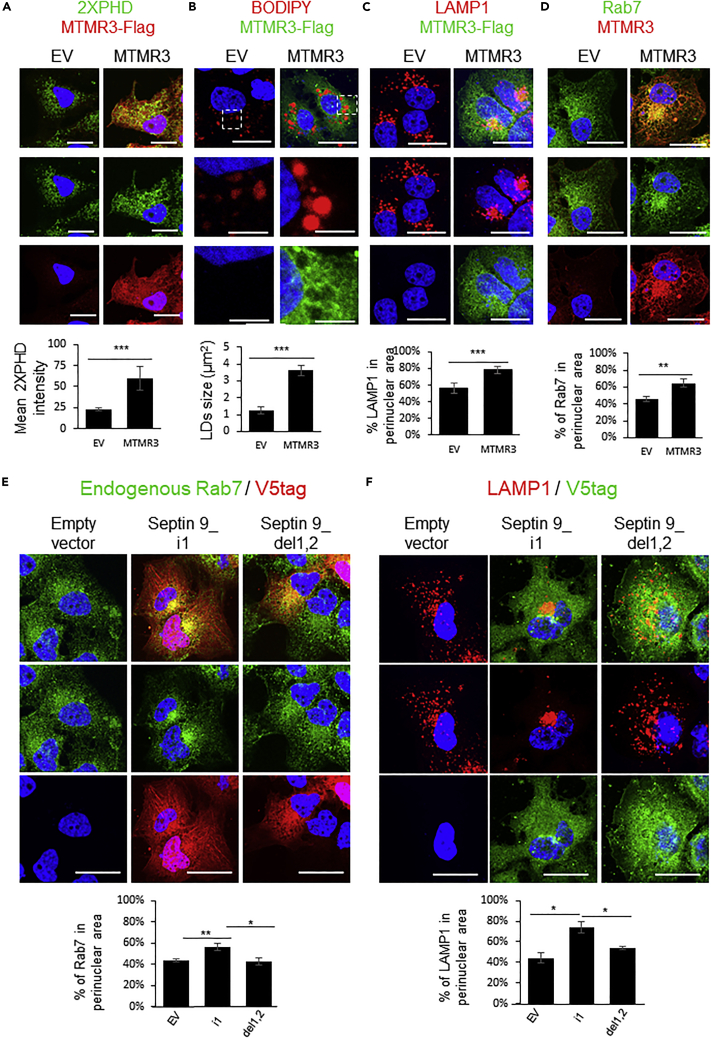
To validate the effects of PtdIns(5)P and PtdIns(3,5)P2, we performed experiments using MTMR3 (Myotubularin Related Protein 3), which is a phosphatase that converts the PtdIns(3,5)P2 into PtdIns(5)P. As expected, overexpression of MTMR3 increased PtdIns(5), which was visualized in the cytoplasm using the 2XPHD probe (Figure 8A). Furthermore, overexpression of MTMR3 increased LD size and also promoted LAMP1 and endogenous Rab7 perinuclear clustering (Figures 8A–8D). The effects PtdIns(3,5)P2 and PtdIns(5)P on MTs and septin 9 co-localization were also evaluated (Figure S11A). Data showed that MTs and septin 9 are enriched and colocalized at MTOC in cells treated with PtdIns(5)P, and not in those treated with PtdIns(3,5)P2 (Figure S11A). Consistent with these data, MTMR3 overexpression enhanced both septin 9 and MTs at MTOC (Figures S11B and S11C).
Figure 8.
Overexpression of MTMR3 recapitulates the cellular effects of PtdIns5P
(A) Huh7 cells were transfected with EV or MTMR3 and were grown for 48 h. Then, cells were stained for 2XPHD (green)/Flag (red). Bar graphs show the mean intensity of 2XPHD from two independent experiments performed as described. Scale bar, 20μm.
(B) Huh7 cells were transfected with EV or MTMR3 and were grown for 48 h. Then, cells were stained for BODIPY (red)/Flag (green). Bar graphs show the average size of LDs from two independent experiments performed as described. Scale bar, 20μm. The dot squares indicate the zoom area. Scale bar, 5μm.
(C) Huh7 cells were transfected with EV or MTMR3 and were grown for 48h. Then, cells were stained for LAMP1 (red)/Flag (green). Bar graphs show the percentage of LAMP1 in perinuclear area from two independent experiments performed as described. Scale bar, 20μm.
(D) Huh7 cells were transfected with EV or MTMR3 and were grown for 48 h. Then, cells were stained for Rab7 (green)/MTMR3 (red). Bar graphs show the percentage of Rab7 in perinuclear area from 2 independent experiments performed as described. Scale bar, 20μm.
(E) Huh7 cells were transfected with EV, septin 9_i1 or Septin 9_del1,2 for 48 h and then stained for Rab7 (green) and V5-tag (red). Bar graphs show the percentage of Rab7 in the perinuclear area from three independent experiments performed as described.
(F) Huh7 cells were transfected with EV, septin 9_i1 or Septin 9_ del1,2 for 48 h and then stained for LAMP1 (red) and V5-tag (green). Bar graphs show the percentage of LAMP1 in the perinuclear area from three independent experiments performed as described. Data information: Bar graphs present Mean ± SEM. Student’s t test was used. ∗p < 0.05, ∗∗p < 0.001, ∗∗∗p < 0.0001.
We reported that septin 9 binds PIs via two polybasic domains (PB1 and PB2s) and deletion of these domains impaired septin 9 filaments formation and their interaction with MTs (Omrane et al., 2019). Thus, we wondered if those PI-binding domains are important for septin 9 induced-lysosome clustering. Therefore, we transfected cells with septin 9_i1 and the septin 9_I1 mutant deleted from its two polybasic domains (septin 9_del1, 2). Expression of septin 9_i1 promoted Rab7 and LAMP1 clustering in the perinuclear area compared to cells transfected with the empty vector (Figures 8E and 8F). By contrast, septin 9_del1,2 expression blocked the clustering of both Rab7 and LAMP1, which are dispersed throughout the cytoplasm (Figures 8E and 8F). Together, these results indicated that the presence of specific PIs and their interaction with septin 9 through its PI-binding domains are crucial for the intracellular distribution of LDs and lysosomes which association is required for LD degradation.
Discussion
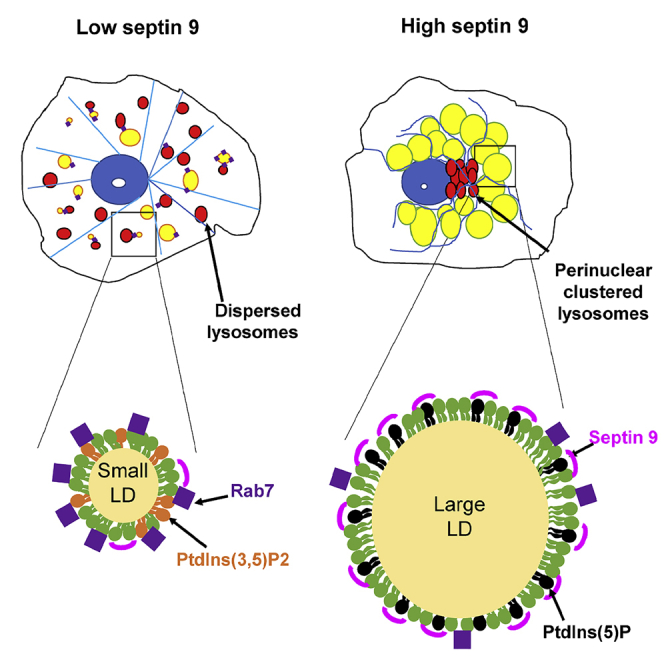
The lipids stored in LDs within cells are controlled by two major metabolic pathways that involved the synthesis of lipids to increase the lipid content of LDs and the catabolism pathway that results in a reduction in the lipid content through β-oxidation, lipolysis by lipases and lipophagy. The latter is dependent on lysosomes and the export of lipids via secretory pathways (Ballabio and Bonifacino, 2019; Olzmann and Carvalho, 2019). We had already provided evidence regarding the role of septin 9 in the growth and perinuclear accumulation of LDs (Akil et al., 2016). In the present study, we showed that septin 9 impaired LD catabolism, and particularly we demonstrated that septin 9 regulated the intracellular localization of lysosomes and the interaction of lysosomal protein Rab7 with LDs. We also showed that there is a shift between septin 9 or Rab7 localization with LDs. Their specific localization on LDs is controlled by PIs and influences the LD behaviors (Figure 8). Indeed septin 9 was associated with large LDs in cells with clustered lysosomes, whereas rab7 is found with small LDs that co-localized with dispersed lysosomes.
The perinuclear clustering and growth of LDs often occur simultaneously. For example, in adipocytes, LDs arise from the peripheral lamellipodia and grow as they move toward the cell center (Nagayama et al., 2007). Conversely, when lipolysis is chronically stimulated in adipocytes, their large perinuclear LDs fragment into small droplets dispersed throughout the cell (Marcinkiewicz et al., 2006). The dispersed to clustered redistribution of LDs may also result from infection by diverse intracellular pathogens (Cocchiaro et al., 2008; Toledo et al., 2016) such as hepatitis C virus (HCV) (Akil et al., 2016). Furthermore, it has been reported that these changes in the cellular distribution of LDs also contributed to their interactions with other organelles including the endoplasmic reticulum, mitochondria, peroxisomes, and autophagic lysosomes (Herms et al., 2015; Jin et al., 2021). A recent study demonstrated that interfering with the perinuclear accumulation of lysosomes could impair LD turnover (Tapia et al., 2019). When we increased oleate concentration or overexpressed septin 9, both LDs and lysosomes were clustered in the perinuclear region, where they appeared to occupy distinct regions. The clustered lysosomes were co-localized with the Golgi apparatus, which occupied the core region, whereas the LDs seem to be tightly packed around this core region (Figures 1 and 4). The Rab7 protein, which is a lysosomal component also found in LDs plays an important role in initiating lysosome-dependent lipophagy. Under the conditions such as 72h after treatment of cells with oleate 100μM and knockdown of septin 9, the LDs and lysosomes behaved more dispersed in the cytoplasm; besides, we observed more co-localization between Rab7 and LDs (Figure 6), which could further permit LD degradation. Conversely, we observed that increased oleate concentration or overexpression of septin 9 promoted Rab7 clustered in the core region with Golgi and then the contact between Rab7 and LDs was reduced. Although our study showed that Rab7 accumulated on the surface of LDs, the data did not allow us to determine whether this is because of a redistribution of Rab7 from lysosomes to LDs or if this rather indicates closeness, proximity of LDs and lysosomes. Nevertheless, we speculated that the interaction of Rab7 with LDs for their degradation was impaired by septin 9 expression. In addition, a high level of septin 9 was found associated with the large clusters of LDs surrounding the core region formed with lysosomal proteins and Golgi. Interestingly, a recent study showed that in the liver, the process involved in LD degradation is dependent on LD size, and indeed, large LDs were not preferentially degraded through lysosomes and remained the targets of cytoplasmic lipases (Schott et al., 2019). Thus, in line with our data, we could speculate that large perinuclear clustered LDs, which have more association with septin 9, have less chance to contact lysosomes and therefore could not be degraded via the lipophagy pathway. In addition, we also observed more LC3B accumulated in oleate treated cells than septin 9 knockdown cells (Figure S9). Those results, at least, confirmed that high septin 9 inhibits autophagy.
We also demonstrated that MTs played a crucial role in lysosome perinuclear clustering and its association with Golgi (Figure 4). Furthermore, we showed that perinuclear LDs are surrounded by organized MT filaments and MT polymerization is enhanced particularly in the MTOC where LAMP is clustered (Figure 5). The strong MT network might form a barrier to block contacts between LDs and lysosomes and thus impair LD degradation and promote their perinuclear growth. Septin 9 controls MTs and therefore an increase of septin 9 could also contribute to lysosome clustering by affecting the MTs dynamics as reported recently (Kesisova et al., 2020). Furthermore, the composition of the monolayer of phospholipids surrounding the core of LDs plays an important role in the regulation of the interaction of LDs with proteins or other organelles (Thiam and Dugail, 2019). Indeed, we have shown that PtdIns5P was able to promote the interaction between LD and septin 9 (Akil et al., 2016). Here, our results revealed that the addition of PtdIns5P on Huh7 cells promoted the perinuclear clustering of Rab7 and reduced the Rab7/LD interactions. On the contrary, PtdIns(3,5)P2 which is enriched in lysosomes promoted the dispersion of LDs through the cytoplasm and increased their interaction with Rab7. In addition, MTMR3 which converts PtdIns(3,5)P2 to PtdIns5P recapitulates the effects of PtdIns5P on the perinuclear distribution of both LDs and lysosomes. These data strongly highlighted the critical role of PI metabolism in the regulation of LD behavior and septin 9 functions.
LDs are dynamic organelles that are essential for cell homeostasis and participate in the processes that maintain cell structure and energy homeostasis. To perform their functions, the interactions of LDs with multiple organelles such as the ER Golgi, mitochondria, peroxisomes, and lysosomes are required. Therefore, to fully understand the biological function of LDs and their alterations during the pathogenesis of LD-associated diseases, their impacts on other organelles needed detailed analysis. Our results have uncovered the role of septin 9 and PIs in LDs behavior and their interaction with lysosomes. To conclude, we have identified molecular machinery that links lipid metabolism to organelle homeostasis. Indeed, understanding how LDs and lysosomes are coordinated both spatially and temporally might help in gaining a clearer understanding of the pathogenesis of LD-associated diseases such as steatosis, cirrhosis, or even cancer.
Limitations of the study
Although our study reveals that septin 9 and phosphoinositides regulate lysosome localization and their association with lipid droplets, there are still questions which need to be addressed. For example, septin 9 activities are often associated with its oligomerization with other septins. Therefore, we could ask about the potential contribution of other members of the septin family in the regulation of lysosome localization.
In this study, we showed the contribution of septin 9 in the regulation of microtubules is involved in both LDs and lysosomes localizations. However, further studies are needed to determine whether septin 9 could also directly bind to lysosome or if other adaptor proteins are required.
The roles of septin 9 and phosphoinositides in the regulation of lysosome and LDs were demonstrated by in vitro studies. Future research based on in vivo studies will help to understand the relevance of our study in pathological conditions such as NAFLD, NASH, and tumorigenesis.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| LAMP1 | Abcam | Cat# ab25630, RRID: AB_470708 |
| TGN46 | Novus | Cat# NBP1-49643, RRID: AB_10011762 |
| alpha Tubulin | Abcama | Cat# ab15246, RRID: AB_301787 |
| β-Tubulin | Sigma-Aldrich | Cat# T4026, RRID: AB_477577 |
| MSF | Abcam | Cat# ab114099, RRID: AB_10863530 |
| Rab7 | Cell Signaling Technology | Cat# 9367, RRID: AB_1904103 |
| GM130 | BD Biosciences | Cat# 610822, RRID: AB_398141 |
| p62/SQSTM1 | Sigma-Aldrich | Cat# P0068, RRID: AB_1841066 |
| LC3B | Sigma-Aldrich | Cat# L7543, RRID: AB_796155 |
| V5 tag | Abcam | Cat# ab9116, RRID: AB_307024 |
| V5 tag | Abcam | Cat# ab27671, RRID: AB_471093 |
| FLAG | Sigma-Aldrich | Cat# F7425, RRID: AB_439687 |
| FLAG | Sigma-Aldrich | Cat# F3165, RRID: AB_259529 |
| MTMR3 | Santa Cruz Biotechnology | sc-398353 |
| Mouse IgG HRP-linked whole Ab | Sigma-Aldrich | NA931V |
| Rabbit IgG HRP Linked Whole Ab | Sigma-Aldrich | NA934V |
| Alexa Fluor® 488 | Thermo Fisher Scientific | Cat# A-11001, RRID: AB_2534069 |
| Alexa Fluor® 488 | Thermo Fisher Scientific | Cat# A-11008, RRID: AB_143165 |
| Alexa Fluor® 546 | Thermo Fisher Scientific | Cat# A10040, RRID: AB_2534016 |
| Alexa Fluor® 546 | Thermo Fisher Scientific | Cat# A-11003, RRID: AB_2534071 |
| Streptavidin, Alexa Fluor™ 488 Conjugate | Thermo Fisher Scientific | S322354 |
| Chemicals, peptides, and recombinant proteins | ||
| LysoTracker™ Red DND-99 | Thermo Fisher | L7528 |
| BODIPY™ 493/503 | Thermo Fisher | D3922 |
| Sodium oleate | Sigma-Aldrich | O7501 |
| Bafilomycin A1 Ready Made | Sigma-Aldrich | SML1661-1ML |
| Albumin, Bovine Serum, Fraction V, Fatty Acid-Free | Sigma-Aldrich | 126575 |
| Nocodazole | Sigma-Aldrich | 487928 |
| Paclitaxel | Sigma-Aldrich | T7191 |
| BODIPY™ 576/589 | Thermo Fisher | D2225 |
| PI(5)P diC8 | Echelon | P-5008 |
| PI(3,5)P2 | Echelon | P-3508 |
| Hoechst 33342 | Thermo Fisher | H3570 |
| Critical commercial assays | ||
| RNeasy Mini Kit | QIAGEN | 74104 |
| RevertAid First Strand cDNA Synthesis Kit | Thermo Fisher | K1621 |
| X-tremeGENE™ 9 DNA Transfection Reagent | Roche | 6365779001 |
| X-tremeGENE™ siRNA Transfection Reagent | Roche | 4476093001 |
| QuantiTect SYBR® Green PCR Kits | QIAGEN | 204141 |
| Experimental Models: Cell Lines | ||
| Huh7 | ATCC | NA |
| Oligonucleotides | ||
| LIPA Forward CTGACAAAGGTCCCAAACCAG LIPA Reverse GCAAGAATGAAGCCCAGGC |
This paper | Invitrogen |
| GNS Forward CCCATTTTGAGAGGTGCCAGT GNS Reverse TGACGTTACGGCCTTCTCCTT |
This paper | Invitrogen |
| GBA Forward GGGCAAAGGTGGTACTGACAG GBA Reverse GCATGGTGTTGGGGAACAG |
This paper | Invitrogen |
| MCOLN1 Forward TTGCTCTCTGCCAGCGGTACTA MCOLN1 Reverse GCAGTCAGTAACCACCATCGGA |
This paper | Invitrogen |
| TMEM55B Forward GTTCGATGCCCCTGTAACTGTC TMEM55B Reverse CCCAGGTTGATGATTCTTTTGC |
This paper | Invitrogen |
| ATP6V1E1 Forward AAAGGTCGGCTTGTGCAAACCC ATP6V1E1 Reverse GGTCATCTCTTGCTCTGAGGAC |
This paper | Invitrogen |
| GAPDH Forward CCACATCGCTCAGACACCAT GAPDH Reverse CCAGGCGCCCAATACG |
This paper | Invitrogen |
| LAMP1 Forward ACGTTACAGCGTCCAGCTCAT LAMP1 Reverse TCTTTGGAGCTCGCATTGG |
This paper | Invitrogen |
| Recombinant DNA | ||
| cDNA GFP-Rab7 | Dr Goud | Institute Curie, PSL Research University, CNRS UMR 144, Paris, France |
| cDNA MTMR3 | Dr Tronchère | INSERM U1048, I2MC and Université Paul Sabatier, 31432 Toulouse, France. |
| cDNA septin 9 isoform 1 | Dr Russell | CCRCB, Queens University Belfast |
| Software and algorithms | ||
| G:BOX Chemi | SYNGENE | https://www.syngene.com/software/ |
| LAS X | Leica | https://www.leica-microsystems.com/products/microscope-software/ |
| LightCycler® 96 SW 1.1 | Roche | https://lifescience.roche.com/en_fr/products/lightcycler-software-4-1.html |
| FIJI | ImageJ | https://imagej.net/software/fiji/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ama Gassama-Diagne (ama.gassama@inserm.fr).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Cell line and culture conditions
Human hepatocarcinoma cells Huh7 was used. Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) containing 1 g/L glucose and supplemented with 10% heat-inactivated fetal bovine serum, 1% nonessential amino acids (GibcoBRL) and 1% penicillin/streptomycin (GibcoBRL). Cells were tested for mycoplasma contamination weekly.
Method details
Cell treatments
Cell treatment with oleate: complexes of sodium oleate with BSA were prepared as previously reported. Briefly, the solution containing 20 mM sodium oleate and 2.4 mM bovine serum albumin-fatty acid free (BSA FFA) was heated to 55°C. The obtained complex was diluted in a pre-warmed culture medium at indicated concentration before addition to the cells. The treated cells were maintained in culture and collected either for immunoblot or immunofluorescence experiments depends on the treated time.
Cells treatment with PIs: PIs as a lyophilized powder were solubilized at 500 mM in water by vortex. The solution was then diluted in Dulbecco’s phosphate-buffered saline (DPBS) to the concentration indicated in the legends of the experiment. Cells were washed with DPBS and the PIs solution was added to the cell for indicated time.
Cell treatment with Bafilomycin A1: The Bafilomycin A1 solution was diluted in cell culture medium to the concentration indicated in the legends of the experiment. Cells were washed with fresh medium twice and the Bafilomycin A1 containing medium was added to the cell then cells were cultured for indicated time. After treatment, the cells were collected either for immunoblot or fixed for immunofluorescence experiments following the methods below.
Reverse transcription and real-time PCR analysis
Total RNA was isolated using RNeasy Mini Kit 50 (Cat# 74104 QIAGEN) and applied to reverse transcription using RevertAid First Strand cDNA Synthesis Kit (Cat#K1612 Thermo Scientific). The cDNA was analyzed by qPCR using QuantiTect SYBR Green PCR Kit (Cat#204143 QIAGEN) and a Light Cycler 480 Real-Time PCR System (Roche). Reaction parameters were Preincubation 10 min at 95°C, followed by 45 cycles of 15 s at 95°C, 30 s at 55°C and 15 s at 72°C. The triplicate mean values were calculated using GAPDH gene transcription as reference for normalization. Used RT–PCR primers sequences are presented in the Table 4.
Cell transfection
The transfection of cDNA and siRNA were performed using X-tremeGENE 9 DNA Transfection Reagent (Roche Diagnostics) following the manufacturer’s protocol. The transfection was performed for 24 h unless indicated otherwise in the text.
Immunofluorescence
Cells were grown on coverslips, fixed with paraformaldehyde 4% for 20 min and permeabilized for 30 min at 37°C using the permeabilizing buffer (PFS): DPBS containing saponin (Cat#10294440 Fisher scientific) 0.025% m.v-1, gelatin from cold water fish skin (Cat#G7041 Sigma 0.7% m.v-1). Then cells were incubated with primary antibody for 2 h washed three times for 5min with PFS and incubated with the appropriate secondary antibodies or with the dye for 90 min. The coverslips were mounted using Prolong Gold (Cat#P36934 Invitrogen). For Lysotracker staining, the cells on coverslips were treated, then, cells were cultured in the same medium contains 75 nM Lysotracker for 1h, after incubation, cells were fixed and performed as described above.
Immunoblot
Cells were washed with ice-cold DPBS and lysed on ice using the following buffer: 20mM Tris, HCl, 100mM NaCl, 1% Triton X-100 at PH 7.4 containing protease inhibitors (cOmplete ULTRA Cat#05892970001 Roche). The proteins were separated on SDS–PAGE and electro-transferred onto nitrocellulose membrane. After transfer, the membrane was saturated in DPBS containing 0.1% Tween 20 and 5% milk. Primary antibodies were added overnight at 4C or for 2 h at room temperature depending on the antibody. The membranes were washed with DPBS and incubated for 1 h at room temperature with appropriate secondary antibody coupled with peroxidase. ECL plus kit (Cat#32132 Thermo Scientific) was used for protein detection. Chemiluminescent signal was detected by G:BOX Chemi Fluorescent & Chemiluminescent Imaging System from SYNGENE. Blot quantification was done using ImageJ software. The uncropped scans are supplied in Figure S12.
Tubulin polymerization assay
Fraction of soluble and polymerized tubulin were performed as described before. Briefly, soluble tubulin extraction buffer A (1mM MgCl2, 2 mM EGTA, 0.5% NP40, 2 mM PMSF, 20 mM Tris-HCl [pH = 6.8], protease inhibitor cocktail) was added to cells at 4°C for 5 min, plates were gently swirled two to three times, and buffer was removed and saved as soluble fraction. Immediately after, polymerized tubulin extraction buffer B (A + 1% SDS) was added for 2 min, cells were scraped, and the polymerized fraction was sonicated briefly and incubated on ice for 30 min. Soluble and polymerized fractions were loaded on a gel and transferred to nitrocellulose membranes to probe with specific antibody as indicated. Cells treated with nocodazole or Taxol alone served as control for efficient extraction of soluble and polymerized tubulin. ImageJ was used to quantify the intensity of beta-tubulin bands from each blot. Further, the percentage of polymerized tubulin was determined by dividing the densitometry value of polymerized tubulin by the total tubulin content (the sum of the densitometry values of soluble and polymerized tubulin).
Images acquisition and analysis
Images were acquired with a Leica TCS SP5 AOBS tandem confocal microscope. For co-localization analysis, images were treated by ImageJ software, the plugin ‘Intensity Correlation Analysis’ was used to generate the Pearson’s correlation coefficient (Rr) which range from −1 (perfect exclusion) to +1 (perfect correlation).
To calculate the total intensity of LAMP1 and LDs, images obtained by confocal microscopy were processed cell ROI by free hand selection tool according to thresholding image, then total intensity of LAMP1 and LDs were measured by ImageJ analyses. The line plot for LAMP1 and BODIPY was generated by Fiji software function ‘Plot profile’.
To determine LAMP1, LDs and Rab7 distribution in cells, the plugin ‘Radial profile Angle’ was used. For analysis, the individual cell total area was generated by free hand selection tool according the thresholding image, then a circle which center is located in the center of the cell nucleus was defined at the periphery of each cell. Then, the plugin produces a profile plot of normalized integrated intensities around concentric circles as a function of distance from a point in the image, where considered as the center of the cell. The concentric circles were assembled in three circle bands, the first corresponding to the area of the nuclei and the rest corresponding to the cytoplasm was divided in two equal bands (the band near the nuclei is considered as the ‘perinuclear’ and the other the ‘periphery’. The intensity in each band was calculated from the total integrated intensities around concentric circles present in the band. A diagram of this analysis was present in Figure S13A.
To determine the number of p62 or LC3B positive puncta in cells, the Fiji function ‘Analyze Particles’ was used. For analysis, the individual cell was generated by free hand selection tool according the thresholding image. After, the ‘Threshold’ function with Default mask was applied on the target channel, then the function ‘Analyze Particles’ was used with ‘Size (microˆ2)’ parameter setting as 0.50-Infinity. Then, the mean count of P62 or LC3B positive puncta in cells was analyzed.
To measure the mean intensity of Rab7 or septin 9 associated with LD, the Fiji function ‘Analyze particles’ were performed on the LDs image to obtain the areas occupied by LDs and marked as area1, then enlarge those areas by 0.5μm, and marked this new area as area 2. Apply those two areas on the target signal image and analyze the total area value and intensity of the target signal. Then calculate. A diagram of this analysis was present in Figure S13B.
Quantification and statistical analysis
Unpaired Student’s t-tests were used, and statistical significance was determined at ∗p < 0.05; ∗∗p < 0.001, ∗∗∗p < 0.0001.
Acknowledgments
We thank B. Goud and S.Miserey-Lenkei (CNRS U144, Institut Curie, Paris) for advice on intracellular trafficking and providing the GFP-Rab7 reagents. We thank H.Tronchère (INSERM U U1048, I2MC, and Université Paul Sabatier) for providing the cDNAs of MTMR3, 2xPDH, and advices on PtdIns(5)P. P.X.S. was supported by a scholarship from the Chinese Scholarship Council. This work was supported by Inserm and University of Paris-Saclay.
Author contributions
P.X.S. contributed to the design and performed most of the experiments, analyzed the data, and wrote the manuscript. J.P. and M.O. analyzed and discussed the data. D.S discussed the data. A.G-D. conceived and supervised the project, designed the experiments, and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: May 20, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104288.
Supplemental information
Data and code availability
-
•
All data are included in the published article and the supplemental information files or are available from the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Akil A., Peng J., Omrane M., Gondeau C., Desterke C., Marin M., Tronchère H., Taveneau C., Sar S., Briolotti P., et al. Septin 9 induces lipid droplets growth by a phosphatidylinositol-5-phosphate and microtubule-dependent mechanism hijacked by HCV. Nat. Commun. 2016;7 doi: 10.1038/ncomms12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelis D., Spiliotis E.T. Septin mutations in human cancers. Front. Cell Dev. Biol. 2016;4:122. doi: 10.3389/fcell.2016.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X., Bowen J.R., Knox T.K., Zhou K., Pendziwiat M., Kuhlenbäumer G., Sindelar C.V., Spiliotis E.T. Novel septin 9 repeat motifs altered in neuralgic amyotrophy bind and bundle microtubules. J. Cell Biol. 2013;203:895–905. doi: 10.1083/jcb.201308068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballabio A., Bonifacino J.S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 2019:1–18. doi: 10.1038/s41580-019-0185-4. [DOI] [PubMed] [Google Scholar]
- Barbosa A.D., Savage D.B., Siniossoglou S. Lipid droplet-organelle interactions: emerging roles in lipid metabolism. Curr. Opin. Cell Biol. 2015;35:91–97. doi: 10.1016/j.ceb.2015.04.017. [DOI] [PubMed] [Google Scholar]
- Barbosa A.D., Siniossoglou S. Function of lipid droplet-organelle interactions in lipid homeostasis. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:1459–1468. doi: 10.1016/j.bbamcr.2017.04.001. [DOI] [PubMed] [Google Scholar]
- Bechmann L.P., Hannivoort R.A., Gerken G., Hotamisligil G.S., Trauner M., Canbay A. The interaction of hepatic lipid and glucose metabolism in liver diseases. J. Hepatol. 2012;56:952–964. doi: 10.1016/j.jhep.2011.08.025. [DOI] [PubMed] [Google Scholar]
- Benedict M., Zhang X. Non-alcoholic fatty liver disease: an expanded review. World J. Hepatol. 2017;9:715–732. doi: 10.4254/wjh.v9.i16.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersuker K., Olzmann J.A. Establishing the lipid droplet proteome: mechanisms of lipid droplet protein targeting and degradation. Biochim. Biophys. Acta. 2017;1862:1166–1177. doi: 10.1016/j.bbalip.2017.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessone F., Razori M.V., Roma M.G. Molecular pathways of nonalcoholic fatty liver disease development and progression. Cell. Mol. Life Sci. 2019;76:99–128. doi: 10.1007/s00018-018-2947-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blümer J., Rey J., Dehmelt L., Mazel T., Wu Y.-W., Bastiaens P., Goody R.S., Itzen A. RabGEFs are a major determinant for specific Rab membrane targeting. J. Cell Biol. 2013;200:287–300. doi: 10.1083/jcb.201209113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges A.A., Jentzsch M.S., Oakes P.W., Occhipinti P., Gladfelter A.S. Micron-scale plasma membrane curvature is recognized by the septin cytoskeleton. J. Cell Biol. 2016;213:23–32. doi: 10.1083/jcb.201512029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci C., Thomsen P., Nicoziani P., McCarthy J., van Deurs B. Rab7: a key to lysosome biogenesis. Mol. Biol. Cell. 2000;11:467–480. doi: 10.1091/mbc.11.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabukusta B., Neefjes J. Mechanisms of lysosomal positioning and movement. Traffic. 2018 doi: 10.1111/tra.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan N., Visram M., Cristobal-Sarramian A., Sarkleti F., Kohlwein S.D. Morphogenesis checkpoint kinase Swe1 is the executor of lipolysis-dependent cell-cycle progression. Proc. Natl. Acad. Sci. U S A. 2015;112:E1077–E1085. doi: 10.1073/pnas.1423175112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu B.-B., Liao Y.-C., Qi W., Xie C., Du X., Wang J., Yang H., Miao H.-H., Li B.-L., Song B.-L. Cholesterol transport through lysosome-peroxisome membrane contacts. Cell. 2015;161:291–306. doi: 10.1016/j.cell.2015.02.019. [DOI] [PubMed] [Google Scholar]
- Cocchiaro J.L., Kumar Y., Fischer E.R., Hackstadt T., Valdivia R.H. Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc. Natl. Acad. Sci. U S A. 2008;105:9379–9384. doi: 10.1073/pnas.0712241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolat L., Spiliotis E.T. Septins promote macropinosome maturation and traffic to the lysosome by facilitating membrane fusion. J. Cell Biol. 2016;214:517–527. doi: 10.1083/jcb.201603030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung K.Y.Y., Dai L., Trimble W.S. In: International Review of Cell and Molecular Biology. Jeon K.W., editor. Academic Press; 2014. Chapter seven - cell and molecular biology of septins; pp. 289–339. [DOI] [PubMed] [Google Scholar]
- Gassama-Diagne A., Yu W., Beest M.T., Martin-Belmonte F., Kierbel A., Engel J., Mostov K. Phosphatidylinositol-3,4,5-trisphosphate regulates the formation of the basolateral plasma membrane in epithelial cells. Nat. Cell Biol. 2006;8:963–970. doi: 10.1038/ncb1461. [DOI] [PubMed] [Google Scholar]
- Haemmerle G., Moustafa T., Woelkart G., Büttner S., Schmidt A., van de Weijer T., Hesselink M., Jaeger D., Kienesberger P.C., Zierler K., et al. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-α and PGC-1. Nat. Med. 2011;17:1076–1085. doi: 10.1038/nm.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henzi T., Lannes N., Filgueira L. Septins in infections: focus on viruses. Pathogens. 2021;10 doi: 10.3390/pathogens10030278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herms A., Bosch M., Reddy B.J.N., Schieber N.L., Fajardo A., Rupérez C., Fernández-Vidal A., Ferguson C., Rentero C., Tebar F., et al. AMPK activation promotes lipid droplet dispersion on detyrosinated microtubules to increase mitochondrial fatty acid oxidation. Nat. Commun. 2015;6:1–14. doi: 10.1038/ncomms8176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inami Y., Yamashina S., Izumi K., Ueno T., Tanida I., Ikejima K., Watanabe S. Hepatic steatosis inhibits autophagic proteolysis via impairment of autophagosomal acidification and cathepsin expression. Biochem. Biophys. Res. Commun. 2011;412:618–625. doi: 10.1016/j.bbrc.2011.08.012. [DOI] [PubMed] [Google Scholar]
- Jean S., Kiger A.A. Coordination between RAB GTPase and phosphoinositide regulation and functions. Nat. Rev. Mol. Cell Biol. 2012;13:463–470. doi: 10.1038/nrm3379. [DOI] [PubMed] [Google Scholar]
- Jin Y., Ren Z., Tan Y., Zhao P., Wu J. Motility plays an important role in the lifetime of mammalian lipid droplets. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22083802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongsma M.L.M., Berlin I., Wijdeven R.H.M., Janssen L., Janssen G.M.C., Garstka M.A., Janssen H., Mensink M., van Veelen P.A., Spaapen R.M., Neefjes J. An ER-associated pathway defines endosomal architecture for controlled cargo transport. Cell. 2016;166:152–166. doi: 10.1016/j.cell.2016.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo E., Surka M.C., Trimble W.S. Mammalian SEPT2 is required for scaffolding nonmuscle myosin II and its kinases. Dev. Cell. 2007;13:677–690. doi: 10.1016/j.devcel.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Kesisova I.A., Robinson B.P., Spiliotis E.T. A septin GTPase scaffold of dynein-dynactin motors triggers retrograde lysosome transport. bioRxiv. 2020 doi: 10.1101/2020.06.01.128488. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.S., Froese C.D., Estey M.P., Trimble W.S. SEPT9 occupies the terminal positions in septin octamers and mediates polymerization-dependent functions in abscission. J. Cell Biol. 2011;195:815–826. doi: 10.1083/jcb.201106131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M., Field C.M., Coughlin M.L., Straight A.F., Mitchison T.J. Self- and actin-templated assembly of mammalian septins. Dev. Cell. 2002;3:791–802. doi: 10.1016/S1534-5807(02)00366-0. [DOI] [PubMed] [Google Scholar]
- Koo S.-H. Nonalcoholic fatty liver disease: molecular mechanisms for the hepatic steatosis. Clin. Mol. Hepatol. 2013;19:210–215. doi: 10.3350/cmh.2013.19.3.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer N., Hilger M., Kory N., Wilfling F., Stoehr G., Mann M., Farese R.V., Walther T.C. Protein correlation profiles identify lipid droplet proteins with high confidence. Mol. Cell Proteomics. 2013;12:1115–1126. doi: 10.1074/mcp.M112.020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokowski S., Lobato-Márquez D., Chastanet A., Pereira P.M., Angelis D., Galea D., Larrouy-Maumus G., Henriques R., Spiliotis E.T., Carballido-López R., Mostowy S. Septins recognize and entrap dividing bacterial cells for delivery to lysosomes. Cell Host Microbe. 2018;24:866–874.e4. doi: 10.1016/j.chom.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurat C.F., Wolinski H., Petschnigg J., Kaluarachchi S., Andrews B., Natter K., Kohlwein S.D. Cdk1/Cdc28-dependent activation of the major triacylglycerol lipase Tgl4 in yeast links lipolysis to cell-cycle progression. Mol. Cell. 2009;33:53–63. doi: 10.1016/j.molcel.2008.12.019. [DOI] [PubMed] [Google Scholar]
- Kwanten W.J., Martinet W., Michielsen P.P., Francque S.M. Role of autophagy in the pathophysiology of nonalcoholic fatty liver disease: a controversial issue. World J. Gastroenterol. 2014;20:7325–7338. doi: 10.3748/wjg.v20.i23.7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcinkiewicz A., Gauthier D., Garcia A., Brasaemle D.L. The phosphorylation of serine 492 of perilipin A directs lipid droplet fragmentation and dispersion. J. Biol. Chem. 2006;281:11901–11909. doi: 10.1074/jbc.M600171200. [DOI] [PubMed] [Google Scholar]
- Michelotti G.A., Machado M.V., Diehl A.M. NAFLD, NASH and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 2013;10:656–665. doi: 10.1038/nrgastro.2013.183. [DOI] [PubMed] [Google Scholar]
- Miyagawa K., Oe S., Honma Y., Izumi H., Baba R., Harada M. Lipid-induced endoplasmic reticulum stress impairs selective autophagy at the step of autophagosome-lysosome fusion in hepatocytes. Am. J. Pathol. 2016;186:1861–1873. doi: 10.1016/j.ajpath.2016.03.003. [DOI] [PubMed] [Google Scholar]
- Mostowy S., Cossart P. Septins: the fourth component of the cytoskeleton. Nat. Rev. Mol. Cell Biol. 2012;13:183–194. doi: 10.1038/nrm3284. [DOI] [PubMed] [Google Scholar]
- Nagayama M., Uchida T., Gohara K. Temporal and spatial variations of lipid droplets during adipocyte division and differentiation. J. Lipid Res. 2007;48:9–18. doi: 10.1194/jlr.M600155-JLR200. [DOI] [PubMed] [Google Scholar]
- Nishihama R., Onishi M., Pringle J.R. New insights into the phylogenetic distribution and evolutionary origins of the septins. Biol. Chem. 2011;392:681–687. doi: 10.1515/BC.2011.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann J.A., Carvalho P. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 2019;20:137–155. doi: 10.1038/s41580-018-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omrane M., Camara A.S., Taveneau C., Benzoubir N., Tubiana T., Yu J., Guérois R., Samuel D., Goud B., Poüs C., et al. Septin 9 has two polybasic domains critical to septin filament assembly and Golgi integrity. iScience. 2019;13:138–153. doi: 10.1016/j.isci.2019.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliuso A., Tham T.N., Stevens J.K., Lagache T., Persson R., Salles A., Olivo-Marin J.-C., Oddos S., Spang A., Cossart P., Stavru F. A role for septin 2 in Drp1-mediated mitochondrial fission. EMBO Rep. 2016;17:858–873. doi: 10.15252/embr.201541612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera R.M., Zoncu R. The lysosome as a regulatory hub. Annu. Rev. Cell Dev. Biol. 2016;32:223–253. doi: 10.1146/annurev-cellbio-111315-125125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiborg C., Wenzel E.M., Pedersen N.M., Olsvik H., Schink K.O., Schultz S.W., Vietri M., Nisi V., Bucci C., Brech A., et al. Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature. 2015;520:234–238. doi: 10.1038/nature14359. [DOI] [PubMed] [Google Scholar]
- Rambold A.S., Cohen S., Lippincott-Schwartz J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell. 2015;32:678–692. doi: 10.1016/j.devcel.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott M.B., Weller S.G., Schulze R.J., Krueger E.W., Drizyte-Miller K., Casey C.A., McNiven M.A. Lipid droplet size directs lipolysis and lipophagy catabolism in hepatocytes. J. Cell Biol. 2019;218:3320–3335. doi: 10.1083/jcb.201803153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder B., Schulze R.J., Weller S.G., Sletten A.C., Casey C.A., McNiven M.A. The small GTPase Rab7 as a central regulator of hepatocellular lipophagy. Hepatology. 2015;61:1896–1907. doi: 10.1002/hep.27667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M., Bohnert M. A different kind of love - lipid droplet contact sites. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2017;1862:1188–1196. doi: 10.1016/j.bbalip.2017.06.005. [DOI] [PubMed] [Google Scholar]
- Sellin M.E., Sandblad L., Stenmark S., Gullberg M. Deciphering the rules governing assembly order of mammalian septin complexes. Mol. Biol. Cell. 2011;22:3152–3164. doi: 10.1091/mbc.e11-03-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Fraldi A., Medina D.L., Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 2013;14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C., Dolat L., Angelis D., Forgacs E., Spiliotis E.T., Galkin V.E. Septin 9 exhibits polymorphic binding to F-actin and inhibits myosin and cofilin activity. J. Mol. Biol. 2015;427:3273–3284. doi: 10.1016/j.jmb.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Russo G., Krauss M. Septins as modulators of endo-lysosomal membrane traffic. Front. Cell Dev. Biol. 2016;4:124. doi: 10.3389/fcell.2016.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Takiguchi Y., Kinoshita M., Takiguchi K. Septin-mediated uniform bracing of phospholipid membranes. Curr. Biol. 2009;19:140–145. doi: 10.1016/j.cub.2008.12.030. [DOI] [PubMed] [Google Scholar]
- Tang T., Abbott M.J., Ahmadian M., Lopes A.B., Wang Y., Sul H.S. Desnutrin/ATGL activates PPARδ to promote mitochondrial function for insulin secretion in islet β cells. Cell Metab. 2013;18:883–895. doi: 10.1016/j.cmet.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia D., Jiménez T., Zamora C., Espinoza J., Rizzo R., González-Cárdenas A., Fuentes D., Hernández S., Cavieres V.A., Soza A., et al. KDEL receptor regulates secretion by lysosome relocation- and autophagy-dependent modulation of lipid-droplet turnover. Nat. Commun. 2019;10:735. doi: 10.1038/s41467-019-08501-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam A.R., Dugail I. Lipid droplet–membrane contact sites – from protein binding to function. J. Cell Sci. 2019;132 doi: 10.1242/jcs.230169. [DOI] [PubMed] [Google Scholar]
- Toledo D.A.M., D’Avila H., Melo R.C.N. Host lipid bodies as platforms for intracellular survival of Protozoan parasites. Front. Immunol. 2016;7 doi: 10.3389/fimmu.2016.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Hong W. Interorganellar regulation of lysosome positioning by the Golgi apparatus through Rab34 interaction with rab-interacting lysosomal protein. Mol. Biol. Cell. 2002;13:4317–4332. doi: 10.1091/mbc.E02-05-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhang X., Chu E.S.H., Chen X., Kang W., Wu F., To K.-F., Wong V.W.S., Chan H.L.Y., Chan M.T.V., et al. Defective lysosomal clearance of autophagosomes and its clinical implications in nonalcoholic steatohepatitis. FASEB J. 2018;32:37–51. doi: 10.1096/fj.201601393R. [DOI] [PubMed] [Google Scholar]
- Zhang J., Kong C., Xie H., McPherson P.S., Grinstein S., Trimble W.S. Phosphatidylinositol polyphosphate binding to the mammalian septin H5 is modulated by GTP. Curr. Biol. 1999;9:1458–1467. doi: 10.1016/S0960-9822(00)80115-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data are included in the published article and the supplemental information files or are available from the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.