Abstract
Retinal ganglion cells (RGCs) die after optic nerve trauma or in degenerative disease. However, acute changes in protein expression that may regulate RGC response to injury are not fully understood, and detailed methods to quantify new protein synthesis have not been tested. Here, we develop and apply a new in vivo quantitative measure of newly synthesized proteins to examine changes occurring in the retina after optic nerve injury. Azidohomoalanine, a noncanonical amino acid, was injected intravitreally into the eyes of rodents of either sex with or without optic nerve injury. Isotope variants of biotin-alkyne were used for quantitative BONCAT (QBONCAT) mass spectrometry, allowing identification of protein synthesis and transport rate changes in more than 1000 proteins at 1 or 5 d after optic nerve injury. In vitro screening showed several newly synthesized proteins regulate axon outgrowth in primary neurons in vitro. This novel approach to targeted quantification of newly synthesized proteins after injury uncovers a dynamic translational response within broader proteostasis regulation and enhances our understanding of the cellular response to injury.
SIGNIFICANCE STATEMENT Optic nerve injury results in death and degeneration of retinal ganglion cells and their axons. The specific cellular response to injury, including changes in new protein synthesis, is obscured by existing proteins and protein degradation. In this study, we introduce QBONCAT to isolate and quantify acute protein synthesis and subsequent transport between cellular compartments. We identify novel candidate protein effectors of the regenerative response and uncover their regulation of axon growth in vitro, validating the utility of QBONCAT for the discovery of novel regulatory and therapeutic candidates after optic nerve injury.
Keywords: neurodegeneration, optic nerve, protein synthesis, proteomics, retina
Introduction
Most adult CNS neurons, such as retinal ganglion cells (RGCs), do not survive or regenerate their axons after injury. As a result, RGC damage or degeneration is irreversible and often leads to apoptotic cell death after traumatic injury or in diseases like glaucoma that affect the optic nerve. Furthermore, although RGCs are the cells directly injured by optic nerve trauma, other retinal neurons including amacrine and bipolar cells, and non-neuronal cells including Mueller glia, microglia, and invading immune cells, all contribute to the overall acute retinal response after injury (Ramírez et al., 2010; Li et al., 2017; Barış and Tezel, 2019).
Understanding retinal response to axon trauma requires a detailed understanding of the dynamic cellular processes occurring from the time of injury, including changes in transcription and translation, which together are major contributors to net changes in protein abundance. Deep sequencing is very effective at characterizing changes in the RNA transcriptome and has been applied to identify key mediators of survival and regenerative failure after optic nerve injury (Yasuda et al., 2014; Tran et al., 2019), but the dissociation between mRNA and protein levels (Liu et al., 2016) may limit interpretation of these data. Identifying the subset of transcripts undergoing active translation using axon-TRAP-RiboTag and related methods (Shigeoka et al., 2016) has moved us closer to understanding protein expression changes after injury but still fails to directly measure changes in protein abundance. Mass spectrometry (MS) allows direct identification of the proteome but provides low quantitative sensitivity to detect small changes relative to the total protein complement.
The ability to detect changes in the proteome and thereby better detect retinal responses to injury in vivo would be greatly aided by methods to specifically detect newly synthesized proteins. This has been aided by two closely related methods using incorporation of noncanonical amino acids (ncAAs) with azide groups that facilitate labeling and enrichment—fluorescent noncanonical amino acid tagging (FUNCAT) and bio-orthogonal noncanonical amino acid tagging (BONCAT; Schiapparelli et al., 2014; Shen et al., 2014). Azide groups are commonly used for copper-catalyzed azide-alkyne cycloaddition, a set of reactions known as click chemistry (Dieterich et al., 2007; Best, 2009; Speers and Cravatt, 2009; Schiapparelli et al., 2014; Shen et al., 2014; Alvarez-Castelao et al., 2017). We have previously shown that intravitreal injection of one such azide-bearing ncAA, azidohomoalanine (AHA), which functionally approximates methionine, efficiently labels proteins in the mammalian retina and allowed identification of more than 1000 AHA-labeled proteins that were newly synthesized over a 24 h period in the retina (Schiapparelli et al., 2014).
Here, we establish a novel, quantitative BONCAT (QBONCAT), using isotopic variants of biotin in the AHA click chemistry reaction. We demonstrate the ability of QBONCAT to measure rapid and sustained protein synthesis dynamics in the retina and subsequent transport of newly synthesized proteins to the optic nerve before and after optic nerve injury. We then determine the cell-autonomous effects of manipulating these candidates through a neurite outgrowth assay, identifying several genes that significantly modulate neurite growth in vitro. Together, these data demonstrate the power of QBONCAT for identifying candidate protein regulators of the degenerative and regenerative response.
Materials and Methods
Animal surgeries
Animal experiments were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee and the Institutional Biosafety Committee of University of California, San Diego; the Scripps Research Institute; and Stanford University, and complied with the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research.
Animal lines
C57BL/6 mice or Sprague Dawley rats 30–45 d old and of either sex were used for all experiments.
Intravitreal injection
A microinjector pressure system (Picosprizer II) with a pulled glass micropipette in a micromanipulator was used to inject reagents intravitreally. For in vivo AHA experiments, we injected each eye with ∼5 μl of 400 mm AHA in PBS, as described previously (Schiapparelli et al., 2014). The injections were given twice over 24 h or once a day for 5 d under deep anesthesia with either 0.5 mg/kg Medetomidine and 75 mg/kg ketamine, i.p., or 2% isofluorane under nose cone. After injections, eyes were treated with topical antibiotics and analgesics.
Optic nerve injury and tissue collection
Under deep anesthesia, the optic nerve was exposed and crushed using fine forceps (Dumont no. 5) at 1.5 mm behind the optic nerve head for 5 s, as described previously (Wang et al., 2015; Cameron et al., 2020). Care was taken to avoid damaging the blood supply to the retina. Sham surgeries exposed the optic nerve, but no crush was given. Postoperatively, animals were allowed to recover on a heating pad and were given subcutaneous injections of buprenorphine hydrochloride, 0.1 mg/kg, twice a day for 3 consecutive days to minimize discomfort.
For tissue collection for mass spectrometry, rodents were killed with CO2 and decapitated for brain removal. The tissue was used immediately for coimmunoprecipitation studies or snap frozen in isopentane in dry ice and stored at −80°C for biochemistry studies. For tissue collection for FUNCAT staining, rodents were deeply anesthetized and transcardially perfused with 4% PFA in PBS. All optic nerve samples include the entire nerve segment from optic head to chiasm to ensure normalization of total tissue volume during downstream sample preparation.
FUNCAT
For FUNCAT, PFA-fixed eyes were cryopreserved by sequential incubation in 15% and 30% sucrose at 4°C for 24 h each before mounting in optimal cutting temperature compound (Tissue-Tek O.C.T, Sakura Finetek), freezing, and sectioning to 10 µm thickness for click chemistry according to the modified protocol in Dieterich et al. (2010) and Hinz et al. (2012). Sections were transferred to Eppendorf tubes or six-well plates with a reaction mixture composed of 100 μm Tris [(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA; Sigma-Aldrich) dissolved in 4:1 tBuOH/DMSO (Sigma-Aldrich), 100 μm CuSO4 (Sigma-Aldrich), 1.25 μm Alexa Fluor 647 alkyne (Invitrogen), and 250 μm Tris(2-carboxyethyl)phosphine (TCEP, Sigma-Aldrich). The reaction proceeded overnight at room temperature. Retinal sections were imaged using an 880 Zeiss confocal microscope.
BONCAT
Freshly dissected optic nerve or retinal tissues were lysed in 0.5% SDS in PBS plus a cocktail of protease inhibitors (Complete Protease Inhibitor Cocktail Tablets, Roche) by homogenizing and sonicating with 10 pulses using a tip sonicator (Sonic Dismembrator, model no. 100, Fisher Scientific). AHA incorporated into proteins was labeled with PEG4 carboxamide-propargyl biotin (Biotin Alkyne, Invitrogen) by click chemistry reaction performed in the total protein suspension as described previously (Speers and Cravatt, 2009; Hulce et al., 2013). For QBONCAT, click reactions were done using biotin-alkyne labeled with heavy stable carbon and nitrogen isotopes, Biotin-β-Alanine-13C3,15N-Alkyne [Biotin Propargyl amide] or light isotope forms of the alkyne (catalog #7884 and #7889, respectively, Setareh Biotech) in the different experimental groups. The following reagents were added in sequence, vigorously vortexing after each addition: 30 μl of 1.7 mm TBTA (Sigma-Aldrich) dissolved in 4:1 tert-butanol/DMSO (Sigma-Aldrich), 8 μl of 50 mm CuSO4 dissolved in ultrapure water (Sigma-Aldrich), 8 μl of 5 mm of biotin-alkyne (in light or heavy form) dissolved in DMSO, and 8 μl of 50 mm TCEP (Sigma-Aldrich) dissolved in water. The click reactions were vortexed and incubated at room temperature overnight with gentle rotation at 4°C. After the completion of the cycloaddition reaction, 10 μl of each click reaction were separated for analyzing biotin-alkyne incorporation to proteins by Western blot using anti-biotin antibody (Pierce). The click reactions of both experimental groups were mixed 1:1, vortexed, and proteins were precipitated with methanol/chloroform. Proteins were precipitated by adding three volumes of methanol, one volume of chloroform, and three volumes of water, vortexed, and centrifuged at 15,000 × g for 2 min at room temperature. The aqueous and organic phases were removed carefully from the tube without disturbing the protein disk at the interface. Protein pellets were washed once by adding three volumes of methanol and centrifuging at 15,000 × g for 2 min. Pellets containing biotinylated proteins were air dried for 10 min. Protein precipitates were resuspended by adding 200 μl 8 m Urea and 200 μl 0.2% ProteaseMax surfactant (Promega) dissolved in 50 mm NH4HCO3. The protein suspension was reduced by adding TCEP to 5 mm final concentration and incubated at 55°C with vigorous orbital shaking using a ThermoMixer (Eppendorf). Protein alkylation was done by adding iodoacetamide (Sigma-Aldrich) to 10 mm final concentration and incubating with vigorous shaking in the dark for 20 min.
To digest the proteins, we added the following in the following order: 150 μl of 50 mm NH4HCO3, 2.5 μl of 1% ProteaseMAX dissolved in 50 mm NH4HCO3, and 1:100 (enzyme/protein, w/w) sequencing grade trypsin (Promega) to a final reaction volume of 500 μl. The digestion reactions were incubated for 3.5 h at 37°C with vigorous orbital shaking. The protein digestion reactions from biotin-AHA labeled proteins were stopped by adding trifluoroacetic acid (TFA; Sigma-Aldrich) to 0.1% final concentration. Samples were centrifuged at 20,000 × g for 20 min at room temperature to remove undigested insoluble material, and supernatant containing the peptide mixture was collected. Any remaining peptides in the insoluble pellet were extracted by adding 0.5 ml of 0.1% TFA in water, resuspending the pellet by pipetting, and centrifuging again for 20 min. The supernatant was pooled with the previous before desalting using Sep-Pak tC18 solid-phase extraction cartridges (Waters). Before loading the mixture of peptides, the cartridges were washed sequentially with 3 ml of acetonitrile, 3 ml of 0.5% acetic acid, 50% acetonitrile in water, and 3 ml of 0.1% TFA in water. After loading the peptide mixtures, the cartridges were washed with 3 ml of 0.1% TFA and then with 0.250 ml of 0.5% acetic acid in water. The peptides were eluted into a clean tube with 1 ml of 0.5% acetic acid, 80% acetonitrile in water, and dried in Eppendorf tubes in a SpeedVac (Thermo Fisher Scientific). Ten milligrams of dried peptide pellet were solubilized in 1 ml of PBS and incubated with a 200 μl slurry of NeutrAvidin beads (Pierce) for 1 h at room temperature. The beads were precipitated by centrifugation at 1000 × g for 5 min, and flow through was collected for MS analysis of unbound peptides. Beads were washed three times by adding 1 ml of PBS with 1 ml of 5% acetonitrile in PBS and a last wash in ultrapure water. Excess liquid was completely removed from the beads using a micropipette, and biotinylated peptides were eluted by adding 0.3 ml of solution containing 0.2% TFA, 0.1% formic acid, and 80% acetonitrile in water. The beads were centrifuged at 1000 × g, and the first elution of biotinylated peptides was collected. A second elution of 0.3 ml was boiled for 5 min for maximum release of peptides from the beads.
Mass spectrometry
Samples were analyzed using a modified MudPIT methodology with a Velos mass spectrometry as described previously (Schiapparelli et al., 2014; McClatchy et al., 2015). Tandem mass spectrometry (MS/MS) spectra remaining after filtering were searched with the Prolucid Software against the UniProt_rat_03-25–2014 concatenated to a decoy database in which the sequence for each entry in the original database was reversed. All searches were parallelized and performed on a Beowulf computer cluster consisting of 100 1.2 GHz Athlon central processing units. No enzyme specificity was considered for any search. The following modifications were searched: a static modification of 57.02,146 on cysteine for all analyses, a differential modification of 523.2749 on methionine for AHA to detect the standard biotin-alkyne, and a differential modification of 351.1774 (heavy) and 347.1702 (light) on methionine to detect the different AHA isotope biotin-alkynes. Prolucid results were assembled and filtered using the DTASelect (version 2.0) program. DTASelect 2.0 uses a linear discriminant analysis to dynamically set XCorr and DeltaCN thresholds for the entire dataset to achieve a user-specified false discovery rate (FDR). In addition, the modified peptides were required to be fully tryptic, <5 ppm deviation from peptide match, and an FDR at the spectra level of 0.01. The FDRs are estimated by the program from the number and quality of spectral matches to the decoy database. For all datasets, the protein FDR was <1%, and the peptide FDR was <0.5%.
In vitro neurite assay
Embryonic day 17 hippocampal neurons were seeded at a density of 200,000 cells/well in six-well tissue culture dishes (Falcon) precoated with poly-d-lysine (0.1 mg/ml; Sigma-Aldrich) and were cultured in defined medium of Neurobasal, l-glutamine (2 mm), penicillin-streptomycin, and B27 supplement (1:50; all from Thermo Fisher Scientific). After 3 d in vitro, candidate gene plasmid vectors (Extended Data Fig. 4-1) were transfected into hippocampal neurons with Lipofectamine LTX (Invitrogen). In general, plasmids contained either a cDNA expression construct or an shRNA against a candidate gene as well as a cytoplasmic fluorescent protein reporter. The amount of DNA per well ranged from 0.5 to 2 µg, depending on transfection efficiency, to yield sparse enough labeling to unambiguously identify individual positively transfected cells in a background of the higher density neuronal culture. After 6 d in vitro (on day 3 after transfection), live fluorescent images of labeled cells were obtained using the 10× objective of an inverted microscope (Carl Zeiss) with cell culture enclosure set at 37°C with 5% CO2.
Neurite quantification
Images were converted into TIF files and analyzed in ImageJ with the Simple Neurite Tracer plug-in, scaled to 1.5385 µm/pixel. Neurons with a neurite of at least 50 µm were traced from the center of the cell body to the end of the longest neurite by an investigator masked to condition. Dead neurons were identified as a cluster of small bright spots with DAPI staining in the cell body representing karyorrhexis and excluded. Faintly labeled neurons were not traced if it was not possible to identify where the neurite ended.
Statistical analysis
Data were analyzed using R version 3.5.0 (R Foundation for Statistical Computing) or GraphPad Prism 7 software. For correlation between proteomic samples, significance testing used Pearson's correlation coefficient in the package GGally. Quantitative proteomic comparisons of across replicates were made using a Stouffer's Z-score. Overexpression plasmids were compared using a Kruskal–Wallis test with the two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli, controlling the false discovery rate to 0.05 (Benjamini et al., 2006). Knock-down plasmids were similarly compared with the scramble control.
Data availability
Data that support the findings of this study are available in this article and the extended data files; all raw data are available from the corresponding author on reasonable request.
Results
AHA-mediated identification of newly synthesized retinal proteins for quantitative mass spectrometry
To label newly synthesized proteins in the retina and transported down optic nerve axons, AHA was injected intravitreally in adult rats (Fig. 1a). AHA is charged onto methionine tRNAs by endogenous tRNA synthetase and incorporated into proteins in place of methionine (Yuet and Tirrell, 2014) and detected after click chemistry conjugation to biotin. In preparation for comparative QBONCAT, we generated biotin beta alanine alkynes with heavy and light carbon and nitrogen isotopes that can be distinguished from endogenous methionine by mass spectrometry (Fig. 1b). We collected retinas and optic nerves 1 or 5 d after daily AHA injection and performed either FUNCAT for localization of newly synthesized proteins or BONCAT for Western blot and mass-spectrometry-based identification and quantification (Fig. 1c).
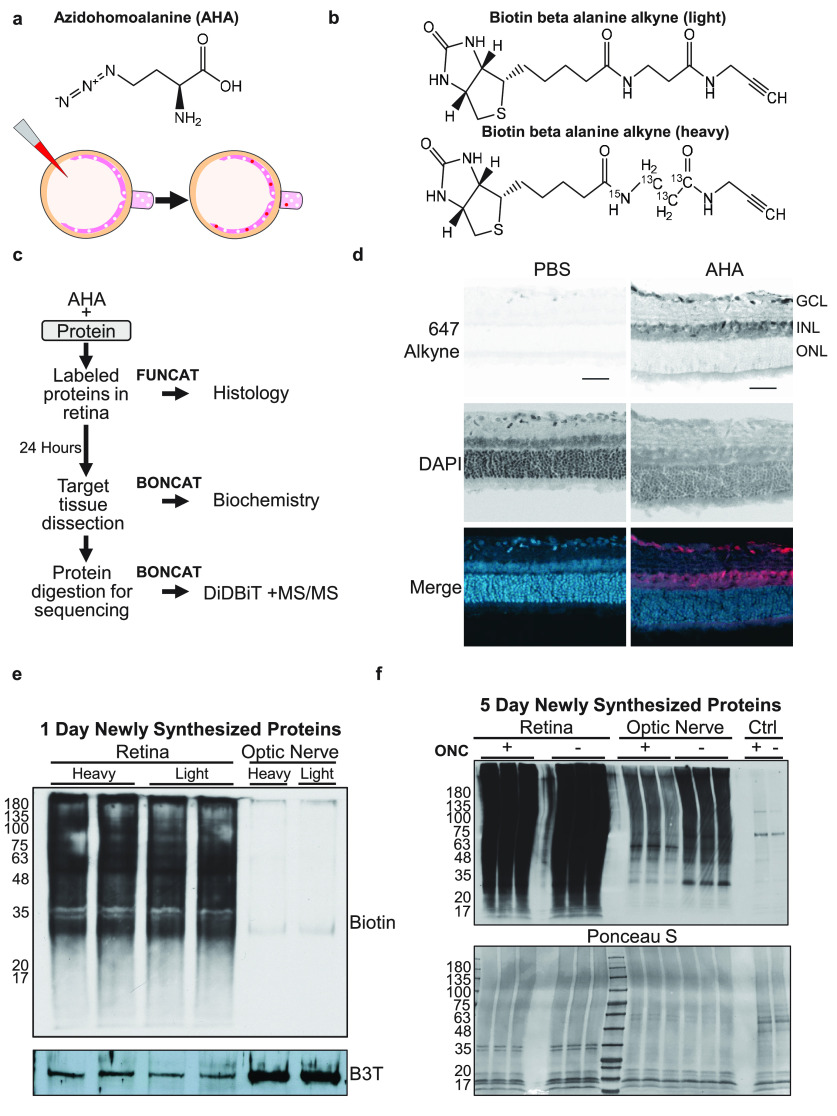
Figure 1.
AHA labeling detects newly synthesized proteins in the retina. a, AHA chemical structure (top) and diagram (bottom) of AHA incorporation into newly translating proteins in the retina, and their transport into the optic nerve, after intravitreal AHA injection. White dots represent unlabeled proteins present in retina. After intravitreal AHA injection (red), labeled proteins (red) are synthesized in the retina and transport to the optic nerve. b, Schematic of the two isotopes of biotin beta alanine used for quantitative QBONCAT proteomics. Light (top) and heavy (bottom) biotin beta alanine attached to AHA have a combined molecular mass of 347 and 351, respectively. c, Protocol for visualization of newly synthesized proteins by FUNCAT or biotin tagging by BONCAT for biochemistry and the enrichment and quantification by Direct Detection of Biotin-Containing Tags (Schiapparelli et al., 2014) and tandem mass spectrometry. d, Click reaction to tag AHA-labeled proteins with Alexa 647 alkyne shows distribution of newly synthesized proteins in the retinal ganglion cell layer (GCL) and inner nuclear layer (INL), with no detectable AHA-labeled protein in the outer nuclear layer (ONL). Cell nuclei are labeled with DAPI. Retinas injected with PBS show minimal background labeling. Scale bar, 50 µm. e, Western blot of biotin after click chemistry with either heavy or light biotin-alkyne shows equivalent labeling in retinal tissue with either isotope of biotin, but little to no labeling in optic nerve samples 1 d after AHA labeling. Two replicates of retinal samples are shown. Beta III tubulin (B3T) used as a loading control. Left, Molecular weights. f, Top, Western blot of biotin after click chemistry with AHA-labeled retinas and optic nerves, with (+) and without (−) optic nerve crush injury (ONC) after 5 d of AHA labeling. Samples were run in triplicate. The retinas are extensively labeled and biotin-labeled proteins are detected after transport into the optic nerve. Control (Ctrl) lanes are optic nerve samples after AHA injection without click reaction showing minimal background endogenous biotin signal. Bottom, Ponceau S stain of the samples showing comparable protein loading across samples. Left, Molecular weights.
We first asked whether intravitreal injection of AHA would result in incorporation of AHA into newly synthesized proteins in retinal cells within 24 h. By FUNCAT conjugation of AHA to a fluorescently tagged alkyne in cryosections of acutely dissected retinas, we observed labeled cell bodies in the ganglion cell and inner nuclear layers of samples after AHA injection compared with control (Fig. 1d). We confirmed AHA incorporation into retinal proteins using BONCAT and Western blot, and also confirmed that isotopic heavy and light variants of biotin-alkynes react equivalently with AHA in click chemistry reactions (Fig. 1e), suggesting these mass tags could be used for quantification and comparison of newly synthesized proteins between different samples. At 24 h, only low levels of biotinylated proteins were detected in the optic nerve using either isotopic biotin (Fig. 1e), suggesting detectable new protein transport down RGC axons is delayed compared with new protein synthesis detected in the retina, further corroborating a mechanism of new protein synthesis in the retina followed by transport to axonal targets. Together, both FUNCAT and BONCAT approaches showed rapid incorporation of AHA into newly synthesized proteins by 24 h in the retina and confirmed comparable labeling by heavy and light biotin isotopes in preparation for QBONCAT.
Following optic nerve injury, injured axons retract from the crush site between 6 h and 1 d after injury, and initial axon sprouting and regrowth from surviving RGCs occurs in the first week (Sellés-Navarro et al., 2001). To characterize the protein synthesis responses to both phases of injury, we injected AHA into the vitreous either twice in the 24 h following optic nerve injury or once every day for 5 d starting 24 h after optic nerve injury. Retinal and optic nerve tissue were collected and analyzed by click chemistry, Western blot, and MS/MS proteomics. In addition to the strong biotin labeling seen in Western blots of the retinal tissue, similar to the 1 d samples (Fig. 1e), we detected newly synthesized proteins transported into the optic nerve after 5 d (Fig. 1f), consistent with greater protein labeling and transport over time. The decrease of newly synthesized protein detected in optic nerve samples after optic nerve injury (Fig. 1f) confirms that the signals detected in the optic nerve samples are from transported proteins labeled in the retina and are not confounded by diffusion of the AHA reagent down the nerve.
Retinal protein synthesis dynamics over 1 and 5 d after optic nerve injury
To quantify changes in the retinal newly synthesized proteome over these two time points of interest, we first compared proteins detected in uninjured retinas collected after 1 d and 5 d of AHA labeling to total proteins in a separate naive retina sample (Fig. 2a). We detected 1910 proteins in the total retinal proteome and 1792 proteins that were newly synthesized at either 1 or 5 d. We next examined how these proteins overlapped between the three sample types. More than 750 proteins were detected in all three groups (Fig. 2a, blue bar; Extended Data Fig. 2-1). Although a majority of the newly synthesized proteins detected at 1 or 5 d were also detected in the naive total retinal protein samples, identifying them as proteins with relatively high constitutive levels of synthesis, 643 proteins were only detected by AHA in the newly synthesized samples. Furthermore, there were 268 and 153 proteins found only in 1 d or 5 d retinas, respectively (Fig. 2a, purple and green bars). These protein identifications highlight the importance of specifically studying newly synthesized, lower abundance proteins for a more comprehensive analysis of the proteomic response.
Figure 2.
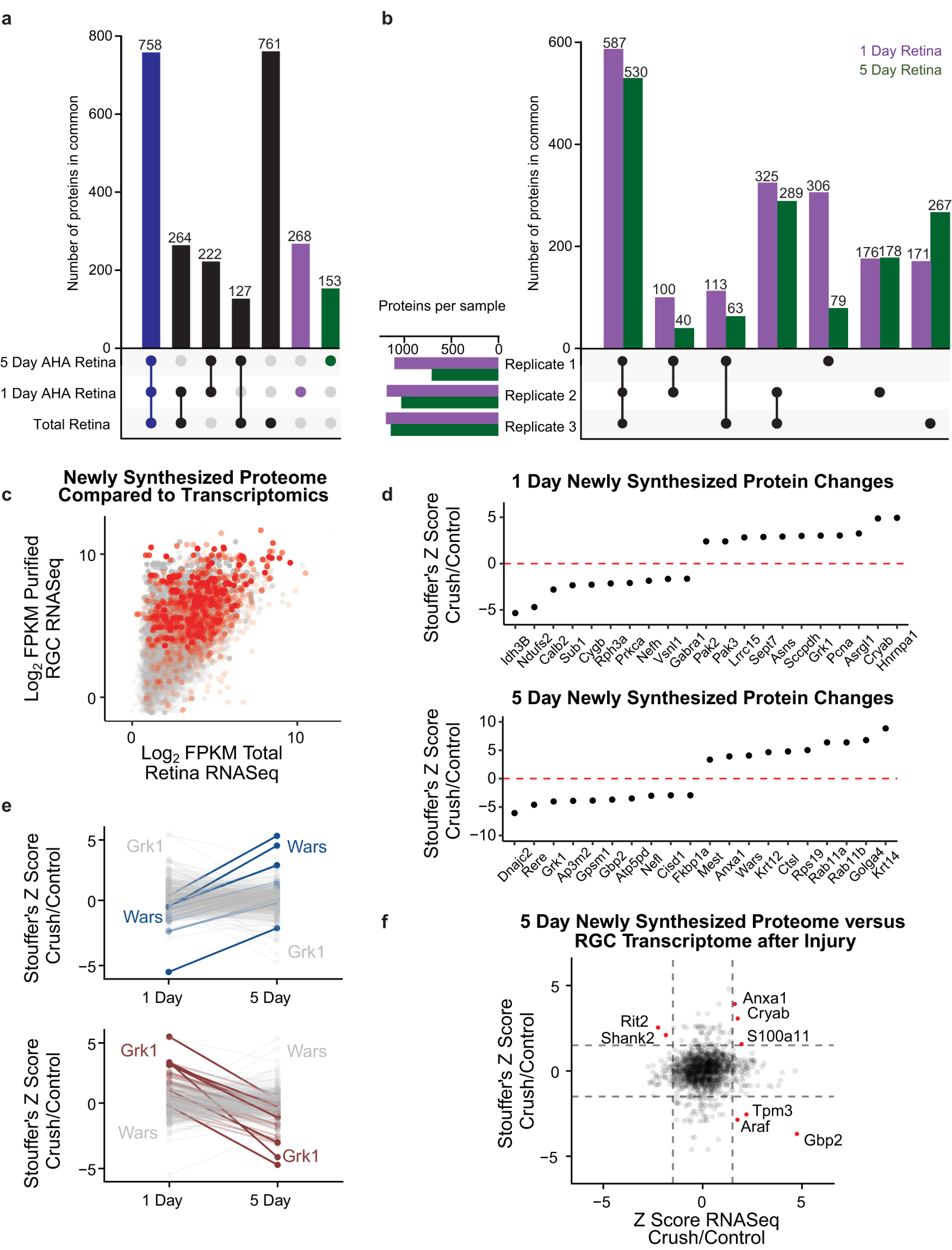
Comparison of retinal new protein synthesis 1 and 5 d after optic nerve injury. a, Comparison of numbers of proteins identified in one or more of each of the following types of retinal sample: 1 d AHA-labeled new protein synthesis, 5 d AHA-labeled new protein synthesis, and total retinal proteome without injury. Bottom left, Sample types. Filled dots indicate overlap between samples, and bar graphs above show number of proteins in group. Seven hundred fifty-eight proteins were found to be in common between all three sample types (blue bar). Two hundred sixty-eight and 153 proteins were found only in the 1 d and 5 d AHA-labeled samples and not in the total retina sample, respectively (purple and green bars, respectively). Full data are available in Extended Data Figure 2-1. b, Comparisons of numbers of AHA-labeled newly synthesized proteins identified in one or more experimental replicates (Replicate 1, 2, 3) after 1 d (purple) or 5 d (green) of AHA labeling, combining analysis of proteomes detected with and without injury. Full data are available in Extended Data Figure 2-2. c, Total RNA-sequencing data from uninjured retinas (Brooks et al., 2011) were compared with purified RGC transcriptomics (Williams et al., 2017) to develop an understanding of transcript enrichment in RGCs. The mRNAs that have corresponding newly synthesized proteins, detected by our proteome study with 5 d AHA labeling, are highlighted in red. This analysis indicates that the newly synthesized proteome captures more of an RGC-related phenotype than non-RGC phenotype. d, Proteins with the largest differences in AHA-biotin labeling 1 d or 5 d after optic nerve injury, quantified by Stouffer's Z-score across three replicates. e, Comparison of AHA-biotin labeling differences from 1 d to 5 d after injury. Left, The proteins with the largest increases labeled in progressively darker blues. Wars showed the largest fold-change increase from 1 to 5 d after optic nerve injury. Right, The proteins with the largest decreases are labeled in progressively darker reds. Grk1 showed the largest fold-change decrease from 1 d to 5 d after optic nerve injury. Full data are available in Extended Data Figure 2-3. f, Comparison of changes in 5 d AHA-labeled proteins after optic nerve injury compared with changes in RGC transcriptomics 5 d after optic nerve injury obtained from previously deposited datasets (Gene Expression Omnibus, GSE142881), both normalized by Z-score. Dashed gray lines represent Z-score of ±1.5 for illustrative purposes. Proteins identified as outliers with either increased protein and RNA expression, increased protein and decreased RNA expression, or decreased protein and increased RNA expression after injury are labeled with red dots.
Protein IDs found in 1 day control samples, 5 day control samples, and/or naive total retina. Download Figure 2-1, XLSX file (73.6KB, xlsx) .
All replicates of AHA retinas from either 1 day or 5 day injection experiments. Download Figure 2-2, XLSX file (100.6KB, xlsx) .
Stouffer's Z-score changes of new protein synthesis in the retina between 1 and 5 days. Download Figure 2-3, XLSX file (46.3KB, xlsx) .
With this increased resolution of newly synthesized proteins, we turned to the novel application of QBONCAT to identify changes in new protein synthesis early (1 d) and late (5 d) after optic nerve injury. We collected eight AHA-injected retinas per optic nerve crush or sham groups and conjugated those protein samples with heavy or light biotin, respectively. These heavy and light biotin-conjugated protein samples were combined for a single MS run. We repeated this approach in three replicates (Replicate 1, Replicate 2, Replicate 3). We first assessed reproducibility of newly synthesized retinal proteomics by exploring the number of unique proteins identified in each replicate for 1 d and 5 d proteomes. We identified more than 700 newly synthesized retinal proteins in each replicate, with more than 1000 proteins in three of three 1 d samples and two of three 5 d samples (Fig. 2b, bottom left graph). When compared with each other, we identified more than 500 proteins in all three replicates in the 1 d and in the 5 d retinal samples (Fig. 2b, first set of bars; Extended Data Fig. 2–2). Five hundred thirty-eight and 392 proteins were found in two of three replicates in the 1 d and 5 d samples, respectively. Interestingly, there was remarkable consistency between the numbers of proteins detected in both 1 and 5 d samples, whether specific to one experimental replicate or detected across two or three experimental replicants (Fig. 2b, compare purple and green bars). Together, these data suggest mass-modified AHA-biotin alkynes can reproducibly identify and quantify newly synthesized retinal proteins after injury.
We next asked whether the newly synthesized proteins identified with AHA labeling preferentially labeled RGCs compared with total retinal cells, as suggested by histology (Fig. 1d). We used previously published transcriptomic datasets from uninjured retinas (Brooks et al., 2011) and uninjured purified RGCs (Williams et al., 2017) to compare gene expression for enrichment in RGCs. We then identified those transcripts with corresponding newly synthesized proteins detected after 5 d of protein labeling (Fig. 2c, red). First, the funnel-shaped rise to the right in Figure 2c confirms that most proteins more highly expressed in RGCs are more likely highly expressed in whole retina. Second, the proteins identified by mass spectrometry (red dots) skewed heavily toward those with higher RGC gene expression but were found among both low- and high-expressed retinal genes. This analysis indicates that the newly synthesized QBONCAT proteome correlated more with RGC gene expression than with retinal gene expression.
To compare changes in specific newly synthesized proteins between control and optic nerve injury conditions, we analyzed AHA-labeled retinal MS/MS datasets using Stouffer's Z-score to separate proteins with significant changes in synthesis between optic nerve crush and control retinas at 1 and 5 d after injury (Fig. 2d). We isolated the proteins that decreased or increased the most in synthesis after injury across three proteomic replicates, shown in the left and right halves of the graphs in Figure 2d, respectively. At 1 d (Fig. 2d, top graph), the protein with the highest increase in synthesis after optic nerve injury is Hnrnpa1, an RNA-binding protein associated with stress granules in neurodegenerative diseases like ALS (Kim et al., 2013). At 5 d after injury (Fig. 2d, bottom graph), Ctsl, a protein involved in protein degradation, is increased in synthesis, coincident with increased degeneration of RGCs (Sánchez-Migallón et al., 2016). Additionally, synthesis of Rab11a and Rab11b, which are known to regulate local protein turnover (Howes et al., 2010), was also increased at 5 d, consistent with a role previously identified in integrin trafficking and in vitro axon growth (Eva et al., 2010), and with RGC growth cone sprouting observed in vivo at this time point (Koseki et al., 2017). This analysis indicates that quantification of newly synthesized proteins by QBONCAT identifies temporally relevant cellular changes after optic nerve injury.
We next asked how retinal protein synthesis dynamics differ at 1 and 5 d after optic nerve injury. After filtering for proteins quantified in the retina at both time points after injury (996 proteins and 563 proteins at 1 and 5 d, respectively, 480 proteins at both time points; Extended Data Fig. 2-3), we analyzed either those that increased or decreased relative to control over time (Fig. 2e). Wars (tryptophanyl tRNA-synthetase), increased the most relative to control at 5 d compared with 1 d after injury. The family of aminoacyl-tRNA synthetases (AARS) play multiple roles beyond canonical translation including regulating cell proliferation, inflammation, and RNA processing (Park et al., 2005). In fact, at least two members of the AARS family affect peripheral nerve degeneration and regeneration (Park et al., 2015), positioning Wars as a candidate to manipulate in CNS injury response. In contrast, synthesis of Grk1 (rhodopsin kinase) decreased in the retina over time after optic nerve injury. As Grk1 is part of the phototransduction cascade, we hypothesize that its decrease is because of the loss of functional vision after optic nerve injury. Together, these findings demonstrate the power of QBONCAT for quantification of dynamic changes to new protein synthesis along a time course in response to injury.
As protein translation only partially correlates with transcription, we next analyzed how our newly synthesized proteome compared with the RGC transcriptome following injury. We compared our 5 d AHA proteomics data to a deposited dataset of transcriptional profiling of RGCs following optic nerve injury (Gene Expression Omnibus, GSE142881). As expected, there was limited global correlation between the two methodologies. There were only very few proteins we identified as outliers with either increased protein and RNA expression, increased protein and decreased RNA expression, or decreased protein and increased RNA expression after injury (Fig. 2f, red dots). Outside these few outliers, the general noncorrelation of mRNA and protein changes after injury points to the importance of proteomic studies using techniques like QBONCAT.
Protein synthesis and transport to the optic nerve after injury
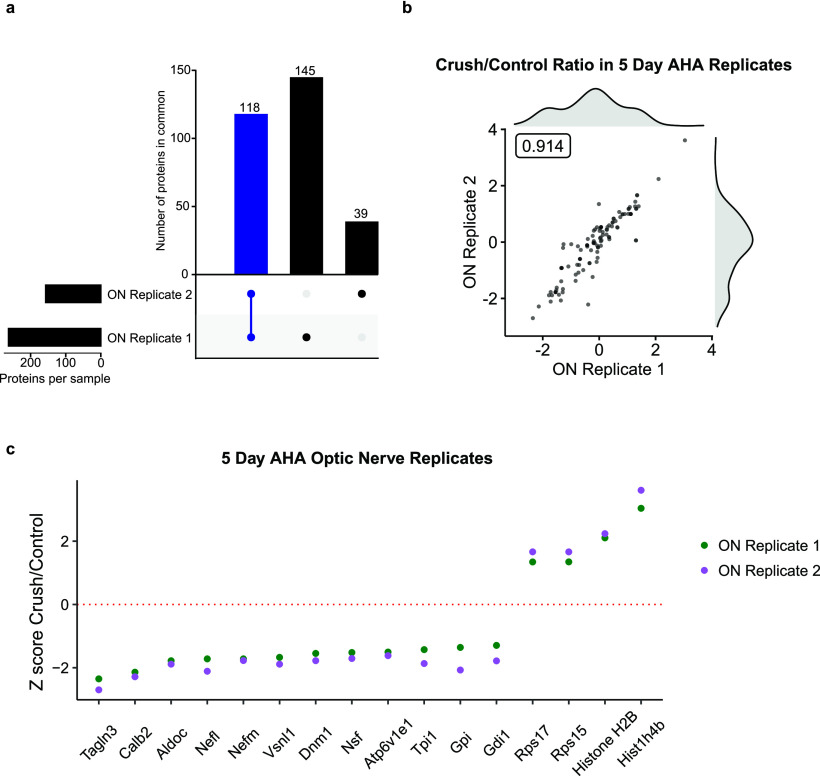
We next asked what newly synthesized proteins were transported to the optic nerve and quantified how this changed after injury. The 5 d AHA Western blot shown in Figure 1f indicated that biotinylated proteins transported to the optic nerve decreased after optic nerve injury. We first compared replicates of the newly synthesized RGC transportome in the optic nerve before and after injury and found 118 proteins in common (Fig. 3a, blue bar). Further analysis of these replicates demonstrated that the quantitative change in newly synthesized transportome after injury was highly correlated (Pearson's r = 0.914; Fig. 3b; Extended Data Fig. 3-1). We next filtered these data for optic nerve transportome proteins whose average normalized ratio between crush and control, expressed as a Z-score, across both replicates changed >1.5 SDs after injury in either direction (Fig. 3b). We found several newly synthesized, transported proteins with large changes 5 d after injury. Surprisingly, we found the presence of histone proteins that increased in the optic nerve after injury. Although previous work has identified histone translation in developing retinal axons in vitro (Cagnetta et al., 2018), their transport to the axonal compartment in vivo in adult RGC axons has only recently been detected (Schiapparelli et al., 2019).
Figure 3.
Changes in new protein synthesis and transport to the optic nerve at 5 d after injury. a, Comparison of proteins in optic nerve samples shows 118 of 302 (39%) AHA-labeled proteins detected in both experimental replicates. b, Scatter plot of peptide ratios of crush/control of two optic nerve (ON) replicates shows tight correlation between samples (Pearson's r = 0.914). Histograms of crush/control ratios of replicate 1 (above) and replicate 2 (right) shows distribution of synthesized and transported proteome. Full data are available in Extended Data Figure 3-1. c, Z-score comparison of crush compared with control, highlighting the largest changes in transport (average Z-score > 1.5) of newly synthesized, AHA-labeled proteins into the optic nerve 5 d after injury. Gene Ontology analysis of compartmentalization of optic nerve proteins is available in Extended Data Figure 3-2.
Replicates of changes to AHA-labeled proteins transported to optic nerve with and without optic nerve crush (Z-score normalized). Download Figure 3-1, XLSX file (15.2KB, xlsx) .
AHA-labeled proteins transported to optic nerve characterized as part of nuclear cellular component by Gene Ontology. Download Figure 3-2, XLSX file (12.8KB, xlsx) .
In fact, analysis of these transported proteins using Gene Ontology reveals several canonically nuclear proteins found to be localized to the optic nerve compartment (Extended Data Fig. 3-2). This unexpected localization of multiple proteins confirms prior observations on the need to expand ontology labeling in the face of new compartmentalized “omics” approaches (Kar et al., 2018); delineating the differential functions of proteins based on context and compartment is a point for future research. Further validation and manipulation of axonal histones will provide insight into possible noncanonical roles in injury response.
Quantification of neurite outgrowth of proteomic candidates in vitro
We hypothesized that proteins whose synthesis changes after injury may play a role in regulating RGC survival or axon growth response. We asked whether any candidate proteins identified from 1 d QBONCAT retinal proteomics affected neurite growth using an in vitro neurite outgrowth assay. We selected candidate proteins as different between optic nerve injury and controls based on meeting any of three criteria for change: Stouffer's Z-score >1.5, presence versus absence in all replicates, or a large fold-change difference in at least one replicate. As it was unclear whether early retinal protein synthesis changes in response to injury are adaptive or degenerative (i.e., pro-axon or antiaxon growth), we generated both overexpression and shRNA knock-down vectors for all candidates, along with overexpression and knock-down plasmids for Kruppel-like factor 9 (KLF9), a transcription factor previously shown to modulate neurite outgrowth in vitro and in vivo (Moore et al., 2009; Apara et al., 2017), as positive controls for outgrowth promotion or inhibition, respectively, and used GFP and a scrambled shRNA plasmid as negative controls (Extended Data Fig. 4-1).
Cultured primary neurons were transfected with plasmids 3 d after plating and imaged for longest neurite quantification 3 d later, with representative images of three such neurons displayed (Fig. 4a,b). We next compared the medians and 95% confidence intervals of all candidate genes with the negative control plasmids medians (Fig. 4c). Although plasmids were designed from known targets before this assay, we relied on functional effects for validation, acknowledging the possibility of false negative results. We found significant regulation of neurite outgrowth by 4 of the 15 candidate genes: knock down of Gtf2i, PCNA (proliferating cell nuclear antigen), and Asrgl1 increased neurite outgrowth significantly, and overexpression of Hnrnpa1 decreased neurite outgrowth significantly (Fig. 4d). These data demonstrate the power and validity of quantifying changes in protein synthesis for candidate selection of relevant regulatory proteins in these data regulating neurite growth.
Figure 4.
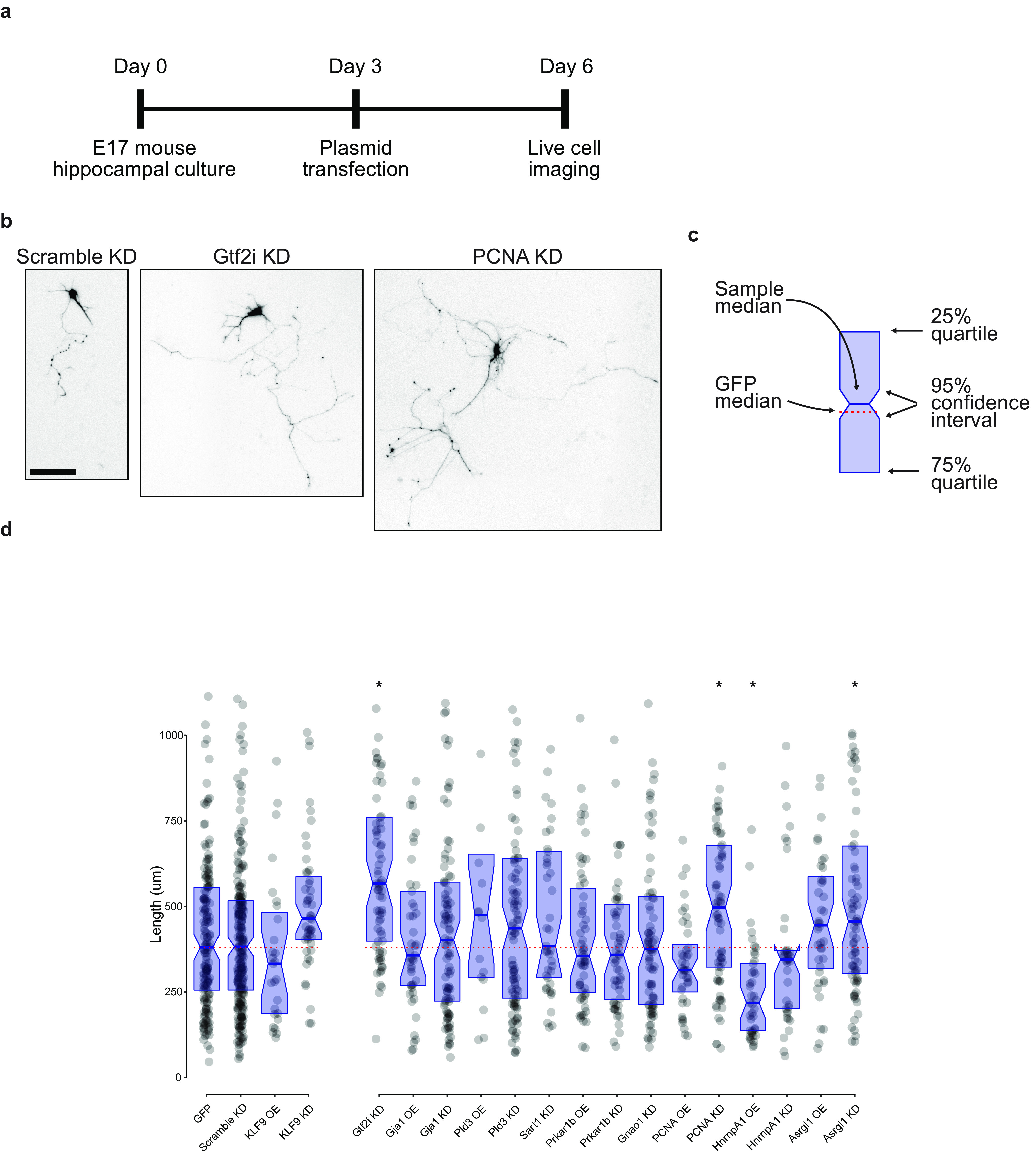
Quantification of neurite outgrowth with manipulation of proteomic candidates. a, Timeline of in vitro experimental protocol. b, Representative images of neurite length after transfection with three shRNA plasmids, as shown. Scale bar, 100 µm. c, Legend for neurite outgrowth graph. Blue rectangle defines the 25% and 75% interquartile range, with the center blue line as the median. Notches on the rectangle define the 95% confidence interval around the median. The dotted red line is the GFP median to aid in visual comparison across conditions. Raw data are also shown in d as gray dots. d, Comparison of longest neurite lengths with each transfection condition. GFP overexpression and a scrambled shRNA construct served as negative controls, and KLF9 overexpression and knock-down plasmids were used as positive controls. Data points above 1100 µm were excluded from graph but included in statistics and comparisons. Overexpression plasmids were compared using a Kruskal–Wallis test with two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli controlling the false discovery rate to 0.05 (Benjamini et al., 2006). Knock-down plasmids were similarly compared with the scramble control (Hnrnpa1 OE, *p < 0.01; Gtf2i KD, *p < 0.01; PCNA KD, *p < 0.01; Asrgl1 KD, *p = 0.02) OE, Overexpression; KD, knock-down. Total neurons traced, 1632. Plasmid design data are available in Extended Data Figure 4-1.
Plasmids used in neurite outgrowth assay. Vectors created by VectorBuilder. Maps can be retrieved using IDs at https://en.vectorbuilder.com/. Download Figure 4-1, XLSX file (11.2KB, xlsx) .
Discussion
Better understanding the molecular basis for axon growth and its application to promoting regeneration in the nervous system remains a major goal in neuroscience. In this article, we quantify changes in protein synthesis after optic nerve injury to identify candidate regulators of retinal and optic nerve injury response. We first demonstrate methodological advances in quantitative BONCAT for newly synthesized proteomics using isotope variants of biotin-alkyne along with direct detection of biotinylated peptides. We adapt this improved technique to work in vivo to quantify changes in newly synthesized retinal proteins along a time course after optic nerve injury and quantify changes in transport of newly synthesized proteins to the optic nerve. We then validate several of these proposed proteins as regulators of neurite outgrowth using an in vitro assay.
Methodologically, QBONCAT confers a significant advantage in quantitative proteomics. Previous studies have detailed transcriptomic changes in the retina after injury, but these remain a proxy for the protein-level changes in the cell (Yasuda et al., 2014). Tandem mass tagging or iTRAQ (isobaric tag for relative and absolute quantitation) for quantitative proteomics can provide protein abundance changes in RGCs after axonal injury (Hollander et al., 2012), as used to identify c-myc as a regulator of survival and regeneration after injury (Belin et al., 2015), but both methods have several limitations. First, total protein abundance does not directly measure translational responses to an injury but instead reflects the net of protein synthesis and degradation, both of which may be dynamic in cellular responses (Savitski et al., 2018). Second, and closely related, total protein abundance may change along a faster or slower time course if protein synthesis and protein turnover rates are not matched to each other, thus limiting accurate temporal resolution.
To overcome these barriers to understanding the cellular response to injury, we quantified newly synthesized proteins with ncAAs to quantify acute changes in protein synthesis across multiple time points in vivo. By using isotope variants of biotin- alkyne, we were able to combine proteins from injured and uninjured samples together in the same MS/MS run, allowing for direct and quantitative comparisons in the acute period after optic nerve injury, extending an approach previously used only for long-term labeling of the full proteome (McClatchy et al., 2015). Interestingly, isolating changes in new protein synthesis from the otherwise overwhelming background of total protein abundance led to the identification of 643 proteins in retinal samples at either 1 or 5 d after optic nerve trauma that were not even detected in total retinal lysates.
A limitation of retinal proteomics in this and prior studies is the inclusion of non-RGCs, although histologic identification of ncAAs by FUNCAT suggests intravitreal injection limits nearly all new protein synthesis identification to inner retinal cells over photoreceptors or retinal pigment epithelium (Fig. 1d). Additionally, comparison of our identified proteins to previous retina and RGC-specific transcriptomic data show a bias toward the RGC phenotype compared with total Retina (Fig. 2c). Additionally, we have previously identified the optic nerve transportome, defining a subset of RGC proteins (Schiapparelli et al., 2019).
One drawback to isolating protein quantification to just newly synthesized proteins is the current technical limitations of mass-spectrometry-based approaches. Even with total proteomics, there is undersampling because of the lack of amplification, so that the most abundant proteins are more reliably detected and many low-abundance proteins are not detected at all. With our technique, especially our 1 d dataset, we are specifically measuring proteins that are synthesized in the restricted time period in a small tissue volume, introducing temporal dynamics and working at the detection limits of state-of-the-art mass spectrometry. To this end, QBONCAT is best suited not to replace other methodologies but rather used in conjunction with biochemical techniques to explore cellular changes at the protein level.
Thus, a lack of detection by QBONCAT at 24 h does not mean that there is no protein synthesis present. Some expected proteins that were expected in the newly synthesized proteome were not able to be measured, possibly because of various biophysical and technical restraints. Alternatively, these data may suggest alternative interpretations of prior data, such as increases in protein being due more to stabilization or decreased degradation/protein turnover rather than the assumed mechanism of increased new protein synthesis. This latter interpretation would also suggest a transcription/translation mismatch, which has already been well described (Schwanhäusser et al., 2011). As this is the first use of this technology in the retina in the context of optic nerve injury, we expect a better understanding of sensitivity and reliability in vivo as more groups attempt to isolate and quantify protein synthesis dynamics in more biological systems. Additional methods still need to be developed to quantitatively measure protein turnover/degradation to properly distinguish these two hypotheses.
Nonetheless, the improved depth of quantitative detection by QBONCAT permitted selection of multiple injury-responsive protein candidates, both at individual time points and those dynamically shifting during the first 5 d of the degenerative process. Several of these proteins were strong candidates for injury-relevant response and were overrepresented in our screen as modulators of neurite outgrowth. For example, neurite growth-relevant genes PCNA, Asrgl1, and Ndufs2 were only found at 1 d after injury, Krt14 was only found in the 5 d group, and Gtf2i and Rab11a were found in both time points. These data demonstrate the specificity of newly synthesized proteomics for isolating modulators of acute cellular responses.
Also interesting was the increased transport of nuclear histone proteins, such as Hist1h4b and Hist1h2b, into the optic nerve 5 d after injury. This was an unexpected finding, although not entirely unprecedented (Schiapparelli et al., 2019). In vitro neuronal proteomics localized several nuclear proteins to the growth cone, including histone proteins as well as PCNA (Estrada-Bernal et al., 2012). The time point of 5 d after injury is consistent with previous studies of optic nerve injury as a time point when retracted axons begin to regrow at the injury site (Sellés-Navarro et al., 2001). With the advent of compartmentalized transcriptomics, translatomics, and now proteomics as shown here, context-dependent localization and function of proteins must be explored to more accurately characterize the dynamics of cellular networks. Exploration of novel axonal functions of canonical nuclear proteins will be a focus of future work.
Our in vitro screen identified Gtf2i, PCNA, Hnrnpa1, and Asrgl1 as regulators of neurite outgrowth, and based on these and other data, all four of these genes merit further study. A positive hit rate of ∼25% is in line with or above that of previous studies using neurite extension assays (Sekine et al., 2018). For example, genetic deletion of Gtf2i, a negative regulator of calcium entry, has previously been shown to affect neurite outgrowth in developing cortical cultured neurons (Deurloo et al., 2019). As electrophysiologic activity regulates RGC axon extension in vitro through mechanisms including calcium-mediated signaling pathways (Goldberg et al., 2002; Corredor et al., 2012), increasing calcium entry by knocking down Gtf2i is a plausible mechanism for a potential therapeutic after neuronal injury. We found that Hnrnpa1 overexpression inhibited neurite outgrowth; we hypothesize that Hnrnpa1 expression is detrimental to RGC survival after injury, similar to its role in concentration-dependent stress granule formation in other neurodegenerative diseases like ALS (Kim et al., 2013; Molliex et al., 2015; Deshaies et al., 2018). Although little is known about Asrgl1 function in RGCs, it has been identified as specifically enriched in human-specific hippocampal gene evolution (Konopka et al., 2012). Overall, newly synthesized proteomics identified several validated gene targets for neurite outgrowth, and future work to test these survival and regeneration candidates in vivo after optic nerve injury may clarify their functions.
Together, these data suggest that quantitative proteomic analysis of newly synthesized proteins highlights the dynamic injury response with better sensitivity than global proteomics and greater reliability to protein expression than transcriptomics. Combing new protein synthesis with QBONCAT and studies that subsequently isolate rates of protein degradation (McClatchy et al., 2015, 2018) through the time course of neurodegeneration and in response to candidate regenerative therapies will greatly increase our understanding of degenerative disease and of neuronal survival and regeneration.
Footnotes
This work was supported by the National Institutes of Health (NIH) Grants EY011261, EY027437, P30 EY019005, and R01MH103134; the Hahn Family Foundation (H.T.C.); NIH Grants P41 GM103533 and R01MH067880 to J.Y.; NIH Grant P30 EY026877, the Glaucoma Research Foundation Grant 100001641, Medical Technology Enterprise Consortium, and Research to Prevent Blindness Grant 100001818 to J.L.G.; and NIH Grant U01EY027261 to J.L.G., J.Y., and H.T.C.
The authors declare no competing financial interests.
References
- Alvarez-Castelao B, Schanzenbächer CT, Hanus C, Glock C, tom Dieck S, Dörrbaum AR, Bartnik I, Nassim-Assir B, Ciirdaeva E, Mueller A, Dieterich DC, Tirrell DA, Langer JD, Schuman EM (2017) Cell-type-specific metabolic labeling of nascent proteomes in vivo. Nat Biotechnol 35:1196–1201. 10.1038/nbt.4016 [DOI] [PubMed] [Google Scholar]
- Apara A, Galvao J, Wang Y, Blackmore M, Trillo A, Iwao K, Brown DP, Fernandes KA, Huang A, Nguyen T, Ashouri M, Zhang X, Shaw PX, Kunzevitzky NJ, Moore DL, Libby RT, Goldberg JL (2017) KLF9 and JNK3 interact to suppress axon regeneration in the adult CNS. J Neurosci 37:9632–9644. 10.1523/JNEUROSCI.0643-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barış M, Tezel G (2019) Immunomodulation as a neuroprotective strategy for glaucoma treatment. Curr Ophthalmol Rep 7:160–169. [PMC free article] [PubMed] [Google Scholar]
- Belin S, Nawabi H, Wang C, Tang S, Latremoliere A, Warren P, Schorle H, Uncu C, Woolf CJ, He Z, Steen JA (2015) Injury-induced decline of intrinsic regenerative ability revealed by quantitative proteomics. Neuron 86:1000–1014. 10.1016/j.neuron.2015.03.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Krieger AM, Yekutieli D (2006) Adaptive linear step-up procedures that control the false discovery rate. Biometrika 93:491–507. [Google Scholar]
- Best MD (2009) Click chemistry and bioorthogonal reactions: unprecedented selectivity in the labeling of biological molecules. Biochemistry 48:6571–6584. 10.1021/bi9007726 [DOI] [PubMed] [Google Scholar]
- Brooks MJ, Rajasimha HK, Roger JE, Swaroop A (2011) Next-generation sequencing facilitates quantitative analysis of wild-type and Nrl(−/−) retinal transcriptomes. Mol Vis 17:3034–3054. [PMC free article] [PubMed] [Google Scholar]
- Cagnetta R, Frese CK, Shigeoka T, Krijgsveld J, Holt CE (2018) Rapid cue-specific remodeling of the nascent axonal proteome. Neuron 99:29–46.e4. 10.1016/j.neuron.2018.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron E, Xia X, Galvao J, Ashouri M, Kapiloff M, Goldberg J (2020) Optic nerve crush in mice to study retinal ganglion cell survival and regeneration. Bio-Protocol 10:e3559. 10.21769/BioProtoc.3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corredor RG, Trakhtenberg EF, Pita-Thomas W, Jin X, Hu Y, Goldberg JL (2012) Soluble adenylyl cyclase activity is necessary for retinal ganglion cell survival and axon growth. J Neurosci 32:7734–7744. 10.1523/JNEUROSCI.5288-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies JE, et al. (2018) TDP-43 regulates the alternative splicing of hnRNP A1 to yield an aggregation-prone variant in amyotrophic lateral sclerosis. Brain 141:1320–1333. 10.1093/brain/awy062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deurloo MHS, Turlova E, Chen WL, Lin YW, Tam E, Tassew NG, Wu M, Huang YC, Crawley JN, Monnier PP, Groffen AJA, Sun HS, Osborne LR, Feng ZP (2019) Transcription factor 2I regulates neuronal development via TRPC3 in 7q11.23 disorder models. Mol Neurobiol 56:3313–3325. 10.1007/s12035-018-1290-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich DC, Lee JJ, Link AJ, Graumann J, Tirrell DA, Schuman EM (2007) Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Protoc 2:532–540. 10.1038/nprot.2007.52 [DOI] [PubMed] [Google Scholar]
- Dieterich DC, Hodas JJL, Gouzer G, Shadrin IY, Ngo JT, Triller A, Tirrell DA, Schuman EM (2010) In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci 13:897–905. 10.1038/nn.2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-Bernal A, Sanford SD, Sosa LJ, Simon GC, Hansen KC, Pfenninger KH (2012) Functional complexity of the axonal growth cone: a proteomic analysis. PLoS One 7:e31858. 10.1371/journal.pone.0031858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eva R, Dassie E, Caswell PT, Dick G, ffrench-Constant C, Norman JC, Fawcett JW (2010) Rab11 and its effector Rab coupling protein contribute to the trafficking of 1 integrins during axon growth in adult dorsal root ganglion neurons and PC12 cells. J Neurosci 30:11654–11669. 10.1523/JNEUROSCI.2425-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg JL, Espinosa JS, Xu Y, Davidson N, Kovacs GT, Barres BA (2002) Retinal ganglion cells do not extend axons by default. Neuron 33:689–702. 10.1016/S0896-6273(02)00602-5 [DOI] [PubMed] [Google Scholar]
- Hinz FI, Dieterich DC, Tirrell DA, Schuman EM (2012) Non-canonical amino acid labeling in vivo to visualize and affinity purify newly synthesized proteins in larval zebrafish. ACS Chem Neurosci 3:40–49. 10.1021/cn2000876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander A, D'Onofrio PM, Magharious MM, Lysko MD, Koeberle PD (2012) Quantitative retinal protein analysis after optic nerve transection reveals a neuroprotective role for hepatoma-derived growth factor on injured retinal ganglion cell. Invest Ophthalmol Vis Sci 53:3973–3989. 10.1167/iovs.11-8350 [DOI] [PubMed] [Google Scholar]
- Howes MT, et al. (2010) Clathrin-independent carriers form a high capacity endocytic sorting system at the leading edge of migrating cells. J Cell Biol 190:675–691. 10.1083/jcb.201002119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulce JJ, Cognetta AB, Niphakis MJ, Tully SE, Cravatt BF (2013) Proteome-wide mapping of cholesterol-interacting proteins in mammalian cells. Nat Methods 10:259–264. 10.1038/nmeth.2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar AN, Lee SJ, Twiss JL (2018) Expanding axonal transcriptome brings new functions for axonally synthesized proteins in health and disease. Neuroscientist 24:111–129. 10.1177/1073858417712668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, et al. (2013) Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495:467–473. 10.1038/nature11922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka G, Friedrich T, Davis-Turak J, Winden K, Oldham MC, Gao F, Chen L, Wang GZ, Luo R, Preuss TM, Geschwind DH (2012) Human-specific transcriptional networks in the brain. Neuron 75:601–617. 10.1016/j.neuron.2012.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koseki H, Donegá M, Lam BYH, Petrova V, van Erp S, Yeo GSH, Kwok JCF, Ffrench-Constant C, Eva R, Fawcett JW (2017) Selective rab11 transport and the intrinsic regenerative ability of CNS axons. Elife 6:e26956. 10.7554/eLife.26956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Andereggen L, Yuki K, Omura K, Yin Y, Gilbert HY, Erdogan B, Asdourian MS, Shrock C, De Lima S, Apfel UP, Zhuo Y, Hershfinkel M, Lippard SJ, Rosenberg PA, Benowitz L (2017) Mobile zinc increases rapidly in the retina after optic nerve injury and regulates ganglion cell survival and optic nerve regeneration. Proc Natl Acad Sci U S A 114:E209–E218. 10.1073/pnas.1616811114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Beyer A, Aebersold R (2016) On the dependency of cellular protein levels on mRNA abundance. Cell 165:535–550. 10.1016/j.cell.2016.03.014 [DOI] [PubMed] [Google Scholar]
- McClatchy DB, Ma Y, Liu C, Stein BD, Martínez-Bartolomé S, Vasquez D, Hellberg K, Shaw RJ, Yates JR (2015) Pulsed azidohomoalanine labeling in mammals (PALM) detects changes in liver-specific LKB1 knockout mice. J Proteome Res 14:4815–4822. 10.1021/acs.jproteome.5b00653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClatchy DB, Ma Y, Liem DA, Ng DCM, Ping P, Yates IIJ (2018) Quantitative temporal analysis of protein dynamics in cardiac remodeling. J Mol Cell Cardiol 121:163–172. 10.1016/j.yjmcc.2018.07.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163:123–133. 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DL, Blackmore MG, Hu Y, Kaestner KH, Bixby JL, Lemmon VP, Goldberg JL (2009) KLF family members regulate intrinsic axon regeneration ability. Science 326:298–301. 10.1126/science.1175737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, Jo HW, Jung J (2015) Expression profile of aminoacyl-tRNA synthetases in dorsal root ganglion neurons after peripheral nerve injury. J Mol Histol 46:115–122. 10.1007/s10735-014-9601-4 [DOI] [PubMed] [Google Scholar]
- Park SG, Ewalt KL, Kim S (2005) Functional expansion of aminoacyl-tRNA synthetases and their interacting factors: new perspectives on housekeepers. Trends Biochem Sci 30:569–574. 10.1016/j.tibs.2005.08.004 [DOI] [PubMed] [Google Scholar]
- Ramírez AI, Salazar JJ, de Hoz R, Rojas B, Gallego BI, Salinas-Navarro M, Alarcón-Martínez L, Ortín-Martínez A, Avilés-Trigueros M, Vidal-Sanz M, Triviño A, Ramírez JM (2010) Quantification of the effect of different levels of IOP in the astroglia of the rat retina ipsilateral and contralateral to experimental glaucoma. Invest Ophthalmol Vis Sci 51:5690–5696. 10.1167/iovs.10-5248 [DOI] [PubMed] [Google Scholar]
- Sánchez-Migallón MC, Valiente-Soriano FJ, Nadal-Nicolás FM, Vidal-Sanz M, Agudo-Barriuso M (2016) Apoptotic retinal ganglion cell death after optic nerve transection or crush in mice: delayed RGC loss with BDNF or a caspase 3 inhibitor. Invest Ophthalmol Vis Sci 57:81–93. 10.1167/iovs.15-17841 [DOI] [PubMed] [Google Scholar]
- Savitski MM, et al. (2018) Multiplexed proteome dynamics profiling reveals mechanisms controlling protein homeostasis resource multiplexed proteome dynamics profiling reveals mechanisms controlling protein homeostasis. Cell 173:260–274.e25. 10.1016/j.cell.2018.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiapparelli LM, Mcclatchy DB, Liu H, Sharma P, Yates JR, Cline HT (2014) Direct detection of biotinylated proteins by mass spectrometry. J Proteome Res 13:3966–3978. 10.1021/pr5002862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiapparelli LM, Shah SH, Ma Y, McClatchy DB, Sharma P, Li J, Yates JR, Goldberg JL, Cline HT (2019) The retinal ganglion cell transportome identifies proteins transported to axons and presynaptic compartments in the visual system in vivo. Cell Rep 28:1935–1947.e5. 10.1016/j.celrep.2019.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M (2011) Global quantification of mammalian gene expression control. Nature 473:337–342. 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- Sekine Y, Lin-Moore A, Chenette DM, Wang X, Jiang Z, Cafferty WB, Hammarlund M, Strittmatter SM (2018) Functional genome-wide screen identifies pathways restricting central nervous system axonal regeneration. Cell Rep 23:415–428. 10.1016/j.celrep.2018.03.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellés-Navarro I, Ellezam B, Fajardo R, Latour M, McKerracher L (2001) Retinal ganglion cell and nonneuronal cell responses to a microcrush lesion of adult rat optic nerve. Exp Neurol 167:282–289. 10.1006/exnr.2000.7573 [DOI] [PubMed] [Google Scholar]
- Shen W, Liu HH, Schiapparelli L, McClatchy D, He H. y, Yates JR, Cline HT (2014) Acute synthesis of CPEB is required for plasticity of visual avoidance behavior in Xenopus. Cell Rep 6:737–747. 10.1016/j.celrep.2014.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka T, Jung H, Jung J, Turner-Bridger B, Ohk J, Lin JQ, Amieux PS, Holt CE (2016) Dynamic axonal translation in developing and mature visual circuits. Cell 166:181–192. 10.1016/j.cell.2016.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speers AE, Cravatt BF (2009) Activity-based protein profiling (ABPP) and click chemistry (CC)-ABPP by MudPIT mass spectrometry. Curr Protoc Chem Biol 1:29–41. 10.1002/9780470559277.ch090138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran NM, Shekhar K, Whitney IE, Jacobi A, Benhar I, Hong G, Yan W, Adiconis X, Arnold ME, Lee JM, Levin JZ, Lin D, Wang C, Lieber CM, Regev A, He Z, Sanes JR (2019) Single-cell profiles of retinal ganglion cells differing in resilience to injury reveal neuroprotective genes. Neuron 104:1039–1055.e12. 10.1016/j.neuron.2019.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cameron EG, Li J, Stiles TL, Kritzer MD, Lodhavia R, Hertz J, Nguyen T, Kapiloff MS, Goldberg JL (2015) Muscle A-kinase anchoring protein-α is an injury-specific signaling scaffold required for neurotrophic- and cyclic adenosine monophosphate-mediated survival. EBioMedicine 2:1880–1887. 10.1016/j.ebiom.2015.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PA, Harder JM, Foxworth NE, Cochran KE, Philip VM, Porciatti V, Smithies O, John SWM (2017) Vitamin B3 modulates mitochondrial vulnerability and prevents glaucoma in aged mice. Science 355:756–760. 10.1126/science.aal0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda M, Tanaka Y, Ryu M, Tsuda S, Nakazawa T (2014) RNA sequence reveals mouse retinal transcriptome changes early after axonal injury. PLoS One 9:e93258. 10.1371/journal.pone.0093258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuet KP, Tirrell DA (2014) Chemical tools for temporally and spatially resolved mass spectrometry-based proteomics. Ann Biomed Eng 42:299–311. 10.1007/s10439-013-0878-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protein IDs found in 1 day control samples, 5 day control samples, and/or naive total retina. Download Figure 2-1, XLSX file (73.6KB, xlsx) .
All replicates of AHA retinas from either 1 day or 5 day injection experiments. Download Figure 2-2, XLSX file (100.6KB, xlsx) .
Stouffer's Z-score changes of new protein synthesis in the retina between 1 and 5 days. Download Figure 2-3, XLSX file (46.3KB, xlsx) .
Replicates of changes to AHA-labeled proteins transported to optic nerve with and without optic nerve crush (Z-score normalized). Download Figure 3-1, XLSX file (15.2KB, xlsx) .
AHA-labeled proteins transported to optic nerve characterized as part of nuclear cellular component by Gene Ontology. Download Figure 3-2, XLSX file (12.8KB, xlsx) .
Plasmids used in neurite outgrowth assay. Vectors created by VectorBuilder. Maps can be retrieved using IDs at https://en.vectorbuilder.com/. Download Figure 4-1, XLSX file (11.2KB, xlsx) .
Data Availability Statement
Data that support the findings of this study are available in this article and the extended data files; all raw data are available from the corresponding author on reasonable request.