Abstract
A facultatively filamentous bacterium was isolated from eutrophic lake water and was identified as Flectobacillus sp. strain MWH38 (a member of the Cytophaga-Flavobacterium-Bacteroides phylum) by comparative 16S rRNA gene sequence analysis. Filament formation by Flectobacillus sp. strain MWH38 and filament formation by Flectobacillus major, the closest known relative of strain MWH38, were studied in chemostat cultures under grazing pressure by the bacterivorous flagellate Ochromonas sp. strain DS and without predation at several growth rates. The results clearly demonstrated that filament formation by the two flectobacilli is growth rate controlled and thus independent of the presence of a predator. However, flagellate grazing positively influenced bacterial growth rates by decreasing bacterial biomass and thus indirectly stimulated filament formation. The results of investigations of cell elongation and filament formation by Comamonas acidovorans PX54 (a member of the β subclass of the class Proteobacteria) supported the recent proposal that in this species the mechanism of filament formation is growth rate controlled. The finding that the grazing defense mechanism consisting of filament formation is growth rate controlled in the flectobacilli investigated and C. acidovorans PX54 (i.e., in bacteria belonging to divergent evolutionary phyla) may indicate that this mechanism is a phylogenetically widely distributed defense strategy against grazing.
Bacteria are important players in the global cycles of carbon, nitrogen, phosphorus, and other elements. In aquatic ecosystems, the substrate supply, phages, bacterivorous nanoflagellates, and other predators influence bacteria. Grazing pressure by bacterivorous flagellates can control bacterial abundance (2, 31) and may potentially influence the species composition of bacterial communities (12, 14, 15, 34). Thus, bacterivorous predators may theoretically influence the overall role of bacteria in nutrient cycles. However, bacterial defense mechanisms against grazing may diminish the influence of bacterivorous predators.
A number of field and experimental reports on the occurrence and presence of filamentous bacteria under strong protozoan grazing pressure conditions (13, 17–20, 28, 32, 35, 36) have suggested that this bacterial morphotype represents an ecologically important bacterial defense strategy against predation by bacterivorous protozoans. It is known that the size of filamentous bacteria exceeds the cell size range that most bacterivorous protists can feed on (4, 9, 33), which protects the bacteria from the protozoan predators. Several field studies and experiments have shown that filamentous bacteria occur after a significant increase in protozoan grazing pressure (15, 19, 20, 29, 34). However, most of these studies did not reveal whether the morphology of nonfilamentous bacteria changed or permanently filamentous bacteria became more abundant. However, quick changes in bacterial communities consisting of small or medium-sized bacteria to communities dominated by filamentous bacteria after a significant increase in protistan grazing pressure have been observed several times (18, 20, 29, 34), and the data suggest that such changes are primarily the result of changes in the morphology of normal-sized bacteria. Pernthaler et al. (29) observed that medium-sized bacteria belonging to the β subclass of the class Proteobacteria (β-Proteobacteria) responded to the addition of a bacterivorous flagellate by developing inedible filaments. These authors speculated that chemical stimuli released by the predator might have triggered the filament formation. We observed the formation of morphologically similar filaments by Comamonas acidovorans PX54 (β-Proteobacteria) after the onset of grazing by a bacterivorous flagellate (15). Experiments without grazing pressure revealed that the formation of filaments by C. acidovorans PX54 was growth rate dependent and thus independent of a chemical stimulus released by the predator.
In this paper we describe on the interaction of a facultatively filamentous, grazing-protected freshwater bacterium, Flectobacillus sp. strain MWH38 (a member of the Cytophaga-Flavobacterium-Bacteroides phylum), with the bacterivorous flagellate Ochromonas sp. strain DS under chemostat conditions. The parameters that controlled filament formation and thus the parameters that controlled the grazing protection mechanism of strain MWH38 were analyzed and compared to the grazing defense strategies of the closest known relative of strain MWH38, Flectobacillus major, and of a filament-forming strain belonging to a different phylum, C. acidovorans PX54 (β-Proteobacteria).
MATERIALS AND METHODS
Microbial strains.
A surface water sample from eutrophic Lake Heidbergsee (Braunschweig, Germany) was taken on 1 January 1995 and adapted, in 2°C steps over a period of 24 h, from the in situ temperature (5.5°C) to room temperature (21°C). Fifty milliliters of the sample was filtered through a 3.0-μm-pore-size polycarbonate filter (Nuclepore) and mixed with 200 ml of an axenic culture (containing no living bacteria) of the bacterivorous nanoflagellate Ochromonas sp. strain DS (15). The mixed culture, which initially contained 4.8 × 104 flagellates ml−1, was incubated at room temperature without shaking. At 24-h intervals 50% of the enrichment culture was exchanged with sterile NSY medium (15) containing 20 mg of complex substrate (equal amounts of nutrient broth, soyotone, and yeast extract) per liter. In a second enrichment step, 24 subcultures (1:1 mixtures of NSY medium and axenic flagellate culture) were established. Each subculture received an inoculum containing ca. 15 bacteria. After 48 h of incubation, samples from nine filament-containing subcultures were plated onto NSY agar, and colonies were inspected for filamentous bacteria by performing a microscopic analysis. The isolate obtained from a single colony was identified as Flectobacillus sp. strain MWH38 (see below).
Flectobacillus sp. strain MWH38, F. major DSMZ 103T (= ATCC 29496T = GromovT), C. acidovorans PX54 (7, 15), and Vibrio sp. strain CB5 (15) were grown on NSY medium (15). The bacterivorous nanoflagellate Ochromonas sp. strain DS was maintained in axenic culture (15) and was used as a model organism for bacterivorous flagellates.
Use of the term filament.
In this paper we use the term filament for both threadlike bacteria (individual strongly elongated cells [e.g., C. acidovorans PX54 cells]) and thin bacterial chains (e.g., Flectobacillus sp. strain MWH38 chains) that are >5 μm long. In addition, we refer to filaments that are 5 to 10 μm long as short filaments and filaments that are >10 μm long as long filaments.
Determination and analysis of the 16S rRNA gene sequence of Flectobacillus sp. strain MWH38.
The genomic DNA of strain MWH38 was extracted and the 16S rRNA gene sequence of this strain was determined and analyzed as described previously (25).
Chemostat studies.
Two different types of chemostat experiments were performed (Table 1). In the first type of experiments the influence of grazing on the morphology of bacteria was examined, and in the second type the influence of growth rate on the morphology of bacteria was examined. In predation experiments, the bacterial strain was cultured initially in the absence of the flagellate Ochromonas sp. strain DS and then in the presence of this flagellate. During these experiments, the chemostat dilution rate was maintained at 0.5 day−1, and thus the growth rate in the flagellate-free phase of the experiments was 0.5 day−1 (doubling time, 33.3 h). In the growth rate chemostat experiments, bacterial strains were grown in flagellate-free chemostat cultures at several growth rates. In the case of Flectobacillus sp. strain MWH38, the two types of experiments were carried out with different types of chemostat equipment. The predation experiment was performed in a single 2-liter reactor (15), and the growth rate experiment was performed in two parallel 1.5-liter reactors, each of which had a working volume of 500 ml. The two experiments with F. major and the one experiment with C. acidovorans PX54 were carried out by using the chemostat system with two parallel reactors. All chemostat cultures were grown at 15°C and were mixed by aeration with sterile air. In most experiments NSY medium containing 9 mg of a complex substrate per liter was used; in the F. major growth rate experiment 4.5 mg of the complex substrate per liter was used (15). Both reactors of the chemostat system were fed from the same reservoir but with separate pumps, which allowed us to use different dilution rates for the two reactors. Samples were taken at 24-h intervals, fixed with formaldehyde (final concentration, 2%), and stored at 4°C until they were analyzed.
TABLE 1.
Overview of the chemostat experiments
| Expt | No. of reactors | Working vol of reactor(s) (liters) | Sterilization methoda | Bacteria | Treatmentb |
|---|---|---|---|---|---|
| Flectobacillus sp. strain MWH38, predation | 1 | 2.0 | Filter sterilized | Flectobacillus sp. strain MWH38 | Ochromonas sp. strain DS |
| Flectobacillus sp. strain MWH38, growth rate | 2c | 0.5 | Autoclaved | Flectobacillus sp. strain MWH38 | Dilution rate |
| F. major, predation | 2c | 0.5 | Autoclaved | F. major DSMZ 103T | Ochromonas sp. strain DS |
| F. major, growth rate | 2c | 0.5 | Autoclaved | F. major DSMZ 103T | Dilution rate |
| C. acidovorans PX54, growth rate | 2c | 0.5 | Autoclaved | C. acidovorans PX54 | Dilution rate |
The same medium was used in all experiments, but two methods of sterilization were used.
In the first phase of all experiments the bacteria were grown at growth rate of 0.5 day−1, and then the flagellate Ochromonas sp. strain DS was introduced or the dilution rate was increased.
Two parallel reactors were fed from the same reservoir.
Batch culture experiments.
The growth of the three bacterial species in batch cultures was monitored to study the dependence of filament formation on growth stage. Bacteria were grown in 250-ml flasks containing 100 ml of NSY medium. Different substrate concentrations were used; the substrate concentration used for flectobacilli was 1 g liter−1, and the substrate concentration used for C. acidovorans PX54 was 9 mg liter−1. The culture flasks were shaken at 100 rpm.
Grazing experiments.
Grazing of Ochromonas sp. strain DS on two Flectobacillus sp. strain MWH38 populations that were pregrown in chemostats with different doubling times (33.3 and 16.6 h) and grazing on a Vibrio sp. strain CB5 population from the stationary phase of a batch culture were studied in batch culture experiments. An axenic flagellate culture was divided into subcultures. The subcultures were each enriched with one of the bacterial populations (three replicates per species, 21°C, no shaking). Samples were taken at zero time and after 0.5, 1.5, 3.5, and 6.5 h and were fixed with formaldehyde (final concentration, 2%). Decreases in bacterial numbers were determined by epifluorescence microscopy.
Determination of microbial abundance and cell size.
To determine the total bacterial abundance, the percentage of bacterial filaments, and the flagellate abundance, fixed subsamples were stained with 0.1% (wt/vol) DAPI (4′,6-diamidino-2-phenylindole), filtered onto 0.2-μm-pore-size polycarbonate filters, and enumerated by epifluorescence microscopy. The lengths of single cells and filaments were measured by using digitized images produced with an epifluorescence microscope (Zeiss Axioplan 2) equipped with a charge-coupled device camera (Sony model MC-3215/PI). Sizes were determined by using the analySIS Pro software (Soft-Imaging Software GmbH, Münster, Germany).
Nucleotide sequence accession number.
The nearly full-length 16S rRNA gene sequence of Flectobacillus sp. strain MWH38 has been deposited in the EMBL database under accession no. AJ011917.
RESULTS
Taxonomy.
A facultatively filamentous strain, identified as Flectobacillus sp. strain MWH38 (see below), was isolated from eutrophic lake water. Flectobacillus sp. strain MWH38 formed smooth, convex, pink colonies on NSY agar. The colonies were similar to those of F. major DSMZ 103T, although the cell morphology of strain MWH38 was different from that of F. major grown under similar conditions in batch or chemostat culture. The cells of isolate MWH38 were straight rods that were 2 to 10 μm long and 0.4 to 0.6 μm wide. This isolate could grow as single cells and as long filaments that were at least 50 μm long. The filaments were chainlike and consisted of a variable number of cells. No motility was observed in samples of colonies from agar plates or samples from batch and chemostat cultures.
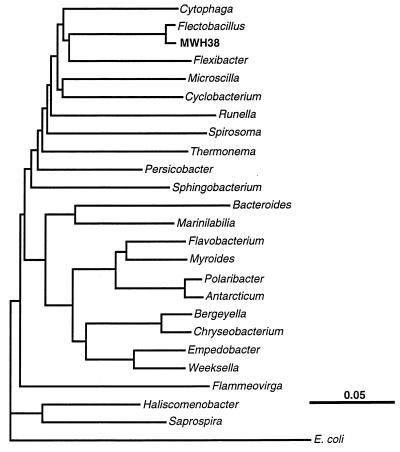
PCR amplification and sequencing of the 16S rRNA gene of strain MWH38 allowed us to determine approximately 97% of the complete sequence. Sequence comparisons (23, 37) demonstrated that strain MWH38 is related to bacteria belonging to the Cytophaga-Flavobacterium-Bacteroides phylogenetic lineage (27) and is most closely related to F. major (Fig. 1), the type species of the genus Flectobacillus (22). Although 16S ribosomal DNA sequence comparisons alone do not define taxonomic relationships between bacteria, the level of 16S ribosomal DNA sequence similarity (95.4%) between strain MWH38 and F. major is within the range of sequence similarity values that are characteristic for different species of a genus. It is likely that strain MWH38 is a member of a new species of the genus Flectobacillus or of a closely related genus, and for the purposes of this study, it was considered Flectobacillus sp. strain MWH38.
FIG. 1.
Dendrogram showing the estimated phylogenetic position of Flectobacillus sp. strain MWH38 within the Cytophaga-Flavobacterium-Bacteroides evolutionary lineage. The dendrogram was generated by using the FITCH algorithm of the PHYLIP package (8) and evolutionary distances (16) calculated from 16S rRNA (and rRNA gene) sequence dissimilarities. The positions of the genera of the Cytophaga-Flavobacterium-Bacteroides evolutionary lineage were calculated from the sequences of the type strains of the type species of the genera.
Influence of grazing and growth rate on morphology of Flectobacillus sp. strain MWH38.
Two chemostat studies (Table 1 and Fig. 2 and 3) and one batch culture study were done to investigate the influence of flagellate grazing and growth rate on the morphology of Flectobacillus sp. strain MWH38.
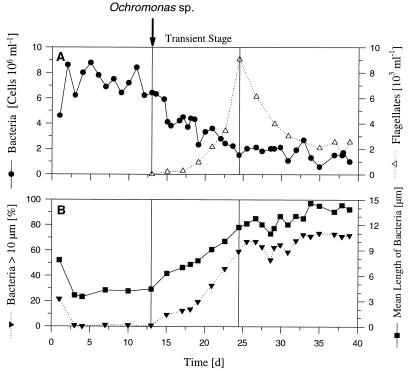
FIG. 2.
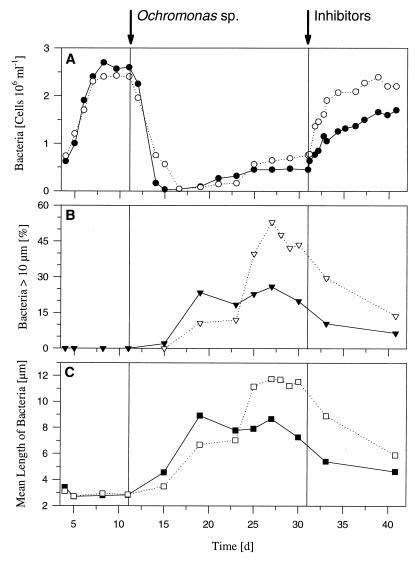
Results of the predation chemostat experiment performed with Flectobacillus sp. strain MWH38 and the bacterivorous flagellate Ochromonas sp. strain DS (introduced on day 13). (A) Influence of flagellate grazing on bacterial numbers. (B) Influence of the predator on the mean length of bacteria (including single cells and filaments) and the percentage of long filaments (length, >10 μm). d, days.
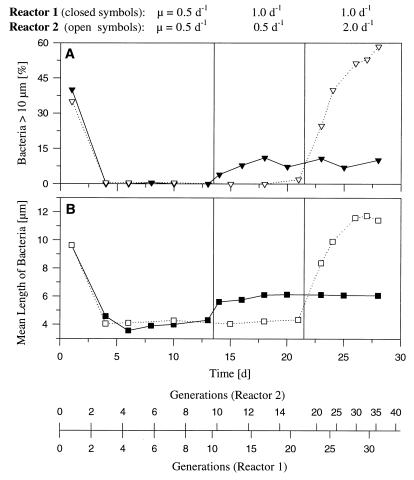
FIG. 3.
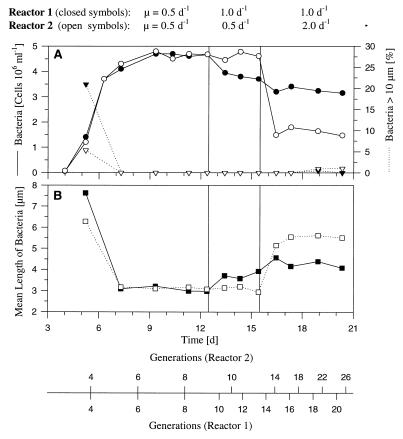
Results of chemostat experiment performed with Flectobacillus sp. strain MWH38, showing the influence of growth rate on the morphology of the strain. Initially, bacteria were grown in both reactors at a growth rate of 0.5 day−1. On day 13, the growth rate of the bacteria in reactor 1 was increased to 1.0 day−1, and 9 days later the growth rate of the bacteria in reactor 2 was increased to 2.0 day−1. (A) Influence of growth rate on the percentage of long filaments (length, >10 μm) in the population. (B) Influence of growth rate on the mean bacterial length (including single cells and filaments). d, days.
(i) Predation experiment.
The chemostat study performed to determine the influence of grazing was initiated with a flagellate-free phase in which the bacteria grew at a rate of 0.5 day−1 (Fig. 2). During this phase, most of the bacteria were single celled (89.1% of the cells) or components of short filaments consisting of two cells (10.9% of the cells) (Fig. 4). After the bacterivorous flagellate Ochromonas sp. strain DS was introduced, the total bacterial cell number decreased, most bacterial cells were elongated, and the fraction of the cells that were components of short and long filaments increased (Fig. 2 and 4). Bacteria that were longer than 10 μm occurred due to this filament formation. After the mean length (single cells and filaments) increased to approximately 12 μm, a steady state was established, in which the bacterial cell number was more or less constant and the mean bacterial length was constant. During the transient phase, flagellate grazing reduced the bacterial cell concentration by 74% to a mean value of 1.9 × 106 ± 0.5 × 106 cells ml−1 (flagellate-controlled steady state). However, the bacterial biomass decreased by only 56% because the decrease in cell number was partially compensated for by the increase in the mean cell length of Flectobacillus sp. strain MWH38 cells.
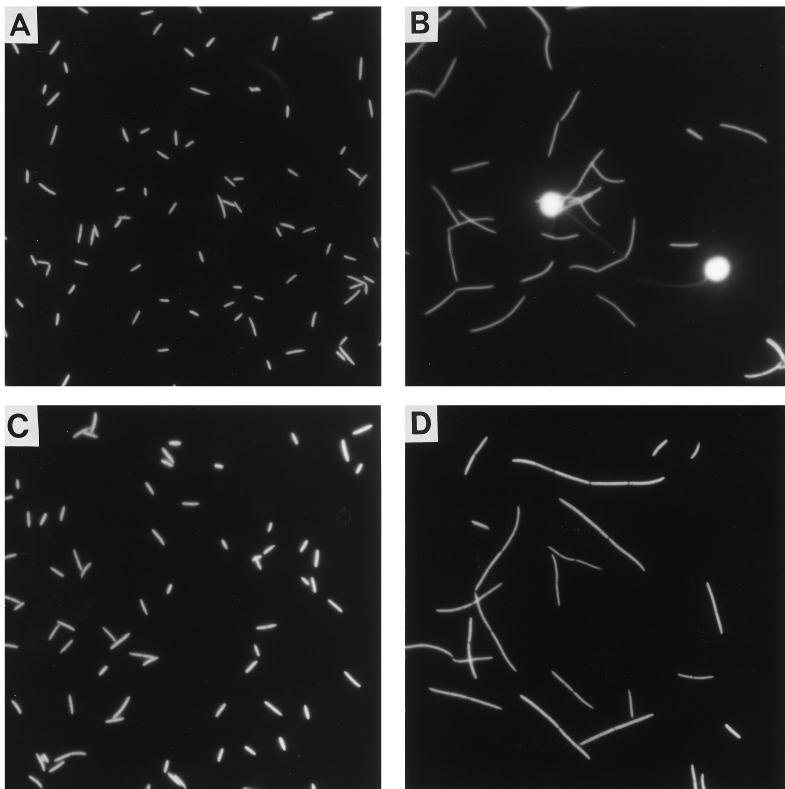
FIG. 4.
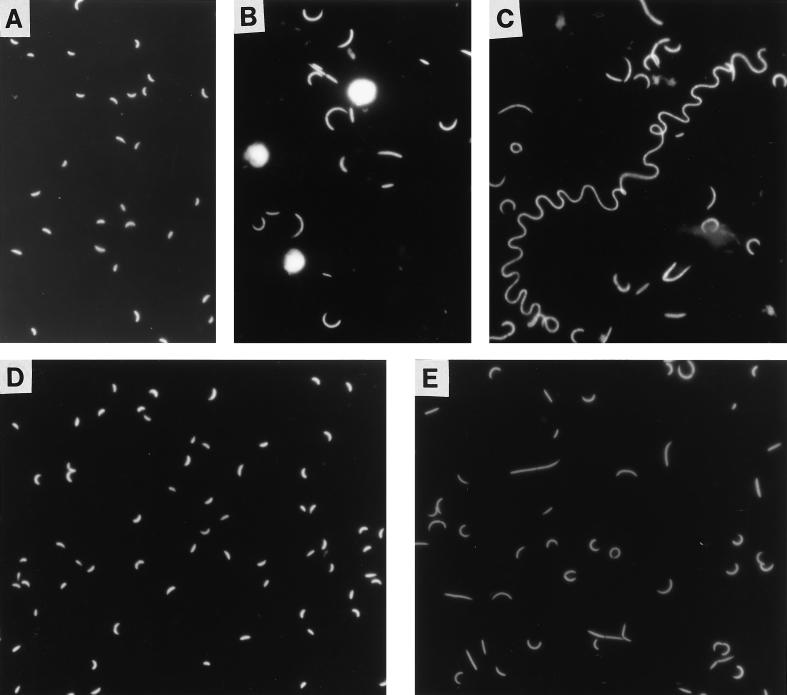
Photomicrographs of Flectobacillus sp. strain MWH38 populations grown in chemostats with and without predation and at different growth rates. (A and B) Flectobacillus sp. strain MWH38 from the predation experiment before (A) and after (B) inoculation with the flagellate. Before the start of predation, the bacteria were cultured at a growth rate of 0.5 day−1. (C and D) Flectobacillus sp. strain MWH38 from the growth rate chemostat experiment at growth rates of 0.5 day−1 (C) and 2.0 day−1 (D). Flagellates were not included in this experiment.
(ii) Growth rate experiment.
Marked changes in the morphology of Flectobacillus sp. strain MWH38 cultured in the absence of the flagellates were observed after the growth rate was increased from 0.5 day−1 to 1.0 or 2.0 day−1 (Fig. 3 through 5). When Flectobacillus sp. strain MWH38 was cultured without flagellates at a growth rate of 2.0 day−1, the bacterial morphology (Fig. 4), the cell size distribution (Fig. 5B and D), the mean bacterial cell length (Fig. 3), and the percentage of bacteria longer than 10 μm (Fig. 3) were very similar to data observed in the flagellate-controlled steady state in the predation experiment (Fig. 2). The bacterial size distributions differed slightly for the classes of cells between 1.5 and 5.5 μm long (Fig. 5B and D). The flagellate-grazed population contained lower percentages of cells in this size range; these findings may indicate the maximum size of bacteria which are potentially edible by Ochromonas sp. strain DS.
FIG. 5.
Cell size distributions of chemostat-grown Flectobacillus sp. strain MWH38, F. major, and C. acidovorans PX54 with and without flagellate predation and at different growth rates in flagellate-free culture. Most of the distributions shown were based on three to eight chemostat samples; the exceptions were the distributions shown in panels K (two samples) and L (one sample). Data for the latter two distributions were obtained from a chemostat experiment described by Hahn and Höfle (15). This experiment was carried out by using the same experimental conditions as those used for the chemostat experiments performed in this study. Note that the graph in panel F represents only the last 5 days of the flagellate-controlled phase of the grazing experiment (Fig. 7), while the graph in panel H represents the first 4 days after a steady state was established in the growth rate experiment (Fig. 8). d, day.
(iii) Batch culture.
In the batch culture experiment performed to determine the effect of growth stage on the morphology of Flectobacillus sp. strain MWH38, a high degree of phenotypic plasticity was observed. During exponential growth, the percentage of the cells in filament form ranged from 80 to 90%. The chainlike filaments started to decay after the exponential phase, and the percentage of single cells increased to 100% during the stationary phase. The mean length of the bacterial cells increased during the exponential phase and reached its highest value at the end of this phase. After this, the mean cell length decreased, as did the percentage of cells in filaments.
Grazing of Ochromonas sp. strain DS on Flectobacillus sp. strain MWH38 populations precultivated at different growth rates.
The two Flectobacillus sp. strain MWH38 populations that were pregrown at growth rates of 0.5 and 1.0 day−1 differed in the degree of filamentation (25.5 and 65.0% of the cells were in filaments that were >5 μm, long, respectively). The decreases in cell numbers due to flagellate grazing were different for the two populations during the first 1.5 h of the experiment. On average, the Flectobacillus sp. strain MWH38 population that was pregrown at a higher growth rate was grazed upon at a 3.2-fold-lower rate during the first 1.5 h of the experiment. After this, the filaments in both populations decayed rapidly, and the cell numbers in the two populations decreased at similar rates. The population of Vibrio sp. strain CB5 consisted exclusively of medium-sized and thus readily edible cells. The number of Vibrio sp. strain CB5 cells decreased during the first 1.5 h of the experiment 8 and 26 times faster than the number of Flectobacillus sp. strain MWH38 cells in the two populations.
Influence of grazing and growth rate on the abundance and morphology of F. major. (i) Predation experiment.
Almost all cells of F. major grew in the first flagellate-free phase of the chemostat experiment as single curved rods (Fig. 6). Only a few cells (<0.1%) grew as straight rather than curved rods, and very few (≪0.1%) formed short filaments consisting of two to five cells. Flagellate grazing reduced the total cell numbers in both reactors by 98 to 99%, to extremely low concentrations (approximately 4 × 104 cells ml−1) (Fig. 7). After the marked reduction in bacterial cell numbers, larger curved and straight cells occurred. The mean length of the bacteria increased slowly, and short filaments (lengths, 5 to 10 μm) occurred. During this phase, most cells had a C-shaped morphology, which has been described as typical for F. major (22). In addition, high percentages of long filaments occurred in both reactors at later stages (Fig. 6 and 7). Along with the occurrence of such filaments, the cell numbers increased rapidly. The numbers of bacteria, the mean lengths of the bacteria (including single cells and filaments), and the percentages of long filaments (lengths, >10 μm) were different in the two reactors, although the bacterial population with the larger forms contained more cells (Fig. 7). After the flagellates were eliminated by specific inhibitors, a further increase in cell number was observed, and the percentage of filaments decreased.
FIG. 6.
Photomicrographs of F. major cells grown in chemostats with and without predation and at different growth rates. (A through C) F. major from the predation experiment before inoculation with the flagellate (A) and 4 days (B) and 15 days (C) after inoculation with the flagellate. Before the start of predation, the bacteria were cultured at a growth rate of 0.5 day−1. Three Ochromonas sp. strain DS cells are shown in panel B. (D and E) F. major from the growth rate chemostat experiment at growth rates of 0.5 day−1 (D) and 2.0 day−1 (E). Flagellates were not included in this experiment.
FIG. 7.
Results of the predation chemostat experiment performed to determine the influence of grazing by the bacterivorous flagellate Ochromonas sp. strain DS on the morphology of F. major. Bacteria were grown in two parallel reactors, reactor 1 (solid symbols) and reactor 2 (open symbols), at the same growth rate, 0.5 day−1. Both reactors were inoculated on day 11 with the flagellate, and on day 31 flagellates were eliminated from both reactors by treatment with specific inhibitors (cycloheximide and colchicine). In the period between introduction and elimination of flagellates, the predators established populations (data not shown) comparable to the populations in other chemostat experiments. (A) Influence of flagellate grazing on bacterial numbers. (B) Influence of flagellate grazing on the percentage of long filaments (length, >10 μm) in the population. (C) Influence of flagellate grazing on the mean length of bacteria (including single cells and filaments). d, days.
(ii) Growth rate experiment.
In the chemostat study, F. major reacted to the increase in the growth rate from 0.5 day−1 to 1.0 or 2.0 day−1 with an increase in cell length (Fig. 8). Although at the higher growth rates most cells grew as single cells (Fig. 6), the cell morphology changed slightly. At the higher growth rates, more straight cells were observed, although cells with more pronounced curves and short filaments occurred. In total, the F. major population grown at a rate of 2.0 day−1 resembled the population observed in the grazing chemostat experiment during the period between the marked reduction in cell number and the occurrence of large filaments (see above). In contrast to what was observed in the late stages of the grazing experiment, only 1% of the cells were in long filaments in the growth rate experiment (Fig. 5H). However, the lack of formation of long filaments may have been a result of a period of growth at a rate of 2.0 day−1 that was too short. In both chemostat experiments, F. major needed 7 days from inoculation to reach the first steady state (Fig. 7 and 8). Such a long incubation period was not observed in any of the other 15 chemostat experiments inoculated in the same way and performed under similar experimental conditions (14, 15). This suggests that the conditions used for the experiments were not optimal for the growth of the strain.
FIG. 8.
Influence of growth rate on the cell morphology of F. major in two parallel chemostat reactors, reactor 1 (solid symbols) and reactor 2 (open symbols). (A) Changes in bacterial cell number and percentage of long filaments (length, >10 μm) in the population with changes in growth rate. (B) Influence of growth rate on the mean length of bacteria (including single cells and filaments). d, days.
(iii) Batch culture.
In contrast to Flectobacillus sp. strain MWH38, only short filaments were observed in batch cultures of F. major. During the exponential phase, the cell shape ranged from half-closed circles to nearly fully closed circles (diameters, 4 to 6 μm). Most cells had shapes that were similar to the letter C and resembled the cells cultured in the chemostat at the higher growth rate. A minority of cells formed short filaments consisting of two or three cells (often with S-like shapes). During the stationary phase, the cell lengths and degrees of curvature decreased. Fifteen hours after the end of the exponential phase, weakly curved rods that were 2 to 3 μm long dominated.
Influence of growth rate on the morphology of C. acidovorans PX54 studied in chemostat culture.
At dilution rates ranging from 0.5 to 3.5 day−1 the mean length of C. acidovorans PX54 cells increased with the growth rate. This resulted in an increase in the percentage of cells that were more than 2.4 μm long to a maximum of approximately 50% (Fig. 9). Bacteria of this size are assumed to be protected by their size from predation by bacterivorous flagellates (28). In contrast to populations grown in the same medium but in batch culture (see below), only a small percentage of short filaments occurred in the chemostat culture. Additional stepwise increases in the growth rate to 4.0 and 4.5 day−1 resulted in decreases in the mean cell length and slight decreases in the percentage of cells longer than 2.4 μm. An additional increase in the growth rate to 5.0 day−1 resulted in washing out of the bacteria.
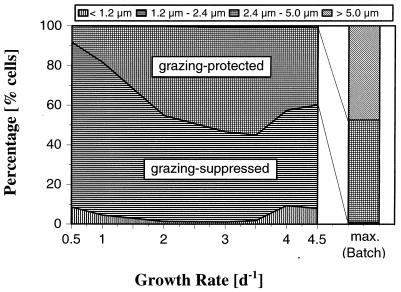
FIG. 9.
Size class distribution of C. acidovorans PX54 cells cultured in chemostats at different growth rates (0.5, 1.0, 2.0, 3.0, 3.5, 4.0, and 4.5 day−1) and in batch culture at the maximum growth rate (bar on the right). The same medium with the same substrate concentration was used for chemostat and batch cultures. The bacterial populations were divided into size classes with different sensitivities to grazing by bacterivorous flagellates (28). Because no C. acidovorans PX54 cells were smaller than 0.4 μm, all cells smaller than 1.2 μm were considered edible bacteria. In chemostat cultures the percentage of short filaments (length, 5 to 10 μm) increased with growth rate from 0% (growth rate, 0.5 day−1) to 1% (growth rate, 4.5 day−1) and was thus much smaller than the percentage observed in samples obtained from the exponential stage of batch cultures. d, day.
For comparison, C. acidovorans PX54 was grown in batch cultures with the same medium used for chemostat experiments and at the same temperature. So that there could be very long exponential growth phases, the cultures were inoculated with 5-μl samples from a preculture (chemostat culture with a growth rate of 3.5 day−1). In the late exponential growth phase (36 h after inoculation), the mean length of cells was twofold greater than the maximum value observed in a chemostat culture. In addition, batch cultures contained much higher percentages of short filaments, although long filaments also were present (Fig. 9).
Comparison of the influence of growth rate on Flectobacillus spp. and C. acidovorans PX54.
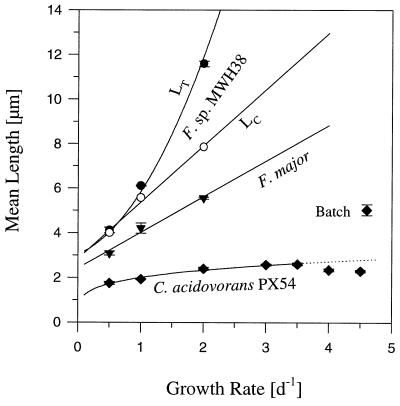
The mean cell lengths (Lc) of Flectobacillus sp. strain MWH38 and F. major increased linearly as the growth rate (μ) increased (Fig. 10). The increase in the Lc of Flectobacillus sp. strain MWH38 (Lc = 2.53 × μ + 2.86; n = 3; r = 0.97) was much more marked than the increase in the Lc of F. major (Lc = 1.6 × μ + 2.42; n = 3; r = 0.99). The mean total length (LT) of Flectobacillus sp. strain MWH38 single cells and filaments increased exponentially as the growth rate increased (LT = e(0.7 × μ + 1.07); n = 3; r = 0.999). The Lc of C. acidovorans PX54 increased as the growth rate increased in the range from 0.5 to 3.5 day−1, but in contrast to the Flectobacillus species the increase was not linear (Lc = 2.01 × μ0.22; n = 5; r = 0.99). In addition, the Lc of C. acidovorans PX54 decreased slightly at higher growth rates.
FIG. 10.
Influence of growth rate on the length of Flectobacillus sp. strain MWH38, F. major, and C. acidovorans PX54 cells and filaments in flagellate-free chemostat cultures. C. acidovorans PX54 never formed chainlike filaments, and only a small percentage (1 to 3%) of F. major cells formed chainlike filaments consisting of two or three cells. High percentages of Flectobacillus sp. strain MWH38 cells formed chainlike filaments at high growth rates (Fig. 3 and 4), and the data for this species distinguish between the influence of growth rate on cell length (Lc) (○) and the influence of growth rate on the total length of bacteria (single cells and filaments (LT) (•). The curves are the result of a regression analysis (see text). In addition, the mean lengths of C. acidovorans PX54 cells observed in the exponential phase of batch cultures are shown. d, day.
DISCUSSION
We performed chemostat and batch culture experiments and demonstrated that the defense mechanism consisting of filament formation is growth rate controlled in the two flectobacilli examined and C. acidovorans PX54. We concluded that in the experiments in which there was grazing pressure (Fig. 2 and 7), filament formation was not a direct reaction to the presence of the flagellate (e.g., filament formation was not induced by a chemical trigger released by the flagellate) but rather was a reaction to an increase in the growth rate caused by the flagellate grazing. In these experiments, flagellate grazing reduced the bacterial cell numbers and thus bacterial biomass. In chemostat cultures, such reductions resulted in increases in the growth rates of the remaining bacteria (15).
In the case of Flectobacillus sp. strain MWH38, we observed very similar size distributions in the flagellate-grazed population and the population grown at a high growth rate (Fig. 4 and 5). With the other two organisms investigated we observed differences between the size distributions for the two experimental conditions (Fig. 5). However, these differences could have been caused by other influences. While grazing stimulated filament formation by increasing the bacterial growth rate (an indirect influence), it may also have influenced the observed size distributions through the elimination of remaining smaller, and thus edible, cells (a direct influence). On the other hand, F. major cultured in the presence of flagellates may have reached higher growth rates than the highest growth rate (2.0 day−1) used for the population in the growth rate experiment, which may have contributed to the more marked shift in the size distribution in the predation experiment.
In contrast to the two flectobacilli, C. acidovorans PX54 did not respond to increases in the growth rate and predation with a shift of nearly the whole population to a larger size. In the predation experiment (15) (Fig. 5I and J), the size of the majority of the cells increased slightly, and only a minority of the cells increased markedly in size, thus expanding the size distribution over a wide range (Fig. 5J). The prerequisite for the observed formation of at least 50-μm-long threadlike filaments by a population of 1- to 4-μm-long C. acidovorans PX54 cells (15) was a 12- to 50-fold increase in biomass without cell division. This required the suppression of cell division for several generation times. In chemostat systems, filament formation by this mechanism is observable only if the growth rate of the bacterial population is significantly higher than the dilution rate of the system. If the dilution and growth rates were equal, cells with suppressed cell division would be washed out before filament formation was recognizable. In steady-state chemostat cultures, such as the chemostat cultures used in the growth rate experiment performed with C. acidovorans PX54 (Fig. 9 and 10), the dilution rate and growth rate are the same. This is not true in chemostat experiments with flagellate predation, in which the bacterial growth rate depends on the dilution rate plus the grazing loss rate (15), and thus the growth rate is higher than the dilution rate. This means that chemostats select against filament formation in growth rate experiments performed with species which do not respond to increases in the growth rate with uniform elongation of cells. However, in predation experiments, selection against filament formation is compensated for by an increase in the bacterial growth rate compared with the dilution rate, due to grazing. Thus, differences in the bacterial growth rate in relation to the chemostat dilution rates in experiments with and without flagellate grazing explain the differences in filament formation by C. acidovorans PX54 observed under the two experimental conditions used (Fig. 5J and L). However, the expansion of the size distribution of C. acidovorans PX54 and the occurrence of short filaments with increasing growth rate (Fig. 5 and 9) confirmed that growth rate control of filament formation occurred.
Comparison of experimental and environmental conditions.
Chemostat experiments were carried out by using low substrate concentrations (9 mg liter−1). Assuming that the carbon content of the complex substrate used was 50%, the dissolved organic carbon concentration of the chemostat medium was within the range of concentrations (4 to 10 mg liter−1) reported for eutrophic Lake Plußsee (26, 39), the natural habitat of C. acidovorans PX54 (7, 39). The bacterial concentrations observed in chemostats without flagellates (2.5 × 106 to 10 × 106 cells ml−1) were within the typical range (106 to 107 cells ml−1) reported for meso- to eutrophic lakes (2, 31). Moreover, the bacterium-to-flagellate ratios observed in our experiments (200 to 1,200 bacteria flagellate −1 fell into the typical range (102 to 104 bacteria flagellate−1; mean, 103) found in marine and freshwater habitats (2, 31). The growth rates of C. acidovorans PX54 in Lake Plußsee were determined by dilution cultures in a single study to be 2.6 to 3.1 day−1 (15°C) (39), and in a seasonal study of the same lake the growth rates of the entire bacterial community ranged from ca. 0.2 to 2.4 day−1 (3). Thus, the range of growth rates used for our investigations covered the range of rates found in the natural habitat of one of the species tested.
General considerations on grazing defense by filament formation.
The filamentous morphology of bacteria may function in different ways in the ecology of bacteria. For example, filamentous morphology may play an important role in the gliding motility of some species (5), or it may be involved in the phage resistance of some bacteria (38). On the other hand, the increasing number of reports of the occurrence of high numbers of filamentous bacteria in situations where there is strong protistan grazing pressure indicates that this morphology plays a role in grazing defense (13, 18–20, 28, 32, 35, 36). However, the mechanism that controls a grazing-mediated change from single-celled bacterial communities to filament-dominated bacterial communities is presently under discussion (15, 34). It is conceivable that permanently filamentous species replace smaller, single-celled species or that external signals induce filament formation by single-celled strains. One possible mechanism is indirect induction of filament formation by grazing-mediated enhanced growth rates, as observed for Flectobacillus spp. and C. acidovorans PX54. Direct induction of filament formation by chemical triggers released by the predator is an alternative mechanism. Theoretically, both types of induction mechanisms provide advantages and disadvantages for the bacteria. If a chemical trigger is synthesized by a predator (predator kairomone), the signal may be limited to interactions of the prey bacteria with a small phylogenetic group to which the predator belongs. Thus, induction may fail if a different phylogenetic group of protozoan predators is the cause of strong grazing pressure. If the trigger is synthesized by the prey bacterium (alarm substance) and is released during the digestion of grazed cells, the signal may be independent of the prey species, but the function of the induction mechanism would depend upon the signal strength and thus on the population density of the prey bacteria. If the population density is too low, a signal that is released during digestion of prey cells may not reach the other members of the population. In contrast to these mechanisms based on chemical triggers, the observed indirect induction by the growth rate is independent of the species to which the predators belong or the population density of the prey bacteria. Such a mechanism should work in any case with a predation-caused increase in growth rate. In the natural environment, such an increase in growth rate may occur if grazing decreases the total bacterial biomass or the biomass of unprotected competitors. Furthermore, grazing may positively influence substrate supply via regeneration of nutrients necessary for nutrient-limited phytoplankton. On the other hand, bacterial growth rates may increase in some situations independent of protozoan grazing pressure. For example, strong grazing pressure by Daphnia spp. may increase the bacterial growth rates by reducing the total bacterial biomass. The formation of filaments under such conditions should be disadvantageous because Daphnia grazing on larger bacteria is more efficient than Daphnia grazing on smaller bacteria. However, perhaps there are mechanisms that suppress filament formation in such situations. However, growth rate-controlled filament formation requires higher growth rates, which may limit this grazing defense mechanism to ecosystems at trophic levels which permit higher bacterial growth rates.
Phylogenetic position of isolate MWH38 and other filamentous bacteria.
Flectobacillus sp. strain MWH38 belongs to a genus which thus far contains a limited number of described species. The closest known relative of strain MWH38 is F. major DSMZ 103T (Fig. 1), which is the type strain of the type species of the genus Flectobacillus (22). Until recently, the genus Flectobacillus included two other species (22, 24), both from marine habitats, but these species have been recognized as members of distinct genera (1, 6, 10, 30). Thus, the genus Flectobacillus is presently a monotypic genus containing only F. major. Only two strains of this species have been described, and both of these strains were isolated from freshwater (30). The most closely related lineages, represented by the genera Cytophaga and Flexibacter (Fig. 1), also contain filament-forming strains (5, 11). However, filamentous bacteria, which have been observed in situations where there is high flagellate grazing pressure, belong to a wide phylogenetic spectrum. In enclosure experiments performed with eutrophic pond water, Jürgens et al. observed that filamentous bacteria belonging to the Cytophaga-Flavobacterium-Bacteroides phylum and the α- and the β-Proteobacteria were present after an increase in grazing pressure (21). Šimek et al. reported that threadlike members of the β-Proteobacteria were present after a bacterivorous flagellate was introduced into a mixed bacterial community cultured in a chemostat (34). We investigated two representatives of the Cytophaga-Flavobacterium-Bacteroides phylum and one representative of the β-Proteobacteria and observed that in these three organisms filament formation is controlled by the growth rate. This may indicate that this type of control is phylogenetically widely distributed among bacteria which protect themselves against protistan grazing by filament formation. However, the number of species investigated thus far is too small to propose a generalization. Other mechanisms are conceivable, and we cannot exclude the possibility that different mechanisms control filament formation in other species of bacteria.
Implications.
A common feature of Flectobacillus sp. strain MWH38, F. major, and C. acidovorans PX54 is that these organisms form only partially grazing-protected populations under strong grazing pressure. In all three cases, most cells were fully protected, but a minor part of each population remained subject to grazing by the flagellate used (Fig. 5 and 9). Each population had a continuous size distribution ranging from smaller, edible cells to very large (lengths, ≥50 μm), inedible filaments. Such broad size distributions may provide the potential for more flexible and rapid reactions by the populations to changes in environmental conditions. This may be advantageous if substrate or growth conditions become rapidly worse or if there is a rapid increase in metazoan predation pressure. Yet it is not known if there are bacterial species which respond to grazing pressure with a shift to fully grazing-protected populations. Nevertheless, in several publications, authors have considered filamentous bacteria to be an independent, fully grazing-protected part of the total bacterial community. This ignored the possibility that the filamentous bacteria might be part of a population of the same species that ranged in size from edible medium-sized cells to filamentous cells. The differences in interpretation have consequences for considerations of carbon flow in microbial food webs if filamentous bacteria are involved.
ACKNOWLEDGMENTS
We acknowledge Carsten Strömpl for technical assistance with the nucleic acid sequencing, Alma K. Steinbach for support in batch culture studies, and Brigitte Albrecht for photographic laboratory work. The comments of two referees improved the manuscript.
This study was supported by grant BEO-0319433B from the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie.
REFERENCES
- 1.Bernardet J-F, Segers P, Vancanneyt M, Berthe F, Kersters K, Vandamme P. Cutting a Gordian knot: emended classification and description of the genus Flavobacterium, emended description of the family Flavobacteriaceae, and proposal of Flavobacterium hydatis nom. nov. (basonym, Cytophaga aquatilis Strohl and Tait 1978) Int J Syst Bacteriol. 1996;46:128–148. [Google Scholar]
- 2.Berninger U-G, Finlay B F, Kuuppo-Leinikki P. Protozoan control of bacterial abundances in freshwater. Limnol Oceanogr. 1991;36:139–147. [Google Scholar]
- 3.Chróst R J, Rai H. Bacterial secondary production. In: Overbeck J, Chróst R J, editors. Microbial ecology of Lake Plußsee. New York, N.Y: Springer; 1994. pp. 93–117. [Google Scholar]
- 4.Chrzanowski T H, Šimek K. Prey-size selection by freshwater flagellated protozoa. Limnol Oceanogr. 1990;35:1424–1436. [Google Scholar]
- 5.Costenbader C J, Bruchard R P. Effect of cell length on gliding motility of Flexibacter. J Bacteriol. 1978;133:1517–1519. doi: 10.1128/jb.133.3.1517-1519.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobson S J, Colwell R R, McMeekin T A, Franzmann P D. Direct sequencing of the polymerase chain reaction-amplified 16S rRNA gene of Flavobacterium gondwanense sp. nov. and Flavobacterium salegens sp. nov., two new species from a hypersaline antarctic lake. Int J Syst Bacteriol. 1993;43:77–83. doi: 10.1099/00207713-43-1-77. [DOI] [PubMed] [Google Scholar]
- 7.Faude U C, Höfle M G. Development and application of monoclonal antibodies for in situ detection of indigenous bacterial strains in aquatic ecosystems. Appl Environ Microbiol. 1997;63:4534–4542. doi: 10.1128/aem.63.11.4534-4542.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 9.González J M, Sherr E B, Sherr B B. Size-selective grazing on bacteria by natural assemblages of estuarine flagellates and ciliates. Appl Environ Microbiol. 1990;56:583–589. doi: 10.1128/aem.56.3.583-589.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gosink J J, Woese C R, Stanley J T. Polaribacter gen. nov., with three new species, P. irgensii sp. nov., P. franzmannii sp. nov. and P. filamentus sp. nov., gas vacuolate polar marine bacteria of the Cytophaga-Flavobacterium-Bacteroides group and reclassification of ‘Flectobacillus glomeratus’ as Polaribacter glomeratus comb. nov. Int J Syst Bacteriol. 1998;48:223–235. doi: 10.1099/00207713-48-1-223. [DOI] [PubMed] [Google Scholar]
- 11.Güde H. Grazing by protozoa as selection factor for activated sludge bacteria. Microb Ecol. 1979;5:225–237. doi: 10.1007/BF02013529. [DOI] [PubMed] [Google Scholar]
- 12.Güde H. The role of grazing on bacteria in plankton succession. In: Sommer U, editor. Plankton ecology. Succession in plankton communities. Berlin, Germany: Springer-Verlag KG; 1989. pp. 337–369. [Google Scholar]
- 13.Güde H, Haibel B, Müller H. Development of planktonic bacterial populations in a water column of Lake Constance (Bodensee-Obersee) Arch Hydrobiol. 1985;105:59–77. [Google Scholar]
- 14.Hahn M W. Experimentelle Untersuchungen zur Interaktion von bakterivoren Nanoflagellaten mit planktischen Bakterien. Ph.D. thesis. Braunschweig, Germany: University of Braunschweig; 1996. [Google Scholar]
- 15.Hahn M W, Höfle M G. Grazing pressure by a bacterivorous flagellate reverses the relative abundance of Comamonas acidovorans PX54 and Vibrio strain CB5 in chemostat cocultures. Appl Environ Microbiol. 1998;64:1910–1918. doi: 10.1128/aem.64.5.1910-1918.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 17.Jürgens K, Güde H. The potential importance of grazing-resistant bacteria in planktonic systems. Mar Ecol Prog Ser. 1994;112:169–188. [Google Scholar]
- 18.Jürgens K, Stolpe G. Seasonal dynamics of crustacean zooplankton, heterotrophic nanoflagellates and bacteria in a shallow, eutrophic lake. Freshwater Biol. 1995;33:27–38. [Google Scholar]
- 19.Jürgens K, Arndt H, Zimmermann H. Impact of metazoan and protozoan grazers on bacterial biomass distribution in microcosm experiments. Aquat Microb Ecol. 1997;12:131–138. [Google Scholar]
- 20.Jürgens K, Arndt H, Rothhaupt K O. Zooplankton-mediated changes of bacterial community structure. Microb Ecol. 1994;27:27–42. doi: 10.1007/BF00170112. [DOI] [PubMed] [Google Scholar]
- 21.Jürgens, K., J. Pernthaler, S. Schalla, and R. Amann. Morphological and compositional changes in a planktonic bacterial community in response to enhanced protozoan grazing. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 22.Larkin J M, Borrall R. Family I. Spirosomaceae. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams & Wilkins Co.; 1984. pp. 125–132. [Google Scholar]
- 23.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGuire A J, Franzmann P D, McMeekin T A. Flectobacillus glomeratus sp. nov., a curved, nonmotile, pigmented bacterium isolated from antarctic marine environment. Syst Appl Microbiol. 1987;9:265–272. [Google Scholar]
- 25.Moore E R B, Mau M, Arnscheidt A, Bittger E C, Hutson R A, Collins M D, Van de peer Y, De Wachter R, Timmis K N. The determination and comparison of the 16S rRNA gene sequences of species of the genus Pseudomonas (sensu stricto) and estimation of the natural intrageneric relationships. Syst Appl Microbiol. 1996;19:478–492. [Google Scholar]
- 26.Münster U. Concentrations and fluxes of organic carbon substrates in the aquatic environment. Antonie Leeuwenhoek. 1993;63:243–274. doi: 10.1007/BF00871222. [DOI] [PubMed] [Google Scholar]
- 27.Paster B J, Ludwig W, Weisburg W G, Stackebrandt E, Haspell R B, Hahn C M, Reichenbach H, Stetter K O, Woese C R. A phylogenetic grouping of the bacteroides, cytophagas and certain flavobacteria. Syst Appl Microbiol. 1985;6:34–42. [Google Scholar]
- 28.Pernthaler J, Sattler B, Šimek K, Schwarzenbacher A, Psenner R. Top-down effects on the size-biomass distribution of a freshwater bacterioplankton community. Aquat Microb Ecol. 1996;10:255–263. [Google Scholar]
- 29.Pernthaler J, Posch T, Šimek K, Vrba J, Amann R, Psenner R. Contrasting bacterial strategies to coexist with a flagellate predator in an experimental microbial assemblage. Appl Environ Microbiol. 1997;63:596–601. doi: 10.1128/aem.63.2.596-601.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raj H D, Maloy S R. Proposal of Cyclobacterium marinus gen. nov., comb. nov. for a marine bacterium previously assigned to the genus Flectobacillus. Int J Syst Bacteriol. 1990;40:337–347. [Google Scholar]
- 31.Sanders R W, Caron D A, Berninger U-G. Relationships between bacteria and heterotrophic nanoplankton in marine and fresh waters: an inter-ecosystem comparison. Mar Ecol Prog Ser. 1992;86:1–14. [Google Scholar]
- 32.Schmaljohann R, Pollingher U, Berman T. Natural populations of bacteria in Lake Kinneret: observations with scanning electron and epifluorescence microscopy. Microb Ecol. 1987;13:1–12. doi: 10.1007/BF02014959. [DOI] [PubMed] [Google Scholar]
- 33.Šimek K, Chrzanowski T H. Direct and indirect evidence of size-selective grazing on pelagic bacteria by freshwater nanoflagellates. Appl Environ Microbiol. 1992;58:3715–3720. doi: 10.1128/aem.58.11.3715-3720.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Šimek K, Vrba J, Pernthaler J, Posch T, Hartman P, Nedoma J, Psenner R. Morphological and compositional shifts in an experimental bacterial community influenced by protists with contrasting feeding modes. Appl Environ Microbiol. 1997;63:587–595. doi: 10.1128/aem.63.2.587-595.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sime-Ngando T, Bouredier G, Amblard C, Pinel-Alloul B. Short-term variations in specific biovolumes of different bacterial forms in aquatic ecosystems. Microb Ecol. 1991;21:211–226. doi: 10.1007/BF02539155. [DOI] [PubMed] [Google Scholar]
- 36.Sommaruga R, Psenner R. Permanent presence of grazing-resistant bacteria in a hypertrophic lake. Appl Environ Microbiol. 1995;61:3457–3459. doi: 10.1128/aem.61.9.3457-3459.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van de Peer Y, Jansen J, De Rijk P, De Wachter R. Database on the structure of small ribosomal subunit RNA. Nucleic Acids Res. 1997;25:111–116. doi: 10.1093/nar/25.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weinbauer M G, Höfle M G. Distribution and life strategies of two bacterial populations in a eutrophic lake. Appl Environ Mikrobiol. 1998;64:3776–3783. doi: 10.1128/aem.64.10.3776-3783.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weinbauer M G, Höfle M G. Size-specific mortality of lake bacterioplankton by natural virus communities. Aquat Microb Ecol. 1998;15:103–113. [Google Scholar]