Abstract
Recent studies have highlighted that the proteome can be used to identify potential biomarker candidates for Alzheimer’s disease (AD) in diverse cohorts. Furthermore, the racial and ethnic background of participants is an important factor to consider to ensure the effectiveness of potential biomarkers for representative populations. A promising approach to survey potential biomarker candidates for diagnosing AD in diverse cohorts is the application of machine learning to proteomics datasets. Herein, we leveraged six existing bottom-up proteomics datasets, which included non-Hispanic White, African American/Black, and Hispanic participants, to study protein changes in AD and cognitively unimpaired participants. Machine learning models were applied to these datasets and resulted in the identification of amyloid-β precursor protein (APP) and heat shock protein β-1 (HSPB1) as two proteins that have high ability to distinguish AD; however, each protein’s performance varied based upon the racial and ethnic background of the participants. HSPB1 particularly was helpful for generating high areas under the curve (AUCs) for African American/Black participants. Overall, HSPB1 improved the performance of the machine learning models when combined with APP and/or participant age, and it is a potential candidate that should be further explored in AD biomarker discovery efforts.
Keywords: Alzheimer’s disease, proteomics, machine learning, African American, AUCs, heat shock protein, APP, amyloid-β precursor protein
Graphical Abstract
Introduction
African American/Black and Hispanic adults are more likely to develop Alzheimer’s disease (AD) than other racial groups,1–2 which is a result of complex and interconnected factors related to structural and systemic racism, “lived experiences”, social determinants of health, comorbidities, and genetics.3–10 Reducing these disparities partially requires better understanding of molecular changes in AD. Neuropathological differences in AD hallmarks (amyloid-beta (Aβ) plaques and tau tangles) have not been reported in African American/Black and non-Hispanic White participants.2, 11–13 Some studies have observed that African American/Black participants are more likely to present with both AD and other dementia pathologies11, 14–16; however, this may be dependent on the sampling of participants in the study in terms of community dwelling versus research centers.17–19 Moreover, potential molecular differences between African American/Black and non-Hispanic White participants have recently been reported, particularly in cerebrospinal fluid (CSF) levels of tau biomarkers for AD.17, 20–22 CSF levels of total tau and tau phosphorylated at position 181 (p-tau181) were lower overall in African American/Black participants than non-Hispanic White participants regardless of cognitive status,17, 20–22 and furthermore, smaller changes in tau levels occurred in African American/Black participants with cognitive decline.20 These differences, however, were related to apolipoprotein E (APOE) ε4 status.17 While such studies have to be replicated, potential differences in biomarker levels based on racial and ethnic background would impact biomarker discovery efforts and biomarker utility.
One promising route forward for establishing AD diagnostic biomarker panels is through incorporating machine learning. In this paradigm, a mathematical model is systematically generated to classify new data based on previous examples of known data.23–24 Machine learning can be used in conjunction with protein biomarkers from diseased patients and healthy controls to predict disease status.24 Machine learning has been previously used for disease classification in AD research.25–26 For example, various machine learning algorithms including XGBoost,27 Support Vector Machine (SVM),28 and the Aristotle Classifier29 were able to classify brain proteomics data from AD and cognitively normal (CN) groups30 across two brain regions with high accuracy,25 and proteins from CSF were also useful in distinguishing the disease.36 In those studies, however, the impact of racial diversity on the model was not ascertainable because the datasets were relatively small, and the vast majority of the samples were from non-Hispanic White participants.
Racial bias is a common problem facing machine learning, particularly when racial subgroups are underrepresented,31 so investigations that address the extent to which machine learning models of AD are effective for all racial groups are necessary. Our laboratories have demonstrated that racial bias in machine learning is relevant to AD proteomics studies. An SVM model trained with proteomics analysis of non-Hispanic White patients’ plasma was effective at discriminating AD in multiple datasets, but only for the racial group used to train the model.26 For African American/Black participants, that specific model was ineffective in distinguishing AD, suggesting that proteomic biomarkers should be established using diverse cohorts. Overall, these studies demonstrate an urgent need for understanding AD-related proteomic changes and ensuring potential biomarkers are evaluated and validated in diverse racial and ethnic participants.
The brain’s direct involvement in AD makes it a valuable tissue in which to initially characterize proteomic changes. Analyses in postmortem brain tissue could identify important target proteins that could later be measured in more accessible biological samples such as plasma or CSF. Proteomics has been widely used to study molecular changes in the AD brain, and many proteins have different abundances between AD and CN groups across spatial brain regions.30, 32–43 However, many existing brain proteomics datasets derived in the United States have included primarily non-Hispanic White participants,30, 32–36 such that characterization of proteomic changes in AD brain in other racial and ethnic groups has been very limited. Availability of postmortem brain tissue from African American/Black participants is significantly limited due to difficulties around recruitment into AD studies,44 particularly related to organ donation.45 Some studies have worked to develop effective strategies for recruiting African American/Black participants into AD research, such as culturally informed storytelling materials, community engagement and AD education, and making CSF and/or organ donation optional instead of required.46–49
Recently, we used proteomics to analyze postmortem brain tissue from a cohort that included African American/Black and non-Hispanic White participants. In those studies, despite most proteins changing similarly in both racial groups we identified a subset of proteins with race-specific changes in AD.37 Others have reported that markers of inflammation and neurodegeneration were increased in AD from the middle temporal gyrus region in African American/Black participants compared to non-Hispanic White participants.50–51 These studies suggest that there is heterogeneity in protein changes in the brain from AD participants, though these studies have had relatively small sample sizes and require replication.
Herein, we combined multiple, recently-published brain proteomics datasets that are described in the literature32, 36, 52 with machine learning to accomplish three goals: 1) identify protein features that strongly correlate to AD across multiple datasets, 2) determine the impact of racial demographic on the utility of these features for discriminating AD from CN, and 3) evaluate the degree to which machine learning models successfully distinguish AD from CN samples within specific studies. Our findings show that proteins expressed in the brain can differentiate AD from CN groups across AD brain proteomics datasets, yet the utility of each of the selected proteins for distinguishing AD depends on the cohort diversity. These studies also highlight the need for further studies of heat shock protein β-1 (HSPB1), which we have identified herein as showing particular promise in discriminating AD in African American/Black participants. Finally, the studies point to an urgent need for enhanced diversity in future brain proteomics analyses in AD.
Methods
Cohort details and proteomics dataset selection
Available proteomics datasets of postmortem brain tissue from CN and AD participants were included in this study. Datasets were limited to those analyzed using Tandem Mass Tags (TMT), an isobaric tagging strategy that allows multiplexing of up to 18 samples in a single experiment, for protein quantification.53–54 This criterion was necessary to ensure that the proteomics sample preparation and analysis process was largely similar for all datasets and resulted in inclusion of six datasets: (1) dorsolateral prefrontal cortex (DLPFC) from the Religious Orders Study and Rush Memory and Aging Project (ROSMAP; N = 192; diagnosis based on Emory strict criteria in 2019)32; (2–3) parahippocampal gyrus (PHG; Brodmann area 36) from the Mount Sinai/JJ Peters VA Medical Center Brain Bank (MSBB-Bai, N = 6236 and MSBB-Full, N = 19052); (4–6) hippocampus, inferior parietal lobule (IPL), and globus pallidus (GP) from the University of Pittsburgh Alzheimer Disease Research Center (Pitt ADRC; N = 20).37 We note that the samples in the MSBB-Bai dataset are also part of the larger MSBB-Full dataset. All cohorts included participants from multiple self-reported racial groups (Table 1). Only findings from AD and CN participants from each dataset were included for machine learning analyses. Participants with asymptomatic AD or mild cognitive impairment were excluded.
Table 1.
Demographics of patients in proteomics datasets.
| CN | AD | CN | AD | CN | AD | CN | AD | CN | AD | CN | AD | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | 84 | 108 | 23 | 39 | 63 | 127 | 8 | 10 | 9 | 10 | 9 | 9 |
| Age at deathb | 85 ± 5 | 88 ± 3 | 81 ± 10 | 84 ± 10 | 81 ± 8 | 80 ± 7 | 65 ± 12 | 82 ± 8 | 67 ± 13 | 82 ± 8 | 67 ± 13 | 81 ± 8 |
| Sex (M/F) | 34/50 | 26/82 | 12/11 | 12/27 | 29/34 2 ND |
41/84 2 ND |
5/3 | 5/5 | 6/3 | 5/5 | 6/3 | 5/4 |
| Racec | 83 W 1 B |
106 W 2 B |
17 W 2 B 4 H |
34 W 3 B 2 H |
44 W 7 B 10 H 2 ND |
102 W 16 B 7 H 2 ND |
5 W 3 B |
5 W 5 B |
5 W 4 B |
5 W 5 B |
5 W 4 B |
5 W 4 B |
ND indicates no data was available for N participants in the designated variable.
Reported as mean ± standard deviation.
Race was self-reported.
W = non-Hispanic White; B = African American/Black; H = Hispanic. Abbreviations: CN, cognitively normal; AD, Alzheimer’s disease; ROSMAP, Religious Orders Study and Rush Memory and Aging Project; MSBB, Mount Sinai Brain Bank; Pitt ADRC, University of Pittsburgh Alzheimer Disease Research Center; IPL, inferior parietal lobule; GP, globus pallidus.
TMT protein intensity data for all quantified proteins from each dataset were used for these analyses. Quantified proteins from the ROSMAP dataset had < 50% missing TMT intensities. Data for these proteins were normalized to pools (samples containing equal amounts of protein from all samples included in each batch).32 Both MSBB datasets included TMT quantification at the peptide spectral match (PSM) level, which involved removing PSMs with low intensities prior to normalizing to the median intensity across all PSMs and mean-centering the data. PSMs were averaged per protein to provide protein-level quantification, which was batch-corrected based on the pools.36, 52 Quantified proteins in the Pitt ADRC dataset were identified across both TMT batches of samples and required that proteins were present for ≥ 80% of channels including the pool channels. Data was normalized to the pool channels.37 The number of quantified proteins from each dataset were as follows: ROSMAP = 8,812, MSBB-Bai = 12,148, MSBB-Full = 12,148, Pitt ADRC-hippocampus = 1,414, Pitt ADRC-IPL = 1,487, Pitt ADRC-GP = 1,173.
Univariate analysis
Data analysis was performed in RStudio, using R version 4.0.3. For all univariate analyses, the predictor variable was disease status (control or AD), and the response variable was protein abundance for the protein of interest. All receiver-operating characteristic (ROC) curves and area under the curve (AUC) calculations were calculated directly from the MS data using the package, pROC.55 Protein identities were matched between datasets using UniProt accession numbers, which were included in the first column or row of each dataset. When the AUC of the entire cohort was specified, all samples in the dataset were used, without regard to racial demographics. When AUCs or fold changes were reported by racial subgroup, the racial identities of the samples along with disease status (CN or AD) were used to stratify samples prior to calculating fold change or AUC. In all cases, fold change was calculated using mean intensity for the group, using the embedded function in R. P-values were calculated using two-tailed t-tests.
Machine learning
Supervised classification was primarily performed with AC.2021, using leave-one-out cross-validation as described previously.25 This classifier recently has been demonstrated to show enhanced classification performance over Support Vector Machine (SVM) and extreme gradient boosting (XGBoost) on a variety of proteomics datasets classifying AD.25 Only two hyperparameters are adjustable in the classifier: the number of Repeats, and X, a variable that influences the weighting of each feature. To tune these hyperparameters, the MSBB-Full dataset was used: the full set of patient samples and the three features of interest (APP, HSPB1, and age) were included in the model. First, the Repeats value was set by starting at Repeats = 500 and increasing the value, from 500 to 1000 to 2000, until both the number of misclassified samples and the AUC (to two decimal places) remained constant for three consecutive classifications. To achieve this standard of reproducibility, Repeats set to 2000 was sufficient. Next, the parameter X was tuned, by starting at 1 and increasing through 10. In this case, the optimal value was that which provided the highest AUC (to two decimal places). In cases where two X values provided equivalent AUCs, the better X value was the one that misclassified the fewest samples. This procedure resulted in an X value of 2 being optimal. These hyperparameters (Repeats = 2000; X = 2) were used for all subsequent classifications on all datasets and all feature sets. Hyperparameters were not re-tuned for different datasets, since the tuning of hyperparameters on small datasets could lead to overtraining, and the goal of these studies is to compare results across feature combinations and datasets when a consistent set of machine learning parameters is used.
For each dataset and feature set, a unique model was developed, using AC.2021, X=2, and Repeats=2000, with the resulting Results vector reporting the outcome of a leave-one-out cross-validation. The Results vector shows the strength of the association between each unlabeled sample and its class assignment, and it was used to determine the percent misclassified for each racial group and to calculate the AUC.
In a secondary test, all the datasets and feature sets previously classified using AC.2021 were reclassified using XGBoost, again using leave-one-out cross-validation. In this case, the package, xgboost, was used. The parameters, eta and max_depth, were tuned to optimize the AUC of the MSBB-Full dataset when all three features (age, HSPB1, APP) were included. The optimized parameters were those that produced the highest AUC for the model. The resulting parameters (eta=1; max_depth=3) were used on all subsequent classifications on all feature sets and all datasets. Other parameters included standard choices that were unoptimized, namely: booster = “gbtree”, objective = “binary:logistic”, nrounds =30, eval_metric=“auc”. This set of experiments was completed as a quality control measure to demonstrate that the major outcomes of the original experiments were not influenced by the selection of the classifier. In cases where only one feature was included in the model, for both XGBoost and AC.2021, the feature of interest was listed twice, since at least two features are required for learning with these tools, but the features do not need to be unique.
Results and Discussion
The overall goal of this study was to understand how inclusion of racial and ethnic diversity in brain proteomics datasets impacts the development of a universal diagnostic biomarker panel for AD. The key challenge to achieve this goal is that a dataset containing large numbers of brain proteomics samples from multiple racial groups does not yet exist. Additionally, use of a single dataset with limited sample size also leads to statistical limitations, is more susceptible to biological noise, and requires further replication. To mitigate these challenges, we employed an experimental design in which multiple existing datasets were leveraged to allow for broader applicability of the findings. We included in this study six publicly available brain proteomics datasets that (1) were analyzed using TMT labeling and (2) contained at least 15% African American/Black participants or at least 50 samples from AD or CN groups (Table 1). The MSBB-Full dataset was chosen as a reference to identify the proteins because it contains N = 190 samples, and the composition of participants was ~12% of African American/Black and ~9% Hispanic.
Each of the 12,148 quantified proteins in the MSBB-Full dataset52 was assessed for its ability to discriminate AD from CN participants. The performance metric selected was the area under the ROC curve, and only proteins whose AUC was at least 0.8 were considered as highly predictive of the disease state. A total of 13 proteins (Table 2) met this criterion and were further interrogated. We note that the AUC, as a performance metric, ranks proteins somewhat differently than p-values or fold changes. We chose AUC as the primary performance metric because 1) each dataset in this study has different numbers of samples and AUC is independent of study size, and 2) AUC directly measures a protein’s ability to discriminate disease state.
Table 2.
List of proteins discriminating AD from CN participants in MSBB-Full dataset.a
| UniProt accession number | Protein name | AUCs | p-value | Fold changeb |
|---|---|---|---|---|
| P21741 | Midkine | 0.93 | 1.78E-17 | 3.53 |
| P05067-108 | Amyloid-β precursor protein | 0.91 | 6.39E-16 | 3.35 |
| Q9H4F8 | SPARC-related modular calcium-binding protein 1 | 0.90 | 3.37E-16 | 1.66 |
| O95631 | Netrin-1 | 0.85 | 3.23E-11 | 1.54 |
| Q96CG8 | Collagen triple helix repeat-containing protein 1 | 0.84 | 2.74E-13 | 1.79 |
| Q8N474 | Secreted frizzled-related protein 1 | 0.84 | 6.03E-13 | 1.55 |
| P04792 | Heat shock protein β-1 | 0.82 | 9.32E-12 | 1.46 |
| P05362 | Intercellular adhesion molecule 1 | 0.82 | 4.88E-10 | 1.70 |
| E7EMC7 | Sequestosome-1 | 0.81 | 5.07E-10 | 1.30 |
| F8WE04 | N/A | 0.81 | 1.87E-11 | 1.44 |
| Q96AQ6 | Pre-B-cell leukemia transcription factor-interacting protein 1 | 0.81 | 5.94E-10 | 1.33 |
| P17948-2 | Vascular endothelial growth factor receptor 1 isoform 2 | 0.81 | 4.72E-11 | 1.21 |
| Q9NPD7 | Neuritin | 0.80 | 3.66E-12 | 0.74 |
Proteins discriminating AD and CN participants with AUCs > 0.80.
Mean fold change value of AD/CN participants.
Of the 13 proteins selected, two were present in each of the five remaining datasets: amyloid-β precursor protein (APP) and HSPB1, which are the subject of the remainder of this investigation. To first assess whether these proteins effectively discriminate AD across all the datasets present in the study, ROC curves were evaluated (Figure 1). Both APP and HSPB1, to varying degrees, could be used to differentiate samples from AD and CN participants. We note that the relative proportion of samples from African American/Black participants in the datasets appears to influence the AUC values obtained for the proteins. The dataset with the lowest proportion of African American/Black participants (ROSMAP) has the largest gap between the ROC curve for APP, which has an AUC of 0.91, and HSPB1, with an AUC of 0.68. By contrast, in the three datasets, Pitt ADRC-hippocampus, -IPL, and -GP, with African American/Black participants comprising at least 40% of the samples, the AUCs of APP and HSPB1 are more similar; in fact, the AUC of HSPB1 (AUC 0.89) in the Pitt ADRC-hippocampus dataset is higher than that of APP (AUC 0.88). APP is noticeably higher than HSPB1 in both MSBB datasets, which have an intermediate number of samples from African American/Black participants. In summary, HSPB1 appears to be a useful indicator of AD, and relative to APP, HSPB1’s utility appears to be associated with the number of African American/Black participants in the dataset.
Figure 1. APP and HSPB1 receiver-operating characteristic (ROC) curves in brain datasets as a function of the percentage of African American/Black participants.
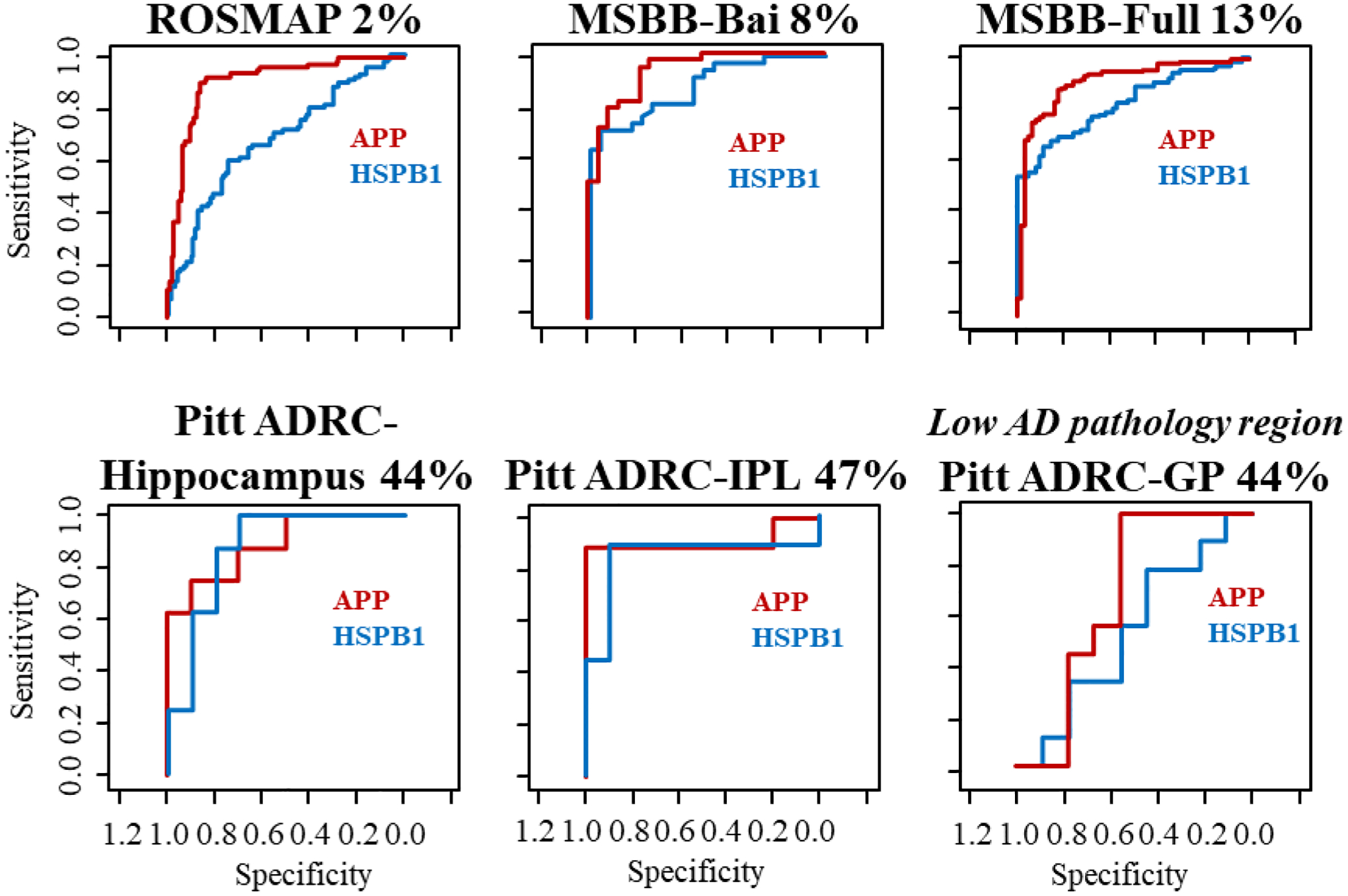
The AUCs for APP and HSPB1, respectively, in each dataset are: ROSMAP (0.91, 0.68); MSBB-Bai (0.93, 0.88); MSBB-Full (0.91, 0.82); Pitt ADRC-hippocampus (0.88, 0.89); Pitt ADRC-IPL (0.91, 0.84); Pitt ADRC-GP (0.67, 0.53). Note: the GP is a brain region with low AD pathology.
We next considered whether HSPB1 is, in fact, a more useful marker of AD in African American/Black participants than it is in non-Hispanic White participants. If this were to be true, then one would expect to see this difference more readily when the samples in each group are first stratified by racial demographic. We tested this hypothesis by comparing the AUCs for APP and HSPB1 in each dataset within the samples from a given racial group (non-Hispanic White or African American/Black). As predicted based on the data in Figure 1, HSPB1 is a better indicator of AD in African American/Black participants than it is in non-Hispanic White participants (Table 3). In six of six datasets, the AUC for this protein is higher for African American/Black participants. The fold change for HSPB1 is higher in all six datasets for the African American/Black participants. Three of the datasets show fold changes > 20% higher; however, we note that the fold change values are dataset-dependent and a reflection of details of inherent changes in the samples, sample preparation, processing, mass spectrometry acquisition, and data normalization.
Table 3.
Performance metrics of APP and HSPB1 by racial group across six datasets.
| HSPB1a | APPa | ||||
|---|---|---|---|---|---|
| NHW | AA/Black | NHW | AA/Black | ||
| AUCs | ROSMAP | 0.68 | 1.00 | 0.91 | 0.50 |
| MSBB-Full | 0.81 | 0.86 | 0.92 | 0.84 | |
| MSBB-Bai | 0.88 | 1.00 | 1.00 | 1.00 | |
| Pitt ADRC-Hippocampus | 0.84 | 0.93 | 0.88 | 0.93 | |
| Pitt ADRC-IPL | 0.76 | 1.00 | 1.00 | 0.85 | |
| Pitt ADRC-GP | 0.32 | 0.75 | 0.52 | 0.88 | |
| FCs | ROSMAP | 1.20 | 1.23 | 1.81 | 1.08 |
| MSBB-Full | 1.46 | 1.52 | 3.40 | 2.24 | |
| MSBB-Bai | 1.4 | 1.70 | 6.93 | 6.35 | |
| Pitt ADRC-Hippocampus | 1.45 | 1.75 | 6.03 | 2.58 | |
| Pitt ADRC-IPL | 1.12 | 1.30 | 7.80 | 4.40 | |
| Pitt ADRC-GP | 0.84 | 1.29 | 1.12 | 1.41 | |
Bold indicates the higher AUC or fold change value between the two racial groups.
Abbreviations: APP, amyloid-β precursor protein; HSPB1, heat shock protein β-1; AUC, area under the curve; NHW, non-Hispanic White; AA, African American; ROSMAP, Religious Orders Study and Rush Memory and Aging Project; MSBB, Mount Sinai Brain Bank; Pitt ADRC, University of Pittsburgh Alzheimer Disease Research Center.
By contrast, there is no discernable race-associated difference in performance when evaluating the AUC for APP. In half of the datasets studied, APP has a higher AUC for the samples originating from non-Hispanic White participants (AUC > 0.90) than from those in the African American/Black participants (AUC 0.5–0.85). Yet, in the other half of the datasets, the AUC is higher for the African American/Black participants. APP does appear to have greater changes in expression level in AD for non-Hispanic White participants than in African American/Black participants. In four of six datasets, the fold change for APP is substantially larger (60–80%) in non-Hispanic White participants. Considering the two datasets that did not show a large difference in fold change, one is the “low AD pathology” dataset (Pitt ADRC-GP), where both racial groups show rather slight fold changes. In the other, the MSBB-Bai dataset, only two samples are present in the African American/Black CN group. A fold change value calculated from more samples would likely result in a different outcome. Overall, it is clear that APP has the ability to distinguish AD from CN in both racial groups; however, the level of its discriminating ability varied in each group.
Interestingly, HSPB1 appears to be a better discriminator of AD in African American/Black participants and in pathological regions. HSPB1 shows particular promise as another protein which can potentially help confirm AD diagnosis. It is necessary, however, to evaluate HSPB1 in the context of larger sample sizes from African American/Black adults and also other racial and ethnic groups. The overall findings for HSPB1 and APP, including which differences are significant (P < 0.05), are shown as bar graphs in Figure 2. Here, all five datasets from high-pathology brain regions are included, and a paired t-test was used to determine if race-associated differences are present, based on fold change or AUC. The results reiterate the findings already stated above.
Figure 2. Assessment of HSPB1 and APP for discriminating AD in five datasets obtained from brain regions with high AD pathology.
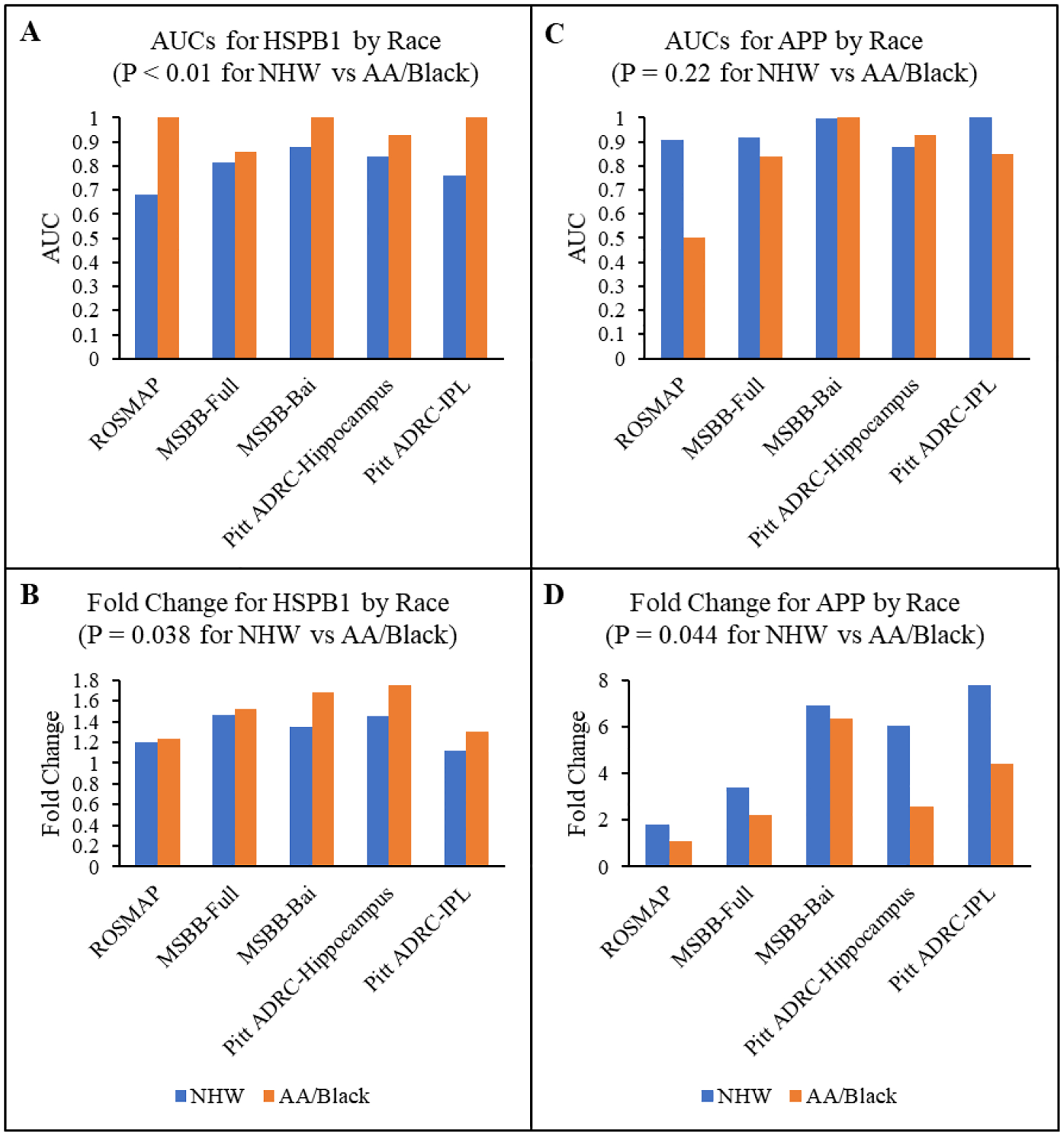
(A) AUCs for HSPB1; (B) fold change for HSPB1; (C) AUCs for APP; (D) fold change for APP. P-values were calculated using paired t-tests. Abbreviations: AUC, are under the curve; HSPB1, heat shock protein β-1; NHW, non-Hispanic White; AA, African American; ROSMAP, Religious Orders Study and Rush Memory and Aging Project; MSBB, Mount Sinai Brain Bank; Pitt ADRC, University of Pittsburgh Alzheimer Disease Research Center; IPL, inferior parietal lobule; APP, amyloid-β precursor protein.
HSPB and APP biomarker panels.
Brain proteomics data are invaluable in the quest to identify useful candidate biomarkers for AD. APP has a well-established role in AD pathogenesis, as APP is cleaved to produce 40- and 42-amino acid Aβ peptides that accumulate in AD’s characteristic amyloid plaques.56–57 While APP has peptides that serve as CSF AD biomarkers, its measurement alone is insufficient to diagnose the disease with high accuracy.58–59 This study has identified an additional protein, HSPB1, that is highly associated with AD across multiple datasets and shows particular promise for improving the diagnosis of AD in African American/Black participants. HSPB1 has been shown to localize to both amyloid plaques60 and tau fibrils61–62 in the brain, and to interact directly with both Aβ63 and tau64 in vitro, leading to prevention or delay of respective fibril formation. Furthermore, neurons from mice lacking HSPB1 were more sensitive to Aβ toxicity,60 while APPswe/PS1dE9 mice overexpressing HSPB1 had improved spatial learning and fewer amyloid plaques in the brain,65 all of which are consistent with HSPB1 having protective effects against AD in the brain.66
To determine the potential benefit of adding HSPB1 into a biomarker panel with APP, machine learning was used to classify four of the datasets (MSBB-Full, ROSMAP, Pitt ADRC-Hippocampus, Pitt ADRC-IPL). Note: these studies were not completed on the Pitt ADRC-GP dataset because this brain region is known to have low AD pathology; also, since the MSBB-Bai dataset is fully represented in MSBB-Full, it was not included in the machine learning studies. The supervised classification relied on different feature combinations, including: APP alone, APP with HSPB1, APP with age, and all three features: APP, HSPB1, age (see Methods). The inclusion of age as an alternative third feature provided an opportunity to assess the relative benefit of HSPB1, as age is the largest risk factor for AD.
In all four datasets, the overall AUC (including all samples/all racial backgrounds) improves when HSPB1 is included in the model, versus a model with APP alone (Table 4). The improvement in AUC is greater in the Pitt ADRC datasets, which have a higher percentage of African American/Black participants. We also assessed the diagnostic accuracy among the racial subgroups. In the MSBB-Full dataset, a total of 23 samples are present in the African American/Black group, and the classification improved from 30% to 22% misclassified when HSPB1 is included as a feature. This difference corresponds to seven vs five samples being misclassified, respectively, so larger studies are still warranted. The inclusion of HSPB1 also substantially improves the classification, from 24% to 6% misclassified, for the Hispanic participants in the MSBB-Full dataset (Table 4).
Table 4.
Classification and AUC metrics for APP, HSPB1, and age features.
| Datasets | % Non-Hispanic White | APP | APP + HSPB1 | APP + Age | All 3 |
|---|---|---|---|---|---|
| MSBB-Full | 76.8% | ||||
| AUC | 0.89 | 0.93 | 0.90 | 0.93 | |
| NHW % error | 15 | 13 | 13 | 14 | |
| AA % error | 30 | 22 | 30 | 22 | |
| H % error | 24 | 6 | 18 | 6 | |
| ROSMAP | 98.4% | ||||
| AUC | 0.89 | 0.91 | 0.92 | 0.93 | |
| NHW % error | 13 | 11 | 12 | 12 | |
| AA % errora | 33 | 33 | 33 | 33 | |
| Pitt ADRC-Hippocampus | 55.6% | ||||
| AUC | 0.76 | 0.89 | 0.90 | 0.91 | |
| NHW # error | 2 | 3 | 3 | 3 | |
| AA # error | 1 | 1 | 1 | 0 | |
| Pitt ADRC-IPL | 52.6% | ||||
| AUC | 0.89 | 0.98 | 0.89 | 0.96 | |
| NHW # error | 0 | 2 | 0 | 1 | |
| AA # error | 1 | 1 | 1 | 1 |
33% of the African American/Black participants is equal to one sample. The AUCs from classification with APP alone and APP + HSPB1 were compared using DeLong’s test, resulting in the following p-values: MSBB-Full, p = 0.01; ROSMAP, p = 0.09; Pitt ADRC-hippocampus, p = 0.20; Pitt ADRC-IPL, p = 0.39.
Abbreviations: AA, African American/Black; APP, amyloid-β precursor protein; H, Hispanic; HSPB1, heat shock protein β-1; MSBB, Mount Sinai Brain Bank; NHW, non-Hispanic White; ROSMAP, Religious Orders Study and Rush Memory and Aging Project; Pitt ADRC, University of Pittsburgh Alzheimer Disease Research Center; IPL, inferior parietal lobule.
The utility of HSPB1 for improving classification of samples from non-Hispanic White participants was also studied in MSBB-Full and ROSMAP datasets. The models that include HSPB1 show consistent improvements in classification: 15 to 13% misclassified in the MSBB- Full dataset and 13 to 11% misclassified in the ROSMAP dataset. These two datasets have 146 and 189 non-Hispanic White participants, respectively. Improvements of classification were difficult to discern in the Pitt ADRC datasets likely because of the small sample sizes.
Finally, we assessed the relative impact of including HSPB1 into the model for predicting AD vs. adding in the participants’ age or adding in both the age and HSPB1. Table 4 shows that including age in the model instead of HSPB1 results in a modest classification improvement for non-Hispanic White participants, no improvement for African American/Black participants, and only minimal improvement for Hispanic participants. We note that more samples or more datasets with Hispanic adults represented are needed to support findings about this racial subgroup. Adding in age as a third feature with the set of APP and HSPB1 does not show any improvement in the model over the two protein-only model for the larger datasets (ROSMAP and MSBB-Full). These results, and particularly the results from the MSBB-Full dataset (largest sample size), demonstrate that HSPB1’s inclusion is important for distinguishing AD in brain samples from non-White participants, particularly for African American/Black adults.
Finally, to verify that the machine learning outcomes are independent of the classifier used, the entire set of supervised classifications was repeated with XGBoost as the classifier, instead of AC.2021. These results can be found in Supplementary Table 1. Overall, all the key outcomes were replicated using this alternative classifier. One notable difference in comparing the reclassification data to the original AC.2021 results is that the AUCs were lower for 14 of the 16 classifications using XGBoost, and the remaining two classifications had equal AUCs to those generated using AC.2021. This comparison between the classifiers further demonstrates that AC.2021 is a better choice for classifying these data.
Strengths and limitations of the study.
The key strength of this study is leveraging datasets from multiple laboratories to better understand how AD is manifested at the protein level and which proteins are important to consider as biomarker candidates for the disease. Each of the datasets used in this study had a similar TMT quantitation strategy, which helps to minimize potential sample processing- and mass spectrometry-related differences. The detection of both APP and HSPB1 in all six datasets is an asset to this study. The general agreement of magnitude of protein change in AD across laboratories and determined by the TMT quantitation strategy supports the reliability of the proteomics results. Finally, the application of machine learning, following the univariate analysis, is a strength, as it identifies the most robust potential protein candidates that can determine AD. The analysis of each protein individually and as a panel demonstrated that the two proteins combined gave more diagnostic utility than using either individually.
The major limitation of this study is the lack of larger and/or more diverse proteomics datasets to conduct machine learning analyses. Even though two of the datasets in this study had ~200 samples, the vast majority of those samples were from non-Hispanic White participants. Assessing the value of biomarkers in sub-populations of participants introduces the challenge of potentially working with low sample sizes. Yet, as this work shows, this type of analysis is critical for generating biomarker panels that are effective for the full population, since some proteins have differential diagnostic utility in sub-populations. While this study focused on racial subgroups, similar subpopulations should be investigated across other parameters, including socioeconomic status, comorbidities, education, lifestyle factors, and other social determinants of health. This study, in addition to others that attempt to undertake the challenging problem of assessing racial sub-populations, would benefit from larger datasets containing a greater representation of African American/Black, Hispanic, and other racial and ethnic participants.
This study focused on two proteins, APP and HSPB1, that were detected in six publicly available datasets but selected from a set of 13 proteins in the MSBB-Full dataset, which served as a reference in this study. Whether there are other shared proteins that could be useful biomarkers would require further analysis and selection from different reference datasets and those with larger numbers and diverse participants.
Conclusions
Leveraging six different brain proteomics datasets, we identified potential AD biomarkers that could serve a racially diverse American population. This combination of datasets and attention to racial subgroup analysis allowed us to identify HSPB1 as a protein that correlates strongly to AD in multiple studies and does so with higher accuracy in samples from African American/Black than non-Hispanic White participants. Furthermore, combining APP and HSPB1 does a better job of discriminating AD than APP alone for all racial groups. HSPB1 should be considered as a potential biomarker candidate for other tissues such as CSF and plasma. Future efforts to identify other potential biomarker candidates using the machine learning strategies presented herein and to replicate the combined value of HSPB1 and APP as diagnostic biomarkers should be explored.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health (R35GM130354 and RF1AG072760, HD), the Alzheimer’s Association (AARGD-17-533405, RASR), pilot funds from the University of Pittsburgh Alzheimer Disease Research Center funded by the National Institutes of Health and National Institute on Aging (P50AG005133, RASR), the Vanderbilt Interdisciplinary Training Program in Alzheimer’s Disease (T32-AG058524, KES), and Vanderbilt University Institutional Funds.
References
- (1).Alzheimer’s Association. 2021 Alzheimer’s disease facts and figures. Alzheimers Dement. 2021, 17 (3), 327–406. [DOI] [PubMed] [Google Scholar]
- (2).Chin AL; Negash S; Hamilton R Diversity and disparity in dementia: the impact of ethnoracial differences in Alzheimer disease. Alzheimer Dis. Assoc. Disord 2011, 25 (3), 187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Wilkins CH; Schindler SE; Morris JC Addressing health disparities among minority populations: Why clinical trial recruitment is not enough. JAMA Neurol 2020, 77 (9), 1063–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Hill CV; Pérez-Stable EJ; Anderson NA; Bernard MA The National Institute on Aging Health Disparities Research Framework. Ethn. Dis 2015, 25 (3), 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Stepler KE; Robinson RAS The potential of ‘omics to link lipid metabolism and genetic and comorbidity risk factors of Alzheimer’s disease in African Americans. In Reviews on Biomarker Studies in Psychiatric and Neurodegenerative Disorders, Guest PC, Ed. Springer International Publishing: Cham, 2019; pp 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Gilmore-Bykovskyi A; Croff R; Glover CM; Jackson JD; Resendez J; Perez A; Zuelsdorff M; Green-Harris G; Manly JJ Traversing the aging research and health equity divide: Toward intersectional frameworks of research justice and participation. Gerontologist 2021, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Mudrazija S; Vega WA; Resendez J; Monroe S Place & brain health equity: Understanding the county-level impacts of Alzheimer’s; UsAgainstAlzheimer’s and the Urban Institute: Washington, DC, 2020. https://www.usagainstalzheimers.org/sites/default/files/2020-11/Urban_UsA2%20Brain%20Health%20Equity%20Report_11-15-20_FINAL.pdf (accessed 2021-09-16). [Google Scholar]
- (8).Zahodne LB; Sharifian N; Kraal AZ; Zaheed AB; Sol K; Morris EP; Schupf N; Manly JJ; Brickman AM Socioeconomic and psychosocial mechanisms underlying racial/ethnic disparities in cognition among older adults. Neuropsychology 2021, 35 (3), 265–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Coogan P; Schon K; Li S; Cozier Y; Bethea T; Rosenberg L Experiences of racism and subjective cognitive function in African American women. Alzheimers Dement. (Amst.) 2020, DOI: 10.1002/dad2.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Pohl DJ; Seblova D; Avila JF; Dorsman KA; Kulick ER; Casey JA; Manly J Relationship between residential segregation, later-life cognition, and incident dementia across race/ethnicity. Int. J. Environ. Res. Public Health 2021, 18 (21), 11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Barnes LL; Leurgans S; Aggarwal NT; Shah RC; Arvanitakis Z; James BD; Buchman AS; Bennett DA; Schneider JA Mixed pathology is more likely in black than white decedents with Alzheimer dementia. Neurology 2015, 85 (6), 528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Gottesman RF; Schneider AC; Zhou Y; Coresh J; Green E; Gupta N; Knopman DS; Mintz A; Rahmim A; Sharrett AR; Wagenknecht LE; Wong DF; Mosley TH Association between midlife vascular risk factors and estimated brain amyloid deposition. JAMA 2017, 317 (14), 1443–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Wilkins CH; Grant EA; Schmitt SE; McKeel DW; Morris JC The neuropathology of Alzheimer disease in African American and white individuals. Arch. Neurol 2006, 63 (1), 87–90. [DOI] [PubMed] [Google Scholar]
- (14).Graff-Radford NR; Besser LM; Crook JE; Kukull WA; Dickson DW Neuropathological differences by race from the National Alzheimer’s Coordinating Center. Alzheimers Dement 2016, 12 (6), 669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Gottesman RF; Fornage M; Knopman DS; Mosley TH Brain aging in African-Americans: the Atherosclerosis Risk in Communities (ARIC) experience. Curr. Alzheimer Res 2015, 12 (7), 607–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Brickman AM; Schupf N; Manly JJ; Luchsinger JA; Andrews H; Tang MX; Reitz C; Small SA; Mayeux R; DeCarli C; Brown TR Brain morphology in older African Americans, Caribbean Hispanics, and whites from northern Manhattan. Arch. Neurol 2008, 65 (8), 1053–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Morris JC; Schindler SE; McCue LM; Moulder KL; Benzinger TLS; Cruchaga C; Fagan AM; Grant E; Gordon BA; Holtzman DM; Xiong C Assessment of racial disparities in biomarkers for Alzheimer disease. JAMA Neurol 2019, 76 (3), 264–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Barnes LL Biomarkers for Alzheimer dementia in diverse racial and ethnic minorities—a public health priority. JAMA Neurol 2019, 76 (3), 251–253. [DOI] [PubMed] [Google Scholar]
- (19).Schneider JA; Aggarwal NT; Barnes L; Boyle P; Bennett DA The neuropathology of older persons with and without dementia from community versus clinic cohorts. J. Alzheimers Dis 2009, 18 (3), 691–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Howell JC; Watts KD; Parker MW; Wu J; Kollhoff A; Wingo TS; Dorbin CD; Qiu D; Hu WT Race modifies the relationship between cognition and Alzheimer’s disease cerebrospinal fluid biomarkers. Alzheimers Res. Ther 2017, DOI: 10.1186/s13195-017-0315-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Garrett SL; McDaniel D; Obideen M; Trammell AR; Shaw LM; Goldstein FC; Hajjar I Racial disparity in cerebrospinal fluid amyloid and tau biomarkers and associated cutoffs for mild cognitive impairment. JAMA Netw. Open 2019, 2 (12), e1917363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Chaudhry A; Rizig M Comparing fluid biomarkers of Alzheimer’s disease between African American or Black African and white groups: A systematic review and meta-analysis. J. Neurol. Sci 2021, 421, 117270. [DOI] [PubMed] [Google Scholar]
- (23).Kelchtermans P; Bittremieux W; De Grave K; Degroeve S; Ramon J; Laukens K; Valkenborg D; Barsnes H; Martens L Machine learning applications in proteomics research: How the past can boost the future. Proteomics 2014, 14 (4–5), 353–366. [DOI] [PubMed] [Google Scholar]
- (24).Swan AL; Mobasheri A; Allaway D; Liddell S; Bacardit J Application of machine learning to proteomics data: Classification and biomarker identification in postgenomics biology. Omics 2013, 17 (12), 595–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Hua D; Desaire H Improved discrimination of disease states using proteomics data with the updated Aristotle Classifier. J. Proteome Res 2021, 20 (5), 2823–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Khan MJ; Desaire H; Lopez OL; Kamboh MI; Robinson RAS Why inclusion matters for Alzheimer’s disease biomarker discovery in plasma. J. Alzheimers Dis 2021, 79 (3), 1327–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Chen T; Guestrin C, XGBoost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Association for Computing Machinery: San Francisco, California, USA, 2016, pp 785–794. [Google Scholar]
- (28).Boser BE; Guyon IM; Vapnik VN, A training algorithm for optimal margin classifiers. In Proceedings of the Fifth Annual Workshop on Computational Learning Theory, Association for Computing Machinery: Pittsburgh, Pennsylvania, USA, 1992, pp 144–152. [Google Scholar]
- (29).Hua D; Patabandige MW; Go EP; Desaire H The Aristotle Classifier: Using the whole glycomic profile to indicate a disease state. Anal. Chem 2019, 91 (17), 11070–11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Ping L; Duong DM; Yin L; Gearing M; Lah JJ; Levey AI; Seyfried NT Global quantitative analysis of the human brain proteome in Alzheimer’s and Parkinson’s Disease. Sci. Data 2018, 5, 180036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Ntoutsi E; Fafalios P; Gadiraju U; Iosifidis V; Nejdl W; Vidal M-E; Ruggieri S; Turini F; Papadopoulos S; Krasanakis E; Kompatsiaris I; Kinder-Kurlanda K; Wagner C; Karimi F; Fernandez M; Alani H; Berendt B; Kruegel T; Heinze C; Broelemann K; Kasneci G; Tiropanis T; Staab S Bias in data-driven artificial intelligence systems—An introductory survey. WIREs Data Min. Knowl 2020, 10 (3), e1356. [Google Scholar]
- (32).Johnson ECB; Dammer EB; Duong DM; Ping L; Zhou M; Yin L; Higginbotham LA; Guajardo A; White B; Troncoso JC; Thambisetty M; Montine TJ; Lee EB; Trojanowski JQ; Beach TG; Reiman EM; Haroutunian V; Wang M; Schadt E; Zhang B; Dickson DW; Ertekin-Taner N; Golde TE; Petyuk VA; De Jager PL; Bennett DA; Wingo TS; Rangaraju S; Hajjar I; Shulman JM; Lah JJ; Levey AI; Seyfried NT Large-scale proteomic analysis of Alzheimer’s disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat. Med 2020, 26, 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Ping L; Kundinger SR; Duong DM; Yin L; Gearing M; Lah JJ; Levey AI; Seyfried NT Global quantitative analysis of the human brain proteome and phosphoproteome in Alzheimer’s disease. Sci. Data 2020, 7 (1), 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Andreev VP; Petyuk VA; Brewer HM; Karpievitch YV; Xie F; Clarke J; Camp D; Smith RD; Lieberman AP; Albin RL; Nawaz Z; El Hokayem J; Myers AJ Label-free quantitative LC-MS proteomics of Alzheimer’s disease and normally aged human brains. J. Proteome Res 2012, 11 (6), 3053–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Seyfried NT; Dammer EB; Swarup V; Nandakumar D; Duong DM; Yin L; Deng Q; Nguyen T; Hales CM; Wingo T; Glass J; Gearing M; Thambisetty M; Troncoso JC; Geschwind DH; Lah JJ; Levey AI A multi-network approach identifies protein-specific co-expression in asymptomatic and symptomatic Alzheimer’s disease. Cell Syst 2017, 4 (1), 60–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Bai B; Wang X; Li Y; Chen P-C; Yu K; Dey KK; Yarbro JM; Han X; Lutz BM; Rao S; Jiao Y; Sifford JM; Han J; Wang M; Tan H; Shaw TI; Cho J-H; Zhou S; Wang H; Niu M; Mancieri A; Messler KA; Sun X; Wu Z; Pagala V; High AA; Bi W; Zhang H; Chi H; Haroutunian V; Zhang B; Beach TG; Yu G; Peng J Deep multilayer brain proteomics identifies molecular networks in Alzheimer’s disease progression. Neuron 2020, 105 (6), 975–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Stepler KE; Mahoney ER; Kofler J; Hohman TJ; Lopez OL; Robinson RAS Inclusion of African American/Black adults in a pilot brain proteomics study of Alzheimer’s disease. Neurobiol. Dis 2020, 146, 105129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Manavalan A; Mishra M; Feng L; Sze SK; Akatsu H; Heese K Brain site-specific proteome changes in aging-related dementia. Exp. Mol. Med 2013, 45, e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Begcevic I; Kosanam H; Martinez-Morillo E; Dimitromanolakis A; Diamandis P; Kuzmanov U; Hazrati LN; Diamandis EP Semiquantitative proteomic analysis of human hippocampal tissues from Alzheimer’s disease and age-matched control brains. Clin. Proteomics 2013, DOI: 10.1186/1559-0275-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).McKetney J; Runde R; Hebert AS; Salamat S; Roy S; Coon JJ Proteomic atlas of the human brain in Alzheimer’s disease. J. Proteome Res 2019, 18 (3), 1380–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Wang Z; Yu K; Tan H; Wu Z; Cho J-H; Han X; Sun H; Beach TG; Peng J 27-plex Tandem Mass Tag mass spectrometry for profiling brain proteome in Alzheimer’s disease. Anal. Chem 2020, 92 (10), 7162–7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Minjarez B; Calderon-Gonzalez KG; Rustarazo ML; Herrera-Aguirre ME; Labra-Barrios ML; Rincon-Limas DE; Del Pino MM; Mena R; Luna-Arias JP Identification of proteins that are differentially expressed in brains with Alzheimer’s disease using iTRAQ labeling and tandem mass spectrometry. J. Proteomics 2016, 139, 103–121. [DOI] [PubMed] [Google Scholar]
- (43).Johnson ECB; Dammer EB; Duong DM; Yin L; Thambisetty M; Troncoso JC; Lah JJ; Levey AI; Seyfried NT Deep proteomic network analysis of Alzheimer’s disease brain reveals alterations in RNA binding proteins and RNA splicing associated with disease. Molec. Neurodegen 2018, 13 (1), 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Stahl SM; Vasquez L Approaches to improving recruitment and retention of minority elders participating in research: examples from selected research groups including the National Institute on Aging’s Resource Centers for Minority Aging Research. J. Aging Health 2004, 16 (5 Suppl), 9s–17s. [DOI] [PubMed] [Google Scholar]
- (45).Boulware LE; Ratner LE; Cooper LA; Sosa JA; LaVeist TA; Powe NR Understanding disparities in donor behavior: race and gender differences in willingness to donate blood and cadaveric organs. Med. Care 2002, 40 (2), 85–95. [DOI] [PubMed] [Google Scholar]
- (46).Robinson RAS; Williams IC; Cameron JL; Ward K; Knox M; Terry M; Tamres L; Mbawuike U; Garrett M; Lingler JH Framework for creating storytelling materials to promote African American/Black adult enrollment in research on Alzheimer’s disease and related disorders. Alzheimers Dement. (N. Y.) 2020, 6 (1), e12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Barnes LL; Shah RC; Aggarwal NT; Bennett DA; Schneider JA The Minority Aging Research Study: ongoing efforts to obtain brain donation from African Americans without dementia. Curr. Alzheimer Res 2012, 9 (6), 734–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Williams MM; Meisel MM; Williams J; Morris JC An interdisciplinary outreach model of African American recruitment for Alzheimer’s disease research. Gerontologist 2011, 51 (Suppl 1), S134–S141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Gilmore-Bykovskyi AL; Jin Y; Gleason C; Flowers-Benton S; Block LM; Dilworth-Anderson P; Barnes LL; Shah MN; Zuelsdorff M Recruitment and retention of underrepresented populations in Alzheimer’s disease research: A systematic review. Alzheimers Dement. (N. Y.) 2019, 5, 751–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Ferguson SA; Panos JJ; Sloper D; Varma V Neurodegenerative markers are increased in postmortem BA21 tissue from African Americans with Alzheimer’s disease. J. Alzheimers Dis 2017, 59, 57–66. [DOI] [PubMed] [Google Scholar]
- (51).Ferguson SA; Varma V; Sloper D; Panos JJ; Sarkar S Increased inflammation in BA21 brain tissue from African Americans with Alzheimer’s disease. Metab. Brain Dis 2020, 35 (1), 121–133. [DOI] [PubMed] [Google Scholar]
- (52).AD Knowledge Portal. https://www.synapse.org/#!Synapse:syn21347564 (accessed 06/03/2021).
- (53).Viner R; Bomgarden R; Blank M; Rogers J Increasing the multiplexing of protein quantitation from 6- to 10-plex with reporter ion isotopologues; Thermo Scientific: 2013. (accessed [Google Scholar]
- (54).Li J; Cai Z; Bomgarden RD; Pike I; Kuhn K; Rogers JC; Roberts TM; Gygi SP; Paulo JA TMTpro-18plex: The expanded and complete set of TMTpro reagents for sample multiplexing. J. Proteome Res 2021, 20 (5), 2964–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Robin X; Turck N; Hainard A; Tiberti N; Lisacek F; Sanchez J-C; Müller M pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011, 12 (1), 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).O’Brien RJ; Wong PC Amyloid precursor protein processing and Alzheimer’s disease. Ann. Rev. Neurosci 2011, 34, 185–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Zhang Y.-w.; Thompson R; Zhang H; Xu H APP processing in Alzheimer’s disease. Molec. Brain 2011, 4, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Jack CR Jr.; Bennett DA; Blennow K; Carrillo MC; Dunn B; Haeberlein SB; Holtzman DM; Jagust W; Jessen F; Karlawish J; Liu E; Molinuevo JL; Montine T; Phelps C; Rankin KP; Rowe CC; Scheltens P; Siemers E; Snyder HM; Sperling R; Elliott C; Masliah E; Ryan L; Silverberg N NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement 2018, 14 (4), 535–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Khoury R; Ghossoub E Diagnostic biomarkers of Alzheimer’s disease: A state-of-the-art review. Biomarkers in Neuropsychiatry 2019, 1, 100005. [Google Scholar]
- (60).Ojha J; Masilamoni G; Dunlap D; Udoff RA; Cashikar AG Sequestration of toxic oligomers by HspB1 as a cytoprotective mechanism. Mol. Cell. Biol 2011, 31 (15), 3146–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Sahara N; Maeda S; Yoshiike Y; Mizoroki T; Yamashita S; Murayama M; Park JM; Saito Y; Murayama S; Takashima A Molecular chaperone-mediated tau protein metabolism counteracts the formation of granular tau oligomers in human brain. J. Neurosci. Res 2007, 85 (14), 3098–3108. [DOI] [PubMed] [Google Scholar]
- (62).Nemes Z; Devreese B; Steinert PM; Van Beeumen J; Fésüs L Cross-linking of ubiquitin, HSP27, parkin, and alpha-synuclein by gamma-glutamyl-epsilon-lysine bonds in Alzheimer’s neurofibrillary tangles. FASEB J 2004, 18 (10), 1135–1137. [DOI] [PubMed] [Google Scholar]
- (63).Wilhelmus MM; Boelens WC; Otte-Höller I; Kamps B; de Waal RM; Verbeek MM Small heat shock proteins inhibit amyloid-beta protein aggregation and cerebrovascular amyloid-beta protein toxicity. Brain Res 2006, 1089 (1), 67–78. [DOI] [PubMed] [Google Scholar]
- (64).Abisambra JF; Blair LJ; Hill SE; Jones JR; Kraft C; Rogers J; Koren J; Jinwal UK; Lawson L; Johnson AG; Wilcock D; O’Leary JC; Jansen-West K; Muschol M; Golde TE; Weeber EJ; Banko J; Dickey CA Phosphorylation dynamics regulate Hsp27-mediated rescue of neuronal plasticity deficits in tau transgenic mice. J. Neurosci 2010, 30 (46), 15374–15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Tóth ME; Szegedi V; Varga E; Juhász G; Horváth J; Borbély E; Csibrány B; Alföldi R; Lénárt N; Penke B; Sántha M Overexpression of Hsp27 ameliorates symptoms of Alzheimer’s disease in APP/PS1 mice. Cell Stress Chaperones 2013, 18 (6), 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Vendredy L; Adriaenssens E; Timmerman V Small heat shock proteins in neurodegenerative diseases. Cell Stress Chaperones 2020, 25 (4), 679–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


