Abstract
We developed an endophenotype disease module-based methodology for Alzheimer’s disease (AD) drug repurposing and identified sildenafil as a potential disease risk modifier. Based on retrospective case-control pharmacoepidemiologic analyses of insurance claims data for 7.23 million individuals, we found that sildenafil usage was significantly associated with a 69% reduced risk of AD (hazard ratio = 0.31, 95% confidence interval 0.25–0.39, P<1.0×10−8). Propensity score stratified analyses confirmed that sildenafil is significantly associated with a decreased risk of AD across all four drug cohorts we tested (diltiazem, glimepiride, losartan and metformin) after adjusting age, sex, race, and disease comorbidities. We also found that sildenafil increases neurite growth and decreases phospho-tau expression in AD patient-induced pluripotent stem cells-derived neuron models, supporting mechanistically its potential beneficial effect in Alzheimer’s disease. The association between sildenafil use and decreased incidence of AD does not establish causality or its direction, which requires a randomized clinical trial approach.
Introduction
Alzheimer’s disease (AD) is the most prevalent form of dementia, anticipated to affect 16 million Americans by 20501. This disease has two prominent cellular hallmarks that have thus far received the greatest attention in drug development efforts: amyloid-beta plaques and neurofibrillary tangles comprised of hyperphosphorylated tau protein2,3. The aggregation of amyloid-beta protein (Aβ) is hypothesized to trigger a cascade of disease-causing processes such as inflammation and synapse dysfunction, as well as contribute to tau tangle formation4. The hypothesis that accumulation of Aβ and consequent tau pathology represents the main cause of AD has dominated the field for more than 25 years5,6. However, anti-tau or anti-amyloid selective drug discovery approaches have lack of clinical benefits for AD patients7. Although the majority of AD cases are sporadic, genetic studies have demonstrated multiple etiologies of familial AD2. However, the predisposition to AD is complex, polygenic, and pleiotropic. Simply identifying susceptibility genes has not led to new therapies8, and in many cases genetic findings must be translated into target identification in order to accelerate drug discovery9–11.
Understanding AD from the point of view of how cellular systems and molecular interactome perturbations underlie the disease is critical for therapeutic development in AD12. This process must incorporate analysis of endophenotypes, which are intermediate genetic traits reflecting the function of a discrete biological system that can further develop into multiple genetically related diseases13. Notably, amyloid and tau pathology in AD have commonly shared endophenotypes14,15. Systematic identification of underlying AD-related endophenotype modules shared by amyloid and tau may provide a foundation for generating predictive models to characterize pathogenesis and therapeutic development for AD16–18. Existing data resources, including genomics, transcriptomics, proteomics, and interactomics (e.g., protein-protein interactions [PPIs]) have not previously been fully utilized and integrated into AD drug discovery19. As drug targets do not operate in isolation from the complex system of proteins that comprise the molecular environment of the cell, each drug-target interaction must be examined in an appropriate integrative context20. These observations may be used to discover the effects of approved drugs that can be repurposed rationally for treatment of diseases other than their original indication21–24.
Several network-based methodologies, such as drug-disease proximity25, shed light on the relationship between drugs (i.e., drug-drug-targets) and diseases (i.e., molecular determinants in disease modules within the human interactome network). In order to prioritize drug candidates identified by network-based approaches, rigorous validation is mandatory. Network-based drug repurposing approaches focus on drugs that are already in clinical practice, and hypothesis testing with this approach is possible using large-scale real-world patient data collected during routine health care delivery21,26.
In this study, we utilized an endophenotype network-based methodology for in silico drug repurposing in AD. We posited that systematic identification of underlying AD-related endophenotype modules, shared by amyloid and tauopathy, would provide a foundation for generating predictive models to characterize AD pathogenesis for efficacious therapeutic development16,18. Specifically, we proposed an integrated network-based framework by incorporating systems pharmacology and network medicine strategies to identify candidate drugs for AD. In doing so, we discovered that sildenafil is associated with a reduced incidence of AD in real-world patient data. In vitro mechanistic observations in human microglia cells and AD patient-induced pluripotent stem cells (iPSC)-derived neurons support a possible mechanism for sildenafil’s beneficial effect in AD.
Results
Alzheimer’s endophenotype disease modules
In this study, we utilized state-of-the-art network-based methodologies to identify AD endophenotype disease modules, and applied these modules to screen FDA-approved drug candidates. We based our studies on the following concepts: 1) genetically supported targets are estimated to double clinical development success rates9,11; 2) proteins that associate with and functionally govern a disease phenotype are localized in the corresponding disease module (molecular determinants of disease pathobiology/physiology) within the comprehensive human protein-protein interaction (PPI) network; and 3) proteins that serve as drug targets for a specific disease may be suitable drug targets for another disease owing to common functional targets and pathways elucidated by the PPIs23,27.
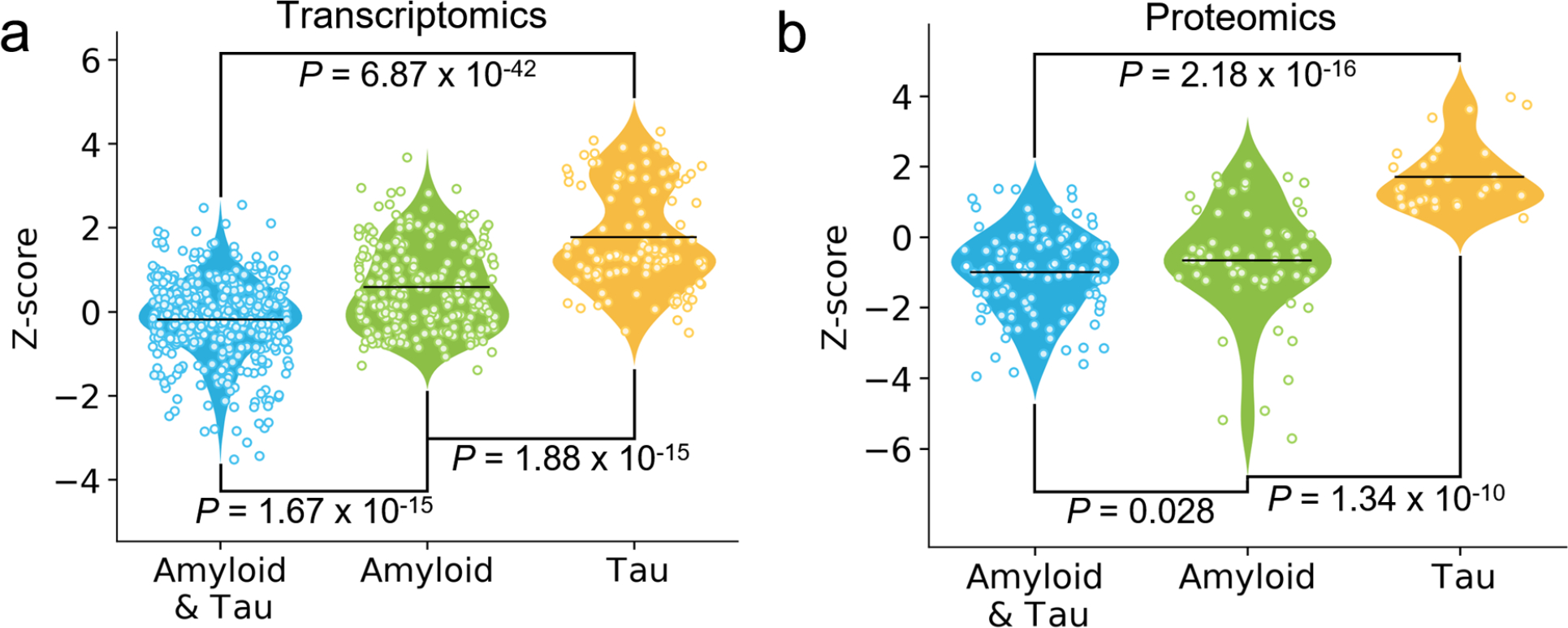
We inspected endophenotype disease modules for AD using three types of disease genes/proteins: (a) seed genes (experimentally validated AD genes), (b) differentially expressed genes from transcriptomics across 7 types of well-established transgenic mouse models with amyloid or tau pathology, and (c) differentially expressed proteins from proteomic data generated from 3 types of transgenic mouse models with well-studied amyloid or tau pathology (see Methods). Functional enrichment analysis revealed that AD seed genes were significantly enriched in all the 13 gene sets related to amyloidosis or tauopathy in AD (P ranging from 3.28 × 10−23 to 2.59 × 10−3, two-tailed Fisher’s exact test, Supplementary Table 1). We also found that AD seed genes formed significant disease modules (see Methods). Out of the 144 AD seed genes, 102 proved to be significantly connected through the human interactome to form a subnetwork (P < 0.001, permutation test, Extended Data Fig. 1 and Supplementary Fig. 1). To test the clinical efficacy of these disease modules in AD therapeutic discovery, we next examined the network relationships between drug-targets and disease modules using network proximity approaches described previously21,23,24. In total, we collected 21 existing drugs (originally FDA-approved with non-AD indications) in new AD clinical trials from the ClinicalTrials.gov database or literatures. Based on the available target and pathway information (Supplementary Table 2), we grouped these drugs into three categories of mechanism-of-action: i) anti-amyloid, (ii) anti-tau, and (ii) dual targeting both amyloid and tau16–18. To examine drug effects on AD, we used a network proximity measure that quantifies the relationship between AD endophenotype disease modules and drug targets in the human interactome network (see Methods). We found that drugs targeting both amyloid and tau have significantly smaller network proximity with AD disease modules, compared with those targeting either amyloid or tau alone (Extended Data Fig. 2). These network observations support a proof-of-concept of endophenotype module-based drug discovery: systematic identification of underlying AD-related endophenotype modules, shared by both amyloid and tau, will generate predictive models to characterize AD likely causal pathways.
We further inspected the endophenotype disease modules generated from large-scale transcriptomics (including DNA microarray and bulk RNA-sequencing) and proteomics data in AD transgenic mouse models, including both amyloid and tau mouse models (see Methods). We found the same network configuration that drugs targeting both amyloid and tau pathways reveal stronger network proximity with AD disease modules built from transcriptomics and proteomics data, compared to drugs targeting amyloid or tau pathways alone (Extended Data Fig. 3). In addition, these drugs have smaller network proximities with AD disease modules built from proteomics data compared to transcriptomics-derived endophenotype modules, suggesting the importance of proteomics data in AD therapeutics discovery. We therefore turned to AD drug repurposing by utilizing amyloid and tau endophenotype module findings, reasoning that drugs targeting the network intersection of amyloid and tau endophenotypes have the greatest potential for success in randomized clinical trials16–18.
Endophenotype network-based Alzheimer’s drug repurposing
We next proposed an endophenotype network-based drug repurposing framework for AD: (1) construction and validation of AD endophenotype module (Fig. 1a); (2) In silico drug repurposing via network proximity analysis (Fig. 1b); (3) population-based validation to test the drug user’s AD-related outcomes (Fig. 1c); (4) network-based in vitro mechanistic observations in human microglia cells and AD patient iPSC-derived neurons (Fig. 1d). In total, we compiled 1,608 FDA-approved drugs by pooling the reported experimental drug-target binding affinity data: EC50, IC50, Ki, or Kd, each ≤ 10 μM as a cutoff. We calculated a z-score for quantifying the significance of the network proximity (the shortest path lengths) between drug-targets and proteins in the endophenotype modules within the human interactome network. We screened all FDA-approved drugs based on their network proximity to the AD seed module (built from AD seed genes), and selected the top 100 candidates with the lowest z-score. We further computed the network proximity of the top 100 candidates with the other 12 AD modules, including 2 modules derived from experimentally validated seed genes related to amyloid and tau, and 10 modules derived from proteomics data across 3 types of AD genetic mouse models with amyloid and tau pathology (Fig. 2).
Figure 1. A diagram illustrating an endophenotype network-based drug repurposing framework for Alzheimer’s disease (AD).
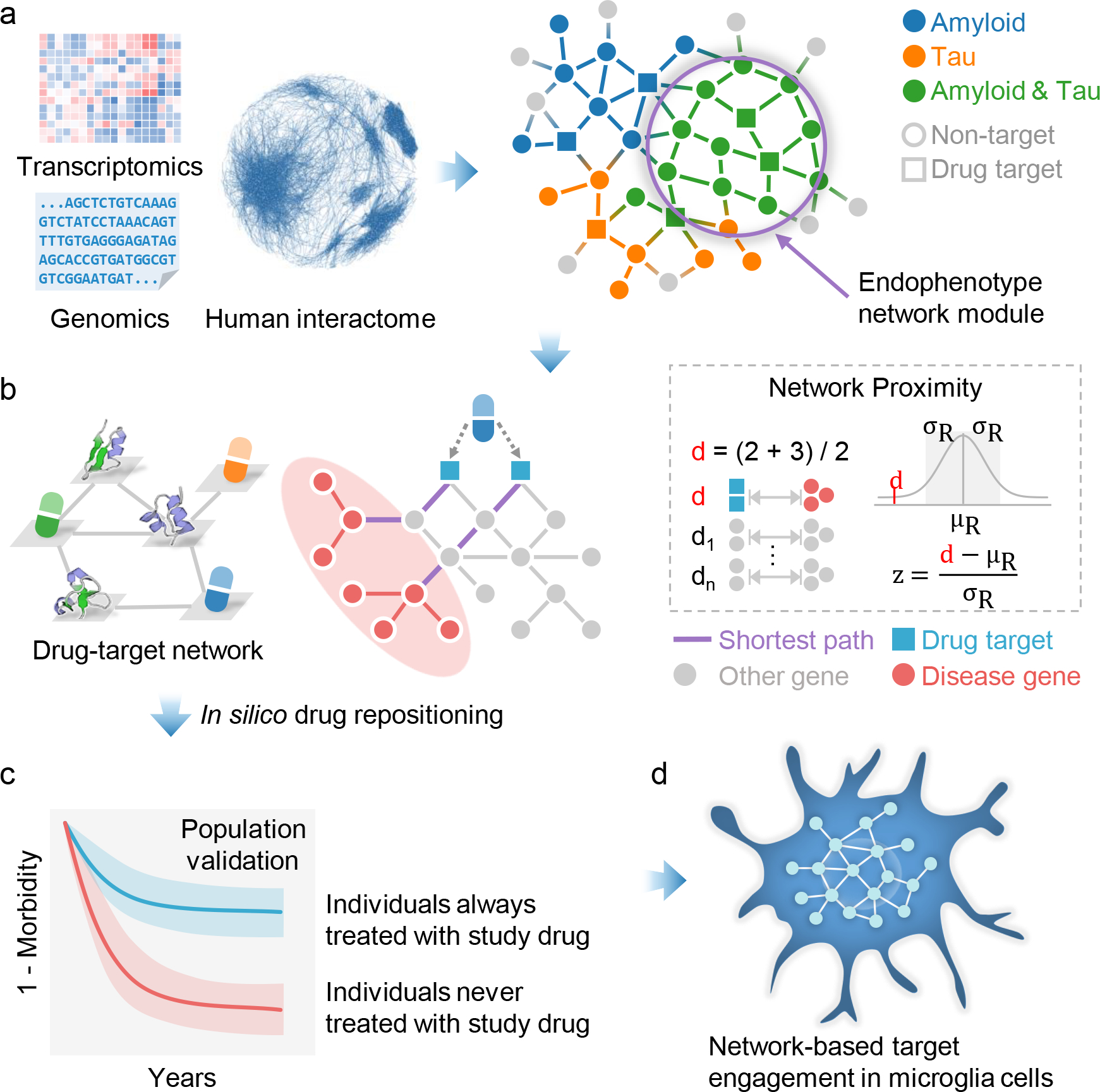
(a) Construction and validation of endophenotype disease modules for AD in the human protein-protein interactome network. (b) In silico drug repurposing by network proximity analysis (see Methods). (c) Population-based validation to test the drug user’s relationship with AD outcomes. (d) Network-based mechanistic observations in human microglia cells and AD patient induced pluripotent stem cells (iPSC)-derived neurons.
Figure 2. Network-based in silico drug repurposing for Alzheimer’s disease (AD).
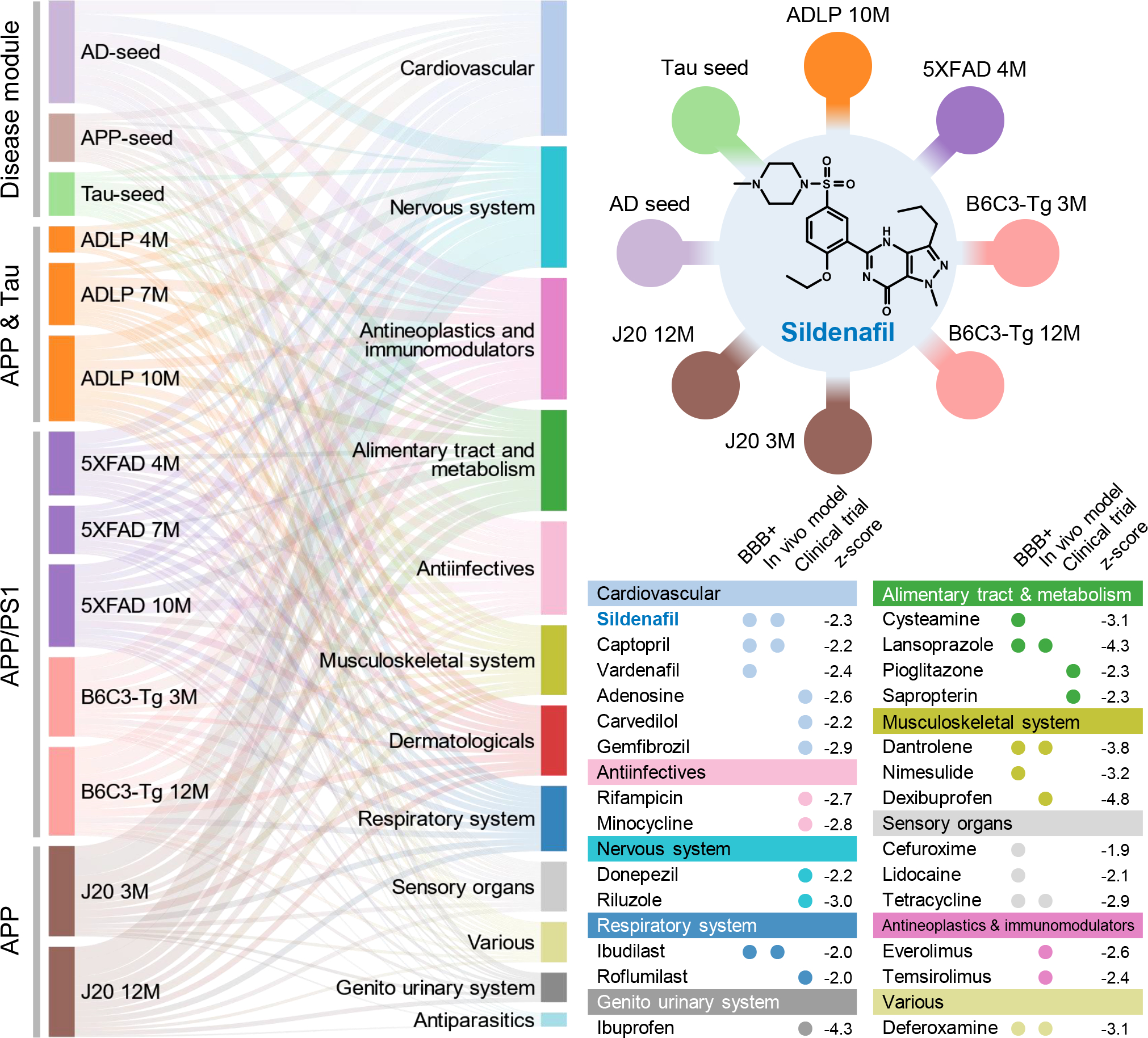
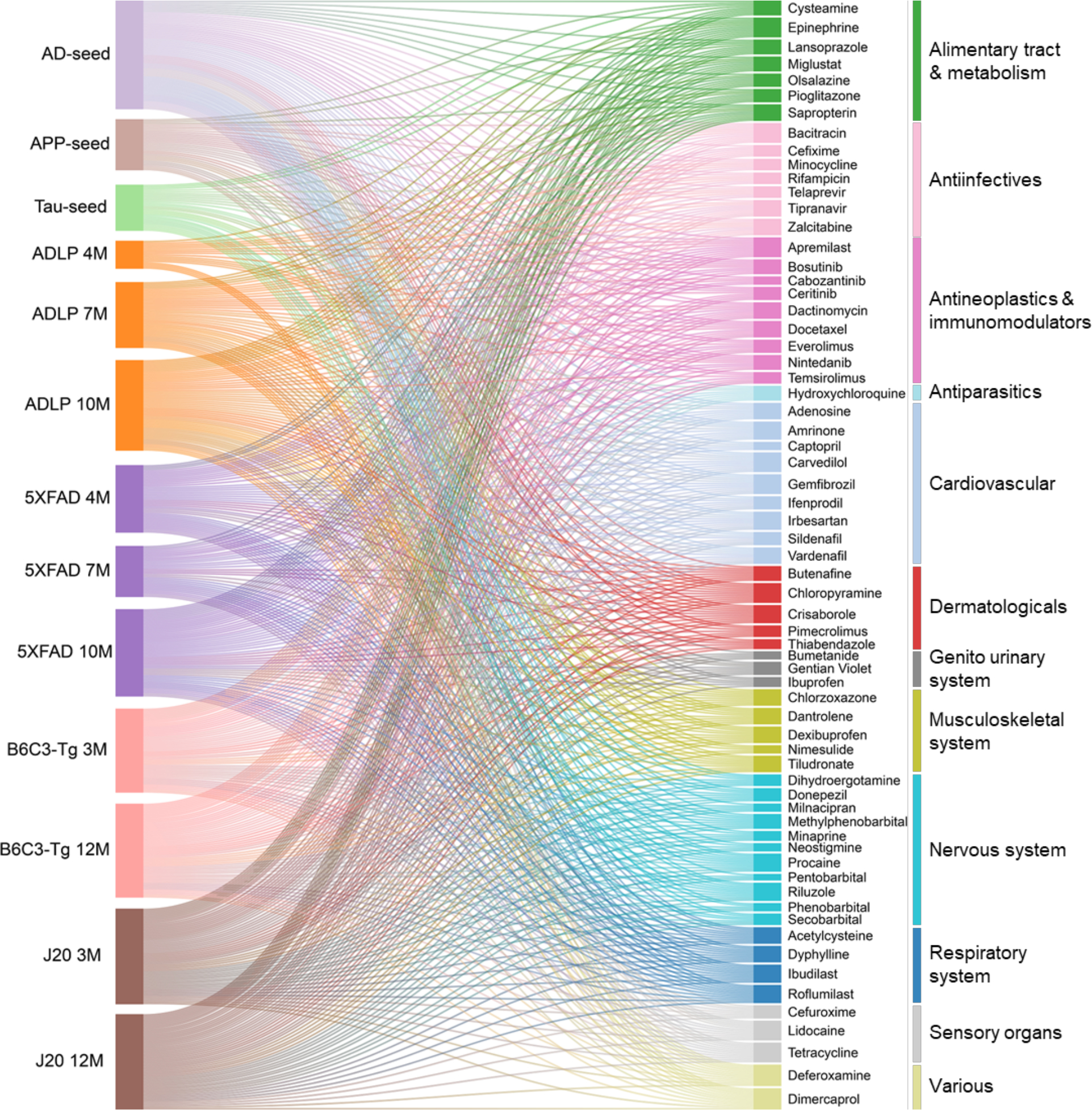
In total, 13 endophenotype disease modules, built by 3 experimentally validated (seed) gene sets in amyloidosis (Amyloid), tauopathy (Tau), and AD, as well as 10 differentially expressed protein from proteomics data generated in AD genetic mouse models, were evaluated to screen FDA-approved drugs using a network proximity measure (Methods). A Sankey diagram illustrates a global view of 66 high-confidence drug candidates, identified by network proximity analysis. Network proximity analysis measures the interplay between endophenotype disease modules (proteins) and drug targets in the human interactome. Drugs are grouped by their first-level Anatomical Therapeutic Chemical Classification (ATC) codes. The drugs with known anti-AD clinical trial status, in vivo animal model, and blood–brain barrier (BBB) properties data are highlighted. The z-score with AD seed module by network proximity is given for each drug. Subject matter expertise criterion-based prioritization resulted in sildenafil as the best candidate (z = −2.30), which has significant close network distance with eight endophenotype disease modules.
We ranked the 100 drugs by counting the number of disease modules that had a z-score less than −1.5 to the drug. Drug candidates that were predicted by at least 4 out of 13 endophenotype modules were kept. After excluding the nutraceutical drugs, metal drugs (copper, zinc), and radioligand diagnostic agents, we identified 66 candidate drugs. We further grouped these 66 drugs (Fig. 2 and Extended Data Fig 4.) into 12 categories according to the first level of the Anatomical Therapeutic Chemical Classification (ATC) code. Among them, drugs affecting the cardiovascular system (n=14) have the largest number of predicted drugs (Fig. 2), followed by nervous system therapeutics (n=11), antineoplastic and immunomodulating agents (n=9), and alimentary tract and metabolism drugs (n=7). Previous studies have suggested that therapeutic strategies aimed at reducing cardiovascular diseases may reduce the prevalence of AD28. Indeed, among the 14 FDA-approved cardiovascular drugs identified, there are currently 9 drugs under preclinical or clinical trials for AD (clinicaltrials.gov database as of July 31, 2019).
Discovery of sildenafil as a new candidate drug in AD
We systematically retrieved anti-AD clinical, in vivo, and blood–brain barrier (BBB) properties from publicly available databases for the 66 candidate drugs (Fig. 2 and Supplementary Table 3). Among the 66 drugs, 21 network-predicted drugs have reported clinical or preclinical evidence in AD (Fig. 2), suggesting reasonable accuracy of our network proximity approach. Among the 21 drugs, 11 drugs (including an approved AD drug donepezil) are in ongoing AD clinical trials. To test model performance, we prioritized the same 100 candidate drugs with the lowest z-score by AD seed modules using a previous proximal pathway enrichment analysis approach (PxEA)29, and found our human interactome-based network proximity measure slightly outperformed PxEA (Supplementary Table 4). In addition, we further collected 120 FDA-approved drugs with reported clinical or preclinical evidence relevant to AD from the AlzGPS database30 (Supplementary Table 5). We found that network-predicted drug candidates are significantly enriched in the 120 collected drugs with known preclinical and clinical evidence related to AD (P = 2.08 × 10−9, two-tailed Fisher’s test). To avoid bias of network proximity scores, we evaluated these drugs again via generating random disease modules of the same size (n=144) as the AD seed module. Heatmap illustration (Supplementary Fig. 2) shows that the z-scores are quite random, which further suggests the robustness of the used network proximity measure. Yet, we found that the incompleteness of drug-target networks may influence the z-score (Supplementary Table 6) after manually checking the network proximity calculation for 12 drugs (dually targeting amyloid & tau) under ongoing AD trials.
We next used subject matter expertise based on a combination of factors: (i) strength of network proximity measures (z-score < −1.5 across 13 AD endophenotype disease modules; (ii) availability of sufficient patient data for meaningful evaluation (exclusion of infrequently used medications); (iii) published positive in vivo experimental data in AD mouse models; (iv) novelty of the predicted associations through exclusion of drugs currently being tested in AD clinical trials; (v) ideal brain penetration from literatures or predicted by admetSAR31, a computational tool for evaluation of drug pharmacokinetic properties (see Supplementary Methods).
Applying these criteria resulted in identifying several top drug candidates, including dantrolene, deferoxamine, lansoprazole and sildenafil. We excluded lansoprazole since its usage was associated with increased risk of AD and AD-related dementia in a pharmacoepidemiologic study32. In addition, dantrolene and deferoxamine (infrequently used medications) were not selected as both of them had an insufficient amount of patient data for population-based validation. Thus, sildenafil (z = −2.30 on AD seed module), an FDA-approved drug for erectile dysfunction, was selected as the best drug candidate. Vascular dementia, often occurring with AD, is one of the leading causes of age-related cognitive impairment and sildenafil has been shown to significantly improve cognition and memory in a rat model of vascular dementia33. In addition, sildenafil treatment also attenuates the magnitude of tauopathy in AD Tg2576 transgenic mice33. Thus, we selected sildenafil to test the drug user’s relationship with AD outcomes using population-based observational studies.
Pharmacoepidemiologic analyses point to an association between sildenafil use and AD incidence
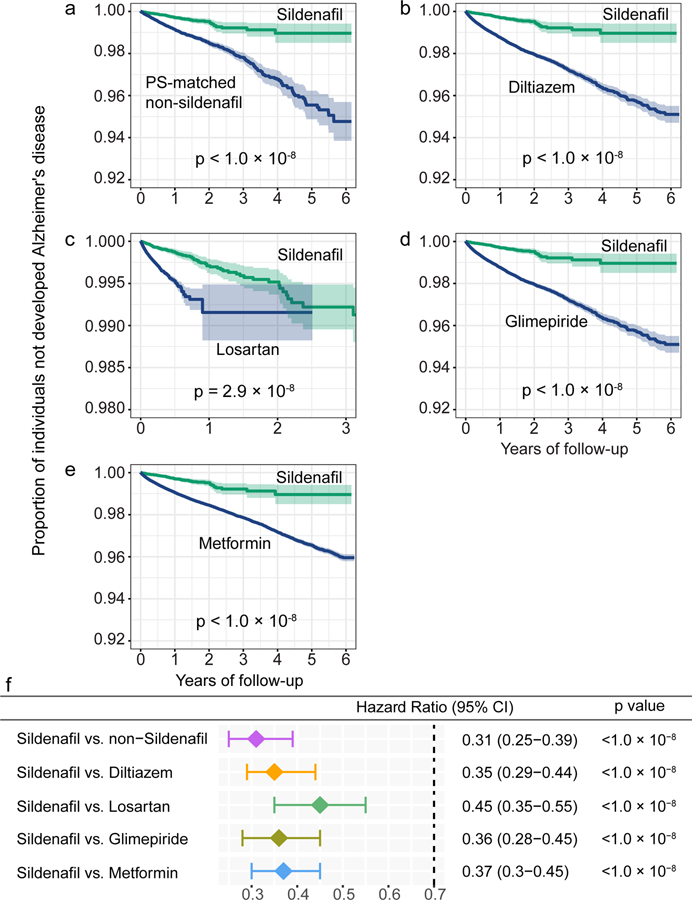
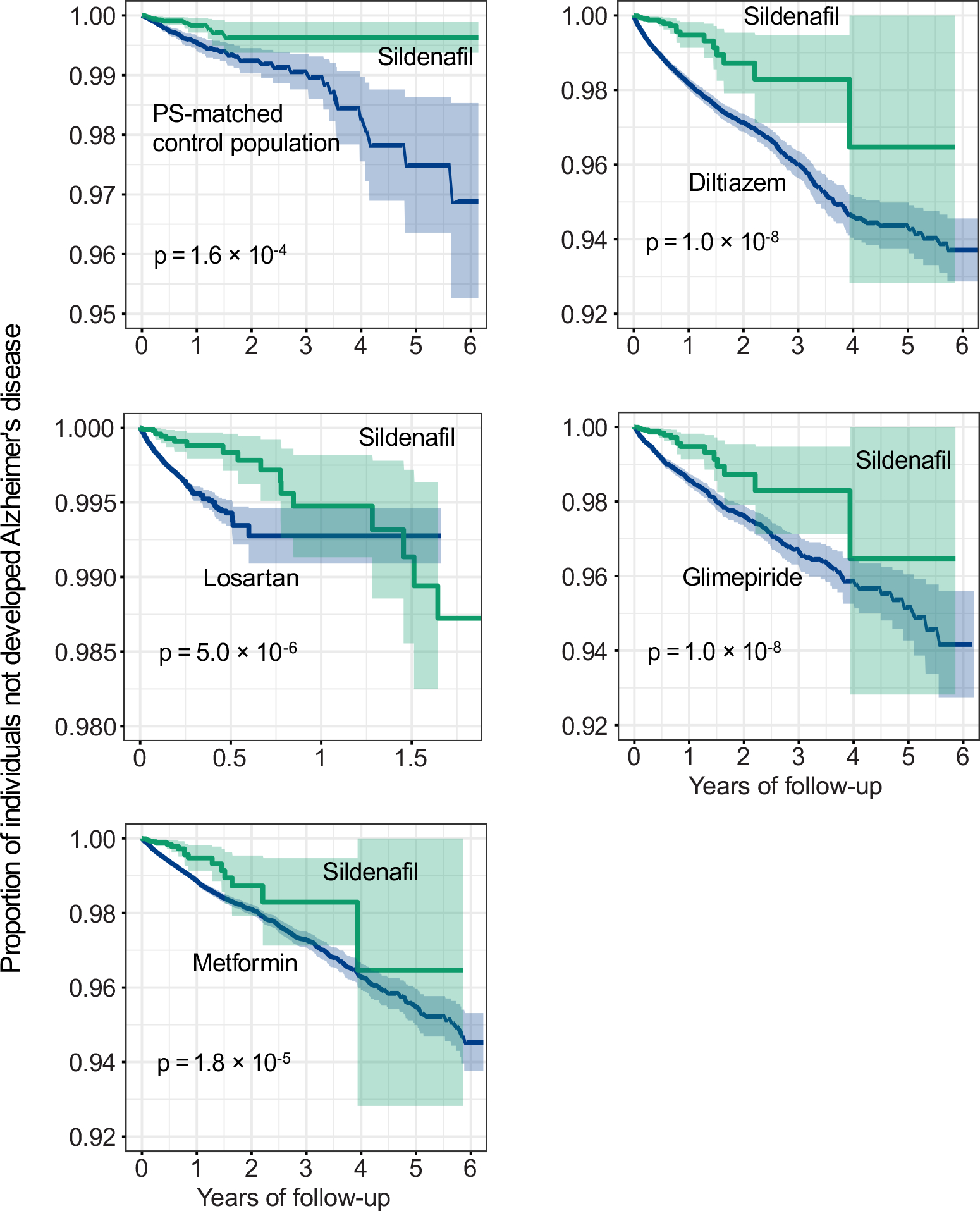
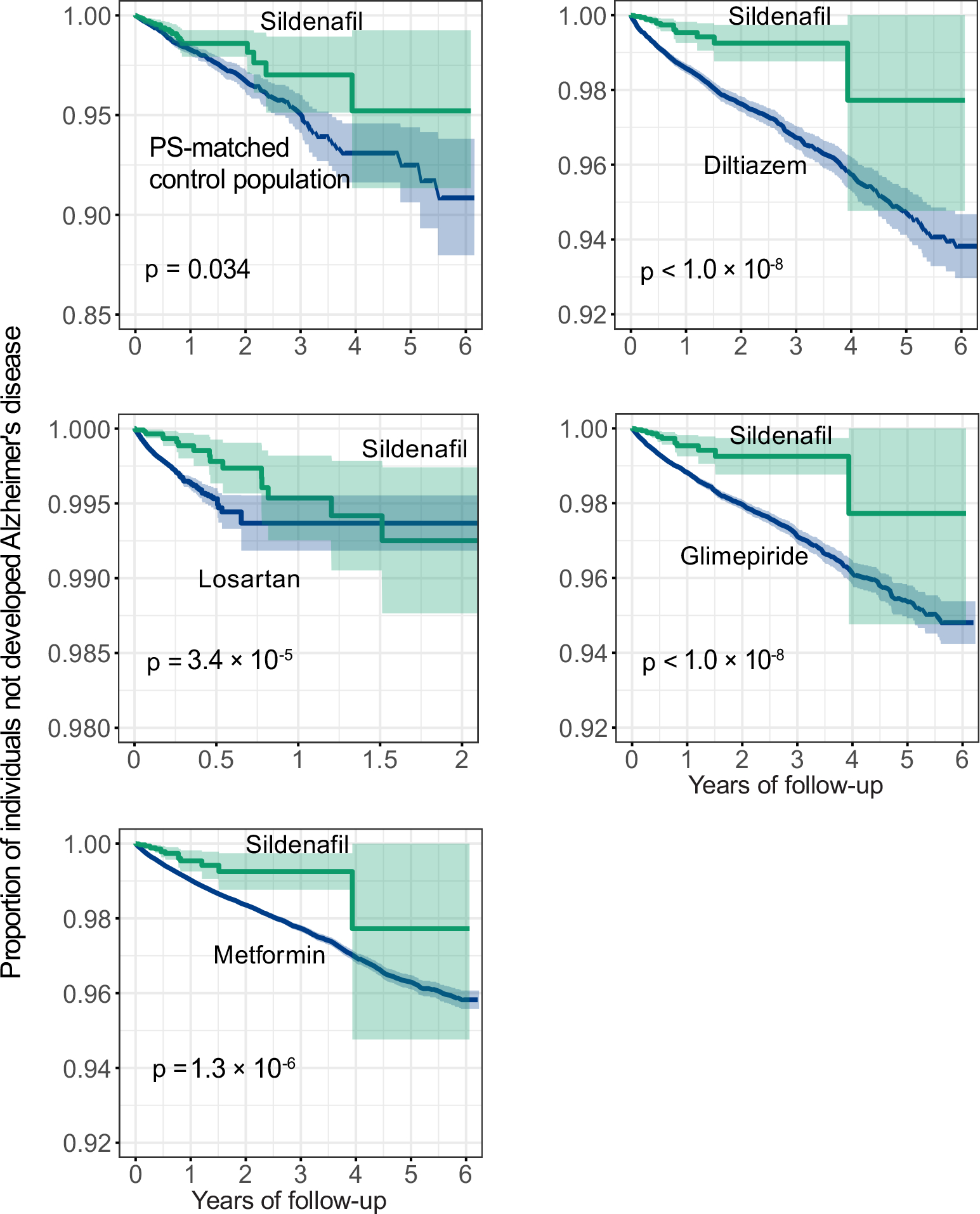
We evaluated the sildenafil user’s relationship with AD outcomes by analyzing 7.23 million U.S. commercially insured individuals. In order to minimize confounding factors, we conducted three types of drug cohort-based observational studies: 1) sildenafil users vs. non-sildenafil users, 2) sildenafil vs. a comparator drug (diltiazem or glimepiride) without reported preclinical and clinical evidence of AD relevance, and 3) sildenafil vs. a comparator drug (losartan or metformin) in active AD clinical trials using a new-user active comparator design21. We conducted five comparison analyses to evaluate the predicted association based on individual level longitudinal patient data and state-of-the-art pharmacoepidemiologic methods. These included: (i) sildenafil (n = 116,412, z = −2.30 with AD seed module) vs. a matched control population (n = 460,356), (ii) sildenafil vs. diltiazem (an anti-hypertensive drug, n = 251,360, z = 0.57), (iii) sildenafil vs. losartan (another anti-hypertensive drug in an AD clinical trial [ClinicalTrials.gov Identifier: NCT02913664], n = 664,265, z = −0.86), (iv) sildenafil vs. glimepiride (an anti-diabetic drug, n= 159,597, z = −0.57), and (v) sildenafil vs. metformin (an anti-diabetic drug in an AD clinical trial [NCT00620191], n = 723,082, z = −0.73). Table 1 summarizes the patient data for state-of-the-art pharmacoepidemiologic analyses. For each comparison, we estimated the un-stratified Kaplan-Meier curves and conducted propensity score stratified (n strata = 10) log-rank tests and the Cox regression analysis. As shown in Fig. 3a, after 6 years of follow-up, sildenafil usage was significantly associated with a 69% reduced risk of AD, compared with matched non-sildenafil users (hazard ratio [HR] =0.31, 95% confidence interval [CI] 0.25–0.39, P < 1 × 10−8) by the Cox regression model (Fig. 3f). Importantly, propensity score-stratified cohort studies reveal that sildenafil usage is significantly associated with a 65% reduced risk of AD compared to diltiazem (HR 0.35, 95% CI 0.29–0.44, P < 1 × 10−8, Figs. 3b and 3f), a 55% reduced risk compared to losartan (HR 0.45, 95% CI 0.35–0.55, P <1 × 10−8, Figs. 3c and 3f), a 64% reduced risk compared to glimepiride (HR 0.36, 95% CI 0.28–0.45, P <1 × 10−8, Figs. 3d and 3f), and a 63% reduced risk compared to metformin (HR 0.37, 95% CI 0.30–0.45, P <1 × 10−8, Figs. 3e and 3f). In summary, these five pharmacoepidemiologic analyses support our network-based prediction of the potential utility of sildenafil in reducing the risk of AD.
Table 1.
Description of the patient data for pharmacoepidemiologic analysis.
| Clinical characteristics | Sildenafil | Diltiazem | Losartan | Metformin | Glimepiride | Control |
|---|---|---|---|---|---|---|
|
| ||||||
| N | 116,412 | 251,360 | 664,265 | 723,082 | 159,597 | 460,356 |
| # of AD | 93 | 5,046 | 8,998 | 8,050 | 2,237 | 1,175 |
| Female (%) | 2.0 | 59.7 | 58.0 | 47.7 | 45.5 | 2.0 |
| Mean age (SD) | 71.0 (5.8) | 76.5 (8.3) | 74.2 (7.8) | 72.1 (6.8) | 73.7 (7.5) | 74.2 (7.2) |
| Geographics (%) | ||||||
| Northeast | 30.8 | 23.9 | 21.0 | 21.3 | 20.4 | 23.0 |
| North central | 18.9 | 29.0 | 27.4 | 27.5 | 30.8 | 31.0 |
| South | 29.9 | 31.3 | 33.7 | 34.2 | 37.8 | 29.8 |
| West | 19.4 | 15.0 | 16.9 | 15.9 | 10.0 | 15.1 |
| Not available | 1.0 | 0.6 | 0.8 | 1.0 | 1.0 | 1.0 |
| Disease comorbidities (%) | ||||||
| T2D | 18.9 | 20.0 | 23.5 | 50.9 | 62.8 | 20.0 |
| HT | 42.6 | 46.9 | 51.2 | 38.0 | 46.3 | 46.0 |
| CAD | 16.4 | 20.6 | 16.8 | 12.7 | 18.8 | 17.3 |
We estimated the un-stratified Kaplan-Meier curves, conducted propensity score stratified (n strata = 10) two-sided log-rank test and Cox model. The AD frequency (1,175/460,356 [0.255%]) in control group was not cumulative AD incidence in the whole life for non-sildenafil users. In fact, these patients were diagnosed with AD during their follow up times (6 years), which had same length as the matched sildenafil exposure windows. AD: Alzheimer’s disease; T2D: Type 2 diabetes; HT: Hypertension; CAD: Coronary artery disease.
Figure 3. Longitudinal analyses reveal that sildenafil usage is significantly associated with reduced likelihood of AD in a longitudinal patient database with 7.23 million subjects.
Five comparator analyses were conducted: (a) sildenafil (n = 116,412) vs. sex and comorbidities (hypertension, diabetes, and coronary artery disease) matched (ratio 1:4) non-sildenafil exposure population (n = 460,356), (b) sildenafil vs. diltiazem (an anti-hypertensive drug, n = 251,360), (c) sildenafil vs. losartan (an anti-hypertensive drug in AD clinical trial [ClinicalTrials.gov Identifier: NCT02913664], n = 664,265), (d) sildenafil vs. glimepiride (an anti-diabetic drug, n= 159,597), and (e) sildenafil vs. metformin (an anti-diabetic drug under an AD clinical trial [ClinicalTrials.gov Identifier: NCT00620191], n = 723,082). First, for each comparator, we estimated the propensity score by using the variables described in Table 1. We estimated the un-stratified Kaplan-Meier curves, conducted propensity score stratified (n strata = 10) two-sided log-rank test and applied a Cox model. (f) Hazard ratios (HR) and 95% confidence interval (CI) across five cohort studies. Propensity score stratified Cox-proportional hazards models were used for statistical inference of the hazard ratios (sildenafil n = 116,412; non-sildenafil n = 460,356; diltiazem n = 251,360; losartan n =664,265; glimepiride n = 159,597; metformin n = 723,082).
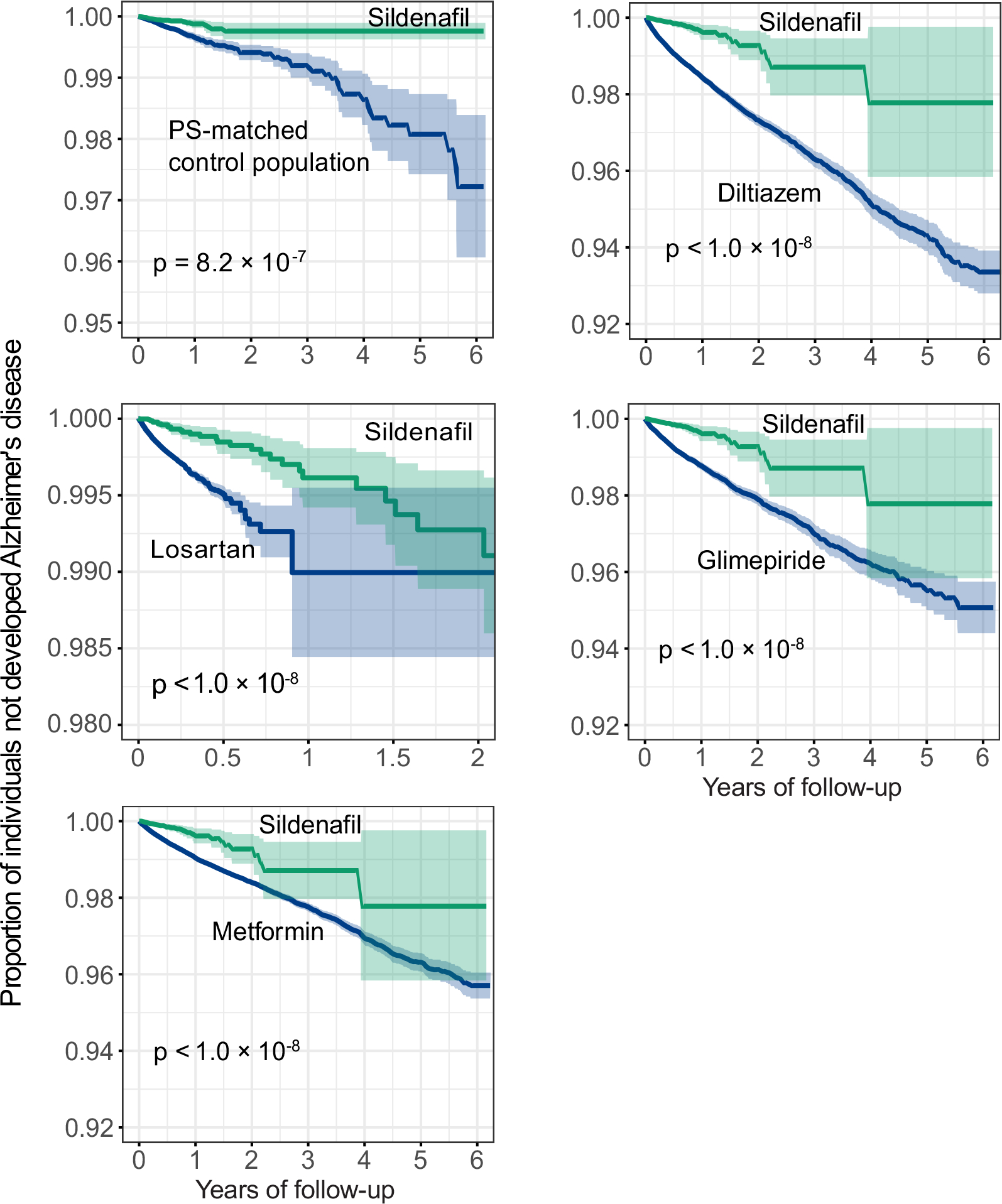
Disease comorbidities, including coronary artery disease (CAD), hypertension (HT), and type 2 diabetes (T2D), are significantly associated with risk of AD34,35. We found that sildenafil usage was significantly associated with reduced risk of AD across five drug cohorts in individuals after we excluded CAD, HT, and T2D as well (Fig. 4a). In addition, sildenafil usage was significantly associated with reduced likelihood of AD across all five drug cohorts in individuals with CAD (Fig. 4b, Extended Data Fig. 5), HT (Fig. 4c, Extended Data Fig. 6), and T2D (Fig. 4d, Extended Data Fig. 7), respectively, although it showed a reduced effect size. In addition, individuals with cognitive impairment and dementia may be less likely to be prescribed sildenafil. After adjusting mild cognitive impairment (MCI), we found that sildenafil usage is significantly associated with reduced likelihood of AD across all five drug cohorts as well (Supplementary Table 7), thus reducing the likelihood of confounds due to selection bias by physicians. Using the commonly used Charlson comorbidity index, we found that sildenafil users have the similar Charlson comorbidity index with comparator treatment groups after PS-matching (Supplementary Fig. 3). Thus, the sildenafil users have the equal health/disease conditions on average with those receiving comparator drugs in the PS-matching patient data observations.
Figure 4. Subgroup analyses of five drug cohort studies to evaluate confounding by disease comorbidities.
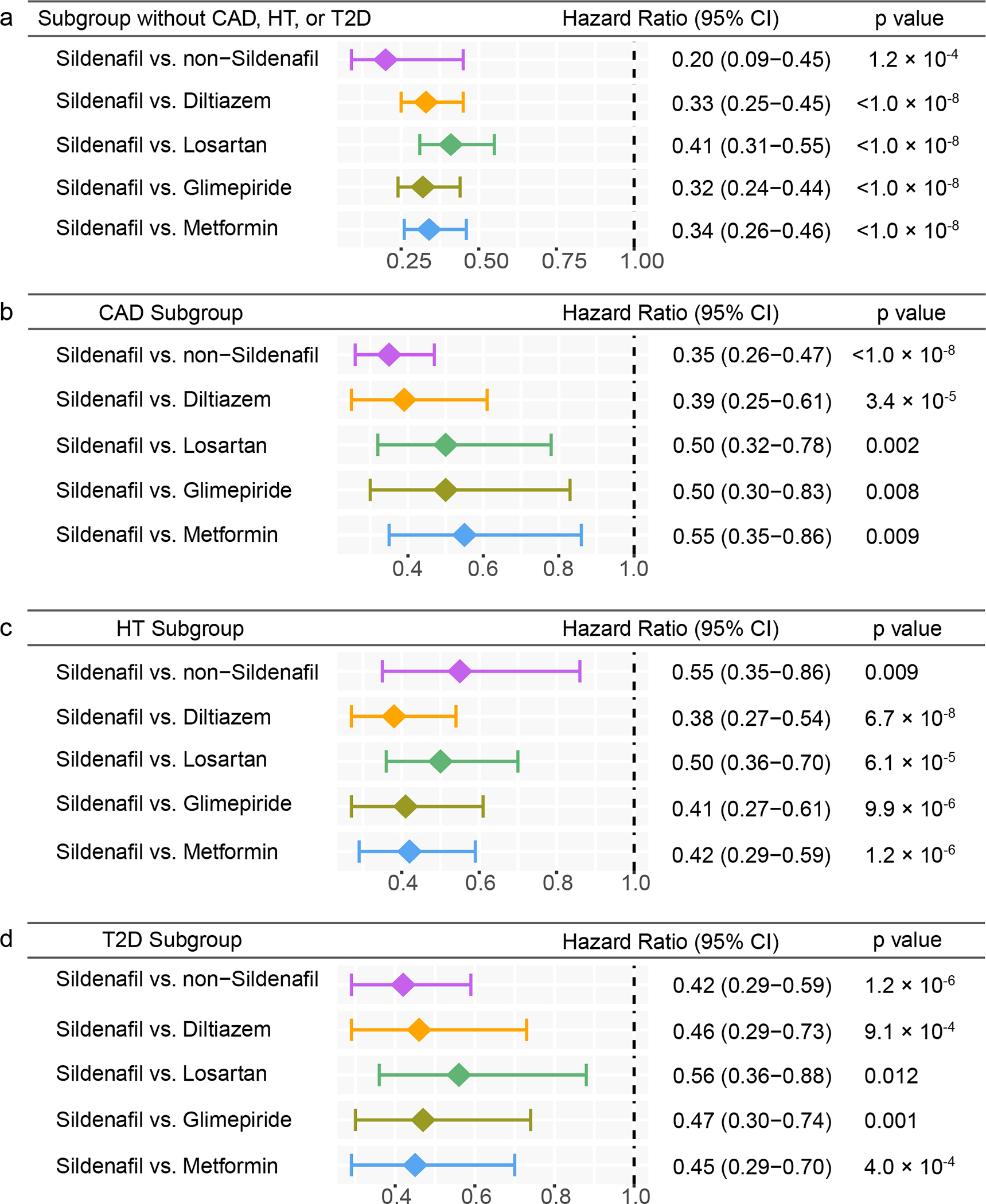
(a) Hazard ratios (HR) and 95% confidence intervals (CI) across five cohort studies after exclusion of individuals with coronary artery disease (CAD), hypertension (HT), and type-2 diabetes (T2D) (sildenafil n = 56,518; diltiazem n = 113,600; losartan n = 275,116; glimepiride n = 52,623; metformin n = 303,008). (b-d) HR and 95% CI plots across five cohort studies in individuals with CAD (b) (sildenafil n = 19,093; diltiazem n = 51,771; losartan n = 111,592; glimepiride n = 30,083; metformin n = 91,705), HT (c) (sildenafil n = 49,541; diltiazem n = 119,097; losartan n = 339,940; glimepiride n = 74,018; metformin n = 275,328), or T2D (d) (sildenafil n = 21,978; diltiazem n = 51,300; losartan n = 156,308; glimepiride n = 100,298; metformin n = 367,754). Non-sildenafil exposure population were matched to the exposures (ratio 4:1) by adjusting the initiation time of sildenafil, enrollment history, sex, and disease comorbidities (CAD, T2D, and HT). Propensity score stratified Cox-proportional hazards models and two-sided log-rank test were used to conduct statistical inference for the hazard ratios.
We also performed sex- and age-specific subgroup analysis. Although slight differences in age distributions were observed across five drug cohorts (Supplementary Fig. 4), we found that sildenafil was significantly associated with reduced likelihood (Fig. 5b, Supplementary Table 8) of AD across all five drug cohorts in both mid-older (65–74 years) and older individuals (75–100 years). Sex-specific subgroup analysis revealed that sildenafil usage is more significantly associated with a reduced risk of AD in males compared to females (Fig. 5a, Supplementary Table 9). Since the majority of sildenafil users are males receiving treatment for erectile dysfunction (ED) and male ED patients (75.9 ± 32.4 [standard deviation] mg) received a higher daily dosage of sildenafil compared to females (22.1 ± 15.0 mg) with pulmonary hypertension (p = 0.03, t-test, Supplementary Table 10), these factors may explain the more significantly reduced risk by sildenafil in male individuals (Fig. 5a). In addition, we found that sildenafil usage was associated with reduced likelihood of AD in both ED and pulmonary hypertension (off-label indication) populations as well (Supplementary Table 11 and Supplementary Fig. 5); yet, this trend lacks statistical significance because of the small sample size. In addition to sex and age, we didn’t observe a significant difference on annual total cost between sildenafil users and non-sildenafil users (p = 0.36, t-test, Supplementary Table 12), further reducing confounding risk of social economic factors. Altogether, these comprehensive subgroup analyses reveal that sildenafil is a candidate drug for prevention or treatment of AD (Figs. 3–5).
Figure 5. Sex-specific and age-specific subgroup analyses across five drug cohort studies.
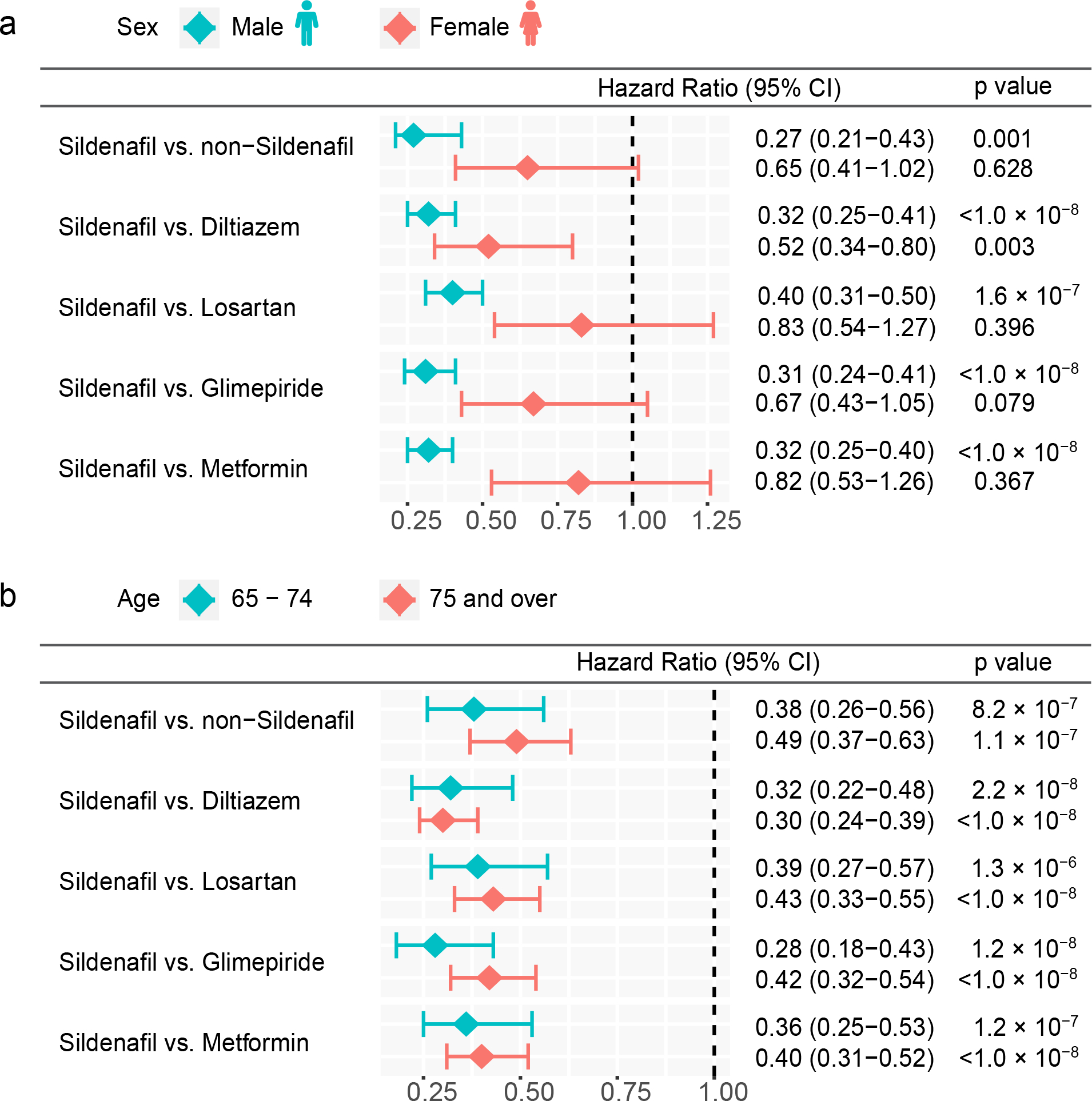
(a) Hazard ratios (HR) and 95% confidence intervals (CI) across five drug cohort studies for males and females respectively (sildenafil female n = 2,280 and male n = 114,132; non-sildenafil female n = 8,182 and male n = 452,174; diltiazem female n = 150,137 and male n = 101,223; losartan female n = 385,454 and male n = 278,811; glimepiride female n = 72,689 and male n = 86,908; metformin female n = 344,944 and male n = 378,138). (b) HR and 95% CI plots across five cohort studies for mild older individuals (65–74 years) and older individuals (75–100 years) (sildenafil 65–74 n = 89,875 and ≥75 n = 26,537; non-sildenafil 65–74 n = 264,209 and ≥75 n = 196,147; diltiazem 65–74 n = 115,892 and ≥75 n = 135,468; losartan 65–74 n = 389,084 and ≥75 n = 275,181; glimepiride 65–74 n = 97,243 and ≥75 n = 62,354; metformin 65–74 n = 505,210 and ≥75 n = 217,872). Non-sildenafil exposure population were matched to the exposures (ratio 4:1) by adjusting the initiation time of sildenafil, enrollment history, sex, and disease comorbidities (hypertension, type 2 diabetes, and coronary artery disease). Propensity score stratified Cox-proportional hazards models and two-sided log-rank test were used to conduct statistical inference for the hazard ratios.
In vitro observation of sildenafil’s mechanism-of-action
To examine the possible mechanism-of-action for sildenafil in AD, we next performed network analysis via an integration of drug targets and AD seed genes into the brain-specific PPI network (see Methods). RNA-sequencing data from the GTEx database36 suggest that several proteins (GSK3β, CDK5 and FYN) are highly expressed in human brain tissues. Network analysis shows that these three proteins have closer network distance to sildenafil’s targets than other seed genes in the human interactome (Fig. 6a). We didn’t find supporting evidence between sildenafil and FYN from the literatures. Importantly, recent studies have shown that GSK3β and CDK5 play crucial roles in AD pathobiology37 and microglia-mediated neuroinflammation38,39. We therefore selected GSK3β and CDK5 to investigate mechanism-of-action of sildenafil using human microglia HMC3 cells. First, HMC3 cells were treated with sildenafil at various concentrations (0.03 μM to 100 μM) for 48 h, and cell viability was determined by MTT assay (see Methods). As presented in Supplementary Fig. 6, sildenafil did not affect cell viability at any concentration, revealing low toxicity. Thus, the optimized concentration of sildenafil (30 and 100 μM) was used in the following experiments. As shown in Fig. 6b, phosphorylation levels of GSK3β and CDK5 were very low in the control vehicle and significantly increased (P = 0.036 for GSK3β and P = 0.007 for CDK5, two-tailed student’s t-test) by LPS treatment (1 μg/mL for 30 min) in HMC3 cells. Of note, pre-treating with sildenafil (30 and 100 μM) reduced phosphorylated GSK3β (P = 0.10 [30 μM] and P = 0.023 [100 μM], two-tailed student’s t-test, Fig. 6c) and CDK5 (P = 0.045 [30 μM] and 0.008 [100 μM], two-tailed student’s t-test, Fig. 6d) in a dose dependent manner (Supplementary Fig. 7).
Figure 6. Experimental validation of sildenafil’s likely mechanism-of-action in Alzheimer’s disease (AD).
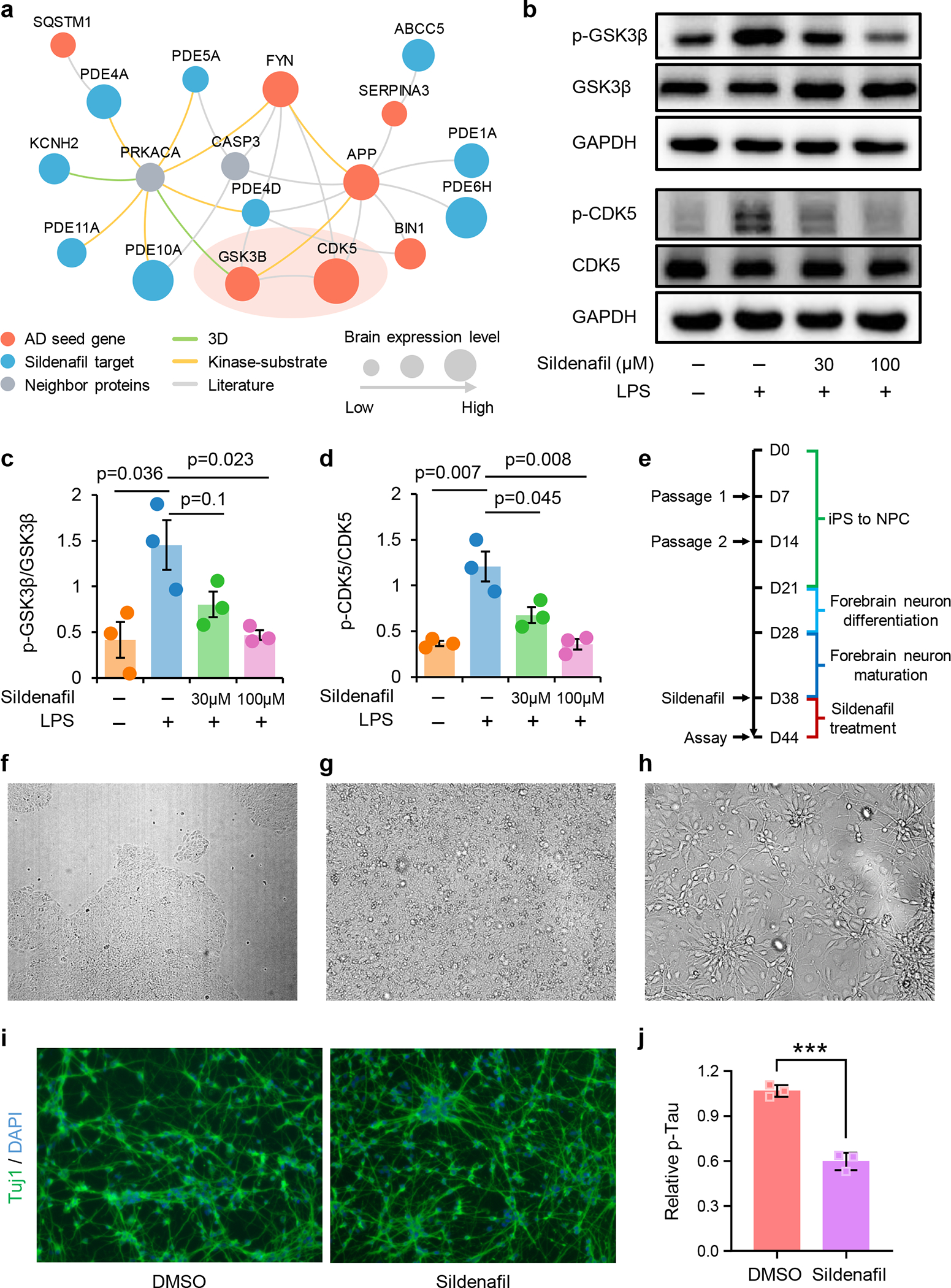
(a) Network analyses highlighting the inferred mechanism-of-action for sildenafil in AD. The molecular mechanisms of sildenafil were investigated via integration of known drug targets and experimentally validated AD seed genes into brain-specific human protein-protein interactome network (see Methods). Node size indicates the protein-coding gene expression level in brain compared with other 31 tissues from GTEx database. Larger size highlighting the high expression level in brain compared with other tissues. (b) Effects of sildenafil on LPS-induced activation of glycogen synthase kinase 3 beta (GSK3β) (c) and cyclin-dependent kinase 5 (CDK5) (d) in human microglia HMC3 cells. HMC3 cells were pretreated with sildenafil and followed LPS treatment (1 μg/mL, 30 min). The total cell lysates were collected and subjected to Western blot analysis. (e) Timeline of differentiation of AD iPSCs into forebrain neurons and downstream drug treatment. (f) iPSC HVRDi001-A-1 colonies, 5× magnification. Scale bar, 300 μm. (g) NPCs at day 7, 20× magnification. Scale bar, 100 μm. (h) Neural precursors at day 27, 20× magnification. Scale bar, 100 μm. (i) Anti-Tuj1 immunostaining of AD neurons after 6 days of treatment with DMSO or sildenafil (30 μM). Scale bar, 100 μm. (j) Decreased level of p-tau (181) after sildenafil treatment in neuronal lysates. Values are normalized to DMSO treatment group. Quantification data represent mean ± standard deviation (SD) of three independent experiments (n=3). P-value was computed by two-tailed student’s t-test. ***p < 0.001.
Sildenafil’s effect on AD patient iPSC-derived neurons
To further test the effect of sildenafil on AD, iPSCs that derived from AD patient were differentiated into forebrain neurons (see Methods). These AD neurons were treated with sildenafil at day 38 (Fig. 6e), when the process of differentiation and maturation are complete. At first, iPSC was differentiated into neural progenitor cells, and then neural precursors (Fig. 6g–6h). Neural precursors were further matured for 10 days before treatment with sildenafil or DMSO control. After 6 days of treatment, the AD neurons were immune-stained with Tuj1, a neuronal marker. The sildenafil treated group had elevated neurite growth comparing to the DMSO control (Fig. 6i). Phosphorylated-tau181 (p-tau 181) is an early biomarker of AD pathology and correlates with cognitive declines40. To investigate whether sildenafil reduces p-tau accumulation in AD neurons, cells were lysed and an ELISA assay was performed to measure the level of p-tau 181. We found that sildenafil significantly reduced the accumulation of p-tau 181 in AD patient iPSC-derived neurons (Fig. 6j). In summary, these data mechanistically support sildenafil’s potential beneficial effect in AD, complementing our endophenotype-based prediction and patient population-based analyses.
Discussion
We demonstrated that endophenotype-informed drug discovery offers a valuable and complementary approach to promote AD therapeutic development, which optimizes the usefulness of available data and adds new knowledge relevant to potential treatments for AD. Specifically, we showed proof-of-concept of an AD endophenotype module, shared by amyloid and tau, in both transcriptomic and proteomic data across multiple types of AD transgenic mouse models. Of note, recent clinical and experimental studies have also demonstrated that synergistic interaction of amyloid and tau is a greater contributor to AD than either amyloid or tau alone16–18. To further prove our AD endophenotype hypothesis, we examined network relationships of drugs under the AD ongoing trials and found that drugs targeting both amyloid and tau pathways had significantly stronger network relationships with amyloid and tau endophenotype modules (Extended Data Fig. 3), compared with those targeting amyloid or tau pathways alone. This prompted us to develop network-based methodologies for AD drug repurposing via quantifying the network distance between AD modules and drug targets in the human interactome.
In silico drug repurposing identified 66 potential drug-AD associations across diverse pathways (Fig. 2). Nervous system drugs for non-AD indications, such as Parkinson’s disease or amyotrophic lateral sclerosis (ALS), may offer potential candidates for AD drug repositioning due to their safety and ability to penetrate the brain. For example, riluzole is an FDA-approved drug for ALS and is now in Phase II trials for mild AD patients (ClinicalTrials.gov Identifier: NCT01703117). In addition, cardiovascular diseases (e.g., hypertension) and metabolic diseases (e.g., diabetes) have been reported as risk factors for AD and share possible pathological mechanisms with AD41,42. Carvedilol (z = −2.23 with AD seed module), an adrenergic receptor blocker for hypertension, significantly attenuates brain oligomeric Aβ levels and cognitive deterioration in two mouse models43.
Sildenafil is a PDE5 inhibitor, and PDEs are broadly expressed in human brain36. Recent studies have shown that PDE inhibitors have beneficial therapeutic effects in preclinical models of AD by modulating neuronal plasticity, reducing tau phosphorylation, improving cognitive impairment, decreasing Aβ plaque accumulation, and enhancing the level of brain-derived neurotrophic factor (BDNF)44. Based on the available data, we selected sildenafil to test for possible associations with AD using a large-scale patient longitudinal database.
Using Cox regression matching, we found that sildenafil usage was significantly associated with a decreased risk of AD (Fig. 3). Importantly, propensity score-stratified cohort studies confirmed that sildenafil usage was significantly associated with a decreased likelihood of AD compared to comparator drugs (Figs. 3b–3e), including diltiazem (Fig. 3b), losartan (Fig. 3c), glimepiride (Fig. 3d) and metformin (Fig. 3e). Since sildenafil is a drug prescribed primarily to male individuals, we performed sex-specific subgroup analyses. This analysis showed that sildenafil usage was associated with a decreased risk of AD across all five comparators for male individuals; in females, sildenafil usage was associated with a reduced AD only when compared to diltiazem (Fig. 5a and Supplementary Table 9). There are two possible explanations. First, as sildenafil is primarily prescribed to male individuals with ED, there are no enough female patients (Table 1) to appropriately power a study for statistical significance. Indeed, there are few female individuals who received sildenafil for treatment of pulmonary hypertension. Second, we found that male individuals with ED had a higher average daily dosage of sildenafil than females (Supplementary Table 10); yet, we found that average daily dosage of sildenafil was not associated with incidence of AD (HR 0.99, 95% CI 0.99–1.0, P = 0.19, log-rank test). Sildenafil usage was associated with reduced likelihood of AD in individuals with ED or pulmonary hypertension (Supplementary Table 11 and Supplementary Fig. 5), although this trend lacks statistically significance by the small sample size. Specifically, there are 10 AD cases in ED population and 7 cases (Supplementary Table 11) in pulmonary hypertension population compared to in total 93 AD cases (Table 1) in all sildenafil users. Taken together, the male-specific, reduced risk by sildenafil must be investigated in sex-balanced populations and sex-matched randomized clinical trials in the future. In addition, sildenafil may have male-specific benefits in reducing the incidence of AD, the basis of which remains to be discovered.
We showed that sildenafil usage was associated with a reduced AD risk in both the general population and subgroup analyses after adjustment of sex, race, age, T2D, HT and CAD (Figs. 3–5). To reduce the likelihood of confounding by indication (such as diabetes, heart disease, and hypertension), we conducted comprehensive subgroup analyses. We found that sildenafil usage was significantly associated with reduced risk of AD across all 5 drug cohorts in either individuals without CAD, HT and T2D (Fig. 4a), or individuals with CAD (Fig. 4b), HT (Fig. 4c), or T2D (Fig. 4d). Importantly, age-specific subgroup analyses revealed that sildenafil usage was significantly associated with reduced risk of AD in the older group as well (Fig. 5b and Supplementary Table 8).
To investigate the mechanism-of-action of sildenafil against AD, we further integrated drug targets of sildenafil and known AD seed genes into the brain-specific human PPI network. Network analysis prompted us to explore whether sildenafil targets GSK3β and CDK5 (Fig. 6a). Hyperphosphorylation of the microtubule-associated protein tau is a hallmark of AD45, and GSK3β and CDK5 are involved in aberrant phosphorylation of tau46. Microglial cells are a specific population of macrophages in the central nervous system proposed to be involved in the tau pathology of AD47. Although previous studies have revealed that sildenafil reverses cognitive impairment in Tg2576 mice via inhibiting overall brain activity of GSK3β and CDK533, it has not previously been tested whether this occurs in human microglial cells. GSK3β and CDK5 are involved in the LPS-induced proinflammatory cytokine expression and innate immune response in macrophages48. We therefore exposed human microglia to LPS to evaluate the effects of sildenafil on GSK3β and CDK5 activation. We found that sildenafil downregulated GSK3β and CDK5 in human microglia cells, supporting mechanistically its potential beneficial effect in AD. We further test the effect of sildenafil using AD patient iPSC-derived neurons. We found that sildenafil increased neurite growth and decreased p-tau 181 expression in AD patient iPSC neurons, further supporting a potential mechanism for its beneficial effect in AD (Fig. 6c–6j). Previous in vivo studies revealed that sildenafil improved memory, amyloid plaque load, inflammation, and neurogenesis in APP/PS1 mice49,50. Sildenafil also improved memory, tau hyperphosphorylation, and GSK3β phosphorylation in J20 mice51, which is consistent with our in vitro mechanistic observation in AD patient-derived iPSC neuron models (Fig. 6). Two pilot studies also showed potential benefits of sildenafil in treatment of AD: (1) a single dosage of 50 mg sildenafil decreased spontaneous neural activity in right hippocampus in 10 AD patients52; and (2) a single dosage of 50 mg sildenafil increased cerebral metabolic rate of oxygen and cerebral blood flow in 12 AD patients, and decreased cerebral vascular reactivity in 8 AD patients53. Altogether, we believe that these data provide a potential mechanism-of-action for the protective efficacy of sildenafil in AD, complementing our endophenotype-based prediction and population-based validation.
There are several strengths of our approach compared to previous studies21. First, we demonstrated a proof-of-concept of an endophenotype-based network methodology for AD drug discovery using a synergistic endophenotype between amyloidosis and tauopathy16–18. We performed multi-omics data integration by incorporating various transcriptomics and proteomics data from AD transgenic mouse models to improve our network-based predictions. Second, our population-based validation attempts were based on a large patient-level longitudinal database and state-of-the-art pharmacoepidemiologic analyses. The massive longitudinal data allows access to real-world patient populations, while maintaining adequate statistical power. Covariates within the data were adjusted for potential confounds of AD and erectile dysfunction.
Limitations of this work should also be acknowledged. Although we have assembled large-scale, experimentally-validated drug-target interactions and the human interactome, potential literature bias and data incompleteness could affect the findings54. For example, we found that the incompleteness of drug-target networks may influence the poor z-score for certain drugs (Supplementary Table 6). Second, detailed clinical information is not available from health insurance claims data although we adjusted age, sex, and disease comorbidities (including CAD, HT, T2D, and MCI). In addition, pharmacoepidemiologic analysis may not be suitable for testing drugs that have fewer patient data. We were able to perform population-based validation for sildenafil, as it has a large amount of patient data available. For drugs with less patient data, replication of the associations using multiple large population-based cohorts is suggested.
Moreover, pharmacoepidemiologic studies are subject to bias due to unmeasured confounding factors. The MarketScan Medicare supplementary database does not contain detailed clinical information with respect to the enrollees’ education level or social economic status, and lacks genotyping information, such as the apolipoprotein E (APOE) status. APOE genotyping status have been associated with treatment responses of AD, including aducanumab55. Yet, we cannot inspect relationship between sildenafil-AD outcome and APOE genotyping as currently used insurance claims don’t have APOE genotyping information for individuals. We may investigate sildenafil’s mechanism-of-action in AD patient-derived iPSC models with specific APOE genotypes56. We defined AD outcomes using the International Classification of Disease codes and accuracy of phenotyping codes may influence our findings as well. Combining biomarker-based confirmation of AD diagnosis and phenotyping codes should be used in the future when those biomarker information (such as imaging and blood biomarkers) are available at the electronic health records.
Sildenafil may be more likely to be prescribed for wealthy individuals and increased wealth was associated with a reduced risk of developing AD and AD-related dementia57,58. We didn’t observe a significant difference on annual total cost between sildenafil users and non-sildenafil users (Supplementary Table 12). Charlson comorbidity index analysis reveals that the sildenafil users have the equal health/disease conditions on average with those receiving comparator drugs after PS-matching (Supplementary Fig. 3). However, social economic status and health inequalities between sildenafil users and comparator drug users should be investigated further in future studies. We found that average daily dosage of sildenafil was not associated with incidence of AD (Supplementary Table 10); yet, dosage and timing of exposure are important confounding factors and they should be investigated in future placebo-controlled trials as well. Finally, population-based observational studies cannot build causal relationship between sildenafil use and beneficial clinical response of AD. Future causal methods, such as Mendelian randomization studies should be utilized further59,60. Taken together, the association between sildenafil usage and decreased incidence of AD does not establish causality or its direction, and our results therefore warrant rigorous clinical trial testing of the treatment efficacy of sildenafil in AD patients, inclusive of both sexes and placebo-controlled.
In summary, this study offers an integrated, network-based approach that incorporates endophenotype module findings, in vitro mechanistic observations, and large-scale patient data analysis for the discovery of effective therapeutics to protect patients from developing AD or reduce their risk. This approach can be applied to other neurodegeneration diseases by exploiting other endophenotypes, such as inflammation and metabolic dysfunction. Other repurposable agents were also identified in this study that could be interrogated in greater detail to determine their anti-AD potential.
Methods
Construction of the human protein–protein interactome
To construct a high-quality and comprehensive human protein-protein interactome, we assembled 15 commonly used data sources with five types of experimental evidence: (1) binary PPIs tested by high-throughput yeast-two-hybrid (Y2H) systems via integrating two publicly available high-quality Y2H datasets21,61,62; (2) kinase-substrate interactions from literature-derived low-throughput and high-throughput experiments; (3) database integration-based PPIs identified by affinity purification followed by mass spectrometry (AP-MS) and literature-derived low-throughput experiments; (4) binary, physical PPIs from protein three-dimensional structures; and (5) signaling networks derived from low-throughput experiments as annotated in SignaLink 2.063. Genes were annotated to their Entrez ID using the NCBI database as well as their official gene symbols from GeneCards (http://www.genecards.org/). Inferred data, such as evolutionary analysis, gene expression data, and metabolic associations, were excluded. The updated human interactome consisted of 351,444 PPIs (edges or links) linked to 17,706 unique proteins (nodes). The detailed descriptions of building the human interactome are given in our recent studies23,24.
Building endophenotype models for Alzheimer’s disease
In order to build disease modules for AD, we first collected experimentally validated (seed) genes in amyloidosis (amyloid) and tauopathy (tau) respectively. These genes satisfied at least one of the following conditions: i) gene validation in large-scale amyloid or tauopathy GWAS studies; ii) in vivo experimental model evidence that knockdown or overexpression of the gene leads to AD-like amyloid or tau pathology. Based on these criteria, we obtained 54 seed genes related to amyloid and 27 seed genes related to tauopathy (Supplementary Table 13). We further constructed a list of AD seed genes for building the AD endophenotype (see module characterized by both amyloid and tau) via assembling multiple data sources as below: i) 54 amyloid seed genes and 27 tauopathy seed genes; ii) 47 late-onset AD common risk genes identified by large-scale genetic studies; iii) 35 genes with at least two AD-causing mutations from the Human Gene Mutation Database (HGMD)64; iv) 39 disease genes curated in at least 2 of 4 following disease gene databases: HGMD (n=63), DisGeNET (n=23, score≥0.2)65, MalaCards (n=79)66, and Open targets (n=81, overall score ≥ 0.7 and literature score > 0)67. After removing the duplicates, 144 AD seed genes were obtained (Supplementary Table 13).
We used a network approach, termed the largest connected component (LCC) approach, to build a disease module. Specifically, we rebuilt the new network modules by searching the LCC formed by the list of AD seed genes. We further performed the permutation testing as below to test the significance of disease modules:
| (1) |
A nominal P was computed by counting the number of permutations (Sm (p)) greater than the number of observed LCC (Sm) formed by the randomly selected proteins with a similar connectivity distribution in the human interactome network. We performed the permutations (1,000 times) to calculate the statistical significance.
Transcriptomics-based endophenotype models
We collected a total of 34 transcriptomic datasets consisting of original brain microarray data (n=17) and bulk RNA-seq data (n=17) from 7 types of transgenic mouse models, including 5XFAD, APP/PS1, HO-TASTPM, Tg2576, TgCRND8, Tau P301L, and rTg4510. The detailed mutations and neuropathology for these models are provided in Supplementary Table 14. All the datasets are composed of samples from total brain, specific brain regions (including hippocampus, cortex, and cerebellum), and brain-derived microglial cells. The original brain microarray datasets were obtained from Gene Expression Omnibus (GEO)68, and detailed information of these GEO datasets is provided in Supplementary Table 14. Expression data were subjected to R statistical processing using the limma/Bioconductor package (v3.13)69. Differentially expressed genes (DEGs) were then identified using a conservative statistical threshold of False Discovery Rate (FDR) < 0.05 and |fold change (FC)| > 1.5 for differential expression (see Supplementary Materials and Methods). The DEGs identified from RNA-seq data were collected from four previous studies70–73 across four types of AD genetic mouse models (5XFAD, Tg2576, TgCRND8, and rTg4510). For 7 RNA-seq transcriptomic profiles from 5XFAD70,71 and rTg451072, the threshold for significance of differential expression was set to FDR < 0.05 using the Benjamini-Hochberg’s method74. Given the larger number of DEGs identified in Tg2576 and TgCRND8 by a threshold P of 0.05, we used a stricter criterion (T test P<0.01) to determine DEGs compared with corresponding wildtype (WT) mice72. All DEGs identified in mouse models were further mapped to unique human-orthologous genes using the Mouse Genome Informatics (MGI) database75. All statistical analyses were performed using R software.
Proteomics-based endophenotype models
In total, 10 proteomic datasets were assembled from 3 types of AD transgenic models76,77. For instance, these AD transgenic models included proteome-wide differential analysis from frontal cortical, hippocampal, and cerebellar extracts in hAPP (3M [month] and 12 M) and hAPP-PS1 (3M and 12M) mice. We obtained four differentially-expressed protein sets (hAPP_3M, hAPP_12M, hAPP-PS1_3M and hAPP-PS1_12M) after merging the differentially expressed proteins from different brain regions (Supplementary Table 14). In addition, we performed differential protein analyses using proteomics data generated in hippocampus across two types of transgenic mice: (i) ADLPAPT mice (4M, 7M, and 10M) carrying three human transgenes (APP, PS1 and MAPT) and (ii) hAPP-PS1 (4M, 7M, 10M) mice. After mapping them to human-orthologs75, we obtained six sets of differentially-expressed proteins, including ADLPAPT_4M, ADLPAPT_7M, ADLPAPT_10M, hAPP-PS1_4M, hAPP-PS1_7M, and hAPP-PS1_10M (see Supplementary Materials and Methods).
Construction of drug-target network
We assembled physical drug-target interactions for U.S. Food and Drug Administration (FDA)-approved drugs via integrating six commonly used data sources21. We defined a physical drug-target interaction using reported binding affinity data: inhibition constant/potency (Ki), dissociation constant (Kd), median effective concentration (EC50), or median inhibitory concentration (IC50) ≤ 10 μM. First, we acquired drug-target interactions from the DrugBank database (v4.3)78, the Therapeutic Target Database (TTD, v4.3.02)79, and the PharmGKB database80. We then extracted bioactivity data related to FDA-approved drugs from three commonly used databases: ChEMBL(v20)81, BindingDB82, and IUPHAR/BPS Guide to PHARMACOLOGY83. We retained only those items satisfying the following criteria: (i) binding affinities, including Ki, Kd, IC50, or EC50, of ≤10 μM; (ii) protein targets having a unique UniProt accession number; (iii) protein targets marked as “reviewed” in the UniProt database; and (iv) proteins present in Homo sapiens. In total, we obtained 15,367 drug–target interactions connecting 1,608 FDA-approved drugs and 2,251 unique human targets/proteins.
Network proximity analysis
Given a set of disease proteins (A), a set of drug targets (B), the closest distance dAB measured by the average shortest path length of all the nodes to the other module in the human protein-protein interactome is defined as:
| (2) |
where d(a, b) indicates the shortest path length between protein a and drug target b. We conducted a permutation test to calculate the significance of the network distance between a given drug and the AD endophenotype disease module. We further constructed a reference distance distribution corresponding to the expected distance between two randomly selected groups of proteins of the same size and degree distribution as the original disease proteins and drug targets in the network. The randomization was performed by first binning all the proteins in the interactome by their degree, then a number of genes were drawn from each bin based on the degree distribution of the AD seed genes21,25. This procedure was repeated 1,000 times. As we do permutation testing for each drug individually, there are no multiple comparisons for drugs in network proximity measures. Details for network proximity measure are provided in previous studies21,25.
Pharmacoepidemiologic validation
Study cohorts.
The pharmacoepidemiology study utilized the MarketScan Medicare Claims database from 2012 to 2017 based on the Medicare Advantage and Fee for services. The dataset included individual-level diagnosis codes, procedure codes and pharmacy claim data for 7.23 million patients. Pharmacy prescriptions of sildenafil, diltiazem, losartan, glimepiride and metformin were identified by using RxNorm and National Drug Code (NDC).
Outcome measurement.
For a subject exposed to the aforementioned drugs, a drug episode is defined as from drug initiation to drug discontinuation (Supplementary Fig. 8). Specifically, drug initiation is defined as the first day of drug supply (i.e. 1st prescription date). Drug discontinuation is defined as the last day of drug supply (i.e. last prescription date + days of supply) and without drug supply for the next 90 days. In another word, gaps of less than 90-day of drug supply were allowed within a drug episode. The sildenafil cohort included the first sildenafil episode for each subject, as well as the diltiazem cohort, the losartan cohort, the glimepiride cohort and the metformin cohort. Further, we excluded observations that started within 180-day of insurance enrollment. For the final cohorts, demographic variables including age, sex and geographical location were collected. Additionally, diagnoses of hypertension (HT), type 2 diabetes (T2D), coronary artery disease (CAD), and Mild cognitive impartment (MCI) (the International Classification of Disease [ICD] codes are given in Supplementary Table 15) before drug initiation were collected. Herein, we focused on AD outcomes only using the well-established ICD9/ICD10 codes by excluding other types of dementia (i.e., vascular dementia)84. All AD ICD9/10 codes were further ascertained the validity and reliability of all claims data85 before use. These variables were specifically selected to address potential confounding biases. Last, a control cohort was selected from patients who were not exposed to sildenafil. Specifically, non-exposures were matched to the exposures (ratio 4:1) by initiation time of sildenafil (Supplementary Fig. 9), enrollment history, sex, HT diagnosis, T2D diagnosis and CAD diagnosis. The geographical location, diagnoses of HT and T2D were collected for the control cohort as well. The primary outcome in pharmacoepidemiologic studies is time from drug initiation to AD diagnosis or drug discontinuation for sildenafil, as well as the comparator drugs. For the sildenafil, diltiazem, losartan, glimepiride and metformin cohorts, observations without diagnosis of AD were censored at the end of drug episodes. For the control cohort, the corresponding sildenafil episode starting date was used as the starting time. Observations without diagnosis of AD were censored at the corresponding sildenafil episode’s end date (Supplementary Fig. 8).
Covariate adjustment.
The covariates include age, sex, geographic location, HT diagnosis, T2D diagnosis, and CAD diagnosis. All variables were specifically selected to address clinical scenarios evaluated in each study.
Charlson comorbidity.
The Charlson comorbidity index predicts the one-year mortality for a patient who may have a range of comorbid conditions86–88. Collected comorbidities contain chronic cardiac disease, chronic respiratory disease (excluding asthma), chronic renal disease (estimated glomerular filtration rate ≤30), mild to severe liver disease, dementia, chronic neurological conditions, connective tissue disease, diabetes mellitus (diet, tablet, or insulin controlled), human immunodeficiency virus (HIV) or acquired immunodeficiency syndrome (AIDS), and malignancy. These conditions were selected a priori by a global consortium to provide rapid, coordinated clinical investigation of patients presenting with any severe or potentially severe acute infection of public interest and enabled standardization. In this study, we used the clinical diagnosis record within 6 months prior the drug episode starting date to determinate comorbidity score.
Statistical analysis.
The survival curves for time to AD were estimated using a Kaplan-Meier estimator approach. Additionally, propensity score stratified survival analyses were conducted to investigate the risk of AD between sildenafil users and non-sildenafil users, sildenafil users and diltiazem users, sildenafil users and losartan users, sildenafil users and glimepiride users, as well as sildenafil users and metformin users (Table 1). Specifically, for each comparison, the propensity score of taking sildenafil was estimated by using a logistic regression model, in which the covariates included age, sex, geographical location, T2D diagnosis, HT diagnosis and CAD diagnoses (Supplementary Fig. 9). Propensity score stratified Cox-proportional hazards models were used to conduct statistical inference for the hazard ratios (HR) of developing AD between cohorts.
Brain-specific subnetwork analysis
We collected RNA-seq data (RPKM value) of 32 tissues from GTEx v6 release36. For each tissue (e.g., brain), we regarded those genes with reads per kilobase millions (RPKM) ≥ 1 in over 80% of samples as tissue-expressed genes, and the rest as tissue-unexpressed. To quantify the expression significance of tissue-expressed gene i in tissue t, we calculated the average expression E(i) and the standard deviation δE(i) of a gene’s expression across all considered tissues89. The significance of gene expression in tissue t is calculated:
| (3) |
Then, we performed a brain-specific co-expressed PPI network analysis by comparing genome-wide expression profiles of brain to 31 other tissues from the GTEx database41.
In vitro mechanistic observation
Reagents.
Sildenafil citrate (purity > 98%) was purchased from Topscience. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), and Lipopolysaccharides (LPS) (Cat# L2880) was purchased from Sigma-Aldrich. Antibodies against CDK5 (Cat# A5730), GSK3β (Cat# A2081), and phospho-GSK3B-Y216 (Cat# AP0261) were obtained from ABclonal Technology. CDK5-Phospho-Tyr15 (Cat# YP0380) was purchased from Immunoway (Plano, Texas, USA). All other reagents were obtained from Sigma-Aldrich unless otherwise specified. The validation of all primary antibodies for the species are available from the manufacturer’s website.
Cell viability.
Human microglia HMC3 cells were obtained from American Type Culture Collection (ATCC, Manassas, VA). Cell viability was analyzed by the MTT method as described previously22. Briefly, 5,000 cells/well were plated in 96-well plates for 12 h, then treated with sildenafil for 48 h. After treatment, MTT solution was added to the cells to a final concentration of 1 mg/mL and the mixture was allowed to incubate at 37 °C for 4 h. The supernatant was removed, and the precipitates were dissolved in DMSO. Absorbance was measured at 570 nm using a Synergy H1 microplate reader (BioTek Instruments, Winooski, VT, USA).
Western blot analysis.
Human microglial HMC3 cells were pre-treated with DMSO (control vehicle) or sildenafil, and followed with 1 μg/mL LPS for 30 min. The concentration (30 μM or 100 μM) of sildenafil used in this study was based on data from previous studies90. Cells were harvested, washed with cold PBS, and then lysed with RIPA Lysis Buffer with 1% Protease Inhibitor (Cat# P8340, Sigma-Aldrich). Total protein concentration was measured using a standard BCA protein assay kit (Bio-Rad, CA, USA) according to the manufacturer’s manual. Samples were electrophoresed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), then blotted onto a polyvinylidene difluoride (PVDF; EMD Millipore, Darmstadt, Germany) membrane. After transferring, the membranes were probed with specific primary antibodies (1:1000) at 4 °C overnight. Specific protein bands were detected using a chemiluminescence reagent after hybridization with a horseradish peroxidase (HRP)-conjugated secondary antibody (Rabbit anti-Human IgG Secondary Antibody (Cat # PA1–28587)) (1:3000).
Differentiation of iPSCs into forebrain neurons
An AD patient-derived iPSC line HVRDi001-A-1 (WiCell) was previously cultured in mTeSR™1 medium. At day 0, they were broken up into single cells and plated onto tissue culture-treated 6-well Matrigel-coated plate with a density of 2 – 2.5 × 105 cells/cm2. Cells were cultured in neural induction medium (05839, STEMCELL Technologies) in a 37°C, 5% CO2 incubator. Monolayer neural progenitors were then generated and passaged at day 7 and day 14. At day 21, cells were dissociated and cultured in forebrain neuron differentiation medium (08600, STEMCELL Technologies). At day 28, cells were differentiated into neuronal precursors and plated in 96 well poly-L-ornithine/laminin-coated plates with a density of 6 × 104 cells/cm2 in forebrain neuron maturation medium (08605, STEMCELL Technologies). After 10 days of maturation, neurons were treated with either DMSO or Sildenafil.
Immunofluorescence analysis of iPSC-derived neuron cultures
Neurons were fixed in 4% paraformaldehyde and permeabilized in 0.1 Triton X-100. Afterward, neurons were incubated with 10% serum and stained with Tuj1 (ab18207, abcam). The secondary antibody was IgG-conjugated Alexa Fluor 488 (111–545-003, Jackson ImmunoResearch). Nuclei were counterstain with DAPI (4′,6-diamidino-2-phenylindole). Images were taken with Leica inverted microscope (DMI6000SD, Leica Microsystems).
p-tau measurement in iPSC-derived neuron lysis
After six days of DMSO or Sildenafil treatment, cell culture media was aspirated and neurons were lysed in 50 μL Neuronal Protein Extraction Reagent (87792, Thermo Fisher Scientific) with protease and phosphatase inhibitors (78442, Halt). Lysates were then diluted into 50 μL and run on a tau (Phospho) [pT181] Human ELISA Kit (KHO0631, Invitrogen). The plate was read at 450 nm absorbance in a microtiter plate reader (Biotek). A 4-parameter standard curve was generated and the concentration of each sample was read from the standard curve. The dilute factor was multiplied to correct the sample dilution and obtain the original p-tau 181 concentration. To make a comparison among different treatments, the total protein concentration of each cell lysis sample was determined by running a BCA protein assay (Thermo Scientific). The final p-tau 181 concentration was given by the original concentration divided by the total protein concentration.
Data availability:
Data supporting the findings of this study are available within Supplementary Table Files. Human protein-protein interactome and drug-target network can be downloaded from https://github.com/ChengF-Lab/endoAD. Tissue/brain-specific expression data were downloaded from GTEx database (https://www.gtexportal.org/home/). The expression datasets used in this study were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) with accession IDs GSE65067, GSE74437, GSE74438, GSE64398, GSE53480, GSE56772, and GSE57583. The health insurance claims data are available from the MarketScan Medicare Claims database (2012 to 2017) based on the Medicare Advantage and Fee for services from IBM MarketScan Research Databases.
Code availability:
Source codes for network proximity analysis and disease module identification are available: https://github.com/ChengF-Lab/GPSnet.
Extended Data
Extended Data Fig 1. Proof-of-concept of network module of Alzheimer’s disease (AD).
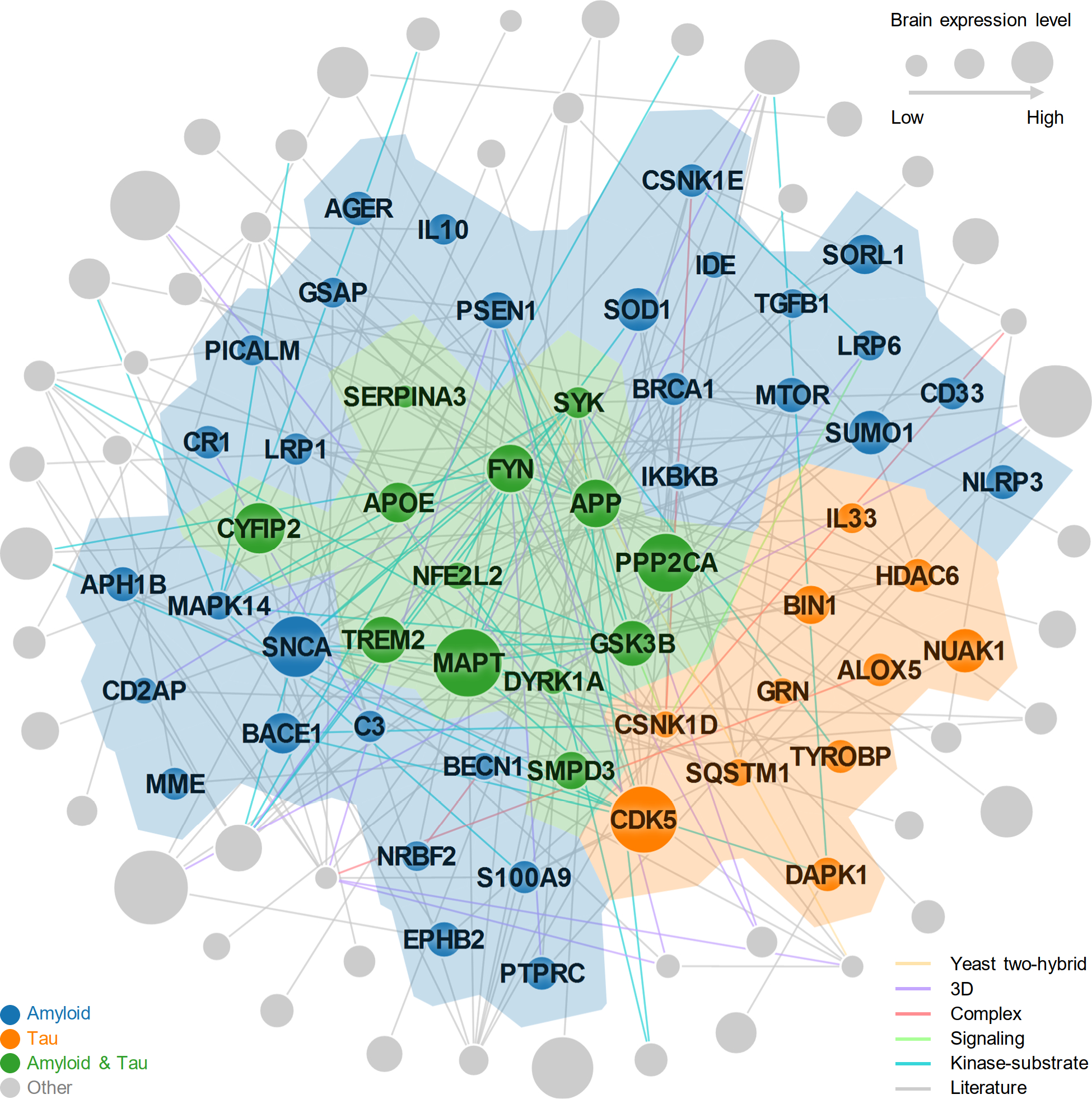
A subnetwork highlighting disease module (AD seed module) characterized by both amyloidosis and tauopathy under the human protein-protein interactome model. The AD seed module includes 227 protein–protein interactions (PPIs) (edges or links) connecting 102 unique proteins (nodes).
Extended Data Fig 2. Heatmap illustrates the network proximity between molecular targets of 21 ongoing repurposable AD drugs and 37 AD disease modules.
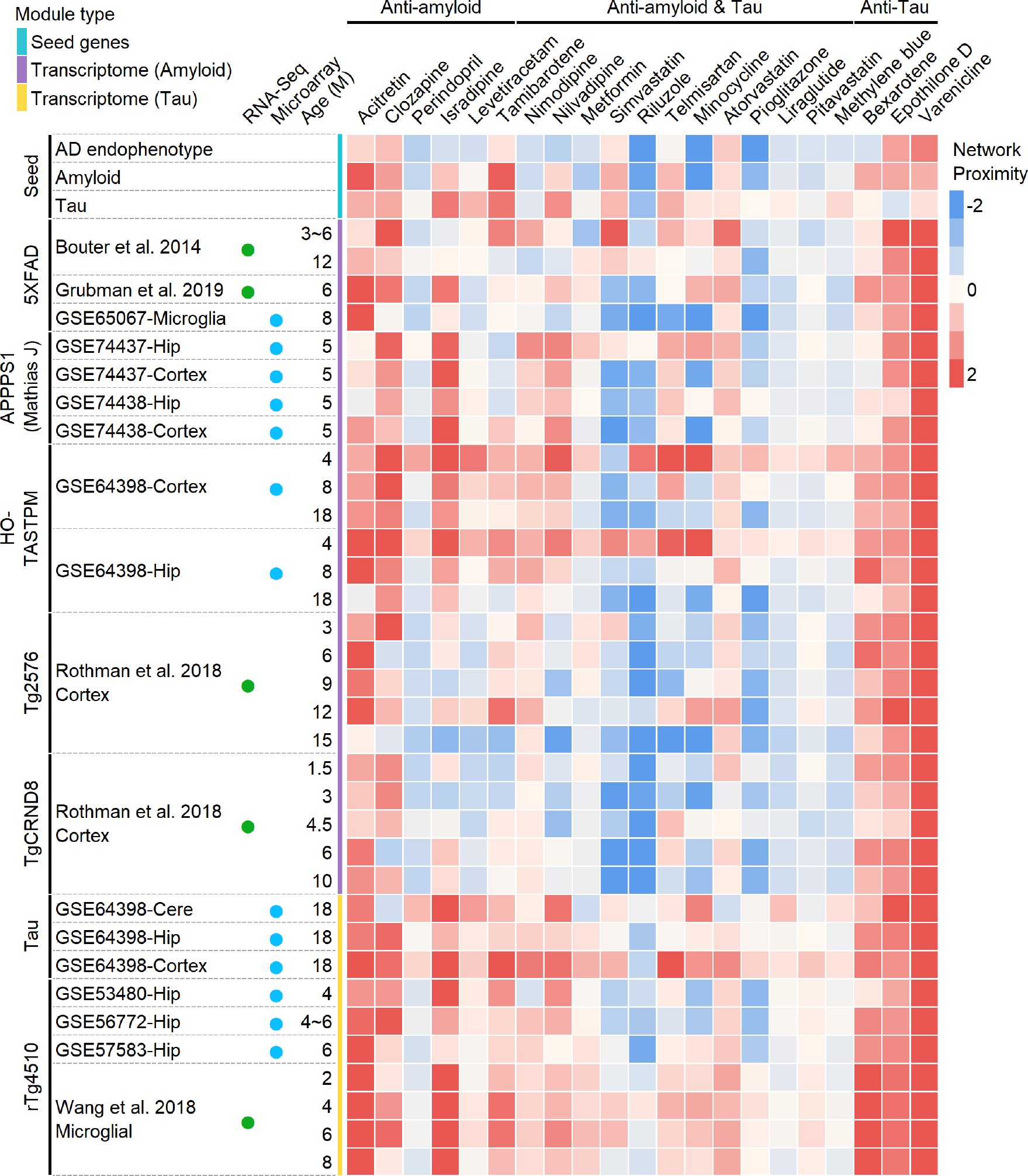
These drugs could ameliorate amyloid, tau or both of them (amyloid & tau) pathology or target amyloid, tau or amyloid & tau related pathways in vitro, in vivo mouse models or in patients with AD. We built three AD network modules by assembling experimentally validated (seed) genes in amyloidosis (amyloid), tauopathy (tau), and AD (characterized by both amyloid and tau). In addition, we also built disease modules from 34 differentially expressed gene (DEG) sets derived from transcriptomics data (including microarray, and bulk RNA-sequencing) from AD genetic mouse.
Extended Data Fig 3. The efficacy of endophenotype-based drug repurposing in Alzheimer’s disease (AD).
Molecular targets of 21 ongoing repurposable AD drugs that target both Amyloid and Tau have significantly closer network distance with AD network modules built from transcriptomics (a) and proteomics (b) data from AD genetic mouse, in comparison to drugs targeting amyloid or tau alone. In a, we built AD modules from 34 differentially expressed gene sets derived from transcriptomics data (including microarray, and bulk RNA-sequencing) from AD genetic mouse. In b, we built AD modules for 10 differentially expressed protein sets from proteomics data in AD genetic mice. P values were calculated by Wilcoxon test (one-side) in a and b.
Extended Data Fig 4. Network-based in silico drug repurposing for Alzheimer’s disease (AD).
13 AD disease modules, including 3 AD seed genes and 10 differentially expressed proteins (DEPs) sets from AD mouse proteomics, were used to screen FDA-approved drugs for AD. A sankey plot illustrates a global view of 66 drug candidates identified by network proximity. Drugs are grouped by their first-level Anatomical Therapeutic Chemical Classification (ATC) codes.
Extended Data Fig 5. Longitudinal analyses reveal that sildenafil usage is significantly associated with reduced likelihood of AD in individuals with coronary artery disease (CAD).
Five comparator analyses were conducted including: (a) sildenafil vs. matched non- sildenafil, (b) sildenafil vs. diltiazem (an anti-hypertensive drug), (c) sildenafil vs. losartan (an anti-hypertensive drug candidate in an AD clinical trial [ClinicalTrials.gov Identifier: NCT02913664]), (d) sildenafil vs. glimepiride (an anti-diabetic drug), and (e) sildenafil vs. metformin (an anti-diabetic drug in an AD clinical trial [ClinicalTrials.gov Identifier: NCT00620191]). For each comparator, we estimated the propensity score by using the variables described in Table 1. Then, we estimated the un-stratified Kaplan-Meier curves, conducted propensity score stratified (n strata = 10) log-rank test and Cox model.
Extended Data Fig 6. Longitudinal analyses reveal that sildenafil usage is significantly associated with reduced likelihood of AD in individuals with hypertension (HT).
Five comparator analyses were conducted including: (a) sildenafil vs. matched non- sildenafil, (b) sildenafil vs. diltiazem (an anti-hypertensive drug), (c) sildenafil vs. losartan (an anti-hypertensive drug candidate in an AD clinical trial [ClinicalTrials.gov Identifier: NCT02913664]), (d) sildenafil vs. glimepiride (an anti-diabetic drug), and (e) sildenafil vs. metformin (an anti-diabetic drug in an AD clinical trial [ClinicalTrials.gov Identifier: NCT00620191]). For each comparator, we estimated the propensity score by using the variables described in Table 1. Then, we estimated the un-stratified Kaplan-Meier curves, conducted propensity score stratified (n strata = 10) log-rank test and Cox model.
Extended Data Fig 7. Longitudinal analyses reveal that sildenafil usage is significantly associated with reduced likelihood of AD in individuals with type-2 diabetes (T2D).
Five comparator analyses were conducted including: (a) sildenafil vs. matched non- sildenafil, (b) sildenafil vs. diltiazem (an anti-hypertensive drug), (c) sildenafil vs. losartan (an anti-hypertensive drug candidate in an AD clinical trial [ClinicalTrials.gov Identifier: NCT02913664]), (d) sildenafil vs. glimepiride (an anti-diabetic drug), and (e) sildenafil vs. metformin (an anti-diabetic drug in an AD clinical trial [ClinicalTrials.gov Identifier: NCT00620191]). For each comparator, we estimated the propensity score by using the variables described in Table 1. Then, we estimated the un-stratified Kaplan-Meier curves, conducted propensity score stratified (n strata = 10) log-rank test and Cox model.
Supplementary Material
Acknowledgments
This work was primarily supported by the National Institute of Aging (NIA) of the National Institutes of Health (NIH) under Award Number R01AG066707 to F.C. This work was supported in part by U01AG073323, 3R01AG066707–01S1, 3R01AG066707–02S1, and 1R56AG074001–01 to F.C. This work was supported in part by the Translational Therapeutics Core of the Cleveland Alzheimer’s Disease Research Center (NIH/NIA: P30AG072959) to F.C., A.A.P. and J.C. This work was supported in part by the Brockman Foundation, Project 19PABH134580006-AHA/Allen Initiative in Brain Health and Cognitive Impairment, the Elizabeth Ring Mather & William Gwinn Mather Fund, S. Livingston Samuel Mather Trust, and the Louis Stokes VA Medical Center resources and facilities to A.A.P. This work was supported in part by the NIA grant R35AG071476 and the Alzheimer’s Disease Drug Discovery Foundation (ADDF) to J.C.
Footnotes
Competing interests. Dr. Cummings has provided consultation to Acadia, Actinogen, Alkahest, Alzheon, Annovis, Avanir, Axsome, Biogen, BioXcel, Cassava, Cerecin, Cerevel, Cortexyme, Cytox, EIP Pharma, Eisai, Foresight, GemVax, Genentech, Green Valley, Grifols, Karuna, Merck, Novo Nordisk, Otsuka, Resverlogix, Roche, Samumed, Samus, Signant Health, Suven, Third Rock, and United Neuroscience pharmaceutical and assessment companies. Dr. Cummings has stock options in ADAMAS, AnnovisBio, MedAvante, and BiOasis. The other authors have declared no competing interest.
Supplementary information is available in the online version of the paper.
References
- 1.Alzheimer’s A 2016 Alzheimer’s disease facts and figures. Alzheimers Dement. 12, 459–509 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Espay AJ et al. Revisiting protein aggregation as pathogenic in sporadic Parkinson and Alzheimer diseases. Neurology 92, 329–337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voorhees JR et al. (−)-P7C3-S243 protects a rat model of Alzheimer’s disease from neuropsychiatric deficits and neurodegeneration without altering Amyloid deposition or reactive Glia. Biol. Psychiatry. 84, 488–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long JM & Holtzman DM Alzheimer disease: An update on pathobiology and treatment strategies. Cell 179, 312–339 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makin S The amyloid hypothesis on trial. Nature 559, S4–s7 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Selkoe DJ & Hardy J The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 8, 595–608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cummings JL, Morstorf T & Zhong K Alzheimer’s disease drug-development pipeline: few candidates, frequent failures. Alzheimers Res. Ther. 6, 37 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tasaki S, Gaiteri C, Mostafavi S, De Jager PL & Bennett DA The molecular and neuropathological consequences of genetic risk for Alzheimer’s dementia. Front. Neurosci. 12, 699 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson MR et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 47, 856–860 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Wang Q et al. A Bayesian framework that integrates multi-omics data and gene networks predicts risk genes from schizophrenia GWAS data. Nat. Neurosci. 22, 691–699 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang H, De Wolf H, Knezevic B & Burnham KL A genetics-led approach defines the drug target landscape of 30 immune-related traits. Nat. Genet. 51, 1082–1091 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang J, Pieper AA, Nussinov R, Lee G, Bekris L, Leverenz JB, Cummings J & Cheng F Harnessing endophenotypes and network medicine for Alzheimer’s drug repurposing. Med. Res. Rev. 40, 2386–2426 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghiassian D et al. Endophenotype network models: Common core of complex diseases. Sci. Rep. 6, 27414 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swarup V et al. Identification of evolutionarily conserved gene networks mediating neurodegenerative dementia. Nat. Med. 25, 152–164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung J et al. Genome-wide association study of Alzheimer’s disease endophenotypes at prediagnosis stages. Alzheimers Dement. 14, 623–633 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pascoal TA et al. Synergistic interaction between amyloid and tau predicts the progression to dementia. Alzheimers Dement. 13, 644–653 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Benbow SJ et al. Synergistic toxicity between tau and amyloid drives neuronal dysfunction and neurodegeneration in transgenic C. elegans. Hum. Mol. Genet. 29, 495–505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pickett EK et al. Amyloid beta and Tau cooperate to cause reversible behavioral and transcriptional deficits in a model of Alzheimer’s disease. Cell Rep. 29, 3592–3604.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greene JA & Loscalzo J Putting the patient back together - Social medicine, network medicine, and the limits of reductionism. N. Engl. J. Med. 377, 2493–2499 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Goh KI et al. The human disease network. Proc. Natl. Acad. Sci. USA, 104, 8685–8690 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng F et al. Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat. Commun. 9, 2691 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Y et al. A systems pharmacology approach uncovers Wogonoside as an angiogenesis inhibitor of triple-negative breast cancer by targeting Hedgehog signaling. Cell Chem. Biol. 26, 1143–1158 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng F, Kovacs IA & Barabasi AL Network-based prediction of drug combinations. Nat. Commun. 10, 1197 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng F et al. A genome-wide positioning systems network algorithm for in silico drug repurposing. Nat. Commun. 10, 3476 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guney E et al. Network-based in silico drug efficacy screening. Nat. Commun. 7, 10331 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mittal S et al. beta2-Adrenoreceptor is a regulator of the alpha-synuclein gene driving risk of Parkinson’s disease. Science 357, 891–898 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bayik D et al. Myeloid-derived suppressor cell subsets drive glioblastoma growth in a sex-specific manner. Cancer Discov. 10, 1210–1225 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Claassen JA New cardiovascular targets to prevent late onset Alzheimer disease. Eur. J. Pharmacol. 763, 131–134 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Aguirre-Plans J et al. Proximal Pathway Enrichment Analysis for Targeting Comorbid Diseases via Network Endopharmacology. Pharmaceuticals 11, 61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y et al. AlzGPS: A Genome-wide Positioning Systems Platform to Catalyze Multi-omics for Alzheimer’s Therapeutic Discovery. Alzheimer’s Res. Ther. 13, 24 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng F et al. admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inf. Model. 52, 3099–3105 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Gomm W et al. Association of Proton Pump Inhibitors With Risk of Dementia: A Pharmacoepidemiological Claims Data Analysis. JAMA Neurol. 73, 410–416 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Cuadrado-Tejedor M et al. Sildenafil restores cognitive function without affecting beta-amyloid burden in a mouse model of Alzheimer’s disease. Br. J. Pharmacol. 164, 2029–2041 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ding J et al. Antihypertensive medications and risk for incident dementia and Alzheimer’s disease: a meta-analysis of individual participant data from prospective cohort studies. Lancet Neurol. 19, 61–70 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xue M et al. Diabetes mellitus and risks of cognitive impairment and dementia: A systematic review and meta-analysis of 144 prospective studies. Ageing Res. Rev 55,100944 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Consortium TG The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Y, Mucke L Alzheimer mechanisms and therapeutic strategies. Cell. 148,1204–1222 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuskaitis CJ, Jope RS Glycogen synthase kinase-3 regulates microglial migration, inflammation, and inflammation-induced neurotoxicity. Cell Signal. 21, 264–273 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Na YR et al. The early synthesis of p35 and activation of CDK5 in LPS-stimulated macrophages suppresses interleukin-10 production. Sci. Signal. 8, ra121 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Karikari TK et al. Diagnostic performance and prediction of clinical progression of plasma phospho-tau181 in the Alzheimer’s Disease Neuroimaging Initiative. Mol. Psychiatry. 26, 429–442 (2021). [DOI] [PubMed] [Google Scholar]
- 41.Su M et al. Mechanisms associated with type 2 diabetes as a risk factor for Alzheimer-related pathology. Mol. Neurobiol. 56, 5815–5834 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Gabin JM, Tambs K, Saltvedt I, Sund E & Holmen J Association between blood pressure and Alzheimer disease measured up to 27 years prior to diagnosis: the HUNT Study. Alzheimers Res. Ther. 9, 37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J et al. Carvedilol as a potential novel agent for the treatment of Alzheimer’s disease. Neurobiol Aging. 32, 2321.e2321–2312 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heckman PRA, Blokland A & Prickaerts J From age-related cognitive decline to Alzheimer’s disease: A translational overview of the potential role for phosphodiesterases. Adv. Neurobiol 17, 135–168 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Li C & Gotz J Tau-based therapies in neurodegeneration: opportunities and challenges. Nat. Rev. Drug Discov. 16, 863–883 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Plattner F, Angelo M & Giese KP The roles of cyclin-dependent kinase 5 and glycogen synthase kinase 3 in tau hyperphosphorylation. J. Biol. Chem. 281, 25457–25465 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Katsumoto A, Takeuchi H, Takahashi K & Tanaka F Microglia in Alzheimer’s disease: Risk factors and inflammation. Front. Neurol. 9, 978 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du J et al. Inhibition of CDKS by roscovitine suppressed LPS-induced *NO production through inhibiting NFkappaB activation and BH4 biosynthesis in macrophages. Am. J. Physiol. Cell Physiol. 297, C742–749 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J et al. Phosphodiesterase-5 inhibitor sildenafil prevents neuroinflammation, lowers beta-amyloid levels and improves cognitive performance in APP/PS1 transgenic mice. Behav. Brain Res. 250, 230–237 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Puzzo D et al. Phosphodiesterase 5 inhibition improves synaptic function, memory, and amyloid-beta load in an Alzheimer’s disease mouse model. J. Neurosci. 29, 8075–8086 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.García-Barroso C et al. Tadalafil crosses the blood-brain barrier and reverses cognitive dysfunction in a mouse model of AD. Neuropharmacology. 64,114–123(2013). [DOI] [PubMed] [Google Scholar]
- 52.Samudra N et al. A Pilot Study of Changes in Medial Temporal Lobe Fractional Amplitude of Low Frequency Fluctuations after Sildenafil Administration in Patients with Alzheimer’s Disease. J. Alzheimers Dis. 70, 163–170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sheng M et al. Sildenafil improves vascular and metabolic function in patients with Alzheimer’s disease. J. Alzheimers Dis. 60, 1351–1364 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Menche J et al. Uncovering disease-disease relationships through the incomplete interactome. Science 347, 1257601 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cummings J et al. Aducanumab: Appropriate use ecommendations. J. Prev. Alzheimers Dis. 8, 398–410 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sienski G et al. APOE4 disrupts intracellular lipid homeostasis in human iPSC-derived glia. Sci. Transl. Med. 13, eaaz4564 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cadar D et al. Individual and area-based socioeconomic factors associated with dementia incidence in England: Evidence from a 12-year follow-up in the English Longitudinal Study of Ageing. JAMA Psychiatry 75, 723–732 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghofrani HA et al. Sildenafil: from angina to erectile dysfunction to pulmonary hypertension and beyond. Nat. Rev. Drug Discov. 5, 689–702 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Andrews SJ et al. Mendelian randomization indicates that TNF is not causally associated with Alzheimer’s disease. Neurobiol. Aging 84, 241.e1–241.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Williams DM et al. Lipid lowering and Alzheimer disease risk: A mendelian randomization study. Ann. Neurol. 87, 30–39 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rolland T et al. A proteome-scale map of the human interactome network. Cell 159, 1212–1226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rual JF et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Csabai L, Olbei M, Budd A, Korcsmaros T & Fazekas D SignaLink: Multilayered Regulatory Networks. Methods Mol. Biol. 1819, 53–73 (2018). [DOI] [PubMed] [Google Scholar]
- 64.Stenson PD et al. The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 136, 665–677 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pinero J et al. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 45, D833–d839 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rappaport N et al. MalaCards: an amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 45, D877–d887 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koscielny G et al. Open Targets: a platform for therapeutic target identification and validation. Nucleic Acids Res. 45, D985–d994 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Edgar R, Domrachev M & Lash AE Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ritchie ME et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bouter Y et al. Deciphering the molecular profile of plaques, memory decline and neuron loss in two mouse models for Alzheimer’s disease by deep sequencing. Front. Aging Neurosci. 6, 75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Grubman A et al. Mouse and human microglial phenotypes in Alzheimer’s disease are controlled by amyloid plaque phagocytosis through Hif1α. bioRxiv, doi: 10.1101/639054 (2019). [DOI] [Google Scholar]
- 72.Rothman SM et al. Human Alzheimer’s disease gene expression signatures and immune profile in APP mouse models: a discrete transcriptomic view of Abeta plaque pathology. J. Neuroinflammation. 15, 256 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang H et al. Genome-wide RNAseq study of the molecular mechanisms underlying microglia activation in response to pathological tau perturbation in the rTg4510 tau transgenic animal model. Mol. Neurodegener. 13, 65 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Benjamini Y & Yekutieli D The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165–1188 (2001). [Google Scholar]
- 75.Eppig JT et al. Mouse Genome Informatics (MGI): Resources for mining mouse genetic, genomic, and biological data in support of primary and translational research. Methods Mol. Biol. 1488, 47–73 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Savas JN et al. Amyloid accumulation drives proteome-wide alterations in mouse models of Alzheimer’s disease-like pathology. Cell Rep. 21, 2614–2627 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim DK et al. Molecular and functional signatures in a novel Alzheimer’s disease mouse model assessed by quantitative proteomics. Mol. Neurodegener. 13, 2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wishart DS et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–d1082 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yunxia W et al. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 48, D1031–d1041 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barbarino JM, Whirl-Carrillo M, Altman RB & Klein TE PharmGKB: A worldwide resource for pharmacogenomic information. Wiley Interdiscip Rev. Syst. Biol. Med. 10, e1417 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gaulton A et al. The ChEMBL database in 2017. Nucleic Acids Res. 45, D945–d954 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gilson MK et al. BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 44, D1045–d1053 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harding SD et al. The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucleic Acids Res. 46, D1091–d1106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Desai RJ et al. Targeting abnormal metabolism in Alzheimer’s disease: The Drug Repurposing for Effective Alzheimer’s Medicines (DREAM) study. Alzheimers Dement (N Y). 6, e12095 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee E et al. Evaluation of Medicare Claims Data as a Tool to Identify Dementia. J Alzheimers Dis. 67,769–778 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Charlson ME et al. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 40, 373–383 (1987). [DOI] [PubMed] [Google Scholar]
- 87.Lash TL et al. Methodology, design, and analytic techniques to address measurement of comorbid disease. J Gerontol A Biol Sci Med Sci. 62, 281–285 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ye J et al. Three Data-Driven Phenotypes of Multiple Organ Dysfunction Syndrome Preserved from Early Childhood to Middle Adulthood. AMIA Annu Symp Proc. 2020,1345–1353 (2021). [PMC free article] [PubMed] [Google Scholar]
- 89.Kitsak M et al. Tissue specificity of human disease module. Sci. Rep. 6, 35241 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Son Y, Kim K & Cho H-R Sildenafil protects neuronal cells from mitochondrial toxicity induced by β-amyloid peptide via ATP-sensitive K channels. Biochem. Biophys. Res. Commun. 500, 504–510 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are available within Supplementary Table Files. Human protein-protein interactome and drug-target network can be downloaded from https://github.com/ChengF-Lab/endoAD. Tissue/brain-specific expression data were downloaded from GTEx database (https://www.gtexportal.org/home/). The expression datasets used in this study were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) with accession IDs GSE65067, GSE74437, GSE74438, GSE64398, GSE53480, GSE56772, and GSE57583. The health insurance claims data are available from the MarketScan Medicare Claims database (2012 to 2017) based on the Medicare Advantage and Fee for services from IBM MarketScan Research Databases.