Abstract
Flow cytometry (FCM) was successfully used to enumerate viruses in seawater after staining with the nucleic acid-specific dye SYBR Green-I. The technique was first optimized by using the Phaeocystis lytic virus PpV-01. Then it was used to analyze natural samples from different oceanic locations. Virus samples were fixed with 0.5% glutaraldehyde and deep frozen for delayed analysis. The samples were then diluted in Tris-EDTA buffer and analyzed in the presence of SYBR Green-I. A duplicate sample was heated at 80°C in the presence of detergent before analysis. Virus counts obtained by FCM were highly correlated to, although slightly higher than, those obtained by epifluorescence microscopy or by transmission electron microscopy (r = 0.937, n = 14, and r = 0.96, n = 8, respectively). Analysis of a depth profile from the Mediterranean Sea revealed that the abundance of viruses displayed the same vertical trend as that of planktonic cells. FCM permits us to distinguish between at least two and sometimes three virus populations in natural samples. Because of its speed and accuracy, FCM should prove very useful for studies of virus infection in cultures and should allow us to better understand the structure and dynamics of virus populations in natural waters.
Quantification of the different biological components present in an ecosystem is one of the first tasks of any ecological investigation. Much effort has been devoted to developing methods for detecting, recognizing, and enumerating different microorganisms such as protozoa, microalgae, bacteria, and viruses that thrive in natural waters. Viruses infecting algae and bacteria are abundant and active components of aquatic ecosystems. The occurrence of bacteriophages has been known for many years (14, 24, 25), but they have not been really investigated until recently. In the last decade, a number of studies have pointed out the ecological importance of viruses in the marine microbial food web (2, 22) and their potential impact on genetic exchange between marine organisms (1, 22).
Diverse culturing and microscopic techniques have been used to estimate the number of viruses in natural waters. The culturing techniques for algal viruses and bacteriophages include plaque counts and most-probable-number techniques (6, 8, 29, 31). The advantage of these assays is that they detect only viruses that are infective for a specific host. This is, however, also a major limitation, because the virus susceptibility of the strains used as assay hosts may be different from the virus susceptibility of the field strains from the same species (7, 28, 31).
The microscopic techniques include transmission electron microscopy (TEM) (1, 4, 33) and epifluorescence microscopy (EFM) combined with fluorescent staining of the viruses (11, 12, 19, 23). For TEM, viruses are harvested directly onto electron microscopy grids by centrifugation (3) or are concentrated by ultrafiltration and transferred to grids (21, 22). Analysis by TEM is time-consuming and requires expensive and bulky equipment, two drawbacks which are not compatible with routine field studies (1, 5, 21, 33). For EFM, the virus samples are usually collected on 0.02-μm-pore-size filters, stained with a DNA-specific fluorescent dye such as DAPI, YO-PRO-1, YOYO-1, POPO-1, or SYBR Green, and viewed under a standard epifluorescence microscope. EFM renders virus detection accessible to field analysis (11, 12, 23, 26), but it is also laborious.
Flow cytometry (FCM) has been routinely used for the analysis of microorganisms in marine samples for the last decade and is now commonly accepted as a reference technique in oceanography. Initially used to discriminate and enumerate the different phytoplanktonic populations (20), it has been more recently applied to the analysis of the heterotrophic bacterial community (15, 18). The major advantage of FCM resides in the possibility of rapidly analyzing a large number of cells and providing statistically significant data. This technique is also amenable to shipboard analysis (20). The increase in sensitivity of recent flow cytometers and the introduction of a new generation of nucleic acid-specific stains like TOTO-1, YOYO-1, PicoGreen, or SYBR Green-I (SYBR-I) have considerably improved the limit of detection of FCM (15–17).
In this paper, we report the use of FCM to detect marine viruses stained with the novel nucleic acid stain SYBR-I and compare this method with TEM and EFM. The method was first optimized with algal virus cultures and then was applied to field samples from different oceanic regions, such as the English Channel, the Equatorial Pacific, and the Mediterranean Sea.
MATERIALS AND METHODS
Cultures.
The algal-host–virus system used in this study was Phaeocystis pouchetii AJ01, obtained from the culture collection at the University of Bergen, Bergen, Norway, and the lytic virus PpV-01 (130 to 160 nm in diameter), isolated from Raunefjorden, Western Norway (13). The algal culture was made axenic after treatment with the antibiotics carbenicillin (0.01 g ml−1; Sigma Chemical Co.) and cefotaxime (0.01 g ml−1; Sigma). The virus culture was made axenic after double filtration over 0.2-μm-pore-size Supor 200 filters (Gelman Sciences). Phaeocystis was grown in f/2 (9), based on aged seawater, with the modifications that KH2PO4 and NaNO3 were added at 5 and 80 μM respectively. The temperature of the cultures was maintained at 8.0 ± 0.5°C, and light was supplied as a light-dark cycle of 16 and 8 h at a photon flux density of 100 ± 20 μmol m−2 s−1.
For the infection experiment, a culture at an algal concentration of 3.5 × 105 cells ml−1 was split into two subcultures of 300 ml each (in sterile 1-liter Erlenmeyer flasks). One of the subcultures was infected with 40 ml of virus lysate, while the other subculture served as control. Samples for algal cell and virus counts were taken at regular intervals of generally 2 to 6 h. After fixation with 0.5% (final) glutaraldehyde (25%, grade I; Sigma), subsamples were either stored directly at 4°C or quickly frozen in liquid N2 and stored at −80°C.
Natural seawater samples.
Equatorial Pacific (150°W, 16°S) samples were collected with Niskin bottles during the OLIPAC cruise onboard the N.O. l’Atalante in November 1994. In the Mediterranean Sea (18°E, 34°N), vertical profiles were collected during the MINOS cruise in June 1996 onboard the N.O. Suroit. Surface seawater samples from the English Channel (Station Estacade, Roscoff, France) were collected with a bucket in February and March 1998. Phytoplanktonic cells were enumerated on fresh samples, onboard for the Mediterranean Sea and Pacific Ocean samples and in the laboratory for the English Channel samples. For bacterial and viral enumeration, an aliquot (1.5 ml) was fixed for 15 min with glutaraldehyde at 0.5 and 0.1% (final concentrations) for the English Channel and Equatorial Pacific samples, respectively, or with a mixture of 1% paraformaldehyde and 0.05% glutaraldehyde for the Mediterranean Sea samples. The samples were then deep frozen in liquid nitrogen and preserved at −80°C until analysis. Bacteria were counted onboard for the Mediterranean Sea samples and in the laboratory for the Pacific Ocean and English Channel samples.
EFM.
During the viral infection experiment with Phaeocystis, 50 μl of fixed sample (4°C) was diluted with autoclaved medium filtered onto 0.2-μm-pore-size Anodisc 25 (Whatman) and stained for 5 min with a 0.2-μm-pore-size-filtered 4′,6-diamidino-2-phenylindole (DAPI) solution (1:1 [vol/vol] of a 10-μg-ml−1 stock solution [Sigma]). After filtration of 400 μl of the solution onto a 0.2-μm-pore-size black membrane filter (Poretics) or a 0.02-μm-pore-size Anodisc 25 filter (Whatman), viruses were counted immediately on a Olympus BH-2 microscope at ×1,000 with Citifluor (phosphate-buffered saline solution AF 3; Citifluor Ltd., London, England) as an antifading agent. Filtered subsamples were also stained with SYBR-I (Molecular Probes Inc.), and the filters were mounted onto a glass slide with 0.1% p-phenylenediamine in phosphate-buffered saline-glycerol (1:1) as described by Noble and Fuhrman (19). The optical density at 495 nm of the SYBR-I stock solution diluted 1,000-fold in distilled water (lot 0561-3) was 0.682.
TEM.
A 3-ml sample of the fixed virus samples was centrifuged onto grids (Ni, 400 mesh) for 30 min at 36,000 rpm in a Beckman SW 41 swingout rotor (3). The grids were positively stained with 2% uranyl acetate and viewed in a JEOL 100S transmission electron microscope at ×20,000 to ×100,000 magnification.
FCM.
All experiments were performed with a FACSort flow cytometer (Becton Dickinson, San Jose, Calif.) equipped with an air-cooled laser providing 15 mW at 488 nm and with the standard filter setup. For photosynthetic-cell enumeration, samples were run for 4 min at a rate of 90 μl min−1 and the discriminator was set on red fluorescence. Bacterial samples were stained with SYBR-I at a final concentration of 10−4 of the commercial stock solution (17). The samples were incubated for 15 min in the dark, the discriminator was set on green fluorescence, and the samples were analyzed for 1 min at a rate of 50 μl min−1.
For virus enumeration, dilutions from 1:100 to 1:2,000 for culture samples and from 1:10 to 1:200 for natural samples were performed in TE buffer (10 mM Tris, 1 mM EDTA [pH 7.5]), to avoid coincidence on the flow cytometer and to minimize the error due to low-volume pipetting. Dilutions were divided into two aliquots of 500 μl each. One was incubated for 15 min in the dark in presence of SYBR-I at a final concentration of 0.5 × 10−4. The second was incubated for 10 min at 80°C in presence of SYBR-I (0.5 × 10−4) and Triton X-100 (0.1%). The latter treatment was not necessary for fixed-frozen samples but essential for samples that were not frozen (see Results). We chose to use this treatment as a reference for virus enumeration. Samples were analyzed by flow cytometry for 1 to 4 min at a delivery rate of 50 μl min−1. The discriminator was set to green fluorescence, which is proportional to the nucleic acids–SYBR-I complex, and the detection threshold was progressively decreased until viruses could be detected (see below), but care was taken to avoid detecting more than 1,000 events per s, a threshold above which coincidence occurs, resulting in the underestimation of particle abundance. Parameters were collected on logarithmic scales and analyzed with the custom-designed software CYTOWIN (30; available freely at http://www.sb-roscoff.fr/Phyto/cyto.html) that discriminates cell populations by using combinations of all recorded parameters.
RESULTS
Optimization of the fixation and staining procedures.
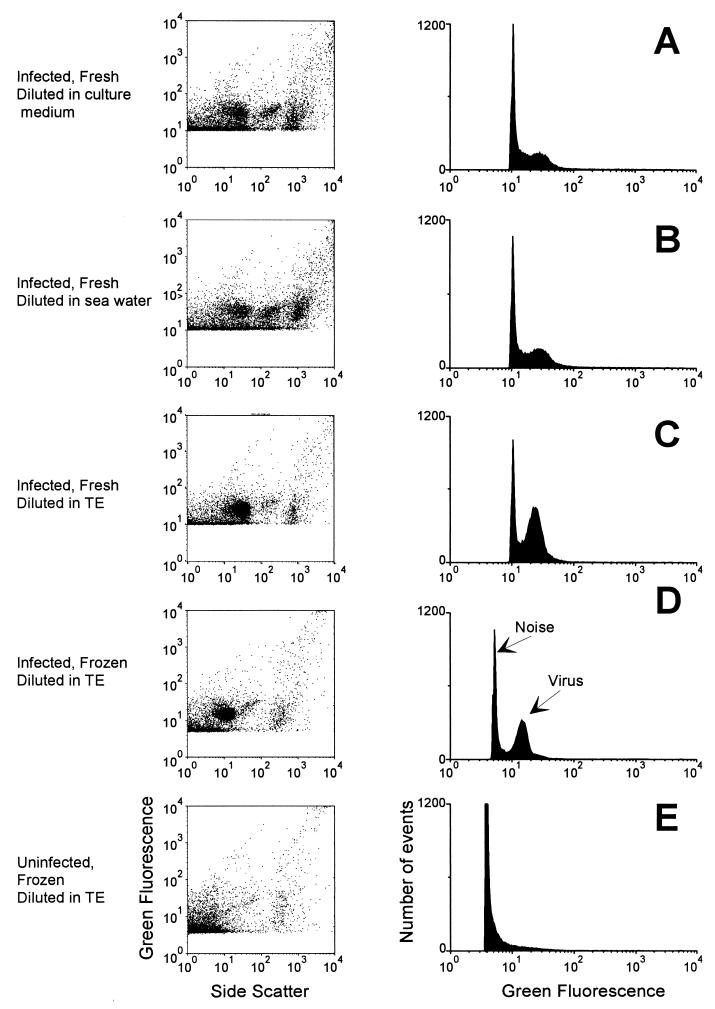
Preliminary tests with Phaeocystis virus samples stained with SYBR-I indicated that FCM could detect a population of fluorescent particles clearly above background but with much lower fluorescence than that usually displayed by bacteria (Fig. 1). For unfrozen Phaeocystis virus samples fixed with 0.5% glutaraldehyde, dilution in culture medium (Fig. 1A) or in 0.2-μm-pore-size-filtered seawater (Fig. 1B) resulted in counts five- to sixfold lower than those obtained after dilution in TE buffer (Fig. 1C). Samples diluted in culture medium (Fig. 1A) displayed higher background than those diluted in seawater (Fig. 1B). FCM analysis of unfrozen samples diluted in TE (Fig. 1C) still displayed virus counts 3- to 10-fold lower than those obtained by EFM. Fixation with different concentrations of glutaraldehyde ranging from 0.1 to 1% had no effect on virus enumeration. Aliquots of the virus suspension were heated for 10 min at 70°C after fixation with different concentrations of paraformaldehyde or glutaraldehyde. Virus counts obtained after this treatment and after staining by SYBR-I were highly correlated with counts obtained by EFM. Varying the incubation temperature between 50 and 95°C or adding SYBR-I before or after heating did not affect the quality of the staining. In contrast, addition of 0.1% Triton X-100 before heating improved the signal, inducing a decrease in the coefficient of variation of the viral population and an increase in the intensity of the green DNA fluorescence (data not shown). Furthermore, we observed a global decrease of viral counts (measured by either FCM or TEM) with time for a suspension fixed with either 0.1 to 0.5% glutaraldehyde or 0.5 to 1% paraformaldehyde and stored at 4°C. We observed an excellent correlation between FCM and EFM counts for samples that were frozen in liquid nitrogen after fixation and diluted in TE buffer after thawing (Fig. 1D). Heating such samples (10 min at 80°C) and incubating them with detergent (0.1% Triton X-100) did not change the virus counts. This suggested that heating is necessary only for samples that have not been frozen in liquid N2.
FIG. 1.
FCM analysis of P. pouchetii samples collected 24 h after the beginning of a viral infection experiment. Samples were analyzed after fixation with 0.5% glutaraldehyde and staining with 5 × 10−4 of the commercial solution of SYBR-I. (Left) Side scatter versus green DNA fluorescence (SYBR-I). (Right) Monoparametric green DNA fluorescence distribution of viruses. Each histogram contains 10,000 events. (A to D) Infected cultures; (E) uninfected culture. Samples were analyzed either immediately after fixation (A to C) or following freezing in liquid nitrogen and storage at −80°C (D and E). Samples were diluted in culture medium (A), 0.2-μm-pore-size-filtered seawater (B), or TE buffer (C to E).
At a final concentration of SYBR-I of 10−4 (usually used for bacteria [17]), staining was not very stable at low virus concentrations, the signal tended to decrease a few seconds after the beginning of analysis, and viruses became difficult to separate from background noise due to unspecific staining of cellular debris or particulate matter. We chose a SYBR-I concentration of 0.5 × 10−4, which gave the best results both for the analysis of suspensions containing low concentrations of viruses and for the homogeneity of the staining.
Comparison of different counting techniques.
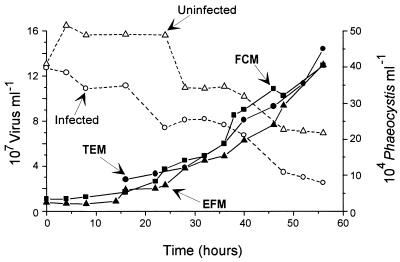
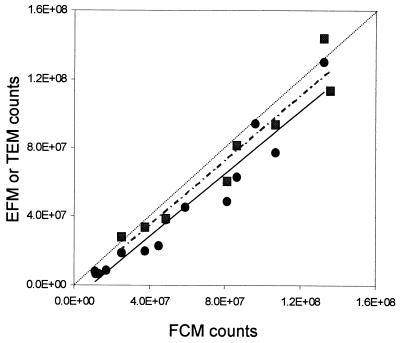
Detailed comparisons between FCM, TEM, and EFM enumeration of viruses were performed during a Phaeocystis infection experiment on samples fixed with 0.5% glutaraldehyde, frozen in liquid nitrogen, and stored at −80°C until analysis. The three techniques gave very similar trends during the experiment (Fig. 2), and counts obtained by FCM were highly reproducible for the different dilutions analyzed (Table 1; Fig. 3). Although a good linear relationship was observed between FCM and EFM and between FCM and TEM (r = 0.97, n = 14, and r = 0.96, n = 8, respectively), the FCM counts were about 30% higher than those obtained by EFM and differences of up to 15% were observed with respect to TEM (Table 1; Fig. 3). At 24 h after the beginning of the experiment, the virus concentration increased in the infected culture (Fig. 2). In both infected and uninfected cultures, the level of debris increased with time but no virus population could be discriminated in the latter cultures (Fig. 1E).
FIG. 2.
Infection experiment of P. pouchetii. Phaeocystis cells were enumerated by FCM for uninfected (open triangles) and infected (open circles) cultures. Virus counts were performed by three different techniques: EFM (solid triangles), TEM (solid circles), and FCM (solid squares). Virus counts reported for FCM correspond to the average value of counts obtained on 0.5% glutaraldehyde-fixed samples, frozen in liquid nitrogen, at different dilutions and analyzed after incubation at room temperature (Table 1) or after heating at 80°C in the presence of Triton X-100.
TABLE 1.
Phaeocystis infection experimenta
| Time (h) | Viral counts (107/ml) by:
|
||||||
|---|---|---|---|---|---|---|---|
| EFM | TEM | FCM at dilution of:
|
|||||
| ×50 | ×100 | ×500 | ×1,000 | ×2,000 | |||
| 0 | 0.80 | 1.09 | 1.13 | 1.14 | |||
| 4 | 0.68 | 1.13 | 1.14 | 1.14 | |||
| 8 | 0.66 | 1.39 | 1.28 | 1.29 | |||
| 14 | 0.88 | 1.73 | 1.66 | 1.80 | |||
| 16 | 1.90 | 2.82 | 2.52 | 2.49 | 2.59 | ||
| 22 | 2.00 | 3.33 | 3.53 | 3.48 | 4.08 | ||
| 24 | 2.30 | 4.64 | 4.33 | ||||
| 28 | 3.80 | 3.86 | 5.12 | 4.64 | |||
| 32 | 4.50 | 5.61 | 6.48 | 5.61 | |||
| 36 | 4.90 | 6.02 | 8.12 | 8.14 | |||
| 40 | 6.30 | 8.10 | 8.91 | 7.98 | 9.00 | ||
| 46 | 7.70 | 9.33 | 11.00 | 10.20 | 10.90 | ||
| 48 | 9.40 | 9.48 | 9.38 | 10.00 | |||
| 52 | 11.30 | 13.00 | 13.80 | 14.00 | |||
| 56 | 13.00 | 14.40 | 13.00 | 13.20 | 13.60 | ||
Samples were fixed with 0.5% glutaraldehyde and immediately frozen in liquid nitrogen. Different dilutions were performed in TE buffer and stained with 5.0 × 10−5 SYBR-I at room temperature.
FIG. 3.
Infection experiment of P. pouchetii. Comparison between FCM, EFM, and TEM virus counts obtained for samples collected every 4 h (Fig. 2; Table 1). FCM versus EFM (solid circles, straight line; EFM = 0.91 × FCM − 8.5 × 106, r = 0.97, n = 14). FCM versus TEM (solid squares, dotted line, FCM = 0.95 × TEM − 3.7 × 106, r = 0.96, n = 8). The dashed line corresponds to a 1:1 relationship.
To make sure that our EFM counts were not biased, we compared different EFM filter and dye combinations (black 0.2-μm-pore-size and white 0.02-μm-pore-size filters; DAPI and SYBR-I) by using different dilutions of a Phaeocystis virus suspension (Table 2). The EFM counts were identical for the different filters and dyes used. They were higher than FCM counts at low dilutions but were 35% lower at the highest dilutions (Table 2). In the latter case, not enough viruses were counted by FCM, leading to statistically insignificant results. At low dilutions, the FACSort began to display coincidence, resulting in an underestimation of virus counts, when there were more than 1,000 events per s. It is therefore critical to adjust the flow rate or sample dilution to remain below this level.
TABLE 2.
Comparison of virus numbers obtained with different dilutions of a Phaeocystis virus suspension fixed with 0.5% glutaraldehyde and immediately frozen in liquid nitrogen and analyzed by FCM after staining with SYBR-I or by EFM after staining with DAPI or SYBR-Ia
| Dilution | Viral counts (105/ml) by:
|
|||
|---|---|---|---|---|
| FCM with SYBR-I | EFM
|
|||
| DAPI, 0.2-μm black filter | DAPI, 0.02-μm white filter | SYBR-I, 0.02-μm white filter | ||
| 1 | 460.0 | 430.0 | 430.0 | |
| 10 | 18.98 | 36.0 | 37.0 | 37.0 |
| 50 | 7.01 | |||
| 100 | 6.40 | 4.40 | 3.70 | 4.90 |
| 250 | 2.71 | |||
| 500 | 1.41 | 1.01 | 1.10 | 1.10 |
| 750 | 0.97 | |||
| 1,000 | 0.74 | 0.48 | 0.48 | |
| 2,000 | 0.39 | |||
| 5,000 | 0.16 | |||
| 10,000 | 0.12 | |||
| 25,000 | 0.03 | |||
| 50,000 | 0.02 | |||
| 100,000 | 0.02 | |||
For EFM, the samples were filtered onto either black 0.2-μm-pore-size or white 0.02-μm-pore-size filters.
Natural seawater samples.
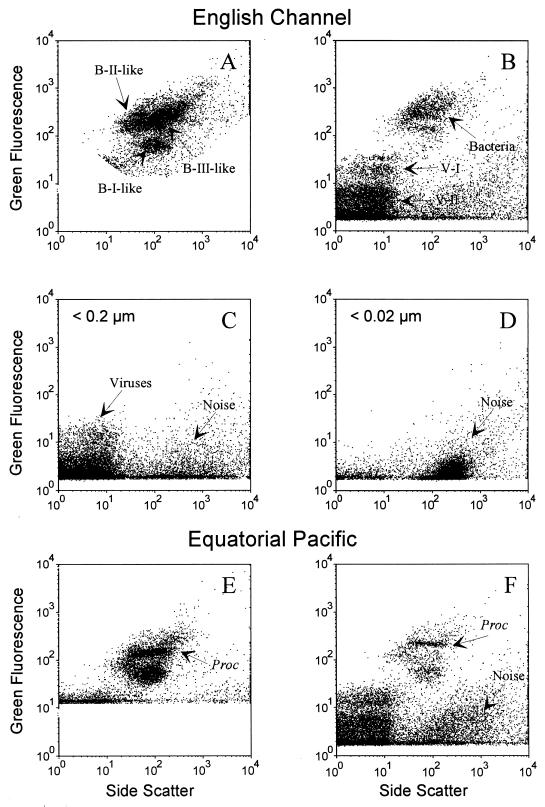
Analysis of natural samples from three different oceanic regions ranging from mesotrophic to oligotrophic conditions revealed two populations of SYBR-I-stained particles that had scatter and DNA fluorescence characteristics similar to those observed for the Phaeocystis viruses (Fig. 4). In this section, these particles are referred to as virus particles (V). The V-I and V-II populations differed in their green emission of the nucleic acid–SYBR-I complex (Fig. 4). The virus populations observed in natural samples by FCM could also be detected by EFM with SYBR-I after filtration onto 0.02-μm-pore-size Anodisc filters. However, the viral counts obtained by EFM were lower than those obtained by FCM (Table 3), due to the difficulty in clearly differentiating virus particles from other abundant detritic particles by microscopy. Virus counts obtained by FCM in the samples from the Mediterranean Sea and the Equatorial Pacific were about 1.5-fold higher those obtained by EFM. In contrast, for English Channel samples, which were analyzed immediately after collection, the EFM and FCM counts differed by only 10% (Table 3). In the English Channel samples, 80% of SYBR-I-stainable virus particles passed through 0.2-μm-pore-size filters (Fig. 4C). The filtrate contained 100% of the V-II population, 20% of the V-I population, and no bacteria. Moreover, the V-I group in the English Channel samples seemed to consist of two subpopulations (Fig. 4). No virus populations could be detected after filtration through 0.02-μm-pore-size filters (Fig. 4D).
FIG. 4.
Side scatter versus green fluorescence (SYBR-I) for surface samples collected in the English Channel and in the Equatorial Pacific and analyzed by FCM. Samples were run undiluted to enumerate bacteria (A and E). Then different dilutions were made to analyze viruses (B and F). Data obtained after filtration of seawater from the English Channel, through 0.2- or 0.02-μm-pore-size filters, and stained by SYBR-I are shown (C and D). Only data from a 1:50 dilution are presented. Proc, Prochlorococcus; B-I, B-II, B-III, bacteria subpopulations; V-I, V-II, virus subpopulations.
TABLE 3.
Concentration of bacteria and viruses, determined by FCM and EFM, for surface samples from the English Channel and the Equatorial Pacific and for samples from a vertical profile collected in the Mediterranean Sea during the MINOS cruise (cast 118)a
| Location | Depth (m) | Bacterial counts (105/ml) | Viral counts (105/ml) by:
|
||
|---|---|---|---|---|---|
| EFM | FCM
|
||||
| Total | V-I/V-II | ||||
| English Channel | 0–2 | 9.1 | 160 | 180 | 0.103 |
| Equatorial Pacific | 5 | 6.3 | 39.0 | 52.9 | 0.216 |
| Mediterranean Sea | 5 | 3.9 | 15.0 | 22.7 | 0.158 |
| Mediterranean Sea | 40 | 4.7 | 43.8 | 0.248 | |
| Mediterranean Sea | 60 | 4.9 | 52.9 | 0.239 | |
| Mediterranean Sea | 100 | 4.1 | 65.1 | 0.276 | |
| Mediterranean Sea | 120 | 3.2 | 58.9 | 0.230 | |
| Mediterranean Sea | 140 | 2.7 | 62.6 | 0.312 | |
| Mediterranean Sea | 200 | 1.9 | 41.2 | 0.249 | |
Samples were stained with 10−4 SYBR-I and analyzed undiluted for bacteria and with 5 × 10−5 SYBR-I and diluted 50- to 200-fold for viruses. EFM counts were performed with SYBR-I as described by Noble and Fuhrman (19).
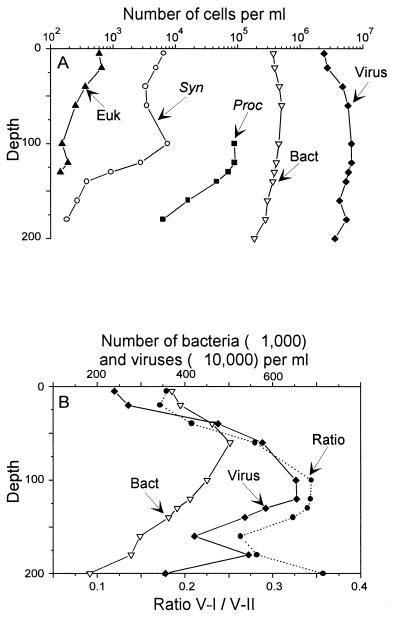
Bacterial concentrations in the English Channel, the Mediterranean Sea, and the Equatorial Pacific were, respectively, 9.1 × 105, 3.6 × 105, and 6.3 × 105 cells ml−1 (Table 3). In the mesotrophic English Channel, 20 times more viruses than bacteria and 10-fold more V-II than V-I viruses were found. The ratio between viruses and bacteria was significantly lower in the more oligotrophic regions (Mediterranean Sea and Equatorial Pacific [Table 3]).
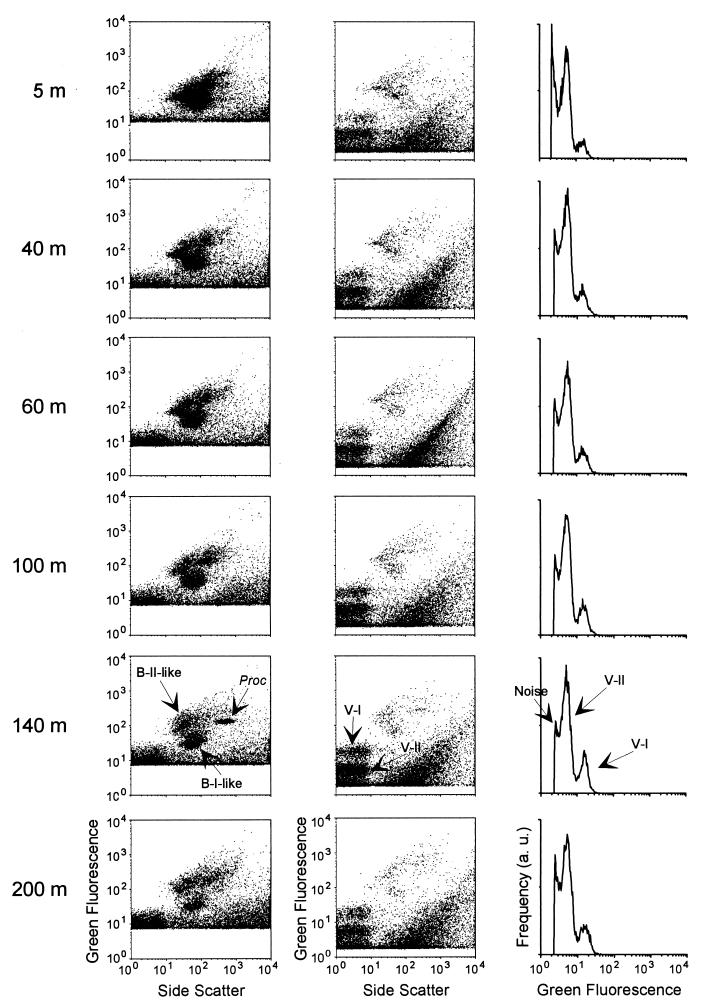
The analysis of a depth profile in the Mediterranean Sea (Fig. 5 and 6) revealed that the abundance of virus particles displayed the same vertical trend as that of bacteria and phytoplanktonic cells. Viral abundance was minimal near the surface, increased down to 100 and 120 m, where phytoplanktonic cells were maximal, and then decreased with depth (Fig. 6A). The ratio between the V-I and V-II populations followed the total number of viruses (Fig. 6B). The minimum value for this ratio was obtained at the surface, and the maximum was obtained between 100 and 130 m.
FIG. 5.
FCM analysis of seawater samples collected at different depths on a vertical profile in the Mediterranean Sea (18°E, 34°N) on 18 June 1996 (MINOS cruise, cast 118). Six representative depths among 12 sampled are shown. Bacteria (left) were enumerated in undiluted samples stained with 10−4 SYBR-I. For virus analysis (middle), samples were diluted 100-fold and stained with 5 × 10−5 SYBR-I. Monoparametric green fluorescence distributions (right), obtained after computation by CYTOWIN and corresponding to the virus populations, are also shown.
FIG. 6.
Vertical distributions of Prochlorococcus (Proc; solid squares), Synechococcus (Syn; open circles), picoeukaryotes (Euk; solid triangles), bacteria (Bact; open triangles), and viruses (solid diamonds) and ratio between V-II and V-I (stars, dashed line) obtained by FCM for a vertical profile collected in the Mediterranean Sea (18°E, 34°N) on 18 June 1996 (MINOS cruise, cast 111). Samples were collected in Niskin bottles from 200 m up to the surface. Phytoplanktonic cells and bacteria were enumerated onboard. Prochlorococcus was undetectable from the surface down to 80 m because of its weak red chlorophyll fluorescence. Viruses were analyzed on paraformaldehyde-preserved samples, and the numbers result from the average of counts obtained on samples diluted 1:100 and 1:200 in TE buffer. (A) Logarithmic scale. (B) Linear scale.
DISCUSSION
In the past, virus in natural marine samples were detected and enumerated by TEM (see, e.g., reference 22). More recently EFM and nucleic acid-specific fluorescent stains, such as DAPI and YO-PRO-1, have been shown to result in more accurate and more easily performed enumeration (12, 32). In the present study, we have gone one step further, by combining SYBR-I staining with detection by a sensitive and widely available method, FCM. This constitutes a real improvement for the study of viral communities, especially because this method is less time-consuming and less operator dependent than other counting techniques such as TEM and EFM (FCM typically allows around 200 samples per day to be processed during a typical cruise, i.e., about 5 to 10 times as many as by EFM). Nevertheless, TEM and EFM can provide complementary information, such as the detection of viruses that are attached to or internalized into host cells or to differentiate different morphotypes. As with EFM or TEM, FCM analysis of viruses can be performed on glutaraldehyde- or paraformaldehyde-fixed samples permitting delayed analysis. A good correlation was obtained between TEM, EFM, and FCM virus counts (Fig. 3), but viral counts by FCM were generally higher. FCM counts appeared highly reproducible (Table 1), and this technique allows the enumeration of viruses in solution at concentrations too low to be counted by the other methods (Table 2).
Samples analyzed by FCM shortly after fixation showed a low percentage of stained viruses compared to those analyzed by the microscopic techniques. However, heating such samples to 95°C in the presence of detergent or freezing in liquid nitrogen resulted in viral counts comparable to TEM or EFM counts. Our observations suggest that fresh viruses may have a structure that makes the nucleic acids not immediately accessible to SYBR-I or to the other nucleic acid-specific dyes that we also tested, such as PicoGreen, SYTOX, and the intercalary cyanine dye TOTO-1 (data not shown). Detergent or heat treatment may denature the viral capsid, allowing the stain to penetrate. For both paraformaldehyde- and glutaraldehyde-fixed samples, we observed a decreased in virus number over time when the samples were kept at 4°C. This decline of virus concentration at 4°C could be due to aggregate formation or to virus degradation by nucleases, as observed for bacterial samples in seawater (10).
Viruses constitute the most abundant group of nucleic acid-containing particles in the ocean (7), and estimates based on TEM range from 105 to 108 ml−1 (1, 2, 5, 11, 12, 22, 27). In pond water (19) or in estuarine environments (21), viral abundance reach 106 to 107 ml−1 and exceed the bacterial abundance by fivefold, while in oligotrophic waters viral and bacterial abundance have been reported to be quite similar from 2.0 to 4.0 × 105 per ml. Other authors found that virus concentrations exceeded the bacterial abundance by 10-fold in Norwegian Fjords, with bacterial counts between 1.0 × 105 and 4.0 × 105 cell ml−1 (5). In the present work, bacterial abundance measured in three different oceanic areas agreed with the literature values, while viral concentrations were about 10-fold higher than bacterial concentrations, ranging from 0.2 × 107 ml−1 in the oligotrophic waters of the Equatorial Pacific or the Mediterranean Sea to 1.8 × 107 ml−1 in coastal waters of the English Channel.
Bergh et al. (1) observed that viruses in seawater samples appear to be free. They identified four different classes of viruses in terms of their size and found that marine samples are dominated by the smallest class (30 to 60 nm). Our FCM data suggest that at least two virus populations (V-I and V-II) are ubiquitous in oceanic samples from very different environments. A total of 20% of the V-I group, which displays the highest SYBR-I fluorescence intensity, and 100% of the V-II viruses could pass through 0.2-μm-pore-size filters (Fig. 4C), suggesting that V-I viruses are larger. The V-I population displays the same relative green fluorescence intensity after staining with SYBR-I than do viruses affecting eukaryotic algal cells such as P. pouchetii (analyzed in the present study) or Micromonas pusilla (data not shown). Furthermore, the V-II population is 4- to 10-fold more abundant than the V-I population, a ratio commensurate with the heterotroph/phytoplankton ratio, which ranged from 3 to 20 (n = 70) in natural samples from the Mediterranean Sea. This suggests that V-I viruses could be infectious to phytoplanktonic (in particular eukaryotic) cells while V-II viruses could be related to the more numerous heterotrophic bacteria. It is noteworthy that in the Mediterranean Sea depth profile the V-I/V-II ratio peaked at the depth where phytoplanktonic cells were most abundant (Fig. 6B). Although these hypotheses require much more detailed work for confirmation, which is clearly beyond the scope of this study, they underline that, in the future, FCM could be one of the major techniques used for the analysis of viruses and will contribute to the knowledge of the distribution and dynamic of viruses in oceanic environments.
ACKNOWLEDGMENTS
This work was supported by grants from the European Community (MAST CT95-0016-MEDEA; MAS3-CT96-5033[DG12-ASAL], TMR program), from JGOFS-France (EPOPE, MINOS, and PROSOPE programs), and by The Research Council of Norway (project 121425/420). The FACSort flow cytometer was funded in part by CNRS-INSU and the Région Bretagne.
The electron microscopic work was done at the Laboratory for Electron Microscopy University of Bergen. We thank Sandrine Boulben for her technical assistance and Jed Fuhrman for an early discussion on the interest of detecting viruses by FCM.
REFERENCES
- 1.Bergh Ø, Børsheim K Y, Bratbak G, Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–468. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- 2.Bratbak G, Heldal M, Norland S, Thingstad T F. Viruses as partners in spring bloom microbial trophodynamics. Appl Environ Microbiol. 1990;56:1400–1405. doi: 10.1128/aem.56.5.1400-1405.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bratbak G, Heldal M. Total count of viruses in aquatic environments. In: Kemp P F, Sherr B F, Sherr E B, Cole J J, editors. Current methods in aquatic microbial ecology. London, England: Lewis Publishers; 1993. pp. 135–138. [Google Scholar]
- 4.Bratbak G, Egge J K, Heldal M. Viral mortality of the marine alga Emiliania huxleyi (Haptophyceae) and termination of algal blooms. Mar Ecol Prog Ser. 1993;93:39–48. [Google Scholar]
- 5.Børsheim K Y, Bradbak G, Heldal M. Enumeration and biomass estimation of planktonic bacteria and viruses by transmission electron microscopy. Appl Environ Microbiol. 1990;56:352–356. doi: 10.1128/aem.56.2.352-356.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Børsheim K Y. Native marine bacteriophages. FEMS Microb Ecol. 1993;102:141–159. [Google Scholar]
- 7.Cottrell M T, Suttle C A. Wide-spread occurrence and clonal variation in viruses which cause lysis of a cosmopolitan eukaryotic marine phytoplankter, Micromonas pusilla. Mar Ecol Prog Ser. 1991;78:1–9. [Google Scholar]
- 8.Cottrell M T, Suttle C A. Dynamics of a lytic virus infecting the photosynthetic marine picoflagellate Micromonas pusilla. Limnol Oceanogr. 1995;40:730–739. [Google Scholar]
- 9.Guillard R R L. Culture of phytoplankton for feeding marine invertebrates. In: Smith W L, Chanley M H, editors. Culture of marine invertebrate animals. New York, N.Y: Plenum Press; 1975. pp. 29–60. [Google Scholar]
- 10.Gundersen K, Bratbak G, Heldal M. Factors influencing the loss of bacteria in preserved seawater samples. Mar Ecol Prog Ser. 1996;137:305–310. [Google Scholar]
- 11.Hara S, Terauchi K, Koike I. Abundance of viruses in marine waters: assessment by epifluorescence and transmission electron microscopy. Appl Environ Microbiol. 1991;57:2731–2734. doi: 10.1128/aem.57.9.2731-2734.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hennes K P, Suttle C A. Direct counts of viruses in natural waters and laboratory cultures by epifluorescence microscopy. Limnol Oceanogr. 1995;40:1050–1055. [Google Scholar]
- 13.Jacobsen A, Bratbak G, Heldal M. Isolation and characterization of a virus infecting Phaeocystis pouchetii (Prymnesiophyceae) J Phycol. 1996;32:923–927. [Google Scholar]
- 14.Kriss A E, Rukina E A. Bacteriophages in the sea. Dokl Akad Nauk SSSR. 1947;57:833–836. [Google Scholar]
- 15.Li W K W, Jellett J F, Dickie P M. DNA distribution in planktonic bacteria stained with TOTO and TO-PRO. Limnol Oceanogr. 1995;40:1485–1495. [Google Scholar]
- 16.Marie D, Vaulot D, Partensky F. Application of the novel nucleic acid dyes YOYO-1, YO-PRO-1, and PicoGreen for flow cytometric analysis of marine prokaryotes. Appl Environ Microbiol. 1996;62:1649–1655. doi: 10.1128/aem.62.5.1649-1655.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marie D, Partensky F, Jacquet S, Vaulot D. Enumeration and cell-cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic-acid stain SYBR Green I. Appl Environ Microbiol. 1997;63:186–193. doi: 10.1128/aem.63.1.186-193.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monger B C, Landry M R. Flow cytometric analysis of marine bacteria with Hoechst 33342. Appl Environ Microbiol. 1993;59:905–911. doi: 10.1128/aem.59.3.905-911.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noble R T, Fuhrman J A. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat Microb Ecol. 1998;14:113–118. [Google Scholar]
- 20.Olson R J, Vaulot D, Chisholm S W. Marine phytoplankton distributions measured using shipboard flow cytometry. Deep-Sea Res. 1985;32:1273–1280. [Google Scholar]
- 21.Paul J H, Jiang S C, Rose J B. Concentration of viruses and dissolved DNA from aquatic environments by vortex flow filtration. Appl Environ Microbiol. 1991;57:2197–2204. doi: 10.1128/aem.57.8.2197-2204.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Proctor L M, Fuhrman J A. Viral mortality of marine bacteria and cyanobacteria. Nature (London) 1990;343:60–62. [Google Scholar]
- 23.Proctor L M, Fuhrman J A. Mortality of marine bacteria in response to enrichments of the virus size fraction from seawater. Mar Ecol Prog Ser. 1992;87:283–293. [Google Scholar]
- 24.Spencer R. A marine bacteriophage. Nature (London) 1955;175:690. doi: 10.1038/175690a0. [DOI] [PubMed] [Google Scholar]
- 25.Spencer R. Indigenous marine bacteriophages. J Bacteriol. 1960;79:614. doi: 10.1128/jb.79.4.614-614.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suttle C A, Chan A M, Cottrell M T. Infection of phytoplankton by viruses and reduction of primary productivity. Nature. 1990;347:467–469. [Google Scholar]
- 27.Suttle C A, Chan A M, Cottrell M T. Use of ultrafiltration to isolate viruses from seawater which are pathogens of marine phytoplankton. Appl Environ Microbiol. 1991;57:721–726. doi: 10.1128/aem.57.3.721-726.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suttle C A, Chan A M. Marine cyanophages infecting oceanic and coastal strains of Synechococcus: abundance, morphology, cross-infectivity and growth characteristics. Mar Ecol Prog Ser. 1993;92:99–109. [Google Scholar]
- 29.Suttle C A, Chan A M. Viruses infecting the marine Prymnesiophyte Chrysochromulina spp.: isolation, preliminary characterization and natural abundance. Mar Ecol Prog Ser. 1995;118:275–282. [Google Scholar]
- 30.Vaulot D. CYTOPC: processing software for flow cytometric data. Signal Noise. 1989;2:8. [Google Scholar]
- 31.Waterbury J B, Valois F W. Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl Environ Microbiol. 1993;59:3393–3399. doi: 10.1128/aem.59.10.3393-3399.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinbauer M E, Suttle C A. Comparison of epifluorescence and transmission electron microscopy for counting viruses in natural marine waters. Aquat Microb Ecol. 1997;13:225–232. [Google Scholar]
- 33.Wommack K E, Hill R T, Kellel M, Russek-Cohen E, Colwell R A. Distribution of viruses in the Chesapeake Bay. Appl Environ Microbiol. 1992;58:2965–2970. doi: 10.1128/aem.58.9.2965-2970.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]