Abstract
STING (Stimulator of interferon genes) is an adaptor protein in innate immunity against DNA viruses or bacteria1–5. STING-mediated immunity could be harnessed for vaccines or cancer immuno-therapies. STING is a transmembrane (TM) dimeric protein located in the endoplasmic reticulon (ER) or Golgi. STING is activated by binding of its cytoplasmic ligand-binding domain (LBD) to cyclic dinucleotides, produced by the DNA-sensor cyclic-GMP-AMP (cGAMP) synthase (cGAS) or invading bacteria1,6,7. Cyclic dinucleotides induce a conformational change to the STING LBD, leading to high-order oligomerization of STING that is essential for triggering the downstream signaling pathways8,9. However, the cGAMP-induced STING oligomers appeared weak and have not been resolved to high resolution, hampering the understanding of the activation mechanism. Here we show that a small molecular agonist, compound 53 (C53)10, promotes human STING oligomerization and activation through a mechanism orthogonal to that of cGAMP. We determined a cryo-EM structure of STING bound to both C53 and cGAMP, revealing a stable oligomer formed by side-by-side packing with a curled overall shape. Surprisingly, C53 binds to a cryptic pocket in the STING TM domain (TMD), between the two subunits of the STING dimer. This binding induces outward shifts of TM helices in the dimer, promoting them to make inter-dimer interactions to mediate the formation of the high-order oligomer. Our functional analyses show that cGAMP and C53 together induce stronger activation of STING.
Innate immunity provides the first-line defense against infection. A major innate immunity pathway for detecting viruses or bacteria is mediated by cyclic GMP-AMP (cGAMP) synthase (cGAS)1,6,7. cGAS serves as a DNA sensor by directly binding to pathogen DNA in the cytosol and generates the second messenger cGAMP. cGAMP activates STING2–5,11, which initiates several downstream signaling pathways, including the type-I interferon pathway, the NFκB pathway and autophagy, to eliminate the pathogen1. cGAS/STING signaling can be activated by DNA from tumor cells and launch antitumor immunity by promoting the production of type I interferons, cell senescence and the adaptive immunity against tumor cells1,12–15. Targeting cGAS/STING dependent signaling has shown promise in clinical application on anti-tumor treatment. In the past few years, a surge effort has been made in the development of STING agonists as novel anti-cancer therapeutics16.
STING contains four TM helices that forms the TM domain (TMD), which is followed by the cytoplasmic LBD that binds cGAMP8,9. In addition, STING has a C-terminal tail that contains the PXPLRXD (X: any residue) motif that recruits the TANK-binding kinase 1 (TBK1)9,17,18. Binding of TBK1 to this motif promotes the phosphorylation of Ser366 in the STING tail19, which subsequently recruits and promotes the phosphorylation of the transcription factor interferon regulatory factor 3 (IRF3), ultimately leading to the expression of type I interferons. We and others have used structural approaches to elucidate the mechanisms that control these activation steps of STING8,9,17,20. STING exists as a constitutive domain-swapped dimer through interactions mediated by both the TMD and LBD8. cGAMP binds the cleft at the center of the butterfly-shaped LBD dimer, inducing concerted conformational changes that include inward rotation of the two wings and closure of the binding pocket. These conformational changes are coupled to a 180°-rotation of the LBD relative to the TMD, as well as a downward tilt of the LBDα2–LBDα3 loop, which mediates the formation of the side-by-side high-order oligomer of STING that is essential for the recruitment of TBK1 and the subsequent phosphorylation events. STING oligomerization and activation are also coupled to cGAMP-induced translocation from the ER to the Golgi apparatus, for which the mechanism is not well understood21–23.
Despite the high affinity of STING for cGAMP24–27, STING oligomers induced by cGAMP alone appeared weak in solution8, which contradicts the observations that cGAMP induces robust puncta formation of STING on the ER or Golgi membrane2,21. In addition, our previous cryo-EM structure of the chicken STING tetramer at low resolution (6.5 Å) shows inter-dimer interactions in the LBD, but the role of the TMD in the oligomerization remains unclear8. These results together suggest that the stability of the high-order oligomer of STING may involve additional factors interacting with the TMD. In search for such factors, we characterized the interaction of human STING with a recently identified activator compound 53 (C53) 10. We determined a cryo-EM structure of STING bound with both cGAMP and C53, which revealed a novel agonist binding pocket in the TMD of STING. Our analyses further showed that the concurrent bindings of cGAMP and C53 to LBD and TMD of STING, respectively, promote the formation of higher-order STING oligomers, and thereby potently activate STING.
C53 promotes STING oligomerization
To stabilize the cGAMP-induced active oligomer of human STING for mechanistic analyses, we searched for STING agonists that are chemically distinct from cGAMP and less likely act as cGAMP mimetics. Recently, Pryde et al identified a series of small benzothiazinone-like compounds, including C53, that activate human STING selectively and potently10. Unlike cGAMP that is negatively charged, these compounds are mostly hydrophobic. We used a native gel assay to assess the effect of C53 on oligomerization of purified human STING (Extended Data Fig. 1). cGAMP and C53 together induced robust oligomerization of STING, while either one alone failed to do so under the same condition (Extended Data Fig. 1c). Consistently, cryo-EM images of STING in the presence of both C53 and cGAMP showed tetramers and higher-order oligomers formed by side-by-side packing of the individual dimers (Extended Data Fig. 2). These results suggested that C53 binds a different site on STING and acts orthogonally with cGAMP to promote STING oligomerization. Intriguingly, these higher-order STING oligomers display a curved overall shape, suggesting a preference for membrane with positive curvature (Extended Fig. 2a). This preference might concentrate the STING oligomers at the rim of ER and Golgi and facilitate their anterograde transport28, which is known to be important for STING signaling in cells21,22.
cGAMP/C53-bound STING oligomer structure
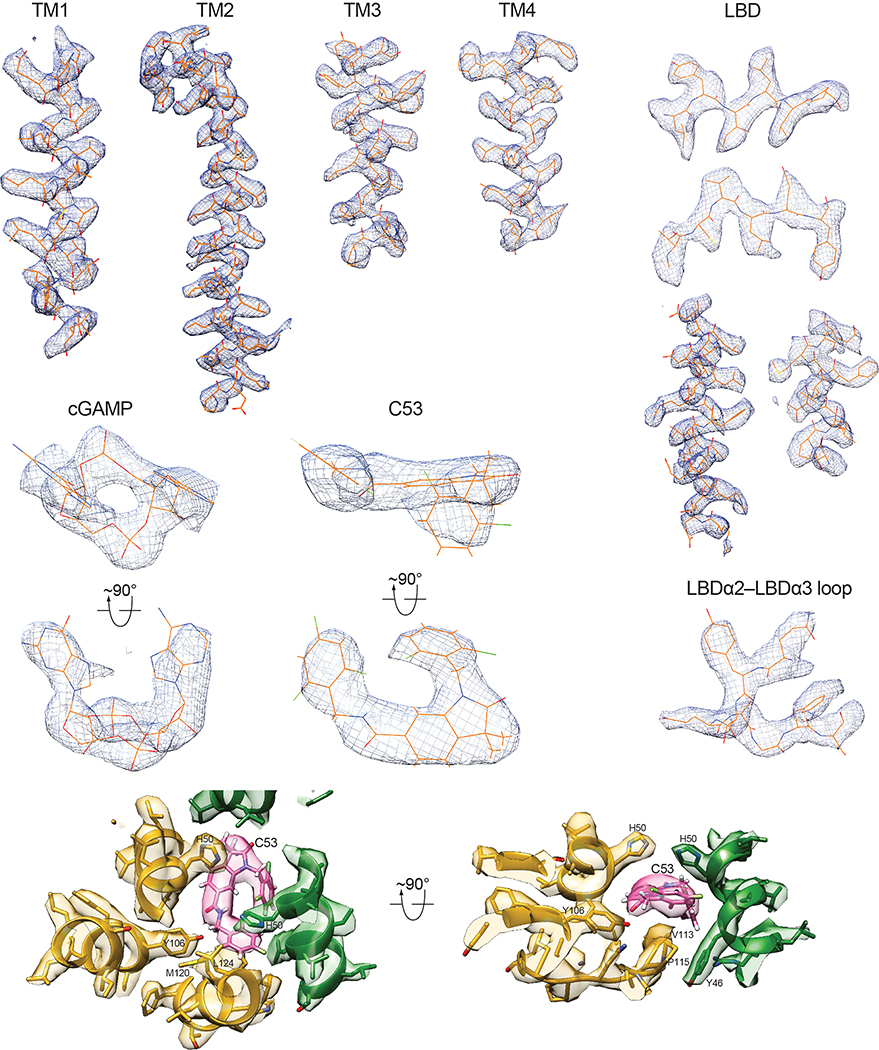
To understand how C53 activates STING, we applied single-particle cryo-EM to determine the structure of the human STING oligomer with both C53 and cGAMP bound (Extended Data Fig. 2). We segmented long oligomers into units containing four dimers for structure determination. This approach resulted in the structure of four STING dimers packed side-by-side in an approximately linear arrangement, which contains all the information for reconstructing higher-order oligomers. The cryo-EM map of the two STING dimers at the center of this structure was further improved to 3.4 Å resolution, enabling us to build a precise model for both the STING tetramer and the bound compounds (Fig. 1).
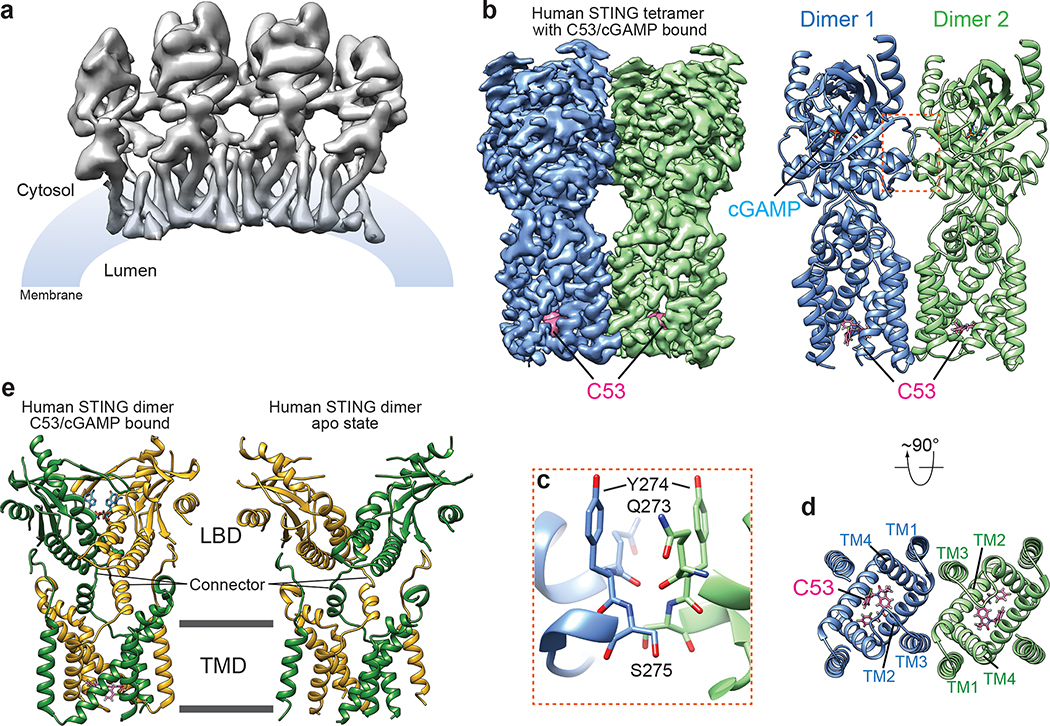
Fig. 1. Structure of the high-order oligomer of human STING bound to both cGAMP and C53.
a, Overall cryo-EM map of the oligomer containing four STING dimers. b, High-resolution cryo-EM map and atomic model of the STING tetramer. c, Expanded view of the inter-dimer interfaced mediated by the LBDα2-LBDα3 loop. The color scheme is the same as in b. d, C53-binding pocket in the TMD of STING viewed from the luminal side of ER/Golgi. The color scheme is the same as in b. e, Comparison of the human STING dimer bound to both cGAMP and C53 with the STING apo-state. The two subunits in the dimer are colored green and yellow respectively.
Our previous structures of chicken STING in both the apo- and cGAMP-bound states have shown that cGAMP induces a 180°-rotation of the LBD with respect to the TMD, converting the two connectors linking the LBD and TMD in the STING dimer from the crossover to parallel configuration8. The human STING dimer bound to both cGAMP and C53 here exhibits the parallel configuration of the connector, in contrast to the crossover configuration in the apo-state solved previously (Fig. 1e)8. These observations together support the notion that the 180°-rotation of LBD is an evolutionarily conserved step in STING activation. cGAMP adopts the typical U-shape and binds to the central pocket in the LBD dimer, consistent with all previous STING/cGAMP complex structures (Extended Data Fig. 3)1.
Our cryo-EM structure of human STING tetramer shows that two STING dimers bound with both C53 and cGAMP assemble in a side-by-side fashion similar to the chicken STING tetramer (Fig. 1). Both the LBD-LBD and TMD-TMD interactions between the two human STING dimers contribute to the assembly of the tetramer. The LBD-LBD contact between the two STING dimers is mediated largely by the loops connecting the α2 and α3 of LBD (LBDα2–LBDα3 loop) (Fig. 1c). This interface was resolved to low resolution in the cryo-EM map of chicken STING tetramer (PDB ID: 6NT8). The high-resolution cryo-EM map of human STING tetramer here allows this interface to be resolved in detail. Interestingly, the interaction of the LBDα2–LBDα3 loops is mostly mediated by the backbone hydrogen bonds, with minimal involvement of sidechains. As a result, this interface appears small and weak, and therefore may not be sufficient for supporting the formation of large STING oligomers. The human STING tetramer is further stabilized by extensive interactions made by the TMD (Fig. 1d), in sharp contrast to the chicken STING tetramer where the TMD barely makes any inter-dimer contact8. C53 induces conformational changes to the TMD of human STING, and thereby promoting the TMD-TMD interactions (See details below). These results explain the additive effects of C53 and cGAMP in promoting the oligomerization of human STING.
C53 binds at a cryptic site in STING-TMD
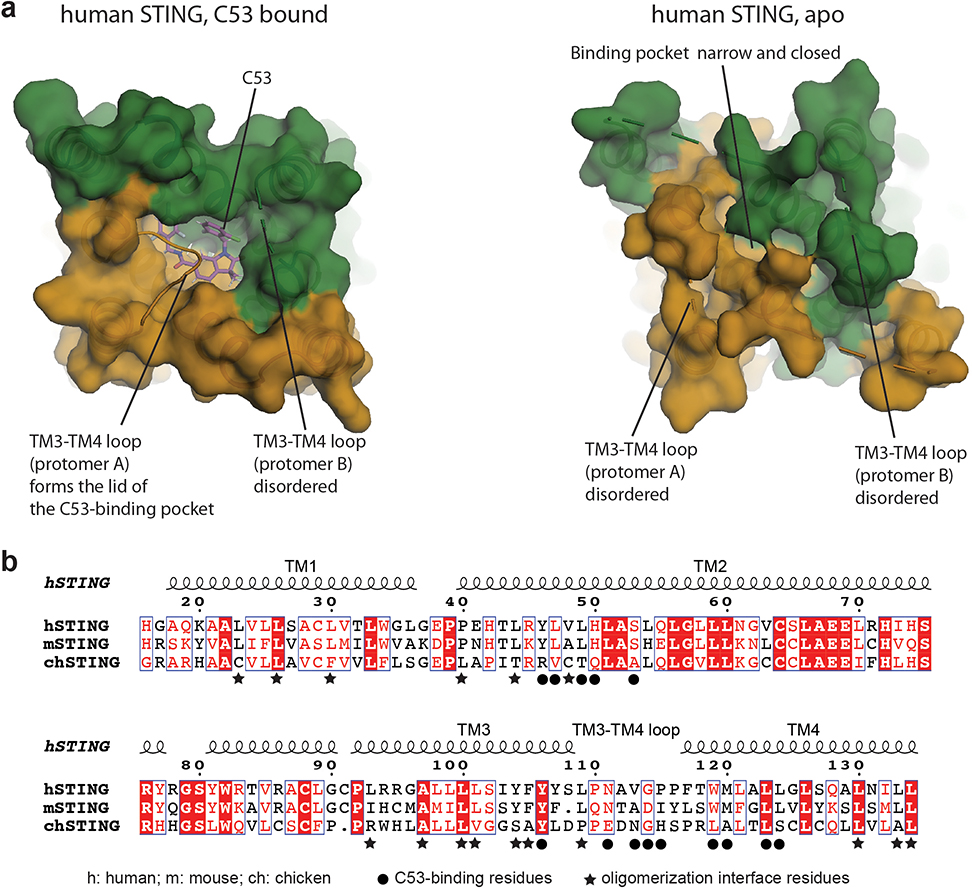
Strikingly, our cryo-EM map shows a strong density peak at the luminal side of the TMD of each STING dimer, which can be assigned to C53 unequivocally based on clear asymmetric C-shape and local chemical environment (Fig. 2 and Extended Data Fig. 3). This agonist binding mode at STING-TMD has not been observed previously. In each STING dimer, a single C-shaped C53 sits approximately at the C2 axis of the STING dimer, in a deep pocket formed among two TM2s and two TM4s. This pocket in the apo-state of human STING is much smaller and cannot accommodate C53 due to tighter packing among these TM helices (Extended Data Fig. 4a). Therefore, C53 binds to the cryptic site in human STING-TMD through an induced-fit mechanism.
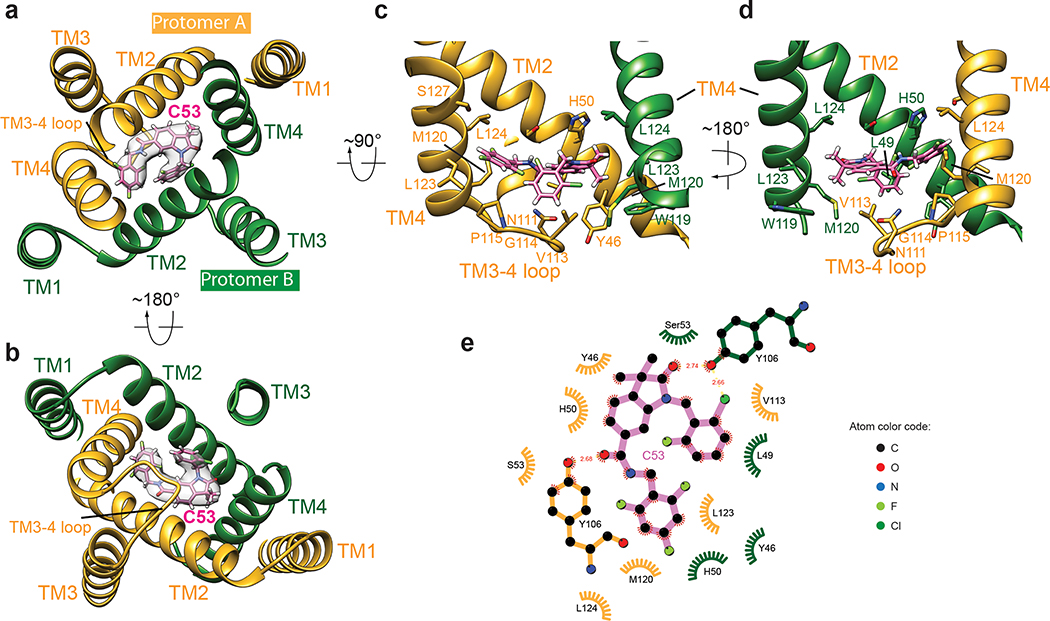
Fig. 2. Interaction between STING and C53.
a, b, Overview of the C53-binding site in the TMD of one STING dimer, viewed from the cytosolic and luminal sides, respectively. Cryo-EM density for C53 is shown in semi-transparent white. c, d, Details of the binding site in two different views. Helices in front of C53 are omitted for clarity. e, Two-dimensional diagram of the interactions between C53 and STING. Hydrogen atoms are omitted.
The 2-Cl-6-F-phenyl ring at one end of C53 is juxtaposed closely with the trifluoro-phenyl ring at the other end, resulting in the overall C-shape (Fig. 2a, b). This compact conformation allows C53 to fit into the narrow rectangular-shaped cavity between TM2 and TM4 from each subunit of the STING dimer. On one side of the binding pocket, the oxindole core makes edge-on packing interactions with TM2 from protomer A of the STING dimer. The outer edge of the oxindole core is accommodated in the space surrounded by Y46, L49, H50 and S53 in TM2 from protomer A (Fig. 2c, d, e). Meanwhile, the two methyl groups on the oxindole core are placed close to W119, M120, L123 and L124 in TM4 from protomer B. On the other side, the 2-Cl-6-F-phenyl and trifluoro-phenyl rings of C53 interact with TM2 from protomer B and TM4 from protomer A. The 2-Cl-6-F-phenyl ring packs its flat face onto a patch composed of Y46, L49, H50 and S53 in TM2 from protomer B. The trifluoro-phenyl wedges between Y46, L47 and H50 of TM2 from protomer B and M120, L123 and L124 of TM4 from protomer A. TM3 contributes to the binding pocket by filling the gap between TM2 and TM4 near the ER/Golgi luminal side. Y106 in TM3 from each protomer makes a hydrogen bond with C53.
The mouth of the binding pocket facing the ER/Golgi lumen is sealed by the TM3-TM4 loop from protomer A of the STING dimer (Fig. 2c, d). V113, G114 and P115 in this loop pack closely with the bottom face of C53. N111 is partially buried in the binding pocket, contributing to the stability of the loop conformation by making hydrogen bonds with the backbone of the loop. The TM3-TM4 loop from protomer B is mostly disordered, while modelling it to the same conformation as that in protomer A leads to clashes between the two loops. Therefore, C53 as an asymmetric molecule induces obvious asymmetry in the TMD of the STING dimer, in contrast to cGAMP, for which its binding site in the LBD is overall symmetric or only shows subtle asymmetry1,29. While most C53-contacting residues from the TM helices are conserved between human and mouse STING, those in the TM3-TM4 loop are not (Extended Data Fig. 4b). This divergence in the TM3-TM4 loop provides an explanation for the observation that C53 activates human STING but not mouse STING10. Similar species specificity has been displayed by DMXAA (5,6-dimethylxanthenone-4-acetic acid), which acts as an agonist for mouse STING by targeting the cGAMP-binding pocket but does not activate human STING30.
To validate this binding mode of C53, we introduced mutations to residues in the binding pocket (H50A, S53L, Y106A, and M120L) and the TM3-TM4 loop (V113Q, G114Q and P115Q) of STING. Our native-gel results showed that, compared to STING wild type (STING-WT), these mutants showed no or greatly reduced high-order oligomerization upon stimulation by C53 and cGAMP (Fig. 3a). Consistently, these mutations also led to significantly decreased C53-induced puncta formation of STING in cells, which reflects the oligomerization of STING on ER/Golgi membrane (Extended Data Fig. 5). In addition, our cell-based functional assay showed that C53 and cGAMP together triggered higher levels of phosphorylation of STING, TBK1 and IRF3 than either agonist alone, suggesting that STING could reach stronger activity when bound to both C53 and cGAMP simultaneously (Fig. 3b and Extended Fig. 6a, b). The mutations in the C53-binding pocket abrogated the phosphorylation of these proteins triggered by C53, confirming that C53 activates STING by targeting this pocket (Fig. 3b). Interestingly, cGAMP-induced phosphorylation was reduced by two mutations in the C53-binding pocket, V113Q (Fig. 3b) and M120L (Extended Fig. 7b), suggesting that this pocket is involved in cGAMP-mediated activation of STING, perhaps through the binding of an endogenous ligand.
Fig. 3. Mutational analyses of the C53-binding site in human STING.
a, Effects of mutations in the C53-binding site on STING oligomerization induced by cGAMP and C53. Native gel results shown are representatives of three biological repeats. b, Effects of mutations in the C53-binding site on phosphorylation of STING, TBK1 and IRF3 induced by cGAMP and C53 in cells. HEK293T cells were transfected with human STING wild type or the mutants. Cells were stimulated with cGAMP, C53 or both and lysates were subjected to western blot analyses. The results shown are representatives of three biological repeats. For gel source data, see Supplementary Figure 1.
C53-induced conformational changes
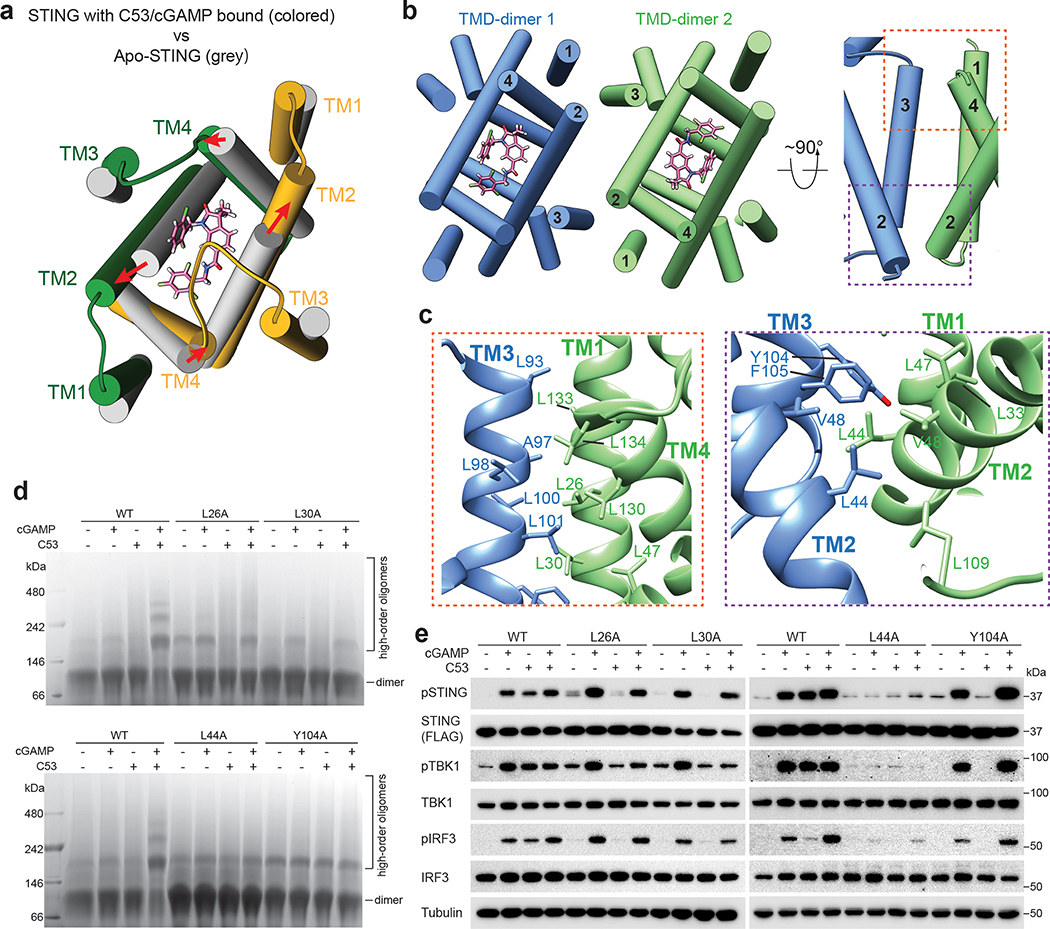
Structural comparison of STING without or with C53 bound revealed that TMs that surround C53 in the STING dimer, especially TM2 and TM4, undergo substantial sideway expansion upon C53 binding (Fig. 4a and Extended Data Fig. 4a). Such conformational rearrangements of TM2 and TM4 are necessary for providing sufficient space for C53 binding. In the tetrameric structure of chicken STING with cGAMP bound, the two STING dimers pack side-by-side mainly through the interaction between the LBDs, while no direct interactions are observed between the TMDs. In contrast, C53-induced dilation of the TMD in human STING allows the TM helices to engage in the TMD-TMD interaction between two STING dimers (Fig. 4b, c). The TMD interface contains two four-helix bundles that are related by the C2-symmetry of the tetramer. Each four-helix bundle is formed by one TM3 from one STING dimer and one set of TM1, TM2 and TM4 from the other dimer (Fig. 4b). Extensive interactions are made by hydrophobic residues from the four helices, including L23, L26 and L30 from TM1, L44 from TM2 as well as L93, A97, L100, L101, Y104, F105 and L109 from TM3 (Fig. 4c). The two four-helix bundles are connected at the ER/Golgi luminal side by the N-terminal residues L44 and V48 from TM2. Most of the residues in the TMD-TMD interaction are conserved between human and mouse STING, but not in chicken STING (Extended Data Fig. 4b), consistent with the observation that chicken STING appears less dependent on the TMD interaction for the high-order oligomerization8. The TMD interface buries ~2600 Å2 solvent accessible area, much larger than the LBD interface (~740 Å2). Notably, the TMDs between the two STING dimers associate more closely on the luminal side than on the cytosolic side, leading to the overall curvature of high-order STING oligomers.
Fig. 4. TMD-mediated interactions contribute to STING oligomerization.
a, C53 induces dilation of the TMD binding pocket in the STING dimer. A comparison with apo-STING shows that the TM helices in the C53-bound structure shift away from each other to enlarge the binding pocket. The structure is viewed from the ER/Golgi luminal side. b, Two orthogonal views of the TMD-TMD interface between the two STING dimers. TM3 from one dimer joins one set of TM1, TM2 and TM4 from the other dimer to form a four-helix bundle. c, Detailed views of the TMD-TMD interface. d, Effects of mutations in the TMD-TMD interface on STING oligomerization induced by cGAMP and C53. Native gel results shown are representatives of three biological repeats. e, Effects of mutations in the TMD-TMD interface on phosphorylation of STING, TBK1 and IRF3 induced by cGAMP and C53 in cells. HEK293T cells were transfected with human STING wild type or the mutants. Cells were stimulated with cGAMP, C53 or both and lysates were subjected to western blot analyses. The results shown are representatives of three biological repeats. For gel source data, see Supplementary Figure 1.
Our tetrameric structure of STING shows an asymmetric binding mode of C53 in each STING dimer, with the side of the STING dimer bound to the closed face of the C-shaped C53 forming the tetramerization interface. The STING tetramers in this configuration represents the dominant state, as no other tetrameric forms were captured from the same cryo-EM dataset. However, the presence of oligomers larger than tetramer in the sample means that both sides of the STING dimer bound to C53 can mediate the oligomerization. We speculate that the STING tetramers assembled by the other side of STING dimer (i.e., the side bound to the open face of C53) are less stable, and therefore could not be determined to high resolution by cryo-EM.
Tetramer and larger oligomers of STING can serve as the signaling platform for efficient recruitment and activation of TBK1 and IRF31,9. The induction of the TMD-TMD interaction therefore likely underlies the activating effect of C53 on STING. To test this model, we introduced mutations to the TMD-TMD interface, including L26A, L30A, L44A and Y104A. Compared to STING-WT, these mutants showed substantially reduced oligomerization in solution in the presence of both C53 and cGAMP (Fig. 4d). Likewise, the mutations led to reduced puncta formation of STING in cells upon stimulation by C53 (Extended Data Fig. 8). The mutations also largely abolished C53-induced phosphorylation of STING, TBK1 and IRF3 in cells (Fig. 4e). In addition, L44A abrogated cGAMP-induced phosphorylation of these proteins (Fig. 4e), whereas L30A reduced STING phosphorylation in cells treated with a lower concentration of cGAMP (100 nM) (Extended Fig. 7c). These results together support the notion that the TMD-TMD interface is important for the activation of STING and downstream signaling by either C53 or cGAMP.
Discussion
Our cryo-EM structure of human STING in complex with both cGAMP and C53 defines a novel agonist binding site in STING-TMD, paving the way for developing more agonists that target this site. Compounds that target the TMD site might be better STING agonists than cGAMP mimetics for therapeutic purposes because they are more hydrophobic and therefore more permeable to the cell membrane (Extended Data Fig. 6c, d)10. In addition, our structural analyses clarify the role of the TMD in the oligomerization and activation of STING. The coupling between the induced opening of the TMD pocket by C53 and the high-order oligomerization of human STING suggests that this conformational change might be an integral part of its activation mechanism in vivo. This notion raises the question how this conformational change is triggered in the cell. It is possible that cGAMP-induced 180°-rotation of the LBD may allosterically induce the open conformation of the TMD on the ER/Golgi membrane, which could not be recapitulated with purified protein in vitro. Alternatively, a cellular endogenous ligand may bind to the TMD pocket and act as a secondary activator to facilitate cGAMP-induced STING activation. Addressing the existence and identity of such a secondary activator of STING will be interesting areas of future studies.
Methods
Protein expression and purification.
The expression constructs and expression and purification procedures for human STING are similar to those described in a previous study8. The coding sequence for human STING residues 1–343, excluding the C-terminal tail that is not a part of the folded structure of the protein, fused to a cleavage site for the human rhinovirus 3C protease and T6SS immunity protein 3 (Tsi3) from Pseudomonas aeruginosa at the C terminus in tandem were inserted into the pEZT-BM vector31. Mutations were introduced by PCR-based mutagenesis. The plasmids were transfected using polyethylenimine (PEI) into HEK293F cells cultured in suspension in FreeStyle293 Expression medium (Gibco, Cat#12338–018), with 1000 μg DNA and 4 ml PEI at 1mg/ml for 1 L cells. These and other cells used were assumed to be authenticated by the commercial sources, and therefore were not authenticated in the study. DAPI (4′,6-diamidino-2-phenylindole) staining and the e-Myco Mycoplasma PCR Detection Kit (Bulldog Bio) were used to ensure cells not contaminated by mycoplasma. Cells were harvested 72 hours after transfection, re-suspended in buffer A (containing 20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM CaCl2, 1% DNase I, 0.2 mM AEBSF and 0.5 mM TECP) and disrupted by French press. The lysates were centrifuged for 10 min at 5000 g to remove debris. Membrane fraction was pelleted by centrifugation at 100,000 g for 1 hour. Proteins in lipid membrane were extracted by 1% n-Dodecyl-B-D-Maltoside (DDM) and 0.2% cholesteryl hemisuccinate tris salt (CHS) in buffer A. The sample was subjected to another round of centrifugation to remove insoluble fraction. The affinity purification step of human STING was based on the high-affinity interaction between the C-terminal Tsi3 tag and the T6SS effector protein Tse38. Detergent solubilized STING was captured by Tse3-conjugated Sepharose 4B resin (GE Healthcare) equilibrated in buffer B (20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM CaCl2, 20 mM imidazole, 0.5 mM TECP, 0.03% DDM and 0.006% CHS). Unbound proteins were removed by extensive wash with buffer B. STING was eluted by cleavage from the Tsi3 tag with the 3C protease on resin at 4 °C for 12 hours. The eluted protein was further purified on a Superdex S200 increase 10/300 column (GE healthcare) in buffer C (25 mM HEPES pH 7.5, 150 mM NaCl, 0.5 mM TECP, 0.03% DDM and 0.006% CHS). Peak fractions were pooled, concentrated and kept at −80 °C before use.
Synthesis of C53.
All solvents for synthesis (N,N’-dimethylformamide (DMF), tetrahydrofuran (THF) and methylene chloride (DCM)) were obtained by passing commercially available pre-dried, oxygen-free formulations through activated alumina columns. All reagents were purchased at high commercial quality (Sigma-Aldrich, Oakwood and AK Scientific) and used without further purification, unless otherwise stated. Reactions were monitored by thin-layer chromatography (TLC) carried out on 0.25 mm E. Merck silica gel plates (60F-254) and visualized under UV light and/or by appropriate staining method (an ethanolic solution of phosphomolybdic acid or cerium sulphate). Flash column chromatography was performed using E. Merck silica gel (60, particle size 0.04–0.063 mm). NMR spectra were recorded on a Bruker Ascend 400 and calibrated using residual not perdeuterated solvent (DMSO-d6: δH =2.50 ppm, δC =39.52 ppm) or using an external reference for 19F NMR [δF=0 (CCl3F) ppm] at 298 K. All 13C NMR spectra were broadband 1H decoupled. The chemical shifts of the peaks of the major rotamer were reported and coupling constants were given in Hz only for the major rotamer. LC-MS was performed on Agilent 1260 Infinity II Single Quadrupole with an Agilent Eclipse XDB-C18 5 μm 4.6 × 150 mm column. Buffer A was 0.1% CF3CO2H in H2O, and buffer B was 0.1% CF3CO2H in MeCN. Analytical gradient (0.0–7.0 min, buffer B from 10% to 60%; 7.0–10.0 min, buffer B from 60% to 100%; 10.0–15.0 min, buffer B maintains 100%) was performed with flow of 0.8 mL/min. For details of the chemical synthesis and characterization, see Supplementary Information.
Native gel analyses of STING oligomerization.
Purified human STING wild type or mutants in buffer C at 50 μM were incubated with DMSO as control, cGAMP at 100 μM, C53 at 100 μM or both at 4°C for 2 hrs. Samples (15 μg protein) were mixed with the Native gel loading buffer (Invitrogen, Cat#BN20032) and resolved using 3–12% gradient native gel (Invitrogen, Cat#BN1003BOX).
Cryo-EM data collection and image processing.
Purified wild type human STING at 50 μM were incubated with cGAMP at 100 μM and C53 at 100 μM for formation of the protein/ligands complex. The complex was purified using a Superose 6 10/300 gel filtration column (GE healthcare) in buffer C. Peak fractions were collected and concentrated to 2.9 mg/ml. Additional cGAMP (100 μM) and C53 (100 μM) were added to ensure saturation of the protein by the ligands. The sample was applied to a glow-discharged Quantifoil R1.2/1.3 300-mesh gold holey carbon grid (Quantifoil, Micro Tools GmbH, Germany), blotted under 100% humidity at 4 °C and plunged into liquid ethane using a Mark IV Vitrobot (FEI).
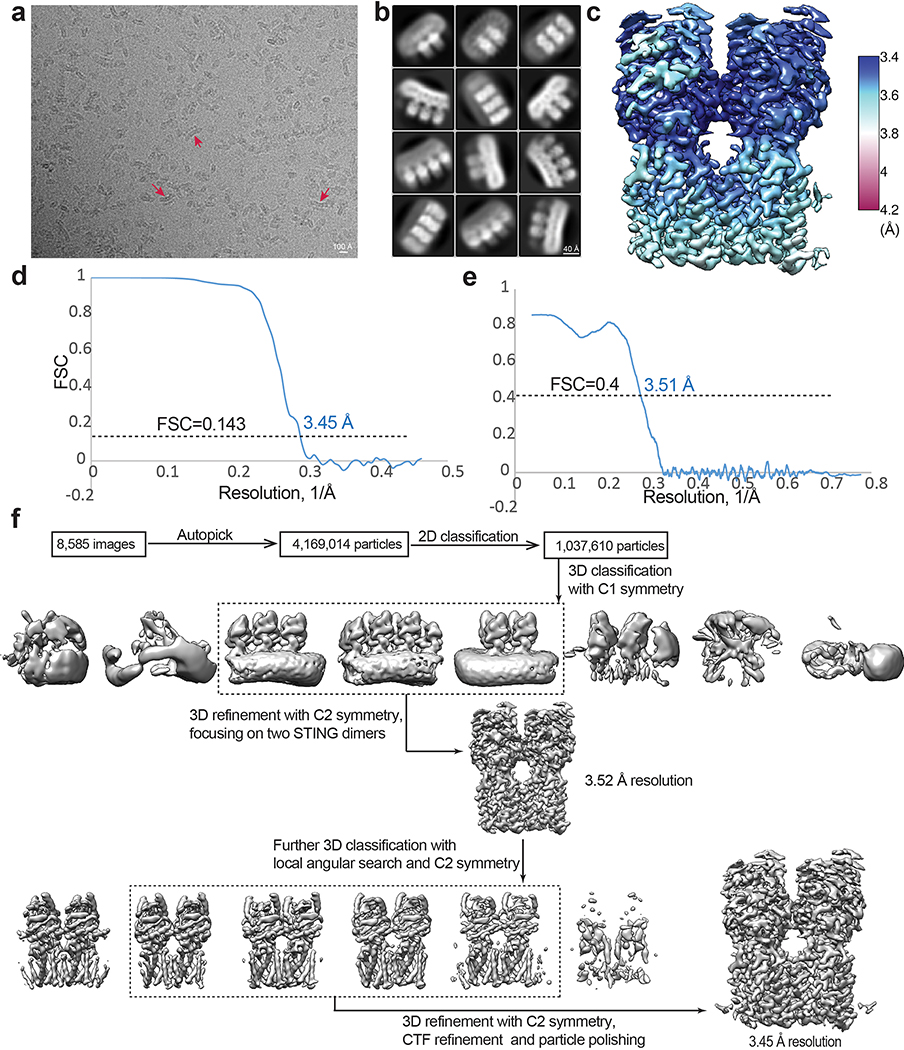
Micrographs were collected on a Titan Krios microscope (FEI) with a K3 Summit direct electron detector (Gatan) in the super-resolution counting mode operated at 300 kV. The slit width of the GIF-Quantum energy filter was set to 20 eV. The nominal magnification was 81,000x and the pixel size of 1.08 Å. Micrographs were dose-fractioned into 36 frames with a total exposure time of 7.2 s at the dose rate of 1.6 e−/Å2/frame in the correlated double sampling (CDS) mode. Movie frames were motion-corrected and dose-weighted using the Motioncorr2 program (version 1.2)32. GCTF 1.06 was used for CTF correction33. Two sets of Particles were picked by using Topaz 0.234 and template-based picking in RELION 3.135, respectively. The two sets were combined with duplicates removed. The rest of the image processing was done in RELION (Extended Data Fig. 2d). Particles were initially extracted with a box size of 160 pixel, which is large enough to accommodate four STING dimers, and binned by a factor of 4 for 2D classification. Particles from good 2D classes were re-extracted with a binning factor 2 and subjected to 3D classification. The initial model was generated with the chicken STING tetramer. A total of 288,021 particles in good 3D classes were selected and re-extracted to the original pixel size for 3D refinement. The 3D reconstructions of the STING tetramer, with C2 symmetry relating the two dimers, reached resolution of ~3.52 Å. An additional round 3D classification with local angular search and the C2 symmetry were performed and two classes showing poor density was removed. The resulting 231,556 particles were subjected to further arounds of 3D refinement, CTF refinement and Bayesian polishing, leading to the final 3D reconstruction with resolution of 3.45 Å. Resolution was estimated by applying a soft mask around the protein density, using the Fourier Shell Correlation (FSC) 0.143 criterion (Extended Data Fig. 2c). Local resolution was calculated in RELION (Extended Data Fig. 2b).
Model building, refinement and analyses.
Model building was initiated by docking the structure of the human STING dimer in the apo state (PDB ID: 6NT5) into the cryo-EM density, followed by manual adjustments in Coot 0.9436. The high quality of the density allows most of the residue sidechains to be clearly identified. C53 and cGAMP were manually fit into the density in Coot. cGAMP is an asymmetric molecule with a 3’−5’ and 2’−5’ phosphodiester bond linking the AMP and GMP moieties24,25,37,38, which could bind to the symmetric STING dimer in two alternative orientations. The subtle asymmetric of cGAMP however do not usually cause obvious asymmetric in the STING LBD1,8. As a result, either way of fitting cGAMP into its binding pocket is equally valid. We therefore chose one orientation arbitrarily to fit cGAMP into the density (Extended Data Fig. 3). In contrast, the asymmetry of C53 is much more pronounced, leading to clear asymmetry in the STING-TMD. The well-defined density for C53 allowed us to dock it to the binding pocket without ambiguity (Extended Data Fig. 3). Real-space refinement of the model was carried out with Phenix 1.1839. Molprobity as a part of the Phenix validation tool was used for assessing the quality of the model40. Statistics of the refined model are summarized in Extended Data Fig. 7. Structural figures were rendered in Coot, PyMOL 2.4 (The PyMOL Molecular Graphics System, Schrödinger) or Chimera 1.1641. Sequence alignment was rendered with ESPript 342. Two-dimensional interaction diagram between C53 and STING was generated with LigPLot+ 2.243. Data collection and model refinement statistics are summarized in Extended data Table 1.
Western blot analyses of phosphorylation of STING, TBK1 and IRF3.
The coding sequence for full-length human STING fused with a C-terminal FLAG-tag was cloned into the pcDNA3.1A(+) vector (Invitrogen). Mutations were introduced by PCR-based mutagenesis. Plasmids were transfected with 500 ng into HEK293T cells in 6-well plates, and cultured for additional 24 hours. Stimulation of cells with cGAMP and C53 was carried out by using digitonin-mediated permeabilization that allows cGAMP to penetrate the cell membrane and enter the cell as described previously44. cGAMP (1 μM), C53 (10 μM) or both in a buffer containing 50 mM HEPES pH7.5, 100 mM KCl, 3mM MgCl2, 0.1 mM DTT, 85 mM sucrose, 0.2% BSA, 1 mM ATP, 0.1 mM GTP and 10 ug/ml digitonin were used to treat cells for 1 hour. For the control experiments in which treatment was carried out without permeabilization as shown in Extended Data Fig. 6 and 7, the same buffer without digitonin was used instead. Cells were lysed in RIPA buffer and lysates were subjected to western blot analyses. Human STING protein was detected by anti- FLAG primary antibody (Bimake, Cat#A5712; 3000X dilution) and anti-Mouse IgG HRP-linked secondary antibody (Cell Signaling Technology, Cat#7076S; 3000X dilution). Phosphorylated STING was detected by Rabbit anti-phospho-STING (S366) antibody (Cell Signaling Technology, Cat#19781S; 1000X dilution) and anti-Rabbit IgG HRP-linked secondary antibody (Cell Signaling Technology, Cat#7074S; 3000X dilution). TBK1 was detected by Mouse anti-TBK1/NAK antibody (Cell Signaling Technology, Cat#51872S; 1000X dilution). Phosphorylated TBK1 was detected by Rabbit anti-phospho-TBK1/NAK antibody (Cell Signaling Technology, Cat#5483S; 1000X dilution). IRF3 was detected by Mouse anti-IRF3 antibody (Abcam, Cat#ab50772; 1000X dilution) and phosphorylated IRF was detected by Rabbit anti-phospho-IRF3 (S386) (Abcam, Cat#ab76493; 1000X dilution).
Fluorescence microscopy of STING oligomerization in cells.
The coding sequence for full-length human STING fused with a C-terminal GFP-tag was cloned into the pmEGFP-N1 vector (Addgene). Mutations were introduced by PRC-based mutagenesis. Plasmids were transfected into Hela cells cultured on glass coverslips in 6-well plates, and cultured for additional 24 hours. Stimulation of cells with cGAMP and C53 was performed in the same manner as described above. One hour after stimulation, cells were washed in PBS and fixed in 4 % paraformaldehyde. STING-GFP was imaged by using a DeltaVision fluorescence microscope with a 40x objective, controlled with the software package SoftWoRx 7.0. Nuclei were stained with DAPI. The experiment for each STING constructs was repeated three times. The percentage of cells containing STING puncta were obtained by counting number of GFP-positive cells with or without STING puncta respectively in images taken from random fields of coverslips. The number of cells counted for each STING construct in each repeat ranged from 60–452. The data was plotted with GraphPad Prism 9.
Data availability.
The atomic coordinates and the cryo-EM map of STING bound to both cGAMP and C53 have been deposited into the RCSB (PDB ID: 7SII) and EMD databases (EMDB ID: EMD-25142).
Extended Data
Extended Data Fig. 1. Purification of human STING and its interaction with C53 and cGAMP.
a, Gel filtration profile of human STING on a Superdex S200 10/300 column. b, SDS-PAGE analyses of fractions indicated by the bracket in a. c, Analyses of STING oligomerization by native gel. C53 and cGAMP together induced robust high-order oligomerization of purified STING solubilized in detergent solution, while either one alone failed to do so under the same condition. The results shown are representatives of three biological repeats.
Extended Data Fig. 2. Image processing procedure of human STING tetramer bound to both cGAMP and C53.
a, Motion corrected micrograph. Red arrows highlight high-order oligomers of STING. The curved overall shape of the oligomers is evident from these examples. b, 2D class averages of high-order oligomers of human STING. Large oligomers were segmented into particles containing four dimers at the maximum. c, Final 3D reconstruction of the tetramer colored based on local resolution. d, Gold-standard FSC curve of the final 3D reconstruction. e, FSC between the final map and the atomic model. f, Image processing procedure.
Extended Data Fig. 3. Sample density maps of various parts of the structure.
Various parts of the structure with the cryo-EM density overlayed are shown. Most of the protein sidechains are clearly visible. The high-quality of the densities for cGAMP and C53 allows unambiguous docking of the compounds.
Extended Data Fig. 4. C53-induced dilation of the binding pocket in the STING-TMD.
a, Comparison of the TMD of human STING in the C53-bound and the apo states. It is evident that the TMD pocket in apo-STING is much smaller and cannot accommodate C53, and binding of C53 induces dilation of this pocket. b, Sequence alignment of the TMD of STING from human (h), mouse (m) and chicken (ch). Black circles highlight residues in the C53-binding pocket. Stars highlight residues in the TMD-TMD interface that contribute to the oligomerization of STING.
Extended Data Fig. 5. Effects of mutations in the C53-binding site on STING oligomerization in cells.
a and c, Representative images of cells expressing STING wild type or the mutants in the binding pocket and the TM3-TM4 loop, respectively. Hela cells were transfected with GFP-tagged human STING wild type or mutants. Cells were stimulated with cGAMP, C53 or both. Localization of STING-GFP in cells was monitored by the fluorescence signal of GFP. Nuclei were stained with DAPI. The experiments were repeated three times. The images are representatives from these experiments. As expected, STING showed a diffuse pattern in cells in the absence of an agonist. Both C53 and cGAMP induced puncta formation of STING wild type. C53-induced puncta formation was reduced or abolished by the mutations. b and d, Quantification of STING puncta formation in cells expressing the wild type or mutants. The bar graph shows the individual data points, mean and s.e.m. of the percentage of cells with STING forming large puncta from the three biological repeats. Statistical significance p-values between the wild type and mutants were calculated by two-tailed Welch’s t-test. Scale bar, 10 μm. Source data of puncta counting results are provided in the source data file.
Extended Data Fig. 6. Further characterization of activation of STING in cells by C53 and cGAMP.
a and b, Activation of STING in cells by C53 and cGAMP at a series of concentrations. Notably, while cGAMP at 10 nM and C53 at 100 nM individually induced minimal phosphorylation of STING, the combination of the two led to robust activation of STING. c and d, Activation of STING by cGAMP or C53 with or without digitonin-mediated cell permeabilization. It is clear from these results that cGAMP-mediated activation of STING is dependent on digitonin-mediated permeabilization, whereas C53 can enter the cell and activate STING on its own. The experiments were carried out in a similar manner as in Fig. 3 and Fig. 4, and the results shown are representatives of three biological repeats. For gel source data, see Supplementary Figure 1.
Extended Data Fig. 7. Additional mutational analyses of STING activation by C53 and cGAMP.
a, C53 could induce phosphorylation of the R238A/Y240A mutant of STING in cells, which has its cGAMP-pocket disrupted and could not be activated by cGAMP. b, M120L, a mutation in the C53-binding pocket, reduced STING phosphorylation when cells were stimulated with cGAMP at a lower concentration (100 nM). c, L30A, a mutation in TMD-TMD oligomerization interface, reduced STING phosphorylation when cells were stimulated with cGAMP at a lower concentration (100 nM). These effects of M120L and L30A were suppressed when cells were treated with cGAMP at 1000 nM. These results suggest that both the TMD pocket and the TMD-TMD interface are important for cGAMP-mediated activation of STING, although high concentrations of cGAMP can overcome the detrimental effects of some mutations in these regions. The experiments were carried out in a similar manner as in Fig. 3 and Fig. 4, and the results shown are representatives of three biological repeats. For gel source data, see Supplementary Figure 1.
Extended Data Fig. 8. Effects of mutations in the TMD-TMD interface on STING oligomerization in cells.
a, Representative images of cells expressing STING wild type or the mutants in the TMD-TMD interface. Hela cells were transfected with GFP-tagged human STING wild type or mutants. Cells were stimulated with cGAMP, C53 or both. Localization of STING-GFP in cells was monitored by the fluorescence signal of GFP. Nuclei were stained with DAPI. The experiments were repeated three times. The images are representatives from these experiments. As expected, STING showed a diffuse pattern in cells in the absence of an agonist. Both C53 and cGAMP induced puncta formation of STING wild type, which was reduced or abolished by the mutants. b, Quantification of STING puncta formation in cells expressing the wild type or mutants. The bar graph shows the individual data points, mean and s.e.m. of the percentage of cells with STING forming large puncta from the three biological repeats. Statistical significance p-values between the wild type and mutants were calculated by two-tailed Welch’s t-test. Scale bar, 10 μm. Source data of puncta counting results are provided in the source data file.
Extended Data Table 1.
Data collection and model refinement statistics.
| Magnification | 81,000 |
| Voltage (kV) | 300 |
| Electron exposure (e−/Å2) | 50 |
| Defocus range (μm) | 1.5–2.5 |
| Pixel size (Å) | 1.08 |
| Symmetry imposed | C2 |
| Initial particle images (no.) | 4,169,014 |
| Final particle images (no.) | 231,556 |
| Map resolution (Å) | 3.45 |
| FSC threshold | 0.143 |
| Initial model used (PDB code) | 6NT5 |
| Model resolution (Å) | 3.5 |
| FSC threshold | 0.4 |
| Map sharpening B factor (Å2) | −100 |
| Model Composition | |
| Non-hydrogen atoms | 10300 |
| Protein residues | 1286 |
| Ligands | cGAMP, 2; C53, 2 |
| B factors (Å2) | |
| Protein | 69.9 |
| Ligand | 65.7 |
| R.m.s. deviations | |
| Bond length (Å) | 0.003 |
| Bond angle (°) | 0.501 |
| Validation | |
| Molprobity score | 1.68 |
| Clashscore | 5.0 |
| Poor rotamers (%) | 2.2 |
| Ramachandran plot | |
| Favored (%) | 97.1 |
| Allowed (%) | 2.9 |
| Outliers (%) | 0 |
Supplementary Material
Acknowledgements.
Cryo-EM data were collected at the University of Texas Southwestern Medical Center (UTSW) Cryo-Electron Microscopy Facility, funded in part by the Cancer Prevention and Research Institute of Texas (CPRIT) Core Facility Support Award RP170644. We thank Daniel Stoddard and Jose Martinez Diaz for facility access. We thank the Structural Biology Lab at UTSW for equipment usage. This work is supported in part by grants from the National Institutes Health (R35GM130289 to X.Z and R01GM143158 to X.-C.B.), the Welch foundation (I-1702 to X.Z. and I-1944 to X.-C.B.) and CPRIT (RP160082 to X.-C.B.). X.-C.B. and X.Z. are Virginia Murchison Linthicum Scholars in Medical Research at UTSW.
Footnotes
Competing interests. The authors declare no competing interests.
References
- 1.Zhang X, Bai XC & Chen ZJ Structures and Mechanisms in the cGAS-STING Innate Immunity Pathway. Immunity 53, 43–53, doi: 10.1016/j.immuni.2020.05.013 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Ishikawa H & Barber GN STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678, doi: 10.1038/nature07317 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhong B et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550, doi: 10.1016/j.immuni.2008.09.003 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Jin L et al. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Molecular and cellular biology 28, 5014–5026, doi: 10.1128/MCB.00640-08 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun W et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci U S A 106, 8653–8658, doi: 10.1073/pnas.0900850106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu J et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830, doi: 10.1126/science.1229963 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun L, Wu J, Du F, Chen X & Chen ZJ Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791, doi: 10.1126/science.1232458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shang G, Zhang C, Chen ZJ, Bai XC & Zhang X Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP. Nature 567, 389–393, doi: 10.1038/s41586-019-0998-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang C et al. Structural basis of STING binding with and phosphorylation by TBK1. Nature 567, 394–398, doi: 10.1038/s41586-019-1000-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pryde DC et al. The discovery of potent small molecule activators of human STING. Eur J Med Chem 209, 112869, doi: 10.1016/j.ejmech.2020.112869 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Burdette DL et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518, doi: 10.1038/nature10429 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dou Z et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550, 402–406, doi: 10.1038/nature24050 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gluck S et al. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nature cell biology 19, 1061–1070, doi: 10.1038/ncb3586 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mackenzie KJ et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465, doi: 10.1038/nature23449 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang H, Wang H, Ren J, Chen Q & Chen ZJ cGAS is essential for cellular senescence. Proc Natl Acad Sci U S A 114, E4612–E4620, doi: 10.1073/pnas.1705499114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Motedayen Aval L, Pease JE, Sharma R & Pinato DJ Challenges and Opportunities in the Clinical Development of STING Agonists for Cancer Immunotherapy. J Clin Med 9, doi: 10.3390/jcm9103323 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao B et al. A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1. Nature 569, 718–722, doi: 10.1038/s41586-019-1228-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka Y & Chen ZJ STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Science signaling 5, ra20, doi: 10.1126/scisignal.2002521 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu S et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630, doi: 10.1126/science.aaa2630 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Ergun SL, Fernandez D, Weiss TM & Li L STING Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition. Cell 178, 290–301 e210, doi: 10.1016/j.cell.2019.05.036 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Dobbs N et al. STING Activation by Translocation from the ER Is Associated with Infection and Autoinflammatory Disease. Cell Host Microbe 18, 157–168, doi: 10.1016/j.chom.2015.07.001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saitoh T et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A 106, 20842–20846, doi: 10.1073/pnas.0911267106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gui X et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 567, 262–266, doi: 10.1038/s41586-019-1006-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X et al. Cyclic GMP-AMP Containing Mixed Phosphodiester Linkages Is An Endogenous High-Affinity Ligand for STING. Molecular cell 51, 226–235, doi: 10.1016/j.molcel.2013.05.022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kranzusch PJ et al. Ancient Origin of cGAS-STING Reveals Mechanism of Universal 2’,3’ cGAMP Signaling. Molecular cell 59, 891–903, doi: 10.1016/j.molcel.2015.07.022 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi H, Wu J, Chen ZJ & Chen C Molecular basis for the specific recognition of the metazoan cyclic GMP-AMP by the innate immune adaptor protein STING. Proc Natl Acad Sci U S A 112, 8947–8952, doi: 10.1073/pnas.1507317112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao P et al. Structure-function analysis of STING activation by c[G(2’,5’)pA(3’,5’)p] and targeting by antiviral DMXAA. Cell 154, 748–762, doi: 10.1016/j.cell.2013.07.023 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ernst AM et al. S-Palmitoylation Sorts Membrane Cargo for Anterograde Transport in the Golgi. Developmental cell 47, 479–493 e477, doi: 10.1016/j.devcel.2018.10.024 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cong X et al. Crystal structures of porcine STING(CBD)-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins. The Journal of biological chemistry 294, 11420–11432, doi: 10.1074/jbc.RA119.007367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conlon J et al. Mouse, but not human STING, binds and signals in response to the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid. J Immunol 190, 5216–5225, doi: 10.4049/jimmunol.1300097 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morales-Perez CL, Noviello CM & Hibbs RE Manipulation of Subunit Stoichiometry in Heteromeric Membrane Proteins. Structure 24, 797–805, doi: 10.1016/j.str.2016.03.004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332, doi: 10.1038/nmeth.4193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang K Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12, doi: 10.1016/j.jsb.2015.11.003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bepler T et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat Methods 16, 1153–1160, doi: 10.1038/s41592-019-0575-8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zivanov J et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, doi: 10.7554/eLife.42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta crystallographica 66, 486–501, doi: 10.1107/S0907444910007493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao P et al. Cyclic [G(2’,5’)pA(3’,5’)p] is the metazoan second messenger produced by DNA-activated cyclic GMP-AMP synthase. Cell 153, 1094–1107, doi: 10.1016/j.cell.2013.04.046 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ablasser A et al. cGAS produces a 2’−5’-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384, doi: 10.1038/nature12306 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica 66, 213–221, doi: 10.1107/S0907444909052925 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica 66, 12–21, doi: 10.1107/S0907444909042073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612, doi: 10.1002/jcc.20084 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Robert X & Gouet P Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42, W320–324, doi: 10.1093/nar/gku316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laskowski RA & Swindells MB LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 51, 2778–2786, doi: 10.1021/ci200227u (2011). [DOI] [PubMed] [Google Scholar]
- 44.Woodward JJ, Iavarone AT & Portnoy DA c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328, 1703–1705, doi: 10.1126/science.1189801 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and the cryo-EM map of STING bound to both cGAMP and C53 have been deposited into the RCSB (PDB ID: 7SII) and EMD databases (EMDB ID: EMD-25142).