Abstract
Population of the world run into several health-related emergencies among mankind and humans as it creates a challenge for the evolution of novel drug discoveries. One such can be the emergence of multidrug-resistant (MDR) strains in both hospital and community settings, which have been due to an inappropriate use and inadequate control of antibiotics that has led to the foremost human health concerns with a high impact on the global economy. So far, there has been application of two strategies for the development of anti-infective agents either by classical antibiotics that have been derived for their synthetic analogs with increased efficacy or screening natural compounds along with the synthetic compound libraries for the antimicrobial activities. However, need for newer treatment options for infectious diseases has led research to develop new generation of antimicrobial activity to further lessen the spread of antibiotic resistance. Currently, the principles aim to find novel mode of actions or products to target the specific sites and virulence factors in pathogens by a series of better understanding of physiology and molecular aspects of the microbial resistance, mechanism of infection process, and gene-pathogenicity relationship. The design various novel strategies tends to provide us a path for the development of various antimicrobial therapies that intends to have a broader and wider antimicrobial spectrum that helps to combat MDR strains worldwide. The development of antimicrobial peptides, metabolites derived from plants, microbes, phage-based antimicrobial agents, use of metal nanoparticles, and role of CRISPR have led to an exceptional strategies in designing and developing the next-generation antimicrobials. These novel strategies might help to combat the seriousness of the infection rates and control the health crisis system.
1. Introduction
Nature has been a rich source for novel finding of potential therapeutic agents that span millions of microbes. Natural products that have been developed are a potential source of antimicrobials and other important metabolites. Microorganisms have been variedly used for the large-scale production of such compounds and have been successful in treatment alternatives that include diseases such as cancer, infectious diseases, diabetes, diarrhoea, atopic dermatitis, and Crohn's disease. They have been a potential source of natural products that include antioxidants, antimicrobials, vitamins, enzymes, immunosuppressants, and enzymes inhibitors. The dawn of antibiotic era has also given a reality check on the emerging antibiotic resistance strains that are dangerous with a disturbing regulatory resistance pattern. The existence of such pathogens came into recognition only in the last 20 years. The escalating MDR pathogens have entitled the researchers for necessitating the postantibiotic era to help in reducing the mortality rate worldwide. Lack of research in developing the newer antibiotics is one of the key factors that have led to the crisis in medical field and denoted elsewhere [1, 2]. The Centre for Disease and Dynamics, Economics and Policy has well documented on the global status of antibiotics policy for better understanding the resistance patterns and need for newer antimicrobials [3]. From 1940 to 1962, there was a rapid development of antibiotics and it is clinically evident from the documentation that about 20 classes had been specified. But in the later years, only two classes have been identified and have been commercialised. However, recently larger pharmaceutical companies have committed towards the drugs treating lifestyle diseases and also yielding high substantial profits [4]. These industry-based partnerships with the smaller companies, academicians, and scientists have helped further in the significant development of antibiotics and its delivery systems.
1.1. Antibiotics: A Wonder Drug for Treating Infections and Its Resistance
Modern medications and technology have been a boon for helping to achieve various aspects for the treatment options related to infectious diseases. One such important key accomplishment has been the discovery of antibiotics that was a boon for human health. However, their misuse has made a very immoral impact on these pathogens in developing resistance factor for various antimicrobials. Thus, it has further complicated and restricted their use in the long-term and its efficacy too. With the discovery of penicillin, other antimicrobials have been elucidated based on the target sites as well as drug molecule modifications. The cell deaths caused by these antimicrobials on bacterial specific target sites involve the cellular structure, biochemical, and molecular levels. The antibiotics induce the death in the pathogens, which is caused due to the breaks in double-stranded DNA followed by the treatment with DNA gyrase inhibitors [5]; following treatment with rifamycin stops the DNA-dependent RNA synthesis [6]; damage caused by cell wall inhibitors on the cell envelop along with the loss of structural integrity [7]; and protein mistranslation, cellular energetics, and ribosomal binding by the protein synthesis inhibitors [8]. Currently, research has proved the mechanism of cell death is due to the drug-induced stress caused by all classes of antimicrobials [9]. Specific treatment options with lethal concentration of antibiotics produce hydroxyl radicals that are harmful through oxidative damage following cellular death pathway by altering the tricarboxylic acid cycle and iron metabolism [10, 11].
Microorganisms over a period have evolved and adjusted according to the environmental changes with subsequent development with various protective mechanisms for their survival strategies in order to reduce their antibiotic susceptibility. Major of these antimicrobials have been losing their efficacy and potency over a period of time with incautious use. Thus, making them useless for the infectious treatment that was once successfully used to treat them. However, with the introduction of new antibiotic class, it has also led to the expansion of resistance levels and it has been utmost evident with their increasing clinical implication shortly after being bought in the market [12]. The cause of MDR in microbes is due to the mutation factor and by the horizontal gene transfer (HGT) between and within the species [13]. It has been observed that there is a rapid and broadly increased dissemination of antimicrobial resistance genes through HGT, especially in Enterobacteriaceae family such as encoding β-lactamases that includes extended spectrum β-lactamases and the metallo-β-lactamases (ESBLs and MBLs, respectively) [14, 15], and it is a serious matter of concern that has reached pandemic proportions in healthcare settings worldwide [16, 17]. These β-lactamases acquired pathogens have evolved as MDR strains, thus limiting to help in treating the infectious diseases, and in worst cases, the bacterial resistance to carbapenems has left us with treatment options using older and more toxic drugs like colistin [18].
Nevertheless, hospitals acquired infections are already accompanied with high antibiotic resistance; however, recent years have also witnessed the gradual increase in the community-associated infections too [19]. Furthermore, globalisation has led the bacterial genes to travel faster than before and spreading the MDR strains worldwide. Henceforth, it becomes primary importance to find newer antibiotics to control these MDR strains. To combat issues related to MDR pathogens recently, antimicrobial science and technology have gained advancement and helped to include strategies in antimicrobial discovery, different types of novel antibiotics, and using newer materials as antimicrobials to target the pathogens to be researched for the purpose.
1.2. Bacterial Target Sites That can Help for Newer Antibiotics
Staphylococcus aureus and Bacillus subtilis encode for the cardiolipin by mutated gene, suggesting that it can be an antibiotic target. A novel antibiotic, teixobactin, prevents the biosynthesis of cell wall by binding the lipid pyrophosphate-sugar motif of lipid II precursor in the peptidoglycan and lipid III in the cell wall of teichoic acid in Gram-positive bacteria (Figure 1). LptD, a specific protein inhibitor, inserts to the lipopolysaccharide in the outer membrane called POL70780 in Pseudomonas aeruginosa, which can act as target site [20]. Biosynthetic pathway in Yersinia pestis and P. aeruginosa has been elucidated, and by using “Staphylopine,” a tetrapeptide isolated from S. aureus binds to cobalt, zinc, nickel, and iron can be used to treat with broad-spectrum antibiotics [21]. Peptidoglycan biosynthesis requires lysine and L,L-diaminopimelate aminotransferase (DapL) pathway that leads to the synthesis of lysine, which identified recently in pathogens such as Chlamydia, Leptospira, and Treponema [22]. The distribution of enzyme among these pathogens also has potential DapL inhibitors that can act as a narrow-spectrum antibiotic [23]. Helicobacter pylori and Mycobacterium tuberculosis are regarded as high MDR pathogens that produce enzymes like shikimate kinase and type II dehyroquinase, which have significant roles in shikimate pathway, and inhibitors for these enzymes are being researched and are under development [24]. Many of the proteins such as TrpB, TrpC, and TrpE for tryptophan biosynthetic pathway in M. tuberculosis are studied for their inhibitory molecules that are facilitated by X-ray diffraction structures in Tuberculosis Structural Genomics Consortium (TBSGC) [25, 26]. M. tuberculosis has shown promising results in Lassomycin, a peptide produced by Lentsea spp, and actinomycetes that inhibit an essential protease enzyme, thus killing both the growing and dormant cells [27]. A study also reveals the mechanism of antibiotic action in food items such as raffinose in ginger that effectively helps to inhibit P. aeruginosa biofilm, wherein the intracellular levels of cyclic-di-GMP are decreased to induce motile planktonic cells [28].
Figure 1.
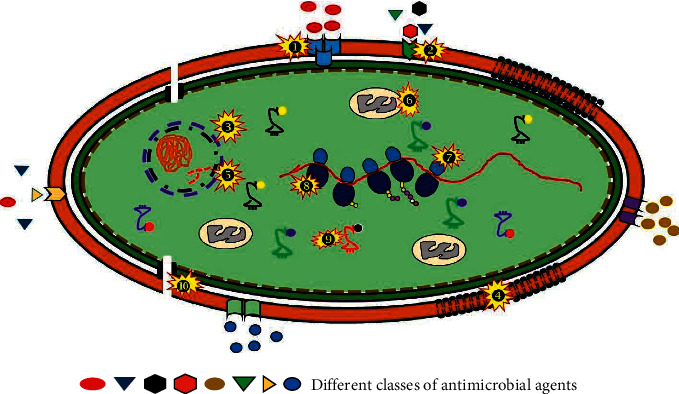
Diagrammatic representation of different target sites for the antimicrobial agent's activity on a bacterial cell. ❶ Alteration in PBP: modified PBP is responsible for the mechanism of action of resistance seen in Gram-positive bacteria as it produces β-lactamases. For example, reduction in affinity of β-lactam antibiotics is due to the mutation on penicillin-binding protein. ❷ Efflux pumps: antibiotics are exported via the membrane proteins inside the cell and maintain their low-intracellular concentrations by pumping them out before they reach their target. Several unrelated antibiotics such as macrolides, tetracyclines, and fluoroquinolones (FQ) are been pumped by cytoplasmic membrane-associated multidrug transporters contributing in the development of MDR strains. ❸ Inhibitors of DNA replication: inhibition of the bacterial division by preventing enzyme DNA gyrase activity (nicks double-stranded DNA and negative supercoils and reseals) is facilitated by quinolones like FQ. ❹ Altered cell wall precursors: Gram-positive bacteria can be inhibited by glycopeptides like vancomycin or teicoplanin as they are involved suppression cell wall synthesis. ❺ DNA mutated-DNA gyrase and topoisomerase IV: quinolone mechanism of resistance involves modification of two enzymes: DNA gyrase and topoisomerase IV. Replication failure is due to mutations in genes gyrA and parC making FQ unable to bind. ❻ Folic acid metabolism inhibitors: combination of few drugs like sulphonamides and trimethoprim acts on biosynthetic pathway, which shows synergy and a reduced mutation rate for resistance. ❼ Alteration in the 30S subunit or 50S subunit: resistance to drugs that affect protein synthesis, that is, macrolides, tetracycline, and chloramphenicol, bind to 30S ribosomal subunit, whereas chloramphenicol, macrolides, lincosamides, and streptogramin B bind to 50S ribosomal subunit to suppress protein synthesis. ❽ Inhibitors of protein synthesis: tetracyclines offer resistance by inhibiting integrity of ribosomal structure and RNA polymerase mutations conferring resistance to rifampicin. ❾ Antibiotic inactivation by enzymes: β-lactamases, aminoglycoside-modifying enzymes, and chloramphenicol acetyltransferases are the three main enzymes that inactivate antibiotics. ❿ Porins: present in the outer membrane. The changed selectivity or decreased in porins stops the antimicrobials to act on the pathogens conferring resistance.
2. Sources of Antimicrobial Agents
2.1. Antimicrobial from Plant Origin
Essential oils derived from the plants are efficient, which depends on their chemical structure, particularly with the hydrophilic functional groups that they possess such as the hydroxyl groups [29]. The compounds of these naturally occurring essential oils (EOs) are the volatile secondary metabolites that are derived from different parts of the plants [30, 31]. The components of essential oils are had been variously studied and have different characteristics like the polypeptides, phenols, tannins, polyphenols, quinones, terpenoids, flavones, lectins, and alkaloids. These metabolites exhibit varied potential activities such as anti-inflammatory, antibacterial, antiviral, antiparasitic, anticancer, antiseptic, antiallergic, and antioxidant. The components of EOs of terpenes such as menthol, geraniol, carvacrol, and thymol have proved with higher antimicrobial activities (Table 1). Relatively, with respect to the individual metabolites, the multiple compounds or the finished EOs are highly efficient and have greater antimicrobial activities [32]. A phenolic compound, Eugenol, is volatile in nature and mainly extracted component from cloves with 70–90% and is accountable for the characteristic clove aroma. It plays a significant role in the oral hygiene and has been used in dental and oral-related preparations, employed as a flavouring agent, sensitiser, irritant, and also as anaesthetic forms. Dental materials adsorbed with eugenol have been employed in clinical dentistry, and it is highly effective against Gram-negative bacteria such as E. coli, Clostridium botulinum, Shigella, Salmonella, and Listeria monocytogenes [33].
Table 1.
List of various plant compounds used for their antimicrobial properties [37].
| Scientific name | Common name | Compound | Antimicrobial activity |
|---|---|---|---|
| Berberis vulgaris | Barberry | Berberine | Bacteria and protozoa |
| Piper nigrum | Black pepper | Piperine | Fungi, Lactobacillus, Micrococcus, E. coli, and E. faecalis |
| Rhamnus purshiana | Cascara sagrada | Tannins | Bacteria, fungi, and viruses |
| Matricaria chamomilla | Chamomile | Anthemic acid | M. tuberculosis, S. typhimurium, and S. aureus |
| Syzygium aromaticum | Clove | Eugenol | Bacteria, fungi, and Trypanosoma cruzi |
| Vaccinium spp. | Cranberry | Fructose | Bacteria |
| Eucalyptus globulus | Eucalyptus | Tannin | Bacteria and viruses |
| Allium sativum | Garlic | Allicin and ajoene | General |
| Hydrastis canadensis | Goldenseal | Berberine and hydrastine | Bacteria, Giardia duodenale, and Trypanosomes |
| Camellia sinensis | Green tea | Catechin | General |
| Glycyrrhiza glabra | Licorice | Glabrol | S. aureus and M. tuberculosis |
| Quercus rubra | Oak | Tannins Quercetin | |
| Allium cepa | Onion | Allicin | Bacteria and Candida |
| Mahonia aquifolia | Oregon grape | Berberine | Plasmodium and Trypanosomes |
| Thymus vulgaris | Thyme | Caffeic acid, Thymol, and Tannins | Viruses, bacteria, and fungi |
| Curcuma longa | Turmeric | Curcumin | Bacteria and protozoa |
One of the foremost important EOs is thymol that is derived from the thymes in the form of monoterpene phenol. It exhibits various activities that include antioxidant, anti-inflammatory, antibacterial, antifungal, antiviral, and anti-immunomodulatory effects. It is highly effective, acts by damaging the membrane integrity, change in pH, homeostasis, and equilibrium of inorganic ions against Salmonella and Staphylococcus spp. [29]. Another phenolic compound derived from vanilla pods has tremendous industrial applications in food, beverages, perfumes, pharmaceuticals, and also used as nutraceuticals [34]. It has inhibitory effects on the food-spoiling microbes, several fungi, and various pathogens such as Escherichia, Klebsiella, Salmonella, Bacillus, Serratia, Staphylococcus, and Listeria (Table 1) [35]. A phenolic compound caffeic acid (3, 4-dihydroxycinnamic acid) is derived from the hydroxycinnamic acid, which has some of the exciting biological properties that include fungicide, antibacterial, and antioxidant. Its antibacterial activity has been effective against different pathogens such as S. epidermidis, S aureus, and K. pneumoniae.
The phenolic metabolites, coumarins, are fabricated from benzene and alpha pyrone ring that are fused together. Even though they have a greater activity against fungi, it also proved well that they can control bacteria [36, 37]. Tannin is a polymeric phenolic substance that may be used to tanning leather and to precipitate gelatin from a solution with astringency. Ellagitannins are also a potent compound that can be used in the medical field. Among several tannins, ellagitannins are present in different parts of the plants and have been an excellent source for inhibiting antimicrobial activities against E coli, S aureus, Salmonella typhimurium, B. subtilis, Shigella sonnei, MDR E. coli, K. pneumonia, and C. albicans. These metabolites bind to the cell wall of the bacteria and thus inhibit their growth [38] (Figure 1).
Nonalcoholic beverages are widely used and consumed by humans with an estimate of over two-third of the population as it acts as mild stimulant, refreshing, and medicinal properties. Tea (Camellia sinensis) is one among the foremost popular nonalcoholic beverages loaded with polyphenols and epigallocatechin-3-gallate. Tea is chargeable for abundant qualities that have health benefits including anti-inflammatory, antimicrobial, antioxidative, antitumor, anti-obesity, defence from disorders, and anti-aging activities [39]. The extract of bioactive components obtained from green tea, EGCG, and flavins shows antibacterial activity over Fusobacterium nucleatum, pathogenic subgingival known to cause destructive periodontitis. This organism is a typical found to be residing in the human gastrointestinal tract and has been related to inflammatory bowel disease too. Consumption of green tea has hassled protection against the damage caused by these bacteria through leakage of membrane and chelating ions preventing biofilm formation [40].
2.1.1. Fruits and Vegetables
Similar to the skin, it acts as the physical barrier to protect us from the invasion of many pathogens in fruits like the peel outer covering. Several investigation evidences show that the fruit peel is mostly disposed as waste that is found to be sustainable resource containing antimicrobial activity [30, 41]. In a study done by Jagtap and Bapat [42], team has shown pomegranate extract reduces the expansion of E. coli due to its great antimicrobial activity as it contains phenols and flavonoids. This study demonstrated similar effect even against other microbes such as S. aureus and B. cereus at a degree of 0.01%. In parallel, several other fruits peel such as lemon grass, citrus, and lime peel extract also exhibited antimicrobial and antibacterial activities over products like meat by inhibiting a wide variety of microbe like S. aureus, E. coli, P. aeruginosa, B. cereus, and S. typhimurium [43]. The antimicrobial activity of peel and capsicum was reported by researchers and was mainly found to be due to the phenolic compounds present in it like the coumaric acid, highly active against S. typhimurium and P. aeruginosa on minced beef [44]. An organic sulphur compound allicin extracted from garlic has antimicrobial activity on both Gram-positive and Gram-negative bacteria such as E. coli, Salmonella, Streptococcus, P. aeruginosa, B. cereus, Klebsiella, Helicobacter pylori, Proteus, and S. typhimurium by inhibiting their growth. It is used as preservative and also has health benefits. Aqueous extracts of garlic have been beneficial for their antimicrobial properties such as S. aureus in hamburger [45]. The antimicrobial effect of onion extract shows high significance with antibacterial activity against E. coli, yeast, and moulds with increased concentrations of the extracts. Antimicrobial efficacy of curcumin, the active metabolite derived from the turmeric plant, has been evaluated against various food pathogens such as S. typhimurium, Listeria monocytogenes, S. aureus, and E. coli showed exceedingly good results in minced meat [46].
2.2. Antimicrobials from Animal Origin
Chitin on partial deacetylation gives chitosan that is a natural polycationic linear polysaccharide obtained basically from the shells of marine crustaceans. Since it has less allergic, nontoxic, and biodegradable, it has been used in wider applications that include antifungal, antimicrobial, antibacterial, and antioxidant activity [47, 48]. It is active against Gram-negative bacteria such as Bacteroides fragilis, Shigella dysenteriae, E. coli, and Vibrio cholerae. Defensin, a small cationic peptide, has been known for their antibacterial and antimycotic activities [49]. Lysozyme, a polypeptide chain consisting of 129 amino acids, is naturally found in tears, saliva, milk, and breast milk with good antimicrobial activity against bacteria causing death by cleaving glycosidic linkages in the cell wall peptidoglycan. It is a crucial defence mechanism with being taken into account the most important element of the innate immune system in mammals [50]. Avidin, a charged glycoprotein, binds to biotin both present in egg that makes unavailable for the microbial growth, and spoilage has been an important diagnostic tool immunoassay using avidin-biotin system, thus controlling growth of several microbes such as Klebsiella pneumoniae, E. coli, P aeruginosa, and Serratia marcescens. An antimicrobial peptide, pleurocidin, is incredibly active against Gram-negative and Gram-positive bacteria. Potentially used in food industries due to their varied applications, heat stability, salt tolerant, and other characteristics favour against pathogens like E. coli, Vibrio parahaemolyticus, and L. monocytogenes [51].
2.3. Antimicrobials from Microbial Origin
Natamycin produced by Streptomyces natalensis is used as food preservative and has been effective against yeast and moulds. However, it does not have effective activity against many of the bacterial pathogens due to its antifungal nature. Nowadays, natamycin has been extensively used due to their effectivity in many food processing industries that include various food products such as meat, juices, and dairy products that are either pasteurised or unpasteurised. Reuterin, a water-soluble antimicrobial derivative obtained from Lactobacillus reuteri, is a nonproteinaceous substance that has been found to have a very broad antimicrobial activity. Importantly, it has been effective against both Gram-positive and Gram-negative bacteria, filamentous moulds, and nonfilamentous fungal organisms. It has shown a very high effectivity to a wide selection and range of pH and immune to various enzymes that include proteolytic and lipolytic. Most importantly, it exhibits bacteriostatic activity particularly against Listeria [52] (Table 2) [53–74].
Table 2.
Representation of various biomolecules derived from bacteria that have different biological activities/applications
| Type | Characteristics | Examples | Producer | Mode of action | References |
|---|---|---|---|---|---|
| Bacteriocin type I | Lantibiotics | Nisin Z and Q, Enterocin W | Lactococcus lactis | Pore formation in the membrane by permeabilisation | [53] |
| Bacteriocin type II | Non Lantibiotics | Enterocin, Leucocin, Munditicin, Lactococcin Q, and Lactocyclicin Q |
Pediococcus pentosaceus,
P. Acidilactici, L. sakei, L. lactis sub sp. Cremoris, L. plantarum, L. gasseri, Enterococcus faecalis, and L. garvieae |
Pore formation in the membrane by permeabilisation | [54–57] |
| Bacteriocin type III | Large peptides | Lysostaphin, Enterolysin A, Helveticin J |
L. crispatus,
L. helveticus, E. faecalis, S. simulans biovar, Staphylolyticus, Enterococcus faecalis, and Lactobacillus helveticus |
Disintegration of the cell-wall imbalance in membrane potential initiating ATP efflux |
[58–60] |
| LPS | Peptides | Surfactin, Iturin, Fengycin |
B. subtilis
B. amyloliquefaciens |
Antifungal, Antimicrobial, Insecticidal, Antimycoplasma, Haemolysis, and Establishment of ion channels in lipid membranes. Acts as bioagents. Immunomodulatory, antitumor enzyme inhibitors, toxins, etc. |
[61–63] |
| Other bioactive compounds | Vinaceuline | Streptomyces spp | Antibacterial activity | [64] | |
| Halocin | Haloferax mediterranei | Alter membrane permeability | [65] | ||
| Sulfolobicins | Sulfolobus islandicus HEN2/2 | Exact mode of action is not known | [66] | ||
| Lactones | Phomopsis spp | Active against A. Niger Botrytis cinerea Fusarium |
[67] | ||
| Jesterone | Pestalotiopsis jesteri | Active against Pythium ultimum, Phytophthora citrophthora, Rhizoctonia solani Sclerotinia sclerotiorum |
[68] | ||
| Diastaphenazines | Streptomyces diastaticus Sub spp. ardesiacus | Antibacterial and antifungal activity | [69] | ||
| Reuterin | Lactobacillus reuteri | Antimicrobial | [70, 71] | ||
| Mollemycin A 20 | Streptomyces spp (CMB-M0244) | Effective against Gram-positive and Gram-negative bacteria, compound shows antimalarial properties | [72] | ||
| Avermectins | Streptomyces avermitilis | Onchocerciasis and lymphatic filariasis | [73] | ||
| Cahuitamycins | Streptomyces gandocaensis | Functional by inhibiting biofilm formation in Gram-negative Acinetobacter baumannii | [74] | ||
2.3.1. Prebiotic and Probiotics Mechanism
One of the most important probiotic microbes, lactic acid bacteria (LAB), helps to confer many health benefits and also acts as preservative. These LABs inhibit the expansion of pathogens by the various antimicrobial agents produced by them that includes the organic acids and bacteriocins that are antimicrobial peptides (Table 2). Different strains of bacteria like L. sakei and L. curvatus are effective against many pathogens like Escherichia O157:H7 and L monocytogenes [75]. Bacteriocins are the antimicrobial compounds that are produced majorly by Gram-positive bacteria, like the LABs, and these metabolites are formed during their growth. These metabolites are polypeptides, wherein the manufacturing microbes gain an advantage to compete with the other microorganisms for their survival [76].
It is well established the advantages of probiotics associated with human health. Mechanisms are possibly being multifactorial, one postulated being antagonistic mechanism on pathogens competing for the nutrients for growth factor, and provide and enhance functions of the gut barrier, competitive adherence to the mucosa and epithelium, defence by the production of antimicrobial substances, and immune system modulation [77, 78]. These probiotic microbes consume most of the monosaccharides in the GI tract making it unavailable for the pathogens such as Clostridium difficile. Lysins have also been improved by bioengineering; for instance, a novel chimeric lysin was constructed by combining the active site present on the lysin protein with a cell wall binding domain [78]. Modulation and enhanced gene expression involved by a number of Lactobacillus spp like the tight junction signalling of E-cadherin and β-catenin has helped to reinforce the integrity of intestinal barrier. Several proteins by these Lactobacillus spp promote the mucosal adhesion by displaying surface adhesins and integrate with complex glycoprotein like mucin, secreted by the intestinal epithelial cells to prevent pathogens [77].
Another mechanism includes modification of microbial flora by the low and high molecular weight compound synthesis like organic acid (acetic and lactic acids) and antimicrobial compounds like bacteriocins [77]. They exhibit strong inhibitory effect by lowering the intracellular pH or accumulating the ionised forms of organic acids that help in disrupting the pH balance and consequently inhibiting growth of pathogens like Helicobacter pylori that causes gastrointestinal disorders and cancers. Bacteriocins produced by probiotics, lactacin B from L. acidophilus, bifidocin B produced by Bifidobacterium bifidum NCFB 1454, plantaricin from L. plantarum, and nisin from Lactococcus lactis [79] have antibacterial activity against the food-borne pathogens by destructing the target cells by pore formation or inhibiting cell wall synthesis [80].
2.4. Miscellaneous Antimicrobial Agents
Antimicrobials are derived from different sources present either in nature or synthesised through artificial and chemical means. As such, today the antimicrobial drugs are of many diverse types based on their source. Microbe-based antibiotics are the most common types, with penicillin from the various Penicillium spp being the first known microbe derived antibiotic to be used by humans. Others include vancomycin derived from Actinobacteria sps, Amycolatopsis orientalis, and Streptomycin from S. griseus. Plant-based antimicrobials are also another class of antimicrobial agents. These are the beneficial nutrients that we attain from plants that help boost our immunity. Apples are a very good example as they are rich in flavonoids, which have strong antioxidant properties, which are harmful to a large number of microbes [81]. Another well-known is aloe vera, whose latex compound is disruptive towards microbes such as Corynebacterium, Salmonella, Streptococcus, and S. aureus [82]. Certain phytochemicals derived from plants have also shown mechanisms to stop microbial proliferation like catechols can cause substrate deprivation in microbes [83], capsaicin initiates membrane disruption [84], and fabatin can form disulphide bridges [85]. Phytochemicals used as antibiotics will majorly contain phenolics and polyphenols that are highly effective against bacteria, fungi, and even viruses. Additionally, they pose significant important group, quinones, which forms complex irreversibly with proteins made of amino acids that are nucleophilic in nature results in their inactivation and loss in the function of proteins [86].
3. Role of CRISPR as an Antimicrobial Agent
Infectious disease continues to pose an enormous burden globally, with the advancement in developing new tools and therapeutics to study the mechanism of host-microbe interactions, and it has not been a tedious job to diagnose and treat infections accurately. Clustered regularly interspaced short palindromic repeats (CRISPR) present in bacteria as well as the members of archaea that act as defence mechanism in protecting them from the invading viruses and bacteriophages. It functions as an adaptive immune system for prokaryotes. It was first discovered in E. coli as a homologous repeat of 29 nucleotides separated by 32 nucleotides spacers [87]. It is used for genome editing and evolutionary analysis of prokaryotic strains and has been identified as a major breakthrough mechanism in the field of science and clinical research. It has been known to regulate stress-related phenomenon in bacteria. One of the global threats contributes to a high mortality rate worldwide in spreading infectious diseases. In order to contain the spread of such pandemics, it is highly crucial to get accurate knowledge of the pathogenesis of bacteria, fungi, viruses, and parasites and, further, know the diagnosis and treatments of such diseases. CRISPR and CRISPR-associated proteins have come into the light for their varied applications in the field of infectious disease, evaluation of host-pathogen interaction, development of diagnostic tests, and prevention of such diseases [88]. CRISPR-Cas9 mechanism has been expansively used across various disciplines, like basic sciences, food and crop development, fuel generation, drug development, and human genome engineering [89]. Rapidly emerging multidrug-resistant strains, it has been tough game to combat diseases. The approach of CRISPR-Cas is recognised as a novel means to control multidrug-resistant bacteria. The rise in antibiotic-resistant strains far exceeds than the development of new antibiotics, which has caused a massive rise in the emergence of diseases. CRISPR-Cas system has also been successful in removing resistant microbiome from the gastrointestinal tract, thus reconstructing a healthy and balanced human microbiome. The CRISPR mechanism is quite comparable to that of RNA interference in eukaryotes, wherein small RNA identifies and neutralises specific DNA sequences, later specifically cleaves DNA encoding the antibiotic resistant genes. This includes the following:
Conjugation-based delivery
Phage-based delivery
Nanoparticle (NP)-based delivery
Antibody-based therapy
The aim of using CRISPR-Cas-mediated methods is to ensure novel ways of developing antibiotics and to combat the MDR pathogens as they use of CRISPR-Cas9 and plasmids to kill bacteria carrying antimicrobial resistance genes (AMR). Alongside, CRISPR-Cas mediated by phages intends to carry plasmids carrying AMR genes leading to the bacterial cell death [90].
Until date on record, there have been two classes of CRISPR-Cas system, further comprising six subtypes.
Class 1 CRISPR—Types I, III, and IV—Multiple Cas proteins
Class 2 CRISPR—Types II, V, and VI—Single Cas protein
CRISPR technology is mainly applied to upregulate or downregulate gene transcription, which is controlled by the activated Cas9 protein through the gene expression. At the molecular level, the genomic locus of CRISPR acts as memory unit wherein the sequences of nucleic acids of the invading pathogens are isolated or hidden away. These particular sequences later help to guide Cas proteins to eliminate the foreign invaders (Figure 2) [91]. Hence, this technology aids to understand regulation of infection causing genes in bacteria, fungi, and viruses. Same knowledge is implicated to combat them by designing accurate therapies and vaccines. The mechanism of CRISPR technology targets nucleic acids upon entry of plasmid DNA or attachment followed by injection of viral genetic material in the host with a progressive step accommodating the Cas gene (Figure 2). When a bacteriophage or plasmid invades a bacterium, a small portion of the DNA that has been invading is cleaved with the help of Cas protein and inserted into CRISPR locus and thus creates memory of sequences [92, 93]. CRISPR ribonucleic acid (crRNA) containing both a single CRISPR and spacer repeat is released into the cytoplasm that manages to bind to the complementary DNA or RNA sequences [94]. Thereafter, a cleavage complex is formed that includes crRNA, its target sequence, Cas nuclease, and a tracer RNA, which cleaves the target at a site adjacent to the crRNA target sequence. As an outcome, this CRISPR-Cas technique has been well contributing in rapid and precise diagnosis of infectious disease. It has allowed enhanced clinical care and manages infection control leading to limited spread of the infection.
Figure 2.
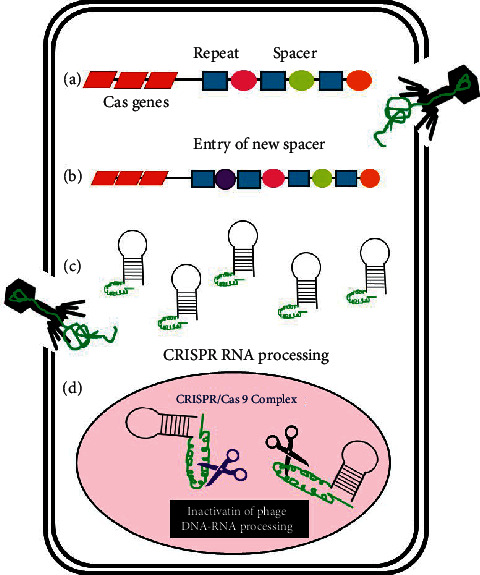
Illustration of the mechanism of action of CRISPR/Cas in a bacterial cell. (a) Entry of genetic material of virus, phage, or plasmid into bacteria. (b) Short segments with genetic information of the invader inserts into CRISPR region. (c) Expression of crRNA in the bacterium. (d) Cleavage of the invading DNA by the formation of a cleavage complex consisting of foreign DNA, crRNA, and Cas9 protein.
In 2016, Winter and team used CRISPR-Cas single-guide ribonucleic acid (sgRNA) to nullify the effect of haemolysins, a toxin produces by S. aureus responsible lysis of red blood cells. Hence, sgRNA can be an efficient tool to elucidate α-haemolysin function in S. aureus virulence factor that induces cytotoxicity [95]. In addition, around seven genes including EMC2, EMC3, SEL1L, DERL2, UBE2G2, UBE2J1, and HRD1 have been identified and reported to inactivate viral particle responsible for West Nile virus known to cause neuronal cell death [96]. These genes have been an important part of the protein degradation pathway and are part of endoplasmic reticulum that has led to the development of novel drug.
Likewise, CRISPR technology has also been employed to mute toxoplasmosis and Chagas disease initiating protozoal parasites like Toxoplasma gondii and Trypanosoma cruzi, respectively [97]. This kind of paralysing the protozoal parasite is due to blockade of paraflagellar rod proteins (PFR), which are required for the motility and the attachment of the parasite flagella to the cell host [98]. Pardee et al. [99] performed an experiment that combined nucleic acid sequence-based amplification (NASBA), a process involving isothermal reactions for amplifying RNA along with CRISPR that help to distinguish closely related Zika virus strains. Upon the cleavage of dsDNA with sgRNA-Cas9 complex, either curtailed or full-length DNA fragments are produced. This activated to trigger sequence to NASBA-amplified viral RNA, which caused colour change on the paper disc that enables the strain differentiation.
Centre for disease and control and prevention had reported about 2 million infections were present worldwide and close to 23,000 deaths in the USA every year due to antibiotic resistant bacteria. Keeping this in challenge CRISPR-based technology aims to prepare therapeutics to target not only drug resistant bacteria but also highly infectious viruses such as hepatitis B (HBV) and human immunodeficiency virus (HIV) [100]. One of the applications of CRISPR is to produce selective antimicrobials for annihilating pathogenic bacteria. One example can be selectively killing specific strains of bacteria like Staphylococcus and Escherichia among pure and mixed cultures by CRISPR-Cas system [101]. Similar practice was also followed by Bikard et al. [102] to selectively kill virulent S. aureus strains with the destruction of plasmids in these bacteria carrying mecA methicillin-resistant gene. By contrary, host bacteria with RNA-guided Cas9 protein remained unaffected.
Nowadays, DNA endonuclease to targeted CRISPR Trans (DETECTR) technique is more frequently being used. It involves the combination of isothermal recombinase polymerase amplification with Cas12a enzymatic activity. This can be used to differentiate between two high-risk strains of human papilloma virus 16 (HPV16) and HPV18. Both are known to cause various types of cancers such as anal, throat, cervical, vaginal, and vulvar mainly in female. A study includes the collection of 25 human anal swab samples followed by testing with DETECTR and PCR analysis. Results showed that all the specimens were correctly identified within an hour of DNA extraction, while the samples that were tested by PCR displayed only twenty-three correctly identified samples signifying the importance of this technique [103].
4. Phage-Based Antimicrobial Agents
The rise of MDR strains in recent years is an ongoing struggle in the medical field, due to the ineffectiveness of antibiotics against them. However, to combat this one effective development was the use of bacteriophages as the active antimicrobial agents. Phage therapy has been around for almost a century and used to treat bacterial infections utilising naturally occurring phages. Recent advances have developed the use of bioengineered phages and the specific use of purified lytic proteins isolated from them. Phages exhibit the use of two major protein classes in their action of lysing bacterial host cells.
These two proteins are as follows:
Transmembrane protein holin
Peptidoglycan cell hydrolase endolysin
Both mechanisms work combined to bring about bacterial lysis leading to death. The cell membrane pores attach to the holing protein causing an opening to appear, which in turn facilitates lysin protein to gain access cell wall and further hydrolysing it [104]. Phages code for numerous lysin and holing proteins, of which some are very specific, and others are effective in a broad-spectrum. Currently, we identified lysins ABgp46. ABgp46 are highly effective to lyse various strains of Gram-negative strains and MDR strains such as A. baumanii, P. aeruginosa, and S. typhimurium [105].
Phage lysins have become a topic of interest as they are capable of even lysing bacterial cells by themselves, while holins cannot act alone. Their use in research has been a boon in treating bacteraemia in mice caused by the strains of A. baumanii [106], S. pneumoniae [107], and methicillin-resistant S aureus (MRSA) [108]. Another study showed that uses of phage lysins with antibiotic combinations are found to be more efficient in action against infections as compared to the antibiotics. The study was performed in vivo and ex vitro in a colon model, with the organism C. difficile [109]. The effectiveness of lysins by a recent study showed that they can cross the epithelial cell barrier and affect hard to treat intercellular infections by S. pyogenes [110]. Furthermore, they disrupt vegetative cells that were illustrated through the B. anthracis lysin PlyG, which exhibited the ability to eliminate endospores of Bacillus. Another advantage of phage lysins is that they can be easily produced in large scale through simple recombinant techniques, as demonstrated by the bacteriophage-derived cysteine gene and histidine-dependent amidohydrolase/peptides that are cloned and inserted in E. coli where it was then overexpressed for further purification [111]. Lysins have also been optimised through bioengineering; were synthesis of a novel chimeric lysin by using a cell wall binding domain to combine the active site present on the lysin protein. This chimeric lysin has shown the ability to treat MRSA bacteraemia in mice [112].
5. Peptides and Peptide-Related Molecules
Biomolecular recognition in signalling steps in biology leads to the importance of nucleic acids, carbohydrates, or lipids in ligand-target interactions, and the effectors of most signal transduction processes are peptides. Peptides can be of various types that can be fragments of proteins, hormones, cytokines, toxins, and antimicrobials [113]. MDR pathogens have spread rapidly causing nosocomial infections increasing mortality and causing concerns on global health issues [114–116]. With the lack of new antibiotics, the discovery of natural structures has been increased drastically for their role in antimicrobial activity. Antimicrobial peptides (AMPSs) are a part of innate immune defence in all class of life that has gained popularity and has been widely studied for their antimicrobial molecule potential [117]. These bioactive peptides have various biological functions and help in contributing towards human health. They have been variedly used for their therapeutics that include antioxidant, antimicrobial, and antithrombotic effects. Recent years has seen the varied applications of these peptides for disease management, peptide vaccines, and toxicity-related issues.
There is a dilemma for the direct administration of the biopeptides and their derivatives with respect to their in vivo stability. However, their therapeutics can be improvised by their coupling with lipids, nonpeptide antibiotics, immunoglobulins, photosensitisers, etc. [118]. Animal models for Neisseria gonorrhoeae and N. meningitidis were found to be susceptible to 2-pyridone nonapeptide, while the Gram-positive gut bacteria were not affected. DNA biosynthesis inhibition has paved way for bactericidal action and it has been suggested that peptides based on pH activity is actively used against the Helicobacter pylori and lipopeptide daptomycin that is been used for treating systemic infections that are caused by MDR Gram-positive bacteria (Figure 3) [119, 120].
Figure 3.
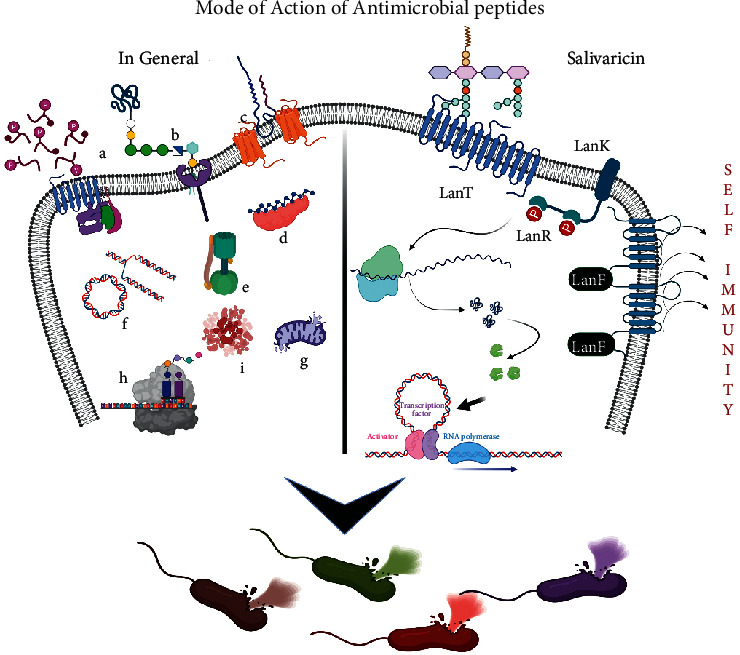
Role of antimicrobial peptides (AMPs): in general, (a), (b), and (c) Different types of AMPs act on the pathogens by penetrating the bacterial membrane through the porins and blocking various cellular and physiological processes of the cells, eventually leading to the death. (d) Inactivation of enzymes by peptide molecules. (e) Acts as ATPase inhibitor. (f) Induces cell death via interacting with intracellular DNAs and RNAs inducing DNA damage. (g) Cationic AMPs interact with organelles as in fungi such as mitochondria, eventually leading to fungal death. (h) Inhibition of the synthesis of protein and also cell wall. (i) Endocytosis.
Gram-positive bacteria produce peptides such as lantibiotics that have rare post-translational modified thioether amino acids like lanthionine and methyl-lanthionine that are effective at low nanomolar concentrations and resistance against bacterial proteases. Recently, lanthionine rings made up of nisin (rings D and E) have been identified, which has been cleaved by the nisin resistance protein of Streptococcus agalactiae, that can help us in synthesising nonhydrolysable nisin analogues [121]. Furthermore, efforts using bioengineering techniques have made possible to modify these lantibiotics producing genes so that newer variants of them can be discovered [122].
Several marine microbes have been studied, and it has been denoted that about 10 different antibiotic metabolites have been found to be synthesised by them. One such example includes species of Entotheonella-producing antibiotic mundticin [123]. Microbes isolated from the gut bacteria of cotton leaf-worm has been fascinating for their production of antimicrobial peptides (AMP), a peptide that helps protect the insect from virulent pathogens. Currently, this has been used as a potential application in microbial coatings and in various food additives [124]. Others include the microbes that are commensals of the human beings like the Staphylococcus lugdunensis and Streptomyces formicae that produce lugdunin and formicamycins, respectively, and are found to be present in the nasal tract [125, 126].
The emerging multidrug resistance pathogen is one of the biggest threats to human beings with getting treatment options for such infectious disease. S. salivarius has been varied studied for its novel bacteriocins that have been a promising antimicrobial activity towards various pathogens. Salivaricins belong to lantibiotics produced by S. salivarius that have selective antimicrobial activity against the oral and upper respiratory tract pathogens (Figure 3). In salivaricin, a lantibiotic peptide synthesises molecules of salivaricin (SalA, SalB, Sal9, SalG32, and SalE) that regulate the immune system. In regulatory system, the LanK and LanR proteins represent sensors that check incidence for the Salivaricin active molecules present in the extracellular environment that induces transcription of lantibiotics structural and immunity genes [127–129]. Synthesis of unmodified inactive linear peptide generated is modified by enzyme LanM. LanT protein transports active peptides across the cell membrane. LanFEG, the self-immunity protein, is the form of ABC transporters that helps to expel the active form of lantibiotics from the cell membrane and thus helps to prevent the pore formation and/or disruption in the cell wall synthesis. In case of nisin and other peptides, encoding through clusters for the transport system involves possibly for the self-protection like lanE, LanF, and LanG (Figure 3). These salivaricins have been in limited use as of now for developing probiotic strains of S. salivarius-producing molecules that have been studied in vivo while colonising the oral cavity. The structure and mechanism of action have been well revised as an important aspect to help enable them in designing newer antimicrobial lantibiotics in the future. Salivaricins are produced by some strains of S. salivarius entirely present in human oral cavity, a polycyclic peptide made of lanthionine, and/or β-methyllanthionine residual molecules. These molecules stem an important role of having antimicrobial affect towards the relevant oral pathogens that has been utilised for the development of probiotic strains that produce salivaricin. In the future, these molecules might help to prove a greater value for developing novel antimicrobial treatment options in the emerging multidrug resistance pathogenic era [127].
6. Nanoparticles Action as Antimicrobials
In recent years, there is a rapid growth in nanotechnology and has new possibilities opened up for controlling microbial infections by using a variety of NPs as drug delivery systems. Usage of conjugated NPs or green synthesized NPs (GSNPs) is becoming more popular from quite a decade because of the dual activity of metallic NPs added with biological origins boost the activity. Therefore, this synergistic effect may be novel-approaching MDR across pathogenic microbes. Various studies exhibit the strength of NPs as a competent antimicrobial agent, possible antifungal, and feasible antiviral with rich anti-inflammatory properties if it is derived from biological basis. Here, the overall mechanism of the NPs or GSNPs showing boundless antimicrobial activity will be due to denaturation microbial outer membrane leading to fragmentation, inference with disulphide or sulfhydryl bonds, or groups of many enzymes, and interruption with the metabolic processes leads to the death of microbial cell by the generation and liberation of reactive oxygen species (ROS) [130–132]. Intensification of the activity is seen when metal oxides are synthesis using plant because biomolecules of various plant sources exhibit a strong reducing forte as well as act as stabilisers in combination with several classes of flavonoids, proteins, tannins, phenols, and terpenoids accompanied during GSNPs. They are also capable of releasing metal ions that can penetrate into the cell and hinder their metabolic processes [133]. To fulfil this, metal NPs such as copper, silver (Ag), silica (Si), zinc, gold, and copper are extensively used in various fields and it poses superb antimicrobial agents to prevent the pathogens with broad-spectrum activity. Charged NPs help to get adhered on the cell surface helping them to penetrate directly into the cell releasing metal ions; further, ions also destabilise the membrane potential leading to cell leakage. Cell wall destabilisation helps to increase bacterial permeability allowing larger NPs to enter cell to have antibiotic like effect [134].
Hurdle lays in the microbes specially bacteria-forming biofilms, which make them to secrete strong diverse polysaccharides and cover up facilitating the rapid multiplication of them. This complex aggregates now protect the bacteria and, on the other hand, prevent to diffusion capacity for the antibiotics and hence make the disease or infection more chronic. Hence, these studies have shown that biofilms and drug-resistant bacteria can effectively killed by SiNPs produced by peppermint oil and cinnamaldehyde against E. coli, P. aeruginosa, Enterobacter cloacae, and S. aureus with a removal rate of 99.9% clearance [135]. High concentration of AgNPs is lethal to bacterial cell resulting extreme levels of oxidative stress (inactivation of enzymes and inhibiting bacterial growth) [136, 137]. To evidence this, a study concluded the effective treatment option using AgNPs, which is used for killing chlorine-resistant apicomplexan parasite Cryptosporidium parvum [138]. Furthermore, AgNPs showed effective results against HIV1 for the antiretroviral activity and aids as a potent viral inhibitory agent. They are also effective against the HBV [139]. Popularisation of GSNPs in the current situation hires usage of Ag in the production of AgNPs by the extracts of lichens to exhibit tremendous antimicrobial activity. Siddiqi et al. [140] reported lichens such as Usnea longissimi, Xanthoria elegans, Cetraria islandica, Usnea Antarctica, and Leptogium puberulum yield good amount of AgNPs, which blocks the growth of pathogens such as S. aureus, Streptococcus mutans, S. pyogenes, S. viridans, Corynebacterium diphtheriae, and C. xerosis [141]. AgNPs cannot directly inhibit BSA and PVP as they are completely encapsulated and there is no exposed surface for AgNPs-virus interactions. Hence, foamy carbon AgNPs have comparatively higher surface of display for the virus attachment, which shows higher cytotoxicity to BSA and PVP resulting in the inhibition [142].
Antimicrobial activities in bacteria and fungi with the use of graphene, graphite graphene oxide, and reduced graphene oxide have been well studied. As graphene (G), NPs gain lot of importance in several carbon-based NPs for multidisciplinary research employed in designing biomedical tools and devices implants, surgical instruments, protein bio-functionalisation, dental accessories, devices used for drug delivery, and many more [143, 144]. The background in this respect has led to the research that GNP derivatives are been found to kill bacteria by membrane disruption and oxidation stress and hence great antimicrobial agent [130–132, 145]. Bactericidal role was detected against E. coli and S. aureus when being treated with GNPs and found to be useful when used in the dental implants [146, 147]. ZnNPs were used in a study by Khan (2016) that displayed significant good blockade in the growth of bacteria like E. coli and B. subtilis along with fungus Candida albicans [148]. Battery of ZnNPs bactericidal activity is seen across several strains includes B. cereus, P. aeruginosa, S. aureus, K. pneumoniae, Salmonella spp, L. monocytogenes, and E. faecium [149] evincing its broader antimicrobial implications. Considerably, CuNPs have been extensively studied for its antimicrobial activity against E. coli, V. cholera, P. aeruginosa, S. typhus, S. aureus, E. faecalis, B. subtilis, and S. faecalis [150]. Cu NPs also have a high antimicrobial effect. They are thought to function against a broad range of bacterial species by interacting with —SH groups and causing proteolysis. These also have a strong affinity for amines and carboxyl groups found on cellular membrane. Cu NPs can disrupt genetic material by linking nucleic acid strands once they penetrate, resulting in bacterial cell death [151]. Low concentration (5 µg/mL) of these CuNPs showed its remedial power by successfully inhibiting the bacteria such as B. cereus, Proteus mirabilis, and Aeromonas caviae [152]. Alavi and team worked with the synthesis of GSNPs using a group of crustose lichen, for example, Protoparmeliopsis muralis, with oxide metals like Ag, Cu, Zno, TiO2, and Fe3O4 checked for different activities that include the antiviral, antimicrobial, antibiofilm, antiquorum sensing, and antioxidant agents against the MDR strains of S. aureus [153]. This study concluded lichen extract generated NPs showed good result by inhibiting the growth of S. aureus. Alongside research has shown that the nanocomposites of Fe3O4 with SiO2 and ZnO/TiO2/SiO2 have effective antifungal properties against Candida spp., like C. albicans, Aspergillus spp., such as A. flavus, A. Niger, and A. terreus [154]. In biological system, magnesium oxide (MgO) assists an excellent natural reducing agent, several studies have used for the GSNPs using plant extracts like aloe vera (A. barbadensis Mill), Pigeon wings plant (Clitoria ternatea), and Indian gooseberry (Emblica Officinalis), and the combination of metal oxide and plant extract boosts the antioxidant property and helps to destroy the harmful pathogens. In a recent investigation, GSMgONPs made by using aqueous juice extract of Indian gooseberry (Phyllanthus emblica) commonly called as amla displayed exceptional antibacterial activity by obstructing the growth of human pathogens, namely, P. aeruginosa, S. aureus, K. pneumoniae, and E. coli [130]. Therefore, these metal NPs can be extensively helpful in biomedical sciences and engineering with their vast applications that include antibacterial, antifungal, and antiviral to help combat various infectious diseases. Fe3O4 in its bulk form is considered inert and lack antimicrobial activity. Surprisingly, these materials can be modified to introduce antimicrobial properties when synthesised in nanosize. Interestingly, microbiological assays have been proved that surface modified using Fe3O4 nanoparticles demonstrates anti-adherent properties and significantly reduces both Gram-positive and Gram-negative bacterial colonisations [155].
7. Nanoparticles for Immunostimulation
NPs as nanomedicines are also a promising approach in immunomodulation, which interact and modify the immune system's activity resulting in immunosuppression or immunostimulation. These modifying effects may have bidirectional action; positive or negative consequences depend on the factors like as composition, size, and surface chemistry, among other NPs [156]. Metals that do not mainly trigger antigen-specific responses or create metal-specific lymphocytes are used to stimulate immune system cells. Many different forms of NPs have been found to create ROS in vitro and in vivo, enhancing immune function and inflammatory responses [157, 158]. Some nanotechnology-based anticancer therapeutics has antitumor qualities in vitro but promotes tumour growth in vivo. Inflammation is a critical immune system response that may be triggered by NPs, as indicated by the production of cytokines and chemokine. The key downstream processes of inflammation have been identified as oxidative stress produced by NPs. In comparison with regular particles, NPs have a vast surface area and significant oxidative capabilities [159]. The immunological imbalance caused by oxidative damage caused by NPs is a major contributor. Pb-based NPs has also been demonstrated to disrupt macrophage-T cell interactions, lowering macrophage capabilities including phagocytosis, cell adhesion, and nitric oxide generation while enhancing antioxidant activity. These reactions might be attributable to Pb's potential to influence the expression of genes linked to innate immunity [160].
8. Antibodies as Treatment Options
With the approval of monoclonal antibodies from past three decades, it has been approved by the United States Food and Drug Administration in 1986. The application of antibodies being engineered has dramatically changed the medical treatment options. Currently, with their high specificity and less adverse effects they make a major class of new drugs for treatment. The market for antibodies as therapeutics has increased drastically over the past five years as the bestselling drug for various treatments that include human diseases like cancers, metabolic disorders, and infectious diseases [161]. Kohler and Milstein produced monoclonal antibodies (mAbs) that are efficient molecules used for therapy for targeting reagents [162, 163]. Antibody like palivizumab has been previously administrated for the prevention of serious lower respiratory tract syncytial virus disease in children documented by the European Medicines Agency, 2013 [164]. The second mAb bezlotoxumab authorised in 2017 to be directed for the use against the infectious disease throughout the European economic area that targets Clostridium difficile toxin B for the prevention of recurring infection as it risked in adults due to the repeated [164, 165].
9. Conclusion
Globally, antibiotic resistance has drastically increased further increasing the health issues and also the economy. Considering very little advancement towards the discovery of novel antibiotics effective against these MDR strains, use of alternative methods could help to resolve the problem. In this grim scenario, every pharmaceutical and scientific community of the world think for alternative treatment modalities, which can improvise their products and cut down the cost. Alternatives can be derived from the natural sources like bioactive compound or secondary metabolites known from many generations coming from plant extracts, and microbial and metal conjugated with bioactive compounds have antimicrobial characteristics. This approach is quite promising and delivers us new class or old class with active variants. The abilities of microbes and plant derivatives can successfully deliver the novel bioactive compounds, which will become useful therapeutic tools. Also, the application of newer molecular tools and techniques will help to contribute for the next generation of antimicrobial compounds and to combat infectious diseases. Comprehensive applications of phytochemical bioactive molecules and microbial-conjugated compounds have marked significant demand and have challenged innovator to think and develop powerful novel bioactive derivatives as an alternative to conventional treatment regime. Accordingly, new biotechnological strategies involving the phage-based therapy, metal oxide-based NPs, GSNPs, or the role of CRISPR have provided access to detect surface active target sites in pathogens and act on them. The need for treating infections would be further enhanced with findings in the structural analysis and molecular docking helping to design the specific antimicrobials with lesser toxicity, high selectivity, and easily biodegradable. Hence, this should be engrossed as the demand for screening of indigenous molecules from different sources, untrapped traditional formulation, and practices across the world reveals the source of them. Reconsidering the facts of these secondary metabolites of biological origin, one should be ready to exploit microbial and plant diversity, with various other techniques to discover novel bioactive molecules or strategies to treat these pathogens that cause life-threatening diseases and to be used as therapeutic tool.
Contributor Information
Shilpa Borehalli Mayegowda, Email: shilpa.siddi@gmail.com.
Fahadul Islam, Email: fahadul29-774@diu.edu.bd.
Data Availability
All data used to support the findings of this study are included within the article.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Walsh C. Antibiotics: Actions, Origins, Resistance . Washington, DC, USA: ASM Press; 2003. [Google Scholar]
- 2.Thornsberry C., Sahm D. F., Kelly L. J., et al. Regional trends in antimicrobial resistance among clinical isolates of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis in the United States: results from the TRUST surveillance program, 1999-2000. Clinical Infectious Diseases . 2002;34(1):S4–S16. doi: 10.1086/324525. [DOI] [PubMed] [Google Scholar]
- 3.Gelband H., Miller P. M., Pant S., et al. The state of the world’s antibiotics 2015. Wound healing southern africa . 2015;8(2):30–34. [Google Scholar]
- 4.Coates A., Hu Y., Bax R., Page C. The future challenges facing the development of new antimicrobial drugs. Nature Reviews Drug Discovery . 2002;1(11):895–910. doi: 10.1038/nrd940. [DOI] [PubMed] [Google Scholar]
- 5.Drlica K., Malik M., Kerns R. J., Zhao X. Quinolone-mediated bacterial death. Antimicrobial Agents and Chemotherapy . 2008;52(2):385–392. doi: 10.1128/AAC.01617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Floss H. G., Yu T.-W. Rifamycin mode of action, resistance, and biosynthesis. Chemistry Review . 2005;105(2):621–632. doi: 10.1021/cr030112j. [DOI] [PubMed] [Google Scholar]
- 7.Tomasz A. The mechanism of the irreversible antimicrobial effects of penicillins: how the beta-lactam antibiotics kill and lyse bacteria. Annual Review of Microbiology . 1979;33(1):113–137. doi: 10.1146/annurev.mi.33.100179.000553. [DOI] [PubMed] [Google Scholar]
- 8.Vakulenko S. B., Mobashery S. Versatility of aminoglycosides and prospects for their future. Clinical Microbiology Reviews . 2003;16(3):430–450. doi: 10.1128/CMR.16.3.430-450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohanski M. A., Dwyer D. J., Hayete B., Lawrence C. A., Collins J. J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell . 2007;130(5):797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 10.Dwyer D. J., Kohanski M. A., Hayete B., Collins J. J. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Molecular Systems Biology . 2007;3(1):p. 91. doi: 10.1038/msb4100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohanski M. A., Dwyer D. J., Wierzbowski J., Cottarel G., Collins J. J. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell . 2008;135(4):679–690. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palumbi S. R. Humans as the world’s greatest evolutionary force. Science . 2001;293(5536):1786–1790. doi: 10.1126/science.293.5536.1786. [DOI] [PubMed] [Google Scholar]
- 13.Thomas C. M., Nielsen K. M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nature Reviews Microbiology . 2005;3(9):711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 14.Chakraborty A., Chanda D. D., Choudhury N., Manjula N. G., Shilpa B. M. A retrospective study on the pyogenic pathogens and their antibiotic susceptibility patterns along with the esßl production. AiM . 2021;11(6):317–326. doi: 10.4236/aim.2021.116024. [DOI] [Google Scholar]
- 15.Nagshetty K., Shilpa B. M., Patil S. A., Shivannavar C. T., Manjula N. G. An overview of extended spectrum beta lactamases and metallo beta lactamases. AiM . 2021;11(1):37–62. doi: 10.4236/aim.2021.111004. [DOI] [Google Scholar]
- 16.Giske C. G., Monnet D. L., Cars O., Carmeli Y. Clinical and economic impact of common multidrug-resistant gram-negative bacilli. Antimicrobial Agents and Chemotherapy . 2008;52(3):813–821. doi: 10.1128/AAC.01169-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hawkey P. M., Jones A. M. The changing epidemiology of resistance. Journal of Antimicrobial Chemotherapy . 2009;64(1):i3–i10. doi: 10.1093/jac/dkp256. [DOI] [PubMed] [Google Scholar]
- 18.Giamarellou H., Poulakou G. Multidrug-resistant gram-negative infections: what are the treatment options? Drugs . 2009;69(14):1879–1901. doi: 10.2165/11315690-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 19.Woodford N., Livermore D. M. Infections caused by gram-positive bacteria: a review of the global challenge. Journal of Infection . 2009;59:S4–S16. doi: 10.1016/S0163-4453(09)60003-7. [DOI] [PubMed] [Google Scholar]
- 20.Srinivas N., Jetter P., Ueberbacher B. J., et al. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science . 2010;327(5968):1010–1013. doi: 10.1126/science.1182749. [DOI] [PubMed] [Google Scholar]
- 21.Ghssein G., Brutesco C., Ouerdane L., et al. Biosynthesis of a broad-spectrum nicotianamine-like metallophore in Staphylococcus aureus. Science . 2016;352(6289):1105–1109. doi: 10.1126/science.aaf1018. [DOI] [PubMed] [Google Scholar]
- 22.Hudson A. O., Gilvarg C., Leustek T. Biochemical and phylogenetic characterization of a novel diaminopimelate biosynthesis pathway in prokaryotes identifies a diverged form of ll-diaminopimelate aminotransferase. Journal of Bacteriology . 2008;190(9):3256–3263. doi: 10.1128/JB.01381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Triassi A. J., Wheatley M. S., Savka M. A., Gan H. M., Dobson R. C. J., Hudson A. O. L,L-diaminopimelate aminotransferase (DapL): a putative target for the development of narrow-spectrum antibacterial compounds. Frontiers in Microbiology . 2014;5 doi: 10.3389/fmicb.2014.00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez-Bello C. Inhibition of shikimate kinase and type II dehydroquinase for antibiotic discovery: structure-based design and simulation studies. CTM . 2015;16(9):960–977. doi: 10.2174/1568026615666150825142527. [DOI] [PubMed] [Google Scholar]
- 25.Lee C. E., Goodfellow C., Javid-Majd F., Baker E. N., Shaun Lott J. The crystal structure of TrpD, a metabolic enzyme essential for lung colonization by Mycobacterium tuberculosis, in complex with its substrate phosphoribosylpyrophosphate. Journal of Molecular Biology . 2006;355(4):784–797. doi: 10.1016/j.jmb.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 26.Czekster C. M., Neto B. A. D., Lapis A. A. M., Dupont J., Santos D. S., Basso L. A. Steady-state kinetics of indole-3-glycerol phosphate synthase from Mycobacterium tuberculosis. Archives of Biochemistry and Biophysics . 2009;486(1):19–26. doi: 10.1016/j.abb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Gavrish E., Sit C. S., Cao S., et al. Lassomycin, a ribosomally synthesized cyclic peptide, kills Mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. Chemistry & Biology . 2014;21(4):509–518. doi: 10.1016/j.chembiol.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim H.-S., Cha E., Kim Y., et al. Raffinose, a plant galactoside, inhibits Pseudomonas aeruginosa biofilm formation via binding to LecA and decreasing cellular cyclic diguanylate levels. Scientific Reports . 2016;6(1) doi: 10.1038/srep25318.25318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lambert G., Fattal E., Couvreur P. Nanoparticulate systems for the delivery of antisense oligonucleotides. Advanced Drug Delivery Reviews . 2001;47(1):99–112. doi: 10.1016/s0169-409x(00)00116-2. [DOI] [PubMed] [Google Scholar]
- 30.Jadimurthy R., Mayegowda S. B., Nayak S. C., Mohan C. D., Rangappa K. S. Escaping mechanisms of ESKAPE pathogens from antibiotics and their targeting by natural compounds. Biotechnology Reports . 2022;34 doi: 10.1016/j.btre.2022.e00728.e00728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prabuseenivasan S., Jayakumar M., Ignacimuthu S. In vitro antibacterial activity of some plant essential oils. BMC Complementary and Alternative Medicine . 2006;6(1):p. 39. doi: 10.1186/1472-6882-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bassolé I. H. N., Lamien-Meda A., Bayala B., et al. Composition and antimicrobial activities of Lippia multiflora Moldenke, Mentha x piperita L. and Ocimum basilicum L. essential oils and their major monoterpene alcohols alone and in combination. Molecules . 2010;15(11):7825–7839. doi: 10.3390/molecules15117825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ceylan E., Fung D. Y. C., Sabah J. R. Antimicrobial activity and synergistic effect of cinnamon with sodium benzoate or potassium sorbate in controlling Escherichia coli O157:H7 in apple juice. Journal of Food Science . 2004;69(4):FMS102–FMS106. doi: 10.1111/j.1365-2621.2004.tb06348.x. [DOI] [Google Scholar]
- 34.Thomas C., Mayegowda Shilpa B., Babu Mythri R. Disease modifying potential of functional foods for neurodegenerative disorders: status update on regulatory compliance. In: Sajid Arshad M., Haseeb Ahmad M., editors. Functional Foods-Phytochemicals and Health Promoting Potential . London, UK: IntechOpen; 2021. [Google Scholar]
- 35.Fitzgerald R. J., Murray B. A. Bioactive peptides and lactic fermentations. International Journal of Dairy Technology . 2006;59(2):118–125. doi: 10.1111/j.1471-0307.2006.00250.x. [DOI] [Google Scholar]
- 36.Tataringa G., Maria Zbancioc A. Coumarin derivatives with antimicrobial and antioxidant activities. In: Rao V., Mans D., Rao L., editors. Phytochemicals in Human Health . London, UK: IntechOpen; 2020. [Google Scholar]
- 37.Khameneh B., Iranshahy M., Soheili V., Fazly Bazzaz B. S. Review on plant antimicrobials: a mechanistic viewpoint. Antimicrobial Resistance and Infection Control . 2019;8(1):p. 118. doi: 10.1186/s13756-019-0559-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saeed F., Afzaal M., Tufail T., Ahmad A. Use of natural antimicrobial agents: a safe preservation approach. In: Var I., Uzunlu S., editors. Active Antimicrobial Food Packaging . London, UK: IntechOpen; 2019. [DOI] [Google Scholar]
- 39.Bansal S., Choudhary S., Sharma M., et al. Tea: a native source of antimicrobial agents. Food Research International . 2013;53(2):568–584. doi: 10.1016/j.foodres.2013.01.032. [DOI] [Google Scholar]
- 40.Ben Lagha A., Haas B., Grenier D. Tea polyphenols inhibit the growth and virulence properties of Fusobacterium nucleatum. Scientific Reports . 2017;7(1):p. 44815. doi: 10.1038/srep44815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar H., Bhardwaj K., Cruz-Martins N., et al. Applications of fruit polyphenols and their functionalized nanoparticles against foodborne bacteria: a mini review. Molecules . 2021;26(11):p. 3447. doi: 10.3390/molecules26113447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jagtap U. B., Bapat V. A. Wines from fruits other than grapes: current status and future prospectus. Food Bioscience . 2015;9:80–96. doi: 10.1016/j.fbio.2014.12.002. [DOI] [Google Scholar]
- 43.Okunowo W. O., Oyedeji O., Afolabi L. O., Matanmi E. Essential oil of grape fruit (Citrus paradisi) peels and its antimicrobial activities. Asian Journal of Plant Science . 2013;4(7):1–9. doi: 10.4236/ajps.2013.47A2001. [DOI] [Google Scholar]
- 44.Careaga M., Fernández E., Dorantes L., Mota L., Jaramillo M. E., Hernandez-Sanchez H. Antibacterial activity of Capsicum extract against Salmonella typhimurium and Pseudomonas aeruginosa inoculated in raw beef meat. International Journal of Food Microbiology . 2003;83(3):331–335. doi: 10.1016/s0168-1605(02)00382-3. [DOI] [PubMed] [Google Scholar]
- 45.El Astal Z. The inhibitory action of aqueous garlic extract on the growth of certain pathogenic bacteria. European Food Research and Technology . 2004;218(5):460–464. doi: 10.1007/s00217-003-0864-3. [DOI] [Google Scholar]
- 46.Sandikci Altunatmaz S., Yilmaz Aksu F., Issa G., Basaran Kahraman B., Dulger Altiner D., Buyukunal S. Antimicrobial effects of curcumin against L. monocytogenes, S. aureus, S. Typhimurium and E. coli O157:H7 pathogens in minced meat. Veterinarni Medicina . 2016;61(5):256–262. doi: 10.17221/8880-VETMED. [DOI] [Google Scholar]
- 47.Chandy T., Sharma C. P. Chitosan—as a biomaterial. Biomaterials, Artificial Cells and Artificial Organs . 1990;18:1–24. doi: 10.3109/10731199009117286. [DOI] [PubMed] [Google Scholar]
- 48.Ngo D.-H., Kim S.-K. Antioxidant effects of chitin, chitosan, and their derivatives. Marine Carbohydrates: Fundamentals and Applications, Part B . 2014;73:15–31. doi: 10.1016/B978-0-12-800268-1.00002-0. [DOI] [PubMed] [Google Scholar]
- 49.Agerberth B., Charo J., Werr J., et al. The human antimicrobial and chemotactic peptides LL-37 and alpha-defensins are expressed by specific lymphocyte and monocyte populations. Blood . 2000;96(9):3086–3093. doi: 10.1182/blood.v96.9.3086. [DOI] [PubMed] [Google Scholar]
- 50.Varahan S., Iyer V. S., Moore W. T., Hancock L. E. Eep confers lysozyme resistance to enterococcus faecalis via the activation of the extracytoplasmic function sigma factor SigV. Journal of Bacteriology . 2013;195(14):3125–3134. doi: 10.1128/JB.00291-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Juneja V. K., Dwivedi H. P., Yan X. Novel natural food antimicrobials. Annual Review of Food Science and Technology . 2012;3(1):381–403. doi: 10.1146/annurev-food-022811-101241. [DOI] [PubMed] [Google Scholar]
- 52.El-Ziney M. G., van den Tempel T., Debevere J., Jakobsen M. Application of reuterin produced by Lactobacillus reuteri 12002 for meat decontamination and preservation. Journal of Food Protection . 1999;62(3):257–261. doi: 10.4315/0362-028x-62.3.257. [DOI] [PubMed] [Google Scholar]
- 53.Garsa A. K., Choudhury P. K., Puniya A. K., Dhewa T., Malik R. K., Tomar S. K. Bovicins: the bacteriocins of streptococci and their potential in methane mitigation. Probiotics and Antimicrobial Proteins . 2019;11(4):1403–1413. doi: 10.1007/s12602-018-9502-z. [DOI] [PubMed] [Google Scholar]
- 54.Gradisteanu Pircalabioru G., Popa L. I., Marutescu L., et al. Bacteriocins in the era of antibiotic resistance: rising to the challenge. Pharmaceutics . 2021;13(2):p. 196. doi: 10.3390/pharmaceutics13020196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meade E., Slattery M. A., Garvey M. B. Potent antimicrobial peptides and the fight against multi drug resistant species: resistance is futile? Antibiotics . 2020;9(1):p. E32. doi: 10.3390/antibiotics9010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Najjari A., Mejri H., Jabbari M., Sghaier H., Cherif A., Ouzari H.-I. Halocins, bacteriocin-like antimicrobials produced by the archaeal domain: occurrence and phylogenetic diversity in halobacteriales. In: Najjari A., Cherif A., Sghaier H., Imene Ouzari H., editors. Extremophilic Microbes and Metabolites-Diversity, Bioprospecting and Biotechnological Applications . London, UK: IntechOpen; 2021. [Google Scholar]
- 57.Ibrahim O. O. Classification of antimicrobial peptides bacteriocins, and the nature of some bacteriocins with potential applications in food safety and bio-pharmaceuticals. EC Microbiology . 2019;15:591–608. [Google Scholar]
- 58.Garcia-Gutierrez E., Mayer M. J., Cotter P. D., Narbad A. Gut microbiota as a source of novel antimicrobials. Gut Microbes . 2019;10:1–21. doi: 10.1080/19490976.2018.1455790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Newstead L. L., Varjonen K., Nuttall T., Paterson G. K. Staphylococcal-produced bacteriocins and antimicrobial peptides: their potential as alternative treatments for Staphylococcus aureus infections. Antibiotics . 2020;9(2):p. E40. doi: 10.3390/antibiotics9020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Negash A. W., Tsehai B. A. Current applications of bacteriocin. International Journal of Microbiology . 2020;2020:7. doi: 10.1155/2020/4374891.4374891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Isa M. H., Shannaq M. A., Mohamed N., Hassan A. R., Al-Shorgani N. K., Hamid A. A. Antibacterial activity of surfactin produced by Bacillus subtilis MSH1. Transactions on Science and Technology . 2017;4(3):402–407. [Google Scholar]
- 62.Calvo H., Mendiara I., Arias E., Blanco D., Venturini M. E. The role of iturin A from B. Amyloliquefaciens BUZ-14 in the inhibition of the most common postharvest fruit rots. Food Microbiology . 2019;82:62–69. doi: 10.1016/j.fm.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 63.Sur S., Romo T. D., Grossfield A. Selectivity and mechanism of fengycin, an antimicrobial Lipopeptide, from molecular dynamics. Journal of Physical Chemistry B . 2018;122(8):2219–2226. doi: 10.1021/acs.jpcb.7b11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang X., Yang Y., Peng T., et al. A new cyclopeptide from endophytic Streptomyces sp. YIM 64018. Natural Product Communications . 2013;8(12):1753–1754. doi: 10.1177/1934578x1300801225. [DOI] [PubMed] [Google Scholar]
- 65.Besse A., Peduzzi J., Rebuffat S., Carré-Mlouka A. Antimicrobial peptides and proteins in the face of extremes: lessons from archaeocins. Biochimie . 2015;118:344–355. doi: 10.1016/j.biochi.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 66.Quehenberger J., Shen L., Albers S.-V., Siebers B., Spadiut O. Sulfolobus–a potential key organism in future biotechnology. Frontiers in Microbiology . 2017;8:p. 2474. doi: 10.3389/fmicb.2017.02474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu S.-H., Chen Y.-W., Shao S.-C., et al. Ten-membered lactones from phomopsis sp., an endophytic fungus of azadirachta indica. Journal of Natural Products . 2008;71(4):731–734. doi: 10.1021/np070624j. [DOI] [PubMed] [Google Scholar]
- 68.Toghueo R. M. K., Sahal D., Boyom F. F. Recent advances in inducing endophytic fungal specialized metabolites using small molecule elicitors including epigenetic modifiers. Phytochemistry . 2020;174 doi: 10.1016/j.phytochem.2020.112338.112338 [DOI] [PubMed] [Google Scholar]
- 69.Li Y., Han L., Rong H., et al. Diastaphenazine, a new dimeric phenazine from an endophytic Streptomyces diastaticus subsp. ardesiacus. Journal of Antibiotics . 2015;68(3):210–212. doi: 10.1038/ja.2014.124. [DOI] [PubMed] [Google Scholar]
- 70.Talarico T. L., Casas I. A., Chung T. C., Dobrogosz W. J. Production and isolation of reuterin, a growth inhibitor produced by Lactobacillus reuteri. Antimicrobial Agents and Chemotherapy . 1988;32(12):1854–1858. doi: 10.1128/AAC.32.12.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gyawali R., Ibrahim S. A. Natural products as antimicrobial agents. Food Control . 2014;46:412–429. doi: 10.1016/j.foodcont.2014.05.047. [DOI] [Google Scholar]
- 72.Blunt J. W., Copp B. R., Keyzers R. A., Munro M. H. G., Prinsep M. R. Marine natural products. Natural Product Reports . 2016;33(3):382–431. doi: 10.1039/c5np00156k. [DOI] [PubMed] [Google Scholar]
- 73.Shen B. A new golden age of natural products drug discovery. Cell . 2015;163(6):1297–1300. doi: 10.1016/j.cell.2015.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Park S. R., Tripathi A., Wu J., et al. Discovery of cahuitamycins as biofilm inhibitors derived from a convergent biosynthetic pathway. Nature Communications . 2016;7(1):p. 10710. doi: 10.1038/ncomms10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rydlo T., Miltz J., Mor A. Eukaryotic antimicrobial peptides: promises and premises in food safety. Journal of Food Science . 2006;71(9):R125–R135. doi: 10.1111/j.1750-3841.2006.00175.x. [DOI] [Google Scholar]
- 76.Ricke S. Perspectives on the use of organic acids and short chain fatty acids as antimicrobials. Poultry Science . 2003;82(4):632–639. doi: 10.1093/ps/82.4.632. [DOI] [PubMed] [Google Scholar]
- 77.Bermudez-Brito M., Plaza-Díaz J., Muñoz-Quezada S., Gómez-Llorente C., Gil A. Probiotic mechanisms of action. Annals of Nutrition & Metabolism . 2012;61(2):160–174. doi: 10.1159/000342079. [DOI] [PubMed] [Google Scholar]
- 78.Khalighi A., Behdani R., Kouhestani S. Probiotics: a comprehensive review of their classification, mode of action and role in human nutrition. In: Rao V., Rao L. G., editors. Probiotics and Prebiotics in Human Nutrition and Health . London, UK: InTech; 2016. [Google Scholar]
- 79.Nielsen D. S., Cho G.-S., Hanak A., Huch M., Franz C. M. A. P., Arneborg N. The effect of bacteriocin-producing Lactobacillus plantarum strains on the intracellular PH of sessile and planktonic Listeria monocytogenes single cells. International Journal of Food Microbiology . 2010;141:S53–S59. doi: 10.1016/j.ijfoodmicro.2010.03.040. [DOI] [PubMed] [Google Scholar]
- 80.Lievin V. Bifidobacterium strains from resident infant human gastrointestinal microflora exert antimicrobial activity. Gut . 2000;47(5):646–652. doi: 10.1136/gut.47.5.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hunter M. D., Hull L. A. Variation in concentrations of phloridzin and phloretin in apple foliage. Phytochemistry . 1993;34(5):1251–1254. doi: 10.1016/0031-9422(91)80010-X. [DOI] [Google Scholar]
- 82.Hernandez-Hernandez A. B., Alarcon-Aguilar F. J., Almanza-Perez J. C., et al. Antimicrobial and anti-inflammatory activities, wound-healing effectiveness and chemical characterization of the latex of Jatropha neopauciflora pax. Journal of Ethnopharmacology . 2017;204:1–7. doi: 10.1016/j.jep.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 83.Ahmad I., Beg A. Z. Antimicrobial and phytochemical studies on 45 Indian medicinal plants against multi-drug resistant human pathogens. Journal of Ethnopharmacology . 2001;74(2):113–123. doi: 10.1016/s0378-8741(00)00335-4. [DOI] [PubMed] [Google Scholar]
- 84.Cichewicz R. H., Thorpe P. A. The antimicrobial properties of Chile peppers (Capsicum species) and their uses in Mayan medicine. Journal of Ethnopharmacology . 1996;52(2):61–70. doi: 10.1016/0378-8741(96)01384-0. [DOI] [PubMed] [Google Scholar]
- 85.Gupta V. K. Bioactive Phytochemicals: Perspectives for Modern Medicine . New Delhi, India: Daya Publishing House; 2012. [Google Scholar]
- 86.Patra A. K. An overview of antimicrobial properties of different classes of phytochemicals. In: Patra A. K., editor. Dietary Phytochemicals and Microbes . Dordrecht, Netherlands: Springer; 2012. pp. 1–32. [DOI] [Google Scholar]
- 87.Ishino Y., Shinagawa H., Makino K., Amemura M., Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. Journal of Bacteriology . 1987;169(12):5429–5433. doi: 10.1128/JB.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mali P., Yang L., Esvelt K. M., et al. RNA-guided human genome engineering via Cas9. Science . 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hsu P. D., Lander E. S., Zhang F. Development and applications of CRISPR-cas9 for genome engineering. Cell . 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pursey E., Sünderhauf D., Gaze W. H., Westra E. R., van Houte S. CRISPR-cas antimicrobials: challenges and future prospects. PLoS Pathogens . 2018;14(6) doi: 10.1371/journal.ppat.1006990.e1006990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shalem O., Sanjana N. E., Zhang F. High-throughput functional genomics using CRISPR–cas9. Nature Reviews Genetics . 2015;16(5):299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Barrangou R., Fremaux C., Deveau H., et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science . 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 93.Barrangou R., Marraffini L. A. CRISPR-cas systems: prokaryotes upgrade to adaptive immunity. Molecular Cell . 2014;54(2):234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Westra E. R., Buckling A., Fineran P. C. CRISPR–Cas systems: beyond adaptive immunity. Nature Reviews Microbiology . 2014;12(5):317–326. doi: 10.1038/nrmicro3241. [DOI] [PubMed] [Google Scholar]
- 95.Virreira Winter S., Zychlinsky A., Bardoel B. W. Genome-wide CRISPR screen reveals novel host factors required for Staphylococcus aureus α-hemolysin-mediated toxicity. Scientific Reports . 2016;6(1):p. 24242. doi: 10.1038/srep24242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ma H., Dang Y., Wu Y., et al. A CRISPR-based screen identifies genes essential for West-Nile-Virus-Induced cell death. Cell Reports . 2015;12(4):673–683. doi: 10.1016/j.celrep.2015.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sidik S. M., Hackett C. G., Tran F., Westwood N. J., Lourido S. Efficient genome engineering of Toxoplasma gondii using CRISPR/Cas9. PLoS One . 2014;9(6) doi: 10.1371/journal.pone.0100450.e100450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lander N., Chiurillo M., Vercesi A., Docampo R. Endogenous C-terminal tagging by CRISPR/Cas9 in trypanosoma cruzi. Bio-Protocol . 2017;7(10) doi: 10.21769/BioProtoc.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pardee K., Green A. A., Takahashi M. K., et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell . 2016;165(5):1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 100.CDC. CDC Works 24/7 [WWW Document]. Centers for Disease Control and Prevention. 2022. https://www.cdc.gov/index.htm .
- 101.Gomaa A. A., Klumpe H. E., Luo M. L., Selle K., Barrangou R., Beisel C. L. Programmable removal of bacterial strains by use of genome-targeting CRISPR-cas systems. mBio . 2014;5(1) doi: 10.1128/mBio.00928-13.e00928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bikard D., Euler C. W., Jiang W., et al. Exploiting CRISPR-cas nucleases to produce sequence-specific antimicrobials. Nature Biotechnology . 2014;32(11):1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen J. S., Ma E., Harrington L. B., et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science . 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Roach D. R., Donovan D. M. Antimicrobial bacteriophage-derived proteins and therapeutic applications. Bacteriophage . 2015;5(3) doi: 10.1080/21597081.2015.1062590.e1062590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Oliveira H., Vilas Boas D., Mesnage S., et al. Structural and enzymatic characterization of ABgp46, a novel phage endolysin with broad anti-gram-negative bacterial activity. Frontiers in Microbiology . 2016;7 doi: 10.3389/fmicb.2016.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lood R., Winer B. Y., Pelzek A. J., et al. Novel phage lysin capable of killing the multidrug-resistant gram-negative bacterium acinetobacter baumannii in a mouse bacteremia model. Antimicrobial Agents and Chemotherapy . 2015;59(4):1983–1991. doi: 10.1128/AAC.04641-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Witzenrath M., Schmeck B., Doehn J. M., et al. Systemic use of the endolysin cpl-1 rescues mice with fatal pneumococcal pneumonia. Critical Care Medicine . 2009;37(2):642–649. doi: 10.1097/CCM.0b013e31819586a6. [DOI] [PubMed] [Google Scholar]
- 108.Schmelcher M., Shen Y., Nelson D. C., et al. Evolutionarily distinct bacteriophage endolysins featuring conserved peptidoglycan cleavage sites protect mice from MRSA infection. Journal of Antimicrobial Chemotherapy . 2015;70(5):1453–1465. doi: 10.1093/jac/dku552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rineh A., Kelso M. J., Vatansever F., Tegos G. P., Hamblin M. R. Clostridium difficile infection: molecular pathogenesis and novel therapeutics. Expert Review of Anti-infective Therapy . 2014;12(1):131–150. doi: 10.1586/14787210.2014.866515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shen Y., Cai J., Davies M. R., et al. Identification and characterization of fluoroquinolone non-susceptible Streptococcus pyogenes clones harboring tetracycline and macrolide resistance in shanghai, China. Frontiers in Microbiology . 2018;9:p. 542. doi: 10.3389/fmicb.2018.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Keary R., Sanz-Gaitero M., van Raaij M. J., et al. Characterization of a bacteriophage-derived murein peptidase for elimination of antibiotic-resistant Staphylococcus aureus. Current Protein & Peptide Science . 2016;17(2):183–190. doi: 10.2174/1389203716666151102105515. [DOI] [PubMed] [Google Scholar]
- 112.Yang H., Zhang H., Wang J., Yu J., Wei H. A novel chimeric lysin with robust antibacterial activity against planktonic and biofilm methicillin-resistant Staphylococcus aureus. Scientific Reports . 2017;7(1):p. 40182. doi: 10.1038/srep40182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Otvos L., Wade J. D. Current challenges in peptide-based drug discovery. Frontiers of Chemistry . 2014;2 doi: 10.3389/fchem.2014.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Willyard C. The drug-resistant bacteria that pose the greatest health threats. Nature . 2017;543(7643):p. 15. doi: 10.1038/nature.2017.21550. [DOI] [PubMed] [Google Scholar]
- 115.Alghoribi M. F., Gibreel T. M., Farnham G., Al Johani S. M., Balkhy H. H., Upton M. Antibiotic-resistant ST38, ST131 and ST405 strains are the leading uropathogenic Escherichia coli clones in riyadh, Saudi arabia. Journal of Antimicrobial Chemotherapy . 2015;70(10):2757–2762. doi: 10.1093/jac/dkv188. [DOI] [PubMed] [Google Scholar]
- 116.Petty N. K., Ben Zakour N. L., Stanton-Cook M., et al. Global dissemination of a multidrug resistant Escherichia coli clone. Proceedings of the National Academy of Sciences of the United States of America . 2014;111(15):5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Torres M. D. T., Sothiselvam S., Lu T. K., de la Fuente-Nunez C. Peptide design principles for antimicrobial applications. Journal of Molecular Biology . 2019;431(18):3547–3567. doi: 10.1016/j.jmb.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 118.Reinhardt A., Neundorf I. Design and application of antimicrobial peptide conjugates. Indian Journal of Management Science . 2016;17(5):p. 701. doi: 10.3390/ijms17050701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tally F. P., DeBruin M. F. D. Development of daptomycin for Gram-positive infections. Journal of Antimicrobial Chemotherapy . 2000;46(4):523–526. doi: 10.1093/jac/46.4.523. [DOI] [PubMed] [Google Scholar]
- 120.Heinis C., Rutherford T., Freund S., Winter G. Phage-encoded combinatorial chemical libraries based on bicyclic peptides. Nature Chemical Biology . 2009;5(7):502–507. doi: 10.1038/nchembio.184. [DOI] [PubMed] [Google Scholar]
- 121.Khosa S., Frieg B., Mulnaes D., et al. Structural basis of lantibiotic recognition by the nisin resistance protein from Streptococcus agalactiae. Scientific Reports . 2016;6(1):p. 18679. doi: 10.1038/srep18679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bierbaum G., Szekat C., Josten M., et al. Engineering of a novel thioether bridge and role of modified residues in the lantibiotic Pep5. Applied and Environmental Microbiology . 1996;62(2):385–392. doi: 10.1128/AEM.62.2.385-392.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wilson M. C., Mori T., Rückert C., et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature . 2014;506(7486):58–62. doi: 10.1038/nature12959. [DOI] [PubMed] [Google Scholar]
- 124.Shao Y., Chen B., Sun C., Ishida K., Hertweck C., Boland W. Symbiont-derived antimicrobials contribute to the control of the Lepidopteran gut microbiota. Cell Chemical Biology . 2017;24(1):66–75. doi: 10.1016/j.chembiol.2016.11.015. [DOI] [PubMed] [Google Scholar]
- 125.Zipperer A., Konnerth M. C., Laux C., et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature . 2016;535(7613):511–516. doi: 10.1038/nature18634. [DOI] [PubMed] [Google Scholar]
- 126.Qin Z., Munnoch J. T., Devine R., et al. Formicamycins, antibacterial polyketides produced by Streptomyces formicae isolated from african tetraponera plant-ants. Chemical Science . 2017;8(4):3218–3227. doi: 10.1039/C6SC04265A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Barbour A., Wescombe P., Smith L. Evolution of lantibiotic salivaricins: new weapons to fight infectious diseases. Trends in Microbiology . 2020;28(7):578–593. doi: 10.1016/j.tim.2020.03.001. [DOI] [PubMed] [Google Scholar]
- 128.Clemens R., Zaschke-Kriesche J., Khosa S., Smits S. H. J. Insight into two ABC transporter families involved in lantibiotic resistance. Frontiers in Molecular Biosciences . 2018;4:p. 91. doi: 10.3389/fmolb.2017.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Smits S. H. J., Schmitt L., Beis K. Self‐immunity to antibacterial peptides by ABC transporters. FEBS Letters . 2020;594(23):3920–3942. doi: 10.1002/1873-3468.13953. [DOI] [PubMed] [Google Scholar]
- 130.Ananda A., Ramakrishnappa T., Archana S., et al. Green synthesis of MgO nanoparticles using Phyllanthus emblica for evans blue degradation and antibacterial activity. Materials Today Proceedings . 2022;49:801–810. doi: 10.1016/j.matpr.2021.05.340. [DOI] [Google Scholar]
- 131.Archana S., Jayanna B. K., Ananda A., Pandiarajan D., Muralidhara H. B., Kumar K. Y. Synthesis of nickel oxide grafted graphene oxide nanocomposites—a systematic research on chemisorption of heavy metal ions and its antibacterial activity. Environmental Nanotechnology, Monitoring & Management . 2021;16 doi: 10.1016/j.enmm.2021.100486.100486 [DOI] [Google Scholar]
- 132.Yadav L. S. R., Shilpa B. M., Suma B. P., Venkatesh R., Nagaraju G. Synergistic effect of photocatalytic, antibacterial and electrochemical activities on biosynthesized zirconium oxide nanoparticles. European Physical Journal-Plus . 2021;136(7):p. 764. doi: 10.1140/epjp/s13360-021-01606-6. [DOI] [Google Scholar]
- 133.Stensberg M. C., Wei Q., McLamore E. S., Porterfield D. M., Wei A., Sepúlveda M. S. Toxicological studies on silver nanoparticles: challenges and opportunities in assessment, monitoring and imaging. Nanomedicine . 2011;6(5):879–898. doi: 10.2217/nnm.11.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Losasso C., Belluco S., Cibin V., et al. Antibacterial activity of silver nanoparticles: sensitivity of different Salmonella serovars. Frontiers in Microbiology . 2014;5:p. 227. doi: 10.3389/fmicb.2014.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Duncan B., Li X., Landis R. F., et al. Nanoparticle-stabilized capsules for the treatment of bacterial biofilms. ACS Nano . 2015;9(8):7775–7782. doi: 10.1021/acsnano.5b01696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Qing Y., Cheng L., Li R., et al. Potential antibacterial mechanism of silver nanoparticles and the optimization of orthopedic implants by advanced modification technologies. International Journal of Nanomedicine . 2018;13:3311–3327. doi: 10.2147/IJN.S165125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gordon O., Vig Slenters T., Brunetto P. S., et al. Silver coordination polymers for prevention of implant infection: thiol interaction, impact on respiratory chain enzymes, and hydroxyl radical induction. Antimicrobial Agents and Chemotherapy . 2010;54(10):4208–4218. doi: 10.1128/AAC.01830-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shahverdi A. R., Fakhimi A., Shahverdi H. R., Minaian S. Synthesis and effect of silver nanoparticles on the antibacterial activity of different antibiotics against Staphylococcus aureus and Escherichia coli. Nanomedicine: Nanotechnology, Biology and Medicine . 2007;3(2):168–171. doi: 10.1016/j.nano.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 139.Lara H. H., Ayala-Nuñez N. V., Ixtepan-Turrent L., Rodriguez-Padilla C. Mode of antiviral action of silver nanoparticles against HIV-1. Journal of Nanobiotechnology . 2010;8(1) doi: 10.1186/1477-3155-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Siddiqi K. S., Rashid M., Rahman A., Tajuddin, Husen A., Rehman S. Biogenic fabrication and characterization of silver nanoparticles using aqueous-ethanolic extract of lichen (Usnea longissima) and their antimicrobial activity. Biomaterials Research . 2018;22(1):p. 23. doi: 10.1186/s40824-018-0135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Baláž M., Goga M., Hegedüs M., et al. Biomechanochemical solid-state synthesis of silver nanoparticles with antibacterial activity using lichens. ACS Sustainable Chemistry & Engineering . 2020;8(37):13945–13955. doi: 10.1021/acssuschemeng.0c03211. [DOI] [Google Scholar]
- 142.Elechiguerra J. L., Burt J. L., Morones J. R., et al. Interaction of silver nanoparticles with HIV-1. Journal of Nanobiotechnology . 2005;3(1):p. 6. doi: 10.1186/1477-3155-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mohammed H., Kumar A., Bekyarova E., et al. Antimicrobial mechanisms and effectiveness of graphene and graphene-functionalized biomaterials: a scope review. Frontiers in Bioengineering and Biotechnology . 2020;8:p. 465. doi: 10.3389/fbioe.2020.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hamzah A. A., Selvarajan R. S., Majlis B. Y. Graphene for biomedical applications: a review. JSM . 2017;46(7):1125–1139. doi: 10.17576/jsm-2017-4607-16. [DOI] [Google Scholar]
- 145.Liu S., Zeng T. H., Hofmann M., et al. Antibacterial activity of graphite, graphite oxide, graphene oxide, and reduced graphene oxide: membrane and oxidative stress. ACS Nano . 2011;5(9):6971–6980. doi: 10.1021/nn202451x. [DOI] [PubMed] [Google Scholar]
- 146.Kumar P., Huo P., Zhang R., Liu B. Antibacterial properties of graphene-based nanomaterials. Nanomaterials . 2019;9(5):p. 737. doi: 10.3390/nano9050737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Radhi A., Mohamad D., Abdul Rahman F. S., Abdullah A. M., Hasan H. Mechanism and factors influence of graphene-based nanomaterials antimicrobial activities and application in dentistry. Journal of Materials Research and Technology . 2021;11:1290–1307. doi: 10.1016/j.jmrt.2021.01.093. [DOI] [Google Scholar]
- 148.Khan M. F., Ansari A. H., Hameedullah M., et al. Sol-gel synthesis of thorn-like ZnO nanoparticles endorsing mechanical stirring effect and their antimicrobial activities: potential role as nano-antibiotics. Scientific Reports . 2016;6(1):p. 27689. doi: 10.1038/srep27689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Vijayakumar S., Krishnakumar C., Arulmozhi P., Mahadevan S., Parameswari N. Biosynthesis, characterization and antimicrobial activities of zinc oxide nanoparticles from leaf extract of glycosmis pentaphylla (retz.) DC. Microbial Pathogenesis . 2018;116:44–48. doi: 10.1016/j.micpath.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 150.Rajendran K., Anwar A., Khan N. A., Siddiqui R. Brain-eating amoebae: silver nanoparticle conjugation enhanced efficacy of anti-amoebic drugs against naegleria fowleri. ACS Chemical Neuroscience . 2017;8(12):2626–2630. doi: 10.1021/acschemneuro.7b00430. [DOI] [PubMed] [Google Scholar]
- 151.Ingle A. P., Duran N., Rai M. Bioactivity, mechanism of action, and cytotoxicity of copper-based nanoparticles: a review. Applied Microbiology and Biotechnology . 2014;98(3):1001–1009. doi: 10.1007/s00253-013-5422-8. [DOI] [PubMed] [Google Scholar]
- 152.Nabila M. I., Kannabiran K. Biosynthesis, characterization and antibacterial activity of copper oxide nanoparticles (CuO NPs) from actinomycetes. Biocatalysis and Agricultural Biotechnology . 2018;15:56–62. doi: 10.1016/j.bcab.2018.05.011. [DOI] [Google Scholar]
- 153.Alavi M., Karimi N., Valadbeigi T. Antibacterial, antibiofilm, antiquorum sensing, antimotility, and antioxidant activities of green fabricated Ag, Cu, TiO2, ZnO, and Fe3O4 NPs via Protoparmeliopsis muralis lichen aqueous extract against multi-drug-resistant bacteria. ACS Biomaterials Science & Engineering . 2019;5(9):4228–4243. doi: 10.1021/acsbiomaterials.9b00274. [DOI] [PubMed] [Google Scholar]
- 154.Abdullah S. M., Kolo K., Sajadi S. M. Greener pathway toward the synthesis of lichen‐based ZnO@TiO2 @SiO2 and Fe3O4 @SiO2 nanocomposites and investigation of their biological activities. Food Sciences and Nutrition . 2020;8:4044–4054. doi: 10.1002/fsn3.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Anghel A., Grumezescu A., Chirea M., et al. MAPLE fabricated Fe3O4@Cinnamomum verum antimicrobial surfaces for improved gastrostomy tubes. Molecules . 2014;19(7):8981–8994. doi: 10.3390/molecules19078981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Jiao Q., Li L., Mu Q., Zhang Q. Immunomodulation of nanoparticles in nanomedicine applications. BioMed Research International . 2014;2014:19. doi: 10.1155/2014/426028.426028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sun J., Xue C. Kupffer cell-mediated hepatic injury induced by silica nanoparticles in vitro and in vivo. International Journal of Nanomedicine . 2013;1129 doi: 10.2147/IJN.S42242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nordberg G., Fowler B. A., Nordberg M. Handbook on the Toxicology of Metals. Volume 1: General Considerations . 4th. Amsterdam, Netherlands: Elsevier, Aademic Press; 2015. [Google Scholar]
- 159.Srinivas A., Rao P. J., Selvam G., Murthy P. B., Reddy P. N. Acute inhalation toxicity of cerium oxide nanoparticles in rats. Toxicology Letters . 2011;205(2):105–115. doi: 10.1016/j.toxlet.2011.05.1027. [DOI] [PubMed] [Google Scholar]
- 160.Pantarotto D., Briand J.-P., Prato M., Bianco A. Translocation of bioactive peptides across cell membranes by carbon NanotubesElectronic supplementary information (ESI) available: details of the synthesis and characterization, cell culture, TEM, epifluorescence and confocal microscopy images of CNTs 1, 2 and fluorescein. Chemical Communications . 2004;16 doi: 10.1039/b311254c. [DOI] [PubMed] [Google Scholar]
- 161.Lu R.-M., Hwang Y.-C., Liu I.-J., et al. Development of therapeutic antibodies for the treatment of diseases. Journal of Biomedical Science . 2020;27:p. 1. doi: 10.1186/s12929-019-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Berger M., Shankar V., Vafai A. Therapeutic applications of monoclonal antibodies. The American Journal of the Medical Sciences . 2002;324(1):14–30. doi: 10.1097/00000441-200207000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Chames P., Van Regenmortel M., Weiss E., Baty D. Therapeutic antibodies: successes, limitations and hopes for the future: therapeutic antibodies: an update. British Journal of Pharmacology . 2009;157(2):220–233. doi: 10.1111/j.1476-5381.2009.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. EMA Zinplava Available Online, 2022, https://www.ema.europa.eu/en/medicines/human/EPAR/zinplava.
- 165.Pelfrene E., Mura M., Cavaleiro Sanches A., Cavaleri M. Monoclonal antibodies as anti-infective products: a promising future? Clinical Microbiology and Infections . 2019;25(1):60–64. doi: 10.1016/j.cmi.2018.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used to support the findings of this study are included within the article.


