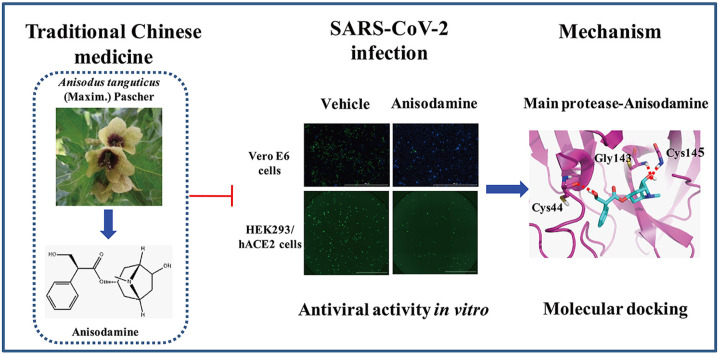
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) provoked a pandemic of acute respiratory disease, namely coronavirus disease 2019 (COVID-19). Currently, effective drugs for this disease are urgently warranted. Anisodamine is a traditional Chinese medicine that is predicted as a potential therapeutic drug for the treatment of COVID-19. Therefore, this study aimed to investigate its antiviral activity and crucial targets in SARS-CoV-2 infection. SARS-CoV-2 and anisodamine were co-cultured in Vero E6 cells, and the antiviral activity of anisodamine was assessed by immunofluorescence assay. The antiviral activity of anisodamine was further measured by pseudovirus entry assay in HEK293/hACE2 cells. Finally, the predictions of crucial targets of anisodamine on SARS-CoV-2 were analyzed by molecular docking studies. We discovered that anisodamine suppressed SARS-CoV-2 infection in Vero E6 cells, and reduced the SARS-CoV-2 pseudovirus entry to HEK293/hACE2 cells. Furthermore, molecular docking studies indicated that anisodamine may target SARS-CoV-2 main protease (Mpro) with the docking score of −6.63 kcal/mol and formed three H-bonds with Gly143, Cys145, and Cys44 amino acid residues at the predicted active site of Mpro. This study suggests that anisodamine is a potent antiviral agent for treating COVID-19.
Keywords: SARS-CoV-2, Anisodamine, Viral infection, Main protease, Molecular docking
Abbreviations: SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; COVID-19, coronavirus disease 2019; FDA, Food and Drug Administration; AHI, hydrobromide injections; ACE2, angiotensin-converting enzyme 2; DMEM, Dulbecco's modified Eagle's medium; FBS, fetal bovine serum; CPEs, cytopathic effects; qRT–PCR, quantitative real-time PCR; BLS-3, biosafety level-3; CCK-8, cell counting kit-8; LDH, lactate dehydrogenase; MOI, multiplicity of infection; IFA, immunofluorescence assay; PBS, phosphate-buffered saline; eGFP, enhance green fluorescent protein; RBD, receptor-binding domain; Mpro, main protease; PLpro, papain-like protease; RdRp, RNA-dependent RNA polymerase; GRP78, glucose-regulated protein 78; MD, molecular dynamics; MOE, Molecular Operating Environment; hpi, post-infection; a7nAChR, nicotinic acetylcholine receptor alpha7
Graphical abstract
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a highly transmissible and pathogenic coronavirus that has caused a pandemic of acute respiratory disease, named coronavirus disease 2019 (COVID-19) [1]. By April 26, 2022, there have been over 500 million confirmed cases of COVID-19, including over 6 million deaths globally (WHO Coronavirus (COVID-19) Dashboard) [2]. High-coverage vaccination is believed to be an effective way to prevent SARS-CoV-2 transmission [3], and over 11 billion vaccine doses have been administered worldwide (WHO Coronavirus (COVID-19) Dashboard) [2]. In addition to vaccines, the development of novel effective therapeutic drugs for the prevention and treatment of COVID-19 is urgently needed, especially for people unsuitable for vaccination (e.g., allergic to the components of Covid-19 vaccines) [4] or unwilling to receive a COVID-19 vaccine.
To date, there are no effective therapeutic drugs to treat patients with COVID-19 [5]. However, the Food and Drug Administration (FDA) has approved remdesivir as antiviral drug for the treatment of COVID-19 [6]. The repurposing of old drugs with potential efficacy against SARS-CoV-2 provides a costless and effective therapeutic solution [7]. Fortunately, numerous traditional Chinese medicine recipes such as Jinhua Qinggan granules and Lianhua Qingwen capsules have demonstrated to be effective treatments for COVID-19 while battling the disease in China [8].
Anisodamine is a type of belladonna alkaloid extracted from the roots of the Chinese medical herb Anisodus tanguticus (Maxim.) Pascher [9]. Similar to atropine and scopalamine, anisodamine is considered a non-specific muscarinic cholinergic antagonist [9]. It has attracted worldwide attention for its therapeutic use in improving the microcirculation in septic shock in China [10]. Furthermore, anisodamine has fewer and less severe adverse effects than atropine [11]. In 2003, anisodamine was used to treat patients with hypoxemia caused by the severe acute respiratory syndrome coronavirus (SARS-CoV) in China [12]. By using network pharmacology integrated molecular docking technology, Su et al. predicted that anisodamine hydrobromide injections (AHI) might indirectly limit the expression of angiotensin-converting enzyme 2 (ACE2), the main target of SARS-CoV-2 [13]. According to its pharmacological properties, Zhen et al. hypothesized that anisodamine might be a feasible treatment for cytokine storm, sticky phlegm in the lung, abnormal coagulation, acute respiratory distress syndrome, and multi-organs damage in COVID-19 infection [14]. Nevertheless, further experiments are required to support anisodamine as an effective therapeutic drug for treating COVID-19.
Herein, we aimed to investigate the antiviral activity and crucial targets of anisodamine on SARS-CoV-2 infection. It was observed that anisodamine suppressed SARS-CoV-2 infection in Vero E6 cells and reduced SARS-CoV-2 pseudovirus entry to HEK293/hACE2 cells. The molecular docking studies indicated that the main protease (Mpro) of SARS-CoV-2 might be a therapeutic target of anisodamine. To our knowledge, our results demonstrated for the first time that anisodamine exhibited antiviral activity against SARS-CoV-2 and might be a promising antiviral agent with an acceptable safety profile to treat COVID-19.
2. Materials and methods
2.1. Chemical
Anisodamine (CAS No. 55869-99-3) was purchased from Hangzhou Fuma Chemical Co., Ltd (Hangzhou, China) with a purity of higher than 98% (HPLC). Anisodamine was dissolved in water at a concentration of 100 mg/mL and stored at −80 °C.
2.2. Cell culture
African green monkey kidney Vero E6 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS), 100 units/mL penicillin, and 100 μg/mL streptomycin (Gibco, Carlsbad, USA). Cells were cultured in a humidified incubator at 37 °C with 5% CO2.
2.3. Virus isolation
A clinical strain of SARS-CoV-2 was isolated from a COVID-19 patient from Changhai Hospital Affiliated with the Naval Medical University/Second Military Medical University (Shanghai, China) in January 2020. In short, the nasopharyngeal aspirate specimen of COVID-19 patient was inoculated in Vero E6 cells, and the latter were monitored daily for cytopathic effects (CPEs) by light microscopy. The SARS-CoV-2 in the cellular supernatant was clarified by centrifugation at 6000×g for 5 min and confirmed via quantitative real-time PCR (qRT–PCR). One-ml aliquots of the supernatant were transferred to tubes and stored at −80 °C. Infection experiments were performed at the biosafety level-3 (BLS-3) laboratory of the Naval Medical University.
2.4. Viral RNA extraction and quantitative real-time PCR (qRT–PCR)
Viral RNA in the cellular supernatant was extracted using an Auto-Pure 96 automated nucleic acid extraction system (Allsheng, Hangzhou, China). The specific ORF1ab and N genes of SARS-CoV-2 were tested by a commercial quantitative PCR assay using the Novel Coronavirus (2019-nCoV) Nucleic Acid Detection Kit (PCR-Fluorescence Probing) (BioGerm, Shanghai, China) and performed on an ABI 7500 real-time PCR system (Life Technologies, Carlsbad, USA) as per manufacturer's instructions.
2.5. Cell viability assay and LDH release assay
Vero E6 cells were seeded into 96-well plates at 4 × 103 cells/well. When 80–90% confluence was reached, cells were treated with different concentrations of anisodamine (50, 100, 200, 400, 800, and 1600 mg/L) for 24 h. Then, cell viability was assessed using the cell counting kit-8 (CCK-8) assay (Dojindo Laboratories, Shanghai, China) and lactate dehydrogenase (LDH) release was detected using the LDH assay kit (GlpBio, Montclair, USA) according to the manufacturer's instructions. 2.6 Immunofluorescence assay.
Vero E6 cells (4.0 × 103 cells/well) were pre-treated with different concentrations of anisodamine (50, 100, and 200 mg/L) for 1 h in 96-well plates, and SARS-CoV-2 at a multiplicity of infection (MOI) of 0.01 was subsequently added to allow infection for 24 h. Viral infection was detected by immunofluorescence assay (IFA). In short, the cells were washed and inactivated with methanol at −20 °C for 30 min and fixed in 4% paraformaldehyde for 10 min. Next, the cells were incubated with sera (1:100 dilution) (from a convalescent COVID-19 patient from Affiliated 3rd People's Hospital of Jiangsu University, Zhenjiang, China) at 37 °C for 30 min, washed with 1 × PBS (phosphate-buffered saline) three times, and incubated with rabbit anti-human IgG (Alexa Fluor® 488)-conjugated secondary antibody (1:1000 dilution, Thermo Fisher Scientific, Rockford, USA) at 37 °C for 30 min. After washing with 1 × PBS three times, the cells were incubated with Hoechst 33,342 (Beyotime Biotechnology, Hangzhou, China) at 37 °C for 5 min. Lastly, images of the cells were captured by Cytation 5 Imaging Reader (BioTek, Winooski, USA).
2.6. Pseudovirus entry assay
HEK293/hACE2 cells (HEK293 cells stably overexpressing human ACE2 receptor; Sino Biological Inc., Beijing, China) were pre-treated with different concentrations of anisodamine (50, 100, and 200 mg/L) for 1 h at 37 °C, then inoculated with SARS-CoV-2 Spike pseudovirus with enhance green fluorescent protein (eGFP) (BPS Bioscience, San Diego, USA). After 48 h of exposure, the fluorescent images of eGFP in HEK293/hACE2 cells were captured by Cytation 5 Imaging Reader (BioTek, Winooski, USA).
2.7. Molecular docking and molecular dynamics simulation
The crystal structures of SARS-CoV-2 Spike RBD (receptor-binding domain) (PDB ID: 6VXX) [15], main protease (Mpro, also referred to as 3CLpro) (PDB ID: 6LZE) [16], papain-like protease (PLpro) (PDB ID: 7CMD) [17], RNA-dependent RNA polymerase (RdRp) (PDB ID: 6M71) [18], and human ACE2 with Spike RBD (PDB ID: 6M0J) [19], glucose-regulated protein 78 (GRP78) (PDB ID: 3LDP) [20] were downloaded from the RCSB Protein Data Bank (http://www.rcsb.org/). A molecular dynamics (MD) simulation study was carried out for anisodamine to target SARS-CoV-2 Spike RBD, Mpro, PLpro, RdRp, human ACE2 with Spike RBD and GRP78 using standard default parameter settings in the Molecular Operating Environment (MOE) software [21].
2.8. Statistical analysis
Statistical analyses were performed using the GraphPad Prism ver. 501 statistical software (GraphPad Software, San Diego, USA) by the two-tailed unpaired Student's t-test. A p value of less than 0.05 was considered statistically significant.
3. Results
3.1. Cytotoxic potential of anisodamine in Vero E6 cells
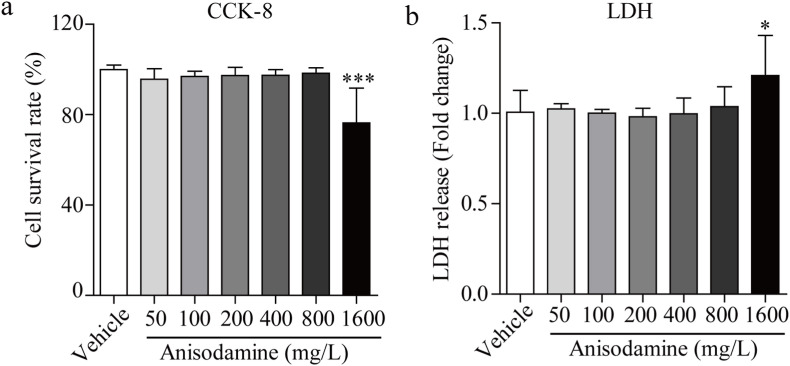
The Vero E6 cell line has been investigated as a potential cell model for SARS-CoV-2 infection. Anisodamine was tested for its cytotoxic effects using the CCK-8 and LDH release assays. Anisodamine did not reduce cell viability (Fig. 1 a) or induce LDH production (Fig. 1b) at concentrations up to 800 mg/L, implying that anisodamine is a safe drug even at high concentration.
Fig. 1.
Cytotoxicity of anisodamine in Vero E6 cells. Vero E6 cells were pre-incubated with anisodamine (0–1600 mg/L) for 24 h, and cell cytotoxicity was analyzed by CCK-8 assays (a), and LDH release assays (b). Experiments were performed in triplicate. Data were presented as the mean ± SD (n = 5). ∗p < 0.05 vs. vehicle, determined with Student's t-test.
3.2. Anisodamine potently inhibited SARS-CoV-2 infection
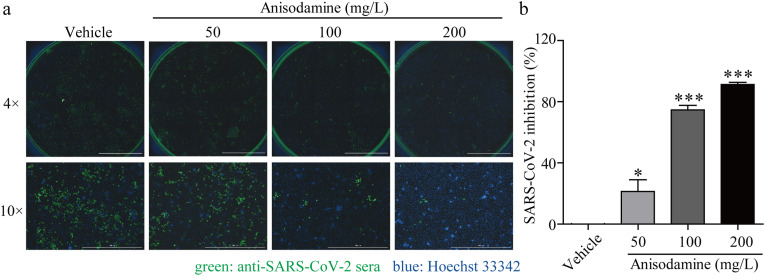
The Vero E6 cells were infected with SARS-CoV-2 and treated with different concentrations of anisodamine. Afterward, anti-SARS-CoV-2 activity was evaluated by quantification of viral copy numbers in the cells via immunofluorescence at 48 h post-infection (hpi). After infection, the number of SARS-CoV-2 increased significantly in Vero E6 cells. However, SARS-CoV-2 replication was potently inhibited after pretreatment with anisodamine in a dose-dependent manner, especially at concentrations of 100 mg/L and 200 mg/L, with over 70% and 90% inhibition of SARS-CoV-2 replication, respectively (Fig. 2 ).
Fig. 2.
Antiviral activity of anisodamine against SARS-CoV-2 in vitro. a. Representative fluorescence images of Vero E6 cells without or with anisodamine (50, 100, and 200 mg/L) under the infection of SARS-CoV-2 at a MOI of 0.01 for 72 h. The green color represents the viral proteins while the blue indicates the nuclei (4 × objective field, scale bar: 2000 μm; 10 × objective field, scale bar: 1000 μm). b. Quantification of inhibition in viral yield. Experiments were performed in triplicate. Data were presented as the mean ± SD (n = 3). ∗p < 0.05, ∗∗∗p < 0.001 vs. vehicle, determined with Student's t-test. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Anisodamine inhibited SARS-CoV-2 pseudovirus infection
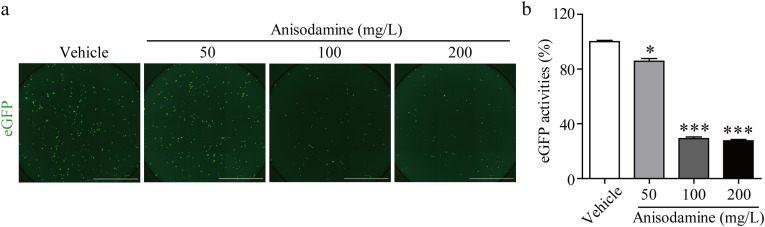
Pseudoviruses are typically regarded as safe and effective virological tools and have been utilized in viral entry mechanistic studies [22] or for identifying entry inhibitors of SARS-CoV-2 [23]. In this study, we measured the entry inhibition of the SARS-CoV-2 spike pseudovirus with anisodamine at different concentrations in HEK293/hACE2 cells. As illustrated in Fig. 3 , the entry of the SARS-CoV-2 pseudovirus was effectively inhibited by anisodamine at concentrations of 100 mg/L and 200 mg/L. As expected, the inhibitory efficacy of anisodamine measured by the pseudovirus infection assay was similar to the immunofluorescence results of SARS-CoV-2.
Fig. 3.
Inhibition of SARS-CoV-2 spike pseudo-virion entry by anisodamine. a. Representative fluorescence images of HEK293/hACE2 cells without or with anisodamine (50, 100, and 200 mg/L) under the infection of SARS-CoV-2 pseudo-virions for 48 h. Transduction efficiency was measured according to eGFP activities (green; scale bar: 2000 μm). b. Quantification of inhibition in eGFP activities. Data were presented as the mean ± SD (n = 3). ∗p < 0.05, ∗∗∗p < 0.001 vs. vehicle, determined with Student's t-test. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.4. The binding affinities of anisodamine into active sites of primary proteins for the SARS-CoV-2 infection
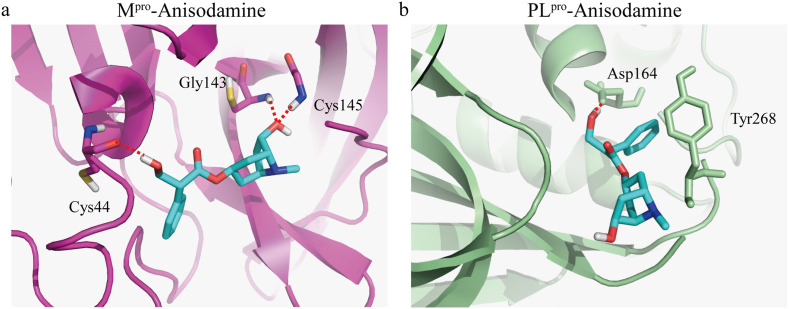
SARS-CoV-2 Spike RBD, Mpro, PLpro and RdRp, and ACE2 and GRP78 in mammalian cells are the druggable targets for the infection of SARS-CoV-2 [24,25]. Docking simulation studies were applied to investigate the binding sites of anisodamine and these proteins. As illustrated in Table 1 , anisodamine demonstrated the highest binding affinity to the predicted active site of Mpro (Docking score of −6.63 kcal/mol) and formed three hydrogen (H)-bonds with Gly143, Cys145, and Cys44 amino acid residues at the predicted active site of Mpro (Fig. 4 a). Likewise, anisodamine displayed considerable binding affinity to the predicted active site of PLpro (Docking score of −5.521 kcal/mol) and formed one H-bond with the Asp164 amino acid residue and one π–π interaction with the Tyr268 amino acid residue at the predicted active site of PLpro (Fig. 4b). In contrast, anisodamine displayed lower binding affinities to the predicted active site of GRP78 (Docking score of −4.801 kcal/mol), Spike RBD (Docking score of −3.898 kcal/mol), RdRp (Docking score of −3.768 kcal/mol) and ACE2 (Docking score of −3.723 kcal/mol), respectively (Table 1). These results suggest that anisodamine can potentially inhibit SARS-CoV-2 invasion through Mpro.
Table 1.
Docking results of anisodamine with the druggable targets for the infection of SARS-CoV-2.
| Target | PBD ID | Docking score (kcal/mol) | Number of H-bonds | H-binding sites |
|---|---|---|---|---|
| Mpro/3CLpro | 6LZE | −6.630 | 3 | Gly145, Gly143, Cys44 |
| PLpro | 7CMD | −5.521 | 1 | Asp164 |
| RdRp | 6M71 | −3.768 | 2 | Phe793, Asp161 |
| Spike RBD | 6VXX | −3.898 | 2 | Tyr489, Gln493 |
| ACE2 with Spike RBD | 6M0J | −3.723 | 1 | Glu310 |
Fig. 4.
Molecular docking of anisodamine with the active sites of SARS-CoV-2 Mpro and PLpro. a. Stereoview of the docked conformation of the Mpro (PDB ID: 6LZE)–anisodamine complex. b. Stereoview of the docked conformation of the PLpro (PDB ID: 7CMD)–anisodamine complex. The dotted red lines indicate hydrogen binding. The carbons of anisodamine are colored in light blue, nitrogen atom in dark blue, and oxygen atoms in red. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Anisodamine has been widely used in multiple diseases such as septic shock since 1965. Little clinical information about its toxicity in humans has been reported so far. It involves a wide range of safe dosing that even 500 mg per day for the treatment of venomous snake bites did not trigger any severe adverse effects [11]. In this study, anisodamine at a dose of up to 800 mg/L showed no toxicity in Vero E6 cells. These studies signify that anisodamine is a very safe drug.
As a fundamental traditional Chinese medicine for improving microcirculation, anisodamine displayed a therapeutic effect on acute respiratory distress syndrome, has been used to treat patients with severe acute respiratory syndrome (SARS), and significantly decreased mortality in 2003 [12]. Both SARS-CoV and SARS-CoV-2 are coronaviruses that belong to the subfamily Coronavirinae and can cause respiratory diseases in humans [26], so it was hypothesized that anisodamine might produce a potential therapeutic effect on patients with COVID-19 caused by SARS-CoV-2. Furthermore, anisodamine could activate the nicotinic acetylcholine receptor alpha7 subunit (a7nAChR) and the cholinergic anti-inflammatory pathway (CAP) and showed an anti-inflammatory effect that is presumed to prevent cytokine storms in patients with COVID-19 [14].
By now, whether anisodamine directly affects the virus infection remains to be elucidated. In this study, anisodamine is convinced to inhibit SARS-CoV-2 infection in Vero E6 cells or effectively prevented entry efficiency of SARS-CoV-2 pseudovirus into HEK293/hACE2 cells, indicating that anisodamine might directly influence the viral entry in human cells.
Undoubtedly, there are several proteins on SARS-CoV-2 and human cells for successfully assisting viral entry in cells. Among them, the spike protein and ACE2 binding is the main responsive for initiation of viral entry [19,27]. In our study, anisodamine inhibited spike pseudovirus entry into HEK293/hACE2 cells. The primary mechanism is the inhibition of the ACE2 expression in HEK293 cells, given that previous research has revealed that anisodamine could indirectly inhibit ACE2 expression [13]. Rayner et al. reported that AR12 suppressed the production of infectious virions via autophagosome formation, which was also associated with the degradation of GRP78 [28]. Another mechanism is that anisodamine may inhibit the spike protein and ACE2 affinity. The molecular docking assay revealed that anisodamine had a low docking score of spike protein and ACE2, reflecting that anisodamine did not directly influence spike protein-ACE2 interactions. Therefore, the mechanism of anisodamine's antiviral activity may be related to the targeting of ACE2 expression. Additionally, anisodamine had a high docking score of inhibiting Mpro which playing essential role in CoV-encoded polyproteins processing [29], indicating anisodamine might play a potential role in inhibiting SARS-CoV-2 infection by affecting the activity of SARS-CoV-2 Mpro.
In conclusion, this study identified anisodamine as a potent antiviral drug against SARS-CoV-2 infection in vitro. Anisodamine with doses up to 800 mg/L showed no toxicity to Vero E6 cells, indicating that anisodamine is a very safe drug. Herein, we observed that anisodamine indirectly disturbed the interaction between ACE2 and the viral spike protein, and SARS-CoV-2 Mpro was a therapeutic target of anisodamine. Nonetheless, further investigations are warranted in order to elucidate the antiviral efficacy of anisodamine in animal model and clinical evaluation in vivo. To conclude, anisodamine may be a promising drug for the treatment of COVID-19 patients.
Ethics statement
The experiments involving clinical samples were approved by the ethical committee for animal experiments of the Navy Military Medical University/Second Military Medical University (SCXK2017-0010). All research participants provided informed consent to participate in this study.
Author contributions
W.W. contributed data analysis and drafted the initial manuscript, N.K. and M.Z.L. performed the experiments, T.H. contributed reagents/materials, J.F.X. revised the manuscript, and C.L. conceived and designed the experiments, and revised the manuscript. All authors read and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank associate professor Yahui Huang for his coordination and support of molecular docking analysis. This study was supported by the Shanghai Biomedical Science and Technology Support Program (20S190280). We thank the Home for Researchers for its linguistic assistance during the preparation of this manuscript.
References
- 1.Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO coronavirus (COVID-19) dashboard. https://covid19.who.int/
- 3.Pablo M., De Salazar N.B.L., Lamarca Karuna, Santillana Mauricio. High coverage COVID-19 mRNA vaccination rapidly controls SARS-CoV-2 transmission in long-term care facilities. Commun. Med. 2021;1:16. doi: 10.1038/s43856-021-00015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cabanillas B., Novak N. Allergy to COVID-19 vaccines: a current update. Allergol. Int. 2021;70:313–318. doi: 10.1016/j.alit.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu R., Wang L., Kuo H.D., Shannar A., Peter R., Chou P.J., et al. An update on current therapeutic drugs treating COVID-19. Curr Pharmacol Rep. 2020:1–15. doi: 10.1007/s40495-020-00216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saul S., Einav S. Old drugs for a new virus: repurposed approaches for combating COVID-19. ACS Infect. Dis. 2020;6:2304–2318. doi: 10.1021/acsinfecdis.0c00343. [DOI] [PubMed] [Google Scholar]
- 8.Huang K., Zhang P., Zhang Z.H., Youn J.Y., Wang C., Zhang H.C., Cai H. Traditional Chinese Medicine (TCM) in the treatment of COVID-19 and other viral infections: efficacies and mechanisms. Pharmacol. Ther. 2021;225:107843. doi: 10.1016/j.pharmthera.2021.107843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenkraft A., Falk A. Possible role for anisodamine in organophosphate poisoning. Br. J. Pharmacol. 2016;173:1719–1727. doi: 10.1111/bph.13486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poupko J.M., Baskin S.I., Moore E. The pharmacological properties of anisodamine. J. Appl. Toxicol. 2007;27:116–121. doi: 10.1002/jat.1154. [DOI] [PubMed] [Google Scholar]
- 11.Li Q.B., Pan R., Wang G.F., Tang S.X. Anisodamine as an effective drug to treat snakebites. J. Nat. Toxins. 1999;8:327–330. [PubMed] [Google Scholar]
- 12.Yang G.D. Patients of severe acute respiratory syndrome with hypoxemia treated by anisodamine. Zhongguo Wei Zhong Bing Ji Jiu Yi Xue. 2003;15:452. [PubMed] [Google Scholar]
- 13.Su J.S., Liu Z.X., Liu C., Li X.H., Wang Y., Zhao J., Wu Q.J., Zheng S.C., Zhang Y. Network pharmacology integrated molecular docking reveals the mechanism of anisodamine hydrobromide injection against novel coronavirus Pneumonia. Evid Based Complement Alternat Med. 2020;2020:5818107. doi: 10.1155/2020/5818107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin Z., Xiang K.F., Su D.F., Sun Y., Liu X. Activation of the cholinergic anti-inflammatory pathway as a novel therapeutic strategy for COVID-19. Front. Immunol. 2021;11:595342. doi: 10.3389/fimmu.2020.595342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai W., Zhang B., Jiang X.M., Su H., Li J., Zhao Y., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao X., Qin B., Chen P., Zhu K., Hou P., Wojdyla J.A., et al. Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharm. Sin. B. 2021;11:237–245. doi: 10.1016/j.apsb.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 20.Macias A.T., Williamson D.S., Allen N., Borgognoni J., Clay A., Daniels Z., et al. Adenosine-derived inhibitors of 78 kDa glucose regulated protein (Grp78) ATPase: insights into isoform selectivity. J. Med. Chem. 2011;54:4034–4041. doi: 10.1021/jm101625x. [DOI] [PubMed] [Google Scholar]
- 21.Khelfaoui H., Harkati D., Saleh B.A. Molecular docking, molecular dynamics simulations and reactivity, studies on approved drugs library targeting ACE2 and SARS-CoV-2 binding with ACE2. J. Biomol. Struct. Dyn. 2020:1–17. doi: 10.1080/07391102.2020.1803967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanders D.A. No false start for novel pseudotyped vectors. Curr. Opin. Biotechnol. 2002;13:437–442. doi: 10.1016/s0958-1669(02)00374-9. [DOI] [PubMed] [Google Scholar]
- 23.Basu A., Mills D.M., Bowlin T.L. High-throughput screening of viral entry inhibitors using pseudotyped virus. Curr. Protoc. Pharmacol. 2010 doi: 10.1002/0471141755.ph13b03s51. 13B.3.1-13B.3.17. [DOI] [PubMed] [Google Scholar]
- 24.Faheem Kumar BK., Sekhar K.V.G.C., Kunjiappan S., Jamalis J., Balaña-Fouce R., et al. Druggable targets of SARS-CoV-2 and treatment opportunities for COVID-19. Bioorg. Chem. 2020;104:104269. doi: 10.1016/j.bioorg.2020.104269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ha D.P., Krieken R.V., Carlos A.J., Lee A.S. The stress-inducible molecular chaperone GRP78 as potential therapeutic target for coronavirus infection. J. Infect. 2020;81:452–482. doi: 10.1016/j.jinf.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Y.C., Xiao Z.Q., Ye K.Y., He X.E., Sun B., Qin Z.R., et al. SARS-CoV-2: characteristics and current advances in research. Virol. J. 2020;17:117. doi: 10.1186/s12985-020-01369-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salvatori G., Luberto L., Maffei M., Aurisicchio L., Roscilli G., Palombo F., et al. SARS-CoV-2 SPIKE PROTEIN: an optimal immunological target for vaccines. J. Transl. Med. 2020;18:222. doi: 10.1186/s12967-020-02392-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rayner J.O., Roberts R.A., Kim J., Poklepovic A., Roberts J.L., Booth L., et al. AR12 (OSU-03012) suppresses GRP78 expression and inhibits SARS-CoV-2 replication. Biochem. Pharmacol. 2020;182:114227. doi: 10.1016/j.bcp.2020.114227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]