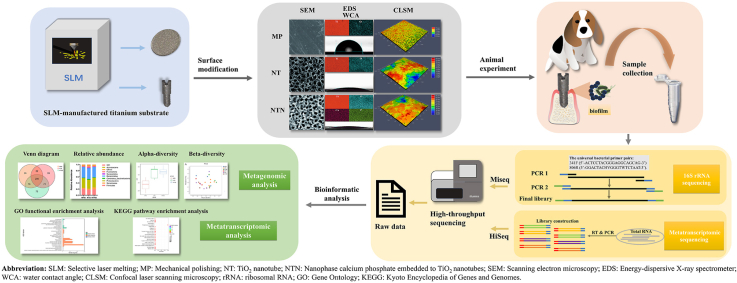
Abstract
Peri-implantitis, the major cause of implant failure, is an inflammatory destructive disease due to the dysbiotic polymicrobial communities at the peri-implant sites. Therefore, it is highly warranted to develop the implant materials with antimicrobial properties and investigate their effects on oral microbiota. However, most of the relevant studies were performed in vitro, and insufficient to provide the comprehensive assessment of the antimicrobial capacity of the implant materials in vivo. Herein, we introduce an innovative approach to evaluate the in vivo antibacterial properties of the most commonly used implant materials, titanium with different nanostructured surfaces, and investigate their antibacterial mechanism via the next-generation sequencing (NGS) technology. We firstly prepared the titanium implants with three different surfaces, i) mechanical polishing (MP), ii) TiO2 nanotubes (NT) and iii) nanophase calcium phosphate embedded to TiO2 nanotubes (NTN), and then characterized them using scanning electron microscopy (SEM), energy-dispersive X-ray spectrometer (EDS), X-ray photoelectron spectroscopy (XPS), confocal laser scanning microscopy (CLSM) and surface hydrophilicity analysis. Afterwards, the implants were placed in the beagle dogs’ mouths to replace the pre-extracted premolar and molar teeth for eight weeks through implant surgery. The supra- and sub-mucosal plaques were collected and subjected to 16S rRNA gene/RNA sequencing and data analysis. It was found that the nanostructured surfaces in NT and NTN groups showed significantly increased roughness and decreased water contact angles compared to the MP group, while the XPS data further confirmed the successful modifications of TiO2 nanotubes and the subsequent deposition of nanophase calcium phosphate. Notably, the nanostructured surfaces in NT and NTN groups had limited impact on the diversity and community structure of oral microbiota according to the 16S rRNA sequencing results, and the nanostructures in NTN group could down-regulate the genes associated with localization and locomotion based on Gene Ontology (GO) terms enrichment analysis. Moreover, the differentially expressed genes (DEGs) were associated with microbial metabolism, protein synthesis and bacterial invasion of epithelial cells. Taken together, this study provides a new strategy to evaluate the antibacterial properties of the biomedical materials in vivo via the high-throughput sequencing and bioinformatic approaches, revealing the differences of the composition and functional gene expressions in the supra- and sub-mucosal microbiome.
Keywords: TiO2 nanotubes, Oral biofilm, 16S, Metatranscriptomics, Dental implants, Surface modification
Graphical abstract
Abbreviations
- MP
Mechanical polishing
- NT
TiO2 nanotube
- NTN
Nanophase calcium phosphate embedded to TiO2 nanotubes
1. Introduction
With the advancement of oral implantology, dental implant has become a common method to rehabilitate partial and full edentulous jaws [1]. In recent years, there is an increasing number of patients with peri-implant diseases, including peri-implant mucositis and peri-implantitis. A recent meta-analysis estimated that the weighted mean prevalence of peri-implant mucositis and peri-implantitis were 43% and 22%, respectively [2]. It is noted that peri-implantitis are the biological complications of dental implantation mainly caused by biofilms [3,4], with symptoms of inflammation in peri-implant mucosa and subsequent progressive loss of supporting bone [5]. Therefore, it is essential to develop dental implants with antibacterial properties to arrest the progression of peri-implant diseases.
Titanium implants with nanostructured surfaces have attracted many attentions on account of their remarkable versatility. It has been reported that titanium nanostructures could increase the osseointegration by promoting the adhesion and proliferation of osteoblasts [6,7], enhancing the differentiation of stem cells [8], and increasing strength of osteoblasts adhesion [9] and mineralization rate [10]. Notably, titanium oxide (TiO2) nanotubes also showed favorable antimicrobial properties [[11], [12], [13], [14]]. Peng et al. found that TiO2 nanotubes coating significantly decreased the initial adhesion and colonization of Staphylococcus epidermidis compared to the acid-etched or polished Ti surfaces in vitro [11]. Moreover, additively manufactured porous titanium biomaterials covered with nanotubular surfaces had less adherent Staphylococcus aureus compared to the porous titanium surface on day one, suggesting that nanotubes alone could be effective in preventing biofilm formation within the first few hours to the first day after the implant surgery [12]. Moreover, antibacterial coatings are important to prevent biofilm formation and infectious diseases [15,16]. This property keeps in line with the reported bactericidal capacity attributed to the high aspect ratio nano-topographies [17,18].
Meanwhile, calcium phosphate coatings have also been applied to promoted the bioactive and biocompatible properties of titanium implants for many years [19], since they show good clinical performance due to the superior osseointegration rate [20]. Moreover, the unique topography, roughness and chemical compositions result in the inherent and stable antibacterial properties of the calcium phosphate coatings against Staphylococcus aureus and Porphyromonas gingivalis [21,22]. Therefore, the nano calcium phosphate together with the TiO2 nanotubes coatings are the two most popular surface modifications on titanium implants.
The selective laser melting (SLM)-processed titanium substrates exhibit great clinical potential and flexibility of constructing titanium implants for the biomedical application. It has been reported that SLM as a manufacturing technique could benefit the clinical individual-based treatments by quickly producing the titanium substrates with intricate shapes [23]. Our previous study has developed a microrough selective laser melting (SLM) titanium substrates with surface modification of nanophase calcium phosphate embedded TiO2 nanotubes [24]. The nanotubes with nanophase calcium phosphate on TiO2 nanotubes showed fewer Streptococcus mutans and Streptococcus sanguinis bacteria colonies after 8 h and 24 h cultivation compared with TiO2 nanotubes in vitro [24]. In consistence with our study, a recent study reported that a significant decrease of Staphylococcus aureus adhesion after 3 h and 22 h was detected on calcium phosphate-coated TiO2 nanotubes compared with those on the TiO2 nanotubes, Ag-coated TiO2 nanotubes and Ag/calcium phosphate-coated TiO2 nanotubes [25]. However, the antibacterial activity of calcium phosphate deposited on TiO2 nanotubes has not been fully exploited, while the investigations of the composition and functional gene expression of oral microbiota presenting on the calcium phosphate-coated TiO2 nanotubes versus TiO2 nanotubes in vivo may facilitate the understanding of antibacterial surface coating and benefit clinical outcome.
So far, the antibacterial properties of the biomaterials as oral implants were usually evaluated by various in vitro experiments, yet the results may solely reflect the influence of materials on specific tested pathogens. These experiments were insufficient to reveal the actual biological effects of the oral implant materials toward the whole oral microbiomes. Moreover, the comprehensive assessment of their in vivo antimicrobial activities remains challenging due to the oral biofilms with complex compositions and structures alongside the host-biofilm interactions. As such, it is urgent to develop an approach to address the aforementioned research gaps.
The advancement of the next-generation sequencing (NGS) technology makes it possible to rapidly generate sequences and provide extensive information of the inhabitant microorganisms in oral cavity [26]. Metagenomics reveal the taxonomical profile of a microbial community, whereas metatranscriptomics could inform us the functional profile of a community [27]. The retrieved information can be used for detecting microbiome-based biomarkers for early diagnosis of oral diseases [26]. Yet, neither metagenomic nor metatranscriptomic sequencing technology has been utilized for in vivo evaluation of the effect of titanium materials on the antibacterial properties.
Taken together, in this study, we innovatively applied the NGS technology to investigate dental implant surface modification, aiming to provide a new strategy to evaluate the antibacterial properties of biomedical materials in vivo. Specifically, we evaluated the in vivo antibacterial properties of titanium with the nanostructured surface from a new perspective, and explored the antibacterial mechanism of the nanostructure through metagenomic and metatranscriptomic sequencing to characterize the diversity and community structure of oral microbiota.
2. Materials and methods
2.1. Specimen preparation and treatment
Specimen preparation and treatment of the mechanical polishing group (MP) and TiO2 nanotube group (NT) have been reported in our previous studies [24]. Briefly, screw-type titanium implants were designed using SolidWorks® 12.0 (SolidWorks Corp, Concord, MA, USA) and manufactured using a selective laser melting (SLM) machine (SLM125HL, SLM solutions GmbH, Lubeck, Germany). A total of two types of specimen shapes were prepared. Titanium round discs with a thickness of 1 mm and a diameter of 10 mm were used for titanium surface characterization. Titanium implant screws were fabricated for the implant surgery in vivo. Each implant body was 13 mm in length and 4.5 mm in diameter as shown in Fig. S1. Then SLM-processed titanium specimens were immersed in a cleaning solution (0.05 mol/L Na2SiO3, 0.1 mol/L Na3PO4 and 0.2 mol/L Na2CO3) for 15 min followed by sonication in deionized water for 15 min. A total of three groups of specimens were fabricated in this study, which were 1) mechanical polishing group (MP, n = 5), specimens were mechanically polished successively by P80, P220, P400, P600, P800, P1000 sandpapers; 2) TiO2 nanotube group (NT), SLM-processed titanium specimens were first sandblasted with 250 μm ZrO2 particles and then chemically polished in a solution of 5% hydrofluoric acid (HF) for 1 min TiO2 nanotube arrays was fabricated on the SLM-processed titanium substrate using anodic oxidation methods (voltage of 20 V in a 0.5 wt% HF electrolyte for 5 min) followed by annealing at 500 °C for 120 min; 3) nanophase calcium phosphate embedded to TiO2 nanotubes groups (NTN), calcium phosphate nanoparticles were in or between the TiO2 nanotubes by electrochemical deposition. Nanotube samples were anchored onto a platinum cathode and subjected to a constant voltage of −3V for 5 min with the counter-electrode platinum using an electrochemical workstation (CHI-660D, USA). The solution for electrochemical deposition contained 0.20 mmol/L of Ca(NO3)2, 0.12 mmol/L of (NH4)2HPO4 and 0.05 mol/L NaNO3 in an 80 °C water bath environment. The duration of electrochemical deposit ion in this study was shortened compared with our previous protocol [24] in order to maintain the topography of nanotubes.
2.2. Surface characterization
The surface topography and elemental compositions were analyzed by field-emission scanning electron microscopy (FE-SEM; Hitachi, S-4800, Tokyo, Japan) equipped with an energy-dispersive X-ray spectrometer (EDS), and X-ray photoelectron spectroscopy (XPS; Thermo Scientific ESCALAB 250Xi, USA), respectively. The average pore diameters and pore size distribution were identified based on SEM micrographs by using Image-Pro Plus 6.0 (Rockville, MD, United States).
In addition, the average roughness (Sa) and root mean square roughness (Sq) measurements were determined via confocal laser scanning microscopy (CLSM; LSM 700, Carl Zeiss AG, Oberkochen, Germany). For surface hydrophilicity analysis, water contact angle measurements were investigated by using sessile drop method from an optical contact angle measuring device (OCA Pro 15 Dataphysics, Germany).
2.3. Animal experiment
This study was ethically approved by the Ethics Committee for Animal Experiments of Sun Yat-Sen University (No. SYSU-IACUC-2018-000207). A total of 3 adult male beagle dogs weighing 11.5–13.5 kg were used. All surgical procedures were performed under general anesthesia using pentobarbital sodium (30 mg/kg/i.m., Pelltobarbitalum, Sigma, USA) and xylazine hydrochloride (0.10 mg/kg/i.m., Sumianxin, Guilin Huamei, China). The third and fourth premolar and the first molar teeth in both mandibular quadrants were extracted gently using a rotational movement after hemi-sectioning. The sockets were curetted, and then interrupted suturing was performed. After a 3-month healing period, implant surgery was performed. A carbon blade no. 15 was used to make a full-thickness crestal incision along the edentulous ridge, and the mucoperiosteal flap was elevated. Three implant holes were drilled in the edentulous alveolar ridge of each mandibular quadrant in No. 1 and No. 2 beagles and of the left mandibular quadrant in No. 3 beagle. Profuse irrigation with saline water was used for cooling during drilling. Five titanium implants of each group were used in this experiment. A total of 6 implants (2 implants of each group) were placed in the implant holes of No. 1 and No. 2 beagles respectively, and 3 implants (1 implant of each group) were placed in the left edentulous alveolar ridge of No. 3 beagle. Then interrupted suturing was performed. The animals received prophylaxis penicillin administration (80,000 IU/kg/i.m., Reyoung, China) during the first three days after surgery. After 8 weeks, clinical parameters were recorded, and supra-mucosal and sub-mucosal plaque were collected. All the animals were euthanized by the administration of a sodium pentobarbital overdose.
2.4. Clinical evaluation and sampling methods
All the residual implants were recruited in this experiment (MP = 3, NT = 5, NTN = 4). The following clinical parameters were examined to characterize the status of peri-implant lesions: modified Plaque Index (mPlI) [28], modified Bleeding Index (mBI) [28], bleeding on probing (BOP) scores [29] and probing pocket depth (PPD) [30].
Prior to sampling, the clinical sites were firstly isolated and dried with sterile cotton rolls. Supra-mucosal plaque samples were collected using sterile Gracey or universal curettes and placed into labeled sterile Eppendorf tubes with 1 mL phosphate buffered saline (PBS) (0.01 M phosphate, 137 mM NaCl, 2.7 mM KCl; pH 7.4). After complete removal of supra-mucosal plaque, sub-mucosal plaque was collected using the same method as the supra-mucosal plaque collection. One supra-mucosal sample and one sub-mucosal sample were collected from each implant for the subsequent experiment. Supra-mucosal samples collected from MP, NT and NTN groups were labeled as MPP, NTP and NTNP, respectively. Sub-mucosal samples acquired from MP, NT and NTN groups were denoted as MPB, NTB and NTNB, respectively. In the laboratory, the Eppendorf tubes were briefly vortexed before being centrifuged (5000 g, 10 min), and the PBS buffer was carefully discarded to obtain the washed plaque samples. Then, all the samples were stored at −80 °C until further analysis.
2.5. 16S rRNA gene sequencing and data analysis
Total bacterial genomic DNA was extracted using QIAamp DNA Mini kits (Qiagen, CA, USA) according to the manufacturer's instructions. PCR amplification of the hypervariable V3–V4 region of the 16S ribosomal RNA (rRNA) genes was performed using the universal bacterial primer pairs, 341F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). PCR amplification and library construction were performed according to Illumina's standard protocol for 16S Metagenomic Sequencing Library Preparation (Part # 15044223 Rev. B). Paired-end MiSeq sequencing was performed on the amplicon libraries using an Illumina MiSeq system (300 PE) at the Beijing Genomics Institute (BGI Institute, Wuhan, China). The paired-end MiSeq sequences were analyzed using the Quantitative Insights Into Microbial Ecology (QIIME1) package [31] following the MiSeq SOP pipeline. Mothur [32] was also used in the downstream sequence analysis. Briefly, the paired ca. 300-bp forward and reverse MiSeq sequence reads were assembled, followed by the filtering of noisy sequences, chimera checking and Operational Taxonomic Units (OTUs) picking based on 97% identity using the usearch quality filtering script. Taxonomy assignments were made against the RDP databases. The Venn plots in OTUs were plotted with R package. The structure of different peri-implant microbial communities, the relative abundance, alpha diversity and beta-diversity were estimated using Mothur and Qiime. Beta-diversity comparisons were performed by both the unweighted and weighted UniFrac analyses, which were visualized in principal coordinate analysis (PCoA) plots. The comparisons of the relative abundance among different groups were determined by R package based on Kruskal-Test. The p value threshold was set to 0.05.
2.6. Metatranscriptomic sequencing and data analysis
Total RNA was extracted using Promega SV Total RNA Isolation System kit (Promega, USA) following the manufacturer's protocol. After rRNA was removed, mRNA was obtained by oligo dT. Then fragmentation buffer was added to interrupt mRNA to short reads. Taking these short reads as templates, Random hexamer-primer were used to synthesize the first-strand cDNA. The second-strand cDNA was serially synthesized using buffer, dATPs, dGTPs, dCTPs, dUTPs, RNase H and DNA polymerase I after removing dNTPs. Short reads were purified with QiaQuick PCR extraction kit and resolved with EB buffer for end reparation and adding poly(A), then the short reads were connected with sequencing adapters. After that, the UNG enzyme was used to degrade the second-strand cDNA, and the product was purified by MiniElute PCR Purification Kit before PCR amplification. Sequencing of mRNA was carried out at the Beijing Genomics Institute (BGI Institute, Wuhan, China) using an Illumina HiSeq2000 platform.
Clean data was obtained using SOAP2 software [33]. A detailed method was shown below: 1) Remove reads with 10% N; 2) Remove reads contaminated by adapter (default:15 bases overlapped by reads and adapter); 3) Remove reads with 20% low quality (20) bases; 4) Remove 16S rRNA, 18S rRNA contamination reads (consistency of 90%). Short reads were assembled using Trinity software [34]. Based on NOIseq package method [35], differentially expressed genes (DEGs) were screened among groups. Gene Ontology (GO) enrichment analysis of differentially expressed genes was implemented by the GOseq R package [36]. KOBAS software [37] was used to test the statistical enrichment of differential expression genes in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The p values were adjusted using the Benjamini & Hochberg method. GO and KEGG terms with corrected p value less than 0.05 were considered significantly enriched by differential expressed genes.
2.7. Statistical analysis
Statistical analysis was conducted using SPSS software 25 (SPSS Inc., Chicago, IL, USA) and RStudio interface (Team, R., RStudio: Integrated Development Environment for R. RStudio Inc., Boston, MA. In 2015) [38]. One-way ANOVA test followed by a Bonferroni's test was used to identify any clinical parameter between multiple groups.
2.8. Data depository
The sequences generated in this study were deposited in NCBI BioProject: PRJNA776400 (Reviewer link: https://www.ncbi.nlm.nih.gov/sra/PRJNA776400) for 16S rRNA gene sequencing data; and PRJNA776641 (https://www.ncbi.nlm.nih.gov/sra/PRJNA776641) for RNA sequencing data.
3. Results
3.1. Surface characterization
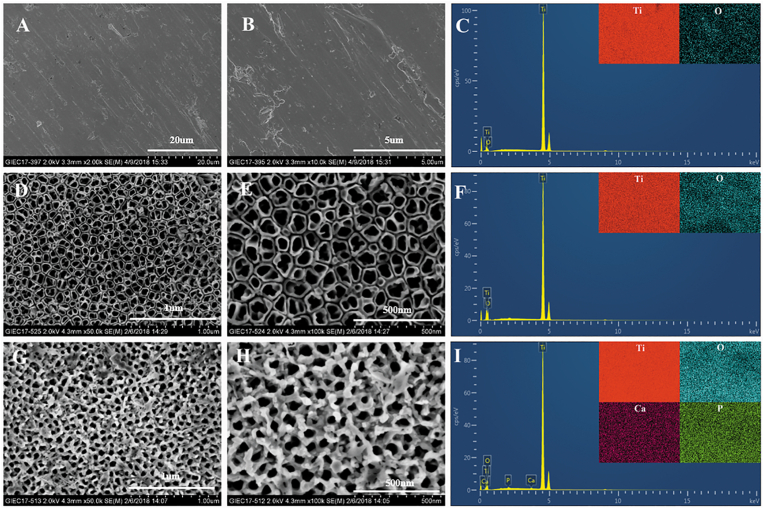
The surface topographies of specimens were shown in Fig. 1. The surface of the MP specimen was smooth with regular scratches resulting from the polishing process (Fig. 1A and B). TiO2 nanotube structures were observed on the surface of NT specimen (Fig. 1D and E). As shown in Fig. 1G and H, the nanophase calcium phosphate was successfully deposited into or between the TiO2 nanotubes on the surface of NTN specimen. According to the pore size distribution in Fig. S2, the average diameter of nanotube in NT group was 61.24 ± 13.82 nm, while the size decreased to 26.63 ± 5.37 nm after the calcium phosphate deposited on the TiO2 nanotubes.
Fig. 1.
Scanning electron microscopy (SEM) images of MP (A & B), NT (D & E) and NTN samples (G & H) at different magnifications. Energy-dispersive X-ray spectrometer (EDS) mapping results showed the elemental compositions of MP (C), NT (F) and NTN (I) samples.
The surface elemental compositions of specimens from different groups were measured by EDS and the data are shown in Table 1 and Fig. 1C, 1F and 1I. It was noted that titanium and oxygen were the major elements in all specimens, while the oxygen proportion increased dramatically in NT and NTN samples, indicating a significant surface elemental change. For NTN specimens, calcium and phosphorus were newly found, suggesting the successful deposition of calcium phosphate nanoparticles.
Table 1.
The surface element compositions of the specimens in different groups (wt%).
| Specimen | Titanium | Oxygen | Calcium | Phosphorus |
|---|---|---|---|---|
| MP | 93.03 | 6.97 | – | – |
| NT | 74.01 | 25.99 | – | – |
| NTN | 78.08 | 21.50 | 0.25 | 0.17 |
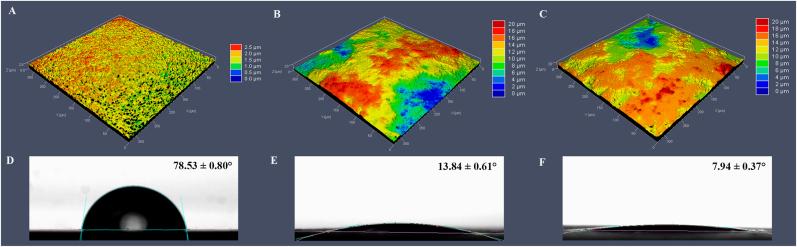
3D images of MP, NT, NTN samples and the representative water contact angle images were presented in Fig. 2. The MP surfaces showed higher mean water contact angle (78.53°) than the NT surfaces (13.84°) and NTN surfaces (7.94°). The surface roughness parameters of specimen surfaces were shown in Table 2. The NT and NTN surfaces showed higher average roughness values (Sa) (2.69 μm and 2.83 μm, respectively) than the MP surfaces (0.26 μm).
Fig. 2.
3D confocal laser scanning microscopy (CLSM) images and water contact angle images of titanium surfaces from MP (A), NT (B), NTN (C) groups. Representative water contact angle images of MP (D), NT (E), NTN (F) samples.
Table 2.
Surface roughness of different specimens.
| Specimen | Sa (μm) | Sq (μm) |
|---|---|---|
| MP | 0.26 ± 0.00 | 0.33 ± 0.01 |
| NT | 2.69 ± 0.04 | 3.30 ± 0.06 |
| NTN | 2.83 ± 0.01 | 3.50 ± 0.09 |
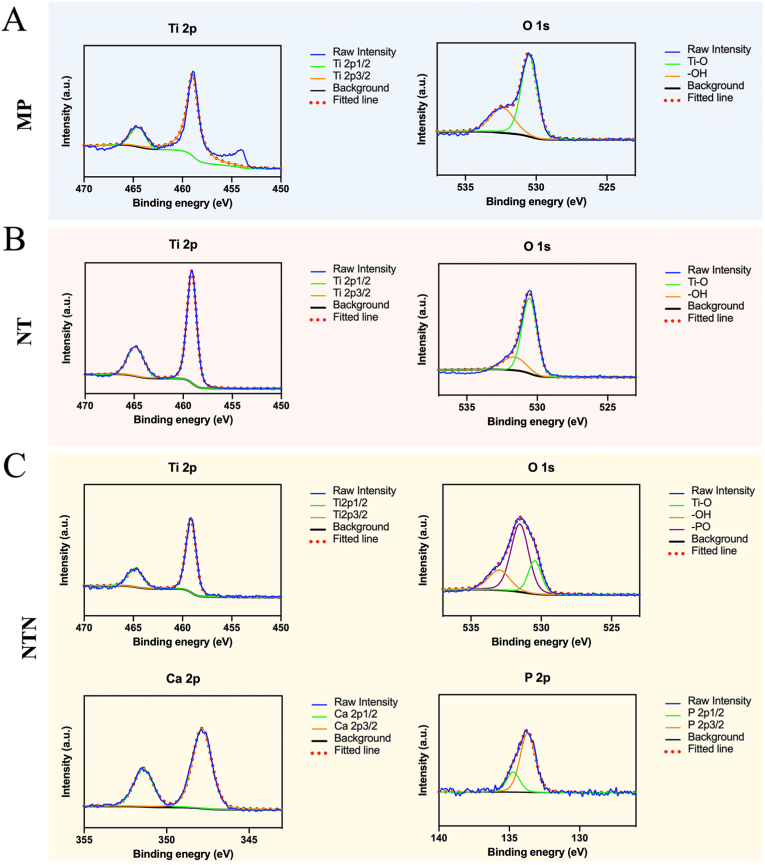
The XPS spectra in Fig. 3 provide more information about the elemental changes resulting from the surface modification. As Ti and O were the major elements found in all the samples, we also analyzed these two elements using XPS. It was found that the O 1s was mainly divided into two bands in MP and NT samples, including Ti–O and –OH, while the surface modification of TiO2 nanotube in NT accounted for the increased intensity of Ti–O with the reference to that in MP samples. After the deposition of calcium phosphate, two new elemental peaks of Ca and P can be identified from the XPS survey spectrum of NTN. Moreover, the O 1s of NTN sample could be differentiated into three peaks, while the new one represented the -PO band.
Fig. 3.
X-ray photoelectron spectroscopy (XPS) spectra (Ti 2p, O 1s, Ca 2p and P 2p) of MP (A), NT (B), and NTN (C).
3.2. Clinical evaluation
Because MP and NTN group lost 2 and 1 implants respectively during the observation period, a total of 12 implants (MP = 3, NT = 5, NTN = 4) were recruited in this study. The clinical characteristics of titanium implants were summarized in Table 3. No significant differences of clinical parameters were detected between the groups.
Table 3.
Clinical characteristics of titanium implants in three groups.
| Clinical parameters | MP | NT | NTN | p value |
|---|---|---|---|---|
| mPlI | 1.67 ± 0.58 | 1.60 ± 0.89 | 1.75 ± 0.50 | 0.953 |
| PPD (mm) | 2.33 ± 0.58 | 2.20 ± 0.57 | 1.88 ± 0.48 | 0.525 |
| mBI | 2.33 ± 0.58 | 2.00 ± 1.41 | 1.00 ± 1.16 | 0.326 |
| BOP (% (frequency of detection)) | 100 | 80 | 50 | 0.366 |
3.3. 16S rRNA gene sequencing results
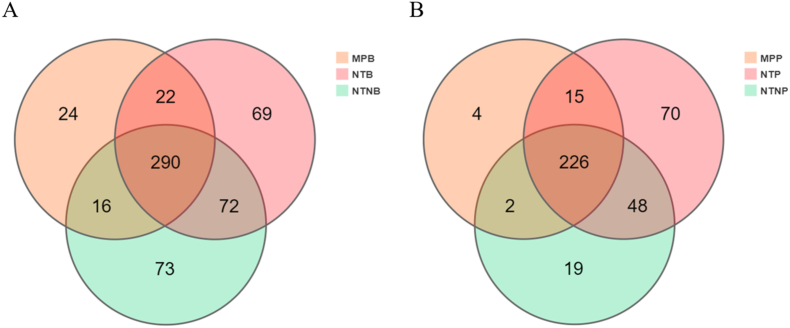
A total of 648,953 assembled quality-filtered sequencing reads were obtained, which were assigned to 572 operational taxonomic units (OTUs) with the identity threshold set at 97%. For sub-mucosal plaque, the three categories shared 290 OTUs in common (i.e. OTUs were detected in all three MP, NT and NTN categories of titanium surfaces). This accounted for nearly half of the total number of OTUs detected, as shown in the Venn diagram (Fig. 4A). Within the supra-mucosal plaque niches, MP, NT and NTN shared 226 OTUs in common (Fig. 4B). As shown in Fig. S3, the rarefaction curves of observed OTUs appeared to approach asymptotes, which indicated high levels of coverage and thus sufficient sampling effort in each biofilm sample.
Fig. 4.
Venn diagrams of Operational Taxonomic Units (OTUs) on the titanium surfaces from three groups. Venn diagrams show the distributions of all 572 OTUs on the titanium surfaces within the sub-mucosal plaque niches (A) and the supra-mucosal plaque niches (B).
3.4. Bacterial community composition at different taxonomic levels
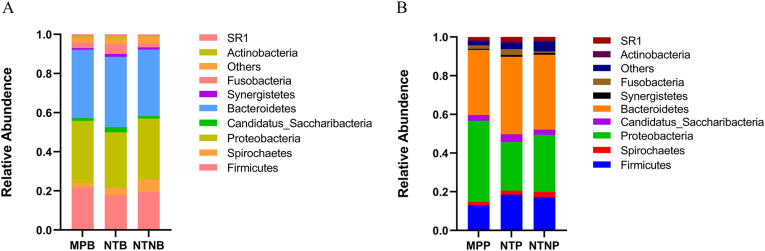
Diverse bacterial taxa were assigned to 14 phyla, 189 genera and 229 species from the clinical samples. As Fig. 5 showed, no significant differences in terms of the relative abundance were detected in the sub-mucosal or supra-mucosal samples at the phylum level. As tabulated in Table S1, a total of 6 genera and 9 species showed significant differences in the sub-mucosal samples among groups. There were no significant differences at the genus level but 3 species showed statistical differences in supra-mucosal samples among groups.
Fig. 5.
The relative abundance of sub-mucosal microbiota (A) and supra-mucosal microbiota (B) at the phylum level. The X-axis indicates different groups; the Y-axis indicates the relative abundance of the phylum annotated. The phyla with the relative abundance of less than 0.5% in the samples are merged into “Others” item.
3.5. Alpha-diversity comparisons
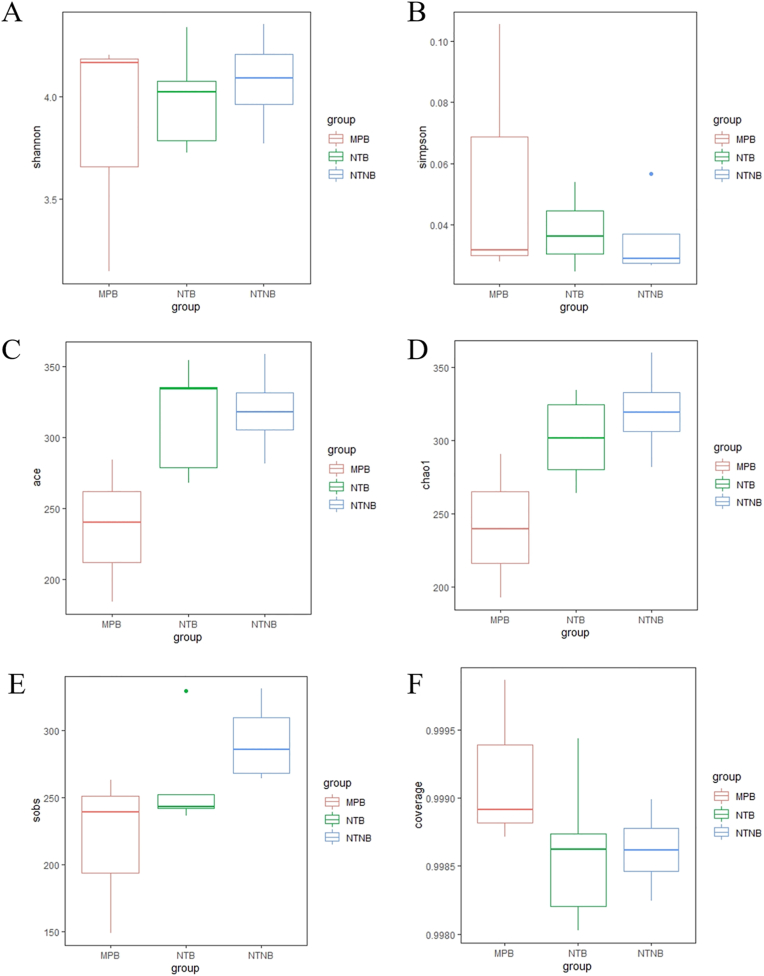
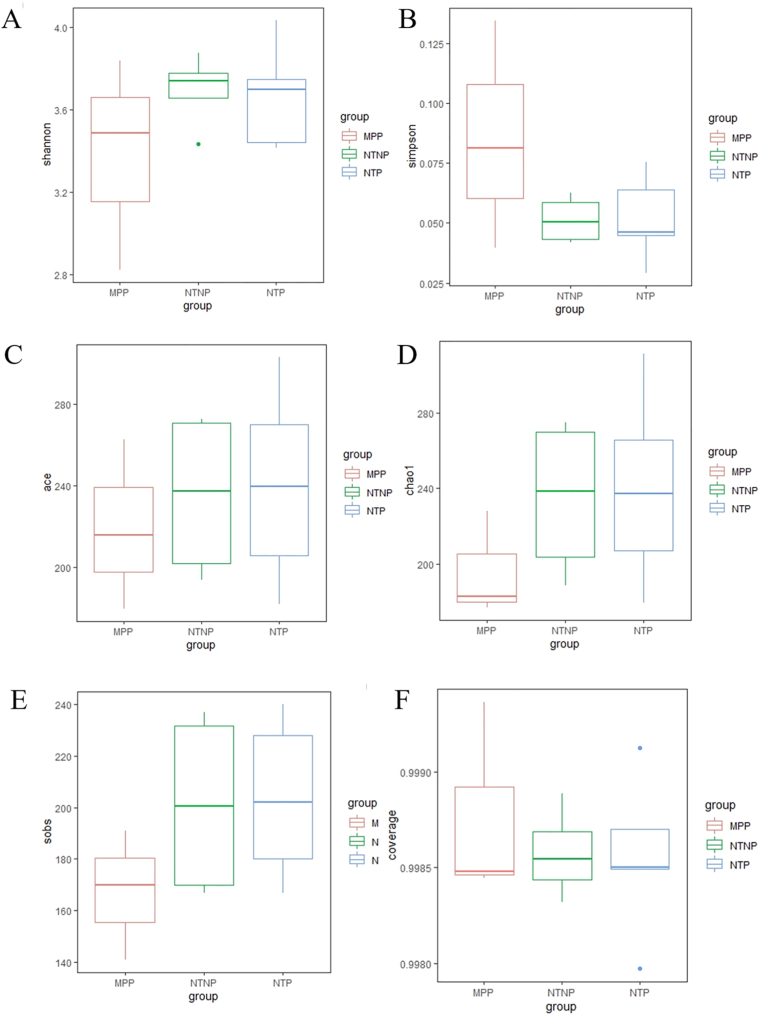
Based on results from alpha-diversity analysis (Fig. 6 and 7), including the Shannon index, Simpson index, ACE index, Chao1 index, sobs index and the coverage index in the sub-mucosal and supra-mucosal samples, no statistically significant difference was found across the three implant surface categories.
Fig. 6.
Boxplots showed the Alpha diversity metrics in the sub-mucosal samples among groups. The comparisons of the Alpha diversity metrics include Shannon (A), Simpson (B), ACE (C), Chao1 (D), sobs (E) and coverage (F). The error bars show the respective minimum and maximum values; the thick line represents the median value; the boxes demarcate the 1st to 3rd quartiles (Q1-Q3), and the abnormal value is shown as ‘o'.
Fig. 7.
Boxplots showed the Alpha diversity metrics in the supra-mucosal samples among groups. The comparisons of the Alpha diversity metrics include Shannon (A), Simpson (B), ACE (C), Chao1 (D), sobs (E) and coverage (F). The error bars show the respective minimum and maximum values; the thick line represents the median value; the boxes demarcate the 1st to 3rd quartiles (Q1-Q3), and the abnormal value is shown as ‘o'.
3.6. Beta-diversity comparisons
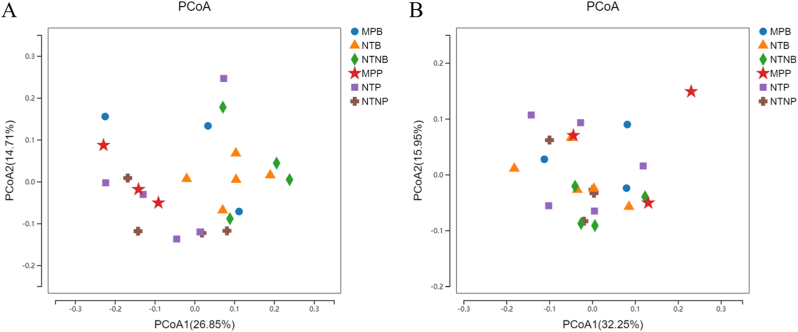
To compare the respective community structures within each titanium surface, the overall bacterial community compositions in each group were compared based on their unweighted and weighted UniFrac distances, and visualized in a PCoA plot. As shown in Fig. 8, a PCoA plot was generated with two principal axes (PCoA1 and PCoA2), which respectively explained 26.85% and 14.71% of the variance based on the unweighted UniFrac distances (32.25% and 15.95% weighted) amongst the bacterial communities on each kind of titanium surface. No clear patterns (clustering arrangements of different samples) were apparent in the PCoA plots colored according to the three titanium surface categories (Permutational multivariate analysis of variance (PERMANOVA): p = 0.31 for weighted UniFrac distances and p = 0.06 for unweighted UniFrac distances).
Fig. 8.
Principal coordinates analysis (PCoA) of the unweighted (A) and weighted (B) UniFrac distances for each of the single site samples. PCoA plots are colored according to different titanium surface categories.
3.7. Differential gene expression
By applying comparative metatranscriptome analysis, we identified major shifts in the overall community activities on different titanium surfaces. Compared with the NTNB group, 10,401 genes were significantly down-expressed and 9497 genes up-expressed in the NTB group (Table S2). Compared with the MPB group, the NTB group showed 9640 up-regulated genes and 9797 down-regulated genes (Table S3). A total of 28,674 genes (14,121 down-regulated genes and 14,553 up-regulated genes) significantly changed their expression levels in NTNB group than in MPB group (Table S4).
3.8. GO functional enrichment analysis
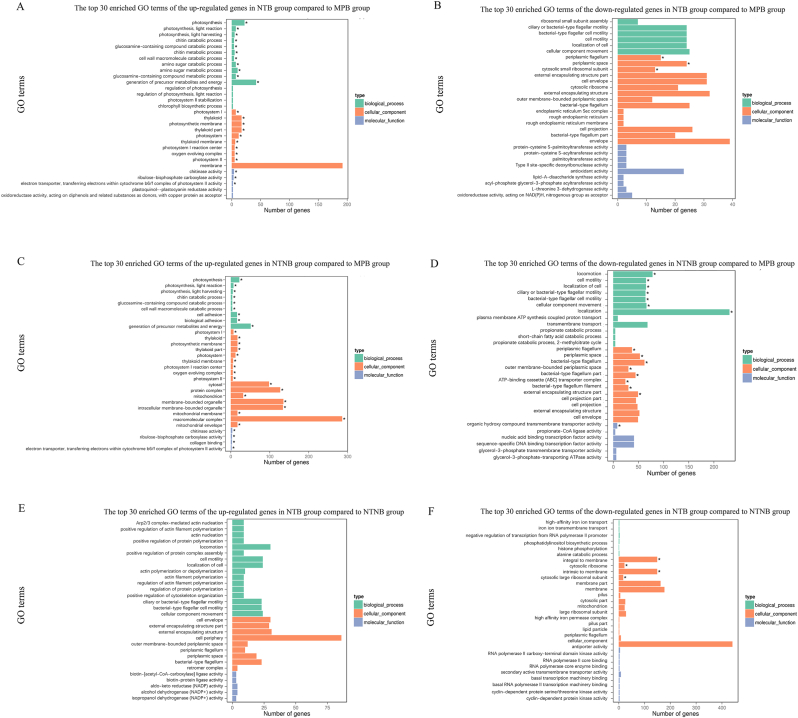
As demonstrated in Fig. 9 and Table S5-S10, a total of 23 and 35 GO terms significantly up-regulated in the NTB group and the NTNB group, respectively, when compared with MPB groups. On the other hand, the NTB group and the NTNB group presented a total of 3 and 16 significantly down-regulated GO terms compared with MPB groups, respectively. The NTB group showed 4 significantly down-regulated GO terms but no significantly up-regulated GO terms when compared to the NTNB group.
Fig. 9.
The top 30 enriched Gene Ontology (GO) terms of the up-regulated (A) and down-regulated (B) differentially expressed genes (DEGs) between NTB and MPB; the top 30 enriched GO terms of the up-regulated (C) and down-regulated (D) DEGs between NTNB and MPB; the top 30 enriched GO terms of the up-regulated (E) and down-regulated (F) DEGs between NTB and NTNB. The Y-axis represents GO terms. The X-axis represents the number of differential genes. The color of the bar represents the type of GO terms. The asterisk (∗) indicates corrected p value < 0.05.
3.9. KEGG pathway enrichment analysis
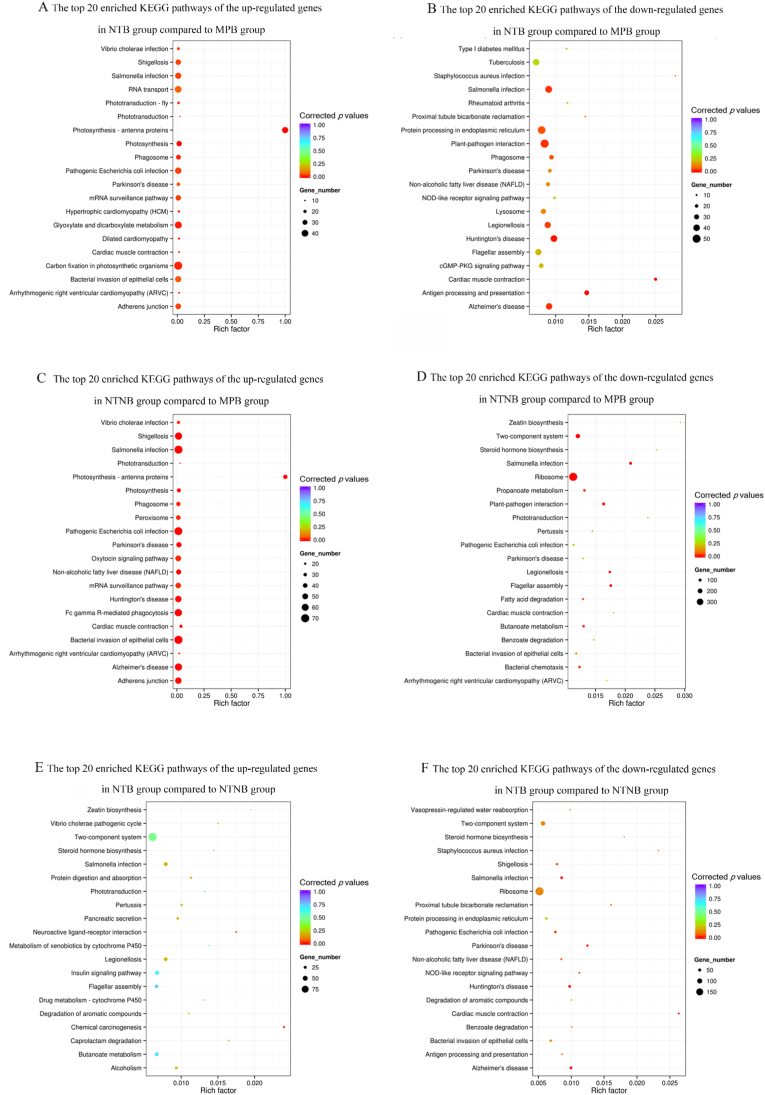
Fig. 10 and Table S11-S16 showed the results of the KEGG pathway enrichment analysis. When compared with the MPB groups, the differentially expressed genes of the NTNB group were enriched in the pathways associated with ribosome, two-component system, Salmonella infection, Shigellosis, bacterial invasion of epithelial cells, pathogenic Escherichia coli infection, oxytocin signaling pathway and mRNA surveillance pathway. The differentially expressed genes of the NTB group compared with the MPB group were mostly enriched in the pathways of glyoxylate and dicarboxylate metabolism, Salmonella infection and protein processing in endoplasmic reticulum. The differentially expressed genes between the NTB group and the NTNB group were significantly enriched in ribosome associated pathway.
Fig. 10.
The top 20 enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) terms of the up-regulated (A) and down-regulated (B) differentially expressed genes (DEGs) between NTB and MPB; the top 20 enriched KEGG terms of the up-regulated (C) and down-regulated (D) DEGs between NTNB and MPB; the top 20 enriched KEGG terms of the up-regulated (E) and down-regulated (F) DEGs between NTB and NTNB.
4. Discussion
The results of this study, together with our previous in vitro experiments [24], have demonstrated that the biological behavior of specific and limited pathogens in the in vitro experiments cannot fully represent the biological performance of the whole sophisticated microbiome established as biofilm on the material surface. Although satisfying results were showed in in vitro study, little is known regarding to the in vivo antibacterial activities of the nanostructured titanium and how nanostructures affected the profiles and functions of the microbial biofilms. In this study, peri-implant microbiota on the different titanium surfaces in the oral cavity of beagle dogs were systematically compared by next-generation sequencing (NGS) technique. We not only described the taxonomy of the oral bacteria sampled, but also profiled and compared the expressed genes, and thus explored the bacterial functions on the different nanostructured surfaces. To the best of our knowledge, it is the first study that applied high-throughput DNA and RNA sequencing technique to evaluate the antibacterial activity of the nanostructured titanium surfaces in vivo.
Based on the results of the 16S rRNA gene sequencing, we found no significant differences in the microbial diversity of the biofilms among the three different titanium surfaces. The α-diversity parameters, indicating both species richness and evenness, in the sub-gingival and supra-gingival microbiota presented on the three kinds of titanium surface were compared (p > 0.05). In addition, the overall composition of the sub-gingival and supra-gingiva microbiota among the three groups was detected by the weighted and unweighted UniFrac metrics. The PCoA plots showed no significant clustering. Together, these results illustrated that the nanostructured titanium had little effect on the community composition of the sub-mucosal and supra-mucosal microbiota established on implant surfaces. No significant difference of diversity and species richness were found among all groups.
The dominant bacterial taxa identified in the sub-mucosal and supra-mucosal niches in this study were in the phyla Bacteroidetes, Proteobacteria and Firmicutes, which were consistent with previous studies in beagles [39,40]. However, a study in human has demonstrated that peri-implantitis microbial communities were enriched with the phyla Bacteroidetes, Spirochetes, and Synergistetes, while Actinobacteria prevailed at healthy sites [41]. This might imply that different hosts or heterogeneity in the methods, such as different sampling techniques, sequencing technique and analysis, may cause the differences of dominant bacterial taxa [42,43].
Despite the fact that the bacterial community composition of peri-implant niches did not show significant differences in our study, DEGs were found between different groups, and were enriched in specific pathways. These results of comparative metatranscriptomic analysis might reveal the underlying mechanisms of antimicrobial effect of implant surface modification. GO functional analysis showed that the differentially expressed genes among the three groups were mainly enriched in the GO terms of biological process and cellular component.
Hydrophilicity and higher surface roughness would promote bacterial adherent and colonization [44]. Although samples of NTN group showed the lowest water contact angle and highest roughness in the current study, the differentially expressed genes were significantly enriched in locomotion and localization in the NTNB group compared to MPB group. Particularly, 231 genes were significantly down-regulated in the GO terms of localization in NTNB group. Meanwhile, there was no significant difference in the expression of genes terms of locomotion and localization between NTB group and MPB group, implying that nanophase calcium phosphate coatings could inhibit bacterial migration and colonization by down-regulating the locomotion and localization-related genes.
Compared to NTB groups, GO terms of integral to membrane, cytosolic ribosome, intrinsic to membrane and cytosolic large ribosomal subunit were significantly up-regulated in the NTNB groups. In our previous study, dead bacteria with deformed and damaged membranes observed by SEM on the surfaces of NTN samples suggested that the sharp nanophase calcium phosphate can affect the integrity of bacterial cell membrane [24]. Taken together, we proposed that the destruction of bacterial cellular membrane on the NTN surface could be one of the vital mechanisms of antibacterial activities of nanostructured titanium surfaces.
KEGG pathway enrichment analysis revealed that over 300 differently expressed genes (DEGs) enriched in the term of ribosome between the NTNB group and MPB group, suggesting that the titanium surface nanostructures affect the gene expression of the microbiota by down-regulating the ribosome-associated pathway. It is noteworthy that several DEGs enriched in the pathway of bacteria-invaded epithelial cells imply that the titanium nanostructures regulated the invasiveness of pathogens. Bacteria invasion into soft tissue is a vital event in the progress of periodontal disease [45,46]. The oral mucosal barrier is exposed to oral commensal microbiome, and a subtle balance is kept through host specific and unspecific immunity. Homeostasis disruption would lead to the periodontal and peri-implant diseases [47]. Healthy peri-implant soft tissue plays a crucial role in maintaining peri-implant health. Collagen fibers in the connective tissue around the implant array in a parallel orientation to the implants, while fibers around teeth perpendicularly and tightly attached to the cementum. Besides, lower density of vascular in the peri-implant connective tissues may make implants more susceptible to peri-implant disease initiation and progression [48]. It can be inferred that titanium nanostructured surface affect the progression of peri-implant diseases through the pathway of bacterial invasion of epithelial cells, whilst the exact mechanism entails further exploration.
From the results of GO functional enrichment analysis, we inferred that the impact on bacterial locomotion and localization, and destruction of cell membrane integrity may be the antimicrobial mechanisms of nanomaterials. KEGG analysis also proved that the metabolic process of bacteria and the ability of bacteria to infect soft tissue may be affected by the nanostructure. In light of these findings, novel nanostructures can be constructed more purposefully to enhance the antibacterial properties of materials in future.
The results of antibacterial activity in vivo were affected by not only the material characteristics but also the condition of the surrounding tissue. Our previous study indicated that calcium phosphate nanoparticles deposited in TiO2 nanotube could enhance the adhesion and proliferation of human gingival epithelial cells (HGECs) and human gingival fibroblasts (HGFs), which indicated that nanostructured titanium surfaces were beneficial to the mucosal barrier [49]. Besides, TiO2 nanotubes can modulate macrophage immune response [50]. Host immune responses and inflammatory state may also influence the complex host-biofilm interaction and therefore influence the composition of the colonizing microbiota and their expressed functions [51].
Although no statistically significant differences in the microbiome composition and diversity were found, differences in metatranscriptomic analysis suggested that the changes in transcriptional level may be prior to changes in the microbiome composition and diversity. The antibacterial mechanism of nanomaterials can be further studied and verified through proteomics. This also suggests that the evaluation and estimation of antibacterial properties of materials should be taken with a more comprehensive examination.
By evaluating the effect of biomaterials on microbiome composition and diversity, and bacterial gene transcription in vivo, this work provides a new perspective of antibacterial feature study. This study estimated the antibacterial abilities by exploring the differences in the bacterial community composition and their biological function within the oral microbiome associated to 3 different surfaces using high-throughput sequencing. Insignificant differences were observed in the microbiome composition and diversity between the three groups; nevertheless, differentially expressed genes of bacteria were detected, which may help us interpret the underlying mechanisms of antibacterial effect of modified nanostructured titanium surfaces.
However, the empirical results of this study have some limitations. The present investigation is a pilot study based on a relatively limited sample size. Future study with a larger sample size is needed to validate and generalize the findings. Meanwhile, there is also a known heterogeneity between different hosts (beagles and humans) of the oral microbiomes. Thus, the results must be interpreted with caution, and should not be directly transferred onto different hosts. Further studies in the relevant hosts are required to establish the antibacterial properties of biomaterials. Nonetheless, this study has important implications for exploring the antibacterial abilities of biomaterials in future studies. Particularly, it highlights the differences between the traditional in vitro antibacterial experiments focusing on limited number of pathogens and the full sequencing-based examination of the in vivo biofilm microbiome established on the biomaterial surfaces.
5. Conclusion
In the current study, high-throughput sequencing analysis has shown that TiO2 nanotube per se and calcium phosphate coating did not change the diversity and community structure of oral microbiota but affected the genes associated with microbial metabolism, protein synthesis, bacterial locomotion, localization and the integrity of organism cellular membranes. In addition, it may affect the antibacterial activity of nanostructures in vivo by influencing the pathways related to bacterial invasion of epithelial cells. Taken together, this study provided a new strategy for evaluating the antibacterial properties of materials in vivo by comparing the composition differences and differential expressed genes of the supra-mucosal and sub-mucosal biofilm on different titanium surfaces with high-throughput sequencing.
Credit author statement
Hanyu Sun: Investigation, Visualization, Writing - original draft, Writing - Reviewing and Editing. Yuki Chan: Investigation, Data curation, Writing - Reviewing and Editing. Xuan Li: Software, Investigation, Writing - Reviewing and Editing. Ruogu Xu: Investigation. Zhengchuan Zhang: Investigation. Xiucheng Hu: Investigation. Fan Wu: Investigation. Feilong Deng: Supervision, Writing - Reviewing and Editing. Xiaolin Yu: Conceptualization, Methodology, Software, Visualization, Writing - original draft, Writing - Reviewing and Editing.
Ethics approval and consent to participate
This study was ethically approved by the Ethics Committee for Animal Experiments of Sun Yat-Sen University (No. SYSU-IACUC-2018-000207) and performed complying to the national and institutional guidelines.
Data availability statement
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Funding
This work was supported by National Natural Science Foundation of China (No. 81801012).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors sincerely acknowledge Ms. Regina Huang for English editing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2022.100275.
Contributor Information
Feilong Deng, Email: dengfl@mail.sysu.edu.cn.
Xiaolin Yu, Email: yuxlin3@mail.sysu.edu.cn.
Appendix ASupplementary data
The following are the Supplementary data to this article:
References
- 1.Fu J.H., Wang H.L. Breaking the wave of peri-implantitis. Periodontol. 2020;84(2000):145–160. doi: 10.1111/prd.12335. [DOI] [PubMed] [Google Scholar]
- 2.Derks J., Tomasi C. Peri-implant health and disease. A systematic review of current epidemiology. J. Clin. Periodontol. 2015;42(Suppl 16):S158–S171. doi: 10.1111/jcpe.12334. [DOI] [PubMed] [Google Scholar]
- 3.Renvert S., Quirynen M. Risk indicators for peri-implantitis. A narrative review. Clin. Oral Implants Res. 2015;26(Suppl 11):15–44. doi: 10.1111/clr.12636. [DOI] [PubMed] [Google Scholar]
- 4.Daubert D.M., Weinstein B.F. Biofilm as a risk factor in implant treatment. Periodontol. 2019;81(2000):29–40. doi: 10.1111/prd.12280. [DOI] [PubMed] [Google Scholar]
- 5.Berglundh T., Armitage G., Araujo M.G., Avila-Ortiz G., Blanco J., Camargo P.M., Chen S., Cochran D., Derks J., Figuero E., Hämmerle C.H.F., Heitz-Mayfield L.J.A., Huynh-Ba G., Iacono V., Koo K.T., Lambert F., McCauley L., Quirynen M., Renvert S., Salvi G.E., Schwarz F., Tarnow D., Tomasi C., Wang H.L., Zitzmann N. Peri-implant diseases and conditions: consensus report of workgroup 4 of the 2017 World Workshop on the classification of periodontal and peri-implant diseases and conditions. J. Periodontol. 2018;89(Suppl 1):S286–S291. doi: 10.1111/jcpe.12957. [DOI] [PubMed] [Google Scholar]
- 6.Oh S., Daraio C., Chen L.H., Pisanic T.R., Finones R.R., Jin S. Significantly accelerated osteoblast cell growth on aligned TiO2 nanotubes. J. Biomed. Mater. Res. 2006;78:97–103. doi: 10.1002/jbm.a.30722. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y., Luo R., Tan J., Wang J., Lu X., Qu S., Weng J., Feng B. Osteoblast behaviors on titania nanotube and mesopore layers. Regen Biomater. 2017;4:81–87. doi: 10.1093/rb/rbw042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oh S., Brammer K.S., Li Y.S., Teng D., Engler A.J., Chien S., Jin S. Stem cell fate dictated solely by altered nanotube dimension. Proc. Natl. Acad. Sci. U. S. A. 2009;106:2130–2135. doi: 10.1073/pnas.0813200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjursten L.M., Rasmusson L., Oh S., Smith G.C., Brammer K.S., Jin S. Titanium dioxide nanotubes enhance bone bonding in vivo. J. Biomed. Mater. Res. 2010;92:1218–1224. doi: 10.1002/jbm.a.32463. [DOI] [PubMed] [Google Scholar]
- 10.Frandsen C.J., Brammer K.S., Noh K., Johnston G., Jin S. Tantalum coating on TiO2 nanotubes induces superior rate of matrix mineralization and osteofunctionality in human osteoblasts. Mater Sci Eng C Mater Biol Appl. 2014;37:332–341. doi: 10.1016/j.msec.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng Z., Ni J., Zheng K., Shen Y., Wang X., He G., Jin S., Tang T. Dual effects and mechanism of TiO2 nanotube arrays in reducing bacterial colonization and enhancing C3H10T1/2 cell adhesion. Int. J. Nanomed. 2013;8:3093–3105. doi: 10.2147/ijn.s48084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amin Yavari S., Loozen L., Paganelli F.L., Bakhshandeh S., Lietaert K., Groot J.A., Fluit A.C., Boel C.H., Alblas J., Vogely H.C., Weinans H., Zadpoor A.A. Antibacterial behavior of additively manufactured porous titanium with nanotubular surfaces releasing silver ions. ACS Appl. Mater. Interfaces. 2016;8:17080–17089. doi: 10.1021/acsami.6b03152. [DOI] [PubMed] [Google Scholar]
- 13.Stolzoff M., Burns J.E., Aslani A., Tobin E.J., Nguyen C., De La Torre N., Golshan N.H., Ziemer K.S., Webster T.J. Decreased bacterial growth on titanium nanoscale topographies created by ion beam assisted evaporation. Int. J. Nanomed. 2017;12:1161–1169. doi: 10.2147/ijn.s119750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y., Yang Y., Li R., Tang X., Guo D., Qing Y., Qin Y. Enhanced antibacterial properties of orthopedic implants by titanium nanotube surface modification: a review of current techniques. Int. J. Nanomed. 2019;14:7217–7236. doi: 10.2147/ijn.S216175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Z., Liu X., Ramakrishna S. Surface engineering of biomaterials in orthopedic and dental implants: strategies to improve osteointegration, bacteriostatic and bactericidal activities. Biotechnol. J. 2021;16 doi: 10.1002/biot.202000116. [DOI] [PubMed] [Google Scholar]
- 16.Djošić M., Janković A., Mišković-Stanković V. Electrophoretic deposition of biocompatible and bioactive hydroxyapatite-based coatings on titanium. Materials. 2021;14:5391. doi: 10.3390/ma14185391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasan J., Crawford R.J., Ivanova E.P. Antibacterial surfaces: the quest for a new generation of biomaterials. Trends Biotechnol. 2013;31:295–304. doi: 10.1016/j.tibtech.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 18.Mahanta U., Khandelwal M., Deshpande A.S. Antimicrobial surfaces: a review of synthetic approaches, applicability and outlook. J. Mater. Sci. 2021;56:17915–17941. doi: 10.1007/s10853-021-06404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eliaz N., Metoki N. Calcium phosphate bioceramics: a review of their history, structure, properties, coating technologies and biomedical applications. Materials. 2017;10:334. doi: 10.3390/ma10040334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Surmenev R.A., Surmeneva M.A., Ivanova A.A. Significance of calcium phosphate coatings for the enhancement of new bone osteogenesis--a review. Acta Biomater. 2014;10:557–579. doi: 10.1016/j.actbio.2013.10.036. [DOI] [PubMed] [Google Scholar]
- 21.Su Y., Cockerill I., Zheng Y., Tang L., Qin Y.X., Zhu D. Biofunctionalization of metallic implants by calcium phosphate coatings. Bioact. Mater. 2019;4:196–206. doi: 10.1016/j.bioactmat.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ge X., Leng Y., Bao C., Xu S.L., Wang R., Ren F. Antibacterial coatings of fluoridated hydroxyapatite for percutaneous implants. J. Biomed. Mater. Res. 2010;95:588–599. doi: 10.1002/jbm.a.32862. [DOI] [PubMed] [Google Scholar]
- 23.Stich T., Alagboso F., Křenek T., Kovářík T., Alt V., Docheva D. Implant-bone-interface: Reviewing the impact of titanium surface modifications on osteogenic processes in vitro and in vivo. Bioeng Transl Med. 2021;7 doi: 10.1002/btm2.10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu X., Xu R., Yu X., Chen J., Wan S., Ouyang J., Deng F. Enhanced antibacterial efficacy of selective laser melting titanium surface with nanophase calcium phosphate embedded to TiO2 nanotubes. Biomed. Mater. 2018;13 doi: 10.1088/1748-605X/aac1a3. [DOI] [PubMed] [Google Scholar]
- 25.Chernozem R.V., Surmeneva M.A., Krause B., Baumbach T., Ignatov V.P., Prymak O., Loza K., Epple M., Ennen-Roth F., Wittmar A., Ulbricht M., Chudinova E.A., Rijavec T., Lapanje A., Surmenev R.A. Functionalization of titania nanotubes with electrophoretically deposited silver and calcium phosphate nanoparticles: structure, composition and antibacterial assay. Mater Sci Eng C Mater Biol Appl. 2019;97:420–430. doi: 10.1016/j.msec.2018.12.045. [DOI] [PubMed] [Google Scholar]
- 26.Verma D., Garg P.K., Dubey A.K. Insights into the human oral microbiome. Arch. Microbiol. 2018;200:525–540. doi: 10.1007/s00203-018-1505-3. [DOI] [PubMed] [Google Scholar]
- 27.Aguiar-Pulido V., Huang W., Suarez-Ulloa V., Cickovski T., Mathee K., Narasimhan G. Metagenomics, metatranscriptomics, and metabolomics approaches for microbiome analysis. Evol Bioinform Online. 2016;12(Suppl 1):5–16. doi: 10.4137/ebo.s36436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mombelli A., Lang N.P. Clinical parameters for the evaluation of dental implants. Periodontol. 1994;4(2000):81–86. doi: 10.1111/j.1600-0757.1994.tb00008.x. [DOI] [PubMed] [Google Scholar]
- 29.Lang N.P., Joss A., Orsanic T., Gusberti F.A., Siegrist B.E. Bleeding on probing. A predictor for the progression of periodontal disease? J. Clin. Periodontol. 1986;13:590–596. doi: 10.1111/j.1600-051x.1986.tb00852.x. [DOI] [PubMed] [Google Scholar]
- 30.Glavind L., Löe H. Errors in the clinical assessment of periodontal destruction. J. Periodontal. Res. 1967;2:180–184. doi: 10.1111/j.1600-0765.1967.tb01887.x. [DOI] [PubMed] [Google Scholar]
- 31.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li R., Yu C., Li Y., Lam T.W., Yiu S.M., Kristiansen K., Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 34.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., Chen Z., Mauceli E., Hacohen N., Gnirke A., Rhind N., di Palma F., Birren B.W., Nusbaum C., Lindblad-Toh K., Friedman N., Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tarazona S., García-Alcalde F., Dopazo J., Ferrer A., Conesa A. Differential expression in RNA-seq: a matter of depth. Genome Res. 2011;21:2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mao X., Cai T., Olyarchuk J.G., Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- 38.Team R. RStudio Inc.; Boston, MA: 2015. RStudio: Integrated Development Environment for R.http://www.rstudio.com [Google Scholar]
- 39.Jiang Q., Yu Y., Xu R., Zhang Z., Liang C., Sun H., Deng F., Yu X. The temporal shift of peri-implant microbiota during the biofilm formation and maturation in a canine model. Microb. Pathog. 2021;158:105100. doi: 10.1016/j.micpath.2021.105100. [DOI] [PubMed] [Google Scholar]
- 40.Qiao S., Wu D., Wang M., Qian S., Zhu Y., Shi J., Wei Y., Lai H. Oral microbial profile variation during canine ligature-induced peri-implantitis development. BMC Microbiol. 2020;20:293. doi: 10.1186/s12866-020-01982-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanz-Martin I., Doolittle-Hall J., Teles R.P., Patel M., Belibasakis G.N., Hämmerle C.H.F., Jung R.E., Teles F.R.F. Exploring the microbiome of healthy and diseased peri-implant sites using Illumina sequencing. J. Clin. Periodontol. 2017;44:1274–1284. doi: 10.1111/jcpe.12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahrmann P., Gilli F., Wiedemeier D.B., Attin T., Schmidlin P.R., Karygianni L. The microbiome of peri-implantitis: a systematic review and meta-analysis. Microorganisms. 2020;8:661. doi: 10.3390/microorganisms8050661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kensara A., Hefni E., Williams M.A., Saito H., Mongodin E., Masri R. Microbiological profile and human immune response associated with peri-implantitis: a systematic review. J. Prosthodont. 2021;30:210–234. doi: 10.1111/jopr.13270. [DOI] [PubMed] [Google Scholar]
- 44.Hao Y., Huang X., Zhou X., Li M., Ren B., Peng X., Cheng L. Influence of dental prosthesis and restorative materials interface on oral biofilms. Int. J. Mol. Sci. 2018;19:3157. doi: 10.3390/ijms19103157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ji S., Choi Y.S., Choi Y. Bacterial invasion and persistence: critical events in the pathogenesis of periodontitis? J. Periodontal. Res. 2015;50:570–585. doi: 10.1111/jre.12248. [DOI] [PubMed] [Google Scholar]
- 46.Bartold P.M., Van Dyke T.E. An appraisal of the role of specific bacteria in the initial pathogenesis of periodontitis. J. Clin. Periodontol. 2019;46:6–11. doi: 10.1111/jcpe.13046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moutsopoulos N.M., Konkel J.E. Tissue-specific immunity at the oral mucosal barrier. Trends Immunol. 2018;39:276–287. doi: 10.1016/j.it.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ivanovski S., Lee R. Comparison of peri-implant and periodontal marginal soft tissues in health and disease. Periodontol. 2018;76(2000):116–130. doi: 10.1111/prd.12150. [DOI] [PubMed] [Google Scholar]
- 49.Xu R., Hu X., Yu X., Wan S., Wu F., Ouyang J., Deng F. Micro-/nano-topography of selective laser melting titanium enhances adhesion and proliferation and regulates adhesion-related gene expressions of human gingival fibroblasts and human gingival epithelial cells. Int. J. Nanomed. 2018;13:5045–5057. doi: 10.2147/ijn.S166661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qiao X., Yang J., Shang Y., Deng S., Yao S., Wang Z., Guo Y., Peng C. Magnesium-doped nanostructured titanium surface modulates macrophage-mediated inflammatory response for ameliorative osseointegration. Int. J. Nanomed. 2020;15:7185–7198. doi: 10.2147/ijn.S239550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Q., Lu H., Zhang L., Yan X., Zhu B., Meng H. Peri-implant mucositis sites with suppuration have higher microbial risk than sites without suppuration. J. Periodontol. 2020;91:1284–1294. doi: 10.1002/jper.19-0634. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.