Abstract
A possible mechanism of resistance to hydrogen peroxide (H2O2) in Vibrio rumoiensis, isolated from the H2O2-rich drain pool of a fish processing plant, was examined. When V. rumoiensis cells were inoculated into medium containing either 5 mM or no H2O2, they grew in similar manners. A spontaneous mutant strain, S-4, derived from V. rumoiensis and lacking catalase activity did not grow at all in the presence of 5 mM H2O2. These results suggest that catalase is inevitably involved in the resistance and survival of V. rumoiensis in the presence of H2O2. Catalase activity was constitutively present in V. rumoiensis cells grown in the absence of H2O2, and its occurrence was dependent on the age of the cells, a characteristic which is observed for the HP II-type catalase of Escherichia coli. The presence of the HP II-type catalase in V. rumoiensis cells was evidenced by partial sequencing of the gene encoding the HP II-type catalase from this organism. A notable difference between V. rumoiensis and E. coli is that catalase is accumulated at very high levels (∼2% of the total soluble proteins) in V. rumoiensis, in contrast to the case for E. coli. When V. rumoiensis cells which had been exposed to 5 mM H2O2 were centrifuged, most intracellular proteins, including catalase, were recovered in the medium. On the other hand, when V. rumoiensis cells were grown on plates containing various concentrations of H2O2, individual cells had a colony-forming ability inferior to those of E. coli, Bacillus subtilis, and Vibrio parahaemolyticus. Thus, it is suggested that when V. rumoiensis cells are exposed to high concentrations of H2O2, most cells will immediately be broken by H2O2. In addition, the cells which have had little or no damage will start to grow in a medium where almost all H2O2 has been decomposed by the catalase released from broken cells.
There are wide varieties of microorganisms which can live under such unusual conditions as low and high pH, low and high temperatures, high salinity, and/or high hydropressure. These organisms possess specific mechanisms to survive in such environments (11). However, there have been no reports on organisms which inhabit environments with hyperoxidative stress caused by factors such as high concentrations of H2O2.
In the course of normal metabolism of O2 in aerobically growing cells, hydrogen peroxide (H2O2), which is toxic to cells, is mostly generated in the respiratory chain through the incomplete reduction of O2 (8, 9). To counteract the potential hazards of intracellular H2O2, which freely diffuses into cells and harms cell membranes, proteins, or DNAs (4, 7, 20), most organisms possess catalase, a high-molecular-weight, heme-containing protein whose primary function is to destroy H2O2, leaving O2 and water as by-products (5). Challenge by reactive oxygen species occurs from extracellular sources as well as from normal aerobic metabolism. Soil pseudomonads are exposed to H2O2 as they contact plant roots (13, 14), and certain soil fungi also produce H2O2 as a mechanism to compete against those organisms (15). Thus, catalase may also be important in overcoming such a challenge (21).
Recently, Yumoto et al. (27) isolated a bacterium which can grow in the presence of relatively high levels of H2O2 from the drain pool of a fish processing factory. This bacterium is regarded as resistant to such hyperoxidative conditions. The bacterium was gram negative, rod shaped, oxidase positive, facultatively psychrophilic, facultatively anaerobic for both fermentative and respiratory metabolism, and sensitive to the vibriostatic compound O/129 (2,4-diamino-6,7-diisopropylpteridine) (27). In the accompanying paper (28), this bacterium was identified as a new species belonging to the genus Vibrio, Vibrio rumoiensis, from its physiological and biochemical characteristics, analysis of its 16S rRNA sequence, and DNA-DNA relatedness. V. rumoiensis S-1 exhibited an extraordinarily high catalase activity compared with other bacteria (27, 28). The catalase activity in cell extracts of this bacterium was 2 orders of magnitude greater than those of Escherichia coli and Bacillus subtilis (27, 28).
Although the mechanisms of adaptation to high oxidative stress by such microorganisms are still unclear, it is postulated that catalase might be involved in eliminating the toxicity of H2O2. In this study, to determine the possible mechanism of adaptation to hyperoxidative stress in V. rumoiensis S-1, the effects of H2O2 on the growth and structure of V. rumoiensis S-1 cells were compared with those on a spontaneous mutant of V. rumoiensis S-1 lacking catalase activity (strain S-4) and other bacterial strains with normal levels of catalase activity. In order to elucidate the molecular properties of catalase involved in the resistance and survival mechanisms in V. rumoiensis S-1, the induction pattern of the catalase was investigated and the partial nucleotide sequence of the catalase gene was determined.
MATERIALS AND METHODS
Bacterial strains and cultivation.
V. rumoiensis was isolated from the drain pool of a fish processing factory; this drain pool usually contains several hundred micromolar H2O2 (27). E. coli XL1-Blue MRA (P2) was purchased from Stratagene (La Jolla, Calif.). B. subtilis IAM 1026 and Vibrio parahaemolyticus JCM 2147 were obtained from type culture collections. V. rumoiensis cells were cultured in PYS medium (pH 7.5) containing 1% polypeptone, 0.5% yeast extract, and 1% NaCl on a rotary shaker at 200 rpm at 27°C until an absorbance of 2.1 at 600 nm, which is equivalent to 1.97 × 108 cells per ml, was obtained. One milliliter of the inoculum was usually transferred to 100 ml of medium. When the inoculum size was changed, 0.1 to 1,000 μl of the inoculum was transferred to 50 ml of medium containing 5 mM H2O2. Strain S-4 was isolated as a spontaneous mutant completely lacking catalase activity during repeated cultivation of V. rumoiensis S-1 (29). S-4 cells were cultured as described above. E. coli, B. subtilis, and V. parahaemolyticus cells were cultured in Luria-Bertani (LB) medium (pH 7.0) containing 0.5% yeast extract, 1% tryptone, and 1% NaCl on a rotary shaker at 200 rpm at 37°C overnight. When V. rumoiensis S-1 cells and strain S-4 cells were cultured on a plate containing H2O2, both strains were grown in PYS medium on a rotary shaker at 200 rpm at 27°C overnight. Cells harvested at the stationary phase were suspended in sterilized LB medium or PYS medium at a concentration of about 2 × 103 cells/ml. Portions (100 μl) of the cell suspensions were spread on plates containing various H2O2 concentrations from 0 to 400 μM. Hydrogen peroxide-containing plates were prepared by adding a 1 M H2O2 solution to the agar plate (25 ml) so as to give the expected concentration.
Preparation of cell extracts and enzyme assays.
Cells of V. rumoiensis S-1 and other strains were harvested by centrifugation at 10,000 × g for 10 min at 4°C and washed three times with 50 mM Tris-HCl buffer (pH 8.0) containing 20 mM MgSO4. The cells, suspended in 1 ml of the same buffer, were sonicated in an ice-water bath for 5 min with a ultrasonic disrupter (type UD-20; Tomy, Tokyo, Japan) at an output setting of 5 and a duty setting of 50. The sonicates were then centrifuged at 10,000 × g for 10 min at 4°C to remove the cell debris and unbroken cells. The supernatants were used as cell extracts.
Catalase activity was assayed in 100 mM potassium phosphate buffer (pH 7.0) containing 30 mM H2O2 as a substrate and 1 to 10 μl of the enzyme preparation in a final volume of 1 ml at 20°C. The reduction of the amounts of H2O2 was monitored by measuring the optical density of the reaction mixture at 240 nm with a spectrophotometer (type U-3210; Hitachi, Tokyo, Japan) (27). One unit was defined as 1 μmol of H2O2 degraded per min. Protein concentrations were measured by the method of Bradford (3) with the Bio-Rad (Hercules, Calif.) protein assay kit II and bovine serum albumin as a standard.
The concentration of H2O2 was measured by the titanium method as follows: 0.8 ml of the sample solution was added to a solution containing 0.25 ml of 20% H2SO4 and 0.15 ml of 1 M TiSO4, and its absorbance at 408 nm was measured (23).
Cloning of the catalase gene from V. rumoiensis S-1. (i) Probe preparation. (a) Determination of the N-terminal amino acid sequence of the V. rumoiensis S-1 catalase.
V. rumoiensis catalase was purified with a DEAE-Sepharose CL-6B anion-exchange column (Pharmacia, Uppsala, Sweden) and a Sephacryl S-300 gel filtration column (Pharmacia) according to the method of Yumoto et al. (29) (details of enzyme purification and characterization will be published elsewhere). To remove minor contaminants, 1.6 mg of the purified V. rumoiensis S-1 catalase was subjected to native polyacrylamide gel electrophoresis with a 5 to 20% gradient gel (type NPG-520L; Atto, Tokyo, Japan) by the standard method of Laemmli (16) and then blotted to a polyvinylidene difluoride membrane (Millipore, Bedford, Mass.) (25). The membrane was stained with 3% trichloroacetic acid containing 0.1% Ponceau S (Wako, Tokyo, Japan). The catalase band was cut out from the membrane and subjected to analysis with a protein autosequencer (Applied Biosystems 491; Perkin-Elmer, Norwalk, Conn.).
(b) Peptide mapping of V. rumoiensis S-1 catalase.
Fifty micrograms of the purified V. rumoiensis S-1 catalase was digested with 0.5 μg of Lysyl-Endopeptidase (Wako) in 1 ml of 10 mM Tris-HCl buffer (pH 9.0) at 37°C overnight. To inactivate the peptidase, 6 M guanidine-HCl was added to the reaction mixture in a final volume of 5 ml and incubated for more than 30 min at room temperature, and then the mixture was loaded onto a TSK gel ODS-80Ts column (0.46 by 15 cm; Toso, Tokyo, Japan) equipped with a reversed-phase high-pressure liquid chromatography system. Peptides were eluted with a liner gradient of 0 to 100% acetonitrile in 0.1% trifluoroacetic acid at a flow rate of 1 ml/min. The eluates were monitored by measuring the absorbance at 214 nm. Each peptide fragment was lyophilized, diluted in trifluoroacetic acid, and then loaded onto the protein autosequencer (Applied Biosystems 491).
(c) Genomic PCR amplification.
Chromosomal DNA was isolated from V. rumoiensis S-1 cells by using the ISOPLANT kit (Nippon Gene, Tokyo, Japan) according to the manufacturer’s instructions. Four oligonucleotides, P-N, P-1, P-3, and P-4.1, were generated according to the N-terminal amino acid sequence and the amino acid sequences of peptide fragments PF-1, PF-3, and PF-4.1, respectively. Genomic PCR amplification was carried out with these primers and V. rumoiensis S-1 genomic DNA as the template. Genomic PCR was carried out as follows. Five microliters of 10× PCR buffer (Takara, Tokyo, Japan), 5 μl of a 2.5 mM deoxynucleoside triphosphate mixture (Takara), 0.7 μg of genomic DNA, 1 pmol of each oligonucleotide primer, 1.25 U of Taq DNA polymerase (Takara), and double-distilled water were mixed in a final volume of 50 μl. The program for PCR was as follows: 93°C for 5 min, 50°C for 2 min, and 72°C for 3 min for one cycle; 94°C for 1 min, 50°C for 2 min, and 72°C for 3 min for 30 cycles, and then 94°C for 1 min, 50°C for 2 min, and 72°C for 5 min for one cycle.
The nucleotide sequence of each PCR product was determined (see below). Among the PCR products a DNA fragment (PP-327) of 327 bp which had been amplified with primers P-3 (ACIGAIGAIGGIAAITGGGCIATG) and P-4.1 (TCICCIGCITCIGCITCIGT) showed high homology with other E. coli HP II-type catalases and especially with the Haemophilus influenzae HktE catalase (2). This PCR product (PP-327) was amplified and thereafter used as a specific probe for cloning the catalase gene (see below).
(ii) Library construction.
Five-microgram portions of chromosomal DNA of V. rumoiensis S-1 were completely digested with EcoRI and electrophoresed on 0.3% low-melting-point agarose gels. DNA fragments of 5 kb which hybridized with PP-327 were recovered from the gels. The recovered DNA fragments were ligated to λgt11 phage DNA at the EcoRI site with ligation kit version 1 (Takara) and packaged into phages by using an in vitro packaging kit (Stratagene). These phages were used to infect E. coli Y1090r− cells. Infected cells were mixed with 5 ml of LB top agar (LB medium containing 0.7% agarose and ampicillin at 20 μg/ml), plated onto LB plates, and then incubated at 37°C for 12 h.
(iii) Library screening.
In the screening of the V. rumoiensis S-1 genomic library, about 10,000 plaques were transferred to Hybond-N+ membranes (Amersham Life Science, Buckinghamshire, United Kingdom) according to the manufacturer’s instructions. PP-327 (10 μg/ml) labeled by use of the ECL direct nucleic acid labeling kit (Amersham) was used as a probe. These membranes were hybridized with PP-327 to detect positive phages (first screening). Positive phages were plated again and screened with PP-327 to obtain single positive clones (second screening). These experiments were performed as described in the manufacturer’s manual (Amersham).
(iv) Nucleotide sequence of the V. rumoiensis S-1 catalase gene.
The insert DNAs of positive phages from the second screening were purified (24). Each positive DNA fragment was ligated at the EcoRI site of the pBluescript SKII(+) plasmid vector by using ligation kit version 2 (Takara), and these plasmids were introduced into E. coli XL1-Blue cells by the heat shock method (24). The transformants were grown in LB medium containing ampicillin at 50 μg/ml. Each plasmid DNA was isolated by the alkaline lysis method (24). The DNA sequencing reaction was performed with sets of M13 forward (CGACGTTGTAAAACGACGGCCAGT) and M13 reverse (GGAAACAGCTATGACCATGATTAC) primers and the PRISM dideoxy terminator cycle sequence kit (Perkin-Elmer), and then the sequence of the candidate clone (gt11A1) was analyzed with a DNA sequencer (model 377; Perkin-Elmer) according to the manufacturer’s manual. The resulting nucleotide sequence of clone gt11A1 was analyzed by a National Center for Biotechnology Information BLAST search.
RESULTS
Effects of H2O2 on the growth of V. rumoiensis S-1 and strain S-4 cells.
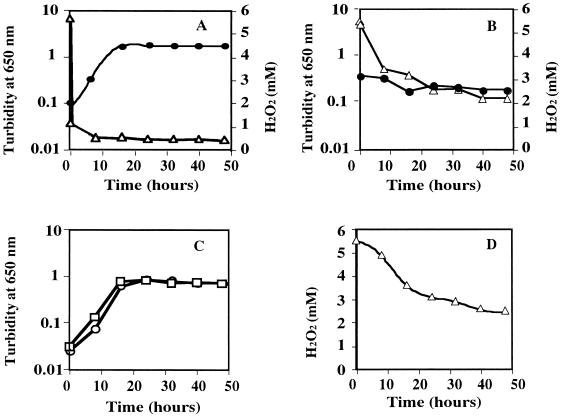
To investigate the effects of H2O2 on growth, V. rumoiensis was cultured either in the presence or in the absence of 5 mM H2O2. As shown in Fig. 1A, V. rumoiensis S-1 cells grew in the presence of 5 mM H2O2, and H2O2 concentration at which E. coli cells cannot grow (6), and the growth reached the stationary phase 16 h after the inoculation. The growth profile of V. rumoiensis S-1 in the absence of H2O2 was almost the same as that in the presence of 5 mM H2O2 (Fig. 1C). When V. rumoiensis S-1 was grown in the presence of 5 mM H2O2, the concentration of H2O2 in the medium decreased immediately after the inoculation of cells, and H2O2 was scarcely detected in the medium 10 min after the inoculation (Fig. 1A). As shown in Fig. 1D, H2O2 spontaneously decomposed to half of the original level within 24 h by shaking without inoculation.
FIG. 1.
Effects of 5 mM H2O2 on growth of V. rumoiensis S-1 and strain S-4. (A) Growth of V. rumoiensis S-1 in 5 mM H2O2 (circles) and concentration of H2O2 in the medium (triangles). (B) Growth of strain S-4 in 5 mM H2O2 (circles) and concentration of H2O2 (triangles). (C) Growth of V. rumoiensis S-1 (squares) and strain S-4 (circles) in medium containing no H2O2. (D) Concentration of H2O2 in medium containing 5 mM H2O2 with no inoculation.
Strain S-4 was obtained as a spontaneous mutant of V. rumoiensis S-1 lacking catalase activity (29). As shown in Fig. 1B, strain S-4 cells could not grow in medium containing 5 mM H2O2, and the H2O2 in the medium was decomposed only spontaneously. V. rumoiensis S-1 and strain S-4 cells showed almost the same growth profile in the absence of H2O2 (Fig. 1C).
Destruction of V. rumoiensis S-1 cells and subsequent release of catalase.
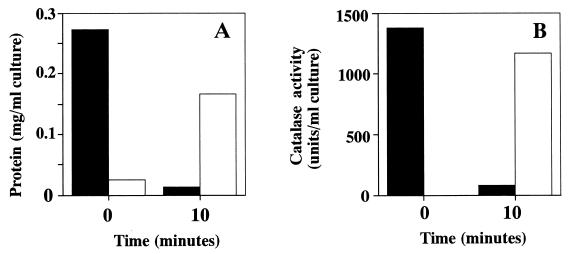
V. rumoiensis S-1 cells accumulate catalase inside the cells, and the catalase is never secreted from the cells (27). To investigate the effects of H2O2 on the cell structure of V. rumoiensis S-1, cells which had been exposed to 5 mM H2O2 were centrifuged, and the localization of the catalase activity was examined. As shown in Fig. 2, most proteins as well as the catalase activity were recovered in the medium after the addition of 5 mM H2O2, whereas there was no catalase activity in the medium before the addition of H2O2. Since H2O2 was scarcely detected in the medium 1 min after the addition of the inoculum (data not shown), H2O2 must be decomposed very quickly by released catalase from disrupted cells.
FIG. 2.
Changes in the amounts of proteins and catalase activity recovered from cells and culture medium of V. rumoiensis S-1 grown in the presence of 5 mM H2O2. (A) Recovered protein in precipitates (solid bars) and in medium (open bars). (B) Recovered catalase activity in precipitates (solid bars) and in medium (open bar).
Effects of the inoculum size of V. rumoiensis S-1 on its growth with H2O2.
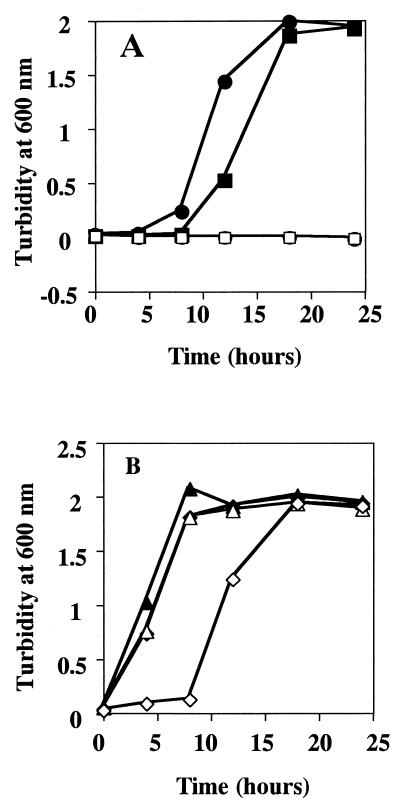
When cells of V. rumoiensis S-1 were grown in the presence or absence of H2O2, their growth profiles were almost the same in some cases (Fig. 1A and 1C), but in other cases a lag time was observed when cells were grown in the presence of H2O2 (data not shown). It was likely that a smaller inoculum size resulted in a much longer lag time in the presence of H2O2 than in its absence. Since the difference in the length of the lag time of growth is considered to be caused by the size of the inoculum, the growth of V. rumoiensis S-1 in 5 mM H2O2 was monitored after inoculation with the various inoculum sizes. As shown in Fig. 3A, a smaller inoculum size brought about a longer lag time. When 1,000-μl (1.97 × 108 cells) and 500-μl (9.85 × 107 cells) portions of the preculture were inoculated into 50 ml of medium, the lag times were 4 and 8 h, respectively. However, no growth was observed when an inoculum of less than 1 μl (1.97 × 105 cells) was used. By contrast, cells grew in medium containing no H2O2 irrespective of the inoculum size (Fig. 3B).
FIG. 3.
Effects of inoculum size on growth of V. rumoiensis S-1 in medium containing 5 mM H2O2. Various volumes of a V. rumoiensis S-1 preculture having an optical density of 2.0 at 600 nm were transferred to 50 ml of medium. (A) A volume of 1,000 μl (closed circles), 500 μl (closed squares), 1 μl (open circles), or 0.1 μl (open squares) of the preculture was transferred to medium containing 5 mM H2O2. (B) A volume of 1,000 μl (closed triangles), 500 μl (closed diamonds), 1 μl (open triangles), or 0.1 μl (open diamonds) of the preculture was transferred to medium containing no H2O2.
Effects of H2O2 concentration on colony formation by V. rumoiensis S-1, strain S-4, and other bacterial strains.
It was considered that the resistance of V. rumoiensis S-1 to H2O2 is primarily due to the endogenously accumulated catalase in cells and/or the released catalase from H2O2-disrupted cells when it was grown in a liquid medium. Thus, the colony-forming ability of individual cells of V. rumoiensis S-1 was compared with that for strain S-4 and other bacterial strains on agar plates containing various concentrations of H2O2. As shown in Table 1, the colony-forming ability of V. rumoiensis S-1 was drastically decreased in the presence of H2O2. At 10 μM H2O2, the colony-forming ability decreased to 68% of the original in V. rumoiensis S-1, whereas 102, 81, and 100% of the original abilities were maintained in V. parahaemolyticus, E. coli, and B. subtilis, respectively. The numbers of colonies on a plate containing 100 μM H2O2 were 35.6, 86.6, 86.1, and 98.5% of the original numbers for V. rumoiensis S-1, V. parahaemolyticus, E. coli, and B. subtilis, respectively. The colony-forming ability of S-4 was very inferior to that of V. rumoiensis S-1. At 100 μM H2O2, strain S-4 completely lost its colony-forming ability (Table 1).
TABLE 1.
Effects of H2O2 concentration on bacterial colony-forming ability
| Species or strain | No. (%) of coloniesa at the following H2O2 concn (μM):
|
||||
|---|---|---|---|---|---|
| 0 | 10 | 100 | 200 | 400 | |
| V. rumoiensis | 315 ± 6 (100) | 215 ± 54 (68.3) | 112 ± 12 (35.6) | 94 ± 5 (29.8) | 10 ± 2 (3.2) |
| Strain S-4 | 179 ± 134 (100) | 75 ± 57 (41.9) | 4 ± 3 (2.2) | 0 ± 0 (0) | 0 ± 0 (0) |
| V. parahaemolyticus | 112 ± 34 (100) | 114 ± 0 (101.8) | 97 ± 8 (86.6) | 21 ± 1 (18.8) | 0 ± 0 (0.0) |
| E. coli | 230 ± 28 (100) | 186 ± 18 (80.9) | 198 ± 17 (86.1) | 149 ± 6 (64.8) | 17 ± 5 (7.4) |
| B. subtilis | 206 ± 5 (100) | 206 ± 7 (100) | 203 ± 18 (98.5) | 184 ± 6 (89.3) | 152 ± 5 (73.8) |
Results are means and standard deviations.
Effects of time of growth on the catalase activity in V. rumoiensis S-1.
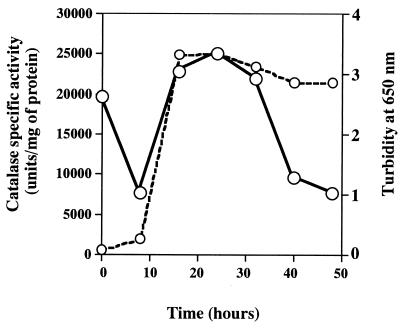
The catalase activity in cell extracts of V. rumoiensis S-1 grown in the absence of H2O2 was assayed. As shown in Fig. 4, the catalase activity of 19,655 units/mg of protein at zero time markedly decreased to 7,627 units/mg of protein at 8 h. The catalase activity then increased and reached its maximum at 25 h (Fig. 4).
FIG. 4.
Changes in the specific activity of catalase in V. rumoiensis cell extracts. Solid line, catalase activity in cell extracts; dashed line, growth curve.
Predicted amino acid sequence of the catalase from V. rumoiensis S-1.
According to the BLAST homology search, the predicted amino acid sequence deduced from the gt11A1 nucleotide sequence showed high homology with the corresponding sequences of E. coli HP II-type catalases and in particular with that of the H. influenzae HktE catalase (Fig. 5). Although clone gt11A1 seemed to contain only a partial sequence of the V. rumoiensis S-1 catalase gene, the predicted amino acid sequence included the same amino acid sequences determined by partial peptide sequencing.
FIG. 5.
Comparison of the partial amino acid sequence of the catalase of V. rumoiensis with the amino acid sequences of the H. influenzae and E. coli HP II-type catalases (2, 26). The partial amino acid sequence of the V. rumoiensis catalase was predicted from the nucleotide sequence of clone gt11A1. Asterisks and dots, identical and similar amino acid residues, respectively. Solid lines, amino acid sequences of the peptide fragments from the peptide mapping experiment.
DISCUSSION
Catalases catalyzing the conversion of H2O2 to water and O2 are found in virtually all aerobic organisms and even in anaerobic organisms. Catalases are thought to be involved in detoxifying H2O2 that has arisen in the cells and also exogenously added H2O2 (1).
As described by Yumoto et al. (27, 28), V. rumoiensis S-1 can survive in environments with high levels of H2O2, and it accumulates a large amount of catalase (levels of 2% of the total soluble proteins inside the cell). As shown in Fig. 1A, when V. rumoiensis S-1 was cultured in a medium where 5 mM H2O2 was present, almost all H2O2 in the medium was quickly decomposed after the inoculation of the cells, and they grew quite normally. On the other hand, strain S-4, a spontaneous mutant of V. rumoiensis S-1 lacking catalase activity, showed no growth in the presence of 5 mM H2O2, and H2O2 in the medium was not decomposed enzymatically (Fig. 1B), suggesting that in this bacterium catalase could play an essential role in decomposing exogenously added H2O2.
Since H2O2 freely diffuses into cells, it was predicted that exogenously added H2O2 could be decomposed by catalase inside the cells and/or by catalase excreted from cells before it diffused into other cells. Although a very small amount of catalase was detected in the culture medium of V. rumoiensis S-1 cells at the stationary phase (28), V. rumoiensis S-1 cells were thought not to excrete catalases under conditions without H2O2 (27). As shown in Fig. 2, most proteins and the catalase activity were recovered in the medium 10 min after the addition of H2O2, suggesting that V. rumoiensis cells were destroyed by H2O2 and catalase was released together with other proteins into the medium. Released catalase could serve to decompose H2O2 in the medium (Fig. 1A). Thus, it is suggested that intact cells and/or less-damaged cells of V. rumoiensis remaining after exposure to high concentrations of H2O2 would begin to grow in this medium, where no H2O2 exists. This scenario implies that individual cells of V. rumoiensis might be rather labile to H2O2.
As shown in Fig. 3, a smaller inoculum size brought about a longer lag time in the growth of V. rumoiensis S-1. This is a very usual and expected phenomenon in bacterial growth. However, when volumes of less than 1 μl (1.95 × 105 cells) of the preculture were inoculated into 50 ml of medium containing 5 mM H2O2, V. rumoiensis S-1 cells exhibited no growth, while the same number of cells of this bacterium did grow, with a lag time of 8 h, in medium containing no H2O2. These results suggest that individual cells of V. rumoiensis S-1 might not have a notable resistance to H2O2 and that they could grow only with low H2O2 concentrations induced by H2O2 decomposition caused by released catalase from cells destroyed by H2O2. This speculation was supported by the findings that individual cells of V. rumoiensis S-1 are significantly inferior in their colony-forming ability to other bacterial strains (see below and Table 1), for which levels of catalase activity were 2 orders of magnitude lower than that for V. rumoiensis S-1.
To exclude the involvement of released catalase from destroyed cells in decomposing exogenous H2O2, V. rumoiensis S-1 cells were grown on agar plates containing various concentrations of H2O2. Thus, the resistance of individual cells to H2O2 could be examined. V. rumoiensis S-1 cells were less resistant to H2O2 than cells of E. coli, B. subtilis, and V. parahaemolyticus (Table 1). Since intracellular levels of catalase in E. coli, B. subtilis, and V. parahaemolyticus were in the range of 10 to 100 U/mg of protein (27), the colony-forming ability on an agar plate would not be associated with the catalase. However, the occurrence of high levels of intracellular catalase seemed to be involved in the colony-forming ability of V. rumoiensis S-1 compared to that of strain S-4 (Table 1). Although the mechanism of disruption of the cell structure is unknown, the bleb structure of the V. rumoiensis S-1 cell envelope (28), which enlarges the surface area of the cell, might be related to the lability of this organism to H2O2. It is likely that V. rumoiensis S-1 and strain S-4 have common structures labile to H2O2 and that the lability of strain S-4 to H2O2 reflects the inherent lability of the cell structure of V. rumoiensis. Some kind of structure unstable to H2O2 and very high-level accumulations of catalase would enable V. rumoiensis S-1 cells to grow in the presence of high concentrations of H2O2. This phenomenon might be designated a self-sacrifice strategy of this bacterium for the maintenance of the species.
In E. coli cells, catalase activity fluctuates markedly depending on the phase of growth (11). For example, the HP I-type catalase, which is induced by H2O2, is produced during the mid-exponential phase, while the HP II-type catalase, a non-H2O2-inducible catalase, is produced during the late exponential to stationary phase (18). As shown in Fig. 4, the total catalase activity dropped at the mid-exponential phase of growth and rapidly increased at the stationary phase in V. rumoiensis S-1 cells grown in the absence of H2O2. A similar profile of change in the HP II-type catalase activity was also found in E. coli cells grown in the absence of H2O2 (10). In E. coli, katE, which encodes the HP II-type catalase, is expressed at high levels in the stationary phase but not in the exponential phase (17, 19) and is controlled by the stationary-phase sigma factor ςs, encoded by rpoS (22). Since V. rumoiensis S-1 produced only one catalase species in the absence of H2O2 (28), it is suggested that the catalase of V. rumoiensis S-1 could be synthesized in a manner similar to that for the HP II-type catalase of E. coli. This suggestion is supported by the fact that V. rumoiensis S-1 has a catalase gene whose predicted amino acid sequence showed high homology with that of the HP II-type catalase of E. coli and, in particular, with that of HktE of H. influenzae (Fig. 5), although a full-length of the catalase gene has not yet been obtained. Whether an H2O2-inducible catalase is present in V. rumoiensis S-1 has never become apparent, because the growth of this bacterium in the presence of H2O2 cannot be examined due to the immediate decomposition of H2O2 in the medium (Fig. 1A).
As described by Yumoto et al. (27), the catalase activity in cell extracts of V. rumoiensis S-1 was 2 orders of magnitude higher than those in E. coli, B. subtilis, and V. parahaemolyticus. These findings suggested that V. rumoiensis S-1 might have multiple copies of the catalase gene and/or a very specific catalytic motif which would enable the catalase to have a highly specific activity. However, V. rumoiensis S-1 had a single copy of the catalase gene on its chromosomal DNA (12). Thus, it is speculated that V. rumoiensis S-1 would have one or more mechanisms for hyperexpression of the catalase gene.
Despite repeated attempts, we have never cloned the complete catalase gene from V. rumoiensis S-1. Only clones lacking 150 to 190 bp at the 5′ region were obtained (Fig. 5), and in some clones, a recombination might have occurred at the same point in the 5′ region of the catalase gene (12). Such an easy rearrangement of the catalase gene in V. rumoiensis S-1 might be related to the occurrence of mutant lacking catalase, like strain S-4.
In conclusion, V. rumoiensis S-1 produced only one catalase species, belonging to the family of the HP II-type catalase of E. coli. The most remarkable difference between the catalase of V. rumoiensis S-1 and other HP II-type catalases is that the catalase in V. rumoiensis S-1 can be synthesized at quite high levels by unknown mechanisms. When V. rumoiensis S-1 cells are exposed to high concentrations of H2O2, cells encountered by H2O2 should be broken and, as a result, released catalase should immediately decompose the exogenous H2O2. Remaining intact cells and/or less-damaged cells of V. rumoiensis S-1 would begin to grow in this medium, where no H2O2 exists. V. rumoiensis S-1 cells apparently are resistant to H2O2. However, individual cells of this bacterium are rather labile to H2O2. Thus, the resistance of V. rumoiensis S-1 to H2O2 and its survival in H2O2 are attributable to high-level accumulation of the intracellular catalase and to the H2O2-labile cell structure of this bacterium.
ACKNOWLEDGMENT
This study was supported by a grant from the Hokkaido Foundation for the Promotion of Scientific and Industrial Technology (Hokscitec).
REFERENCES
- 1.Ames B N, Shigenaga M K, Hagen T M. Oxidants, antioxidants, and the degenerative diseases of aging. Proc Natl Acad Sci USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishai W R, Smith H O, Barcak G J. A peroxide/ascorbate-inducible catalase from Haemophilus influenzae is homologous to the Escherichia coli katE gene product. J Bacteriol. 1994;176:2914–2921. doi: 10.1128/jb.176.10.2914-2921.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;14:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Brot N, Weissbach L, Werth J, Weissbach H. Enzymatic reduction of protein-bound methionine sulfoxide. Proc Natl Acad Sci USA. 1981;78:2155–2158. doi: 10.1073/pnas.78.4.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown S M, Howell M L, Vasil M L, Anderson A J, Hassett D J. Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide. J Bacteriol. 1995;177:6536–6544. doi: 10.1128/jb.177.22.6536-6544.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demple B, Halbrook J. Inducible repair of oxidative DNA damage in Escherichia coli. Nature. 1983;304:466–468. doi: 10.1038/304466a0. [DOI] [PubMed] [Google Scholar]
- 7.Farr S B, Touati D, Kogoma T. Effects of oxygen stress on membrane functions in Escherichia coli: role of HP I catalase. J Bacteriol. 1980;170:1837–1842. doi: 10.1128/jb.170.4.1837-1842.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fridovich I. The biology of oxygen radicals. Science. 1978;201:875–880. doi: 10.1126/science.210504. [DOI] [PubMed] [Google Scholar]
- 9.González-Flecha B, Demple B. Metabolic sources of hydrogen peroxide in aerobically growing Escherichia coli. J Biol Chem. 1995;270:13681–13687. doi: 10.1074/jbc.270.23.13681. [DOI] [PubMed] [Google Scholar]
- 10.Hassan H M, Fridovich I. Regulation of the synthesis of catalase and peroxidase in Escherichia coli. J Biol Chem. 1978;253:6445–6450. [PubMed] [Google Scholar]
- 11.Horikoshi K, Grant W D. Superbugs; microorganisms in extreme environments. Tokyo, Japan: Japan Scientific Societies Press; 1991. [Google Scholar]
- 12.Ichise, N., et al. Unpublished results.
- 13.Katsuwon J, Anderson A J. Characterization of catalase activities in root colonizing isolates of Pseudomonas putida. Can J Microbiol. 1992;38:1026–1032. [Google Scholar]
- 14.Katsuwon J, Zdor R, Anderson A J. Superoxide dismutase activities in a root-colonizing pseudomonads. Can J Microbiol. 1993;39:420–429. doi: 10.1139/m93-061. [DOI] [PubMed] [Google Scholar]
- 15.Kim K K, Fravel D R, Papavizas G C. Identification of a metabolite produced by Talaromyces flavus as glucose oxidase and its role in the biocontrol of Verticillium dahliae. Phytopathology. 1978;78:488–492. [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Loewen P C, Triggs B L. Genetic mapping of katF, a locus that with katE affects the synthesis of second catalase species in Escherichia coli. J Bacteriol. 1984;160:668–675. doi: 10.1128/jb.160.2.668-675.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loewen P C, Switala J, Triggs-Raine B L. Catalases HPI and HPII of Escherichia coli are induced independently. Arch Biochem Biophys. 1985;243:144–149. doi: 10.1016/0003-9861(85)90782-9. [DOI] [PubMed] [Google Scholar]
- 19.Loewen P C. Regulation of bacterial catalase synthesis. In: Scandalios J G, editor. Molecular biology of free radical scanvenging systems. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. pp. 97–115. [Google Scholar]
- 20.McCord J M, Fridovich I. Superoxide dismutase: an enzymatic function for erythrocuprein. J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 21.Miller C D, Kim Y C, Anderson A J. Cloning and mutational analysis of the gene for the stationary-phase inducible catalase (catC) from Pseudomonas putida. J Bacteriol. 1997;179:5241–5245. doi: 10.1128/jb.179.16.5241-5245.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murvey M R, Loewen P C. Nucleotide sequence of katF of Escherichia coli suggests KatF protein is a novel ς transcription factor. Nucleic Acids Res. 1989;17:9979–9991. doi: 10.1093/nar/17.23.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pobiner H. Determination of hydroperoxides in hydrocarbon by conversion to hydrogen peroxide and measurement by titanium complexing. Anal Chem. 1961;33:1423–1426. [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 25.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.von Ossowski I, Mulvey M R, Leco P A, Borys A, Loewen P C. Nucleotide sequence of Escherichia coli katE, which encodes catalase HPII. J Bacteriol. 1991;173:514–520. doi: 10.1128/jb.173.2.514-520.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yumoto I, Yamazaki K, Kawasaki K, Ichise N, Morita N, Hoshino T, Okuyama H. Isolation of Vibrio sp. S-1 exhibiting extraordinarily high catalase activity. J Ferment Bioeng. 1998;85:113–116. [Google Scholar]
- 28.Yumoto I, Iwata H, Sawabe T, Ueno K, Ichise N, Matsuyama H, Okuyama H, Kawasaki K. Characterization of a facultatively psychrophilic bacterium, Vibrio rumoiensis sp. nov., that exhibits high catalase activity. Appl Environ Microbiol. 1999;65:67–72. doi: 10.1128/aem.65.1.67-72.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yumoto, I., et al. Unpublished results.