Abstract
From 24 December 2020 to 8 February 2021, 163 cases of SARS-CoV-2 Alpha variant of concern (VOC) were identified in Chieti province, Abruzzo region. Epidemiological data allowed the identification of 14 epi-clusters. With one exception, all the epi-clusters were linked to the town of Guardiagrele: 149 contacts formed the network, two-thirds of which were referred to the family/friends context. Real data were then used to estimate transmission parameters. According to our method, the calculated Re(t) was higher than 2 before the 12 December 2020. Similar values were obtained from other studies considering Alpha VOC. Italian sequence data were combined with a random subset of sequences obtained from the GISAID database. Genomic analysis showed close identity between the sequences from Guardiagrele, forming one distinct clade. This would suggest one or limited unspecified viral introductions from outside to Abruzzo region in early December 2020, which led to the diffusion of Alpha VOC in Guardiagrele and in neighbouring municipalities, with very limited inter-regional mixing.
Keywords: Abruzzo; SARS-CoV-2, Alpha variant; Epidemiology; Genomic surveillance; Reproduction number
Highlights
-
•
SARS-CoV-2 Alpha VOC has been identified in Guardiagrele (Abruzzo, Italy) starting from late December 2020.
-
•
Epidemiological investigations led to the identification of epi-clusters comprising 163 Alpha VOC cases.
-
•
A reconstructed transmission chain can be used to estimate transmission parameters including Re(t).
-
•
A comparison between sequences in the GISAID database supports limited virus introduction scenario in the area.
1. Introduction
Since late 2020, SARS-CoV-2 variants circulating globally, posing an increased risk to global public health, have been classified as Variants of Interest (VOI) and Variants of Concern (VOC), to prioritize global monitoring and research efforts (World Health Organization WHO, 2021a). These variants are characterised by specific multiple genomic mutations with respect to the Wuhan-Hu strain, that may cause diagnostic detection failures and reduced efficacy of treatments (Centers for Disease Control and Prevention CDC, 2021). In mid-December 2020 a new SARS-CoV-2 variant, belonging to lineage B.1.1.7 (PANGO) and designated as Variant of Concern (VOC) 202012/01 (O’Toole et al., 2021), was detected in the United Kingdom (UK) during routine genomic surveillance (Rambaut et al., 2020). This VOC, re-designated as Alpha (World Health Organization WHO, 2021a), showed a number of mutations, six of which occurring in the spike protein. In particular, the deletion of amino acids 69 and 70 (Δ69–70) on S protein encoding gene was responsible for the so-called spike gene target failure (SGTF), which prevents PCR amplification of the target, thus resulting in a no detectable S-gene when using certain commercial RT-PCR kits (as Thermo Fisher TaqPath COVID-19 assay) (Public Health England, 2021).
The two earliest positive samples to Alpha in the UK had been collected in late September in Kent and Greater London (Rambaut et al., 2020). By 20 December 2020, Alpha had spread geographically in and to London, South East and East of England regions (Public Health England, 2020). In order to contain the spread of the new variant, Italy decided to ground air traffic from the UK from 20 December (Gazzetta Ufficiale della Repubblica Italiana, 2020). As an emergency measure, entrance and transit in Italy was forbidden to people that, in the previous 14 days, had stayed or transited through the UK; whoever had already arrived in Italy in the previous 14 days, and had stayed or transited through the UK before arrival, was obliged to report to the Local Health Authority to be tested (Gazzetta Ufficiale della Repubblica Italiana, 2020).
Alpha showed increased transmissibility and attack rate when compared to non-VOC variants (Buchan et al., 2021, Horby et al., 2021, World Health Organization (WHO), 2021b). A modelling analysis of Alpha cases in the UK, performed on samples collected between 1 October 2020 and 16 January 2021, estimated a growth rate difference of 0.33 per week (95% confidence interval (CI) 0.09–0.62) of this variant with a R0, or reproduction rate, increased by 1.89 (95% CrI 1.43–2.65) (Volz et al., 2021). Moreover, this VOC caused a more severe illness, with an increased risk of hospitalization and death, when compared to non-VOC circulating variants at that time (Bager et al., 2021, Davies et al., 2021b, Horby et al., 2021). A paper concerning the circulation of the Alpha VOC in Abruzzo region (central Italy) suggested that the infection sustained by this variant was characterized by longer persistence and higher viral RNA loads in nasopharyngeal swabs, but no major differences in mortality were observed in the same geographical areas with respect to other circulating variants (Calistri et al., 2021). Nevertheless, immunity from previous infections was overall secured towards Alpha VOC, as secured was the protection against the disease provided by the vaccination (Muik et al., 2021, World Health Organization (WHO), 2021b).
Since the beginning of the pandemic, the Istituto Zooprofilattico Sperimentale of Abruzzo and Molise regions (IZSAM) was appointed by the Italian Ministry of Health to support the molecular diagnosis and genomic characterization of SARS-CoV-2. In addition, IZSAM has been providing epidemiological support to the Local Health Authorities of Abruzzo region (Amato et al., 2021, Cito et al., 2020, Danzetta et al., 2020, Di Giallonardo et al., 2020, Lorusso et al., 2020). From March 2020 onward, IZSAM started to regularly perform whole genome sequencing by Next Generation Sequencing (NGS) on SARS-CoV-2-positive swabs collected in Abruzzo. On 24 December 2020, SARS-CoV-2 Alpha VOC was identified for the first time in four samples collected about one week earlier in Guardiagrele (42°11'26.7"N - 14°13'15.3"E), a town of nearly 9000 inhabitants in the hinterland of Chieti province. Guardiagrele, located on a hill on the foothill of Majella massif, was already in the spotlight of the Local Health Authority because of an unusual increase of COVID-19 cases in December 2020. In the weeks following the first identification of Alpha VOC, the Local Health Authority of Chieti, with the support of IZSAM, performed additional epidemiological investigations in an attempt to identify the possible introduction pathways of the new variant, and to verify the transmission chain. Guardiagrele represented the first Italian cluster of SARS-CoV-2 Alpha VOC to hit the headlines (The Washington Post, 2021). The main epidemiological and genomic findings are addressed in this paper.
2. Materials and methods
2.1. Ethics
The testing and sequencing of suspected SARS-CoV-2 cases and traced contacts in the Abruzzo region was conducted within the official surveillance program established by the Italian Health Authorities, and is exempt from ethical approval.
2.2. Specimens collection and examination
Nasopharyngeal swabs were collected from individuals showing SARS-CoV-2 clinical signs, either hospitalized or not, screened in the framework of contact-tracing activities or monitoring programs for employees of the National Health Care System (Servizio Sanitario Nazionale, SSN). Detection of SARS-Cov-2 RNA in the swabs followed the laboratory procedure and diagnostic methods described in Lorusso et al. (Lorusso et al., 2020). The workflow for SARS-CoV-2 RNA detection followed two main steps: the viral inactivation (PrimeStore® MTM) carried out in a BSL3 biocontainment laboratory, and RNA detection by the TaqManTM 2019-nCoV Assay Kit v2 (Thermo Fisher, qPCR), targeting three different portions of SARS-CoV-2 genome located in the replicase, S and N protein encoding genes. Selected positive samples showing threshold cycle (CT) values less or equal to 25 were further processed by NGS in order to obtain the whole genome sequence of the occurring strains. Genome sequencing was performed as previously described (Di Giallonardo et al., 2020). SARS-CoV-2 lineages were assigned to each sequence using the PANGO COVID-19 Lineage Assigner tool v2.0.7 (Github, 2021). Sequences, once produced, were immediately shared with the GISAID database.
2.3. Study population and epi-clusters
Alpha VOC SARS-CoV-2 confirmed cases detected from 1 December 2020-18 January 2021 in Chieti province were firstly considered in the cluster analysis. When genome sequencing was not possible (i.e. high CT values), spike gene target failure (SGTF) was taken as proxy of Alpha VOC. SGTF is defined as any test with CT < 30 for ORF1ab and N targets but no detectable S gene (Davies et al., 2021b). The presence of SGTF and a link to an Alpha VOC sequenced case were used for assigning these individuals to the Alpha cluster. If PCR results were not available (i.e. sample processed by a different laboratory), the link with the Alpha VOC confirmed case was individually evaluated on the basis of the robustness of the epidemiological information.
Data on confirmed SARS-CoV-2 cases in Chieti province are routinely collected and stored in an electronic database by the Local Health Authority. Personal data (i.e. name, surname, address, municipality, individual fiscal code, date of birth) and epidemiological information (i.e. case unique identification number; date of onset of symptoms; date of sampling and date of diagnosis; date of start of health surveillance; date of recovery; additional remarks like workplace details, school attended, or any other relevant information, and, if known, in-contact case and relationship with the in-contact case) of the study population were retrieved from the Local Health Authority dataset on 8 February 2021.
Data quality was improved by means of clean-up, validation, and update of the original information. Tools and network analysis techniques were used to assist data validation process and rebuild the infectious transmission chains. Cases were grouped into clusters on the basis of epidemiological information (hereafter referred as “epi-clusters”). Links between cases were categorised according to common environments: household/friends, work/occupational, school, health care structures, spatial proximity (households within 5 m), or unknown. A relational spatial-temporal network dataset was created (visNetwork, dplyr and igraph libraries in R environment, version 1.4.1106). To identify all possible transmission chains, a trace forward analysis was performed, using each case as seed and a time window of 66 days, starting from 1 December 2020. The network analysis was conducted to identify sub-network structures providing a greater contribution to the transmission of the infection. Address and municipality information was used to find spatial coordinates through OpenStreetMap and geocoding procedures developed using the R package tidygeocoder (Cambon and Hernangómez, 2021). The detailed contact tracing procedure is described in the Supplementary Information.
Gender, age, presence of symptoms and fatality rate were evaluated. A two-tailed Mann–Whitney test was applied to evaluate differences between the age of symptomatic and asymptomatic cases. Data management was performed by using Microsoft Excel® (Microsoft Corporation, 2013) and Microsoft Access® (Microsoft Corporation, 2013). Statistical analysis was performed by Statistical Software for Excel—XLSTAT (XLSTAT Version 2013.2.04).
2.4. Estimation of the key epidemiological parameters
The effective reproduction number Re(t) represents the number of expected secondary cases deriving from each primary case at time t. Its calculation is used as an indicator of the epidemic trend or to evaluate the effectiveness of interventions. To estimate Re(t), it is essential to quantify the serial interval number or the generation interval (Griffin et al., 2020).
As the dataset of the Alpha VOC cluster contained detailed information on the possible transmission chains, it was used for providing the incidence time series and the corresponding contact chains for the whole outbreak. A procedure to sample plausible exposure times within a reconstructed contact chain was developed. Exposure times, with assumed upper and lower bounds, were calculated from the index case of each epi-cluster. The process was iterated for each epi-cluster. At the end of the iteration 10,000 infection trees were generated. The detailed procedure is described in the Supplementary Information.
2.4.1. Incidence and generation interval
The model described by Cori et al. (Cori et al., 2013) for Re(t) estimation, integrated with the best practice procedures and considerations on Re(t) estimation proposed by Gostic et al. (Gostic et al., 2020), was applied to the obtained infection incidence and generation interval distributions. The average daily infection incidence was calculated across the set of generated trees. The generation interval was determined by first calculating the median of the times for each infector-infected pair. Then, a Bayesian Markov Chain Monte Carlo (MCMC) approach has been used to estimate the distribution parameters, given the median generation intervals and assuming they were Gamma distributed (using the uninformative Uniform(0, 10) as prior for both shape and scale parameters). In this way, the uncertainty of the generation interval parameters is linked to the size of the chain rather than the number of trees generated. The parameters of the final distribution are obtained from the median values of the distributions of the parameters. The BayesTools library was used for the MCMC framework.
2.4.2. Effective reproduction number Re(t)
Re(t) calculation was carried out following two different approaches, one using the infectious transmission chain and the other based on symptoms onset data.
The first approach consists of calculating Re(t) on simulated trees starting from the real infectious transmission chain observed. This method, therefore, allows to determine Re(t) from the date of symptoms onset and to simulate the exposure time. The average case-reproduction number (the number of secondary cases per each case infected on day t) distribution was used to estimate Re(t) from the 10,000 generated trees, similarly to what was done by Hens et al. (Hens et al., 2012).
For comparison to the first approach, it was applied the model described by Cori et al. (Cori et al., 2013), and implemented in R package Epiestim (Cori et al., 2019), which allows estimating Re(t) in a Bayesian framework by having the incidence and the serial interval distribution. Gostic et al., and Knight & Mishra (Gostic et al., 2020; Knight and Mishra, 2020) showed that using infection incidence and generation interval improves the accuracy of the estimations. Thus, estimations derived from generated trees were used to feed the Cori model and to obtain Re(t) on a sliding window of seven days.
2.5. Phylogenetic analysis
A random subset of 1500 Alpha variant sequences was obtained from GISAID database (GISAID, 2021) covering a time span from 1 November 2020 to 10 March 2021. This ‘background’ data was combined with Italian sequence data (n = 608, 18 December 2020–1 March 2021). An alignment was performed in MAFFT applying the L-INS-I algorithm and manually inspected for accuracy using Geneious Prime® 2021.1.1 (Biomatters, 2020). Sequences which did not cover the complete coding region or had > 5% ambiguities were removed. The final alignment consisted of 608 Italian sequences and 1363 global sequences. A P.1 lineage was used as an outgroup (EPI_ISL_833137). A phylogenetic tree was estimated in IQ-TREE using the Hasegawa-Kishino-Yano nucleotide substitution model with a gamma distributed rate variation among sites (HKY+Γ) and an SH-like approximate likelihood ratio test for branch support (1000 replicates). All Italian sequences fell into one node containing 1097 sequences (see Supplementary Information, Fig. S10), and which was used for subsequent analysis. Italian clusters with more than 10 Italian sequences were defined as branches with high node support (>80%) and less than 5% of global sequences.
3. Results
3.1. 163 cases and 14 epi-clusters identified in the outbreak
From 24 December 2020–8 February 2021, 94 cases were confirmed by genome sequencing to be infected by SARS-CoV-2 Alpha VOC. Additional 69 individuals were recognized as suspected to be infected by Alpha VOC because epidemiologically linked to one or more confirmed cases. Of the 69 suspected cases, 64 samples were not suitable for sequencing but showed SGTF. For the remaining five cases the information about PCR CT was not available and they were considered only on the basis of strong epidemiological connections with a confirmed Alpha VOC case.
The overall study population comprised 163 cases: 85 women (52%) and 78 men (47%). The mean age was 46 years, and the most represented age group was 50–59 years old (32 cases, 19.63%). The 76.69% of people (125/163) reported some SARS-CoV-2 clinical symptoms, while 38 cases (23%) were asymptomatic. The median age of symptomatic (51 years, ± 29 years of IQR) and asymptomatic cases (45 years ± 42.75 years of IQR) were not significantly different (p-value = 0.137, two tails Mann—Whitney Test). Two fatalities were recorded within the study population (2/163, 1.23%), in a 63-year-old man and a 41-year-old woman.
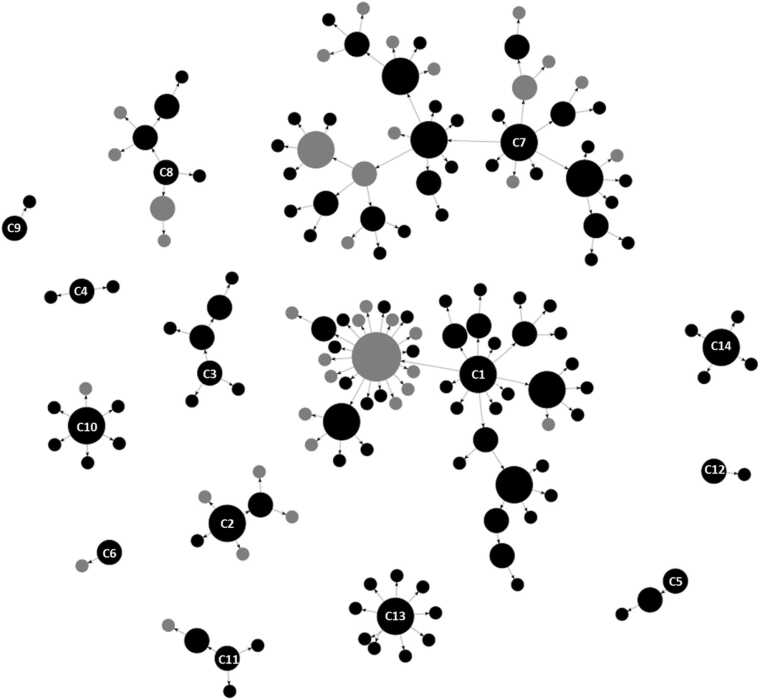
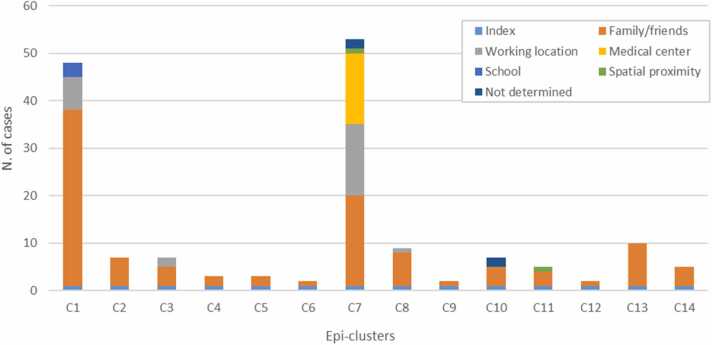
The network comprised 149 transmission links (Fig. 1). Cases were grouped into 14 epi-clusters according to a) reported connections with other cases, b) spatial proximity and/or c) any additional information. About two third of contacts (100/149; 67.11%) were recorded in the family/friends context; 25 connections (16.78%) were related to working locations and 15 connections (10.07%) were linked to a common residential medical centre. The remaining contacts were associated to the same school (3/149; 2.01%), spatial proximity (2/149; 1.34%) or were not determined (4/149; 2.68%) (Fig. 2). Most of the cases (97/163, 59.51%) were residents in the town of Guardiagrele, while the others belonged to 15 other municipalities located in Chieti province and in the bordering provinces of Pescara and L’Aquila.
Fig. 1.
Network based on epidemiological data. 14 apparently unrelated epi-clusters were identified, with 163 cases (circles) and 149 transmission links (arrows). Epi-clusters are numbered (number located on the index case) and circles’ diameter is proportional to the number of secondary cases. Color of the circles represent date of symptoms' onset: recorded (black circles) or not (grey circles). Arrows represent the transmission chain, from the infector to the infected.
Fig. 2.
Epi-clusters and nature of connections between cases. One-hundred contacts happened in the family/friends context; 25 connections were related to working locations, 15 connections were linked to a residential medical center; three cases were associated to a fourth one because of attending the same school; spatial analysis highlighted two connections between cases, and, lastly, the nature of four connections reported by the Local Health Authorities was not specified.
Two large epi-clusters (C1 and C7) were documented. C1, which consists of 48 cases, was the first to occur. The acknowledged index case reported to the Local Health Authority an unspecified contact with people coming from outside of the region. C7 was the largest identified epi-cluster and it was linked to a medical centre. Among the 53 cases of C7, 15 were inpatients of the centre, 15 workers (health personnel and cleaning workers) and 19 related family members and friends. C3-C6, and C8-C14 were mostly family-related epi-clusters (from 2 to maximum 10 cases each). C8 and C3 also included work-related transmission links. It is noteworthy that C2 epi-cluster, apparently unrelated to any other cluster, was directly caused by the return from the UK of a person, who resulted positive to nasopharyngeal swab after arrival at home, with the subsequent infection of other six members of the family. With the exception of C2, although clear direct epidemiological links were not demonstrated between the other 13 epi-cluster, they are all linked to the town of Guardiagrele, because of the epidemiological links of people living or working in the town.
3.2. Re(t) and generation interval estimated using real data
The average and the standard deviation of the median generation intervals (for each infector-infected person pair over the 10,000 trees) resulted in 4.7 days and 2.9 days, respectively.
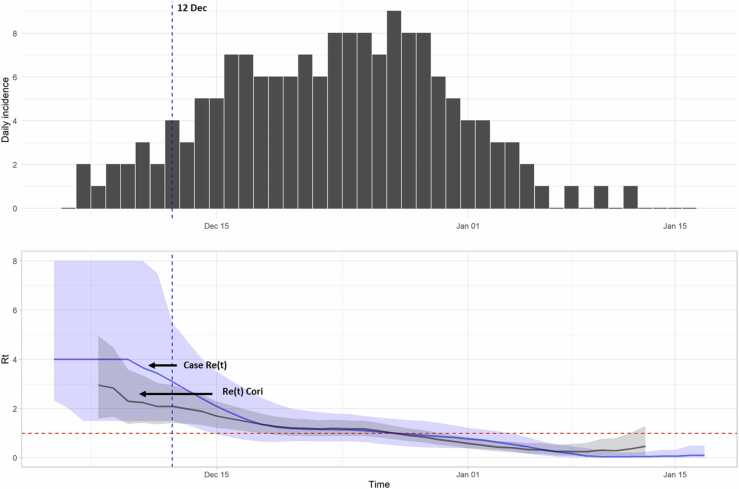
The infection incidence curve estimated by the generated trees is shown in Fig. 3 (upper panel). The overall period considered ranges from 4 December 2020–17 January 2021, resulting in eight days back shifted with respect to the onset of the first symptoms (12 December) and 1 day before the date of the last positive test. The curves of the estimated Re(t) values distribution according to the model described by Cori et al. (Cori et al., 2013), and considering the average case-reproduction number (the number of secondary cases per each case infected on day t), were reported in Fig. 3 (lower panel). Although in the initial growth phase, i.e. during the first 10 days of the epidemic, case Re(t) shows a greater dispersion and higher values than the Re(t) Cori distribution, the Re(t) curves show a similar trend, and both of them reach the value 1 when the infection incidence achieves the peak (maximum number of 9 cases on 27 December).
Fig. 3.
Infection incidence and estimated incidence. The vertical dashed blue line represents the first symptom onset (12 Dec, blue line). Upper panel: infection incidence (bars). Lower panel: Re(t) and 95% CI estimated by Cori model (grey area), and from case reproduction number (blue area); the dashed red line represents Re(t)= 1.
3.3. All sequences from Guardiagrele form one distinct clade
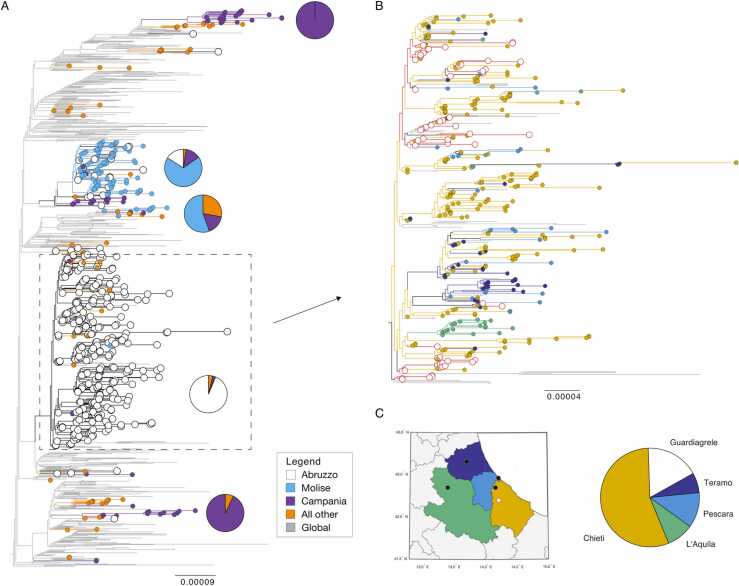
All Italian Alpha VOC sequences fell within one node in the global phylogeny. There was an overrepresentation of 61% of sequences from infections in Abruzzo (n = 372), and 16% and 12% from Molise (n = 99) and Campania regions (n = 70), respectively. The phylogeny reveals limited mixing between the sequences detected in Italy and those abroad (Supplementary Information, Fig. S10). We identified six larger clades with more than 10 Italian sequences and node support > 80%. These contained 26, 129, 19, 383, and 14 sequences, respectively (Fig. 4A). Within these phylogenetic clades most infections were sampled within one region only, but infections from the same region were scattered across different clades (Fig. 4A). We found two clades with infections from Campania (clades 1 and 5), two clades with infections from Molise (clades 2 and 3), and also two clades with infections from Abruzzo (clades 2 and 5). This suggests multiple introductions of Alpha VOC into different regions with very limited subsequent inter-region transmission. Notably, the four sequences linked to C2-epiclusters were not linked to any of these six clades but instead formed independent branches on the phylogeny.
Fig. 4.
Phylogeny of B.1.1.7 in Italy. (A) Maximum likelihood tree showing 1097 full-genome SARS-CoV-2 sequences. Branches are colored according to Italian regions; global data is shown in grey. For the five main Italian clusters the proportion of sequences per region is shown as a pie chart. (B) The largest clusters containing all sequences from Guardiagrele is shown enlarged (n = 383). Branches are colored according to the province in Abruzzo. Sequences from Guardiagrele are marked with white. (C) The provinces in Abruzzo and the proportion of sequences from each province in the large cluster. Branch length shows nucleotide substitutions per site.
All sequences from Guardiagrele form one distinct clade containing 383 infections, including 12 sequences sampled outside of Italy (Fig. 4A, clade 5). This largest clade contained 341 sequences from infections in Abruzzo samples between 18 December 2020 and 1 March 2021, lasting over 67 days. The sequences were labelled according to the Abruzzo province of sampling: L’Aquila, Teramo, Pescara, and Chieti (Fig. 4B). The latter was further subdivided into Guardiagrele (which is part of Chieti) and all other towns in this province. Within this clade, 55% (n = 188) of Abruzzo sequences were from Chieti, and additional 17% (n = 59) were from Guardiagrele only (Fig. 4C). Notably, the first infection was sampled in Guardiagrele on 18 December and only three days later in Chieti (21 December), then on the 28 December in L’Aquila, 29 December in Pescara, and lastly on the 30 December in Teramo. We could not determine the exact number of within and between province transmission due to low node support. However, the phylogeny does not exclude between province transmission as sequences from all four provinces are scattered across the phylogeny. Although, such mixing was limited.
4. Discussion
Following the first detection of the SARS-CoV-2 Alpha VOC in Guardiagrele on 18 December 2020, Italy witnessed the spread of this VOC also in other regions. Indeed, a prevalence study on SARS-CoV-2 variants performed on 4 February 2021 all over the country, demonstrated the progressive spread of Alpha VOC across Italy (Istituto Superiore di Sanità, 2021a) with the 54% of all new cases of infections caused by this variant. This percentage increased up to 86.7% in a second survey performed in mid-March and over 91% in a third survey conducted in late April, supporting for the higher transmissibility of this variant (Istituto Superiore di Sanità, 2021b, Istituto Superiore di Sanità, 2021c). The spread of Alpha VOC coincided with the third-wave of cases in Italy, which hit the country from February to May 2021.
From a certain point of view, the detection of the new Alpha VOC in the town of Guardiagrele was not a surprise. This town, despite not being a commercial hub or located in an industrial district, was already classified as one of the most vulnerable municipalities of Abruzzo region for SARS-CoV-2 spread (Savini et al., 2020), due to the proximity of the town to the most industrial hub of the region, in the Sangro Valley (42°08'41.2"N - 14°26'42.0"E), where several medium and large factories are present.
Noteworthy, social distancing measures were already in place in Abruzzo region when the outbreak in Guardiagrele started. The region was classified as “red area” from mid-November 2020–7 December and from 23 December until 7 January 2021, and “orange area” during the period in between. Red is the highest level of alert and it foresees the ban of people movement, even within the municipalities, apart from those for urgent needs related to health or necessary works (e.g.: linked to food supply production chain, health workers, etc.). The measures foreseen in the orange area include the ban of movements across regions, except for urgent or working reasons. However, the restrictions in place imposed by the partial lockdown appeared not able to stop the spread of the Alpha VOC in a context like Guardiagrele, where the transmission occurred, at least for the two thirds, in family settings.
When the period of time before the official confirmation of the first cases is taken into consideration (before the 12 December 2020), the Re(t) calculated according to the model of Cori et al. (Cori et al., 2013) was higher than 2 (Fig. 3).
A systematic review of 29 studies on the reproductive number for SARS-CoV-2 transmission showed an overall estimation of 2.87 (95% CI: 2.39–3.44), under several different conditions, with the highest values reported for the Diamond Princess Cruise Ship in Japan (14.8), followed by some country-level estimations, as those reported for France (6.32, 95% CI: 5.72–6.99), Germany (6.07, 95% CI: 5.51–6.69) and Spain (3.56, 95% CI: 1.62–7.82) (Billah et al., 2020).
These values seem clearly higher than our estimation. However, when the Alpha VOC is considered and studies more similar to ours are taken into account, the distance appears less important. Using the method described in Abbott (Abbott, 2020) and in Sherratt et al. (2021), and implemented in the EpiNow2 R package (Abbott et al., 2020a), Davies et al. (Davies et al., 2021a) estimated Re(t) values of Alpha VOC, from October 2020 to January 2021, for some regions of England, which varied from 1.01 to 1.04 (Davies et al., 2021a). In Switzerland, between 01 January and 17 January 2021, before the introduction of more severe control measures, the reproductive number for the Alpha VOC was significantly above 1 (1.24 [1.07–1.41], and 1.46 [1.21–1.72] according two different datasets) (Chen et al., 2021).
Nevertheless, considering the specific approach followed in our work, the results of any comparison of Re(t) values estimated by us with those reported in previous published papers should be carefully interpreted. In fact, our estimations are strictly linked to the epidemiological situation observed in the area under study during the time window considered, thus influenced by the control measures in place, the people’s behaviour and the level of immunity of the population.
In our approach, the median generation intervals across the set of generated trees have been used to estimate the distribution parameters, assuming they were Gamma distributed. The uncertainty of the generation interval parameters is linked to the size of the chain rather than to the number of generated trees. However, this approach could be biased by the variance reduction of the estimated generation interval distribution. In the cluster observed in Guardiagrele, social distancing measures, strict trace back, testing and quarantine measures were in place, which probably contained and eventually interrupted the viral transmission. In addition, the lack of epidemiological links between some observed cases in Guardiagrele may have led to underestimate the real Re(t) as well as the assumptions made in the model (e.g. considering the incubation period of 1–15 days) may limit the comparability of our results with those of other authors.
As mentioned, the available dataset did not include the full transmission chain. Therefore, we decided to apply the EpiEstim method to overcome the availability of only incomplete epidemiological data (i.e., chain of transmission, introduction of the virus, case zero, etc.). Also, we attempted to identify a plausible chain of transmission, based on available data. As reported in literature, many models could be applied for the calculation of Re(t) when the chain of transmission is unknown, but the dates of clinical onset of symptoms are available (Abbott et al., 2020a, Abbott et al., 2020b). One of the first described is the one proposed by Cori et al. (Cori et al., 2013), and later improved in the EpiEstim package (Cori et al., 2019). However, the main limits of Cori’s approach have been reported by Gostic et al. (Gostic et al., 2020). The method is indeed distorted as for misspecifications of the generation interval and because of existing lag between infection’s incidence and symptoms’ incidence.
Although the limits of the method presented by Cori and colleagues have been exceeded by other packages (Abbott et al., 2020a), in the present work we applied the EpiEstim package also because, at the time of our study, EpiEstim was the standard method applied by the Italian National Institute of Health when defining restriction areas and/or regions in Italy (Istituto Superiore di Sanità, 2021d). The proposed method aimed at temporally relating the estimated case reproduction number to infections date; in our view, this approach would have been more useful than obtaining estimates based on symptoms’ onset dates. In fact, in this second case, the impact of control measures would have been observed with a temporal delay because of the incubation period distribution. Therefore, we think that our approach could be used to evaluate applied control measures in a timelier manner.
Although a case-by-case comparison between genetic and epidemiological data cannot be done, since sequence data could be used only for 59 of 163 cases belonging to the Guardiagrele cluster, the results of the genetic analyses do not seem to contradict the main findings of the epidemiological investigations. The phylogenetic analysis shows that the sequences obtained from the cases linked to this cluster are closely related to each other, with a limited mixing with other SARS-CoV-2 Alpha VOC sequences from the same province (Fig. 4). The sole exception is represented by the sequences belonging to the epi-cluster C2, falling within a different node, thus confirming the different introduction pathway as already resulted from the epidemiological investigations.
In particular, the results of the genetic analyses do not seem to contradict the hypothesis arising from the epidemiological investigations about the possibility that the observed cluster in Guardiagrele, with the exception of C2 epi-cluster, might derive from a single introduction from outside occurred in early December 2020. Although multiple introductions cannot be excluded, the close similarity of genome sequences would suggest that a single common source of infection caused the Alpha VOC SARS-CoV-2 cluster in Guardiagrele. The observed differences among the sequences might be coherent with a local independent evolution during the outbreak, with no or limited re-introduction from outside the cluster. The limited mixing between geographic regions investigated might be also due to the lockdown restrictions imposed and the reduced number of travels allowed across the country.
However, several factors may have influenced and partially biased the results of our epidemiological investigations. The lack of evidence of epidemiological connections among the observed epi-clusters may indicate the existence of multiple infection sources or, more coherently with the results of the genetic analyses, may be linked to the difficulties of identifying all possible social connections among cases and exposure opportunities, which can easily occur in a small town like Guardiagrele. Likewise, hesitancy in epidemiological interviews may have interfered with the identification of linkages within and between epi-clusters.
5. Conclusions
The methodological approach presented in this study allowed to estimate in a more accurate way the Re(t) and other transmission parameters (generation interval and incubation period), using data from the epidemiological investigations carried out on a cluster of SARS-CoV-2 Alpha VOC infection, identified in a town of Abruzzo region on late December 2020.
The whole genome sequencing of viral RNA present in the nasopharyngeal swabs of cases belonging to the outbreak under investigation partially confirmed that the virus belonged to the same phylogenetic node, suggesting that one or limited unspecified introductions from outside in early December 2020 led to the diffusion of Alpha VOC in Guardiagrele and in the neighbouring municipalities.
Funding
This work was supported by funding from the European Union’s Horizon 2020 Research and Innovation programme (One Health European Joint Programme under grant agreement No 773830, Recipient: Alessio Lorusso) and by funding from the Italian Ministry of Health (Ricerca Corrente 2020, PanCO “Epidemiologia e Patogenesi dei coronavirus umani ed animali”, recipient Alessio Lorusso, and Ricerca Strategica 2020, “Suscettibilità dei mammiferi a SARS-COV-2: rischi di zoonosi inversa e possibilità in medicina traslazionale”, recipient Alessio Lorusso).
CRediT authorship contribution statement
Laura Amato: Methodology, Formal analysis, Data curation, Writing – original draft, Writing – review & editing. Luca Candeloro: Methodology, Software, Formal analysis, Writing – original draft, Writing – review & editing. Arturo Di Girolamo: Conceptualization, Formal analysis, Data curation, Investigation. Lara Savini: Methodology, Software, Formal analysis, Writing – original draft, Writing – review & editing. Ilaria Puglia: Methodology, Data curation. Maurilia Marcacci: Methodology, Data curation. Marialuigia Caporale: Methodology, Data curation. Iolanda Mangone: Methodology, Data curation. Cesare Cammà: Methodology, Data curation. Annamaria Conte: Conceptualization, Methodology, Software, Writing – review & editing. Giuseppe Torzi: Conceptualization, Investigation, Resources, Supervision. Adamo Mancinelli: Conceptualization, Investigation, Resources. Francesca Di Giallonardo: Methodology, Software, Data curation, Writing – review & editing. Alessio Lorusso: Methodology, Data curation, Writing – review & editing, Funding acquisition. Giacomo Migliorati: Supervision, Project administration, Funding acquisition. Thomas Schael: Supervision, Project administration, Resources, Funding acquisition. Nicola D’Alterio: Supervision, Project administration, Funding acquisition. Paolo Calistri: Conceptualization, Methodology, Formal analysis, Writing – original draft, Visualization, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the IZSAM.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.epidem.2022.100578.
Contributor Information
Laura Amato, Email: l.amato@izs.it.
Luca Candeloro, Email: l.candeloro@izs.it.
Arturo Di Girolamo, Email: arturo.digirolamo@asl2abruzzo.it.
Lara Savini, Email: l.savini@izs.it.
Ilaria Puglia, Email: i.puglia@izs.it.
Maurilia Marcacci, Email: m.marcacci@izs.it.
Marialuigia Caporale, Email: mr.caporale@izs.it.
Iolanda Mangone, Email: i.mangone@izs.it.
Cesare Cammà, Email: c.camma@izs.it.
Annamaria Conte, Email: a.conte@izs.it.
Giuseppe Torzi, Email: giuseppe.torzi@asl2abruzzo.it.
Adamo Mancinelli, Email: adamo.mancinelli@asl2abruzzo.it.
Francesca Di Giallonardo, Email: fdigiallonardo@kirby.unsw.edu.au.
Alessio Lorusso, Email: a.lorusso@izs.it.
Giacomo Migliorati, Email: g.migliorati@izs.it.
Thomas Schael, Email: direzione.generale@asl2abruzzo.it.
Nicola D’Alterio, Email: n.dalterio@izs.it.
Paolo Calistri, Email: p.calistri@izs.it.
Appendix A. Supplementary material
Supplementary material
.
References
- Abbott, S., 2020. epiforecasts/EpiNow2: Initial release [WWW Document]. 10.5281/ZENODO.3957490. [DOI]
- Abbott S., Hellewell J., Sherratt K., Gostic K., Hickson J., Badr H.S., DeWitt M., Thompson R., EpiForecasts, Funk S. (2020a). EpiNow2: Estimate Real-Time Case Counts and Time-Varying Epidemiological Parameters. doi: 10.5281/zenodo.3957489.
- Abbott S., Hellewell J., Thompson R.N., Sherratt K., Gibbs H.P., Bosse N.I., Munday J.D., Meakin S., Doughty E.L., Chun J.Y., Chan Y.-W.D., Finger F., Campbell P., Endo A., Pearson C.A.B., Gimma A., Russell T., Flasche S., Kucharski A.J., Eggo R.M., Funk S. Estimating the time-varying reproduction number of SARS-CoV-2 using national and subnational case counts. Wellcome Open Res. 2020;5:112. doi: 10.12688/wellcomeopenres.16006.2. [DOI] [Google Scholar]
- Amato L., Jurisic L., Puglia I., Di Lollo V., Curini V., Torzi G., Di Girolamo A., Mangone I., Mancinelli A., Decaro N., Calistri P., Di Giallonardo F., Lorusso A., D’Alterio N. Multiple detection and spread of novel strains of the SARS-CoV-2 B.1.177 (B.1.177.75) lineage that test negative by a commercially available nucleocapsid gene real-time RT-PCR. Emerging Microbes & Infections. 2021:1148–1155. doi: 10.1080/22221751.2021.1933609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bager P., Wohlfahrt J., Fonager J., Rasmussen M., Albertsen M., Michaelsen T.Y., Møller C.H., Ethelberg S., Legarth R., Button M.S.F., Gubbels S., Voldstedlund M., Mølbak K., Skov R.L., Fomsgaard A., Krause T.G. Risk of hospitalisation associated with infection with SARS-CoV-2 lineage B.1.1.7 in Denmark: an observational cohort study. Lancet Infect. Dis. 2021 doi: 10.1016/s1473-3099(21)00290-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billah M.A., Miah M.M., Khan M.N. Reproductive number of coronavirus: A systematic review and meta-analysis based on global level evidence. PLoS One. 2020;15 doi: 10.1371/journal.pone.0242128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biomatters, 2020. Geneious - Bioinformatics Software for Sequence Data Analysis. Geneious.
- Buchan S.A., Tibebu S., Daneman N., Whelan M., Vanniyasingam T., Murti M., Brown K.A. Increased Household Secondary Attacks Rates With Variant of Concern Severe Acute Respiratory Syndrome Coronavirus 2 Index Cases. Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciab496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calistri P., Amato L., Puglia I., Cito F., Di Giuseppe A., Danzetta M.L., Morelli D., Di Domenico M., Caporale M., Scialabba S., Portanti O., Curini V., Perletta F., Cammà C., Ancora M., Savini G., Migliorati G., D’Alterio N., Lorusso A., D’Alterio N., Lorusso A. Infection sustained by lineage B.1.1.7 of SARS-CoV-2 is characterised by longer persistence and higher viral RNA loads in nasopharyngeal swabs. Int. J. Infect. Dis. 2021;105:753–755. doi: 10.1016/j.ijid.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambon, J., Hernangómez, D., 2021. Tidygeocoder: Geocoding Made Easy. 10.5281/zenodo.3981510. [DOI]
- Centers for Disease Control and Prevention (CDC), 2021. SARS-CoV-2 Variant Classifications and Definitions [WWW Document]. URL 〈https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html〉 (accessed 8.31.21).
- Chen C., Nadeau S.A., Topolsky I., Manceau M., Huisman J.S., Jablonski K.P., Fuhrmann L., Dreifuss D., Jahn K., Beckmann C., Redondo M., Noppen C., Risch L., Risch M., Wohlwend N., Kas S., Bodmer T., Roloff T., Stange M., Egli A., Eckerle I., Kaiser L., Denes R., Feldkamp M., Nissen I., Santacroce N., Burcklen E., Aquino C., de Gouvea A.C., Moccia M.D., Grüter S., Sykes T., Opitz L., White G., Neff L., Popovic D., Patrignani A., Tracy J., Schlapbach R., Dermitzakis E.T., Harshman K., Xenarios I., Pegeot H., Cerutti L., Penet D., Blin A., Elies M., Althaus C.L., Beisel C., Beerenwinkel N., Ackermann M., Stadler T. Quantification of the spread of SARS-CoV-2 variant B.1.1.7 in Switzerland. Epidemics. 2021;37 doi: 10.1016/J.EPIDEM.2021.100480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cito F., Amato L., Di Giuseppe A., Danzetta M.L., Iannetti S., Petrini A., Lorusso A., Bonfini B., Leone A., Salini R., Mancinelli A., Torzi G., Savini G., Migliorati G., Schael T., D’alterio N., Calistri P. A covid-19 hotspot area: Activities and epidemiological findings. Microorganisms. 2020;8:1–11. doi: 10.3390/microorganisms8111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cori A., Ferguson N.M., Fraser C., Cauchemez S. A new framework and software to estimate time-varying reproduction numbers during epidemics. Am. J. Epidemiol. 2013;178:1505–1512. doi: 10.1093/aje/kwt133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cori, A., Kamvar, Z.N., Stockwin, J., Jombart, T., Thompson, R., Dahlqwist, E., 2019. EpiEstim 2.2 [WWW Document]. 10.5281/ZENODO.3333654. [DOI]
- Danzetta M.L., Amato L., Cito F., Di Giuseppe A., Morelli D., Savini G., Mercante M.T., Lorusso A., Portanti O., Puglia I., Monaco F., Casaccia C., Di Gennaro A., Testa L., Migliorati G., D’alterio N., Calistri P. SARS-CoV-2 RNA persistence in naso-pharyngeal swabs. Microorganisms. 2020;8:1–10. doi: 10.3390/microorganisms8081124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., Wenseleers T., Gimma A., Waites W., Wong K.L.M., van Zandvoort K., Silverman J.D., CMMID COVID-19 Working Group, COVID-19 Genomics UK (COG-UK) Consortium, Diaz-Ordaz K., Keogh R., Eggo R.M., Funk S., Jit M., Atkins K.E., Edmunds W.J. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021 doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Jarvis C.I., Edmunds W.J., Jewell N.P., Diaz-Ordaz K., Keogh R.H. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021:1–5. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giallonardo F., Duchene S., Puglia I., Curini V., Profeta F., Cammà C., Marcacci M., Calistri P., Holmes E.C., Lorusso A. Genomic Epidemiology of the First Wave of SARS-CoV-2 in Italy. Viruses. 2020:12. doi: 10.3390/v12121438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzetta Ufficiale della Repubblica Italiana, 2020. Ordinanza 20 dicembre 2020. Ulteriori misure urgenti in materia di contenimento e gestione dell’emergenza epidemiologica da COVID-19.
- GISAID, 2021. GISAID Initiative [WWW Document]. URL 〈https://www.gisaid.org/〉 (accessed 6.28.21).
- Github, 2021. Cov-lineages/pangolin: Software package for assigning SARS-CoV-2 genome sequences to global lineages.
- Gostic K.M., McGough L., Baskerville E.B., Abbott S., Joshi K., Tedijanto C., Kahn R., Niehus R., Hay J.A., De Salazar P.M., Hellewell J., Meakin S., Munday J.D., Bosse N.I., Sherrat K., Thompson R.N., White L.F., Huisman J.S., Scire J., Bonhoeffer S., Stadler T., Wallinga J., Funk S., Lipsitch M., Cobey S. Practical considerations for measuring the effective reproductive number, Rt. PLoS Comput. Biol. 2020;16:1–21. doi: 10.1371/journal.pcbi.1008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin J., Casey M., Collins Á., Hunt K., McEvoy D., Byrne A., McAloon C., Barber A., Lane E.A., More S. Rapid review of available evidence on the serial interval and generation time of COVID-19. BMJ Open. 2020;10:1–9. doi: 10.1136/bmjopen-2020-040263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hens N., Calatayud L., Kurkela S., Tamme T., Wallinga J. Robust reconstruction and analysis of outbreak data: Influenza A(H1N1)v transmission in a school-based population. Am. J. Epidemiol. 2012;176:196–203. doi: 10.1093/aje/kws006. [DOI] [PubMed] [Google Scholar]
- Horby, P., Huntley, C., Davies, N., Edmunds, J., Ferguson, N., Medley, G., Semple, C., 2021. NERVTAG Note on B.1.1.7 Severity.
- Istituto Superiore di Sanità, 2021a. Prevalenza della variante VOC 202012/01, lineage B.1.1.7 in Italia - Studio di prevalenza 4–5 febbraio 2021. Roma.
- Istituto Superiore di Sanità, 2021b. Prevalenza delle varianti VOC (Variant Of Concern) in Italia: lineage B.1.1.7, P.1, P.2, lineage B.1.351, lineage B.1.525 (Indagine del 18/3/2021). Roma.
- Istituto Superiore di Sanità, 2021c. Prevalenza delle VOC (Variant Of Concern) del virus SARS-CoV-2 in Italia: lineage B.1.1.7, P.1 e B.1.351, e altre varianti (Variant Of Interest, VOI) tra cui lineage P.2 e lineage B.1.525 (Indagine del 20/4/2021). Roma.
- Istituto Superiore di Sanità, 2021d. FAQ sul calcolo del Rt [WWW Document]. URL 〈https://www.iss.it/coronavirus/-/asset_publisher/1SRKHcCJJQ7E/content/faq-sul-calcolo-del-rt〉 (accessed 1.3.22).
- Lorusso A., Calistri P., Mercante M.T., Monaco F., Portanti O., Marcacci M., Cammà C., Rinaldi A., Mangone I., Di Pasquale A., Iommarini M., Mattucci M., Fazii P., Tarquini P., Mariani R., Grimaldi A., Morelli D., Migliorati G., Savini G., Borrello S., D’Alterio N. A “One-Health” approach for diagnosis and molecular characterization of SARS-CoV-2 in Italy. One Heal. 2020;10 doi: 10.1016/j.onehlt.2020.100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muik A., Wallisch A.K., Sänger B., Swanson K.A., Mühl J., Chen W., Cai H., Maurus D., Sarkar R., Türeci Ö., Dormitzer P.R., Şahin U. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine–elicited human sera. Science (80-. ) 2021;371:1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole Á., Kraemer M.U.G., Hill V., Pybus O.G., Watts A., Bogoch I.I., Khan K., Messina J.P., Tegally H., Lessells R.R., Giandhari J., Pillay S., Tumedi K.A., Nyepetsi G., Kebabonye M., Matsheka M., Mine M., Tokajian S., Hassan H., Salloum T., Merhi G., Koweyes J., Geoghegan J.L., de Ligt J., Ren X., Storey M., Freed N.E., Pattabiraman C., Prasad P., Desai A.S., Vasanthapuram R., Schulz T.F., Steinbrück L., Stadler T., Parisi A., Bianco A., García de Viedma D., Buenestado-Serrano S., Borges V., Isidro J., Duarte S., Gomes J.P., Zuckerman N.S., Mandelboim M., Mor O., Seemann T., Arnott A., Draper J., Gall M., Rawlinson W., Deveson I., Schlebusch S., McMahon J., Leong L., Lim C.K., Chironna M., Loconsole D., Bal A., Josset L., Holmes E., St. George K., Lasek-Nesselquist E., Sikkema R.S., Oude Munnink B., Koopmans M., Brytting M., Sudha rani V., Pavani S., Smura T., Heim A., Kurkela S., Umair M., Salman M., Bartolini B., Rueca M., Drosten C., Wolff T., Silander O., Eggink D., Reusken C., Vennema H., Park A., Carrington C., Sahadeo N., Carr M., Gonzalez G., de Oliveira T., Faria N., Rambaut A. Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2. Wellcome Open Res. 2021;6 doi: 10.12688/wellcomeopenres.16661.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Public Health England . Sage,; 2021. SARS-CoV-2 variants of concern and variants under investigation in England; pp. 1–50. [Google Scholar]
- Public Health England, 2020. Investigation of novel SARS-COV-2 variant: Variant of Concern 202012/01 - Technical briefing 4, gov.uk.
- Rambaut, A., Loman, N., Pybus, O., Barclay, W., Barrett, J., Carabelli, A., Connor, T., Peacock, T., Robertson, D.L., Volz, E.M., 2020. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations - SARS-CoV-2 coronavirus / nCoV-2019 Genomic Epidemiology - Virological, virological.org.
- Savini L., Candeloro L., Calistri P., Conte A. A municipality-based approach using commuting census data to characterize the vulnerability to influenza-like epidemic: The COVID-19 application in Italy. Microorganisms. 2020;8:1–21. doi: 10.3390/microorganisms8060911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherratt K., Abbott S., Meakin S.R., Hellewell J., Munday J.D., Bosse N., Jit M., Funk S. Exploring surveillance data biases when estimating the reproduction number: With insights into subpopulation transmission of COVID-19 in England. Philos. Trans. R. Soc. B Biol. Sci. 2021:376. doi: 10.1098/rstb.2020.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Washington Post, 2021. A cluster of the coronavirus’s U.K. variant was found in Italy. Four cases grew to 29 before the town was alert [WWW Document]. URL 〈https://www.washingtonpost.com/world/europe/italy-covid-uk-variant/2021/01/16/0732bd24–544e-11eb-acc5–92d2819a1ccb_story.html〉 (accessed 1.28.21).
- Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., O’Toole Á., Amato Robert, Ragonnet-Cronin M., Harrison I., Jackson B., Ariani C.V., Boyd O., Loman N.J., McCrone J.T., Gonçalves S., Jorgensen D., Myers R., Hill V., Jackson D.K., Gaythorpe K., Groves N., Sillitoe J., Kwiatkowski D.P., Koshy C., Ash A., Wise E., Moore N., Mori M., Cortes N., Lynch J., Kidd S., Fairley D.J., Curran T., McKenna J.P., Adams H., Fraser C., Golubchik T., Bonsall D., Hassan-Ibrahim M.O., Malone C.S., Cogger B.J., Wantoch M., Reynolds N., Warne B., Maksimovic J., Spellman K., McCluggage K., John M., Beer R., Afifi S., Morgan S., Marchbank A., Price A., Kitchen C., Gulliver H., Merrick I., Southgate J., Guest M., Munn R., Workman T., Connor T.R., Fuller W., Bresner C., Snell L.B., Patel A., Charalampous T., Nebbia G., Batra R., Edgeworth J., Robson S.C., Beckett A.H., Aanensen D.M., Underwood A.P., Yeats C.A., Abudahab K., Taylor B.E.W., Menegazzo M., Clark G., Smith W., Khakh M., Fleming V.M., Lister M.M., Howson-Wells H.C., Berry Louise, Boswell T., Joseph A., Willingham I., Jones C., Holmes C., Bird P., Helmer T., Fallon K., Tang J., Raviprakash V., Campbell S., Sheriff N., Blakey V., Williams L.A., Loose M.W., Holmes N., Moore Christopher, Carlile M., Wright V., Sang F., Debebe J., Coll F., Signell A.W., Betancor G., Wilson H.D., Eldirdiri S., Kenyon A., Davis T., Pybus O.G., du Plessis L., Zarebski A.E., Raghwani J., Kraemer M.U.G., Francois S., Attwood S.W., Vasylyeva T.I., Zamudio M.E., Gutierrez B., Torok M.E., Hamilton W.L., Goodfellow I.G., Hall G., Jahun A.S., Chaudhry Y., Hosmillo M., Pinckert M.L., Georgana I., Moses S., Lowe H., Bedford L., Moore J., Stonehouse S., Fisher C.L., Awan A.R., BoYes J., Breuer J., Harris K.A., Brown J.R., Shah D., Atkinson L., Lee J.C.D., Storey N., Flaviani F., Alcolea-Medina A., Williams R., Vernet G., Chapman M.R., Levett L.J., Heaney J., Chatterton W., Pusok M., Xu-McCrae L., Smith D.L., Bashton M., Young G.R., Holmes A., Randell P.A., Cox A., Madona P., Bolt F., Price J., Mookerjee S., Ragonnet-Cronin M., Nascimento F.F., Jorgensen D., Siveroni I., Johnson R., Boyd O., Geidelberg L., Volz E.M., Rowan A., Taylor G.P., Smollett K.L., Loman N.J., Quick J., McMurray C., Stockton J., Nicholls S., Rowe W., Poplawski R., McNally A., Nunez R.T.M., Mason J., Robinson T.I., O’Toole E., Watts J., Breen C., Cowell A., Sluga G., Machin N.W., Ahmad S.S.Y., George R.P., Halstead F., Sivaprakasam V., Hogsden W., Illingworth C.J., Jackson C., Thomson E.C., Shepherd J.G., Asamaphan P., Niebel M.O., Li K.K., Shah R.N., Jesudason N.G., Tong L., Broos A., Mair D., Nichols J., Carmichael S.N., Nomikou K., Aranday-Cortes E., Johnson N., Starinskij I., da Silva Filipe A., Robertson D.L., Orton R.J., Hughes J., Vattipally S., Singer J.B., Nickbakhsh S., Hale A.D., Macfarlane-Smith L.R., Harper K.L., Carden H., Taha Y., Payne B.A.I., Burton-Fanning S., Waugh S., Collins J., Eltringham G., Rushton S., O’Brien S., Bradley A., Maclean A., Mollett G., Blacow R., Templeton K.E., McHugh M.P., Dewar R., Wastenge E., Dervisevic S., Stanley R., Meader E.J., Coupland L., Smith L., Graham C., Barton E., Padgett D., Scott G., Swindells E., Greenaway J., Nelson A., McCann C.M., Yew W.C., Andersson M., Peto T., Justice A., Eyre D., Crook D., Sloan T.J., Duckworth N., Walsh S., Chauhan A.J., Glaysher S., Bicknell K., Wyllie S., Elliott S., Lloyd A., Impey R., Levene N., Monaghan L., Bradley D.T., Wyatt T., Allara E., Pearson C., Osman H., Bosworth A., Robinson E., Muir P., Vipond I.B., Hopes R., Pymont H.M., Hutchings S., Curran M.D., Parmar S., Lackenby A., Mbisa T., Platt S., Miah S., Bibby D., Manso C., Hubb J., Ramsay M., Bradshaw D., Thornton A., Schaefer U., Gallagher E., Lee D., Williams D., Ellaby N., Hartman H., Manesis N., Patel V., Bishop C., Chalker V., Ledesma J., Twohig K.A., Holden M.T.G., Shaaban S., Birchley A., Adams A., Davies A., Gaskin A., Plimmer A., Gatica-Wilcox B., McKerr C., Moore Catherine, Williams C., Heyburn D., Lacy E.De, Hilvers E., Downing F., Shankar G., Jones H., Asad H., Coombes J., Watkins J., Evans J.M., Fina L., Gifford L., Gilbert L., Graham L., Perry M., Morgan M., Bull M., Cronin M., Pacchiarini N., Craine N., Jones R., Howe R., Corden S., Rey S., Kumziene-Summerhayes S., Taylor S., Cottrell S., Jones S., Edwards S., O’Grady J., Page A.J., Mather A.E., Baker D.J., Rudder S., Aydin A., Kay G.L., Trotter A.J., Alikhan N.F., de Oliveira Martins L., Le-Viet T., Meadows L., Casey A., Ratcliffe L., Simpson D.A., Molnar Z., Thompson T., Acheson E., Masoli J.A.H., Knight B.A., Ellard S., Auckland C., Jones C.R., Mahungu T.W., Irish-Tavares D., Haque T., Hart J., Witele E., Fenton M.L., Dadrah A., Symmonds A., Saluja T., Bourgeois Y., Scarlett G.P., Loveson K.F., Goudarzi S., Fearn C., Cook K., Dent H., Paul H., Partridge D.G., Raza M., Evans C., Johnson K., Liggett S., Baker P., Bonner S., Essex S., Lyons R.A., Saeed K., Mahanama A.I.K., Samaraweera B., Silveira S., Pelosi E., Wilson-Davies E., Williams R.J., Kristiansen M., Roy S., Williams C.A., Cotic M., Bayzid N., Westhorpe A.P., Hartley J.A., Jannoo R., Lowe H.L., Karamani A., Ensell L., Prieto J.A., Jeremiah S., Grammatopoulos D., Pandey S., Berry Lisa, Jones K., Richter A., Beggs A., Best A., Percival B., Mirza J., Megram O., Mayhew M., Crawford L., Ashcroft F., Moles-Garcia E., Cumley N., Smith C.P., Bucca G., Hesketh A.R., Blane B., Girgis S.T., Leek D., Sridhar S., Forrest S., Cormie C., Gill H.K., Dias J., Higginson E.E., Maes M., Young J., Kermack L.M., Gupta R.K., Ludden C., Peacock S.J., Palmer Sophie, Churcher C.M., Hadjirin N.F., Carabelli A.M., Brooks E., Smith K.S., Galai K., McManus G.M., Ruis C., Davidson R.K., Rambaut A., Williams T., Balcazar C.E., Gallagher M.D., O’Toole Á., Rooke S., Hill V., Williamson K.A., Stanton T.D., Michell S.L., Bewshea C.M., Temperton B., Michelsen M.L., Warwick-Dugdale J., Manley R., Farbos A., Harrison J.W., Sambles C.M., Studholme D.J., Jeffries A.R., Darby A.C., Hiscox J.A., Paterson S., Iturriza-Gomara M., Jackson K.A., Lucaci A.O., Vamos E.E., Hughes M., Rainbow L., Eccles R., Nelson C., Whitehead M., Turtle L., Haldenby S.T., Gregory R., Gemmell M., Wierzbicki C., Webster H.J., de Silva T.I., Smith N., Angyal A., Lindsey B.B., Groves D.C., Green L.R., Wang D., Freeman T.M., Parker M.D., Keeley A.J., Parsons P.J., Tucker R.M., Brown R., Wyles M., Whiteley M., Zhang P., Gallis M., Louka S.F., Constantinidou C., Unnikrishnan M., Ott S., Cheng J.K.J., Bridgewater H.E., Frost L.R., Taylor-Joyce G., Stark R., Baxter L., Alam M.T., Brown P.E., Aggarwal D., Cerda A.C., Merrill T.V., Wilson R.E., McClure P.C., Chappell J.G., Tsoleridis T., Ball J., Buck D., Todd J.A., Green A., Trebes A., MacIntyre-Cockett G., de Cesare M., Alderton A., Amato Roberto, Beale M.A., Beaver C., Bellis K.L., Betteridge E., Bonfield J., Danesh J., Dorman M.J., Drury E., Farr B.W., Foulser L., Goncalves S., Goodwin S., Gourtovaia M., Harrison E.M., Jamrozy D., Johnston I., Kane L., Kay S., Keatley J.P., Kwiatkowski D., Langford C.F., Lawniczak M., Letchford L., Livett R., Lo S., Martincorena I., McGuigan S., Nelson R., Palmer Steve, Park N.R., Patel M., Prestwood L., Puethe C., Quail M.A., Rajatileka S., Scott C., Shirley L., Chapman M.H.S., Thurston S.A.J., Tonkin-Hill G., Weldon D., Rajan D., Bronner I.F., Aigrain L., Redshaw N.M., Lensing S.V., Davies R., Whitwham A., Liddle J., Lewis K., Tovar-Corona J.M., Leonard S., Durham J., Bassett A.R., McCarthy S., Moll R.J., James K., Oliver K., Makunin A., Barrett J., Gunson R.N., Flaxman S., Ratmann O., Bhatt S., Hopkins S., Gandy A., Rambaut A., Ferguson N.M. Vol. 593. 2021. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England; pp. 266–269. (Nature). [DOI] [PubMed] [Google Scholar]
- World Health Organization (WHO), 2021a. Tracking SARS-CoV-2 variants [WWW Document]. Geneva, Switz. URL 〈https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/〉 (accessed 6.28.21).
- World Health Organization (WHO) World Health Organization,; 2021. COVID-19 Weekly Epidemiological Update 45. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material