Abstract
We developed procedures for isolating and characterizing in situ-transcribed mRNA from groundwater microorganisms catabolizing naphthalene at a coal tar waste-contaminated site. Groundwater was pumped through 0.22-μm-pore-size filters, which were then frozen in dry ice-ethanol. RNA was extracted from the frozen filters by boiling sodium dodecyl sulfate lysis and acidic phenol-chloroform extraction. Transcript characterization was performed with a series of PCR primers designed to amplify nahAc homologs. Several primer pairs were found to amplify nahAc homologs representing the entire diversity of the naphthalene-degrading genes. The environmental RNA extract was reverse transcribed, and the resultant mixture of cDNAs was amplified by PCR. A digoxigenin-labeled probe mixture was produced by PCR amplification of groundwater cDNA. This probe mixture hybridized under stringent conditions with the corresponding PCR products from naphthalene-degrading bacteria carrying a variety of nahAc homologs, indicating that diverse dioxygenase transcripts had been retrieved from groundwater. Diluted and undiluted cDNA preparations were independently amplified, and 28 of the resulting PCR products were cloned and sequenced. Sequence comparisons revealed two major groups related to the dioxygenase genes ndoB and dntAc, previously cloned from Pseudomonas putida NCIB 9816-4 and Burkholderia sp. strain DNT, respectively. A distinctive subgroup of sequences was found only in experiments performed with the undiluted cDNA preparation. To our knowledge, these results are the first to directly document in situ transcription of genes encoding naphthalene catabolism at a contaminated site by indigenous microorganisms. The retrieved sequences represent greater diversity than has been detected at the study site by culture-based approaches.
Among the many goals of microbial ecology is identification of the microorganisms and genes that are responsible for catalyzing biogeochemical reactions in soil, water, and sediment (5, 30, 58). Techniques that focus on markers of in situ metabolism that are specific and transient, such as unstable metabolites (2, 71) and mRNA, offer the possibility of measuring activity that is taking place at the time of sampling (71). mRNA transcripts are short-lived and unique nucleic acid sequences that can be detected with a high degree of specificity. When nucleic acid sequence information is available for the analysis of specific transcripts, this approach obviates the need for culturing microorganisms or incubating environmental samples under laboratory-imposed conditions and, therefore, may avoid the biases and limitations of other methods (30, 31). Retrieved transcripts can be used to compare environmental expression of individual members of gene families. Thus, when properly applied to field samples, mRNA-based methods may be useful in determining relationships between environmental conditions prevailing in microbial habitats and particular in situ activities of native microorganisms.
mRNA extraction techniques have been used to examine diverse microbiological processes, including in situ carbon fixation in oceans (48–52) and lakes (73), in situ nitrogen assimilation by the marine cyanobacterium Trichodesmium thiebautii (28, 72, 75), in situ mercury volatilization in freshwater (25, 26, 43), naphthalene metabolism in samples of soil (11, 56, 57), methane monooxygenase transcription in samples of trichloroethylene-contaminated soils (12), and potential biodegradation activities of microorganisms in deep subsurface samples (45).
The metabolism of naphthalene begins with the dioxygenase-mediated formation of 1,2-dihydroxy-1,2-dihydronaphthalene (6, 63). The iron-sulfur protein components of dioxygenases that catalyze this type of reaction are typically composed of two identical large subunits and two identical small subunits (19, 38, 70). The large subunit, thought to confer substrate specificity (14, 47), is encoded by nahAc in the model naphthalene-degrading organism, Pseudomonas putida G7 (74). The sequences of several related subunits have been determined (see below).
The primary objective of this investigation was to use reverse transcription-PCR (RT-PCR) to analyze the expression of the genes encoding naphthalene catabolism in microorganisms residing in a coal tar waste-contaminated site. To achieve this goal, we (i) improved methods for the extraction of in situ-transcribed mRNA from groundwater microorganisms present at low cell concentrations; (ii) incorporated approaches to limit changes in the physiological status of the native cells; (iii) designed a suite of PCR primers, targeted at conserved regions of naphthalene dioxygenases, to maximize the probability of detection and analysis of diverse sequences; (iv) tested the set of primers on diverse naphthalene-degrading isolates to ensure that we could amplify diverse naphthalene dioxygenase sequences; (v) reverse transcribed and amplified specific fragments from mRNA transcripts retrieved from site groundwater; (vi) assessed the diversity of sequences which were retrieved and amplified by hybridization analysis, using the digoxigenin-labeled RT-PCR products as probes and the corresponding PCR products of diverse isolates as targets; and (vii) cloned and analyzed the mRNA sequences amplified from both diluted and undiluted preparations of groundwater-derived cDNA.
MATERIALS AND METHODS
Site.
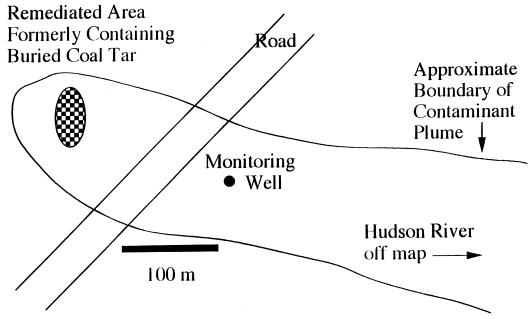
The study site is a rural wooded area in upstate New York, where coal tar waste was buried in a shallow trench in the early 1960s. The source material was removed, and the groundwater plume is undergoing intrinsic bioremediation. This site has been described previously (41) and has been used extensively for a variety of field-oriented microbiological studies (21–23, 33–36, 40, 55, 61, 71). A plan view of the site is presented in Fig. 1.
FIG. 1.
Plan view of the coal tar waste-contaminated site in Glens Falls, N.Y., showing the monitoring well where groundwater was collected.
Groundwater sampling.
Groundwater samples were collected at a flow rate of 300 ml/min after purging 4 well volumes with a peristaltic pump with new polyethylene tubing. Cells in 5 liters of groundwater (collected for ≈17 min) were concentrated onto 142-mm-diameter Millipore Corp. (Bedford, Mass.) Durapore membranes (pore size, 0.22 μm), placed in sterile Whirlpak bags, and frozen immediately by immersion in a dry ice-ethanol bath. Samples remained on dry ice during transport to the laboratory, where they were transferred to a −85°C freezer.
Microorganisms used in this study.
P. putida G7, originally isolated from soil in Berkeley, Calif. (23), was a gift from G. S. Sayler, University of Tennessee, Knoxville, Tenn. Comamonas testosteroni GZ39 and GZ42, originally isolated from sediments of the Passaic River in New Jersey (16), were gifts from G. J. Zylstra, Rutgers University. Genes for the large subunit of the dioxygenases cloned from these organisms have approximately 80% (GZ42) and 55% (GZ39) (15) identity to nahAc at the nucleotide level. P. putida Cg1 and putative Pseudomonas sp. strain Cg7 were previously isolated from a contaminated seep area at the coal tar-contaminated site (23). Sequencing of PCR-amplified nahAc from strains Cg1 and Cg7 indicated about 95% identity to nahAc from P. putida G7 and about 93% mutual identity (21).
Alignment of dioxygenases.
Sixteen bacterial dioxygenase genes retrieved from GenBank (3) and the sequence of C. testosteroni GZ39 phdAc (16), which was kindly provided prior to publication by Gerben Zylstra, Rutgers University, were aligned. The sequences included four dioxygenases from naphthalene-degrading organisms (with GenBank accession numbers in parentheses): nahAc (M83949) from P. putida G7 (60), doxB (M60405) from Pseudomonas sp. strain C18 (9), ndoB (M23914) from P. putida NCIB 9816-4 (60), and nahAc (D16629) from Pseudomonas sp. strain OUS82 (65). The sequences also included 11 dioxygenase sequences from organisms capable of growth on either benzene, chlorobenzene, benzoate, toluene, biphenyl, or chlorinated biphenyls: bedC1 (L04642) from P. putida ML2 (66), benA (M76990) from Acinetobacter calcoaceticus ADP1 (44), todC1 (J04996) from P. putida F1 (76), tcbA (U15298) from Pseudomonas sp. strain P51 (68), xylX (M64747) from P. putida (20), bphA (M83673) from P. pseudoalcaligenes KF707 (64), bphA (D16831) from Pseudomonas sp. strain KKS102 (27), bphA1 (D32142) from Rhodococcus sp. strain RHA1 (37), bphA (M86348) from Pseudomonas sp. strain LB400 (10), bphA1 (X80041) from Rhodococcus globerulus P6 (1), and bpdC2 (U27591) from Rhodococcus sp. strain M5 (69). Sequences were first aligned with the software programs EditSeq and MegAlign (DNASTAR Inc, Madison, Wis.), and the alignments were then adjusted manually.
PCR primers.
Degenerate PCR primers were designed with the aid of the OLIGO 4.0 software package (National Biosciences, Inc., Plymouth, Minn.). To target genes and mRNA transcripts that were specifically involved in naphthalene degradation, the primers were chosen to distinguish nahAc-type dioxygenase genes from all of the other characterized dioxygenases. The numbers used in the primer names refer to the 5′ end of each primer, using the nucleotide sequence of the P. putida G7 nahAc. The forward primers were Ab248F, 5′-CGTGACA(G/C)AGAACATCAAAACATA-3′; Ac114F, 5′-CTGGC(T/A)(T/A)TT(T/C)CTCAC(T/C)CAT-3′; and Ac307F, 5′-TATCACGGCTGG(G/T)(G/C)(C/T)TTCGGCT-3′. The reverse primers were Ac596R, 5′-C(G/A)GGTG(C/T)CTTCCAGTTG-3′; Ac893R, 5′-AGTTGAGGTG(G/C)(G/C)(A/T)GCGATA-3′; and Ac1095R, 5′-TTGTCA(T/C)TGTCGTCG(C/G)(T/C)TTC-3′.
RNA extraction.
RNA extraction was carried out by a modification of the method of Jeffrey et al. (26) and incorporated procedures from the method of Siering and Ghiorse (59). Chemicals and reagents were obtained from Sigma (St. Louis, Mo.) except where otherwise noted and were of molecular biology grade. Standard precautions against the presence of RNases were taken. Filters containing groundwater microorganisms were manually crushed while still frozen in their Whirlpak bags, and the pieces were poured into 40-ml screw-cap centrifuge tubes. Then 5 ml of lysis buffer (1% sodium dodecyl sulfate, 0.1 M NaCl, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA) which had been preheated to 85°C was added, and the tubes were capped, vortexed for 0.5 min, and placed in a boiling-water bath for 5 min. The samples were vortexed again for 0.5 min, and the liquid was decanted from the filter pieces into a new centrifuge tube, which was placed on ice. The filter pieces were extracted three times with 5 ml of pH 5.1 buffer (50 mM sodium acetate, 10 mM EDTA [pH 5.1]) by vortexing for 0.5 min, and all four of the decanted extracts were combined and incubated on ice for 15 min. A 15-ml volume of phenol, equilibrated with pH 5.1 buffer, was added, and the sample was mixed vigorously. After incubation on ice for 5 min, the sample was centrifuged at 10,000 × g at 4°C for 15 min. The aqueous layer was removed to another tube, reextracted with an equal volume of phenol-chloroform-isoamyl alcohol (125:25:1) (pH 4.7), and centrifuged as above to separate the phases. The resultant aqueous layer was extracted with an equal volume of chloroform-isoamyl alcohol (25:1) and centrifuged to separate the phases, and RNA present in the aqueous layer was precipitated overnight at −20°C with 0.5 volume of 7.5 M ammonium acetate, 2 volumes of absolute ethanol, and 2 μl of 20-mg/ml glycogen. The precipitated RNA was pelleted at 12,000 × g at 4°C for 30 min. The supernatant was removed, and 5 ml of 70% ethanol at −20°C was washed along the side of the tube, which was then centrifuged again at 12,000 × g at 4°C for 5 min. The supernatant was removed, and the tube was inverted and allowed to dry for 15 min in a sterile hood. Precipitated RNA was resuspended in 2.1 ml of buffer (10 mM Tris-HCl, 1 mM EDTA [pH 7.5]) containing 20% formamide (the formamide was included, as suggested in reference 7, to facilitate resuspension and to protect the RNA from nucleases) by incubation at room temperature for 30 min and then mixed by gently pipetting up and down. The RNA was reprecipitated in three 1.7-ml tubes (700 μl/tube) overnight at −20°C by the addition of 1/10 volume of 4 M sodium acetate (pH 4.0), 2 volumes of absolute ethanol, and 1.5 μl of 20-mg/ml glycogen. RNA was pelleted at room temperature in a bench top microcentrifuge for 30 min. Supernatants were removed with a pipette, and the pellets washed with 0.5 ml of 70% ethanol at −20°C. The tubes were then centrifuged for 5 min, the supernatants were removed, and the tubes were inverted and allowed to dry for 15 min in a sterile hood. Precipitated RNA was resuspended by adding 30 μl of the 20% formamide buffer to each tube, incubating at room temperature for 30 min, and gently pipetting the buffer up and down. The three samples were combined and digested at 37°C for 3 h with 50 U of RNase-free DNase I (Ambion, Inc., Austin, Tex.) in a 0.5-ml volume containing 2.5 mM CaCl2, 10 mM MgCl2, and 25 mM Tris-HCl (pH 7.5). The reaction was stopped by addition of 0.1 ml of 50 mM EDTA. DNase-digested RNA samples were extracted with an equal volume of phenol-chloroform-isoamyl alcohol and centrifuged at room temperature for 5 min to separate the phases, and the aqueous layer was reserved. The organic layer was extracted twice with 0.5 ml of buffer containing 10 mM Tris-HCl and 1 mM EDTA (pH 7.5). The combined aqueous extracts were extracted with chloroform-isoamyl alcohol (25:1), and the aqueous phase of this extraction (final volume, approximately 1.5 ml) was precipitated for cDNA synthesis as outlined below.
cDNA synthesis.
cDNA synthesis was carried out as suggested previously (4). A 4-pmol sample of reverse primer was added to 0.5 ml of DNase-digested RNA preparation and coprecipitated overnight at −20°C with sodium acetate-ethanol-glycogen as above. The pellet was resuspended by incubation at room temperature for 30 min in 40 μl of buffer containing 80 mM Tris-HCl (pH 8.0) and 80 mM KCl. The RNA was denatured at 70°C for 4 min and incubated at 48°C for 3 h to allow the primers to anneal (this prolonged incubation was critical for successful cDNA synthesis from groundwater mRNA). Then 6 μl of 0.1 mM dithiothreitol, 1.5 μl of 10 mM deoxynucleoside triphosphates (dNTPs), and 12 μl of buffer (375 mM KCl, 15 mM MgCl2, 250 mM Tris-HCl [pH 8.3]) were added to the sample, and the solution was gently mixed and divided into two 30-μl aliquots. These aliquots were incubated at 42°C for 2 min, and 1 μl (200 U) of Superscript II RNase H− reverse transcriptase (Life Technologies, Gaithersburg, Md.) was added to one of the aliquots. The sample with no reverse transcriptase functioned as a negative control. cDNA synthesis was carried out at 42°C for 3 h, after which the cDNA solutions were stored at −20°C.
PCR.
PCR amplifications were carried out in 0.2-ml thin-walled PCR tubes (Lab Product Sales, Rochester, N.Y.), with 50- or 100-μl volumes containing a final concentration of 1 μM each primer, 50 μM each dNTP, 1.5 U of Taq polymerase (Promega Corp., Madison, Wis.), 50 mM KCl, 10 mM Tris-HCl (pH 8.0), 1.5 mM MgCl2, 0.01% nuclease-free bovine serum albumin (New England Biolabs, Beverly, Mass.), and 0.05% Tween 20. For amplification of mRNA sequences, 2 μl of the cDNA reaction mixture was used as the template. For amplification from pure cultures, a sterile platinum wire was touched to the surface of a fresh naphthalene-grown colony and then swirled briefly in 10 μl of sterile deionized water in a PCR tube. The cells were lysed by incubating the PCR tube at 95°C for 5 min and then at 4°C for 5 min (21) before addition of the PCR cocktail and cycling as described below. Reaction mixes were brought to 80°C in a thermal cycler (MJ Research Minicycler, Watertown, Mass.) for the “hot-start” addition of dNTPs, after which a Touchdown PCR program (53) was used, which consisted of 1 cycle of denaturation at 94°C for 1 min, annealing at 57°C for 1 min, and extension at 72°C for 1.5 min; 29 cycles in which the annealing temperature was reduced 0.5°C/cycle from the preceding cycle; and then 15 cycles of 94°C for 1 min, 42°C for 1 min, and 72°C for 1.5 min, with the last cycle followed by a 5-min extension at 72°C. Amplification products were separated in a 1.0% agarose gel and stained with ethidium bromide (54).
Production of digoxigenin-labeled RT-PCR product.
Digoxigenin-labeled RT-PCR product was produced by using primers Ac114F and Ac596R and the above PCR procedure, except that the dTTP concentration was reduced to 35 μM and digoxigenin-11-dUTP (Boehringer Mannheim, Indianapolis, Ind.) was included at 15 μM, as suggested by the manufacturer. Ten PCRs were carried out in a volume of 100 μl, each using 2 μl of undiluted cDNA as template, and the resulting amplified products were combined. Then 500 μl of the PCR product was loaded onto a 1% low-melting-temperature agarose (SeaPlaque GTG Agarose; FMC BioProducts, Rockland, Maine) gel and run at 2.5 V/cm for 90 min. After ethidium bromide staining, the band corresponding to the RT-PCR product was excised from the gel. The labeled probe was quantified with a control digoxigenin-labeled probe as described by Boehringer Mannheim for the Genius system.
Hybridization.
nahAc homologs were PCR amplified from several naphthalene-degrading strains containing diverse naphthalene dioxygenase genes by using primers Ac114F and Ac596R. The strains and the identity which their dioxygenase large-subunit genes shared with nahAc, were as follows: P. putida G7, 100% (60); P. putida Cg1, 95% (23); putative Pseudomonas sp. strain Cg7, 95% (23); and C. testosteroni GZ39, 55%, and GZ42, 80% (15). A 500-ng portion (determined by densitometery with ethidium bromide-stained standards) of each PCR product or control DNA (salmon sperm DNA, and lambda DNA digested with HindIII) was suspended in 10 mM Tris-HCl–1 mM EDTA (pH 7.8) and denatured by addition of 0.1 volume of 3 M NaOH and incubation at 60°C for 60 min. The DNA solutions were allowed to cool to room temperature, and 1 volume of 6× SSC (diluted from 20× SSC stock: 3 M NaCl plus 0.3 M sodium citrate [pH 7.0]) was added. The samples were blotted onto a Magna Graph (MSI, Westborough, Mass.) nylon membrane by using a vacuum-based dot-blot apparatus (Bio-Rad, Hercules, Calif.) as specified by the membrane manufacturer. The blot was dried, and DNA was cross-linked to the membrane by baking at 80°C for 60 min. Hybridization was carried out as suggested by Boehringer Mannheim. Hybridization and prehybridization were carried out at 65°C in SHB/50 buffer, containing 5× SSC, 2% blocking reagent, 0.1% N-lauroylsarcosine, 0.02% sodium dodecyl sulfate, and 50% formamide. The probe was diluted in SHB/50 to 20 ng/ml, denatured by heating to 95°C for 10 min, and cooled to 65°C. Prehybridization in SHB/50 was carried out for 120 min, after which the prehybridization solution was discarded and the preheated probe solution was added. Hybridization was allowed to occur for 18 to 24 h. The membranes were washed twice for 15 min each in 2× wash solution (n× wash solution contains n× SSC and 0.1% sodium dodecyl sulfate) which had been preheated to 65°C. The membranes were washed twice for 15 min each to achieve three stringencies: 0.3× wash solution (ca. 12% mismatch blot), 0.2× wash solution (ca. 9% mismatch blot), or 0.1× wash solution (ca. 5% mismatch blot). The percent mismatch for a given wash buffer was calculated from the formula Tm = (81.5 + 16.6 log[Na+] + 0.41%G+C − 600/l) − (1.25 × percent mismatch), where [Na+] represents the total salt concentration in moles, %G+C represents the percentage of guanine-cytosine pairs in the probe sequence, and l represents the length of the probe (39, 54). Bound probe was detected with the Genius kit and CSPD chemiluminescent substrate for alkaline phosphatase, and the X-ray film was exposed as specified by the manufacturer (Boehringer Mannheim).
Amplification, cloning, and sequencing of RT-PCR products.
The experimental design sought to contrast sequences of transcripts that were abundant (detected in diluted cDNA) and rare (detected in undiluted cDNA) in the groundwater. Although we could not assume that the proportions of different transcripts would be unaltered by the PCR, it is likely that the diluted cDNA preparation contained fewer rare transcripts than the undiluted preparation did. Separate aliquots of a 1:100 dilution of site-retrieved cDNA (a 1:200 dilution was the lowest dilution to produce amplification products) were amplified in each of nine independent PCRs (40-μl volume) with primers Ac114F and Ac596R. Five separate aliquots of undiluted cDNA were processed in an identical manner. After holding at 80°C for the hot-start addition of dNTPs, the thermal cycler carried out 30 cycles of amplification consisting of 1 min at 94°C, 1 min at 43°C, and 1 min at 72°C. A final extension was carried out at 72°C for 7 min, and the reaction mixtures were then brought to 4°C. The PCR products were cloned with the original T/A cloning kit (Invitrogen Corp., Carlsbad, Calif.) as specified by the manufacturer. The nucleotide sequences were determined for one to six of the cloned PCR products that were obtained from each of the 14 independent PCR amplifications from the diluted or undiluted cDNA. Plasmid preparations were carried out with the PlasmidPURE DNA miniprep kit (Sigma Chemical Co.) as specified by the manufacturer. Sequences were determined by using the Taq DyeDeoxy terminator cycle-sequencing procedure on an Applied Biosystems (Foster City, Calif.) 373 DNA sequencer at the Automated Sequencing Facility of the Cornell University Center for Advanced Technology. Nucleotide sequencing of the inserts was carried out in both directions with primers that corresponded to the M13 (5′-CAGGAAACAGCTATGAC-3′) and T7 (5′-TACGACTCACTATAGGG-3′) sites of the pCR2.1 vector.
Analysis of cloned RT-PCR products.
Sequences (401 bp) were aligned and cluster analysis was performed by using the DNASTAR software program MegAlign. For pairwise comparison of sequences, percent dissimilarities were used directly, without any corrections based on models of nucleotide substitution.
Nucleotide sequence accession numbers.
The sequences of the nahAc-type mRNA clones have been deposited in GenBank under accession no. AF099747 to AF099774.
RESULTS
Primer design, development of a touchdown PCR method, and amplification of nahAc homologs from naphthalene-degrading cultures.
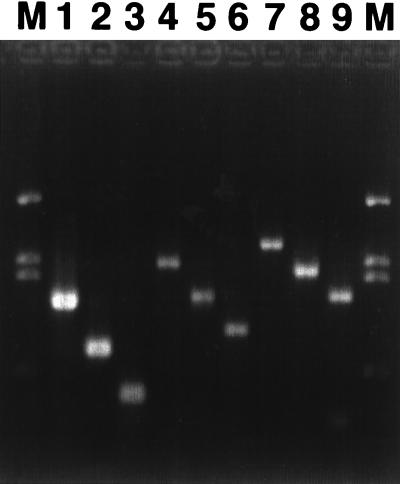
Three forward and three reverse primers were chosen in regions conserved among the dioxygenases from naphthalene-degrading organisms but not conserved among the dioxygenases from organisms metabolizing other aromatic substrates. Four of the five naphthalene dioxygenase sequences were highly conserved, but degeneracy in the primers was needed to accommodate the phdAc sequence from C. testosteroni GZ39. We designed a temperature-cycling regime that allowed amplification of specific nahAc-like sequences from P. putida G7 DNA with each of the nine possible primer combinations (Fig. 2). The sizes of the amplified products corresponded to the sizes predicted from the published nahAb and nahAc sequences (60), and no nonspecific amplification was observed. When C. testosteroni GZ39 DNA was used, five of the nine primer combinations amplified well, with the other four primer pairs exhibiting no or weak amplification (data not shown). Amplifications with primer Ac893R and primer pair Ab248F and Ac1095R were unsuccessful (data not shown). Results for C. testosteroni GZ42 were similar to those for C. testosteroni GZ39: four of the nine primer combinations amplified well, and the other five primer pairs exhibited weak or no amplification (data not shown). The amplifications that included primer Ac893R or primer Ab248F were unsuccessful. PCR products of the successful primer pairs corresponded in size to the PCR amplicons produced from P. putida G7.
FIG. 2.
Amplification of nahAc from P. putida G7 with each of the nine possible combinations of three forward and three reverse primers. A single touchdown PCR temperature-cycling program was used. Lanes: 1 to 3, amplicons with primer Ac596R and primers Ab248F, Ac114F, and Ac307F, respectively; 4 to 6, amplicons with primer Ac893R and primers Ab248F, Ac114F, and Ac307F, respectively; 7 to 9, amplicons with primer Ac1095R and primers Ab248F, Ac114F, and Ac307F, respectively; M, HindIII-digested lambda DNA markers.
RNA isolation.
Initial attempts at extracting RNA from cells present in site groundwater focused on the bead-beating methods of Pichard and Paul (52), which were modifications of the method of Chomzynski and Sacchi (8). These extracts failed to yield detectable rRNA bands on ethidium bromide-stained agarose gels. We therefore attempted to use the method of Jeffrey et al. (26), which involves filters having a diameter of 142 mm rather than 47 mm and thus allows the sampling of larger water volumes. The larger filters also required the use of alternative lysis procedures, because they do not fit into the bead-beating tubes. After a boiling-SDS lysis step, the method of Jeffrey et al. (26) was essentially identical to that of Pichard and Paul (52); both used guanidinium isothiocyanate solutions to denature proteins and thus protect the RNA from nucleases. However, our initial attempts to use this method produced smears on ethidium bromide-stained agarose gels, indicating that RNA was extracted from the larger filters but that nuclease activity was also present. Furthermore, the guanidinium-containing solutions would not reliably separate into an aqueous phase and an organic phase after being mixed with an equal volume of phenol. We therefore shifted to a series of acidic phenol-chloroform extractions, similar to those used with pure cultures by Siering and Ghiorse (59). This resulted in extracts containing undegraded rRNA bands.
RT-PCR amplification of nahAc sequences from RNA extracts of P. putida G7.
The RT-PCR procedure was first tested with RNA derived from naphthalene-grown cells of P. putida G7. Each of the three reverse primers was used in a separate cDNA synthesis reaction, and this cDNA was then amplified with each of the forward primers in conjunction with the reverse primer used for reverse transcription. Amplified products of the predicted sizes were produced with each of the nine possible primer sets (data not shown). Negative controls, treated identically to the test reactions except that reverse transcriptase was left out of the cDNA reaction, did not produce detectable products. Reagent-only negative controls also failed to produce amplified products. Less stringent RT-PCR methods, which omitted the coprecipitation of RNA and primer or substituted a 2-min incubation for the 3-h annealing step, were also found to be successful at achieving RT-PCR amplification of nahAc mRNA transcripts from P. putida G7.
RT-PCR amplification from RNA extracts of microorganisms in contaminated well water.
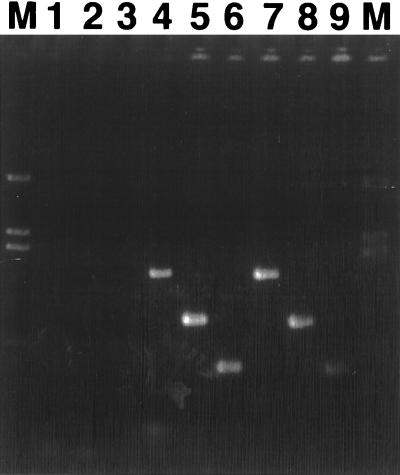
The procedures that successfully detected nahAc transcripts in naphthalene-grown P. putida G7 cells were applied to microorganisms collected from well water in the contaminated study site. RNA was extracted from cells contained in well water, and cDNA was reverse transcribed from the extracted RNA with primer Ac596R. The cDNA was then used as the template in the touchdown PCR assay. Successful amplification was observed when reverse primer Ac596R was included with each of the three forward primers, (Fig. 3, lanes 4 to 6). These products corresponded in size to those produced from P. putida G7 (lanes 7 to 9). Negative controls, identical in all respects except for the omission of reverse transcriptase during the synthesis of cDNA, did not produce amplification products (1 to 3), indicating that the RNA extracts were not contaminated with DNA. Similar experiments in which the site RNA extracts were reverse transcribed with primer Ac1095R were performed, but no amplification was observed. Digesting the cDNA preparations with DNase-free RNase, in an attempt to alleviate interference with amplification that may have been due to RNA-DNA hybrid formation, did not allow successful amplification with this primer. We did not attempt amplification with primer Ac893R, because this primer was not successful at amplifying from DNA isolated from C. testosteroni GZ39 or GZ42. RT-PCR amplification from site-derived RNA was unsuccessful, even with primer Ac596R, when an abbreviated cDNA synthesis method was used that did not have a primer-RNA coprecipitation step and omitted the 3-h annealing step at an elevated temperature.
FIG. 3.
RT-PCR amplification of nahAc mRNA transcripts isolated from cells in groundwater at the coal tar-contaminated site, using PCR reverse primer Ac596R with three forward primers. Reverse transcriptase was omitted from the cDNA synthesis reactions in lanes 1 to 3, demonstrating that products in lanes 4 to 6 (primers Ab248F, Ac114F, and Ac307F, respectively) were not derived from DNA sequences contaminating the RNA preparation. Groundwater-derived amplicons (lanes 4 to 6) were identical in size to those from RNA of naphthalene-grown P. putida G7 cells (lanes 7 to 9; primers Ab248F, Ac114F, and Ac307F, respectively).
Hybridization.
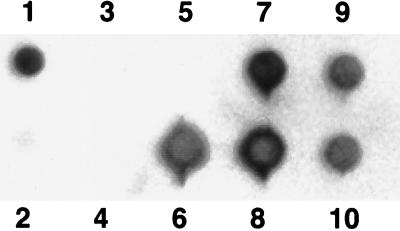
Using primers Ac114F and Ac596R, we produced digoxygenin-labeled PCR products from the site-retrieved RNA. The diversity of these products was assessed by hybridization with a range of diverse dioxygenase sequences obtained from several bacteria: P. putida G7 (60), P. putida Cg1 (23), putative Pseudomonas sp. strain Cg7 (23), and C. testosteroni GZ39 and GZ42 (15). These strains represent the total diversity of naphthalene dioxygenase gene sequences described to date. Primers Ac114F and Ac596R were used to produce amplified products from each of these isolates for use in the hybridization assay. Other target DNAs included the corresponding unlabeled PCR product derived from site RNA, an equivalent volume of the control PCR product which lacked reverse transcriptase during cDNA synthesis, HindIII-digested bacteriophage lambda DNA, salmon sperm DNA, and a buffer control. Results were identical for each of the stringencies examined, and only results for the 5% mismatch conditions are presented in Fig. 4. The probe hybridized, under high-stringency conditions, with the corresponding PCR product of each of the strains tested (Fig. 4, positions 6 to 10), which together represent the total described diversity of pure cultures known to catabolize naphthalene. The probe also hybridized with the unlabeled PCR product derived from site RNA but not the corresponding preparation in which reverse transcriptase was omitted (positions 1 and 2, respectively). No hybridization was observed between the probe and the lambda DNA or salmon sperm DNA or the well to which only buffer was added (positions 3 to 5, respectively).
FIG. 4.
Hybridization of a digoxigenin-labeled probe mixture from groundwater with the corresponding PCR products from diverse naphthalene-degrading organisms. The probe was produced by RT-PCR of groundwater mRNA. Conditions allowed ∼5% base pair mismatch. Target DNA was blotted in positions as follows: 1, unlabeled RT-PCR product from groundwater RNA; 2, negative control RT-PCR product prepared as in position 1, which lacked reverse transcriptase during cDNA synthesis; 3, salmon sperm DNA; 4, HindIII-digested bacteriophage lambda DNA; 5, no DNA; 6, P. putida G7; 7, P. putida Cg1; 8, putative Pseudomonas sp. strain Cg7; 9, C. testosteroni GZ39; 10, C. testosteroni GZ42.
Cloning and sequencing of amplified site transcripts.
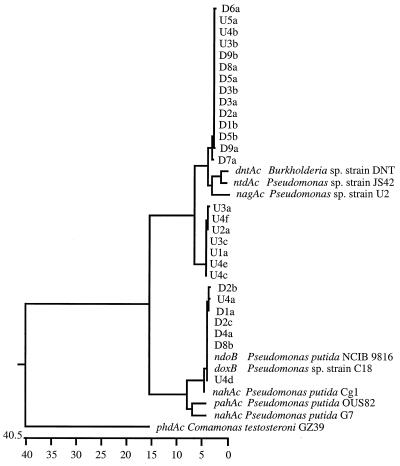
It was hypothesized that clones from the diluted cDNA pool would be of low diversity, representing genes from dominant, actively growing groundwater microorganisms. Conversely, this reasoning would predict that clones from the undiluted cDNA pool might include transcripts from less active, less abundant, and perhaps more diverse microorganisms. Five replicate PCR amplifications were performed with 2 μl of undiluted cDNA. Additionally, nine independent replicates of a 1:100 dilution (1:200 was the extinction dilution) were amplified in separate reaction tubes. PCR products from each of these 14 independent reactions were individually ligated into plasmid vectors and individually transformed, screened, and sequenced. Nucleotide sequences of one to six clones from each of the independent dilutions were determined. The retrieved sequences were aligned, and their relationships to each other and to previously characterized sequences are presented in Fig. 5 as a cluster analysis based on simple percent dissimilarity. Twenty-eight sequences derived from site mRNA were included in this analysis, as well as the corresponding 401-nucleotide region of nahAc from P. putida G7 (60), doxB from Pseudomonas sp. strain C18 (9), ndoB from P. putida NCIB-9816 (60), phdAc from C. testosteroni GZ39 (77), dntAc from Burkholderia sp. strain DNT (62), ntdAc from Pseudomonas sp. strain JS42 (46), nagAc from Pseudomonas sp. strain U2 (13), pahAc from P. putida OUS82 (65), and P. putida Cg1 (23), whose sequence in the corresponding region was determined as part of this study. The sequences of dntAc and ntdAc were not from naphthalene-utilizing bacteria but are reported to be greater than 98% identical at the nucleotide level to the sequence of the gene encoding the large subunit of the dioxygenase cloned from C. testosteroni GZ42 (15). Each of the sequences derived from groundwater mRNA are assigned a three-part name explained in the legend to Fig. 5. For example, clone U4f was found as an insert in the sixth [f] colony picked from the fourth [4] PCR performed on undiluted [U] cDNA.
FIG. 5.
Cluster analysis examining the percent dissimilarity of 28 nahAc-like mRNA sequences derived from well water and the corresponding 401-bp sequence from nine bacteria (see the text). The notation for each sequence specifies if the cDNA was diluted (D) or undiluted (U) before PCR amplification and cloning; which PCR (1 to 5 for U and 1 to 9 for D) produced the clone; and the order in which an insert was sequenced from each reaction (e.g., “a” was the first and “e” was the fifth).
The sequences retrieved from groundwater clustered tightly into three groups shown in the top, middle, and bottom portions of Fig. 5. The top group, obtained from both diluted and undiluted cDNA, was closely related to the sequence from Burkholderia sp. strain DNT (94.8% similar at the nucleotide level and 94% similar at the amino acid level). The middle group, obtained exclusively from undiluted cDNA, was more distant from pure-culture-derived alleles than was any other group of groundwater transcripts (when compared to Burkholderia sp. strain DNT, the central group was 90.3% similar at the nucleotide level and 92.5% similar at the amino acid level). This divergence from previously known sequences may have been because the central group of sequences were cloned from undiluted cDNA, where minor members of the groundwater microbial community may have been better represented. The bottom group of mRNA sequences in Fig. 5 was derived from both diluted and undiluted cDNA and was closely related to the sequences from P. putida NCIB 9816-4 (98% similar at the nucleotide level and 100% identical at the amino acid level).
Despite the fine structure discussed above (three groups), the dendrogram in Fig. 5 reveals that the groundwater naphthalene dioxygenase-type sequences fell into two major branches: one with Burkholderia sp. strain DNT and another with P. putida NCIB 9816-4. The two major branches differed in approximately 21% of the nucleotide positions, while the amino acid sequences differed by approximately 10%. In 11 of these 13 amino acid substitutions, the changes between the two branches were neutral. The two exceptions involve substitution of ionizable amino acids in the ntdAc-like sequences for nonionizable polar amino acids at positions 90 (Asn→His) and 129 (Asn→Lys) of the P. putida G7 nahAc-encoded protein.
DISCUSSION
In situ naphthalene biodegradation has been established previously in our study site by criteria that include (i) metabolic adaptation linked to protozoan predation (35), (ii) contraction of the contaminant plume coupled to oxygen depletion profiles (41), (iii) adaptation of microbial communities resulting in an enrichment of PCR-amplifiable nahAc sequences (22), and (iv) detection of the unique transient intermediary metabolite, 1,2-dihydroxynaphthalene-cis-dihydrodiol (71). In the present study, we developed a RT-PCR protocol for documenting biodegradation gene transcription (mRNA) in naphthalene-grown P. putida G7 cells and then produced specifically amplified products from groundwater whose sizes and sequences were homologous to those of nahAc-type mRNAs. This result indicates that naphthalene dioxygenase genes were being actively transcribed in site groundwater at the time of sampling and that we were able to successfully extract these transcripts and amplify them by PCR. Because this analysis focused on short-lived molecules specifically associated with naphthalene biodegradation and because the samples were obtained in a manner that minimized sampling-imposed artifacts, the results constitute compelling molecular biological evidence for ongoing intrinsic biodegradation activity at the study site.
To our knowledge, the diversity of field-expressed biodegradation genes has not been analyzed before. Thus, the results shown in Fig. 3 provided an opportunity to explore molecular relationships between model bacteria studied in pure culture and processes actually carried out by field microbial communities. With this objective, we characterized the mRNA transcripts which had been retrieved from contaminated well water. The RT-PCR products hybridized, under high-stringency conditions, to the corresponding PCR products derived from the total described diversity of pure cultures known to catabolize naphthalene. This result indicates that a variety of dioxygenase sequences were being simultaneously transcribed when the water was sampled. These results, obtained by a non-culture-based method, differ from results obtained in a recent culture-based study of naphthalene dioxygenase sequences from organisms at this site (23). In that study, a 373-bp portion of nahAc in eight naphthalene-degrading isolates obtained from a contaminated seep area was analyzed, and the sequences were found to be nearly identical (23). Because the shared nahAc sequences were derived from a group of isolates whose 16S rRNA sequences varied significantly, the lack of divergence among the nahAc genes was interpreted as evidence of in situ horizontal transfer of the nah genes (23, 61). The limited range of dioxygenases found in the previous study of site microorganisms (23) may reflect biases implicit in culture-based investigation or differences between the microbial communities present in surface sediment (23) and groundwater (this study).
The sequences of mRNA retrieved from the groundwater clustered into two major phylogenetic branches, one which was closely related to the ndoB sequence from P. putida NCIB 9816 and one which was related to the dntAc sequence from Burkholderia sp. strain DNT (Fig. 5). When more than one clone from a single RT-PCR was sequenced, the individual clones were often not identical, and separate clones clustered with separate branches (Fig. 5). This indicates that mRNA sequence in both of the phylogenetic branches were abundant in RNA extracts from the site; therefore, both types of genes were probably being translated into proteins being utilized by numerically prevalent organisms. These different proteins may have a variety of substrate ranges and specific activities and may thus influence the fate of chemical contaminants at the site in previously undescribed ways. A detailed examination of the dendrogram in Fig. 5 revealed a central cluster of sequences that departed further from pure-culture-derived sequences than did the other clones. Because undiluted cDNA was the source of the disparate sequences, it is likely that they were less predominant in the environmental mRNA pool than were the other two major alleles but amplified at a higher efficiency under our PCR conditions. None of our recovered sequences clustered with the phylogenetically divergent phdAc of C. testosteroni GZ39 (Fig. 5), suggesting that phdAc-like transcripts were present in relatively low concentrations or amplified inefficiently.
Failure to recover detectable RNA by the bead-beating method of Pichard and Paul (52) may have been a consequence of the low total-cell numbers that we were able to collect on the 47-mm-diameter membrane filters required by the use of 2.2-ml bead-beating tubes in this method. Acridine orange direct cell counts (59) indicated that 2 × 104 to 5 × 104 cells/ml were present in the groundwater sampled from the monitoring well. Unsuccessful bead-beating extractions, carried out with 0.5-liter sample volumes collected on 47-mm-diameter filters, had been attempted from about 107 cells. The method of Jeffrey et al. (26) uses filters with a diameter of 142 mm and thus allows the sampling of larger water volumes. The water that Jeffrey et al. used in their studies had cell concentrations of 0.5 × 106 to 2 × 106 cells/ml, and they were able to filter 8 liters of water, for a total of 0.4 × 1010 to 1.6 × 1010 cells/filter (26). We found that at the contaminated site in this study, flow through the 142-mm filters became restricted after 5 liters; therefore, only 1 to 2% of the number of cells used by Jeffrey et al. were obtained. As outlined in Results, we tested several modifications of the extraction procedure before we were able to effectively isolate RNA from the cells contained in groundwater at this site.
RT-PCR amplification of nahAc homologs was not observed in experiments in which primer Ac1095R was used to produce cDNA, even though the sequencing results suggested that some of the retrieved sequences were closely related to sequences known to be complementary to Ac1095R. It is unlikely that primer Ac1095R was not complementary to the retrieved transcripts, implying that the lack of amplification was related to the length of the products. Amplification from site-derived cDNA was also not observed, even with primer Ac596R, when a shortened RT-PCR method that was effective with P. putida G7 RNA was used. This difference may reflect an increase in the nonspecific binding of the primer by RNA extracts that are more heterogeneous and contain a lower target concentration. These variable results reaffirm the utility of using multiple primer sets and methods in analyzing nucleic acids extracted from environmental samples.
nah gene transcription has been the focus of several previous studies. In situ RT-PCR has been used to visualize nah transcript in laboratory cultures of naphthalene-degrading bacteria (24). Using a polycyclic aromatic hydrocarbon-contaminated soil from a manufactured gas plant site, researchers analyzed RNA extracts from uninoculated samples, samples inoculated with pure cultures of bacteria capable of growth on naphthalene, and samples which had been amended with salicylate (an inducer of the nah operon) and incubated for 3 days (67). No nahAb transcripts were detected in the RNA extracted from the uninoculated and unamended soil or the salicylate-induced soil, but hybridization was observed with RNA extracted from inoculated soils. Additional studies examining nah transcripts have been performed at the University of Tennessee (11, 56, 57). These researchers, using an RNase protection assay, were able to detect nahA transcripts in RNA extracts from uninoculated soils. Additionally, they made efforts to quantitate the extracted transcripts and successfully correlated the concentrations of the transcripts with naphthalene mineralization rates and with naphthalene concentrations. The samples that were examined (11, 56, 57) had been removed from their sites up to several years before being analyzed, were sieved, were stored at 4°C, and then were incubated as water slurries for 18 h at 27°C before being subjected to mRNA isolation. Therefore, the data produced could not be interpreted as reflecting the activity of in situ field populations.
The major limitations of mRNA-based analyses concern a lack of knowledge of the sequence variability among environmentally significant organisms, difficulties in relating transcript presence to a specific activity, and the inherently nonquantitative nature of the methods used. While the transient nature of mRNA makes transcripts more useful as real-time indicators of biodegradation activity, it also adds an uncertainty to quantitative RNA extraction analysis. Typical controls for mRNA extraction and purification analyses are in vitro-transcribed partial transcripts or transcripts contained in actively growing pure cultures. Both of these controls are probably unrepresentative of intracellular transcripts in the native microbial populations residing in situ. Alternate quantitation strategies, which attempt to normalize RNA concentrations to their corresponding DNA concentrations (52), must consider possible discrepancies between both DNA and RNA extraction efficiencies and laboratory- and field-grown microorganisms. Additionally, there are difficulties in interpreting measurements of transcriptional activity even when physiological and geochemical data supporting in situ microbial activity are available (35, 41, 71). One cannot easily extrapolate from expression of the first gene in an operon to an entire operon or to enzymatic activity. Difficulties arise because oxygenase enzymes often feature low substrate specificity and because microorganisms may have low levels of constitutive transcriptional activity. Constitutive levels of transcription are likely to vary among different naphthalene-degrading organisms (18), as is the specific activity of different enzymes encoded by different transcripts. Like other approaches, transcriptional analyses must be combined with independent methods aimed at understanding microorganisms in field settings (31, 32, 36, 42).
mRNA approaches also have their unique strengths. By minimizing sampling-induced changes in the microbial community, artifacts which might be created by laboratory incubations or time course assays can be avoided. Methods of nucleic acid extraction allow sampling from almost all of the cells present (17, 40). Because mRNA is very short lived, its detection ensures that the gene and the microorganism to which it belongs were active when sampled. Finally, these methods are characterized by high sensitivity and specificity.
ACKNOWLEDGMENTS
This research was supported by Air Force Office of Scientific Research grant F49620-95-0346 and USDA/Hatch grant 189434.
We are grateful to D. R. Bond for discussions, to R. Garen for producing the gel images, and for P. L. Lisk for expert manuscript preparation.
REFERENCES
- 1.Asturias J A, Diaz E, Timmis K N. The evolutionary relationship of biphenyl dioxygenase from gram-positive Rhodococcus globerulus P6 to multicomponent dioxygenases from gram-negative bacteria. Gene. 1995;156:11–18. doi: 10.1016/0378-1119(94)00530-6. [DOI] [PubMed] [Google Scholar]
- 2.Beller H R, Spormann A M. Anaerobic activation of toluene and o-xylene by addition to fumarate in denitrifying strain T. J Bacteriol. 1997;179:670–676. doi: 10.1128/jb.179.3.670-676.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson D A, Boguski M S, Lipman D J, Ostell J, Ouellette B F F. GenBank. Nucleic Acids Res. 1998;26:1–7. doi: 10.1093/nar/26.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beverley S M. Enzymatic amplification of RNA by PCR. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. New York, N.Y: Greene Publishing Associates and John Wiley & Sons; 1992. pp. 15.13–15.14. [Google Scholar]
- 5.Boschker H T S, Nold S C, Wellsbury P, Bos D, de Graaf W, Pel R, Parkes R J, Cappenburg T E. Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature. 1998;392:801–805. [Google Scholar]
- 6.Cerniglia C E. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation. 1992;3:351–368. [Google Scholar]
- 7.Chomczynski P. Solubilization in formamide protects RNA from degradation. Nucleic Acids Res. 1992;20:3791–3792. doi: 10.1093/nar/20.14.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Denome S A, Stanley D C, Olson E S, Young K D. Metabolism of dibenzothiophene and naphthalene in Pseudomonas strains: complete DNA sequence of an upper naphthalene catabolic pathway. J Bacteriol. 1993;175:6890–6901. doi: 10.1128/jb.175.21.6890-6901.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erickson B D, Mondello F J. Nucleotide sequencing and transcriptional mapping of genes encoding biphenyl dioxygenase, a multicomponent PCB-degrading enzyme in Pseudomonas strain LB400. J Bacteriol. 1992;174:2903–2912. doi: 10.1128/jb.174.9.2903-2912.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleming J T, Sanseverino J, Sayler G S. Quantitative relationship between naphthalene catabolic gene frequency and expression in predicting PAH degradation in soils at town gas manufacturing sites. Environ Sci Technol. 1993;27:1068–1074. [Google Scholar]
- 12.Fleming J T, Zhang D J. Abstracts of the 94th General Meeting of the American Society for Microbiology 1994. Washington, D.C: American Society for Microbiology; 1994. Abstr. Q-161. [Google Scholar]
- 13.Fuenmayor S L, Wild M, Boyes A L, Williams P A. A gene cluster encoding steps in conversion of naphthalene to gentisate in Pseudomonas sp. strain U2. J Bacteriol. 1998;180:2522–2530. doi: 10.1128/jb.180.9.2522-2530.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furukawa K, Hirose J, Suyama A, Zaiki T, Hayashida S. Gene components responsible for discrete substrate specificity in the metabolism of biphenyl (bph operon) and toluene (tod operon) J Bacteriol. 1993;175:5224–5232. doi: 10.1128/jb.175.16.5224-5232.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goyal A K, Zylstra G J. Genetics of naphthalene and phenanthrene degradation by Comomonas testosteroni. J Ind Microbiol Biotechnol. 1997;19:401–407. doi: 10.1038/sj.jim.2900476. [DOI] [PubMed] [Google Scholar]
- 16.Goyal A K, Zylstra G J. Molecular cloning of novel genes for polycyclic aromatic hydrocarbon degradation from Comomonas testosteroni GZ39. Appl Environ Microbiol. 1996;62:230–236. doi: 10.1128/aem.62.1.230-236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gray J P, Herwig R P. Phylogenetic analysis of the bacterial communities in marine sediments. Appl Environ Microbiol. 1996;62:4049–4059. doi: 10.1128/aem.62.11.4049-4059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerin W F, Boyd S A. Maintenance and induction of naphthalene degradation activity in Pseudomonas putida and an Alcaligenes sp. under different culture conditions. Appl Environ Microbiol. 1995;61:4061–4068. doi: 10.1128/aem.61.11.4061-4068.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harayama S, Kok M, Neidle E L. Functional and evolutionary relationships among diverse oxygenases. Annu Rev Microbiol. 1992;46:565–601. doi: 10.1146/annurev.mi.46.100192.003025. [DOI] [PubMed] [Google Scholar]
- 20.Harayama S, Rekik M, Bairoch A, Neidle E L A, Ornston L N. Potential DNA slippage structures acquired during evolutionary divergence of Acinetobacter calcoaceticus chromosomal benABC and Pseudomonas putida TOL pWWO plasmid xylXYZ, genes encoding benzoate dioxygenases. J Bacteriol. 1991;173:7540–7548. doi: 10.1128/jb.173.23.7540-7548.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herrick J B. Ph.D. thesis. Ithaca, N.Y: Cornell University; 1995. [Google Scholar]
- 22.Herrick J B, Madsen E L, Batt C A, Ghiorse W C. Polymerase chain reaction amplification of naphthalene-catabolic and 16S rRNA gene sequences from indigenous sediment bacteria. Appl Environ Microbiol. 1993;59:687–694. doi: 10.1128/aem.59.3.687-694.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herrick J B, Stuart-Keil K G, Ghiorse W C, Madsen E L. Natural horizontal transfer of a naphthalene dioxygenase gene between bacteria native to a coal tar-contaminated field site. Appl Environ Microbiol. 1997;63:2330–2337. doi: 10.1128/aem.63.6.2330-2337.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodson R E, Dustman W A, Garg R P, Moran M A. In situ PCR for visualization of microscale distribution of specific genes and gene products in prokaryotic communities. Appl Environ Microbiol. 1995;61:4074–4082. doi: 10.1128/aem.61.11.4074-4082.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeffrey W H, Nazaret S, Barkay T. Detection of the merA gene and its expression in the environment. Microb Ecol. 1996;32:293–303. doi: 10.1007/BF00183064. [DOI] [PubMed] [Google Scholar]
- 26.Jeffrey W H, Nazaret S, Von Haven R. Improved method for recovery of mRNA from aquatic samples and its application to detection of mer expression. Appl Environ Microbiol. 1994;60:1814–1821. doi: 10.1128/aem.60.6.1814-1821.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimbara K, Hashimoto T, Fukuda M, Koana T, Takagi M, Oishi M A, Yano K. Cloning and sequencing of two tandem genes involved in degradation of 2,3-dihydroxybiphenyl to benzoic acid in the polychlorinated biphenyl-degrading soil bacterium Pseudomonas sp. strain KKS102. J Bacteriol. 1989;171:2740–2747. doi: 10.1128/jb.171.5.2740-2747.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kramer J G, Wyman M, Zehr J P, Capone D G. Diel variability in transcription of the structural gene for glutamine synthetase (glnA) in natural populations of the marine diazotrophic cyanobacterium Trichodesmium thiebautii. FEMS Microbiol Ecol. 1996;21:187–196. [Google Scholar]
- 29.Kurkela S, Lehvaeslaiho H, Palva E T, Teeri T H. Cloning, nucleotide sequence and characterization of genes encoding naphthalene dioxygenase of Pseudomonas putida strain NCIB9816. Gene. 1988;73:355–362. doi: 10.1016/0378-1119(88)90500-8. [DOI] [PubMed] [Google Scholar]
- 30.Madsen E L. A critical analysis of methods for determining the composition and biogeochemical activities of soil microbial communities in situ. In: Stotzky G, Bollag J M, editors. Soil biochemistry. Vol. 9. New York, N.Y: Marcel Dekker, Inc.; 1996. pp. 287–370. [Google Scholar]
- 31.Madsen E L. Epistemology of environmental microbiology. Environ Sci Technol. 1998;32:429–439. [Google Scholar]
- 32.Madsen E L. Methods for determining biodegradability. In: Hurst C J, Knudsen G R, McInerney M J, Stetzenbach L D, Walter M V, editors. Manual of environmental microbiology. Washington, D.C: American Society for Microbiology; 1997. pp. 709–720. [Google Scholar]
- 33.Madsen E L, Bilotta-Best S E, Ghiorse W C. Development of a rapid 14C-based field method for assessing potential biodegradation of organic compounds in soil and sediment samples. J Microbiol Methods. 1995;21:317–327. [Google Scholar]
- 34.Madsen E L, Mann C L, Bilotta S E. Oxygen limitations and aging as explanations for the field persistence of naphthalene in coal tar-contaminated surface sediments. Environ Toxicol Chem. 1996;15:1876–1882. [Google Scholar]
- 35.Madsen E L, Sinclair J L, Ghiorse W C. In situ biodegradation: microbiological patterns in a contaminated aquifer. Science. 1991;252:830–833. doi: 10.1126/science.2028258. [DOI] [PubMed] [Google Scholar]
- 36.Madsen E L, Thomas C T, Wilson M S, Sandoli R L, Bilotta S E. In situ dynamics of aromatic hydrocarbons (AHs) and bacteria capable of AH metabolism in a coal tar waste-contaminated field site. Environ Sci Technol. 1996;30:2412–2416. [Google Scholar]
- 37.Masai E, Yamada A, Healy J M, Hatta T, Kimbara K, Fukuda M, Yano K. Characterization of biphenyl catabolic genes of gram-positive polychlorinated biphenyl degrader Rhodococcus sp. strain RHA1. Appl Environ Microbiol. 1995;61:2079–2085. doi: 10.1128/aem.61.6.2079-2085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mason J R, Cammack R. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu Rev Microbiol. 1992;46:277–305. doi: 10.1146/annurev.mi.46.100192.001425. [DOI] [PubMed] [Google Scholar]
- 39.Meinkoth J, Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984;138:267–272. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- 40.More M I, Herrick J B, Silva M C, Ghiorse W C, Madsen E L. Quantitative cell lysis of indigenous microorganisms and rapid extraction of microbial DNA from sediment. Appl Environ Microbiol. 1994;60:1572–1580. doi: 10.1128/aem.60.5.1572-1580.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murarka I, Neuhauser E, Sherman M, Taylor B B, Mauro D M, Ripp J, Taylor T. Organic substances in the subsurface: Delineation, migration, and remediation. J Hazard Mater. 1992;32:245–261. [Google Scholar]
- 42.National Research Council. In situ bioremediation: when does it work? Washington, D.C: National Academy Press; 1993. [Google Scholar]
- 43.Nazaret S, Jeffrey W H, Saouter E, Von Haven R, Barkay T. merA gene expression in aquatic environments measured by mRNA production and Hg(II) volatilization. Appl Environ Microbiol. 1994;60:4059–4065. doi: 10.1128/aem.60.11.4059-4065.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neidle E L, Hartnett C, Ornston L N, Bairoch A, Rekik M, Harayama S. Nucleotide sequences of the Acinetobacter calcoaceticus benABC genes for benzoate 1,2-dioxygenase reveal evolutionary relationships among multicomponent oxygenases. J Bacteriol. 1991;173:5385–5395. doi: 10.1128/jb.173.17.5385-5395.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogram A, Sun W, Brockman F J, Fredrickson J K. Isolation and characterization of RNA from low-biomass deep-subsurface sediments. Appl Environ Microbiol. 1995;61:763–768. doi: 10.1128/aem.61.2.763-768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parales J V, Kumar A, Parales R E, Gibson D T. Cloning and sequencing of the genes encoding 2-nitrotoluene dioxygenase from Pseudomonas sp. JS42. Gene. 1996;181:57–61. doi: 10.1016/s0378-1119(96)00462-3. [DOI] [PubMed] [Google Scholar]
- 47.Parales R E, Emig M D, Lynch N A, Gibson D T. Substrate specificities of hybrid naphthalene and 2,4-dinitrotoluene dioxygenase enzyme systems. J Bacteriol. 1998;180:2337–2344. doi: 10.1128/jb.180.9.2337-2344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paul J H. Carbon cycling: molecular regulation of photosynthetic carbon fixation. Microb Ecol. 1996;32:231–245. doi: 10.1007/BF00183060. [DOI] [PubMed] [Google Scholar]
- 49.Pichard S L, Campbell L, Kang J B, Tabita F R, Paul J H. Regulation of ribulose bisphosphate carboxylase gene expression in natural phytoplankton communities. I. Diel rhythms. Mar Ecol Prog Ser. 1996;139:257–265. [Google Scholar]
- 50.Pichard S L, Frischer M E, Paul J H. Ribulose bisphosphate carboxylase gene expression in subtropical marine phytoplankton populations. Mar Ecol Prog Ser. 1993;101:55–65. [Google Scholar]
- 51.Pichard S L, Paul J H. Detection of gene expression in genetically engineered microorganisms and natural phytoplankton populations in the marine environment by mRNA analysis. Appl Environ Microbiol. 1991;57:1721–1727. doi: 10.1128/aem.57.6.1721-1727.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pichard S L, Paul J H. Gene expression per gene dose, a specific measure of gene expression in aquatic microorganisms. Appl Environ Microbiol. 1993;59:451–457. doi: 10.1128/aem.59.2.451-457.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roux K H. Optimization and troubleshooting in PCR. In: Dieffenbach C W, Dveksler G S, editors. PCR primer: a laboratory manual. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 53–62. [Google Scholar]
- 54.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 55.Sandoli R L, Ghiorse W C, Madsen E L. Regulation of microbial phenanthrene mineralization in sediment samples by sorbent-sorbate contact time, inocula and gamma irradiation-induced sterilization artifacts. Environ Toxicol Chem. 1996;15:1901–1907. [Google Scholar]
- 56.Sanseverino J, Werner C, Fleming J, Applegate B, King J M H, Sayler G S. Molecular diagnostics of polyaromatic hydrocarbon biodegradation in manufactured gas plant soils. Biodegradation. 1993;4:303–321. doi: 10.1007/BF00695976. [DOI] [PubMed] [Google Scholar]
- 57.Sayler G S, Layton A, Lajoie C, Bowman J, Tschantz M, Fleming J T. Molecular site assessment and process monitoring in bioremediation and natural attenuation. Appl Biochem Biotechnol. 1995;54:277–290. doi: 10.1007/BF02787926. [DOI] [PubMed] [Google Scholar]
- 58.Siering P L. The double helix meets the crystal lattice: the power and pitfalls of nucleic acid approaches for biomineralogical investigations. Am Mineral. 1998;83:1593–1607. [Google Scholar]
- 59.Siering P L, Ghiorse W C. Development and application of 16S rRNA-targeted probes for detection of iron- and manganese-oxidizing sheathed bacteria in environmental samples. Appl Environ Microbiol. 1997;63:644–651. doi: 10.1128/aem.63.2.644-651.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Simon M J, Osslund T D, Saunders R, Ensley B D, Suggs S, Harcourt A, Suen W C, Cruden D L, Gibson D T, Zylstra G J. Sequences of genes encoding naphthalene dioxygenase in Pseudomonas putida strains G7 and NCIB 9816-4. Gene. 1993;127:31–37. doi: 10.1016/0378-1119(93)90613-8. [DOI] [PubMed] [Google Scholar]
- 61.Stuart-Keil K G, Hohnstock A M, Drees K P, Herrick J B, Madsen E L. Plasmids responsible for horizontal transfer of naphthalene catabolic genes between bacteria at a coal tar-contaminated site are homologous to pDTG1 from Pseudomonas putida 9816-4. Appl Environ Microbiol. 1998;64:3633–3640. doi: 10.1128/aem.64.10.3633-3640.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suen W C, Haigler B E, Spain J C. 2,4-Dinitrotoluene dioxygenase from Burkholderia sp. strain DNT: similarity to naphthalene dioxygenase. J Bacteriol. 1996;178:4926–4934. doi: 10.1128/jb.178.16.4926-4934.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sutherland J B, Rafii F, Khan A A, Cerniglia C E. Mechanisms of polycyclic aromatic hydrocarbon degradation. In: Young L Y, Cerniglia C E, editors. Microbial transformation and degradation of toxic organic chemicals. New York, N.Y: John Wiley & Sons, Inc.; 1995. pp. 269–306. [Google Scholar]
- 64.Taira K, Hirose J, Hayashida S A, Furukawa K. Analysis of bph operon from the polychlorinated biphenyl-degrading strain of Pseudomonas pseudoalcaligenes KF707. J Biol Chem. 1992;267:4844–4853. [PubMed] [Google Scholar]
- 65.Takizawa N, Kaida N, Torigoe S, Moritani T, Sawada T, Satoh S, Kiyohara H. Identification and characterization of genes encoding polycyclic aromatic hydrocarbon dioxygenase and polycyclic aromatic hydrocarbon dihydrodiol dehydrogenase in Pseudomonas putida OUS82. J Bacteriol. 1994;176:2444–2449. doi: 10.1128/jb.176.8.2444-2449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tan H, Tang H, Joannou C, Abdel-Wahab N, Mason J. The Pseudomonas putida ML2 plasmid-encoded genes for benzene dioxygenase are unusual in codon usage and low in G+C content. Gene. 1993;130:33–39. doi: 10.1016/0378-1119(93)90343-2. [DOI] [PubMed] [Google Scholar]
- 67.Tsai Y-L, Park M J, Olson B H. Rapid method for direct extraction of mRNA from seeded soils. Appl Environ Microbiol. 1991;57:765–768. doi: 10.1128/aem.57.3.765-768.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van der Meer J R, Zehnder A J, de Vos W M. Identification of a novel composite transposable element, Tn5280, carrying chlorobenzene dioxygenase genes of Pseudomonas sp. strain p51. J Bacteriol. 1991;173:7077–7083. doi: 10.1128/jb.173.22.7077-7083.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y, Garnon J, Labbe D, Bergeron H, Lau P C K. Sequence and expression of the bpdC1C2BADE genes involved in the initial steps of biphenyl/chlorobiphenyl degradation by Rhodococcus sp. M5. Gene. 1995;164:117–122. doi: 10.1016/0378-1119(95)00448-f. [DOI] [PubMed] [Google Scholar]
- 70.Williams P A, Sayers J R. The evolution of pathways for aromatic hydrocarbon oxidation in Pseudomonas. Biodegradation. 1994;5:195–217. doi: 10.1007/BF00696460. [DOI] [PubMed] [Google Scholar]
- 71.Wilson M S, Madsen E L. Field extraction of a transient intermediary metabolite indicative of real time in situ naphthalene biodegradation. Environ Sci Technol. 1996;30:2099–2103. [Google Scholar]
- 72.Wyman M, Zehr J P, Capone D G. Temporal variability in nitrogenase gene expression in natural populations of the marine cyanobacterium Trichodesmium thiebautii. Appl Environ Microbiol. 1996;62:1073–1075. doi: 10.1128/aem.62.3.1073-1075.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu H H, Tabita F R. Ribulose-1,5-bisphosphate carboxylase/oxygenase gene expression and diversity of Lake Erie planktonic microorganisms. Appl Environ Microbiol. 1996;62:1913–1921. doi: 10.1128/aem.62.6.1913-1921.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yen K-M, Serdar C M. Genetics of naphthalene catabolism in psuedomonads. Crit Rev Microbiol. 1988;15:247–268. doi: 10.3109/10408418809104459. [DOI] [PubMed] [Google Scholar]
- 75.Zehr J P, Wyman M, Miller V, Duguay L, Capone D G. Modification of the Fe protein of nitrogenase in natural populations of Trichodesmium thiebautii. Appl Environ Microbiol. 1993;59:669–676. doi: 10.1128/aem.59.3.669-676.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zylstra G J, Gibson D T. Toluene degradation by Pseudomonas putida F1: nucleotide sequence of the todC1C2BADE genes and their expression in E. coli. J Biol Chem. 1989;264:14940–14946. [PubMed] [Google Scholar]
- 77.Zylstra, G. J., and A. K. Goyal. 1996. Personal communication.