Abstract
The bone remodeling process is crucial for titanium (Ti) osseointegration and involves the crosstalk between osteoclasts and osteoblasts. Considering the high osteogenic potential of Ti with nanotopography (Ti Nano) and that osteoclasts inhibit osteoblast differentiation, we hypothesized that nanotopography attenuate the osteoclast-induced disruption of osteoblast differentiation. Osteoblasts were co-cultured with osteoclasts on Ti Nano and Ti Control and non-co-cultured osteoblasts were used as control. Gene expression analysis using RNAseq showed that osteoclasts downregulated the expression of osteoblast marker genes and upregulated genes related to histone modification and chromatin organization in osteoblasts grown on both Ti surfaces. Osteoclasts also inhibited the mRNA and protein expression of osteoblast markers, and such effect was attenuated by Ti Nano. Also, osteoclasts increased the protein expression of H3K9me2, H3K27me3 and EZH2 in osteoblasts grown on both Ti surfaces. ChIP assay revealed that osteoclasts increased accumulation of H3K27me3 that represses the promoter regions of Runx2 and Alpl in osteoblasts grown on Ti Control, which was reduced by Ti Nano. In conclusion, these data show that despite osteoclast inhibition of osteoblasts grown on both Ti Control and Ti Nano, the nanotopography attenuates the osteoclast-induced disruption of osteoblast differentiation by preventing the increase of H3K27me3 accumulation that represses the promoter regions of some key osteoblast marker genes. These findings highlight the epigenetic mechanisms triggered by nanotopography to protect osteoblasts from the deleterious effects of osteoclasts, which modulate the process of bone remodeling and may benefit the osseointegration of Ti implants.
Keywords: histone, nanotopography, osteoblast, osteoclast, titanium
1. Introduction
A focus of biomaterial investigation is the interactions between titanium (Ti) implants and bone tissue, an essential component of osseointegration [1–4]. To improve the integrative ability of Ti, modifications of implant surfaces have been made, such as functionalization with molecules and creation of different topographies. These modifications affect signaling pathways and cellular events involved in cell adhesion and proliferation, as well as osteoprogenitor cell differentiation, which ultimately regulate the formation of mineralized extracellular matrix [5–9].
Chemical conditioning with H2SO4/H2O2 solution creates a Ti surface with nanotopography that reduces the contaminants, increases the oxide layer (TiO2) from 5 to 32–40 nm and generates nano-sized pits of 20–22 nm average diameters [6,10,11]. This nanotopography favors cell adhesion and osteoblast differentiation by modulating several signaling pathways such as integrins, bone morphogenetic proteins (BMPs) and Wnt [6,7,11–14]. To date, there are no investigations on the effects of this nanotopography on the crosstalk between osteoblasts and osteoclasts, which may have a direct impact on bone remodeling and consequently on Ti osseointegration.
Bone remodeling is a physiological process regulated by continuous cycles of bone resorption and formation and depends on complex interactions between osteoblasts and osteoclasts [15]. Osteoblasts are derived from mesenchymal stem cells and osteoclasts are derived from hematopoietic cells through the fusion of macrophages after contact with stromal or osteoblastic cells, thus forming multinucleated cells [16,17]. Osteoblasts synthesize cytokines and growth factors that regulate the formation and activity of osteoclasts, including receptor activator of nuclear factor Kappa B ligand (RANKL) and macrophage colony stimulating factor (M-CSF); osteoclasts secrete sphingosine 1 phosphate (S1P), collagen triple helix repeat containing 1 (CTHRC1) and complement component C (C3) that stimulate while semaphorin 4D (SEMA4D) and sclerostin (SCL) inhibit osteoblast differentiation [16,18–21]. Such fine balance of the interactions between osteoblasts and osteoclasts is regulated by several cell mechanisms, including epigenetic modifications.
Epigenetic modifications comprise changes in response to environmental stimuli without alterations of the DNA sequence. The phenomenon is based on chemical modifications of proteins and DNA resulting in a diversity of chromatin accessibility [22]. Histone methylation, a mechanism that contributes to chromatin dynamics and occurs through enzymes that catalyze the addition of methyl groups to lysine and arginine residues of histones, is associated with changes in the activation and repression of gene expression. Depending on the histone and the residue that will undergo, these changes can participate in the regulation of several tumors and acting on cell differentiation [22–25]. Histones H3K9 and H3K27 methylations, which are associated with a highly compacted and transcriptionally repressed chromatin, inhibit gene transcription and the osteoblast differentiation [26–28]. Such inhibitory effects of histone methylation on osteoblast differentiation are underexplored in the crosstalk between osteoblasts and osteoclasts, especially in the context of Ti osseointegration.
Considering the high osteogenic potential of Ti with nanotopography and that osteoclasts inhibit osteoblast differentiation, we hypothesized that this nanotopography inhibits the deleterious effects of osteoclasts on osteoblasts by regulating histone modifications. To test this hypothesis, we established a co-culture model in which osteoblasts were grown on Ti discs and osteoclasts were cultured into inserts positioned above the discs, thus sharing the same microenvironment. Our results demonstrated that this nanotopography attenuated the disruption of osteoblast differentiation induced by osteoclasts by reducing the accumulation of methylated histones that repress the promoter regions of some osteoblast marker genes.
2. Materials and methods
2.1. Materials and reagents
Discs of commercially pure grade II Ti (1.0 mm thickness and 13 mm in diameter) were purchased from Realum (São Paulo, São Paulo, Brazil) and silicon carbides from Norton (Guarulhos, São Paulo, Brazil). MC3T3-E1 and RAW 264.7 cell lines were obtained from American Type Culture Collection (ATCC) (Manassas, Virginia, USA). Minimum essential medium alpha-modification (α-MEM), Dulbeccco’s modified eagle medium (DMEM), fetal bovine serum (FBS), penicillin-streptomycin, TRIzol reagent and immunoprecipitation kit dynabeads protein G were purchased from Thermo Fisher Scientific (Waltham, Massachusetts, USA). β-glycerophosphate, L-ascorbic acid, bovine serum albumin, Tween® 20, bone morphogenetic protein receptor type 1A antibody (anti-BMPR1A, SAB1302614), Hoechst 33258 (bisBenzimide H 33258 solution), phenylmethanesulfonyl fluoride and ethyl alcohol pure were purchased from Sigma-Aldrich (St. Louis, Missouri, USA). Runt related transcription factor 2 (anti-RUNX2, 8486S), di-methyl-histone H3 (Lys9) (anti-H3K9me2, D85B4), tri-methyl-histone H3 (Lys27) (anti-H3K27me3, C36B11), enhancer of zeste 2 polycomb repressive complex 2 subunit (anti-EZH2, D2C9) and secondary horseradish peroxidase-conjugated goat anti-rabbit IgG (7074S) antibodies were purchased from Cell Signaling (Beverly, Massachusetts, USA). Alkaline phosphatase (anti-ALPL, ab108337) and anti-RUNX2 (ab76956) were purchased from Abcam (Cambridge, Massachusetts, United Kingdom) and glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH, sc-25778) and anti-ALPL (sc-137213) from Santa Cruz Technology (Dallas, Texas, USA). Red- fluorescent Alexa Fluor 594- conjugated goat anti- mouse secondary antibody (A11005) and green-fluorescent Alexa Fluor 488- conjugated goat anti- rabbit secondary antibody (A11008) were purchased from Molecular Probes (Eugene, Oregon, USA). Twenty-four-well ThinCert™ cell culture inserts and 24-well plates were purchased from Greiner Bio-One (Frickenhausen, Baden-Württemberg, Germany), Millex GV filter (0.22 μm), ethanol, formaldehyde, sulfuric acid (95–97%) and hydrogen peroxide (30%) from Merck Millipore (Darmstadt, Hesse, Germany) and RANKL from Peprotech (Rocky Hill, Connecticut, USA). SMARTer® Stranded Total RNA Hi Mammalian Sample Prep Kit was purchased from Takara (Mountain View, California, USA) and Qubit® RNA HS assay kit from Life Technologies (Carlsbad, California, USA). High-capacity cDNA reverse transcription kit and Fast SYBR green master mix reagent was obtained from Applied Biosystems (Foster City, California, USA). Protease inhibitor tablets and MG132 proteasome inhibitor were acquired from Roche Applied Science (Indianapolis, Indiana, USA). Trans-blot turbo PVDF membrane, nonfat powdered milk and Clarity western ECL substrate were purchased from Bio-Rad Laboratories (Hercules, California, USA). MinElute PCR purification kit was purchased from QIAGEN (Hilden, Nordrhein-Westfalen, Germany). UNC1999 was purchased from Cayman Chemical (Ann Arbor, Michigan, USA).
2.2. Ti surface modification and characterization
The discs of Ti (Realum) were polished with silicon carbide (320 and 600 grit) (Norton), cleaned in 70% ethanol (Merck) and washed by sonication in distilled water. The discs were conditioned for 4 hours, at room temperature, with a mixture of 10 N H2SO4 (Merck) and 30% aqueous H2O2 (Merck) to obtain the nanotopography (Ti Nano) [10]. Non-conditioned discs were used as control (Ti Control). Before cell culture experiments, all discs were cleaned by sonication, air-dried and autoclaved. The surfaces were characterized by field emission scanning electron microscopy (SEM) operated at 5 kV (Inspect S50, FEI, Hillsboro, Oregon, USA) and by atomic force microscopy (AFM) using an AFM Bruker Multimode 8 Microscope (Bruker, Santa Barbara, California, USA) operated in air using the contact mode and a contact probe DNP-10 (Bruker) with tip radius of 20 nm and spring constant of 0.35 N/m. The AFM images were treated, and the surface area (n = 5) were calculated using the software NanoScope Analysis 1.5 (Bruker).
2.3. Effect of osteoclasts on osteoblasts grown on Ti Nano
2.3.1. Cell cultures
MC3T3-E1 pre-osteoblastic cells were cultured in α-MEM (Gibco - Thermo Fisher Scientific), supplemented with 10% FBS (Gibco - Thermo Fisher Scientific), 100 U/mL penicillin (Gibco - Thermo Fisher Scientific) and 100 μg/mL streptomycin (Gibco - Thermo Fisher Scientific) and RAW 264.7 macrophage cells were cultured in DMEM (Gibco - Thermo Fisher Scientific), supplemented with 10% FBS (Gibco - Thermo Fisher Scientific), 100 U/mL penicillin (Gibco - Thermo Fisher Scientific) and 100 μg/mL streptomycin (Gibco - Thermo Fisher Scientific), both non-inducing differentiation media, and kept at 37 °C in a humid atmosphere containing 5% CO2 and 95% atmospheric air. After subconfluence, MC3T3-E1 cells were plated on Ti Nano and Ti Control discs and RAW 264.7 cells on 24-well ThinCert (Greiner Bio-One), in 24-well culture plates (Greiner Bio-One) at a density of 1×104 cells/well or insert. MC3T3-E1 cells were cultured in osteogenic medium, constituted by α-MEM (Gibco - Thermo Fisher Scientific), supplemented with 10% FBS (Gibco - Thermo Fisher Scientific), 100 U/mL penicillin (Gibco - Thermo Fisher Scientific), 100 μg/mL streptomycin (Gibco - Thermo Fisher Scientific), 7 mM β-glycerophosphate (Sigma-Aldrich) and 5 μg/mL L-ascorbic acid (Sigma-Aldrich) for 5 days and RAW 264.7 cells were cultured in osteoclastogenic medium, constituted by DMEM (Gibco - Thermo Fisher Scientific), supplemented with 10% FBS (Gibco - Thermo Fisher Scientific), 100 U/mL penicillin (Gibco - Thermo Fisher Scientific), 100 μg/mL streptomycin (Gibco - Thermo Fisher Scientific) and 50 ng/mL of RANKL (Peprotech) also for 5 days, and they were kept separated during this time. On day 5, the inserts containing the osteoclasts were positioned above osteoblast cultures, thereby establishing an indirect co-culture system. The osteoblast-osteoclast co-cultures were kept in α-MEM (Gibco - Thermo Fisher Scientific), supplemented with 10% FBS (Gibco - Thermo Fisher Scientific), 100 U/mL penicillin (Gibco - Thermo Fisher Scientific), 100 μg/mL streptomycin (Gibco - Thermo Fisher Scientific), 7 mM β-glycerophosphate (Sigma-Aldrich), 5 μg/mL L-ascorbic acid (Sigma-Aldrich) and 50 ng/mL of RANKL (Peprotech) for another 2 days, completing 7 days of culture in differentiation media. The co-culture medium was α-MEM supplemented with osteoblast and osteoclast differentiation factors, since DMEM reduced osteoblast differentiation compared to α-MEM, while α-MEM did not affect osteoclast differentiation compared to DMEM (data not shown). The controls were non-co-cultured osteoblasts grown on both Ti surfaces.
2.3.2. RNA isolation and library preparation
On day 7 (2 days of co-culture), the total RNA was isolated from the osteoblasts using TRizol reagent (Invitrogen - Thermo Fisher Scientific), according to the manufacturer’s specifications. RNA integrity was assessed by Bioanalyzer (Agilent, Santa Clara, California, USA). RNA-Seq libraries were built with the SMARTer® Stranded Total RNA Hi Mammalian Sample Prep Kit (Takara) according to the manufacturer’s protocol. The quality and quantification of RNA-Seq libraries were determined by Bioanalyzer (Agilent) and Qubit® (Life Technologies). The RNA-Seq libraries were single-end sequenced considering 75-bp single-end reads using a Hi-Seq 1500 Platform (Illumina, Hayward, California, USA) and three biological replicates (n = 3) of each sample.
2.3.3. RNA-Seq analysis
The dataset analysis was performed using the Galaxy Platform and RStudio [29,30]. The quality control of raw reads was done using FastQC and the alignment to reference genome mm10 using STAR [31,32]. The process data from high-throughput sequencing assays (expression counts) was performed using HTSeq with gene annotations (Gencode m23) [33,34]. Differential expression was analyzed by DESeq2 [35]. The GO enrichment analyses were performed using Gene Ontology (GO Ontology database Released 2019–12-09) and PANTHER Overrepresentation Test (Released 20190711) [36–38]. To consider differential gene expression, the cutoff for significant fold change was > 1.7, and adjusted p-value < 0.05.
2.3.4. mRNA expression by real-time polymerase chain reaction (RT-qPCR)
On day 7 (2 days of co-culture), RT-qPCR was carried out to evaluate the gene expression of runt related transcription factor 2 (Runx2), distal-less homeobox 5 (Dlx5), alkaline phosphatase (Alpl), bone sialoprotein (Ibsp), osteocalcin (Bglap), osteoprotegerin (Opg), bone morphogenetic protein receptor type 1A (Bmpr1a), inhibitor of DNA binding 3 (Id3), jun D proto-oncogene (Jund) and matrix metallopeptidase 13 (Mmp13). The total RNA collected from osteoblasts was quantified and the cDNA was done using high-capacity cDNA reverse transcription kit (Applied Biosystems) following the manufacturer’s instructions. The real-time PCR reactions were done with Fast SYBR green master mix reagent (Applied Biosystems) and the selected primers (Supplementary Material 1, Table s1). The results were normalized to actin-beta (Actb) and calibrated by non-co-cultured osteoblasts grown on Ti Control. The data (n = 3) were calculated according to the comparative 2−ΔΔCt method [39].
2.3.5. Protein expression by western blotting
The protein expression of RUNX2, ALPL and BMPR1A was detected by western blotting on day 7 (2 days of co-culture). Cells were lysed with 150 μL of buffer constituted by 1 × protease inhibitor mixture (Roche Applied Science), 1 mM phenylmethanesulfonyl fluoride (Sigma-Aldrich) and 25 mM MG132 proteasome inhibitor (Roche Applied Science). Briefly, 50 μg of total protein was denatured, separated in 10% SDS polyacrylamide electrophoresis gel and transferred to a trans-blot turbo PVDF membrane (Bio-Rad Laboratories). The membrane was blocked for 1 hour in Tris-buffered saline with 0.1% Tween® 20 (Sigma-Aldrich), containing 5% nonfat powdered milk (Bio-Rad Laboratories), probed with primary antibodies anti-RUNX2 (1:2000, Cell Signaling), anti-ALPL (1:3000, Abcam) and anti-BMPR1A (1:250, Sigma-Aldrich) overnight at 4°C, and incubated with secondary horseradish peroxidase-conjugated antibody goat anti-rabbit IgG (1:2000, Cell Signaling) for 1 hour at room temperature. The proteins were detected using Clarity Western ECL Substrate (Bio-Rad Laboratories) and the images were acquired using G-Box gel imaging (Syngene, Cambridge, UK). The proteins were quantified (n = 3) by counting pixels through Image-J software (National Institutes of Health, Bethesda, Maryland, USA), normalized to GAPDH (Santa Cruz Technology) and calibrated by non-co-cultured osteoblasts grown on Ti Control.
2.3.6. In situ ALPL activity by Fast red staining
The in situ ALPL activity was evaluated on day 7 (2 days of co-culture) using Fast red staining, as previously described [13]. Macroscopic images were digitally obtained with a stereomicroscope coupled to a high-resolution digital camera Leica DC 300F (Leica Biosystem, Wetzlar, Hesse, Germany) and ALPL staining was quantified (n = 4) by counting pixels using LASV 4.0 Image Analysis Software (Leica Biosystem). The data are presented as percentage of the area of the disc.
2.4. Effect of osteoclasts on methylated histones H3K9 and H3K27 accumulation in osteoblasts grown on Ti Nano
2.4.1. Protein expression by western blotting
The protein expression of H3K9me2, H3K27me3 and EZH2 was detected by western blotting on day 7 (2 days of co-culture). The membrane was blocked for 1 hour in Tris-buffered saline with 0.1% Tween® 20 (Sigma-Aldrich), containing 5% bovine serum albumin (Sigma-Aldrich), probed with primary antibodies anti-H3K9me2 (1:1000, Cell Signaling), anti-H3K27me3 (1:1000, Cell Signaling) and anti-EZH2 (1:1000, Cell Signaling) overnight at 4°C and incubated with secondary horseradish peroxidase-conjugated antibody goat anti-rabbit IgG (1:2000, Cell Signaling) for 1 hour at room temperature. The proteins were detected as described above in 2.3.5.
2.4.2. Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was carried out on day 7 (2 days of co-culture) as described elsewhere [40]. The osteoblasts were cross-linked with 1% formaldehyde (Merck) and lysed, and the DNA-protein complexes were immunoprecipitated with anti-H3K9me2 (Cell Signaling), anti-H3K27me3 (Cell Signaling) antibodies and protein G dynabeads kit (Invitrogen - Thermo Fisher Scientific). The DNA was extracted and purified with MinElute PCR purification kit (QIAGEN) after DNA-protein crosslink reversion. The PCR reactions were performed using the sequence primers for Runx2, Alpl, Ibsp, Id3, Bglap and Jund (Supplementary Material 1, Table s2) and the products were analyzed through 7% acrylamide gel, quantified by counting pixels through Image-J software (National Institutes of Health), normalized to positive control (INPUT) and calibrated by non-co-cultured osteoblasts grown on Ti Control.
2.4.3. Protein co-immunolocalization by immunofluorescence
On day 7 (2 days of co-culture), the RUNX2, ALPL and H3K27me3 proteins were evaluated by indirect immunofluorescence with anti- RUNX2 (1:100, Abcam), anti- ALPL (1:50, Santa Cruz Technology) and anti-H3K27me3 (1:200, Cell Signaling) antibodies, followed by a mixture of the red- fluorescent Alexa Fluor 594- conjugated goat anti- mouse secondary antibody (1:200, Molecular Probes) to detect the RUNX2 and ALPL and green-fluorescent Alexa Fluor 488- conjugated goat anti- rabbit secondary antibody to detect anti-H3K27me3 (1:200, Molecular Probes). Nuclei were detected with Hoechst 33258 solution (1:1000, Sigma-Aldrich). The samples were analyzed and randomly photographed in two areas per disc with an epifluorescence microscope ZEISS, ApoTome.2 (Carl Zeiss, Oberkochen, Baden-Württemberg, Germany).
2.5. Statistical analysis
The data of Ti surface area were compared using Student’s t-test while the data of mRNA expression by RT-qPCR, protein expression by western blotting and in situ ALPL activity by Fast red staining were compared by two-way ANOVA followed by the Holm-Sidak post-test when appropriate. All analyses were made using the SigmaPlot software (Systat Software Inc., San Jose, California, USA) and the level of significance was set at 5% (p ≤ 0.05).
3. Results
3.1. Characterization of Ti surfaces
The Ti Control exhibited a polished surface (Figure 1A) while the Ti Nano (Figure 1B) generated by H2SO4/H2O2 treatment presented nanopores distributed over the entire surface as observed under SEM. The surface area of Ti Nano (21.96 ± 0.52) was greater (p = 0.001) compared to Ti Control (14.44 ± 0.89) as determined by AFM (Figure 1 C–F).
Figure 1.
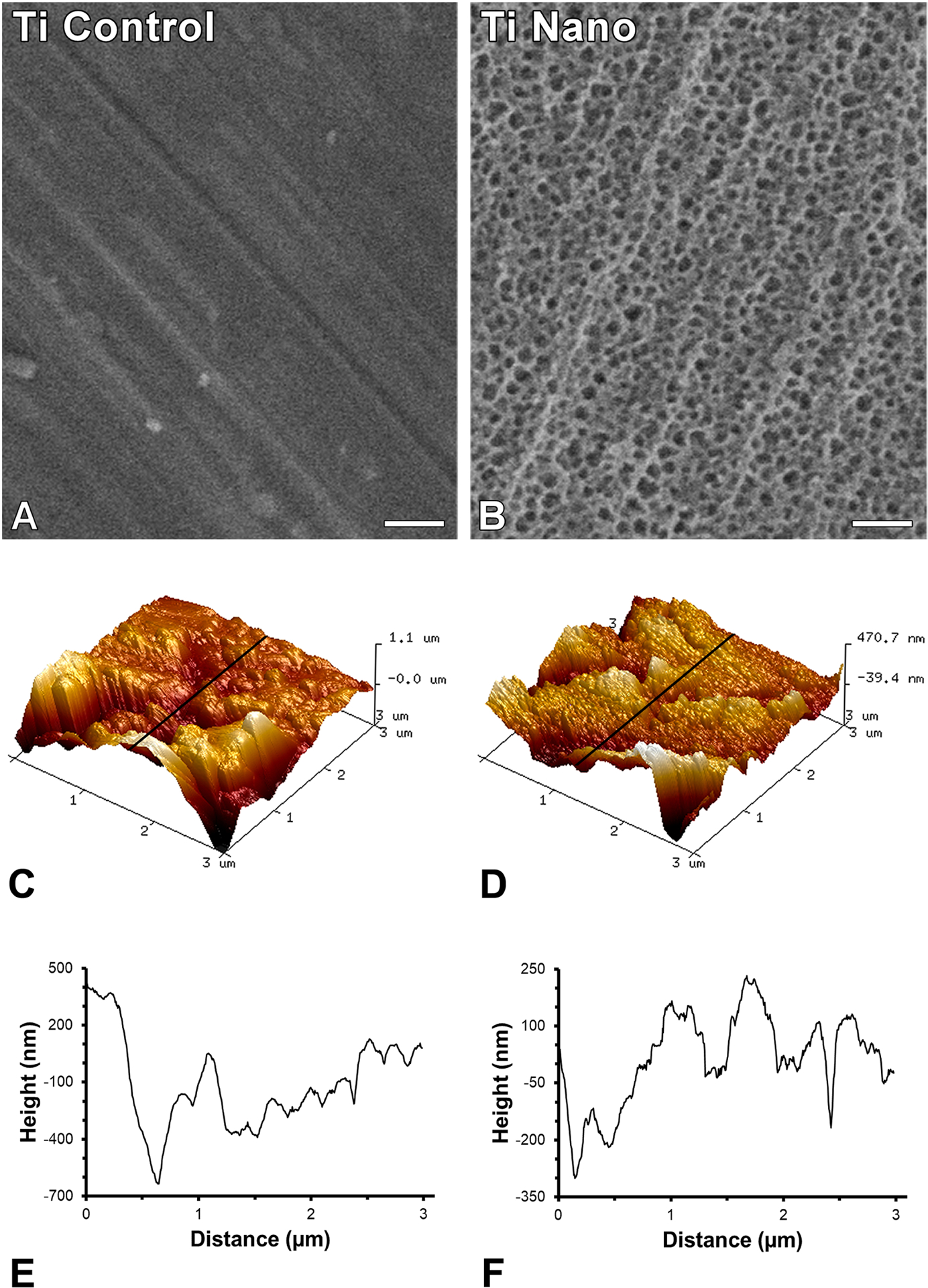
Surface topography of titanium (Ti) discs. Scanning electron micrographies of polished Ti (A, Ti Control) and Ti with nanotopography (B, Ti Nano). Three-dimensional surface topography generated by atomic force microscopy and the corresponding line profile of Ti Control (C, E) and Ti Nano (D, F). Scale bars (A and B) = 100 nm. The reader is referred to the web version of this article for the colour representation of this figure.
3.2. Osteoclasts induce differential expression of genes in osteoblasts grown on Ti Nano
Changes in gene expression of osteoblasts grown on both Ti Control and Ti Nano, either in the presence or absence of osteoclasts, were identified (Figure 2). The relationship between samples during this interaction was evidenced by both principal component analysis (PCA, Figure 2A) and Euclidean distance analysis (Figure 2B). Irrespective of Ti surface, the presence of osteoclasts generated separation from the samples cultured in the absence of osteoclasts in the first principal component (PC1, x-axis, 79% of variance, Figure 2A). The y-axis (PC2) shows 11% of variance and the samples of osteoblasts co-cultured with osteoclasts on Ti Nano were separately clustered from the osteoblasts co-cultured with osteoclasts on Ti Control (Figure 2A). No relevant differences were noted between non-co-cultured osteoblast grown on Ti Nano compared to non-co-cultured osteoblasts grown on Ti Control (Figure 2A). The Euclidean distance analysis (Figure 2B) corroborates the findings of PCA, indicating the great variance between osteoblasts grown in the presence of osteoclasts on both Ti Control and Ti Nano compared to the variance between non-co-cultured osteoblasts.
Figure 2.
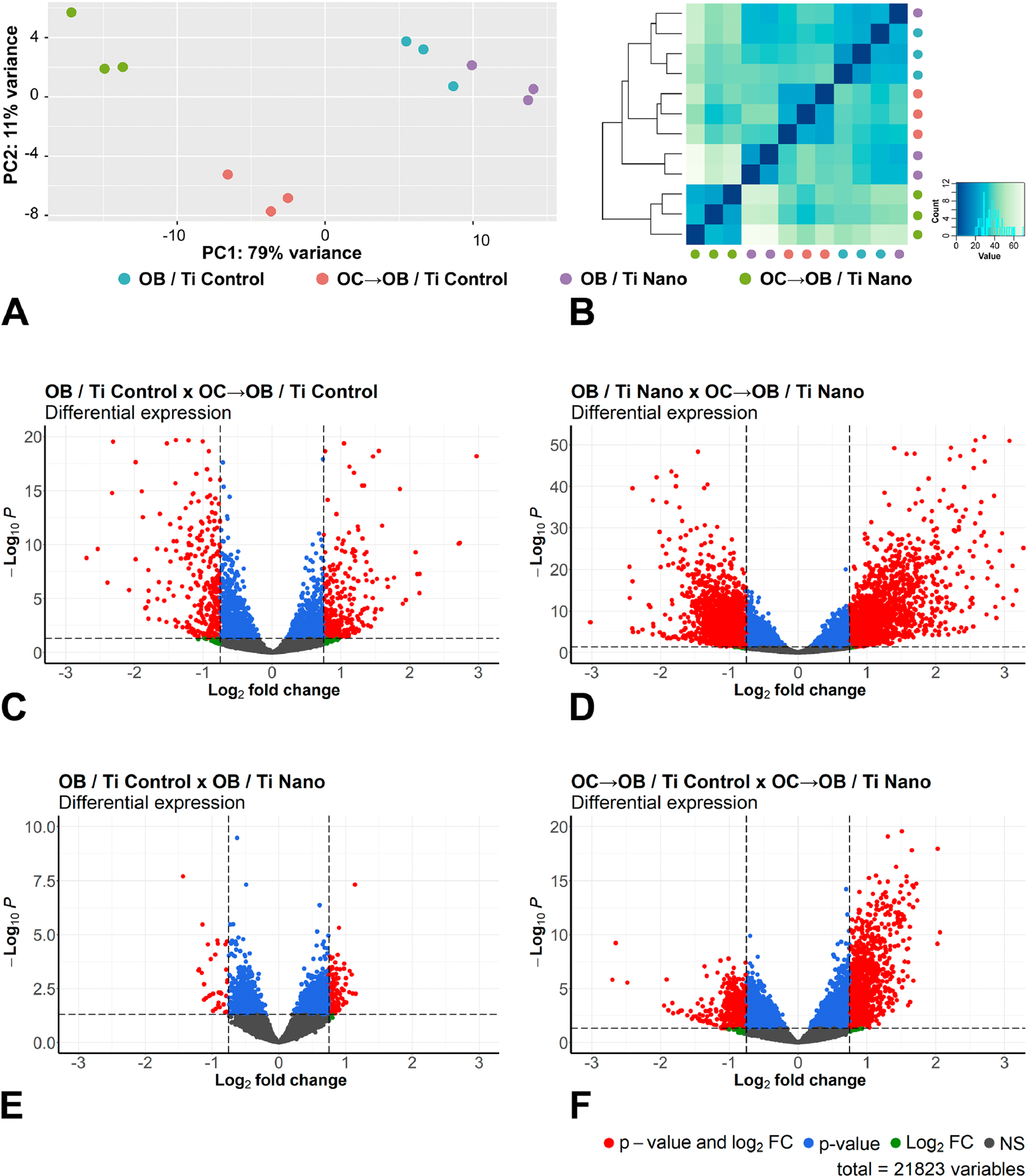
Osteoclasts induce differential expression of genes in osteoblasts grown on titanium with nanotopography (Ti Nano) compared to polished titanium (Ti Control). Principal component analysis (A) and Euclidean distance metric plot (B) with dots in different colors representing osteoblasts grown on Ti Control (OB / Ti Control), osteoblasts grown on Ti Control in the presence of osteoclasts (OC→OB / Ti Control), osteoblasts grown on Ti Nano (OB / Ti Nano) and osteoblasts grown on Ti Nano in the presence of osteoclasts (OC→OB / Ti Nano) (n = 3); the first component (PC1) showed 79% of variance and the second component (PC2) showed 11% (A). Volcano plots of the RNA-Seq data showing the differential gene expression (n = 3) by comparing OB / Ti Control with OC→OB / Ti Control (C), OB / Ti Nano with OC→OB / Ti Nano (D), OB / Ti Control with OB / Ti Nano (E) and OC→OB / Ti Control with OC→OB / Ti Nano (F). The upregulated genes are represented as red dots on the right and the downregulated ones as red dots on the left, which means that these genes presented statistically significant differences (adjusted p-value) and magnitude of fold change (FC). Blue dots indicate genes that only presented statistically significant differences (adjusted p-value), green dots, the genes that only presented magnitude of FC and the grey ones indicate genes without both statistically significant differences and magnitude of FC (C-F). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The Volcano plots showed the patterns of global gene expression and compared osteoblasts grown on Ti Control either in the presence or absence of osteoclasts (Figure 2C), osteoblasts grown on Ti Nano either in the presence or absence of osteoclasts (Figure 2D), osteoblasts grown on Ti Control and Ti Nano (Figure 2E) and osteoblasts grown in the presence of osteoclasts on Ti Control and Ti Nano (Figure 2F). The comparison of osteoblasts grown on Ti Control either in the presence or absence of osteoclasts showed that the presence of osteoclasts upregulated 305 and downregulated 322 genes (Figure 2C). By comparing osteoblasts grown on Ti Nano either in the presence or absence of osteoclasts, we found that the presence of osteoclasts upregulated 1825 and downregulated 1629 genes, quite a large difference from the Ti control (Figure 2D). Osteoblasts grown on either Ti Control or Ti Nano showed that nanotopography upregulated 116 and downregulated 34 genes (Figure 2E). By comparing osteoblasts grown in the presence of osteoclasts on Ti Control and Ti Nano, nanotopography upregulated 934 and downregulated 471 genes (Figure 2F) indicating that osteoblasts grown on Ti Nano are more responsive to osteoclasts than the ones grown on Ti Control. These data are summarized in Supplementary Material 1, Table s3.
The Heatmap shows a total of 4328 genes of osteoblasts that were regulated by both Ti surfaces and the presence or absence of osteoclasts (Figure 3A). Hierarchical clustering of these differentially expressed genes was performed and GO analysis identified biological processes involved in osteoblast differentiation and regulation of chromatin organization (Figure 3A,B). Cluster 1 presented a total of 223 genes highly expressed in non-co-cultured osteoblasts and a complete change in the pattern of gene expression when grown in the presence of osteoclasts, both being more evident on Ti Nano (Figure 3A). Among the regulated genes, GO identified genes involved in an essential biological process termed “regulation of osteoblast differentiation” including Wnt4 and Wnt7b, which are involved in bone formation and enhancement of fracture healing, Id1 and Id3, both targets modulated by the BMP signaling pathway, Jund that stimulates the osteocalcin gene transcription and Gli1, a hedgehog gene directly regulated by Runx2 (Figure 3B) [41–46]. Cluster 3 presented 1388 genes with low expression in osteoblasts grown on Ti Nano, which were upregulated by the presence of osteoclasts, while no relevant regulation was observed in osteoblasts grown on Ti Control irrespective of the presence of osteoclasts (Figure 3A). GO identified genes linked to “regulation of chromatin organization” and “histone modification” that have well-known roles in bone biology, including Sirt1, a regulator of bone mass and Kdm5a that inhibits bone formation [47,48]. The genes encoding the receptors Bmpr1a, Bmpr2 and Fzd5, involved with BMP and Wnt signaling pathways, are included in the GO category “positive regulation of transcription by RNA polymerase II” (Figure 3B). Cluster 4 presented a total of 771 genes that were downregulated by osteoclasts in osteoblasts grown on Ti Nano, while on Ti Control, osteoclasts induced a slight upregulation of these genes (Figure 3A). Among these, GO identified the following genes: Rarg that influences the development of trabecular bone mass and hematopoiesis; Notch1, an essential receptor involved in Notch signaling that regulates skeletal cell proliferation and differentiation; Alpl, a marker of early osteoblast differentiation that is crucial for bone formation; Bglap that encodes the most abundant non-collagenous protein in the bone matrix; Mustn1 that represents a pan-musculoskeletal cell marker that precedes Sox9 in cartilage; and Col5a1, a gene activated by osterix during osteoblast differentiation, that is related to “tube formation”, “alpha-amino acid biosynthetic process”, “tissue development” and “extracellular matrix organization” (Figure 3B) [49–55]. Cluster 5 presented a total of 747 genes that were downregulated by osteoclasts in osteoblasts cultured on both Ti surfaces, with more pronounced effect on Ti Nano (Figure 3A). Xbp1 encodes a protein that targets Osterix and Dot1l that plays an important role in prenatal and postnatal chondrocyte development and trabecular bone maintenance; each of these genes are included in the GO categories “cellular macromolecule biosynthetic process” and “cellular protein metabolic process” respectively (Figure 3B) [56,57,58]. Cluster 6 presented a total of 419 genes that were upregulated by osteoclasts in osteoblasts cultured on both Ti surfaces, with more pronounced effect on Ti Nano (Figure 3A). Among these, GO identified Grem1, a BMP antagonist associated with “regulation of cell growth”, Cav1 that negatively regulates osteoblasts and cementoblasts associated with “inactivation of MAPK activity”, Spry2 that is involved in proliferation and differentiation of osteoblasts, included in “developmental growth”; Mmp13, a marker of bone matrix remodeling and Spp1, a bone sialoprotein involved in osteoclast attachment to mineralized bone matrix, both associated with “Ossification” [59–63].
Figure 3.
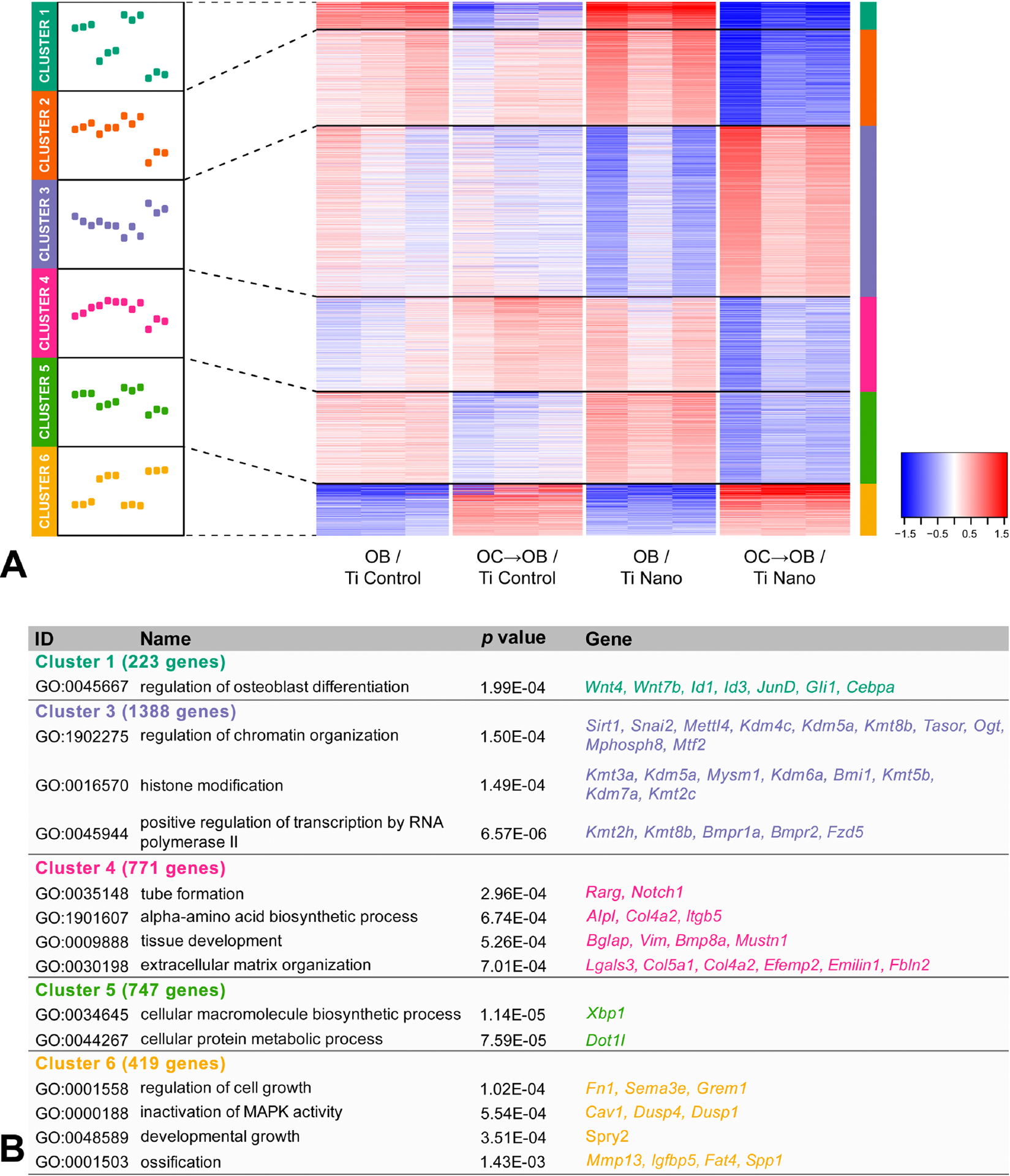
Osteoclasts induce differential expression of genes in osteoblasts grown on titanium with nanotopography (Ti Nano) compared to polished titanium (Ti Control). Heatmap showing the differential gene expression of osteoblasts grown on Ti Control (OB / Ti Control), osteoblasts grown on Ti Control in the presence of osteoclasts (OC→OB / Ti Control), osteoblasts grown on Ti Nano (OB / Ti Nano) and osteoblasts grown on Ti Nano in the presence of osteoclasts (OC→OB / Ti Nano) (n = 3). Six clusters were defined, and the line plots indicate the gene expression dynamics of each cluster (A). The clusters were submitted to GO analysis and selected GO terms and genes are presented in the table (B). The reader is referred to the web version of this article for the colour representation of this figure.
3.3. Ti Nano attenuates the osteoclast-induced disruption of osteoblast differentiation
To investigate if Ti Nano attenuates the negative effect of osteoclasts on osteoblasts, we measured the mRNA and protein expressions of key bone markers and the in situ ALPL activity (Figure 4). The gene expression of Runx2 and Bmpr1a in osteoblasts was upregulated (p = 0.001 for both) by Ti Nano, and the presence of osteoclasts downregulated their expressions on Ti Control (p = 0.001 for both genes) and upregulated on Ti Nano (p = 0.001; p=0.04) (Figure 4A). The gene expression of Dlx5, Ibsp and Opg in osteoblasts was upregulated (p = 0.005; p = 0.001; p = 0.001, respectively) by Ti Nano and the presence of osteoclasts downregulated their expressions (p = 0.001 for all three) in a more pronounced way on Ti Control (Figure 4A). The gene expression of Alpl and Bglap in osteoblasts was inhibited (p = 0.001 for both genes) by Ti Nano and the presence of osteoclasts downregulated their expressions (p = 0.001 for both genes) in a more pronounced way on Ti Control (Figure 4A). The gene expression of Id3 in osteoblasts was not affected (p = 0.990) by Ti surfaces and the presence of osteoclasts downregulated its expression on Ti Control and Ti Nano (p = 0.001 for both surfaces) (Figure 4A). The gene expression of Jund in osteoblasts was not affected (p = 0.319) by Ti surfaces and the presence of osteoclasts downregulated its expression on Ti Control (p = 0.021) and Ti Nano (p = 0.001) (Figure 4A). The gene expression of Mmp13 in osteoblasts was not affected (p = 0.551) by Ti surfaces and the presence of osteoclasts downregulated its expression on Ti Control and upregulated on Ti Nano (p = 0.001 for both surfaces) (Figure 4A). The protein expression of RUNX2 in osteoblasts was upregulated (p = 0.001) by Ti Nano and the presence of osteoclasts downregulated its expression on Ti Control and Ti Nano (p=0.001 for both) in a more pronounced way on Ti Control (Figure 4B). The protein expression of ALPL in osteoblasts was upregulated (p = 0.001) by Ti Nano and the presence of osteoclasts downregulated its expression on Ti Control and upregulated on Ti Nano (p = 0.001 for both) (Figure 4C). The protein expression of BMPR1A in osteoblasts was inhibited (p = 0.001) by Ti Nano and the presence of osteoclasts downregulated its expression on Ti Control and upregulated on Ti Nano (p = 0.001 for both) (Figure 4C). The ALPL activity in osteoblasts was increased (p = 0.001) by Ti Nano and the presence of osteoclasts downregulated its activity on Ti Control and Ti Nano (p = 0.001 for both) in a more pronounced way on Ti Control (Figure 4E).
Figure 4.
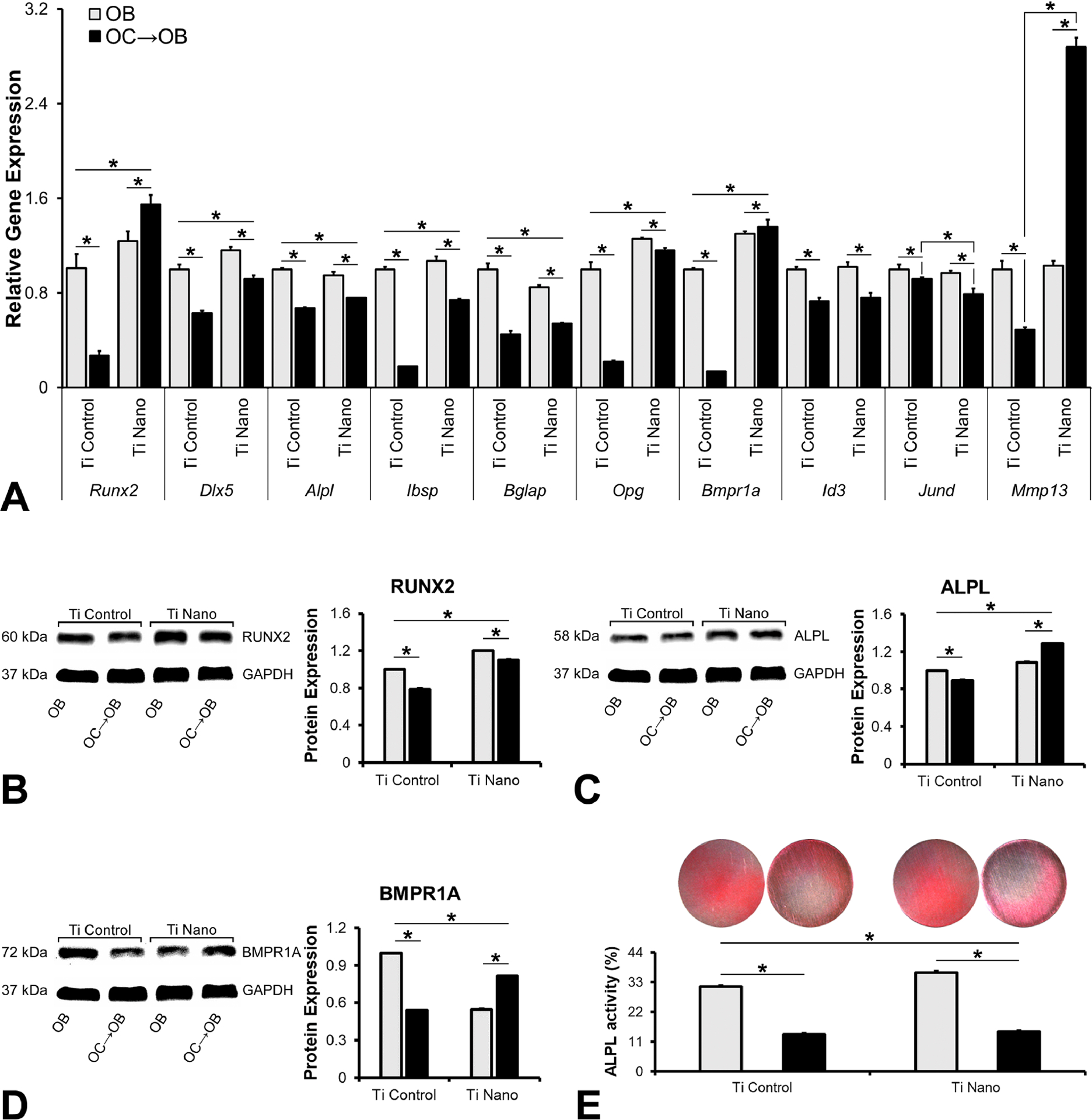
Titanium with nanotopography (Ti Nano) attenuates osteoclast-induced disruption of osteoblast differentiation. Gene expression of the bone markers Runx2, Dlx5, Alpl, Ibsp, Bglap, Opg, Bmpr1a, Id3, Jund and Mmp13 (A), protein expression of RUNX2 (B), ALPL (C) and BMPR1A (D) and in situ ALPL activity (E) of osteoblasts (OB) grown on polished titanium (Ti Control), osteoblasts grown in the presence of osteoclasts (OC→OB) on Ti Control, OB grown on Ti Nano and OC→OB on Ti Nano. The data of gene expression (n = 3), protein expression (n = 3) and in situ ALPL activity (n = 4) are presented as mean ± standard deviation and asterisks (*) indicate statistically significant differences (p ≤ 0.05). The reader is referred to the web version of this article for the colour representation of this figure.
3.4. Ti Nano attenuates the osteoclast-induced disruption of osteoblast differentiation by reducing accumulation of the methylated histones
The RNA-Seq analysis, especially cluster 3, identified genes strongly associated with regulation of chromatin organization and histone modification, and the downregulation of osteoblast markers induced by osteoclasts, which was attenuated by Ti Nano. We used this experimental design to evaluate the mechanisms involved in the regulation of the crosstalk between osteoclasts and osteoblasts that were mediated by the nanotopography. Initially, we evaluated the the expression of H3K9me2 (Figure 5A), H3K27me3 (Figure 5B) and EZH2 (Figure 5C). The protein expression of H3K9me2 in osteoblasts was increased by Ti Nano (p = 0.004) and the presence of osteoclasts did not affect its expression on Ti Control (p = 0.130) and upregulated it on Ti Nano (p = 0.001) (Figure 5A). The protein expression of H3K27me3 in osteoblasts was increased by Ti Nano (p = 0.018) and the presence of osteoclasts upregulated its expression on Ti Control (p = 0.001) and Ti Nano (p = 0.020) (Figure 5B). The protein expression of EZH2 in osteoblasts was increased by Ti Nano (p = 0.004) and the presence of osteoclasts upregulated its expression on Ti Control (p = 0.007) and Ti Nano (p = 0.014) (Figure 5C).
Figure 5.
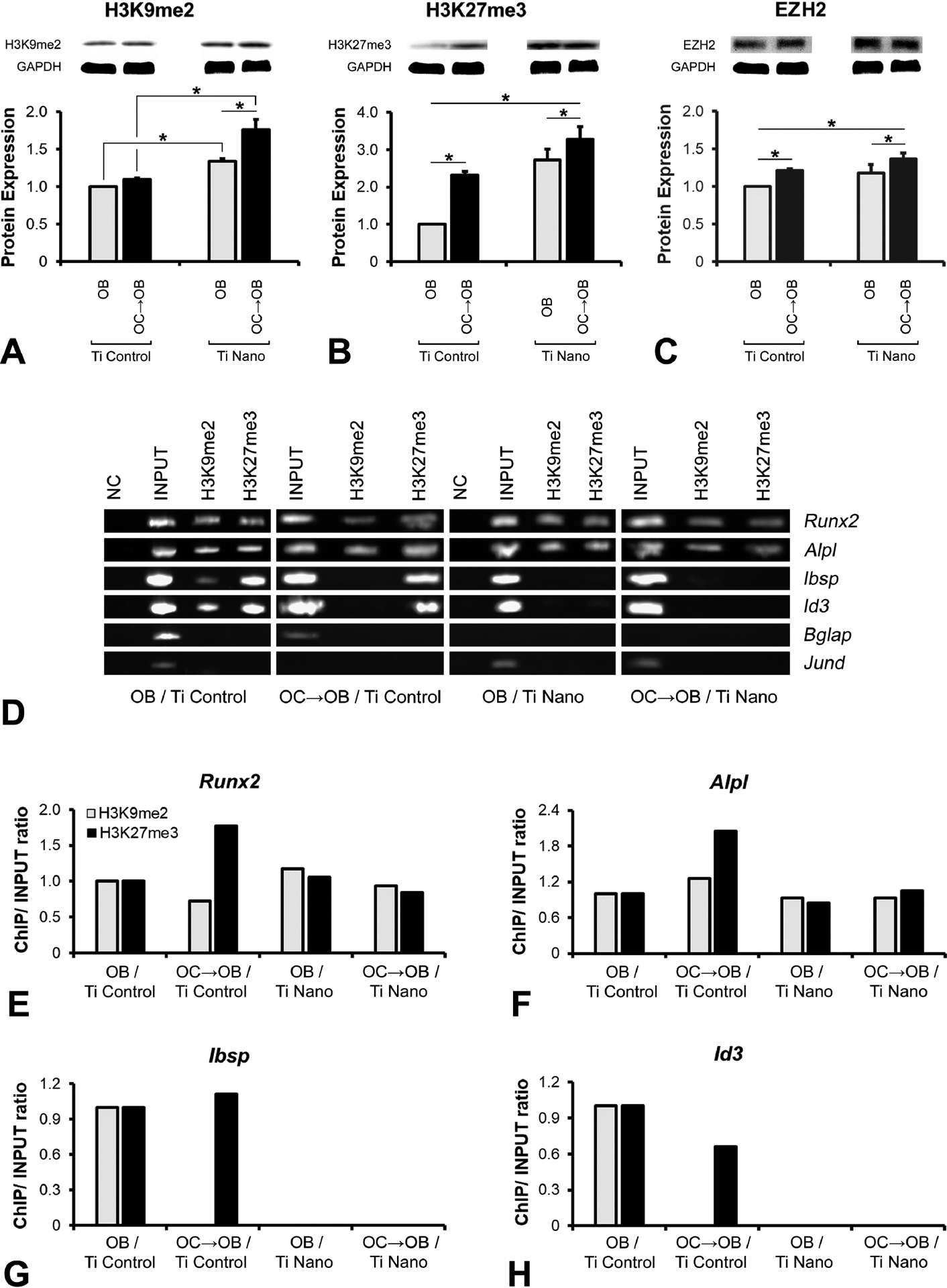
Titanium with nanotopography (Ti Nano) attenuate osteoclast-induced disruption of osteoblast differentiation by reducing the methylated histone accumulation. Protein expression of H3K9me2 (A), H327me3 (B) and EZH2 (C) of osteoblasts (OB) grown on polished titanium (Ti Control), osteoblasts grown in the presence of osteoclasts (OC→OB) on Ti Control, OB grown on Ti Nano and OC→OB on Ti Nano. Binding of the H3K9me2 and H3K27me3 to the promoter regions of the bone markers Runx2, Alpl, Ibsp, Id3, Bglap and Jund in OB grown on Ti Control, OC→OB on Ti Control, OB grown on Ti Nano and OC→OB on Ti Nano (D-H). Representative values from the PCR quantification of the genes, Runx2, Alpl, Ibsp and Id3, regulated by histone methylation (E-H). NC lanes represent samples immunoprecipitated with anti-IgG antibody, INPUT samples consist of total DNA, and H3K9me2 and H3K27me3 lanes refer to DNA immunoprecipitated with anti-H3K9me2 and anti-H3K27me3 antibodies (D). The data of protein expression (n = 3) are presented as mean ± standard deviation and asterisks (*) indicate statistically significant differences (p ≤ 0.05).
Based on these findings, the ChIP assay (Figure 5D–H) was performed to identify if the higher expression of H3K9me2 and H3K27me3 induced by osteoclasts contributes to the downregulation of osteoblast markers through chromatin condensation at the promoters, which prevents gene expression. Osteoclasts induced a high accumulation of H3K27me3, repressing the promoter regions of Runx2 and Alpl of osteoblasts grown on Ti Control compared to Ti Nano (Figure 5D–F). Irrespective of the presence of osteoclasts, the Ti Nano prevented the accumulation of H3K9me2 and H3K27me3 in the promoter regions of Ibsp and Id3 of the osteoblasts (Figure 5D,G,H). On Ti Control, only the accumulation of H3K9me2 in these regions was inhibited by the presence of osteoclasts (Figure 5D,G,H). No methylation of histones H3K9 or H3K27 was identified in the promoter regions of Bglap and Jund (Figure 5D). As H3K27me3 seemed to be more involved in the regulation of osteoblast differentiation by osteoclasts on Ti Nano, we used an immunofluorescence assay that corroborated the ChIP data and showed that osteoclasts increased the expression of H3K27me3 (Fig 6A,B), which reduced the expression of RUNX2 (Figure 6A) and ALPL (Fig 6B) in osteoblasts grown on both Ti surfaces, but in a less pronounced way on Ti Nano. These results indicate the protective role of Ti Nano on osteoblasts, since this nanotopography attenuated the osteoclast-induced disruption of osteoblast differentiation by reducing the accumulation of H3K9 and H3K27, mainly H3K27, methylated histones.
Figure 6.
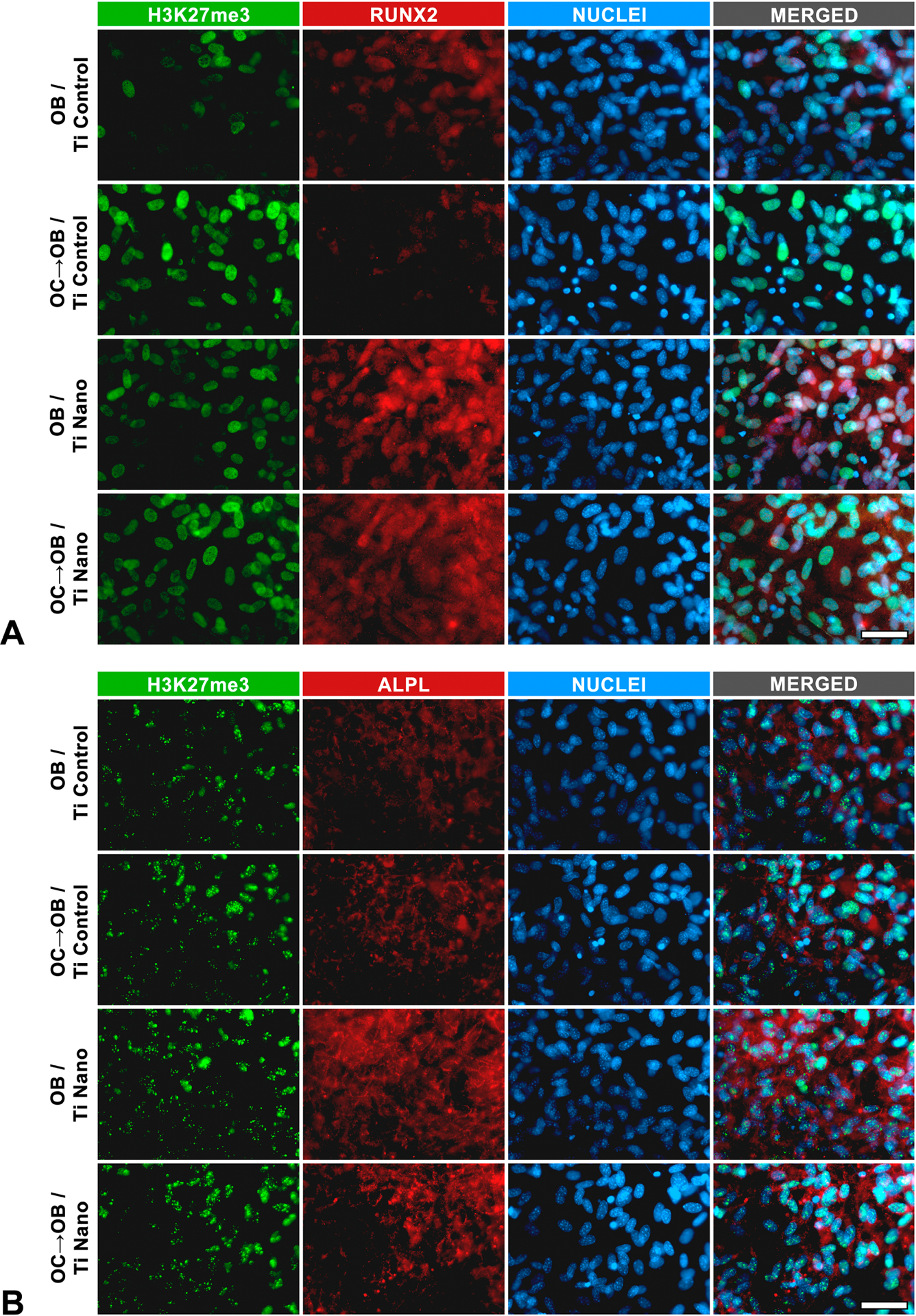
Titanium with nanotopography (Ti Nano) attenuate osteoclast-induced disruption of osteoblast differentiation by reducing the methylated histone accumulation. Co-immunolocalization of H3K27me3 and RUNX2 (A), and H3K27me3 and ALPL (B) in osteoblasts (OB) grown on polished titanium (Ti Control), osteoblasts grown in the presence of osteoclasts (OC→OB) on Ti Control, OB grown on Ti Nano and OC→OB on Ti Nano. Scale bars (A and B) = 50 μm. The reader is referred to the web version of this article for the colour representation of this figure.
4. Discussion
Several studies have shown the relevance of implant surface topography to the process of osseointegration. This event is characterized by the establishment of a direct interface between bone and the biomaterial, resulting in functional stability, load support and decreased risk of failure, thus maintaining the longevity of the implanted material and the health of the adjacent tissues [3,17,64]. Ti has been widely used to produce implants and the nanotopography generated by chemical conditioning with H2SO4/H2O2 favors osteoblast differentiation by modulating several cellular signaling pathways [6,7,11–14]. Considering the importance of bone remodeling to the Ti osseointegration, we demonstrated that nanotopography attenuated the negative effect of osteoclasts on osteoblast differentiation by reducing the accumulation of H3K27me3 in the promoter regions of Runx2 and Alpl (Figure 7), which adds further explanation to the high osteogenic potential of this Ti surface.
Figure 7.
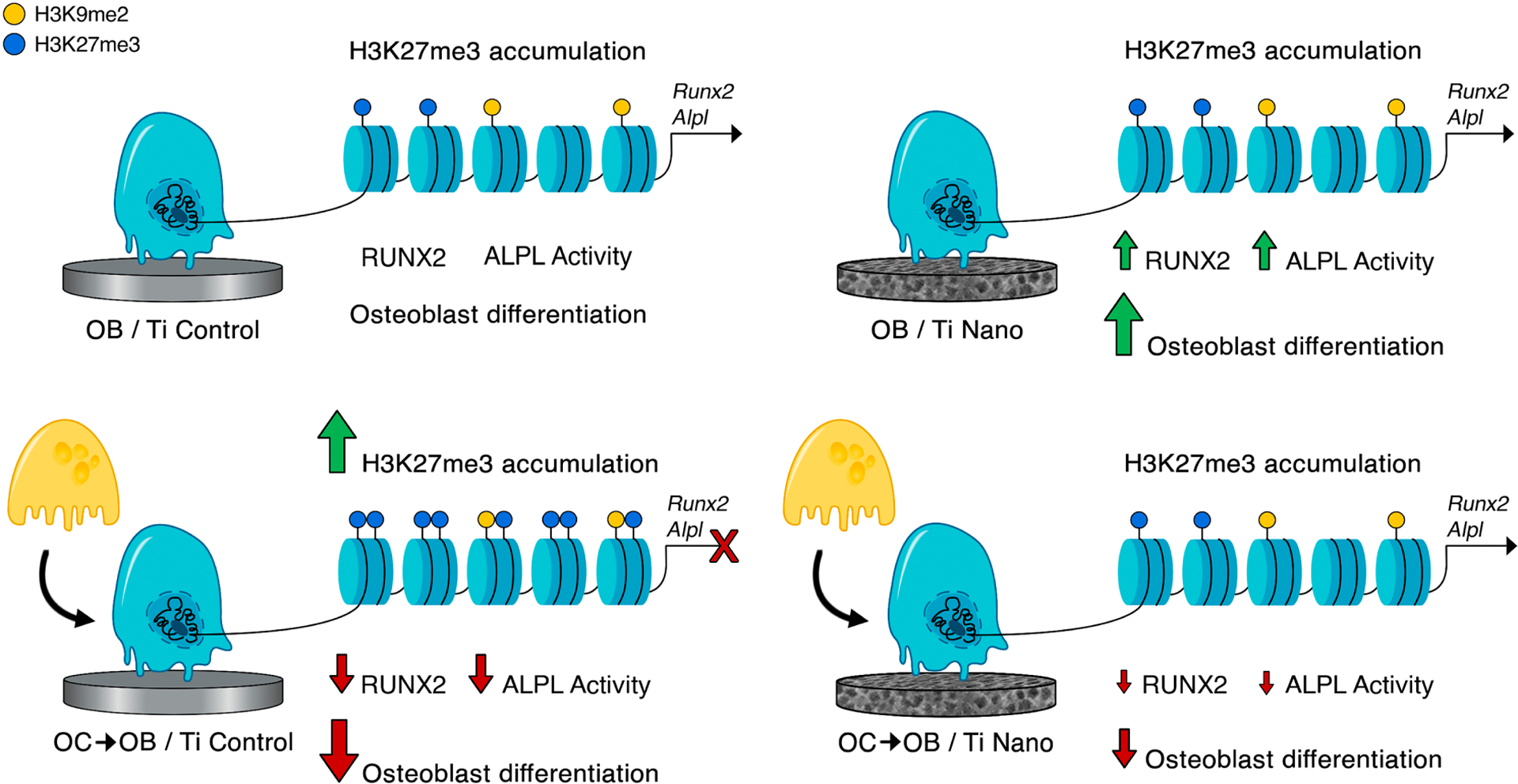
Schematic representation of the main findings of this study. It is demonstrated that osteoclasts inhibit osteoblast differentiation of cells grown on both Ti Control and Ti Nano and that Ti Nano attenuates the osteoclast-induced disruption of osteoblast differentiation by preventing the accumulation of H3K27me3 in the promoter regions of Runx2 and Alpl. The reader is referred to the web version of this article for the colour representation of this figure.
To investigate the impact of the osteoclasts on osteoblasts grown on Ti Nano, we initiated a transcriptome analysis that revealed different patterns of gene expression by osteoblasts depending on Ti surfaces and the presence of osteoclasts. The downregulation of transcripts from the essential biological process “regulation of osteoblast differentiation” was induced by osteoclasts irrespective of the Ti surface. Indeed, some studies have indicated that osteoclasts secrete microRNA-enriched exosomes that are transferred to osteoblasts, inhibiting osteoblast differentiation and activity [65,66]. This mechanism highlights the relevance of exosomes secreted by osteoclasts on bone loss and could be, at least in part, involved in the inhibitory action of osteoclasts on osteoblast differentiation observed here. It was also identified an upregulation of genes involved with “histone modification” and “regulation of chromatin organization” in the presence of osteoclasts that were more pronounced in osteoblasts grown on Ti Nano compared to Ti Control. These results indicate that osteoclasts trigger post-translational modifications of chromatin and histones in osteoblasts, which is corroborated by studies that link epigenetic modifications with bone development [22,26,28].
The Ti Nano favored osteoblast differentiation compared to Ti Control, as evidenced by gene and protein expression of osteoblastic markers as well as ALPL activity, corroborating our previous studies [6,7,12–14]. As expected, osteoclasts inhibited osteoblast differentiation and interestingly this effect was more pronounced in cells grown on Ti Control compared to Ti Nano. It has been shown that exosomes derived from osteoclasts contain miR-23a-5p that suppresses osteoblast activity through a connecting site between this miRNA and Runx2 [67]. These data demonstrate that osteoclasts reduced not only Runx2, but many genes and proteins associated with osteoblast differentiation and activity, again in a more marked way in cells grown on Ti Control than on Ti Nano. It is noteworthy that Bmpr1a, a receptor that mediates the BMP signaling pathway was downregulated by osteoclasts in osteoblasts grown on Ti Control, which was prevented by Ti Nano. Because the knockdown of Bmpr1a generates a striking inhibition of the osteogenic potential, bone quality and bone strength, these results suggest a compensatory mechanism when osteoblasts are grown on Ti Nano in the presence of osteoclasts, as the nanotopography induces osteoblast differentiation, at least in part, by the increase of BMP-2 production and BMP signaling pathway modulation [13,68]. Another interesting finding is the upregulation of Mmp13 induced by osteoclasts in osteoblasts cultured on Ti Nano. MMP13 is a member of the matrix metalloproteinases that acts on bone remodeling and cartilage degradation due to its particular ability to cleave type II collagen. This upregulation suggests that nanotopography could be associated not only with the increased osteoblast differentiation but also with high osteoclast activity leading to an enhancement of the bone remodeling process [69].
As the RNA-Seq and GO results showed differential patterns of expression of genes related to chromatin dynamics in osteoblasts depending on both the Ti surface topography and the presence of osteoclasts, we hypothesized that the inhibition of osteoblast differentiation induced by osteoclasts grown on Ti surfaces, which was attenuated by nanotopography, is due to histone modifications. Indeed, the expression of methylated histones H3K9me2 and H3K27me3, and EZH2, the enzyme that catalyzes the addition of methyl groups to histone H3 at Lys 27 (H3K27), was higher in osteoblasts grown on both Ti surfaces under the influence of osteoclasts. Methylation of histone proteins, a post-translational modification, is a remarkable epigenetic modification involved in bone development by regulating gene expression through chromatin compaction that represses gene transcription [22,27,28]. Despite H3K27me3 global distribution was higher in osteoblasts grown on both Ti surfaces under the influence of osteoclasts, the ChIP data showed that osteoclasts enhanced the accumulation of histone H3K27me3 in the promoter regions of key osteoblast marker genes, mainly Runx2 and Alpl, on Ti Control, suppressing the gene expression of Runx2, Alpl, Ibsp, and Id3, which was attenuated on Ti Nano. The co-immunolocalization confirmed that the increase of H3K27me3 reduced the protein expression of RUNX2 and ALPL in a more pronounced way in osteoblasts grown on Ti Control in the presence of osteoclasts. Attesting that this mechanism of histone modification is involved in the protective effect of nanotopography on osteoblasts, the treatment with UNC1999, an EZH2 inhibitor, reversed both H3K27me3 increase and RUNX2 reduction induced by osteoclasts in osteoblasts grown on Ti Nano (Supplementary Material 2). In agreement with these findings, it has been demonstrated that the increase in the H3K27 trimethylation resulted in a repression of Runx2 and Spp1 through a decrease in EZH2 phosphorylation and that H3K9me2 repressed the regulatory regions of Twist in primary osteogenic mesenchyme from calvaria [26,70]. Considering the relevance of this mechanism to the bone-biomaterial interactions, further studies are needed to decipher which factors secreted by osteoclasts are inducing the accumulation of H3K27me3 in the Runx2 and Alpl promoter regions of osteoblasts.
In conclusion, to the best of our knowledge, this study is the first to show the impact of the Ti Nano on the crosstalk between osteoblasts and osteoclasts. Despite osteoclasts inhibiting osteoblasts grown on both Ti Control and Ti Nano, the nanotopography attenuated the osteoclast-induced disruption of osteoblast differentiation by preventing the increase of H3K27me3 accumulation, which mediate repression of gene expression, in the promoter regions of Runx2 and Alpl. These findings shed light on the epigenetic mechanisms triggered by nanotopography to protect osteoblasts from the deleterious effects of osteoclasts, generating a fine balance of the process of bone remodeling that may benefit the osseointegration of Ti implants.
Supplementary Material
Highlights.
Osteoclasts inhibit osteoblasts grown on titanium (Ti) surfaces
Ti with nanotopography protects osteoblasts from the negative effect of osteoclasts
Nanotopography prevents the increase of H3K27me3 in Runx2 and Alpl promoter regions
Acknowledgments
The authors would like to thank Fabiola S. Oliveira and Roger R. Fernandes for their technical assistance during the experiments. The next-generation sequencing was performed in the Vermont Integrative Genomics Resource Massively Parallel Sequencing Facility and was supported by the University of Vermont Cancer Center, Lake Champlain Cancer Research Organization, UVM College of Agriculture and Life Sciences, and the UVM Larner College of Medicine.
Funding sources
This study was supported by the State of São Paulo Research Foundation (FAPESP, Brazil) [grant numbers 2019/09349-2, 2018/17356-6 and 2017/23888-8]; National Council for Scientific and Technological Development (CNPq, Brazil) [grant number 303464/2016-0]; Coordination of Improvement of Higher Education Personnel (CAPES, Brazil); and National Institutes of Health (USA) [grant numbers R01AR039588 and R01DE029311].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
All authors confirm that there is no conflict of interest.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability:
Data supporting the findings of this study are available upon request to the corresponding author.
References
- [1].Brånemark PI, Osseointegration and its experimental background, J. Prosthet. Dent 50 (1983) 399–410, 10.1016/s0022-3913(83)80101-2. [DOI] [PubMed] [Google Scholar]
- [2].Serre CM, Boivin G, Obrant KJ, Linder L, Osseointegration of titanium implants in the tibia. Electron microscopy of biopsies from 4 patients, Acta Orthop. Scand 65 (1994) 323–327, 10.3109/17453679408995462. [DOI] [PubMed] [Google Scholar]
- [3].Silverwood RK, Fairhurst PG, Sjöström T, Welsh F, Sun Y, Li G, Yu B, Young PS, Su B, Meek RMD, Dalby MJ, Tsimbouri PM, Analysis of osteoclastogenesis/osteoblastogenesis on nanotopographical titania surfaces, Adv. Healthc. Mater 5 (2016) 947–955, 10.1002/adhm.201500664. [DOI] [PubMed] [Google Scholar]
- [4].Lee S, Chang YY, Lee J, Madhurakkat Perikamana SK, Kim EM, Jung YH, Yun JH, Shin H, Surface engineering of titanium alloy using metal-polyphenol network coating with magnesium ions for improved osseointegration, Biomater. Sci 8 (2020) 3404–3417, 10.1039/d0bm00566e. [DOI] [PubMed] [Google Scholar]
- [5].Oliveira PT, Nanci A, Nanotexturing of titanium-based surfaces upregulates expression of bone sialoprotein and osteopontin by cultured osteogenic cells, Biomaterials 25 (2004) 403–413, 10.1016/s0142-9612(03)00539-8. [DOI] [PubMed] [Google Scholar]
- [6].Oliveira PT, Zalzal SF, Beloti MM, Rosa AL, Nanci A, Enhancement of in vitro osteogenesis on titanium by chemically produced nanotopography, J. Biomed. Mater. Res. A 80 (2007) 554–564, 10.1002/jbm.a.30955. [DOI] [PubMed] [Google Scholar]
- [7].Rosa AL, Kato RB, Castro Raucci LMS, Teixeira LN, de Oliveira FS, Bellesini LS, de Oliveira PT, Hassan MQ, Beloti MM, Nanotopography drives stem cell fate toward osteoblast differentiation through α1β1 integrin signaling pathway, J. Cell. Biochem 115 (2014) 540–548, 10.1002/jcb.24688. [DOI] [PubMed] [Google Scholar]
- [8].Liu Y, Wang Y, Cheng X, Zheng Y, Lyu M, Di P, Lin Y, MiR-181d-5p regulates implant surface roughness-induced osteogenic differentiation of bone marrow stem cells, Mater. Sci. Eng. C Mater. Biol. Appl 121 (2021), 111801, 10.1016/j.msec.2020.111801. [DOI] [PubMed] [Google Scholar]
- [9].Tang J, Chen L, Yan D, Shen Z, Wang B, Weng S, Wu Z, Xie Z, Shao J, Yang L, Shen L, Surface functionalization with proanthocyanidins provides an anti-oxidant defense mechanism that improves the long-term stability and osteogenesis of titanium implants, Int. J. Nanomedicine 15 (2020) 1643–1659, 10.2147/IJN.S231339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yi J-H, Bernard C, Variola F, Zalzal SF, Wuest JD, Rosei F, Nanci A, Characterization of a bioactive nanotextured surface created by controlled chemical oxidation of titanium, Surf. Sci 600 (2006) 4613–4621, 10.1016/j.susc.2006.07.053. [DOI] [Google Scholar]
- [11].Guadarrama Bello D, Fouillen A, Badia A, Nanci A, Nanoporosity stimulates cell spreading and focal adhesion formation in cells with mutated paxillin, ACS Appl. Mater. Interfaces 12 (2020) 14924–14932, 10.1021/acsami.0c01172. [DOI] [PubMed] [Google Scholar]
- [12].Kato RB, Roy B, De Oliveira FS, Ferraz EP, De Oliveira PT, Kemper AG, Hassan MQ, Rosa AL, Beloti MM, Nanotopography directs mesenchymal stem cells to osteoblast lineage through regulation of microRNA-SMAD-BMP-2 circuit, J. Cell. Physiol 229 (2014) 1690–1696, 10.1002/jcp.24614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Castro-Raucci LMS, Francischini MS, Teixeira LN, Ferraz EP, Lopes HB, de Oliveira PT, Hassan MQ, Rosa AL, Beloti MM, Titanium with nanotopography induces osteoblast differentiation by regulating endogenous bone morphogenetic protein expression and signaling pathway, J. Cell. Biochem 117 (2016) 1718–1726, 10.1002/jcb.25469. [DOI] [PubMed] [Google Scholar]
- [14].Abuna RPF, Oliveira FS, Lopes HB, Freitas GP, Fernandes RR, Rosa AL, Beloti MM, The Wnt/β-catenin signaling pathway is regulated by titanium with nanotopography to induce osteoblast differentiation 184 (2019), 110513, 10.1016/j.colsurfb.2019.110513. [DOI] [PubMed] [Google Scholar]
- [15].Eriksen EF, Cellular mechanisms of bone remodeling, Rev. Endocr. Metab. Disord 11 (2010) 219–227, 10.1007/s11154-010-9153-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Matsuo K, Irie N, Osteoclast–osteoblast communication, Arch. Biochem. Biophys 473 (2008) 201–209, 10.1016/j.abb.2008.03.027. [DOI] [PubMed] [Google Scholar]
- [17].Young PS, Tsimbouri PM, Gadegaard N, Meek RMD, Dalby MJ, Osteoclastogenesis/osteoblastogenesis using human bone marrow-derived cocultures on nanotopographical polymer surfaces, Nanomedicine 10 (2015) 949–957, 10.2217/nnm.14.146. [DOI] [PubMed] [Google Scholar]
- [18].Kusu N, Laurikkala J, Imanishi M, Usui H, Konishi M, Miyake A, Thesleff I, Itoh N, Sclerostin is a novel secreted osteoclast-derived bone morphogenetic protein antagonist with unique ligand specificity, J. Biol. Chem 278 (2003) 24113–24117, 10.1074/jbc.M301716200. [DOI] [PubMed] [Google Scholar]
- [19].Negishi-Koga T, Shinohara M, Komatsu N, Bito H, Kodama T, Friedel RH, Takayanagi H, Suppression of bone formation by osteoclastic expression of semaphorin 4D, Nat. Med 17 (2011) 1473–1480, 10.1038/nm.2489. [DOI] [PubMed] [Google Scholar]
- [20].Zhang Y, Chen SE, Shao J, van den Beucken JJJP, Combinatorial surface roughness effects on osteoclastogenesis and osteogenesis, ACS Appl. Mater. Interfaces 10 (2018) 36652–36663, 10.1021/acsami.8b10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kim JM, Lin C, Stavre Z, Greenblatt MB, Shim JH, Osteoblast-osteoclast communication and bone homeostasis, Cells 9 (2020) 2073, 10.3390/cells9092073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gordon JAR, Stein JL, Westendorf JJ, van Wijnen AJ, Chromatin modifiers and histone modifications in bone formation, regeneration, and therapeutic intervention for bone-related disease, Bone 81 (2015) 739–745, 10.1016/j.bone.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mu W, Starmer J, Yee D, Magnuson T, EZH2 variants differentially regulate polycomb repressive complex 2 in histone methylation and cell differentiation, Epigenetics Chromatin 11 (2018) 71, 10.1186/s13072-018-0242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li Y, Guo D, Sun R, Chen P, Qian Q, Fan H, Methylation patterns of Lys9 and Lys27 on histone H3 correlate with patient outcome in gastric cancer, Dig. Dis. Sci 64 (2019) 439–446, 10.1007/s10620-018-5341-8. [DOI] [PubMed] [Google Scholar]
- [25].Tellez CS, Picchi MA, Juri D, Do K, Desai DH, Amin SG, Hutt JA, Filipczak PT, Belinsky SA, Chromatin remodeling by the histone methyltransferase EZH2 drives lung pre-malignancy and is a target for cancer prevention, Clin. Epigenetics 13 (2021) 44, 10.1186/s13148-021-01034-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wei Y, Chen YH, Li LY, Lang J, Yeh SP, Shi B, Yang CC, Yang JY, Lin CY, Lai CC, Hung MC, CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells, Nat. Cell Biol 13 (2011) 87–94, 10.1038/ncb2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Deng P, Chen QM, Hong C, Wang CY, Histone methyltransferases and demethylases: regulators in balancing osteogenic and adipogenic differentiation of mesenchymal stem cells, Int. J. Oral Sci 7 (2015) 197–204, 10.1038/ijos.2015.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Montecino M, Carrasco ME, Nardocci G, Epigenetic control of osteogenic lineage commitment, Front. Cell Dev. Biol 8 (2021), 611197, 10.3389/fcell.2020.611197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Eberhard C, Gruning B, Guerler A, Hillman-Jackson J, Von Kuster G, Rasche E, Soranzo N, Turaga N, Taylor J, Nekrutenko A, Goecks J, The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update, Nucleic Acids Res. 44 (2016) W3–W10, 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].RStudio, Integrated development for R. http://www.rstudio.com. (Accessed 14 January 2020).
- [31].Anders S, FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. (Accessed 14 January 2020).
- [32].Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner, Bioinformatics 29 (2013) 15–21, 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigó R, Hubbard TJ, GENCODE: the reference human genome annotation for the ENCODE project, Genome Res. 22 (2012) 1760–1774, 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Anders S, Pyl PT, Huber W, HTSeq–a python framework to work with high-throughput sequencing data, Bioinformatics 31 (2015) 166–169, 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Love MI, Huber W, Anders A, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol. 15 (2014) 550, 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G, Gene ontology: tool for the unification of biology. The gene ontology consortium, Nat. Genet 25 (2000) 25–29, 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD, PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools, Nucleic Acids Res. 47 (2019) D419–D426, 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].The Gene Ontology Consortium, The gene ontology resource: 20 years and still going strong, Nucleic Acids Res. 47 (2019) D330–D338, 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Livak KJ, Schmittgen TD, Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method, Methods 25 (2001) 402–408, 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- [40].Almeida LO, Goto RN, Pestana CR, Uyemura SA, Gutkind S, Curti C, Leopoldino AM, SET overexpression decreases cell detoxification efficiency: ALDH2 and GSTP1 are downregulated, DDR is impaired and DNA damage accumulates, FEBS J. 279 (2012) 4615–4628, 10.1111/febs.12047. [DOI] [PubMed] [Google Scholar]
- [41].Akhouayri O, St-Amaud R, Differential mechanisms of transcriptional regulation of the mouse osteocalcin gene by Jun family members, Calcif. Tissue Int 80 (2007) 123–131, 10.1007/s00223-006-0102-7. [DOI] [PubMed] [Google Scholar]
- [42].Kobaku S, Ohte S, Sasanuma H, Shin M, Yoneyama K, Murata E, Kanomata K, Nojima J, Ono Y, Yoda T, Fukuda T, Katagiri T, Supression of BMP-smad signaling axis-induced osteoblastic differentiation by small C-terminal domain phosphatase 1, a smad phosphatase, Mol. Endocrinol 25 (2011) 474–481, 10.1210/me.2010-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Vrijens K, Lin W, Cui J, Farmer D, Low J, Pronier E, Zeng FY, Shelat AA, Guy K, Taylor MR, Chen T, Roussel MF, Identification of small molecule activators of BMP signaling, PLoS One 8 (2013), e59045, 10.1371/journal.pone.0059045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yu B, Chang J, Liu Y, Li J, Kevork K, Al-Hezaimi K, Graves DT, Park NH, Wang CY, Wnt4 signaling prevents skeletal aging and inflammation by inhibiting nuclear factor-kappaB, Nat. Med 20 (2014) 1009–1017, 10.1038/nm.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Komori T, Regulation of proliferation, differentiation and functions of osteoblasts by runx 2, Int. J. Mol. Sci 20 (2019) 1694, 10.3390/ijms20071694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Song D, He G, Song F, Wang Z, Liu X, Liao L, Ni J, Silva MJ, Long F, Inducible expression of Wnt7b promotes bone formation in aged mice and enhances fracture healing, Bone Res. 8 (2020) 4, 10.1038/s41413-019-0081-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zainabadi K, Liu CJ, Caldwell ALM, Guarente L, SIRT1 is a positive regulator of in vivo bone mass and a therapeutic target for osteoporosis, PLoS One 12 (2017), e0185236, 10.1371/journal.pone.0185236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wang C, Wang J, Li J, Hu G, Shan S, Li Q, Zhang X, KDM5A controls bone morphogenic protein 2-induced osteogenic differentiation of bone mesenchymal stem cells during osteoporosis, Cell Death Dis. 7 (2016), e2335, 10.1038/cddis.2016.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Christenson RH, Biochemical markers of bone metabolism: an overview, Clin. Biochem 30 (1997) 573–593, 10.1016/s0009-9120(97)00113-6. [DOI] [PubMed] [Google Scholar]
- [50].Wu YF, Matsuo N, Sumiyoshi H, Yoshioka H, Sp7/Osterix is involved in the up-regulation of the mouse pro-α1(V) collagen gene (Col5a1) in osteoblastic cells, Matrix Biol. 29 (2010) 701–706, 10.1016/j.matbio.2010.09.002. [DOI] [PubMed] [Google Scholar]
- [51].Green AC, Rudolf-Stringer V, Straszkowski L, Tjin G, Crimeen-Irwin B, Walia M, Martin TJ, Sims NA, Purton LE, Retinoic acid receptor γ activity in mesenchymal stem cells regulates endochondral bone, angiogenesis, and B lymphopoiesis, J. Bone Miner. Res 33 (2018) 2202–2213, 10.1002/jbmr.3558. [DOI] [PubMed] [Google Scholar]
- [52].Hadjiargyrou M, Mustn1: a developmentally regulated pan-musculoskeletal cell marker and regulatory gene, Int. J. Mol. Sci 19 (2018) 206, 10.3390/ijms19010206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Dirckx N, Moorer MC, Clemens TL, Riddle RC, The role of osteoblasts in energy homeostasis, Nat. Rev. Endocrinol 15 (2019) 651–665, 10.1038/s41574-019-0246-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Novak S, Roeder E, Sinder BP, Adams DJ, Siebel CW, Grcevic D, Hankenson KD, Matthews BG, Kalajzic I, Modulation of Notch1 signaling regulates bone fracture healing, J. Orthop. Res 38 (2020) 2350–2361, 10.1002/jor.24650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wehner C, Lettner S, Moritz A, Andrukhov O, Rausch-Fan X, Effect of bisphosphonate treatment of titanium surfaces on alkaline phosphatase activity in osteoblasts: a systematic review and meta-analysis, BMC Oral Health 20 (2020) 125, 10.1186/s12903-020-01089-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B, The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation, Cell 108 (2002) 17–29, 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- [57].Tohmonda T, Miyauchi Y, Ghosh R, Yoda M, Uchikawa S, Takito J, Morioka H, Nakamura M, Iwawaki T, Chiba K, Toyama Y, Urano F, Horiuchi K, The IRE1α-XBP1 pathway is essential for osteoblast differentiation through promoting transcription of osterix, EMBO Rep. 12 (2011) 451–457, 10.1038/embor.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Jo SY, Domowicz MS, Henry JG, Schwartz NB, The role of Dot1l in prenatal and postnatal murine chondrocytes and trabecular bone, JBMR Plus 4 (2019), e10254, 10.1002/jbm4.10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Sanui T, Tanaka U, Fukuda T, Toyoda K, Taketomi T, Atomura R, Yamamichi K, Nishimura F, Mutation of Spry2 induces proliferation and differentiation of osteoblasts but inhibits proliferation of gingival epithelial cells, J. Cell. Biochem 116 (2015) 628–639, 10.1002/jcb.25014. [DOI] [PubMed] [Google Scholar]
- [60].Lee SY, Yi JK, Yun HM, Bae CH, Cho ES, Lee KS, Kim EC, Expression of caveolin-1 in periodontal tissue and its role in osteoblastic and cementoblatic differentiation in vitro, Calcif. Tissue Int 98 (2016) 497–510, 10.1007/s00223-015-0095-1. [DOI] [PubMed] [Google Scholar]
- [61].Hu K, Sun H, Gui B, Sui C, Gremlin-1 suppression increases BMP-2-induced osteogenesis of human mesenchymal stem cells, Mol. Med. Rep 15 (2017) 2186–2194, 10.3892/mmr.2017.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Khayal LA, Grünhagen J, Provazník I, Mundlos S, Kornak U, Robinson PN, Ott CE, Transcriptional profiling of murine osteoblast differentiation based on RNA-seq expression analyses, Bone 113 (2018) 29–40, 10.1016/j.bone.2018.04.006. [DOI] [PubMed] [Google Scholar]
- [63].Zhao H, Chen Q, Alam A, Cui J, Suen KC, Soo AP, Eguchi S, Gu J, Ma D, The role of osteopontin in the progression of solid organ tumour, Cell Death Dis. 9 (2018) 356, 10.1038/s41419-018-0391-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lotz EM, Berger MB, Schwartz Z, Boyan BD, Regulation of osteoclasts by osteoblast lineage cells depends on titanium implant surface properties, Acta Biomater. 68 (2018) 296–307, 10.1016/j.actbio.2017.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Li D, Liu J, Guo B, Liang C, Dang L, Lu C, He X, Cheung HY, Xu L, Lu C, He B, Liu B, Shaikh AB, Li F, Wang L, Yang Z, Au DW, Peng S, Zhang Z, Zhang B, Pan X, Qian A, Shang P, Xiao L, Jiang B, Wong CK, Xu J, Bian Z, Liang Z, Guo D, Zhu H, Tan W, Lu A, Zhang G, Osteoclast-derived exosomal miR-214–3p inhibits osteoblastic bone formation, Nat. Commun 7 (2016) 10872, 10.1038/ncomms10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Sun W, Zhao C, Li Y, Wang L, Nie G, Peng J, Wang A, Zhang P, Tian W, Li Q, Song J, Wang C, Xu X, Tian Y, Zhao D, Xu Z, Zhong G, Han B, Ling S, Chang Y, Li Y, Osteoclast-derived microRNA-containing exosomes selectively inhibit osteoblast activity, Cell Discov. 2 (2016) 16015, 10.1038/celldisc.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Yang JX, Xie P, Li YS, Wen T, Yang XC, Osteoclast-derived miR-23a5p-containing exosomes inhibit osteogenic differentiation by regulating Runx2, Cell. Signal 70 (2020), 109504, 10.1016/j.cellsig.2019.109504. [DOI] [PubMed] [Google Scholar]
- [68].Bao Q, Li A, Chen S, Feng J, Liu H, Qin H, Li J, Liu D, Shen Y, Zong Z, Disruption of bone morphogenetic protein type IA receptor in osteoblasts impairs bone quality and bone strength in mice, Cell Tissue Res. 374 (2018) 263–273, 10.1007/s00441-018-2873-3. [DOI] [PubMed] [Google Scholar]
- [69].Hu Q, Ecker M, Overview of MMP-13 as a promising target for the treatment of osteoarthritis, Int. J. Mol. Sci 22 (2021) 1742, 10.3390/ijms22041742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Higashihori N, Lehnertz B, Sampaio A, Underhill TM, Rossi F, Richman JM, Methyltransferase G9A regulates osteogenesis via twist gene repression, J. Dent. Res 96 (2017) 1136–1144, 10.1177/0022034517716438. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


