Abstract
Contamination of soils with toxic metals is a major problem on military, industrial, and mining sites worldwide. Of particular interest to the field of bioremediation is the selection of biological markers for the end point of remediation. In this microcosm study, we focus on the effect of addition of a mixture of toxic metals (cadmium, cobalt, cesium, and strontium as chlorides) to soil on the population structure and size of the ammonia oxidizers that are members of the beta subgroup of the Proteobacteria (β-subgroup ammonia oxidizers). In a parallel experiment, the soils were also treated by the addition of five strains of metal-resistant heterotrophic bacteria. Effects on nitrogen cycling were measured by monitoring the NH3 and NH4+ levels in soil samples. The gene encoding the α-subunit of ammonia monooxygenase (amoA) was selected as a functional molecular marker for the β-subgroup ammonia oxidizing bacteria. Community structure comparisons were performed with clone libraries of PCR-amplified fragments of amoA recovered from contaminated and control microcosms for 8 weeks. Analysis was performed by restriction digestion and sequence comparison. The abundance of ammonia oxidizers in these microcosms was also monitored by competitive PCR. All amoA gene fragments recovered grouped with sequences derived from cultured Nitrosospira. These comprised four novel sequence clusters and a single unique clone. Specific changes in the community structure of β-subgroup ammonia oxidizers were associated with the addition of metals. These changes were not seen in the presence of the inoculated metal-resistant bacteria. Neither treatment significantly altered the total number of β-subgroup ammonia-oxidizing cells per gram of soil compared to untreated controls. Following an initial decrease in concentration, ammonia began to accumulate in metal-treated soils toward the end of the experiment.
Toxic metal wastes from defense-related activities, industry, and municipal sources have routinely entered the environment through disposal in landfill sites or by accidental release in accidents such as that which occurred at Chernobyl. These practices have resulted in surface contamination problems, transport to groundwater, and/or bioaccumulation of radionuclides and toxic metals (see, e.g., references 8, 9, 21, and 31). Metals such as Cs, Sr, Cd, and, to a lesser extent, Co are prevalent in soils near industrial centers (see, e.g., references 7 and 21) at concentrations up to 50 μg of Cs/g, 350 μg of Cd/g, and 500 μg of Sr/g (31). As cocontaminants, toxic metals are often inhibitory to other bioremediative processes, e.g., hydrocarbon degradation (34).
Due to biotic and abiotic chemical dynamics, microbial metal toxicity is reduced by 1 to 2 orders of magnitude in soils relative to solution, depending on factors such as soil type, aeration conditions, metal speciation, carbon sources, pH, and Eh (2, 12). Microbial communities are of primary importance in bioremediation of metal-contaminated soils and represent a substantial proportion of the in situ biomass and metabolic diversity. The structure and diversity of soil microbial communities have been shown to change in soil in the presence of toxic metals (2, 13, 14, 28). Microorganisms can alter metal chemistry and mobility through reduction, accumulation, mobilization, and immobilization (1, 5, 20, 39). Since metal ion species are generally more readily soluble in acidic environments, acidogenic microbial metabolic activities may contribute to the introduction of metals into groundwater from contaminated soils.
In soils, the ammonia oxidizers that are members of the beta subgroup of the Proteobacteria (β-subgroup ammonia oxidizers) form an important part of the bacterial community, being chiefly responsible for the first step in the oxidation of immobile ammonia to highly mobile nitrate via nitrite (29). This process involves a concomitant release of protons, which can lead to significant soil acidification where the ammonia input is high (4). The coherent phylogenetic and physiological characteristics of the β-subgroup ammonia oxidizers has provided the opportunity to view them as an indicator species for environmental change (16, 18, 33, 37). By using 16S rDNA as a marker, specific changes in β-subgroup ammonia oxidizer populations have been observed to occur with changing pH (37), with addition of swine manure (16) to soil, through effects of salmon farm waste in marine sediments (37), and with proximity to the ocean and aging in coastal sand dunes (18). It is now practical to use a second molecular marker in environmental studies, amoA, the gene encoding the α-subunit of ammonia monooxygenase. Rotthauwe et al. (33) have demonstrated the use of a highly specific set of PCR primers to amplify a fragment of amoA from a variety of pure cultures of β-subgroup ammonia oxidizers and environmental samples.
In this study, we use amoA as a functional molecular marker to assess whether any changes in the population structure of β-subgroup ammonia oxidizers could be induced by an 8-week exposure of an indigenous soil community to high levels of a mixture of toxic metal ions. This study had two aims. First, we wished to determine whether all indigenous species of β-subgroup ammonia oxidizers were equally susceptible to damage by toxic metals through observation of changes in the structure of amoA clone libraries. Observation of unequal susceptibility may provide the basis for using β-subgroup ammonia oxidizers as an indicator group for the end point of metal removal or immobilization, defined as return of the impacted communities to control values. Our initial approach was to assess the natural diversity of amoA genes by PCR amplification, cloning, and sequence analysis of selected clones (33). The second aim of this study was to assess whether inoculation of soil with a group of metal-resistant bacteria could reduce the bioavailability of toxic metal ions to the indigenous microflora. A second set of soil microcosms was created as above, but these were also inoculated with several bacteria which have been considered metal resistant in the literature. Here, the aim was to assess whether any changes to the β-subgroup ammonia oxidizer population would also be observed in the presence of these added bacteria or whether the activities of these bacteria might protect the indigenous β-subgroup ammonia oxidizer community from the toxic effects of the metals.
MATERIALS AND METHODS
Soil microcosms.
Microcosms consisted of 150-ml polypropylene beakers (VWR Scientific) containing 75g (dry weight) of sieved (2-mm) agricultural loam top soil (depth, 0 to 100 mm) from the University of Tennessee Agricultural Experiment Station in Alcoa (Sequatchie series). The soil was slightly acidic (pH 5.5) and contained 0.06% (wt/wt) organic carbon and 0.05% (wt/wt) total nitrogen. Indigenous NH3/NH4+ was 25.0 ppm. Sand (0.05 to 1 mm), silt, and clay were measured at 53.5, 32.9, and 13.6% respectively. After metal or inoculum additions (final water content, 17% [wt/wt]), the microcosms were thoroughly mixed and the soils were compacted to 1.2 g/cm3 and loosely covered with foil for aerobic incubation in the dark at 23°C and high atmospheric humidity (>70%). Metal chlorides were added in aqueous solution (CoCl2 · 6H2O [EM Industries, Inc., Gibbstown, N.J.]; CsCl [Alfa Aesar, Ward Hill, Mass.]; SrCl2 · 6H2O [Fischer Scientific, Co., Fair Lawn, N.J.]; and CdCl2 · 2 1/2 H2O [J. T. Baker Chemical Co., Phillipsburg, N.J.]). The final concentrations of Cd, Co, and Sr in soil were 500 μg/g (dry weight) of soil, and that of Cs was 1,800 μg/g (dry weight) of soil. Triplicate microcosms were sacrificed at 0, 4, and 8 weeks for analysis. Moist soil samples (10 g) were frozen at −20°C for DNA extraction.
Bacterial strains.
The soil inoculum consisted of a five-strain mixture each grown separately to stationary phase in batch culture. Shewanella putrifaciens 200 (oil pipeline isolate) (26), Pseudomonas aeruginosa FRD-1 (cystic fibrosis patient isolate) (27), and Alcaligenes eutrophus CH34 (metal-resistant strain; ATCC 43123) were grown for ∼26 h at 23°C in nutrient broth with shaking. Sphingomonas sp. strain B0695 (contaminated soil isolate) (3) was grown as above for 48 h. Desulfovibrio vulgaris (ATCC 29579) was grown anaerobically for 48 h in an acetate-lactate medium at pH 7.2 containing (grams per liter) sodium acetate, 2.8; sodium lactate, 2.26; yeast extract, 0.5; ascorbic acid, 0.1; MgSO4 · 7H2O, 0.5; Na2SO4, 0.5; K2HPO4, 0.5; NH4Cl, 0.5; FeSO4 · 7H2O, 0.1; NaCl, 7.0; and sodium thioglycolate, 0.1. The strains were then washed twice in 0.85% (wt/vol) NaCl by centrifugation and resuspended together in distilled water to a density of ∼2.5 × 109 cells/ml each for delivery of 3 ml of cell suspension per 75-g microcosm, providing ∼2 × 107 cells of each species per g (dry weight) of soil.
DNA extraction.
The direct nucleic acid extraction was performed by using a bead-beating system adapted from reference 6 with modifications. Soil (0.5 g), 0.12 M sodium phosphate buffer (pH 8.0) (425 μl), chaotropic reagent (CRSR; Bio-101, Vista, Calif.) (175 μl), and 0.17-mm glass beads (0.5 g) were agitated in a 1.5-ml microcentrifuge tube by using a high-speed Crescent WIG-L-BUG bead beater (Crescent Dental Mfg. Co., Lyons, Ill.) for 1.5 min. The sample mixture was centrifuged at 13,000 × g for 5 min, and the supernatant was collected. Chloroform (300 μl) was added to the soil pellet, mixed thoroughly, and centrifuged at 13,000 × g for 5 min. The aqueous supernatant was collected and combined with the first supernatant fraction. DNA was precipitated from the aqueous phase with an equal volume of isopropanol in an ice bath for 30 min. DNA was pelleted by centrifugation at 13,000 × g at 4°C for 15 min, washed with 1 ml of 80% ethanol twice, air dried, and redissolved in 200 μl of Tris-EDTA (TE) buffer (pH 8.0). The DNA extract was purified by extracting twice with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol) followed by a glass milk DNA purification protocol with a Gene Clean kit (Bio-101) as described by the manufacturer.
Construction and analysis of amoA gene fragment libraries.
The amoA gene fragments were amplified as described previously (33) with minor modification as follows. Two ambiguities were created in the forward primer (GGGGHTTYTACTGGTGGT; H = not G; Y = C or T; altered bases are underlined) to improve matching with the target sequences of various cultured representatives of the β-subgroup ammonia oxidizer clade. This was referred to as amoA-1F*. The reverse primer was unaltered; amoA-2R (CCCCTCKGSAAAGCCTTCTTC; K = G or T). The reaction conditions were 35 cycles of 92°C for 60 s, 55°C for 60 s, and 68 C for 45 s with 1.225 U of Expand HF polymerase, the supplied buffer (Boehringer Mannheim, Indianapolis, Ind.), and 10 pmol of each primer in a total volume of 25 μl on a Robocycler 96 thermocycler (Stratagene, La Jolla, Calif.). PCR products were gel purified and extracted with a Gene-Clean kit. Purified fragments were cloned with the vector PCR2.1 TOPO and Escherichia coli TOP10F′ competent cells as specified by the manufacturer (Invitrogen, Carlsbad, Calif.). From each library, 36 to 47 white colonies were randomly selected and the cloned inserts were reamplified with the vector primers M13 reverse and T7 (30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 45 s). A portion (5 μl) of the resulting amplification product was digested with the restriction endonuclease MspI as specified by the manufacturer (Boehringer) and analyzed by separation of fragments on a 2% agarose gel with Tris-acetate-EDTA (TAE) buffer. Representative plasmids from each digestion pattern were selected for sequencing. The remaining portion of the M13 reverse-T7 amplification product was gel purified, extracted with a Gene Clean kit, and subjected to double-strand sequencing with the same primers on an Applied Biosystems automated sequencer (model 373) with Prism dye terminators. The sequences were assembled and aligned with SeqAp version 0.6 (15). Phylogenetic algorithms (DNA-DIST, NEIGHBOR, and SEQBOOT) operated through the PHYLIP package (10) and the ARB sequence management system (38).
Competitive PCR.
Competitive PCR was carried out as described previously (35), using a molar-equivalent conversion factor of 1.11 to account for the size difference between the target and competitor molecules. An amoA internal control standard was generated as follows: 3′ truncation of the amoA sequence carried by clone NAB_8_23 was achieved by amplification with amoA-1F and amoA-2R_DEL (CCCCTCTGCAAAGCCTTCTTCCCTTCACGTAGAAGAAG). The 5′ 20 bases (underlined) correspond to amoA-2R (33), and the 3′ 17 bases correspond to positions 535 to 553 of the coding sequence of Nitrosomonas europaea amoA (23). The amplification product (428 bp) was cloned with the vector PCR2.1 TOPO and E. coli TOP10F′ to generate p428-NAB_8_23. Plasmids were isolated with a Wizard mini-prep kit (Promega Corp., Madison, Wis.) and linearized with EcoRI (Promega), for which the amplified control sequence did not carry a recognition site. Linearized products were gel purified on 0.8% agarose with TAE buffer and extracted with a Gene-Clean kit prior to quantification by using a Hoefer DyNA-Quant 200 fluorometer and Hoechst H33258 dye binding assay (Pharmacia Biotech. Inc, Piscataway, N.J.).
Metal extractions and soil pH.
Metal extraction was performed by shaking soil for 1 h in distilled water at 1:10 (soil dry weight/solution volume). Filtrates were collected after centrifugation (2,500 × g) with a 12-sample filtration manifold (Millipore Corp., Bedford, Mass.), with Whatman no. 40 filter paper and 2 drops of 1% (wt/vol) sodium pyrophosphate per 15 ml of filtrate for stabilization of metals (30). Samples were stored at 4°C for 1 to 4 weeks. Sr, Co, Mn, Fe, and Cd were measured by monitoring inductively coupled argon plasma atomic emission (Plant and Soil Science Department, University of Tennessee, Knoxville, Tenn.), and Cs was determined by flame atomic absorption spectrometry (Galbraith Laboratories Inc., Knoxville, Tenn.). The soil pH was determined by using distilled water (1:1, wt/vol) (22) and a pH combination electrode.
Ammonia and ammonium measurements.
Soil samples (2 g) were assayed for NH3 and NH4+ content after extraction with 1 M KCl by the modified indophenol blue microtiter plate method of Sims et al. (36).
Statistical analysis.
Chi-square and Student t tests were performed with an Excel spreadsheet (Microsoft Office 97, Microsoft Corp.).
Nucleotide sequence accession numbers.
Sequences have been submitted to GenBank under accession no. AF056049 to AF056069.
RESULTS AND DISCUSSION
Bioavailability of metals in soil microcosms.
No significant differences in the solubility of any of the metals added were detected between the inoculated and noninoculated soils. The percent availability of each metal at each time point consistently followed the order Sr > Co > Cs > Cd. The water-soluble fraction of each averaged 324.8 ± 14.7, 284.7 ± 17.1, 560.6 ± 198.1, and 213.6 ± 50.8 ppm, respectively (means ± standard deviations of 30 readings taken over 8 weeks).
Effect of metal addition on NH3 and NH4+ levels in soil.
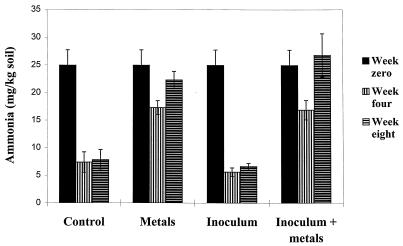
The ammonia and ammonium level in the test soil at time zero was 25.0 ppm (Fig. 1). Ammonia and ammonium levels in the microcosms which were not treated by the addition of metals fell by 80% of this value over the first 4 weeks and then were stable until the end of the sampling period. The ammonia and ammonium levels in the metal-treated microcosms fell by only approximately 30% of the initial level over the first 4 weeks and then accumulated until they had returned to the starting values by the eighth week. There was no significant difference in ammonia and ammonium levels between the inoculated and noninoculated microcosms. The nitrogen turnover in metal-treated microcosms was therefore significantly affected by the addition of the metal chlorides, an imbalance that was not corrected by the addition of metal-resistant bacteria.
FIG. 1.
Ammonia and ammonium levels in soil microcosms. Ammonia and ammonium levels were determined as described previously (36). Week 0 values are given for the combined analysis of all microcosms (n = 12). All others are given for triplicate microcosms. NH3 and NH4+ levels fell rapidly in uncontaminated soils and stabilized by week 4.NH3 and NH4+ levels fell more slowly in metal-contaminated microcosms and then accumulated between weeks 4 and 8.
Recovery of amoA gene fragments from soil.
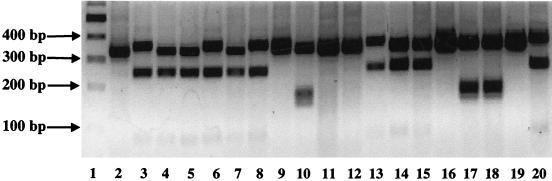
Single PCR products of the predicted size (ca. 490 bp) were recovered from each soil sample taken from week 0 and week 4 microcosms by primary amplification with the primers amoA-1F* and amoA-2R. Amplification products were generated from all week 8 soils, but the rate of individual successful attempts was reduced to 30%. Randomly selected clones were picked from each library, and the inserts were reamplified with vector primers for restriction digestion analysis. Digestion with MspI revealed three major banding patterns (Fig. 2) represented in each library. The total number of clones screened by restriction digestion analysis was 664. Representatives of each of the three major pattern types (a total of 20 clones) and all clones showing “other” banding patterns (a total of 10 clones) were selected for sequence analysis. Neighbor-joining analysis of the 449 bases of amplified sequence (corresponding to the minimum length of submissions in reference 32) (Fig. 3) and BLASTN searches of the GenBank database confirmed that all sequenced clones represented amoA-like sequences.
FIG. 2.
Restriction digestion analysis of amoA clones with MspI. Cloned amoA gene fragments were amplified from the cloning vector by using primers directed at the T7 and M13 reverse RNA polymerase binding sites, producing a fragment with approximately 70 bp of vector sequence on each end. The vector sequence contained no MspI recognition sites. Products were digested with a twofold excess of MspI for 1 h, analyzed by electrophoresis on a 2% agarose gel with TAE buffer, and visualized by ethidium bromide fluorescence. Lanes: 1, molecular size marker (100-bp ladder; Boehringer); 2, 9, and 19 pattern 1 amoA clones; 10, 17, and 18, pattern 2 amoA clones; 3 to 8, 11 to 16, and 20, pattern 3 amoA clones.
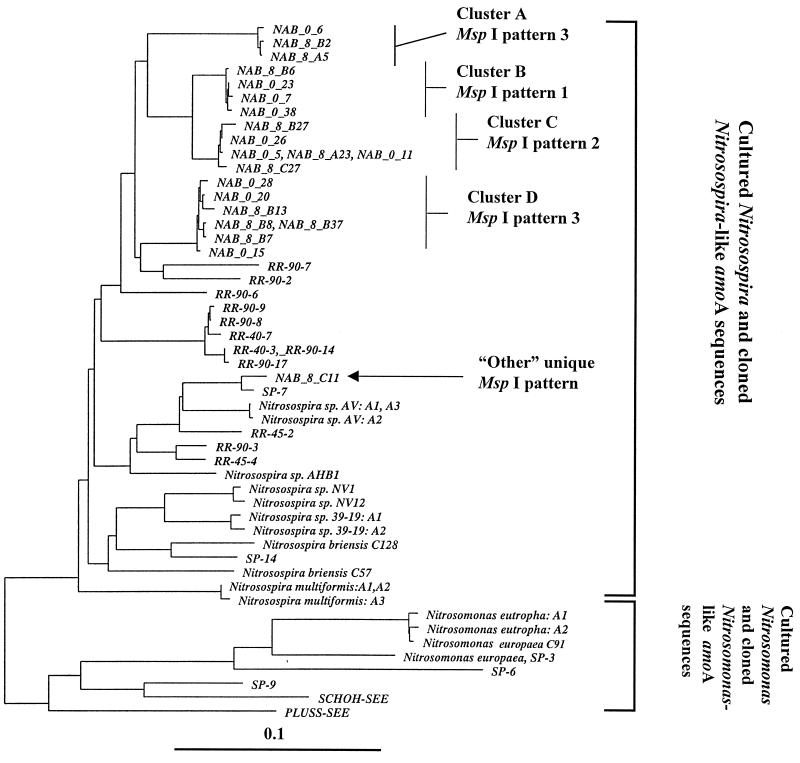
FIG. 3.
Neighbor-joining tree with Fitch-Margoliash correction of amoA sequences recovered from soil microcosms. The tree is derived from a compilation of available amoA sequences spanning the 449 nucleotide bases available for all sequences used. Sequences prefixed with NAB were generated during this study (accession no. AF056049 to AF056069). Clones were selected from libraries on the basis of MspI restriction patterns to provide a survey of sequence diversity in the target amoA sequences. Sequences which are identical over the fragment analyzed are presented on the same branch and separated by commas. Reference sequences: sequences SCHOH-SEE and PLUSS-SEE and sequences prefixed with RR and SP (environmental clones) (33); Nitrosospira briensis C57, Nitrosovibrio tenuis NV1, and Nitrosomonas europaea C-91 (pure cultures) (33); Nitrosospira sp. strain AHB1 (32); Nitrosomonas eutropha copies A1 and A2: (accession no. U51630 and U72670) (25); Nitrosospira multiformis copies A1, A2 and A3 (accession no. U91603, U15733, and U89833 respectively) (25); Nitrosospira briensis C128 (accession no. U76553) (25); Nitrosovibrio tenuis NV12 (accession no. U76552) (25); Nitrosospira sp. strain 39-19 copies A2 and A3 (accession no. AF016002 and AF006692, respectively) (25); Nitrosomonas europaea (23).
Diversity of recovered sequences.
Neighbor-joining analysis revealed four clusters of amoA sequences and 10 single sequences that did not fall within any of the four clusters. Independent analysis of the 3′ 205 bases and the 5′ 244 bases demonstrated poor stability in placement of 9 of the 10 single sequences, which were therefore tentatively regarded as chimeric artifacts of the PCR amplification process (37). Bootstrap analysis (100 replicates) also provided values of less than 50 for placement of these sequences. Bootstrap values of 100% values were recovered for support of each cluster and the grouping of sequence NAB_8_C11 with clone SP-1 (32). Sequence clusters B and C also grouped together with 100% bootstrap support. The only systematic difference between the translated protein sequences of the clusters is the conservative exchange at position 195 of an isoleucine in clusters A, B, and C (typical of published Nitrosomonas-related AmoA primary sequences) with a valine in cluster D sequences (typical of published Nitrosospira-related AmoA primary sequences). Comparison with the available data from cultured species (compiled in reference 33) suggested that all the sequences recovered in this study were derived from members of the genus Nitrosospira (Fig. 3). Despite selection of all variants of MspI restriction digestion patterns from the first 213 randomly selected clones analyzed, no sequences related to the genus Nitrosomonas were recovered. This may reflect the apparently small number of Nitrosomonas cells compared to Nitrosospira cells as previously recorded in agricultural soil (16, 37). Equally, it may suggest that the Nitrosomonas species inferred to exist in soil carry amoA sequences that are not compatible with the primer set chosen. Rotthauwe et al. (33) predicted that this may be the case for some untested lineages of the genus Nitrosomonas.
Although it would be naive to attempt to determine the number of ammonia oxidizer species in these samples from the sequences of a single gene, some points of interest can be made. Multiple copies of amoA are carried by a number of β-subgroup proteobacterium ammonia oxidizers. Nitrosospira multiformis, Nitrosospira sp. strain AV, and Nitrosospira sp. strain 39-19 (24, 25) carry three nearly identical copies of amoA (Fig. 3). This near identity is reflected in the short branch lengths separating the gene copies. The much longer branch lengths separating the four clusters and the one isolated representative of amoA sequences presented here suggest that they have been derived from at least five distinct strains of β-subgroup ammonia oxidizers.
Comparison of the population structures of clone libraries.
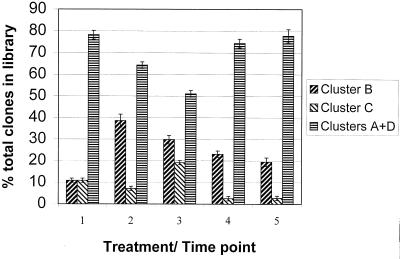
MspI digestion pattern analysis of amoA clones from these microcosms was selected as an analytical tool due to its rapidity and the observation that some phylogenetic information was maintained, inasmuch as two of the four supported clusters could be identified. amoA sequence clusters B (pattern 1) and C (pattern 2) could be distinguished from each other and from clusters A and D (pattern 3) with 100% accuracy as judged from the initial survey of 20 recovered sequences within these groups (Fig. 3). Clusters A and D could not be differentiated by this method; therefore, any changes in the abundance of these groups relative to each other were not detected. Clone libraries were generated from each soil microcosm and between 36 and 47 clones analyzed for each soil sample (Fig. 4). At time zero, clone libraries were dominated by sequences related to clusters A and D (80%), and the remainder were split equally between clusters B and C. After the 8-week microcosm incubation, the structures of all recovered libraries had changed significantly as judged by chi-square analysis. The greatest positive change was a relative increase in the proportion of cluster B (pattern 1) sequences.
FIG. 4.
Proportions of sequence types in clone libraries. The library population structures of all week 8 samples were significantly different from the starting population (chi-square probabilities of <0.05, <0.0002, <0.05, and <0.05, respectively). The only significantly different population within the week 8 clone libraries was between the metal-treated soil without added inoculum and all other week 8 libraries (P = <0.003). Error bars represent standard deviations between libraries generated from duplicate microcosms. Chi-square values were calculated on the combined data.
Effect of toxic metals.
The effect of metal addition after 8 weeks was seen in a comparison of libraries recovered from microcosms treated or not treated with toxic metals. These populations were significantly different, with cluster C (pattern 2) sequences being present in higher relative abundance following metal treatment, suggesting that the source organisms carried a selective advantage over other detectable β-subgroup ammonia oxidizers in the toxic-metal-treated soils.
Effect of the metal-resistant inoculum on metal solubility and changes to the β-subgroup ammonia oxidizer population.
Extraction of metals from soils in water did not indicate that the presence of the inoculated metal-resistant bacteria had any significant affect on the solubilities of any of the added metals (data not shown). The presence of the inoculum did not appear to influence the β-subgroup ammonia oxidizer population structure of noncontaminated microcosms, as seen in the comparison of libraries from 8-week-old untreated soil and untreated soil plus inoculum. Therefore, the inoculated bacteria did not affect the ammonia oxidizer population structure in the absence of toxic metals. A highly significant difference in population structure was, however, seen between amoA libraries recovered from metal-treated soils with and without the presence of the metal-resistant bacteria (P < 0.003). Further, there was no significant difference in the β-subgroup ammonia oxidizer population structure of 8-week-old untreated microcosms and that of metal-contaminated soils treated with the inoculum. These results demonstrated that the source organisms of amoA cluster C sequences gained no selective advantage over other indigenous β-subgroup ammonia oxidizers in the presence of toxic metals when the inoculum was present. We interpret this finding as evidence that the addition of the inoculum had lowered the bioavailability of the toxic metals to the β-subgroup ammonia oxidizer community sufficiently to protect it from specific metal-induced population change. The fact that no differences in metal availability were detected following water extraction may have been due to high standard errors on replicate samples, a weakness in the salt-free water extraction method (30).
Abundance of target amoA sequences in soil microcosms.
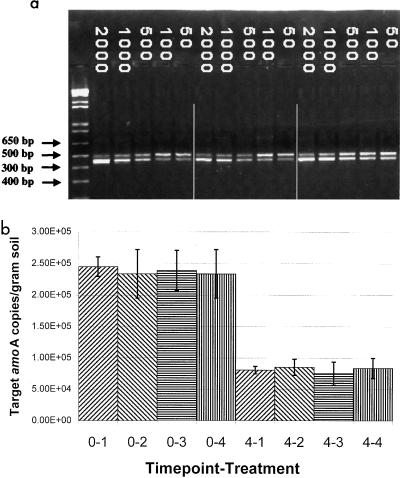
To compare changes in the total number of ammonia oxidizers per gram of toxic-metal-contaminated soil, competitive PCR for amoA sequences was used. This demonstrated that the sequences targeted by these primers dropped from approximately 2.3 × 105 copies per g of soil to 7 × 104 to 8 × 104 copies per g of soil over the first 4 weeks of the experiment in the presence of metals, irrespective of the presence of the inoculum. The ability to retrieve amplification products from week 8 soil samples was too inconsistent to gain an accurate estimate of target numbers, presumably since their numbers had dropped close to detection limits. These values were similar to those for the non-metal-treated soils (data not shown). Thus, the decrease in numbers of amoA target molecules was attributed to incubation under laboratory conditions. The presence of the inoculum did not protect the β-subgroup ammonia oxidizer population from this effect (Fig. 5).
FIG. 5.
(a) Quantification of amoA target sequences by competitive PCR. Results shown are from three soil microcosms at week 0 with added metals and inoculum. Lane 1 is a molecular size marker (1 kb-plus; Boehringer). Numbers indicate the number of linear amoA deletion fragments added to the reaction mixture. This generated the lower band visible in each lane. The relative abundances of amoA and amoA deletion fragments were assayed by ethidium bromide fluorescence and quantification with Alpha Imager software (Alpha Innotech). (b) Changes in amoA target numbers per gram of metal-contaminated soil microcosms. Values are averages of three reactions on DNA samples extracted from triplicate microcosms. Error bars indicate standard deviation. The prefix shows the time point: 0 indicates samples taken at week zero; 4 indicates samples taken at week 4. The suffix shows the treatment designation: 1, no inoculum, no metals; 2, inoculum added, no metals; 3, no inoculum, metals added; 4, inoculum added, metals added. Inoculation with metal-resistant bacteria did not significantly affect the total number of amoA target sequences per gram of soil detected by competitive PCR after 4 weeks.
Conclusions.
Due to the incomplete nature of the available amoA data set with respect to cultured organisms of the 16S phylogeny proposed previously (17), it is impossible to state what proportion of the soil β-subgroup ammonia oxidizer community was targeted by the PCR primers used. This is particularly notable in that phylogenetic interpretation of all available 16S rDNA sequence data from cultures, enrichments, and environmental clones suggests that the amoA sequences available from cultured organisms represent members of only 16S rDNA clusters 3 and 6/7 (32, 37). However, the amoA DNA sequence data presented here strongly suggests that the target organisms in the soil tested are quite distinct from any cultured organisms for which amoA sequence data is available. In this system, the source organisms of cluster C amoA sequences carried a demonstrable selective advantage over the other target organisms within the β-subgroup ammonia oxidizers following exposure to toxic metals. The rapid decline in amoA targets and the change in community structure associated with incubation of the soil under laboratory conditions preclude any strong conclusions on the effect of toxic metals on this group in the field. Nonetheless, sufficient evidence has been gathered to provide a working hypothesis which can now be tested at contaminated sites. Should the β-subgroup ammonia oxidizer community structure, measured as described, show elevated levels of cluster C amoA over neighboring control sites, a rapid and sensitive method will be available for the determination of a defensible end point to toxic-metal bioremediation. Reinstatement of the ratios of amoA sequence types to local control values may become a valuable measure of metal bioavailability.
The finding that the changes induced in the indigenous β-subgroup ammonia oxidizer population by toxic metals were abolished by the addition of exogenous bacterial species also supports the use of metal-resistant bacteria in reducing the bioavailability of metal species at sensitive sites. Determination of which member(s) of the five-species inoculum was responsible for this effect is under investigation.
ACKNOWLEDGMENTS
This work was supported by Department of Energy, Office of Energy Research, grant DE-FC02-96ER62278White as part of the Assessment Component of the Natural and Accelerated Bioremediation Research Program (NABIR), administered by John Houghton to D.C.W.
We thank Werner Liesack for helpful discussion and Julia Brüggemann for thoughtful comments on the manuscript.
REFERENCES
- 1.Avery S V. Cesium accumulation by microorganisms: uptake mechanisms, cation competition, compartmentalization and toxicity. J Ind Microbiol. 1995;14:76–84. doi: 10.1007/BF01569888. [DOI] [PubMed] [Google Scholar]
- 2.Babich H, Stotzky G. Heavy metal toxicity to microbe-mediated ecologic processes: a review and potential application to regulatory policies. Environ Res. 1985;36:111–137. doi: 10.1016/0013-9351(85)90011-8. [DOI] [PubMed] [Google Scholar]
- 3.Balkwill D L, Reeves R H, Drake G R, Reeves J Y, Crocker F H, King M B, Boone D R. Phylogenetic characterization of bacteria in the subsurface microbial culture collection. FEMS Microbiol Rev. 1997;20:201–216. doi: 10.1111/j.1574-6976.1997.tb00309.x. [DOI] [PubMed] [Google Scholar]
- 4.Beiderbeck V O, Campbell C A, Ukrainetz H, Curtin D, Bouman O T. Soil microbial and biochemical properties after ten years of fertilization with anhydrous ammonia. Can J Soil Sci. 1996;76:7–14. [Google Scholar]
- 5.Beveridge T J. Role of cellular design in bacterial metal accumulation and mineralization. Annu Rev Microbiol. 1989;43:147–171. doi: 10.1146/annurev.mi.43.100189.001051. [DOI] [PubMed] [Google Scholar]
- 6.Borneman J, Skroch P W, O’Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bunzl K, Schimmack W. Kinetics of the sorption of 137Cs, 85Sr, 57 Co, 65Zn, and 109Cd by the organic horizons of a forest soil. Radiochim Acta. 1991;54:97–102. [Google Scholar]
- 8.Cornish J E, Golberg W C, Levine R S, Benemann J R. Phytoremediation of soils contaminated with toxic elements and radionuclides. In: Hinchee R E, Means J L, Burris D R, editors. Bioremediation of inorganics. Third International In Situ and On-Site Bioreclamation Symposium, no. 10. Columbus, Ohio: Battelle Press; 1995. pp. 55–62. [Google Scholar]
- 9.Cunningham S D, Berti W R, Huang J W. Remediation of contaminated soils and sludges by green plants. In: Hinchee R E, Means J L, Burris D R, editors. Bioremediation of inorganics. Third International In Situ and On-Site Bioreclamation Symposium, no. 10. Columbus, Ohio: Battelle Press; 1995. pp. 33–45. [Google Scholar]
- 10.Felsenstein J. PHYLIP: phylogeny inference package (version 3.57c). Seattle: Department of Genetics, University of Washington; 1993. http://info.ibb.waw.pl/docs/PHYLIPdoc/index.html . Available from the author at: http://info.ibb.waw.pl/docs/PHYLIPdoc/index.html. [Google Scholar]
- 11.Fitch W M, Margoliash E. Construction of phylogenetic trees. Science. 1967;155:279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- 12.Franzle O. Contaminants in terrestrial environments. Springer Ser Phys Environ. 1993;13:312–315. [Google Scholar]
- 13.Frostegård A, Tunlid A, Bååth E. Phospholipid fatty acid composition, biomass and activity of microbial communities from two soil types experimentally exposed to different heavy metals. Appl Environ Microbiol. 1993;59:3605–3617. doi: 10.1128/aem.59.11.3605-3617.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frostegård A, Tunlid A, Bååth E. Changes in microbial community structure during long-term incubation in two soils experimentally contaminated with metals. Soil Biol Biochem. 1996;28:55–63. [Google Scholar]
- 15.Gilbert D G. SeqApp sequence alignment editor. 1996. Bloomington, Ind. Available from author by ftp ( ftp.bio.indiana.edu). [Google Scholar]
- 16.Hastings R C, Ceccherini M T, Miclaus N, Saunders J R, Bazzicalupo M, McCarthy A J. Direct molecular biological analysis of ammonia oxidizing bacteria populations in cultivated soil plots treated with swine manure. FEMS Microbiol Ecol. 1997;23:45–54. [Google Scholar]
- 17.Head I M, Hiorns W D, Embley T M, McCarthy A J, Saunders J R. The phylogeny of autotrophic ammonia-oxidizing bacteria as determined by analysis of 16S ribosomal RNA gene sequences. J Gen Microbiol. 1993;139:1147–1153. doi: 10.1099/00221287-139-6-1147. [DOI] [PubMed] [Google Scholar]
- 18.Kowalchuk G A, Stephen J R, De Boer W, Prosser J I, Embley T M, Woldendorp J W. Analysis of proteobacteria ammonia-oxidizing bacteria in coastal sand dunes using denaturing gradient gel electrophoresis and sequencing of PCR amplified 16S rDNA fragments. Appl Environ Microbiol. 1997;63:1489–1497. doi: 10.1128/aem.63.4.1489-1497.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liesack W, Janssen P H, Rainey F A, Ward-Rainey N L, Stackebrandt E. Microbial diversity in soil: the need for a combined approach using molecular and cultivation techniques. In: Van Elsas J D, Trevors J T, Wellington E M, editors. Modern soil microbiology. New York, N.Y: Marcel Dekker, Inc.; 1997. pp. 375–439. [Google Scholar]
- 20.Lovley D R. Microbial reduction of iron, manganese and other metals. Adv Agron. 1994;54:175–231. [Google Scholar]
- 21.Lux D, Kammerer L, Rühm W, Wirth E. Cycling of Pu, Sr, Cs, and other longliving radionuclides in forest ecosystems of the 30-km zone around Chernobyl. Sci Total Environ. 1995;173/174:375–384. [Google Scholar]
- 22.McLean E O. Soil pH and lime requirement. In: Page A L, Miller R H, Keeney D R, editors. Agronomy. 2nd ed. Madison, Wis: ASA; 1982. pp. 199–244. [Google Scholar]
- 23.McTavish H, Fuchs J A, Hooper A B. Sequence of the gene coding for ammonia monooxygenase in Nitrosomonas europaea. J Bacteriol. 1993;175:2436–2444. doi: 10.1128/jb.175.8.2436-2444.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norton J M, Low J M, Klotz M G. The gene encoding ammonia monooxygenase subunit A exists in three nearly identical copies in Nitrosospira sp. NpAV. FEMS Microbiol Lett. 1996;139:181–188. doi: 10.1111/j.1574-6968.1996.tb08200.x. [DOI] [PubMed] [Google Scholar]
- 25.Norton, J. M., J. Alzerreca, and M. G. Klotz. Unpublished data.
- 26.Obuekwe C O. Ph.D. thesis. Edmonton, Alberta, Canada: University of Alberta; 1980. [Google Scholar]
- 27.Ohman D E, Chakrabarty A M. Genetic mapping of chromosomal determinants for the production of exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect Immun. 1981;33:124–148. doi: 10.1128/iai.33.1.142-148.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pennanen T, Frostegåard Å, Fritze H, Bååth E. Phospholipid fatty acid composition and heavy metal tolerance of soil microbial communities along two heavy metal-polluted gradients in coniferous forests. Appl Environ Microbiol. 1996;62:420–428. doi: 10.1128/aem.62.2.420-428.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prosser J I. Autotrophic nitrification in bacteria. Adv Microbiol Physiol. 1989;30:125–181. doi: 10.1016/s0065-2911(08)60112-5. [DOI] [PubMed] [Google Scholar]
- 30.Rhoades J D. Soluble salts. Methods of soil analysis. 2. Chemical and microbiological properties. In: Page A L, Miller R H, Keeney D R, editors. Agronomy. 2nd ed. Madison, Wis: ASA; 1982. pp. 167–179. [Google Scholar]
- 31.Riley R G, Zachara J M. Chemical contaminants on DOE lands and selection of contaminant mixtures of subsurface science research. Publication DOE/ER-0547/T. U.S. Washington, D.C: Department of Energy; 1992. [Google Scholar]
- 32.Rotthauwe J-H, de Boer W, Liesack W. Comparative analysis of gene sequences encoding ammonia monooxygenase of Nitrosospira sp. AHB1 and Nitrosolobus multiformis C-71. FEMS Microbiol Lett. 1995;133:131–135. doi: 10.1111/j.1574-6968.1995.tb07873.x. [DOI] [PubMed] [Google Scholar]
- 33.Rotthauwe J-H, Witzel K-P, Liesack W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol. 1997;63:4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Said W A, Lewis D L. Quantitative assessment of the effects of metals on microbial degradation of organic chemicals. Appl Environ Microbiol. 1991;57:1498–1503. doi: 10.1128/aem.57.5.1498-1503.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fitch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Sims G K, Ellsworth T R, Mulvaney R L. Microscale determination of inorganic nitrogen in water and soil extracts. Commun Soil Sci Plant Anal. 1995;26:303–316. [Google Scholar]
- 37.Stephen J R, McCaig A E, Smith Z, Prosser J I, Embley T M. Molecular diversity of soil and marine 16S rDNA sequences related to β-subgroup ammonia-oxidizing bacteria. Appl Environ Microbiol. 1996;62:4147–4154. doi: 10.1128/aem.62.11.4147-4154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strunk O, Ludwig W. ARB. Munich, Germany: Computer program distributed by the Technical of University Munich; 1997. [Google Scholar]
- 39.Valentine N B, Bolton H, Jr, Kingsley M T, Drake G R, Balkwill D L, Blymale A E. Biosorption of cadmium, cobalt, nickel, and strontium by a Bacillus simplex strain isolated from the vadose zone. J Ind Microbiol. 1996;16:189–196. [Google Scholar]