Dear Editor,
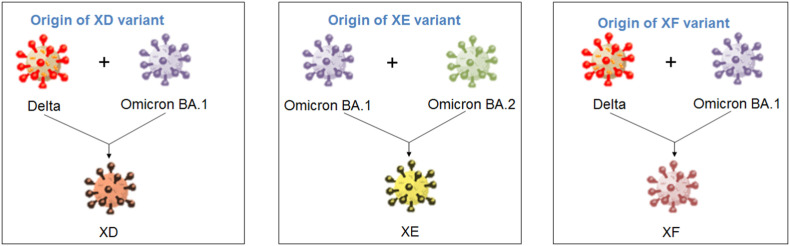
A unique SARS-CoV-2 recombinant variant was detected in France during Jan 2022. Recently, WHO has designated the recombinant variant as XD on March 09, 2022 [1]. The recombinant variant has been positioned in the category of VUM (variants under monitoring) by WHO. The researchers first designated the recombinant variant as “deltacron”, and CDC, China, has published a report about this recombinant variant, providing some scientific insight into the recombinant variant [2,3]. GISAID (Global Initiative on Sharing Avian Influenza Data) offers open-access information on avian influenza genomic data to researchers as a global science initiative. This open access to genomic data has played a significant role in sharing genomic data of SARS-CoV-2 to researchers during the pandemic period and allowed performance of faster research on the virus and its variants [4]. Presently, GISAID has highlighted the unique SARS-CoV-2 recombinant variant “in the focus section” [5]. The XD variant (AY.4/BA.1) is also recognized and confirmed by the Institut Pasteur in France, and the genome sequence is submitted to the GISAID. XD is considered a recombinant lineage developed from the recombination of two VOCs, i.e., the Delta (AY.4) variant and Omicron BA.1 variant. Simultaneously, the variant was reported in Denmark and Belgium and has created concern in these countries and other parts of Europe.
At the same time, the recombinant variant was confirmed by the UK Health security agency. Other than this recombinant variant (XD), this health agency also confirmed another two SARS-CoV-2 recombinant variants: XE and XF [6]. As per the eCDC, XF was first detected in the UK in January 2022 [7]. The eCDC has announced that all these recombinant variants can be positioned in the VUM category. However, all these three recombinant variants (XD, XE, and XF) are being closely monitored by researchers and policymakers throughout the world.
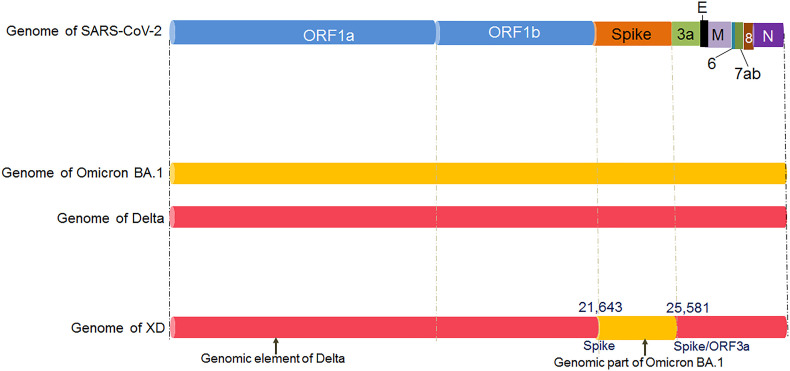
The XD variant contains the genomic elements from the Delta variant and Omicron BA.1 variant (Fig. 1 a). The XD variant contains the genomic elements ORF1a and ORF1b integrated from the Delta variant, while the spike protein is integrated from the Omicron BA.1 variant. The other genomic parts (E, M, ORF6, ORF7a, ORF7b, ORF8, N, 3UTR) of the XD is incorporated from the Delta variant (Fig. 1b). Researchers have noted that approximately 1 bp to 21463 bp of the genomic element is integrated from the Delta variant. Similarly, the genomic element from 21643 bp to 25581 bp is integrated from the Omicron BA.1 variant (spike protein region). Again, from 25581 bp to the endpoint, the genomic element is from the Delta variant. Nevertheless, it has been observed that XD contains the novel mutation (E172D). The E172D mutation is located in the genomic region of NSP2 [6].
Fig. 1.
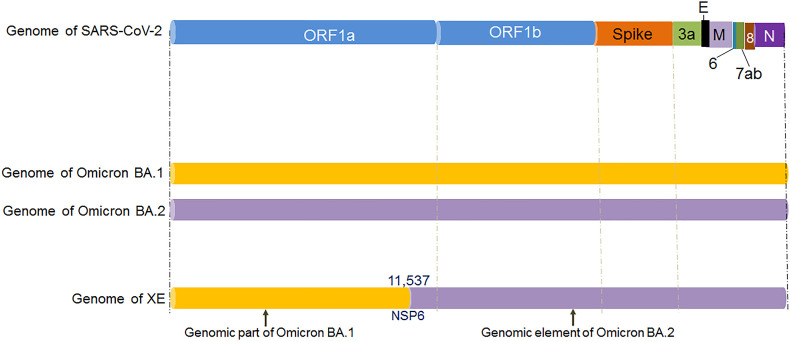
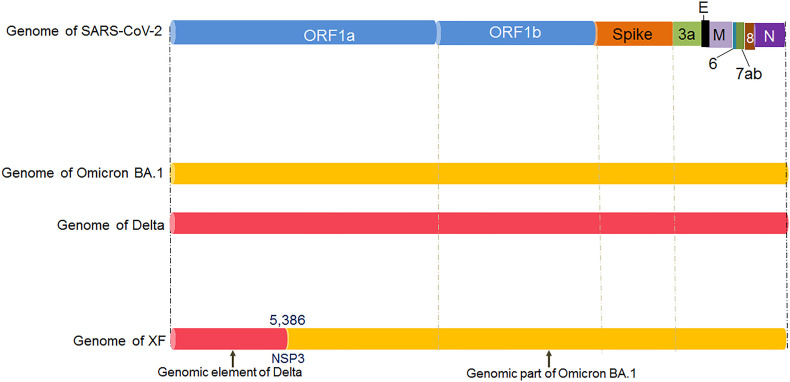
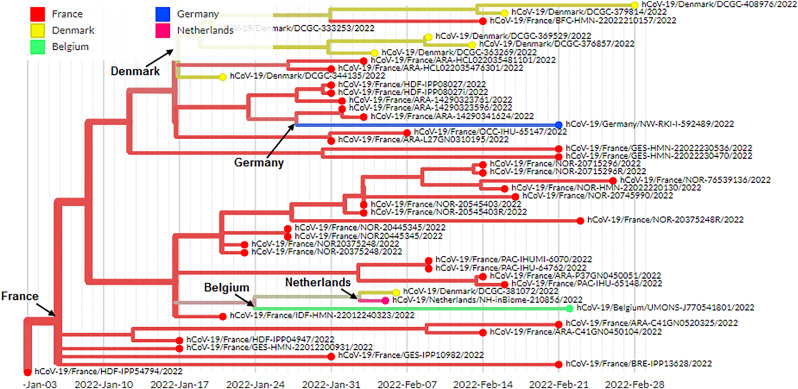
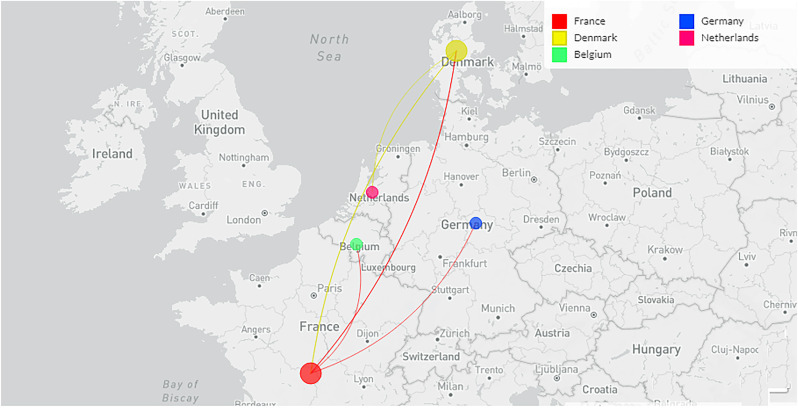
The diagram shows the schematic representation of the origin and genetic elements of recombinant variants XD, XE, and XF. The figure also describes the country-wise molecular phylogenetic of XD and spill over of the recombinant SARS-CoV-2 variants XD (a) Schematic representation of the origin of the recombinant SARS-CoV-2 variants XD, XE, and XF. The diagram illustrated the origin of the XD from Delta (AY.4) and Omicron BA.1 variant, the origin of XE from two Omicron variants (Omicron BA.1 and Omicron BA.2 variant), and the origin of XF from Delta and the Omicron BA.1 variant. (b) The figure represents the genome of XD variant. It also describes the genomic elements of Delta and Omicron BA.1 variants which are participated in recombinant XD variant's genome development. (c) The figure represents the genome of the XE variant. It also illustrates the genomic elements of Omicron BA.1 and Omicron BA.2 variants which are participated in the recombinant XE variant's genome creation. (d) The figure represents the genome of the XF variant. The figure also describes the genomic elements of Delta and Omicron BA.1 variants which are participated in the recombinant XF variant's genome development. (e) A phylodynamics of the XD variant which was detected in France and was developed using GISAID. The phylogenetic tree was depicted by GISAID. To show the country-wise relationship among the recombinant XD variant sequences of SARS-CoV-2. It is showing 47 genomes collected from January 2022 to February 2022 (As per the GISAID server) that have been used to develop the phylogenetic tree. (f) Figure shows the transmission pattern of the XD variant. It shows the variant spill over from France to the neighboring countries in Europe, and the variant is circulated to adjacent countries such as Germany, Belgium, Netherlands, and France. Fig. 1e and f are based on GISAID open data source.
The recombinant XE variant originated from the recombinant genomic elements from the two Omicron variants, the Omicron sister variants: Omicron BA.1 variant and Omicron BA.2 variant (Fig. 1a). However, it has been found that the XE variant contains more genomic elements from the Omicron BA.2 variant compared to the Omicron BA.1 variant. At the same time, the XE variant contains the first portion of the genomic elements from the Omicron BA.1 variant and the next/last segment of the genomic elements from the Omicron BA.2 variant (Fig. 1c). This recombinant variant contains up to 11,537 bp of genomic elements from the Omicron BA.1 variant, and it is in the NSP6 protein-coding genomic region. After the 11,537 bp position of genomic elements, the recombinant XE variant contains genomic elements from the Omicron BA.2 variant.
Similarly, the recombinant XF variant originated from the recombinant genomic elements of the two variants: the Delta variant and the Omicron BA.1 variant (Fig. 1a). However, the XF variant contains the genomic element of NSP1 to NSP3 from the Delta variant. The subsequent portion of the XF variant encloses the genomic element of the Omicron BA.1 variant. The first part of the genomic element of the Delta variant is noted up to 5386 bp, while after the 5386 bp position of genomic elements, the recombinant XF variant contains genomic elements from the Omicron BA.2 variant. Therefore, the XF variant contains more genomic elements from the Omicron BA.1 variant compared to the Delta variant (Fig. 1d). However, a comparative analysis of genomic elements of XD and XF variant shows that the XD variants contains more genomic part of the Delta variant. In contrast, the XF variant contains more genomic parts of the Omicron BA.1 variant.
A country-wise phylogenetic tree has been depicted to show the country-wise relationship among the recombinant XD variant of SARS-CoV-2 from GISAID (Fig. 1e). After identification of the XD variant from France, the variant spill over to the neighboring countries, and the circulation of the XD variant was even noted from the adjacent countries such as Germany, Belgium, Netherlands, and France (Fig. 1f). This phenomenon has raised several questions which require early answers for the properties of these recombinant variants, such as infectivity, re-infectivity, vaccine escape, immune escape, etc. Moreover, it is also an urgent need to unfold an in-depth evaluation of the SARS-CoV-2 recombinant linage.
The emergence of a SARS-CoV-2 recombinant linage is likely, and scientists expected it. However, very little is known about the recombinant variants. Thus, specific primers for the RT-PCR are urgently required to detect the recombinant variants efficiently. Therefore, more research is needed to efficiently design and develop the primers to identify these variants. Concurrently, more research is also needed to understand better the characteristics of these recombinant variants, such as their interaction properties (interaction between RBD of S-glycoprotein and hACE2 receptor to explain the infectivity), molecular determinants involved in the creation of recombinant linage, epidemiological data to develop appropriate prevention and control strategies to stop the pandemic.
Ethical Statement
No applicable.
Funding
None.
Data statement
The data in this correspondence article is not sensitive in nature and is accessible in the public domain. The data is therefore available and not of a confidential nature.
International journal of surgery author disclosure form
The following additional information is required for submission. Please note that failure to respond to these questions/statements will mean your submission will be returned. If you have nothing to declare in any of these categories, then this should be stated.
Research registration Unique Identifying number (UIN)
The World Medical Association's Declaration of Helsinki 2013 states in article 35: ‘Every research study involving human subjects must be registered in a publicly accessible database before recruitment of the first subject’. Editors of IJS require that all types of research studies involving human participants should be registered prospectively and failing that retrospectively. There are many places to register your research, and you can choose which is the most suitable for your needs:
-
•
https://www.clinicaltrials.gov/- for all human studies - free
-
•
http://www.chictr.org.cn/index.aspx - for all human studies - free
-
•
https://www.researchregistry.com/- for all human studies - charge
-
•
https://www.isrctn.com/- for all human studies - charge
-
•
Prospero - for systematic reviews - free
-
•
There are many national registries approved by the UN that can be found here
Elsevier does not support or endorse any registry.
-
1.
Name of the registry: Not applicable
-
2.
Unique Identifying number or registration ID: Not applicable
-
3.
Hyperlink to your specific registration (must be publicly accessible and will be checked): Not applicable
Guarantor
The Guarantor is the one or more people who accept full responsibility for the work and/or the conduct of the study, had access to the data, and controlled the decision to publish. Please note that providing a guarantor is compulsory.
Professor Chiranjib Chakraborty.
Department of Biotechnology, School of Life Science and Biotechnology.
Adamas University, Kolkata, West Bengal 700126, India.
Email: drchiranjib@yahoo.com Tel: +91-9871608125.
Provenance and peer review
Not commissioned, internally peer-reviewed.
CRediT authorship contribution statement
Chiranjib Chakraborty: Conceptualization, Data curation, Investigation, Writing – original draft, Writing – review & editing. Manojit Bhattacharya: Validation, figure preparation. Ashish Ranjan Sharma: Review and validation, discussion, review & editing. Kuldeep Dhama: Validation, reviewing, All authors critically reviewed and approved the final version of the manuscript.
Declaration of competing interest
All authors report no conflicts of interest relevant to this article.
References
- 1.WHO Tracking SARS-CoV-2 variants. 2022. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (Last accessed: April, 5 2022)
- 2.Wang L., Gao G.F. The “Wolf” is indeed coming: recombinant “deltacron” SARS-CoV-2 detected. China CDC Weekly. 2022;4(14):285–287. doi: 10.46234/ccdcw2022.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahase E. British Medical Journal Publishing Group; 2022. Covid-19: what Do We Know about the Delta Omicron Recombinant Variant? [DOI] [PubMed] [Google Scholar]
- 4.Maxmen A. One million coronavirus sequences: popular genome site hits mega milestone. Nature. 2021;593(7857) doi: 10.1038/d41586-021-01069-w. 21-21. [DOI] [PubMed] [Google Scholar]
- 5.GISAID Global initiative on sharing all influenza data. https://www.gisaid.org/ (Last accessed: April, 5 2022) [DOI] [PMC free article] [PubMed]
- 6.UKHSA SARS-CoV-2 variants of concern and variants under investigation in England: Technical briefing 39. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1063424/Tech-Briefing-39-25March2022_FINAL.pdf (Last accessed: April, 5 2022)
- 7.ECDC Data on SARS-CoV-2 variants in the EU/EEA. https://www.ecdc.europa.eu/en/publications-data/data-virus-variants-covid-19-eueea (Last accessed: April, 5 2022)