Abstract
Small nucleolar RNAs (snoRNAs), a type of non-coding RNA, are widely present in the nucleoli of eukaryotic cells and play an important role in rRNA modification. With the recent increase in research on snoRNAs, new evidence has emerged indicating that snoRNAs also participate in tRNA and mRNA modification. Studies suggest that numerous snoRNAs, including tumor-promoting and tumor-suppressing snoRNAs, are not only dysregulated in tumors but also show associations with clinical prognosis. In this review, we summarize the reported functions of snoRNAs and the possible mechanisms underlying their role in tumorigenesis and cancer development to guide the snoRNA-based clinical diagnosis and treatment of cancer in the future.
Subject terms: Tumour biomarkers, Cancer epigenetics
FACTS
SnoRNAs can be mainly divided into three types: H/ACA box snoRNAs, C/D box snoRNAs, and scaRNAs.
SnoRNAs are related to the modification of RNAs, including 2′-O-methylation and pseudouridylation of rRNAs and ac4C of 18 S rRNA. SnoRNAs can also regulate alternative splicing and have a function like miRNAs.
SnoRNAs take part in the occurrence and development of cancers.
OPEN QUESTIONS
How are snoRNAs produced and what are their functions?
How do snoRNAs take part in biological processes?
How do snoRNAs take part in tumorigenesis and cancer development?
Introduction
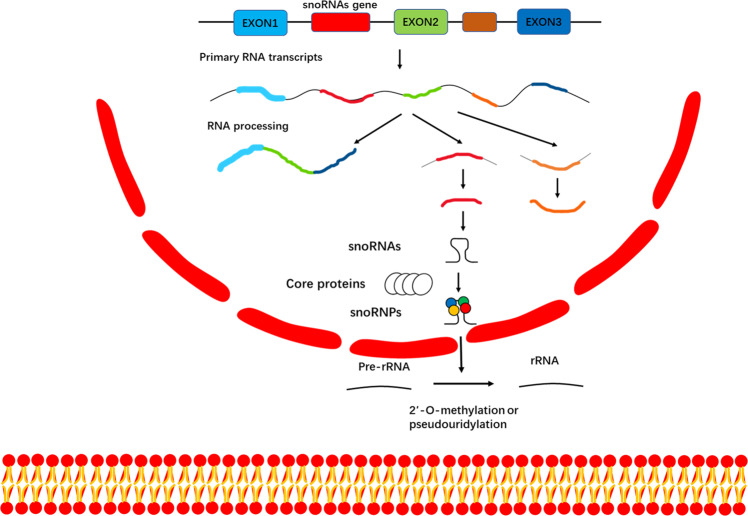
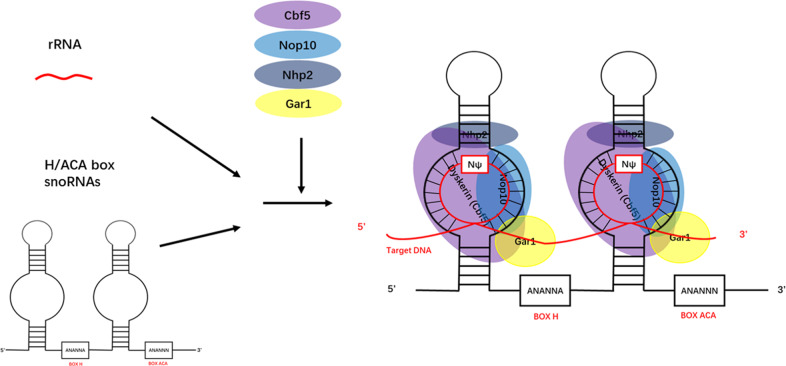
Small nucleolar RNAs (snoRNAs) are small non-coding RNAs widely present in the nucleoli of eukaryotic cells and have a length of 60–300 nt [1]. snoRNAs are mainly encoded by intronic regions of both protein coding and non-protein coding genes [2]. Normally, snoRNAs can be mainly classified into three groups: H/ACA box snoRNAs, C/D box snoRNAs, and small cajal RNAs (scaRNAs) [3]. The former two types of snoRNAs participate in the processing of ribosomal RNA (rRNA) by adding 2′-O-methylation and pseudouridylation modifications to rRNA molecules, respectively. However, a type of snoRNAs are located at Cajal bodies (CBs), so they are called scaRNAs. They also follow C/D-H/ACA classification, but some scaRNAs contain both C/D and H/ACA structures [4]. C/D box snoRNAs bind to four essential proteins—Nop1p, Nop56p, Nop58p, and Snu13p—to generate functional small nucleolar ribonucleoproteins (snoRNPs). Likewise, H/ACA box snoRNAs form functional snoRNPs by binding to Cbf5p, Gar1p, Nhp2p, and Nop10p [5] (Fig. 1).
Fig. 1. The biosynthesis of snoRNAs and snoRNPs.
Originate from the nucleolus, snoRNAs are mainly encoded in the intron region of the gene transcribed by RNA polymerase II. SnoRNAs form functional snoRNPs through binding to core proteins. SnoRNAs stabilize the structure of rRNA through modifying rRNA with 2 ‘- O-methylation and pseudouridylation.
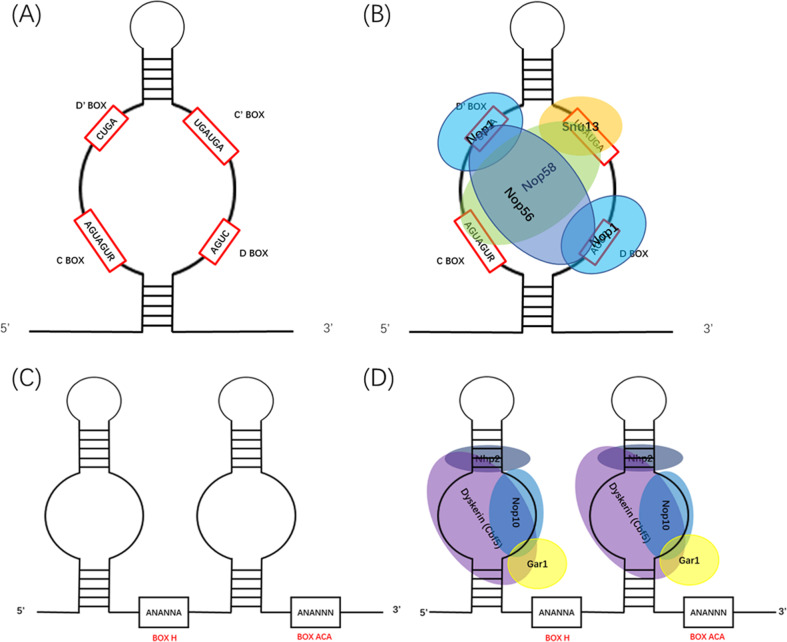
The length of eukaryotic C/D box snoRNAs usually ranges from 70 to 120 nt. These snoRNAs contain two conserved sequences: the C box and the D box. The C box consists of the nucleotides RUGAUGA, which are located at the 5′-end of the snoRNA molecule. In contrast, the D box is located at the 3′-end and consists of the nucleotides CUGA [6]. Together, these elements depend on the base-pairing to fold into a structure called a kink-turn. This structure is recognized by Snu13p, which then recruits Nop1p (also called fibrillarin [FBL]), Nop58p, and Nop56p for 2′-O-methylation modification [5, 7].
H/ACA snoRNAs are usually 60–75 nt in length and contain the region called the pseudouridylation pockets wherein uridine residues on the substrate RNA are isomerized [8]. H/ACA box snoRNPs bind to Cbf5p, Nop10p, Gar1p, and Nhp2p, among which Cbf5p acts as the catalytic protein involved in pseudouridylation [9]. Eukaryotic H/ACA box snoRNAs contain two conserved sequences: the H box and the ACA box, which are located downstream of the first and second hairpin, respectively [10] (Fig. 2).
Fig. 2. Structures of snoRNAs and snoRNPs.
The structure of C/D box snoRNAs A, C/D box snoRNPs B, H/ACA box snoRNAs C, and H/ACA box snoRNPs D.
Besides, some snoRNAs have been found that they have no apparent complementarity with rRNAs at known modified positions and they are called orphan snoRNAs. These indicated that snoRNAs have more function other than 2′-O-methylation and pseudouridylation of rRNAs. Kishore et al. indicated that HBII-52/SNORD115 had no complementarity with known modified positions of canonical snoRNAs. They found that HBII-52 bound to exon Vb of the 5-HT2C receptor and regulated alternative splicing [11, 12]. Another study found that orphan snoRNA SNORA73 inhibits PARP1 auto-PARylation to affect cancer genome stability by forming a small nucleolar ribonucleoprotein with PARP1 and DKC1/NHP2 [13].
SnoRNAs are also reported that they play a significant role in several tumors, such as lung cancer, gastric cancer. colorectal cancer, breast cancer, and so on. This review focuses on the functions of snoRNAs and the possible regulatory mechanisms underlying their role in biological processes, as well as their involvement in cancer pathophysiology.
Possible molecular mechanisms underlying the role of snoRNAs in biological processes
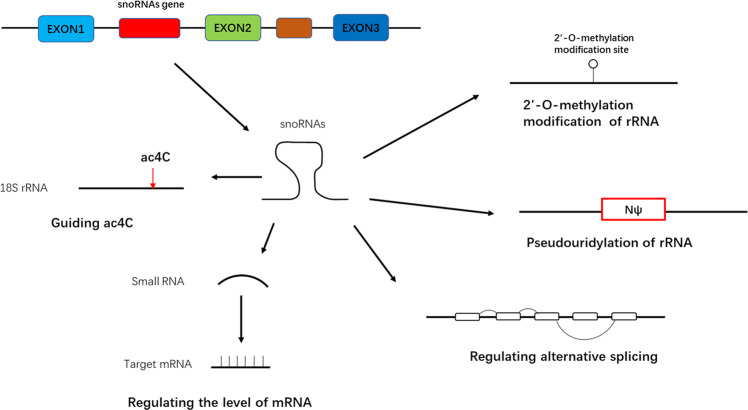
The common actions of snoRNAs include the 2′-O-methylation and pseudouridylation of rRNAs [14]. In recent years, there has been increasing research on snoRNAs, and several studies have confirmed that snoRNAs can also regulate cell physiology by guiding N4-acetylcytidine (ac4C) modifications, regulating alternative splicing (AS), and performing miRNA-like functions (Fig. 3).
Fig. 3. Molecular mechanisms of snoRNAs in biological processes.
The two common mechanisms of snoRNAs contain 2′-O-methylation and pseudouridylation of rRNAs. It is reported that snoRNAs also can regulate alternative splicing, guide N4-acetylcytidine and regulate the level of mRNA like a miRNA.
2′-O-methylation
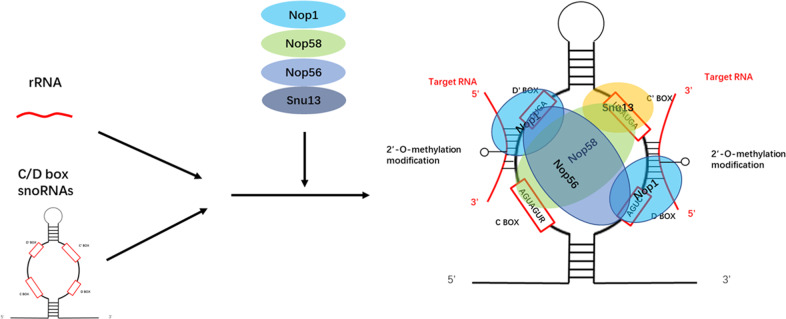
In 1981, Langberg et al. first detected methyltransferase activity in extracts from HeLa cells [15]. In 2000, it was discovered that one member of the C/D box snoRNP complex, i.e., Nop1p, was similar in terms of sequence and structural motifs to methyltransferases [16]. While all C/D box snoRNAs contain a C box and a D box, most of them also contain two additional components: the C′ box and the D′ box. One or two antisense elements (10–21 nt in length) are present upstream of the D box and/or the D′ box. The sequence of these antisense elements is complementary to that of the target rRNAs. Therefore, the snoRNAs bind to target rRNAs through these antisense elements [17–21].
As a methyltransferase, Nop1p is a key component of snoRNPs [22]. It transfers the methyl group from SAM to the 2′-hydroxyl group of ribose molecules in the target RNA [23]. The introduction of the methyl group changes the spatial structure of the target RNA and increases its hydrophobicity, protecting the RNA molecule from nucleolytic attacks [24] (Fig. 4).
Fig. 4. C/D box snoRNPs modifie the target RNA by 2′-O-methylation.
SnoRNAs bind to target RNAs by antisense elements upstream of D box and/or D’ box. SnoRNAs form functional snoRNPs by binding to four core proteins, including Nop1p, Nop56p, Nop58p, and Snu13p. Among them, Nop1p transfers the methyl group on s-adenosine-methionine to the 2′- hydroxyl group of the target RNA ribose.
Pseudouridylation
Pseudouridylation is the most prevalent RNA modification and can be found in all species of cellular RNA [25, 26]. Pseudouridylation can maintain RNA stability and modulate ribosome synthesis. Further, it plays an important role in transforming nonsense codons into sense codons [27–29]. Cohn first found a new nucleoside in 1951, which was named pseudouridine after soon [30, 31]. Currently, there are two known modes of pseudouridylation: RNA-independent and RNA-dependent. RNA-independent pseudouridylation can be achieved by an enzyme called pseudouridine synthase, while RNA-dependent pseudouridylation requires H/ACA box snoRNPs [32, 33]. H/ACA box snoRNAs bind to target RNA and transform the target uridine into pseudouridine, increasing target specificity. This modification occurs in the pseudouridylation pockets of H/ACA box snoRNAs [34]. In addition, Cbf5p can also act as an independent pseudouridine synthase and modify transfer RNA (tRNA) substrates [35] (Fig. 5).
Fig. 5. H/ACA box snoRNPs modifie the target RNA by Pseudouridylation.
The H/ACA box snoRNAs form functional snoRNPs through binding to core proteins, including Cbf5p, Nop10p, Nhp2p and Gar1p. SnoRNPs bind to target RNA by their pseudouridylation pockets. In these pseudouridylation pockets, the target uridine in the target RNA is modified into pseudouridine.
N4-acetylcytidine (ac4C)
As a highly conserved RNA modification, ac4C can be found on both rRNAs, tRNAs, and mRNAs [36]. In 1978, Thomas et al. found that ac4C was present in the ribosomes of rats, chickens, and budding yeast [37]. Then, the acetyltransferases NAT10 and Kre33 were found to catalyze ac4C modifications in humans and yeast, respectively. NAT10 is an ATP-dependent RNA acetyltransferase and is necessary for cytokinesis and nucleologenesis [38, 39]. It has been reported that NAT10 can also regulate DNA damage responses and telomerase function [38, 39]. Eukaryotic 18 S rRNAs contain two acetylated cytidines, one in helix 34 and the other in helix 45. The former is crucial for translation fidelity, whereas the latter is part of the ribosome decoding site. With the help of snoRNAs, NAT10 can catalyze the formation of ac4C on rRNA [40, 41].
Sharma et al. found that Kre33 could bind to rRNAs and tRNAs in yeast. Moreover, they showed that Kre33 could also bind to two orphan snoRNAs, snR4 and snR45. Through quantitative reverse-phase high-performance liquid chromatography (RP-HPLC), they confirmed that low levels of snR45 were related to a decrease in 18 S rRNA acetylation [38]. Tyc et al. discovered that in vertebrates, the C/D box snoRNA U13 is complementary to the 3′ end of 18 S rRNA owing to the presence of two extended complementary base pair regions [42]. Meanwhile, Sharma et al. showed that residue 1842, which is present between these two regions of complementarity, corresponds to the acetylated residue C1773 in yeast. They hypothesized that the C/D box snoRNA U13 could be involved in 18 S rRNA acetylation. Using HPLC, they found that the acetylation levels of 18 S rRNA purified from HCT116 cells were reduced by half after the depletion of the C/D box snoRNA U13 for 72 h [38]. These findings strongly indicated that snoRNAs play a role in 18 S rRNA acetylation.
Regulation of alternative splicing (AS)
AS is a process through which different combinations of splice sites are selected from a pre-messenger RNA (pre-mRNA) to produce variably spliced mRNAs [43]. In eukaryotes, AS significantly enriches the proteomic and transcriptomic output of the coding genome. In addition, AS is important for gene expression [44]. Multiple mRNA subtypes are generated from the same gene through AS in mammals. Due to their different coding capacities, stabilities, and translational efficiencies, these subtypes are translated into proteins with different structures and functions [45]. A primary RNA transcript contains exons, introns, and intervening sequences. Pre-mRNAs are converted to mature mRNAs through the removal of introns and the joining of spliced exons. This intron excision process is catalyzed by the spliceosome [42]. Moreover, AS allows the generation of mRNAs with different structures and functions, and therefore, different encoded proteins. It also influences intracellular localization, protein stability, enzymatic activity, and posttranslational modification of gene products [46]. Some studies show that numerous snoRNAs do not have binding site with rRNAs [47]. These indicate that snoRNAs could have functions beyond the modification of rRNA. Falaleeva et al. demonstrated that SNORD27 is present in nuclear components that lack FBL. The whole genome was searched for potential targets complementary to the SNORD27 sequence, and complementarity was identified between the alternatively spliced exons of the E2F7 gene and SNORD27. Further, low levels of SNORD27 were found to be associated with reduced levels of alternative exon skipping [48]. Moreover, Cavaillé et al. found that the orphan snoRNA SNORD115 plays a role in regulating the AS of serotonin receptor 2c (Htr2c) mRNA [12]. These findings indicate that snoRNAs may be involved in the regulation of AS.
MicroRNA(miRNA)-like functions
miRNAs are short regulatory RNAs that can regulate post-transcriptional gene expression. Lai found that miRNAs inhibit translation by binding to specific sequences in the 3′-untranslated region, thereby performing regulatory functions [49].
Surprisingly, in 2008, Ender et al. used northern blotting and verified that small RNAs could be derived from the snoRNA ACA45. They also found that these ACA45-derived small RNAs had miRNA-like functions, through which they could target CDC2L6 [50]. Additionally, Ono et al. demonstrated that while the C/D box snoRNA HBII-180C contains a 2′-O-methylation site, it also has an M-box region, through which it can act as a miRNA and inhibit the mRNA and protein expression of target genes [51]. Hence, the literature suggests that some snoRNAs can play miRNA-like roles within cells.
SnoRNAs in cancer
SnoRNAs in lung cancer
Non-small-cell lung carcinoma (NSCLC) is a major contributor to cancer-related deaths. NSCLC accounts for more than 75% of all lung cancer cases, and patients are often diagnosed at an advanced stage, which considerably diminishes the probability of complete recovery [52, 53]. Despite the significant progress in treatments for lung cancer, the prognosis of NSCLC continues to remain dismal [54].
Cui et al. found that the levels of NOP10, a component of H/ACA box snoRNPs, are elevated in NSCLC and are related to poor outcomes. Additionally, they found that the reduction of pseudouridylation resulting from the knockout of SNORA7A, SNORA7B, and SNORA65 and the inhibition of NOP10 can decrease the proliferation, invasion, and migration of lung cancer cells. [55] According to Mourksi et al., low levels of SNORA80E can increase the rate of apoptosis and the cleavage of caspase-3 and PARP1 in lung cancer cells, while increased levels of this snoRNA is associated with reduced p53 levels. SNORA80E inhibits apoptosis through a p53-dependent pathway [56]. Zheng et al. showed that low levels of SNORD78 could also inhibit cell proliferation. This effect is likely related to the consequent increase in the proportion of G0/G1 cells. P21 and P16, which are G0/G1 arrest markers, are up-regulated after SNORD78 knockdown. Moreover, the low expression of SNORD78 also increases the proportion of Bax/Bcl-2-positive cells, thus promoting cell apoptosis. Overall, the results indicate that low levels of SNORD78 promote apoptosis and induce cell cycle arrest, thereby inhibiting cell proliferation [57]. In addition, Tang et al. demonstrated that SNORA71A could influence the cell cycle, cell migration, cell invasion, and the epithelial–mesenchymal transition (EMT) via the phosphorylation of MEK and ERK1/2 in MAPK signaling pathway [58]. Taken together, these findings suggest that snoRNAs are involved in the development of NSCLC.
SnoRNAs in colorectal cancer (CRC)
CRC is a common type of cancer with the fourth-largest contributor to cancer mortality [59]. The treatments for CRC include surgery, chemotherapy, radiotherapy, and targeted therapy. However, despite the rapid development of therapeutic strategies against CRC, the prognosis of patients with CRC is still poor [60].
Owing to increasing research on snoRNAs, new evidence supporting the association between snoRNAs and CRC development has been uncovered. Liu et al. found that SNORD1C promotes the development of CRC by regulating β-catenin and TCF7 expression. High levels of SNORD1C are associated with a reduced five-year survival rate in CRC patients [61]. Another study showed that SNORA21 can promote CRC cell proliferation by regulating cancer-related pathways such as Hippo signaling pathway and Wnt signaling pathway and so on, and that high levels of SNORA21 is related to distant metastasis in CRC [62]. Fang’s group showed that in CRC, SNORD126 up-regulates FGFR2, thereby activating the PI3K-AKT pathway. The proteins downstream of this pathway include CREB, P27, MDM2, IKK, mTOR, p70S6K and GSK-3β. The overexpression of SNORD126 promotes the phosphorylation of GSK-3β and p70S6K, and promote the development of CRC via the PI3K-AKT signaling pathway [63]. SNORD12C/78 regulates the expression of target genes EIF4A3 and LAMC2 in a ZFAS1-dependent manner through NOP58-mediated 2’-O-methylation, promoting the development of CRC [64]. Hence, snoRNAs could be viable therapeutic targets for CRC.
SnoRNAs in gastric cancer (GC)
GC, one of the most common malignant tumors, is the second-largest contributor to cancer-associated deaths in the world [65]. Like most tumors, the treatments of GC include surgery, radiotherapy, chemotherapy, and targeted therapy. Notably, early diagnosis is believed to improve the treatment outcomes and prognosis of GC significantly.
The study showed that in GC, SNORD105B could promote tumorigenesis by binding to ALDOA and thereby upregulating the expression of C-myc [66, 67]. In addition, Liu et al. demonstrated that the overexpression of SNORA21 was associated with increased lymph node metastasis and distant metastasis in GC [68]. These studies suggest that snoRNAs may play a significant role in the occurrence and development of GC.
SnoRNAs in breast cancer (BC)
BC is the most common malignant tumor among women and the primary cause of cancer-related death in this group [69]. Hence, there is a great need to find effective prognostic biomarkers and therapeutic targets for BC.
Su et al. found that the levels of FBL were elevated in BC. FBL, a core protein of C/D box snoRNPs, was found to be important for the accumulation of snoRNAs and could affect Myc levels [70]. In turn, Myc also induced FBL expression. Meanwhile, low levels of FBL increased p53 activity, while its overexpression reduced the p53 response. Therefore, the findings indicated that snoRNAs could contribute to the development of BC by modulating the p53 response [71]. Another study found that snoRNA U50 mediates the methylation of C2848 in 28 S rRNA [72], suggesting that it may act as a tumor suppressor-like gene. Additionally, Dong et al. discovered that snoRNA U50, which inhibits cell colony formation, is frequently downregulated in BC [73]. Hence, snoRNA U50 may exert tumor-suppressive effects in BC. SNORD50A/B significantly enhances their interaction by forming a complex between the E3 ubiquitin ligase TRIM21 and its substrate GMPS, thereby promoting GMPS ubiquitination. Deletion of SNORD50A/B in p53 wild-type breast cancer cells releases GMPS and induces GMPS translocation into the nucleus, where GMPS can recruit USP7 and form a complex with p53, thereby reducing p53 ubiquitination, stabilizing p53 protein, and suppressing malignant phenotypes [74]. Kim et al. indicated that SNORA73A、SNORA73B and SNORA74A bound to PARP-1 to activate the catalytic activity of PARP-1 and mediated ADPRylation of DDX21, so as to promote cell proliferation in BC [75]. Moreover, Hu et al. found that in BC, SNORA71A promoted the binding of G3BP1-ROCK2 and increased the expression of ROCK2, promoting EMT process [76]. These evidences together support the association of snoRNAs with the occurrence and development of BC.
SnoRNAs in Hepatocellular carcinoma (HCC)
HCC is common cancer with a high mortality rate [77]. Currently, therapies for HCC include surgery, radiotherapy, chemotherapy, and other comprehensive treatments, which extend life expectancy to a certain extent [78, 79]. However, owing to the unavailability of early screening markers, most patients with HCC are diagnosed in the advanced stage, and the lack of effective treatment leads to high rates of mortality [80].
Fang et al. found that orphan snoRNA SNORD126 promotes cell growth in HCC and binds to hnRNPK protein to up-regulate FGFR2, thus activating the PI3K-AKT pathway. Meanwhile, FGFR2 downregulation suppresses the growth of Huh-7 cells with high levels of SNORD126. Hence, SNORD126 appears to regulate HCC development via the PI3K-AKT pathway [81]. SnoU2_19 participates in the regulation of the Wnt/β-catenin signaling pathway by inducing the translocation of β-catenin between the cytoplasm and nucleus, thereby promoting the progression of hepatocellular carcinoma [82]. SNORD52 upregulated CDK1 by binding and enhancing the stability of CDK1 proteins to promote HCC tumorigenesis [83]. SNORD17 reduces p53 activation by anchoring nucleophosmin 1 and MYB-binding protein 1a in the nucleolus to drive HCC progression [84]. In addition, other studies have shown that the overexpression of SNORD105 can increase cell viability and motility in HCC [85] and SNORA42 can promote the development of HCC by inhibiting p53 signal pathways [86]. Further, SNORD113-1 suppresses tumorigenesis in HCC by regulating the transforming growth factor-β (TGF-β) and mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK-ERK) pathways [87]. These studies indicate that snoRNAs are involved in the development of HCC.
SnoRNAs in ovarian cancer (OC)
Of all gynecological tumors, OC has the highest mortality rate [88]. Despite advancements in surgery and chemotherapeutics, the five‐year survival rate in women diagnosed with OC remains below 30% [89].
Zhang et al. found that SNORA72 influences cell stemness in OC via the Notch1/c-Myc pathway [90]. In addition, SNORD89 can affect cell proliferation, invasion, migration, and self-renewal ability in OC by regulating the Notch1/c-Myc pathway [91]. Therefore, snoRNAs may play a role in the development of OC.
SnoRNAs in leukemia
Leukemia is classified into several types, including acute lymphoblastic leukemia, acute myeloid leukemia (AML), chronic lymphocytic leukemia, and chronic myelogenous leukemia [92]. The main treatment strategies for leukemia include chemotherapy and radiotherapy, although the former causes severe toxicity and adverse effects.
Valleron et al. discovered that SNORD112, SNORD113, and SNORD114 are ectopically expressed at the DLK1-DIO3 locus in acute promyelocytic leukemia. Their study showed that the variants of SNORD114-1 cause cell cycle arrest at G0/G1 and inhibit cell growth [93]. Another study by Pauli et al. demonstrated that the knockout of SNORD42A could reduce 2′-O-methylation levels of U116, causing ribosome activity and protein translation decreased. SNORD42A deficiency could inhibit cell proliferation and colony-forming ability in malignant cells [94]. All in all, snoRNAs may be related to the development of leukemia (Table 1).
Table 1.
The mechanism and function of snoRNAs in several types of cancers.
| Cancer | snoRNA name | Expression | Mechanism/Pathway/Target | Function | Reference |
|---|---|---|---|---|---|
| Lung cancer | SNORA7A | Upregulation | unknown | Promote the cell proliferation, invasion, and migration | [55] |
| SNORA7B | Upregulation | unknown | |||
| SNORA65 | Upregulation | unknown | |||
| SNORA80E | Upregulation | P53 | Inhibit apoptosis and support stemness | [56] | |
| SNORD78 | Upregulation | unknown | promoted the cell proliferation invasion and EMT process | [57] | |
| SNORA71A | Upregulation | MAPK/MEK/ERK | Promote the cell proliferation, invasion, and migration | [58] | |
| SNORA47 | Upregulation | PI3K/Akt/EMT | Promote cell proliferation, migration, invasion and EMT process and inhibit apoptosis | [95] | |
| snoRNA U3 | Upregulation | P53 | Involve in pre-rRNA processing and required for in vitro and in vivo tumorigenesis | [96] | |
| snoRNA U8 | Upregulation | ||||
| SNORA42 | Upregulation | P53 | Promote cell proliferation, migration, invasion and xenograft growth in vivo | [97] | |
| Colorectal cancer | SNORD1C | Upregulation | Wnt/β-catenin | Promote cell proliferation, migration, invasion and inhibit apoptosis and enhance cancer cell stemness | [61] |
| SNORA21 | Upregulation | Hippo signaling pathway and Wnt signaling pathway | Promote cell proliferation | [62] | |
| SNORD78 | Upregulation | Regulate the expression of EIF4A3 and LAMC2 in a ZFAS1-dependent manner | Promote the development of CRC | [63] | |
| SNORD12C | Upregulation | ||||
| SNORD126 | Upregulation | FGFR2, PI3K-AKT/GSK-3β, p70S6K | Promote cell growth | [80] | |
| snoRNA U44 | Upregulation | P53 | Involve in p53-regulated cellular response to DNA damage | [98] | |
| snoRNA U47 | Upregulation | ||||
| Gastric cancer | SNORD105B | Upregulation | ALDOA / C-myc | Promote cell proliferation, migration and invasion | [65, 66] |
| SNORA21 | Upregulation | unknown | Be associated with increased lymph node metastasis and distant metastasis | [67] | |
| Breast cancer | snoRNA U50 | Downregulation | Methylation of C2848 in 28 S rRNA | Inhibit cell colony formation | [71, 72] |
| SNORD50A/B | Upregulation | TRIM21-GMPS/P53 | Enhance malignant phenotypes in p53wt breast cancer cells, while inhibit malignant phenotypes in p53mt breast cancer cells | [73] | |
| SNORA73A/B | Upregulation | PARP-1/mediate ADPRylation of DDX21 | Promote cell proliferation | [74] | |
| SNORA74A | Upregulation | ||||
| SNORA71A | Upregulation | G3BP1/ROCK2 | Promote EMT process | [75] | |
| SNORA71B | Upregulation | Unknown | Promote cell proliferation, migration, invasion and EMT process | [99] | |
| SNORA7B | Upregulation | unknown | Be related to poor prognosis for BC and promote cell proliferation, migration, invasion | [100] | |
| Hepatocellular carcinoma | SNORD126 | Upregulation | FGFR2, PI3K-AKT/GSK-3β, p70S6K | Promote cell growth | [80] |
| snoRNA U2_19 | Upregulation | Wnt/β-catenin | Promote cell proliferation and inhibit apoptosis | [81] | |
| SNORD52 | Upregulation | CDK1 | Be related to poor prognosis and promote cell proliferation | [82] | |
| SNORD17 | Upregulation | P53 | Promote the growth and tumorigenicity of HCC cells | [83] | |
| SNORD105 | Upregulation | unknown | Increase cell viability and motility | [84] | |
| SNORA42 | Upregulation | P53 | Promote cell proliferation, migration, invasion and inhibit apoptosis | [85] | |
| SNORD113-1 | Downregulation | TGF-β、MAPK-ERK pathways | Suppress HCC tumorigenesis | [86] | |
| ACA11 | Upregulation | PI3K/AKT | Promote cell growth, migration, invasion and induce EMT process | [101] | |
| SNORA18L5 | Upregulation | P53 | Promote cell proliferation and inhibit apoptosis | [102] | |
| SNORD126 | Upregulation | hnRNPK, FGFR2, PI3K-AKT | Promote HCC tumorigenesis in vitro and in vivo | [81] | |
| SNORA47 | Upregulation | unknown | Promote cell proliferation, migration, invasion and inhibit apoptosis | [103] | |
| SNORD76 | Upregulation | Wnt/β-catenin | Promote cell growth and induce EMT process | [104] | |
| SNORA42 | Upregulation | P53 | Promote cell proliferation, migration, invasion and inhibit apoptosis | [85] | |
| SNORA23 | Downregulation | Impairing the 2′-O-ribose methylation of 28 S rRNA | Inhibit cell proliferation, migration and invasion | [105] | |
| Ovarian cancer | SNORA72 | Upregulation | Notch1/c-Myc | Induce the stemness of OC | [89] |
| SNORD89 | Upregulation | Notch1/c-Myc | Promote cell proliferation, migration and invasion | [90] | |
| Leukemia | SNORD114-1 | Ectopically expressed at the DLK1-DIO3 | Rb/p16 | Implicate in the G0/G1 to S phase transition | [92] |
| SNORD42A | Upregulation in AML | Increase the 2′-O-methylation level of U116 | Promote cell proliferation and colony-forming ability | [93] |
Conclusion
Previously, snoRNAs were thought only to be involved in 2′-O-methylation and pseudouridylation. However, with an increase in the number of studies, other functions of snoRNAs, including ac4C modification, AS regulation, and microRNA-like actions, have been discovered. Accumulating evidence indicates that the levels of snoRNAs are perturbed in malignant tissues. However, their specific roles in tumors have not been fully elucidated. In the existing studies, snoRNAs mainly bind to proteins, mRNAs, rRNAs, etc. directly or participate in protein regulatory pathways to regulate the modification and stability of proteins and RNAs, regulate protein expression and subcellular localization, and change the activity of proteins and protein complexes, thereby involved in tumorigenesis and cancer progression. According to the previous studies, 2’-O-methylation, pseudouracillation, ac4C modification, AS regulation, and microRNA-like effects also play key roles in tumorigenesis and cancer development. However, there are few reports that snoRNAs participate in tumor regulation by means of the above ways. So the questions whether and how snoRNAs use these pathways to participate in tumor regulation warrants further investigation. Such research could improve our understanding of the link between cancer and snoRNAs and bolster the use of snoRNAs as effective biomarkers and therapeutic targets for various cancers.
Acknowledgements
This study was supported by the National Natural Scientific Foundation of China (No. 82072854) and The Project for Key Medicine Discipline Construction of Guangzhou Municipality (No. 2021-2023-17).
Author contributions
Guarantor of the article: YZ designed and revised the review. Z-HH and Y-PD wrote and revised the manuscript. J-TW and B-FL revised the manuscript. Z-HH constructed and revised the figures. All authors approved the final manuscript and agreed to be responsible for this review.
Data availability
The data used to support the findings of this study are available from the corresponding author upon request.
Competing interests
The authors declare no competing interests.
Ethics statement
Not applicable.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zheng-hao Huang, Yu-ping Du.
References
- 1.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 2.Dieci G, Preti M, Montanini B. Eukaryotic snoRNAs: a paradigm for gene expression flexibility. Genomics. 2009;94:83–8. doi: 10.1016/j.ygeno.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 3.van der Werf J, Chin CV, Fleming NI. SnoRNA in cancer progression, metastasis and immunotherapy response. Biol (Basel) 2021;10:809. doi: 10.3390/biology10080809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bratkovič T, Rogelj B. The many faces of small nucleolar RNAs. Biochim Biophys Acta. 2014;1839:438–43. doi: 10.1016/j.bbagrm.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer? Nat Rev Cancer. 2012;12:84–8. doi: 10.1038/nrc3195. [DOI] [PubMed] [Google Scholar]
- 6.Kiss-László Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85:1077–88. doi: 10.1016/S0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 7.Baldini L, Charpentier B, Labialle S. Emerging data on the diversity of molecular mechanisms involving C/D snoRNAs. Noncoding RNA. 2021;7:30. doi: 10.3390/ncrna7020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bortolin ML, Ganot P, Kiss T. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. The. EMBO J. 1999;18:457–69. doi: 10.1093/emboj/18.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krell er UT. How a single protein complex accommodates many different H/ACA RNAs. Trends Biochem Sci. 2006;31:311–5. doi: 10.1016/j.tibs.2006.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–56. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 11.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–2. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 12.Cavaillé J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, et al. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci USA. 2000;97:14311–6. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han C, Sun LY, Luo XQ, Pan Q, Sun YM, Zeng ZC, et al. Chromatin-associated orphan snoRNA regulates DNA damage-mediated differentiation via a non-canonical complex. Cell Rep. 2022;38:110421. doi: 10.1016/j.celrep.2022.110421. [DOI] [PubMed] [Google Scholar]
- 14.Panse VG, Johnson AW. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem Sci. 2010;35:260–6. doi: 10.1016/j.tibs.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langberg SR, Moss B. Post-transcriptional modifications of mRNA. Purification and characterization of cap I and cap II RNA (nucleoside-2’-)-methyltransferases from HeLa cells. J Biol Chem. 1981;256:10054–60. doi: 10.1016/S0021-9258(19)68740-5. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Boisvert D, Kim KK, Kim R, Kim SH. Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 A resolution. EMBO J. 2000;19:317–23. doi: 10.1093/emboj/19.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bratkovič T, Božič J, Rogelj B. Functional diversity of small nucleolar RNAs. Nucleic Acids Res. 2020;48:1627–51. doi: 10.1093/nar/gkz1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiss-Laszlo Z, Henry Y, Kiss T. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 1998;17:797–807. doi: 10.1093/emboj/17.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tycowski KT, Shu MD, Steitz JA. A mammalian gene with introns instead of exons generating stable RNA products. Nature. 1996;379:464–6. doi: 10.1038/379464a0. [DOI] [PubMed] [Google Scholar]
- 20.Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20:3617–22. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cavaille J, Nicoloso M, Bachellerie JP. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature. 1996;383:732–5. doi: 10.1038/383732a0. [DOI] [PubMed] [Google Scholar]
- 22.Filipowicz W, Pelczar P, Pogacic V, Dragon F. Structure and biogenesis of small nucleolar RNAs acting as guides for ribosomal RNA modification. Acta Biochim Pol. 1999;46:377–89. doi: 10.18388/abp.1999_4171. [DOI] [PubMed] [Google Scholar]
- 23.Stepanov GA, Filippova JA, Komissarov AB, Kuligina EV, Richter VA, Semenov DV. Regulatory role of small nucleolar RNAs in human diseases. Biomed Res Int. 2015;2015:206849. doi: 10.1155/2015/206849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sproat BS, Lamond AI, Beijer B, Neuner P, Ryder U. Highly efficient chemical synthesis of 2′-O-methyloligoribonucleotides and tetrabiotinylated derivatives; novel probes that are resistant to degradation by RNA or DNA specific nucleases. Nucleic Acids Res. 1989;17:3373–86. doi: 10.1093/nar/17.9.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilbert WV, Bell TA, Schaening C. Messenger RNA modifications: form, distribution, and function. Science. 2016;352:1408–12. doi: 10.1126/science.aad8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karijolich J, Yi C, Yu YT. Transcriptome-wide dynamics of RNA pseudouridylation. Nat Rev Mol Cell Biol. 2015;16:581–5. doi: 10.1038/nrm4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kierzek E, Malgowska M, Lisowiec J, Turner DH, Gdaniec Z, Kierzek R. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res. 2013;42:3492–501. doi: 10.1093/nar/gkt1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jack K, Bellodi C, Landry DM, Niederer RO, Meskauskas A, Musalgaonkar S, et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol Cell. 2011;44:660–6. doi: 10.1016/j.molcel.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernández IS, Ng CL, Kelley AC, Wu G, Yu YT, Ramakrishnan V. Unusual base pairing during the decoding of a stop codon by the ribosome. Nature. 2013;500:107–10. doi: 10.1038/nature12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cohn WE. Some results of the applications of ion-exchange chromatography to nucleic acid chemistry. J Cell Physiol Suppl. 1951;38:21–40. doi: 10.1002/jcp.1030380405. [DOI] [PubMed] [Google Scholar]
- 31.Cohn WE. Pseudouridine, a carbon-carbon linked ribonucleoside in ribonucleic acids: isolation, structure, and chemical characteristics. J Biol Chem. 1960;235:1488–98. doi: 10.1016/S0021-9258(18)69432-3. [DOI] [PubMed] [Google Scholar]
- 32.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89:799–809. doi: 10.1016/S0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 33.Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–73. doi: 10.1016/S0092-8674(00)80238-X. [DOI] [PubMed] [Google Scholar]
- 34.Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–34. doi: 10.1016/S0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 35.Kamalampeta R, Kothe U. Archaeal proteins Nop10 and Gar1 increase the catalytic activity of Cbf5 in pseudouridylating tRNA. Sci Rep. 2012;2:663. doi: 10.1038/srep00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sas-Chen A, Thomas JM, Matzov D, Taoka M, Nance KD, Nir R, et al. Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping. Nature. 2020;583:638–43. doi: 10.1038/s41586-020-2418-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas G, Gordon J, Rogg H. N4-Acetylcytidine. A previously unidentified labile component of the small subunit of eukaryotic ribosomes. J Biol Chem. 1978;253:1101–5. doi: 10.1016/S0021-9258(17)38117-6. [DOI] [PubMed] [Google Scholar]
- 38.Sharma S, Langhendries JL, Watzinger P, Kötter P, Entian KD, Lafontaine DL. Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 2015;43:2242–58. doi: 10.1093/nar/gkv075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ito S, Horikawa S, Suzuki T, Kawauchi H, Tanaka Y, Suzuki T, et al. Human NAT10 is an ATP-dependent RNA acetyltransferase responsible for N4-acetylcytidine formation in 18 S ribosomal RNA (rRNA) J Biol Chem. 2014;289:35724–30. doi: 10.1074/jbc.C114.602698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin G, Xu M, Zou M, Duan S. The processing, gene regulation, biological functions, and clinical relevance of N4-acetylcytidine on RNA: a systematic review. Mol Ther Nucleic Acids. 2020;20:13–24. doi: 10.1016/j.omtn.2020.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ito S, Akamatsu Y, Noma A, Kimura S, Miyauchi K, Ikeuchi Y, et al. A single acetylation of 18 S rRNA is essential for biogenesis of the small ribosomal subunit in Saccharomyces cerevisiae. J Biol Chem. 2014;289:26201–12. doi: 10.1074/jbc.M114.593996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tyc K, Steitz JA. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989;8:3113–9. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–63. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leung SK, Jeffries AR, Castanho I, Jordan BT, Moore K, Davies JP, et al. Full-length transcript sequencing of human and mouse cerebral cortex identifies widespread isoform diversity and alternative splicing. Cell Rep. 2021;37:110022. doi: 10.1016/j.celrep.2021.110022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–5. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 46.Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, et al. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 47.Filipowicz W, Pogacić V. Biogenesis of small nucleolar ribonucleoproteins. Curr Opin Cell Biol. 2002;14:319–27. doi: 10.1016/S0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- 48.Falaleeva M, Pages A, Matuszek Z, Hidmi S, Agranat-Tamir L, Korotkov K, et al. Dual function of C/D box small nucleolar RNAs in rRNA modification and alternative pre-mRNA splicing. Proc Natl Acad Sci USA. 2016;113:E1625–34. doi: 10.1073/pnas.1519292113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lai EC. microRNAs: runts of the genome assert themselves. Curr Biol. 2003;13:R925–36. doi: 10.1016/j.cub.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 50.Ender C, Krek A, Friedländer MR, Beitzinger M, Weinmann L, Chen W, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–28. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 51.Ono M, Yamada K, Avolio F, Scott MS, van Koningsbruggen S, Barton GJ, et al. Analysis of human small nucleolar RNAs (snoRNA) and the development of snoRNA modulator of gene expression vectors. Mol Biol Cell. 2010;21:1569–84. doi: 10.1091/mbc.e10-01-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 53.Tan S, Gou Q, Pu W, Guo C, Yang Y, Wu K, et al. Circular RNA F-circEA produced from EML4-ALK fusion gene as a novel liquid biopsy biomarker for non-small cell lung cancer. Cell Res. 2018;28:693–5. doi: 10.1038/s41422-018-0033-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, Chen WJ, Gan TQ, Zhang XL, Xie ZC, Ye ZH, et al. Clinical significance and effect of lncRNA HOXA11-AS in NSCLC: a study based on bioinformatics, in vitro and in vivo verification. Sci Rep. 2017;7:5567. doi: 10.1038/s41598-017-05856-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cui C, Liu Y, Gerloff D, Rohde C, Pauli C, Köhn M, et al. NOP10 predicts lung cancer prognosis and its associated small nucleolar RNAs drive proliferation and migration. Oncogene. 2021;40:909–21. doi: 10.1038/s41388-020-01570-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mourksi NE, Morin C, Fenouil T, Diaz JJ, Marcel V. snoRNAs offer novel insight and promising perspectives for lung cancer understanding and management. Cells. 2020;9:541. doi: 10.3390/cells9030541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng D, Zhang J, Ni J, Luo J, Wang J, Tang L, et al. Small nucleolar RNA 78 promotes the tumorigenesis in non-small cell lung cancer. J Exp Clin Cancer Res. 2015;34:49. doi: 10.1186/s13046-015-0170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang G, Zeng Z, Sun W, Li S, You C, Tang F, et al. Small nucleolar RNA 71A promotes lung cancer cell proliferation, migration and invasion via MAPK/ERK pathway. J Cancer. 2019;10:2261–75. doi: 10.7150/jca.31077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467–80. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 60.Nishihara R, Wu K, Lochhead P, Morikawa T, Liao X, Qian ZR, et al. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N. Engl J Med. 2013;369:1095–105. doi: 10.1056/NEJMoa1301969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Y, Zhao C, Wang G, Chen J, Ju S, Huang J, et al. SNORD1C maintains stemness and 5-FU resistance by activation of Wnt signaling pathway in colorectal cancer. Cell Death Disco. 2022;8:200. doi: 10.1038/s41420-022-00996-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoshida K, Toden S, Weng W, Shigeyasu K, Miyoshi J, Turner J, et al. SNORA21 - an oncogenic small nucleolar RNA, with a prognostic biomarker potential in human colorectal cancer. EBioMedicine. 2017;22:68–77. doi: 10.1016/j.ebiom.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fang X, Yang D, Luo H, Wu S, Dong W, Xiao J, et al. SNORD126 promotes HCC and CRC cell growth by activating the PI3K-AKT pathway through FGFR2. J Mol Cell Biol. 2017;9:243–55. doi: 10.1093/jmcb/mjx043. [DOI] [PubMed] [Google Scholar]
- 64.Wu H, Qin W, Lu S, Wang X, Zhang J, Sun T, et al. Long noncoding RNA ZFAS1 promoting small nucleolar RNA-mediated 2′-O-methylation via NOP58 recruitment in colorectal cancer. Mol Cancer. 2020;19:95. doi: 10.1186/s12943-020-01201-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–64. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 66.Ji S, Zhang B, Liu J, Qin Y, Liang C, Shi S, et al. ALDOA functions as an oncogene in the highly metastatic pancreatic cancer. Cancer Lett. 2016;374:127–35. doi: 10.1016/j.canlet.2016.01.054. [DOI] [PubMed] [Google Scholar]
- 67.Zhang C, Zhao LM, Wu H, Tian G, Dai SL, Zhao RY, et al. C/D-Box Snord105b promotes tumorigenesis in gastric cancer via ALDOA/C-Myc pathway. Cell Physiol Biochem. 2018;45:2471–82. doi: 10.1159/000488265. [DOI] [PubMed] [Google Scholar]
- 68.Liu CX, Qiao XJ, Xing ZW, Hou MX. The SNORA21 expression is upregulated and acts as a novel independent indicator in human gastric cancer prognosis. Eur Rev Med Pharm Sci. 2018;22:5519–24. doi: 10.26355/eurrev_201809_15812. [DOI] [PubMed] [Google Scholar]
- 69.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 70.Watkins NJ, Lemm I, Ingelfinger D, Schneider C, Hossbach M, Urlaub H, et al. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol Cell. 2004;16:789–98. doi: 10.1016/j.molcel.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 71.Su H, Xu T, Ganapathy S, Shadfan M, Long M, Huang TH, et al. Elevated snoRNA biogenesis is essential in breast cancer. Oncogene. 2014;33:1348–58. doi: 10.1038/onc.2013.89. [DOI] [PubMed] [Google Scholar]
- 72.Pacilli A, Ceccarelli C, Treré D, Montanaro L. SnoRNA U50 levels are regulated by cell proliferation and rRNA transcription. Int J Mol Sci. 2013;14:14923–35. doi: 10.3390/ijms140714923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dong XY, Guo P, Boyd J, Sun X, Li Q, Zhou W, et al. Implication of snoRNA U50 in human breast cancer. J Genet Genomics. 2009;36:447–54. doi: 10.1016/S1673-8527(08)60134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Su X, Feng C, Wang S, Shi L, Gu Q, Zhang H, et al. The noncoding RNAs SNORD50A and SNORD50B-mediated TRIM21-GMPS interaction promotes the growth of p53 wild-type breast cancers by degrading p53. Cell Death Differ. 2021;28:2450–64. doi: 10.1038/s41418-021-00762-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim DS, Camacho CV, Nagari A, Malladi VS, Challa S, Kraus WL. Activation of PARP-1 by snoRNAs controls ribosome biogenesis and cell growth via the RNA helicase DDX21. Mol Cell. 2019;75:1270–85. doi: 10.1016/j.molcel.2019.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hu T, Lu C, Xia Y, Wu L, Song J, Chen C, et al. Small nucleolar RNA SNORA71A promotes epithelial-mesenchymal transition by maintaining ROCK2 mRNA stability in breast cancer. Mol Oncol. (2022). 10.1002/1878-0261.13186. [DOI] [PMC free article] [PubMed]
- 77.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 78.Department of Medical Administration, National Health and Health Commission of the People’s Republic of China. Guidelines for diagnosis and treatment of primary liver cancer in China (2019 edition) Zhonghua Gan Zang Bing Za Zhi. 2020;28:112–28. doi: 10.3760/cma.j.issn.1007-3418.2020.02.004. [DOI] [PubMed] [Google Scholar]
- 79.Liu CY, Chen KF, Chen PJ. Treatment of liver cancer. Cold Spring Harb Perspect Med. 2015;5:a021535. doi: 10.1101/cshperspect.a021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen W, Zheng R, Zhang S, Zeng H, Xia C, Zuo T, et al. Cancer incidence and mortality in China, 2013. Cancer Lett. 2017;401:63–71. doi: 10.1016/j.canlet.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 81.Xu W, Wu Y, Fang X, Zhang Y, Cai N, Wen J, et al. SnoRD126 promotes the proliferation of hepatocellular carcinoma cells through transcriptional regulation of FGFR2 activation in combination with hnRNPK. Aging(Albany NY) 2021;13:13300–17. doi: 10.18632/aging.203014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang H, Ma P, Liu P, Chen B, Liu Z. Small nucleolar RNA U2_19 promotes hepatocellular carcinoma progression by regulating Wnt/β-catenin signaling. Biochem Biophys Res Commun. 2018;500:351–6. doi: 10.1016/j.bbrc.2018.04.074. [DOI] [PubMed] [Google Scholar]
- 83.Li C, Wu L, Liu P, Li K, Zhang Z, He Y, et al. The C/D box small nucleolar RNA SNORD52 regulated by Upf1 facilitates Hepatocarcinogenesis by stabilizing CDK1. Theranostics. 2020;10:9348–63. doi: 10.7150/thno.47677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liang J, Li G, Liao J, Huang Z, Wen J, Wang Y, et al. Non-coding small nucleolar RNA SNORD17 promotes the progression of hepatocellular carcinoma through a positive feedback loop upon p53 inactivation. Cell Death Differ (2022). 10.1038/s41418-022-00929-w. [DOI] [PMC free article] [PubMed]
- 85.Chen X, Zhang Q, Yang Z, Yu H, Qu Y, Tan R, et al. An SNP reducing SNORD105 and PPAN expression decreases the risk of hepatocellular carcinoma in a Chinese population. J Clin Lab Anal. 2021;35:e24095. doi: 10.1002/jcla.24095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang G, Li J, Yao Y, Liu Y, Xia P, Zhang H, et al. Small nucleolar RNA 42 promotes the growth of hepatocellular carcinoma through the p53 signaling pathway. Cell Death Disco. 2021;7:347. doi: 10.1038/s41420-021-00740-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu G, Yang F, Ding CL, Zhao LJ, Ren H, Zhao P, et al. Small nucleolar RNA 113-1 suppresses tumorigenesis in hepatocellular carcinoma. Mol Cancer. 2014;13:216. doi: 10.1186/1476-4598-13-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 89.Holmes D. Ovarian cancer: beyond resistance. Nature. 2015;527:S217. doi: 10.1038/527S217a. [DOI] [PubMed] [Google Scholar]
- 90.Zhang L, Ma R, Gao M, Zhao Y, Lv X, Zhu W, et al. SNORA72 activates the Notch1/c-Myc pathway to promote stemness transformation of ovarian cancer cells. Front Cell Dev Biol. 2020;8:583087. doi: 10.3389/fcell.2020.583087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu W, Niu J, He M, Zhang L, Lv X, Liu F, et al. SNORD89 promotes stemness phenotype of ovarian cancer cells by regulating Notch1-c-Myc pathway. J Transl Med. 2019;17:259. doi: 10.1186/s12967-019-2005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 93.Valleron W, Laprevotte E, Gautier EF, Quelen C, Demur C, Delabesse E, et al. Specific small nucleolar RNA expression profiles in acute leukemia. Leukemia. 2012;26:2052–60. doi: 10.1038/leu.2012.111. [DOI] [PubMed] [Google Scholar]
- 94.Pauli C, Liu Y, Rohde C, Cui C, Fijalkowska D, Gerloff D, et al. Site-specific methylation of 18S ribosomal RNA by SNORD42A is required for acute myeloid leukemia cell proliferation. Blood. 2020;135:2059–70. doi: 10.1182/blood.2019004121. [DOI] [PubMed] [Google Scholar]
- 95.Yu H, Tian L, Yang L, Liu S, Wang S, Gong J. Knockdown of SNORA47 inhibits the tumorigenesis of NSCLC via mediation of PI3K/Akt signaling pathway. Front Oncol. 2021;11:620213. doi: 10.3389/fonc.2021.620213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Langhendries JL, Nicolas E, Doumont G, Goldman S, Lafontaine DL. The human box C/D snoRNAs U3 and U8 are required for pre-rRNA processing and tumorigenesis. Oncotarget. 2016;7:59519–34. doi: 10.18632/oncotarget.11148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mei YP, Liao JP, Shen J, Yu L, Liu BL, Liu L, et al. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene. 2012;13:2794–804. doi: 10.1038/onc.2011.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Krell J, Frampton AE, Mirnezami R, Harding V, De Giorgio A, Roca Alonso L, et al. Growth arrest-specific transcript 5 associated snoRNA levels are related to p53 expression and DNA damage in colorectal cancer. PLoS ONE. 2014;9:e98561. doi: 10.1371/journal.pone.0098561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Duan S, Luo X, Zeng H, Zhan X, Yuan C. SNORA71B promotes breast cancer cells across blood-brain barrier by inducing epithelial-mesenchymal transition. Breast Cancer. 2020;27:1072–81. doi: 10.1007/s12282-020-01111-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sun Y, Chen E, Li Y, Ye D, Cai Y, Wang Q, et al. H/ACA box small nucleolar RNA 7B acts as an oncogene and a potential prognostic biomarker in breast cancer. Cancer Cell Int. 2019;19:125. doi: 10.1186/s12935-019-0830-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wu L, Zheng J, Chen P, Liu Q, Yuan Y. Small nucleolar RNA ACA11 promotes proliferation, migration and invasion in hepatocellular carcinoma by targeting the PI3K/AKT signaling pathway. Biomed Pharmacother. 2017;90:705–12. doi: 10.1016/j.biopha.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 102.Cao P, Yang A, Wang R, Xia X, Zhai Y, Li Y, et al. Germline duplication of SNORA18L5 increases risk for HBV-related hepatocellular carcinoma by altering localization of ribosomal proteins and decreasing levels of p53. Gastroenterology. 2018;155:542–56. doi: 10.1053/j.gastro.2018.04.020. [DOI] [PubMed] [Google Scholar]
- 103.Li G, He Y, Liu X, Zheng Z, Zhang M, Qin F, et al. Small nucleolar RNA 47 promotes tumorigenesis by regulating EMT markers in hepatocellular carcinoma. Minerva Med. 2017;108:396–404. doi: 10.23736/S0026-4806.17.05132-1. [DOI] [PubMed] [Google Scholar]
- 104.Wu L, Chang L, Wang H, Ma W, Peng Q, Yuan Y. Clinical significance of C/D box small nucleolar RNA U76 as an oncogene and a prognostic biomarker in hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2018;42:82–91. doi: 10.1016/j.clinre.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 105.Liu Z, Pang Y, Jia Y, Qin Q, Wang R, Li W, et al. SNORA23 inhibits HCC tumorigenesis by impairing the 2’-O-ribose methylation level of 28S rRNA. Cancer Biol Med. 2021;19:104–19. doi: 10.20892/j.issn.2095-3941.2020.0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.