Abstract
The HOM3 gene of Saccharomyces cerevisiae codes for aspartate kinase, which plays a crucial role in the regulation of the metabolic flux that leads to threonine biosynthesis. With the aim of obtaining yeast strains able to overproduce threonine in a controlled way, we have placed the HOM3-R2 mutant allele, which causes expression of a feedback-insensitive enzyme, under the control of four distinctive regulatable yeast promoters, namely, PGAL1, PCHA1, PCYC1-HSE2, and PGPH1. The amino acid contents of strains bearing the different constructs were analyzed both under repression and induction conditions. Although some differences in overall threonine production were found, a maximum of around 400 nmol/mg (dry weight) was observed. Other factors, such as excretion to the medium and activity of the catabolic threonine/serine deaminase, also affect threonine accumulation. Thus, improvement of threonine productivity by yeast cells would probably require manipulation of these and other factors.
Threonine is an essential amino acid that is often limiting in animal feed and human food (14). On the other hand, this amino acid is a precursor of a number of commonly used flavoring agents (31). Threonine has been successfully marketed as a feed additive, with a worldwide demand of more than 10,000 tons per year. The industrial production of this amino acid is carried out mainly by fermentation through the use of overproducing bacterial strains, which yield up to 80 g of threonine per liter (for a review, see reference 17).
The yeast Saccharomyces cerevisiae plays an important role in the food industry in the production of bread, beer, and wine, etc., being considered a GRAS (generally recognized as safe) microorganism. This fact together with its high nutritional value (high protein and vitamin contents) has favored the increasing consumption of yeasts as natural supplements or as flavor enhancers in conventional ready-to-serve food. In this context, the obtaining of yeast strains able to overproduce threonine might be of great interest to the food industry, since this amino acid-enriched biomass could be directly added to food.
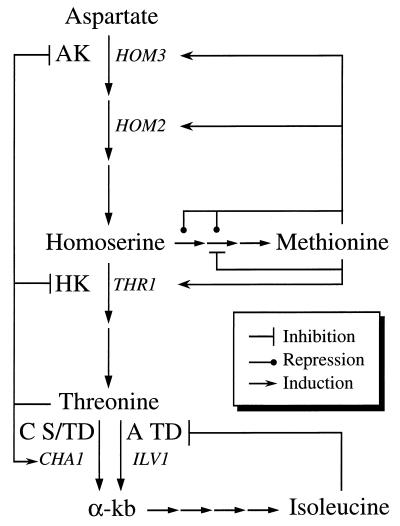
Threonine-overproducing mutants of S. cerevisiae that accumulate up to 40 times more threonine than the wild-type strain have been previously isolated (1, 5, 20, 24, 28). In all cases studied, the overproduction is linked to a mutant HOM3 allele that causes expression of a feedback-insensitive aspartate kinase, the key enzyme in the regulation of the threonine biosynthetic pathway (Fig. 1). Three of these alleles have been cloned in our laboratory, and the analysis of their DNA sequences shows that in each case a different point mutation is responsible for the feedback-insensitive phenotype (references 1 and 21 and unpublished results). The amplification of two of these mutant alleles, namely, HOM3-R2 and HOM3-R6, resulted in a 2.5-fold increase in threonine accumulation with respect to that of the mutant strain (7). However, this overproduction seems to have a deleterious effect on the growth of the strain, probably due to interference with the general cell metabolism.
FIG. 1.
Biosynthesis of aspartate-derived amino acids and its regulation. AK, aspartate kinase; HK, homoserine kinase; C S/TD, catabolic serine/threonine deaminase; A TD, anabolic threonine deaminase; α-kb, α-ketobutyrate.
The aim of this work has been the construction of yeast strains able to overproduce threonine in a controlled way so that threonine accumulation neither impairs other physiological processes nor interferes with the fermentative growth of the industrial strains to which the results of this study can be applied. With that objective, we have placed the HOM3-R2 mutant allele under the control of four distinctive regulatable yeast promoters: two inducible by nutritional factors (presence of either galactose or serine and threonine in the medium) and two others induced by distinct physiological conditions (high temperature or stationary phase). We have analyzed the amino acid contents of strains bearing the different constructs under both repression and induction conditions.
MATERIALS AND METHODS
Strains, plasmids, media, and growth conditions.
S. cerevisiae strains and plasmids used in this work are listed in Table 1. Escherichia coli DH5α (10) was used as the host for plasmid maintenance and recovery; BMH71-18 mutS (16) was used to recover the mutant plasmids after site-directed mutagenesis.
TABLE 1.
Yeast strains and plasmids
| Strain or plasmid | Genotype or relevant features | Source or reference |
|---|---|---|
| S. cerevisiae | ||
| XMP10-7B | MATa ura3 ade1 hom3 | This work |
| GRF167 | MATα ura3-167 his3-Δ200 | 2 |
| SG15 | MATα ura3-52 trp1 gal2 | S. Holmberg |
| SG183 | MATα ura3-52 trp1 cha1 gal2 | S. Holmberg |
| HT2 | MATα ura3-52 trp1 hom3::TRP1 gal2 | I. Velasco |
| Plasmids | ||
| pYH3 | URA3, 2μm, Aps Tcr, HOM3 | 7 |
| pMP1 | URA3, Apr, HOM3 | This work |
| pMR3-R21 | URA3, Apr, HOM3-R2 | 21 |
| pCGS42 | URA3, 2μm, Apr Tcr | F. del Rey |
| YCp50 | URA3, ARS, CEN, Apr Tcr | 25 |
| pYES2 | URA3, 2μm, Apr, PGAL1 | J. M. Pardo |
| pTK120 | URA3, ARS, CEN, Apr, PCHA1 | S. Holmberg |
| YEP/HSE2 | 2μm, Apr PCYC1-HSE2 | A. Rodríguez |
| YEp356, GPH-lacZ | URA3, 2μm, Apr, PGPH1 | J. François |
| pEGAL-H3 or -R2a | pYES2 containing a 1.9-kb HindIII fragment (HOM3 or HOM3-R2) from pMP1 or pMR3-R21, respectively | This work |
| pCGAL-H3 or -R2a | YCp50 containing a 2.4-kb SpeI fragment (PGAL1::HOM3 or HOM3-R2) from pEGAL-H3 or -R2, respectively | This work |
| pCHA-H3 or -R2a | YCp50 containing a 1.9-kb BamHI-EcoRI fragment (HOM3 or HOM3-R2) from pEHSE-H3 or -R2, respectively, and a 0.7-kb BamHI fragment (PCHA1) from pTK120 | This work |
| pEHSE-H3 or -R2a | pCGS42 containing a 0.8-kb SalI-HindIII fragment (PCYC1-HSE2) from YEP/HSE2+Hb and a 1.9-kb HindIII fragment (HOM3 or HOM3-R2) from pMP1 or pMR3-R21, respectively | This work |
| pCHSE-H3 or -R2ac | YCp50 containing a 1.9-kb BamHI-EcoRI fragment (HOM3 or HOM3-R2) from pEHSE-H3 or -R2, respectively, and a 0.8-kb SalI-BglII fragment (PCYC1-HSE2) from YEP/HSE2 | This work |
| pGPH-H3 or -R2a | YCp50 containing a 0.4-kb EcoRI-HindIII fragment (PGPH1) from YEp356, GPH-lacZ, and a 1.9-kb HindIII fragment (HOM3 or HOM3-R2) from pMP1-Hd or pMR3-R21-Hd, respectively | This work |
In these plasmids, the expression of the HOM3 alleles is under the control of the respective inducible promoter; the HindIII site used for cloning is 25 bp upstream from the ATG start codon of the HOM3 gene.
This plasmid was constructed by inserting a linker HindIII in the BamHI site of YEP/HSE2.
In these plasmids an additional ATG start codon from a lac gene present in the original promoter was deleted.
These plasmids are identical to pMP1 and pMR3-R21 except that the internal HindIII site in the HOM3 or HOM3-R2 open reading frame was removed by site-directed mutagenesis, without altering the amino acid sequence.
Standard media for S. cerevisiae (YPD and SD) have been previously described (29). When indicated, SD medium was buffered with NaOH and succinate (pH 5.8) (23). Appropriate supplements were added at standard concentrations (29), except for tryptophan, which was used at a fourfold-higher concentration when indicated. SGal medium is similar to SD but contains 2% galactose as the sole carbon source instead of glucose. Unless otherwise indicated, yeast cells were grown at 30°C with shaking at 200 rpm. E. coli was grown in Luria-Bertani medium (27) at 37°C. When appropriate, ampicillin (100 μg/ml) was added.
DNA techniques.
Standard protocols (27) were used for construction, purification, and analysis of plasmid DNA. Enzymes were purchased from Boehringer GmbH (Mannheim, Germany) and used according to the manufacturer’s instructions. Yeast cells were transformed after lithium acetate treatment as previously described (9).
RNA isolation and analysis.
Yeast RNA preparation was carried out by the hot-phenol extraction method as previously described (15). Northern analysis was performed after size fractionation of 10 μg of RNA and transfer onto nylon filters (Micron Separations Inc., Westborough, Mass.) by conventional procedures (27). A 0.95-kb StyI internal fragment of the HOM3 gene was radiolabeled with [α-32P]dCTP by random oligonucleotide-primed synthesis (8) and used as a probe. Radioactive signals were quantified with a betascope (Instantimager; Packard, Meriden, Conn.). Total rRNA was quantified from methylene blue-stained filters by using the program Bio Image Intelligent Quantifier and used as a loading control.
Amino acid determination.
Intracellular and extracellular amino acids were analyzed as ortho-phthaldialdehyde derivatives by reverse-phase high-pressure liquid chromatography as previously described (7).
Site-directed mutagenesis.
In vitro oligonucleotide-directed mutagenesis was carried out by using the Altered Sites system (Promega Corporation, Falkenberg, Sweden) according to the manufacturer’s instructions. The mutagenic oligonucleotide ΔHIND (5′ CCAGAAGAAGCCTCTGAATTAACA 3′) was used to alter the HindIII site present in the HOM3 open reading frame. The HOM3 gene-bearing phagemid pYH3 was used as the template. The system employs a phagemid vector and is based on the use of a second mutagenic oligonucleotide (Amp Repair) which restores ampicillin resistance to the newly synthesized DNA strand during the mutagenesis reaction. A repair-minus strain (BMH71-18 mutS) is transformed, and ampicillin resistance is selected. A second round of transformation in DH5α ensures proper segregation of mutant and wild-type plasmids.
RESULTS
Expression of HOM3-R2 from the GAL1 promoter.
The GAL1 gene encodes galactokinase, an enzyme involved in galactose fermentation. The regulation of the GAL1 promoter is well known, and its expression under inducing conditions, i.e., the presence of galactose and absence of glucose, is up to 1,000-fold higher than that under repressive conditions (12). Thus, it is an efficient but physiologically complex system that could serve to evaluate the usefulness of other similarly controlled promoters.
Strain XMP10-7B was transformed with the episomic plasmid pEGAL-R2 and with pEGAL-H3 as a control (Table 1). Plasmid stability in SGal- and SD-grown cultures was 80 and 100%, respectively. The intracellular threonine pool and the threonine excreted to the medium were monitored in the different cultures. As expected, the HOM3-R2 allele conferred threonine and homoserine overproduction (Table 2). In the case of the wild-type HOM3 allele, the SD- and SGal-grown cultures accumulated less than 35 and 85 nmol of threonine/mg (dry weight), respectively. Consistent with previous results, the threonine-overproducing strains reached lower final cell densities than the strains bearing the wild-type allele (optical density at 600 nm [OD660] = 3.7 versus 4.3 in SD and 1.7 versus 2.2 in SGal after 38 h of incubation). The maximum growth rates of the strains in each medium were nevertheless similar (0.26 ± 0.03 h−1 in SD and 0.18 ± 0.01 h−1 in SGal).
TABLE 2.
Amino acid production under repression (SD) and induction (SGal) conditions by XMP10-7B transformed with pEGAL-R2
| tb (h) | Intracellular (extracellular) concn (nmol/mg [dry wt])a
|
|||
|---|---|---|---|---|
| Threonine
|
Homoserine
|
|||
| SD | SGal | SD | SGal | |
| 14 | 75 | 154 (95) | 8 | 33 (30) |
| 20 | 50 | 166 (68) | 8 | 84 (48) |
| 24 | 58 (11) | 206 (70) | 14 (2) | 127 (60) |
| 38 | 86 (5) | 355 (72) | 18 (3) | 66 (44) |
| 46 | 115 (4) | 290 (60) | 24 (2) | 26 (35) |
| 64 | 83 (27) | 186 (128) | 17 (12) | 11 (62) |
Results are the mean values from at least two independent experiments. Coefficients of variation were less than 20%.
Cultivation time.
The results presented so far indicate that under repressive conditions (SD), the amino acid accumulation by cells bearing PGAL1::HOM3 constructions is relatively high. Moreover, under induction conditions (SGal), the cultures accumulate only four times more threonine than the repressed cultures. In order to achieve a better regulation of threonine accumulation, we considered it convenient to reduce the copy number of the construction to 1 to 2 per cell.
For that purpose, centromeric plasmids containing PGAL1::HOM3 fusions (pCGAL-H3 and pCGAL-R2 [Table 1]) were constructed. Strain XMP10-7B transformed with pCGAL plasmids was unable to grow in the absence of homoserine even on carbon sources different from glucose if galactose was not present in the medium. In order to avoid the addition of this amino acid to the culture medium, which would interfere with the quantification of this and other related amino acids, we decided to use the Hom3+ strain GRF167, which is genetically related to XMP10-7B. To avoid glucose repression, medium containing raffinose as the sole carbon source (SR) was used in further experiments. In this medium, both transformants had a maximum growth rate of 0.13 h−1 and reached an OD660 of up to 3.5. Table 3 summarizes the results obtained from the estimation of the amino acid production by GRF167 transformed with pCGAL-R2 under noninducing conditions (SR) and after induction at mid-log growth phase (OD660 ∼ 1 to 1.5) by the addition of galactose at a final concentration of 0.2% (SR plus Gal). In this case, both a very low basal threonine content and a high accumulation after induction were obtained. When a similar experiment was carried out with pCGAL-H3 as a control, the maximum amounts of threonine and homoserine accumulated by induced cultures were 16 and 2 nmol/mg (dry weight), respectively.
TABLE 3.
Amino acid production in uninduced (SR) and induced (SR plus Gal) cultures of GRF167 transformed with pCGAL-R2
| tb (h) | Intracellular (extracellular) concn (nmol/mg [dry wt])a
|
|||
|---|---|---|---|---|
| Threonine
|
Homoserine
|
|||
| SR | SR plus Gal | SR | SR plus Gal | |
| 17 | 2 | NDc | ||
| 21 | 1 | 194 (26) | ND | 28 (10) |
| 25 | 5 | 164 (62) | 1 | 21 (26) |
| 41 | 6 | 138 (129) | 1 | 34 (73) |
| 65 | 11 | 199 (199) | 1 | 14 (109) |
Results are the mean values from at least two independent experiments. Coefficients of variation were less than 20%.
Cultivation time.
ND, not detectable.
Expression of HOM3-R2 from the CHA1 promoter.
The CHA1 gene encodes the catabolic serine/threonine deaminase, one of the two enzymes catalyzing the first step in threonine degradation (Fig. 1) and also involved in serine catabolism (24a). Its transcription is induced by serine and/or threonine (23). HOM3-R2 expression from this promoter can be triggered by the addition of serine to the medium, and once a certain amount of threonine has been accumulated, threonine itself would induce its own overproduction.
The transcriptional fusions with this promoter were constructed in a centromeric vector (pCHA plasmids [Table 1]). As in the previous case, to avoid the addition of homoserine to the growth medium, a Hom3+ strain (SG15) was used as the recipient of the plasmid. In order to assess the influence of the CHA1 activity, which is also inducible by serine, on threonine accumulation, a Cha1− strain (SG183) isogenic to SG15 was also used. Both strains were transformed with the pCHA plasmids; the transformants were grown in minimal medium (SD plus Trp), and threonine production was monitored. The results (Table 4) show that the intracellular threonine concentrations in 14-h cultures (OD660 ∼ 0.7) of the Cha1+ and Cha1− strains transformed with pCHA-R2 were similar. However, in 38-h cultures (OD660 ∼ 1) the threonine concentration in the Cha1− strain was twofold higher than that in the Cha1+ strain, probably because of the absence of threonine deaminase activity in the former.
TABLE 4.
Amino acid production under repression and induction conditions by SG15 and SG183 transformed with pCHA-R2
| ta (h) and strain | Intracellular (extracellular) threonine concn (nmol/mg [dry wt]) with the following concn of serine (g/liter) added to the medium:
|
||||
|---|---|---|---|---|---|
| 0 | 1 | 0.01b | 0.1b | 1b | |
| 14 | |||||
| SG15 | 37 (NEc) | 100 (80) | |||
| SG183 | 49 (NDd) | 35 (30) | |||
| 15 | |||||
| SG15 | 41 (NE) | 99 (78) | 40 (NE) | 37 (NE) | 40 (NE) |
| SG183 | 48 (3) | 38 (31) | 48 (4) | 47 (4) | 47 (4) |
| 17 | |||||
| SG15 | 40 (NE) | 118 (79) | 34 (NE) | 35 (NE) | 33 (NE) |
| SG183 | 60 (10) | 49 (34) | 61 (7) | 65 (11) | 68 (8) |
| 38 | |||||
| SG15 | 58 (NE) | 112 (77) | 78 (NE) | 57 (NE) | 70 (NE) |
| SG183 | 99 (27) | 67 (40) | 161 (44) | 194 (55) | 226 (67) |
Cultivation time.
Serine was added to 14-h cultures.
NE, not estimated.
ND, not detectable.
Surprisingly, when the transformants were grown in the presence of 1 g of serine per liter, the Cha1+ strain initially accumulated three times more threonine than the Cha1− strain, and this amount was constant throughout the experiment (Table 4). This effect might be related to the fact that under these conditions, the Cha1− strain also accumulates a large amount of serine (∼300 nmol/mg [dry weight], versus 60 in the Cha1+ strain). A possible explanation might be that serine inhibits homoserine dehydrogenase, as occurs in E. coli (9a), although with a high inhibition constant. Another possibility is that serine might have an effect of trans stimulation on an eventual amino acid carrier that mediates serine uptake down a concentration gradient, and as a consequence, threonine export could take place. The behavior of the Cha1− strain was similar to that without serine, except that now threonine was accumulated and excreted to the medium to the same extent. The same experiments carried out with the wild-type HOM3 allele under control of the CHA1 promoter revealed that serine has no effect on threonine accumulation, which was at most 10 nmol/mg (dry weight) (data not shown).
In another set of experiments, different concentrations of serine were added to 14-h cultures and threonine accumulation was periodically monitored. As shown also in Table 4, in the pCHA-R2-bearing Cha1+ strain, a slight increase in threonine accumulation was observed only at 24 h after serine addition (t = 38 h) and irrespective of the amount of serine added. However, in the Cha1− strain the increase was much higher and was dependent on the concentration of serine, although not proportionally.
An improvement in the culture medium was achieved by buffering the SD medium (pH 5.8) and adding four times more tryptophan than usual. Under these conditions, threonine production at the early states of growth was similar to that described above (data not shown), but as the duration of growth increased more threonine was accumulated, reaching in the case of the Cha1− strain, to which 0.1 g of serine per liter had been added, 320 nmol of threonine/mg (dry weight) at an OD660 of ∼5.
Expression of HOM3-R2 from the CYC1-HSE2 promoter.
CYC1-HSE2 is a hybrid promoter inducible by heat shock (30). It is based on the promoter of the CYC1 gene, which encodes iso-1-cytochrome c, but the upstream activation sequence required for heme and glucose regulation has been replaced by the HSE2 (for heat shock element) region of SSA1, a gene from the Hsp70 family. Thus, in this construct, HOM3-R2 expression could be induced by shifting the culture incubation temperature from 22 to 37°C.
Transcriptional fusions of the HOM3 and HOM3-R2 alleles with the CYC1-HSE2 promoter were constructed and placed in episomic plasmids, generating pEHSE-H3 and pEHSE-R2, respectively (Table 1). Strain XMP10-7B transformed with either of these plasmids was grown in selective minimal medium at 22°C to an OD660 of ∼0.7, and the threonine content was measured. In both cases, the amount of accumulated threonine was quite high (∼25 and ∼75 nmol/mg [dry weight] in the cases of the wild-type and the mutant alleles, respectively), probably due to the high copy number of the plasmid and despite the fact that only 75% of the cells of both cultures retained the plasmids. Moreover, similar amounts of threonine were found when the incubation temperature was shifted to 37°C (data not shown).
On the basis of these results, these constructions were placed in the centromeric vector Ycp50, resulting in plasmids pCHSE-H3 and -R2 (Table 1). In order to quantify the amino acid production, the XMP10-7B transformants with these plasmids were grown in selective minimal medium at 22°C. When the cultures reached an OD660 of ∼1, a portion of each was shifted to 37°C, and threonine and homoserine were periodically measured. In the case of the wild-type allele, the heat shock had no appreciable effect on threonine and homoserine accumulations, which were at most 10 and 1 nmol/mg (dry weight), respectively. Conversely, the results with pCHSE-R2 (Table 5) show that the longer the incubation was maintained at this temperature, the more amino acid was produced. However, a good proportion of the produced amino acid was excreted to the medium.
TABLE 5.
Amino acid production in uninduced (22°C) and induced (transferred to 37°C) cultures of XMP10-7B transformed with pCHSE-R2
| tb (h) | Intracellular (extracellular) concn (nmol/mg [dry wt])a
|
|||
|---|---|---|---|---|
| Threonine
|
Homoserine
|
|||
| 22°C | 37°C | 22°C | 37°C | |
| 0 | 28 ± 15 | 4 ± 2 | ||
| 1 | 40 ± 16 | 159* (5 ± 2) | 7 ± 2 | 43* (3 ± 1) |
| 3 | 63 ± 22 | 254 ± 15 (27 ± 2) | 22 ± 10 | 93 ± 5 (16 ± 3) |
| 24 | 106 ± 9 (16 ± 1) | 151 ± 7 (264 ± 7) | 25 ± 4 (9 ± 2) | 23 ± 2 (91 ± 29) |
Results are the mean values and standard errors from at least two independent experiments, except for those marked with an asterisk, which are the results of a single experiment.
Time after heat shock.
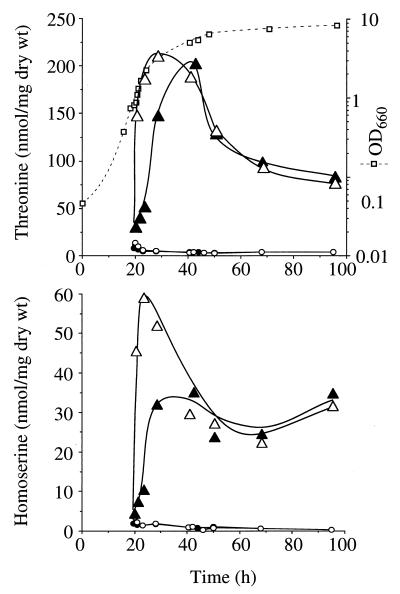
To reduce the effect of excretion, which had been previously reported to be favored at 37°C (24), a similar experiment in which the heat shock lasted for only 30 min was carried out. For this and the subsequent experiments, strain HT2, which was derived from strain SG15 by disruption of the HOM3 gene, was used. Cultures of HT2 transformants in buffered SD medium reached a maximum OD660 of ∼8. When the cultures grown at 22°C reached an OD660 of ∼1, they were split into two: one portion was subjected to a 30-min heat shock, and the other was kept at 22°C; amino acids in both portions were periodically measured. The threonine and homoserine accumulations are shown in Fig. 2; the amounts of these amino acids excreted to the medium were negligible. The results show that a progressive increase in the threonine and homoserine contents in low-temperature-grown cultures occurred, reaching a maximum at the end of the exponential growth phase. No significant threonine production was attained when the pCHSE-H3 plasmid was used. Another heat shock was applied to a sample of the cultures grown at 22°C when they reached an OD660 of ∼5. This resulted in slightly increased amino acid accumulation for the pCHSE-R2-bearing cells (data not shown).
FIG. 2.
Threonine and homoserine accumulation in strain HT2 transformed with plasmid pCHSE-H3 (circles) or pCHSE-R2 (triangles), grown at 22°C (closed symbols) and after a 30-min heat shock at an OD660 of ∼1 (open symbols). Growth of HT2(pCHSE-R2) is also shown (squares). The results are means from two independent experiments, and the coefficients of variation were lower than 20%. wt, weight.
In order to prove that these effects were indeed due to transcriptional induction of the HOM3-R2 allele from the CYC1-HSE2 promoter, a Northern analysis with samples taken at different times after the shift to 37°C and from nonshocked cultures was carried out. As shown in Table 6, a maximum HOM3-R2 transcript level was reached shortly after the temperature shift, followed by a progressive decrease during the next 90 min. In fact, an additional experiment (data not shown) revealed that a 5-min heat shock resulted in threonine production similar to that described above. Similar transcription levels were obtained in the case of the wild-type HOM3 allele fused to the CYC1-HSE2 promoter (Table 6). On the other hand, consistent with the threonine production data, a fourfold increase in the HOM3-R2 transcript level was observed at the end of the exponential growth phase. This could indicate that induction of the construction takes place at this growth state. An explanation for the temporary lack of correlation between the message level and threonine content could be that while mRNAs are rapidly degraded after the heat shock (Table 6), the resulting enzyme and also the threonine pool are more stable. In fact, a lack of correspondence between enzyme activity and transcript levels in the threonine/methionine pathway has also been observed by others (22). Posttranscriptional regulation has been postulated to explain this phenomenon.
TABLE 6.
Quantification of the HOM3 transcript level in HT2 transformed with the pCHSE plasmids
| Time after heat shock | Heat shock | Transcript levela
|
|
|---|---|---|---|
| HOM3-R2 | HOM3 | ||
| 0 | − | NEb | 1 |
| 5 min | + | 30.5 | 24.7 |
| 15 min | + | 14.4 | 17.1 |
| 45 min | + | 0.7 | 0.6 |
| 90 min | + | 0.2 | 0.1 |
| − | 1 | 1.4 | |
| 24 h | + | 3.8 | 2.9 |
| − | 4.9 | 4.5 | |
The values were normalized with respect to the total rRNA.
NE, not estimated because of being below the reliable limit. Instead, the value for the uninduced culture at 90 min was considered the reference value.
Expression of HOM3-R2 from the GPH1 promoter.
GPH1 encodes glycogen phosphorylase, which is involved in glycogen utilization. GPH1 transcription is induced at the late exponential growth phase, almost simultaneously with the onset of intracellular glycogen accumulation but before glucose depletion from the medium (11). The main advantage of the use of this promoter is that no external inducer is needed to trigger its expression.
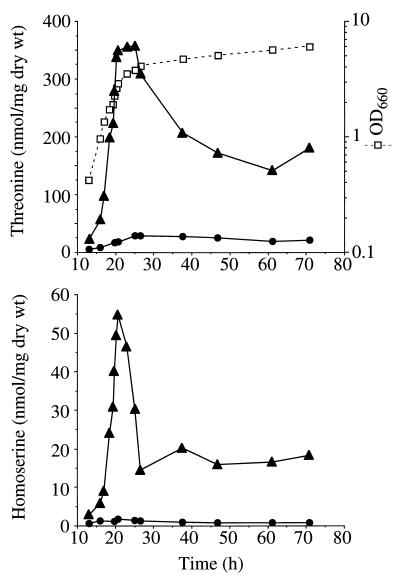
Strain HT2 was transformed with the pGPH plasmids (Table 1). The transformants were grown in SD plus Trp (at a concentration fourfold higher than usual) at 30°C, and threonine and homoserine production was monitored throughout the growth curve until the late stationary phase. The results shown in Fig. 3 reveal that cells with plasmid pGPH-R2 accumulated a considerable amount of threonine and homoserine at the end of the exponential growth phase. In 140-h cultures these amino acid concentrations were the same as those at 70 h (data not shown). The external concentration of threonine was negligible.
FIG. 3.
Threonine and homoserine accumulation in strain HT2 transformed with plasmids pGPH-H3 (circles) and pGPH-R2 (triangles). Growth of HT2(pGPH-R2) is also shown (squares). The results are means from two independent experiments, and the coefficients of variation were lower than 20%. wt, weight.
DISCUSSION
From an energetic viewpoint, accumulation of a metabolite competes with energy production for cell growth and maintenance. Thus, in order to overproduce a metabolite such as threonine, it is convenient to separate the trophophase or growth phase from the production phase. This separation turns out to be necessary in the case of microorganisms coming from fermentative industries that are intended to be used afterwards as enriched biomass.
In this work we show that the control of expression from four different inducible promoters of the yeast HOM3-R2 allele, which confers the ability to overproduce threonine, allows circumvention of the overlap of the two phases. The use of each construction has some advantages and inconveniences derived from their characteristics, which makes the choice of a single promoter difficult. For instance, the initial assumption that the presence of a high copy number of a certain HOM3-R2 construct would favor threonine overproduction is not necessarily true. The results obtained with the episomic plasmids pEGAL-R2 and pEHSE-R2 suggest that a limitation of negative effector molecules may make the basal level of threonine accumulation higher than desirable under repression conditions. Moreover, under induction conditions, the expression might not be optimal due to a limitation of the number of activator molecules. In fact, it has been reported that the level of Gal4p (transcriptional activator of GAL genes) in the cell is very low (3), being insufficient even for the maximum induction of the normal complement of GAL promoters in a wild-type strain (13). To overcome this problem, we decided to place the different constructs in single-copy plasmids, which at the same time would represent more accurately the hypothetical situation in an industrial yeast strain in which the construct would be integrated into the genome, thus increasing the stability of the construction.
The induction of the fusion CYC1-HSE2::HOM3/-R2 observed during the late exponential growth phase is an interesting feature of this construct. In S. cerevisiae there are a number of genes whose expression is induced under stress conditions (18). The stress response is related to the presence of an STRE element in the corresponding promoter (19). In the SSA1 gene there are two STRE elements, although this gene is not induced at the entry into the stationary growth phase (4). One of them is present in the HSE2 region and hence in the CYC1-HSE2 promoter. This together with the fact that cytochrome c-related genes are coordinately induced by glucose exhaustion (6) might be responsible for the observed induction effect. The GPH1 promoter induction observed at the entry into stationary growth phase (11) is consistent with the presence of STRE elements. In this case, the induction has an effect on threonine accumulation, which is congruent with the GPH1 transcription pattern previously described.
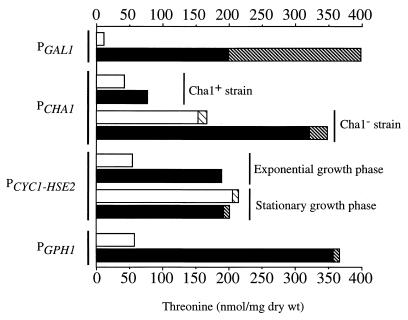
A summary of the maximum threonine production achieved with each of the constructions in centromeric plasmids, under both induction and repression conditions, is shown in Fig. 4. Besides differences in overall production, two general factors need to be considered if maximum accumulation is to be achieved. First, under certain circumstances a large part of the produced threonine is excreted to the culture medium, especially at advanced growth phases. Threonine excretion by a yeast strain seems to be highly dependent on its genetic background (5), although we still do not have a complete picture of the genetic factors involved in that phenomenon. Further investigations in this field are necessary in order for this factor to be controlled. Second, the Cha1p activity lowers threonine accumulation, although its effect is observed mainly at the stationary growth phase. Thus, inactivation of the CHA1 gene in the producer strain should also improve threonine accumulation.
FIG. 4.
Maximum threonine production by strains carrying one copy of the mutant HOM3-R2 allele under the control of different regulatable promoters. The production is expressed as the sum of threonine accumulated in the cells under either repression (open bars) or induction (shaded bars) conditions and that excreted to the medium (hatched bars).
The results presented in this and previous papers (7, 24) show that manipulation of the genes involved in threonine biosynthesis, by both mutation and amplification, leads to a maximum production that in all cases is around 400 nmol/mg (dry weight). Thus, with these kinds of operations, this seems to be the upper limit, which may be influenced by the efflux system for this amino acid. In fact, it has been shown that threonine overproduction also leads to increased formation of homoserine, an intermediate metabolite, and to a lesser extent that of isoleucine and glycine, which are derived from threonine (data not shown). These results are in agreement with those obtained for bacteria (25). Therefore, further improvement of the production should probably be achieved by engineering other factors, such as the input of metabolites into the route or the degradation and excretion of the amino acid. We are presently working on these topics.
ACKNOWLEDGMENTS
This work was supported by the Spanish CICYT (grants BIO93-0423 and ALI96-0938) and by the Junta de Andalucía (grant CVI-0153).
We very much appreciate the collaboration of J. E. Martín-Oar and L. Navas (Grupo Cruzcampo S.A., Seville, Spain) and P. Niederberger and D. Jäger (Nestlé Research Center, Lausanne, Switzerland). We thank S. Holmberg, A. Aguilera, F. del Rey, J. M. Pardo, A. Rodríguez, and J. François for kindly providing the strains and plasmids mentioned in the text; M. Arévalo for many helpful discussions; I. Velasco for constructing strain HT2; and D. Jäger for critical reading of the manuscript.
REFERENCES
- 1.Arévalo M. Aislamiento y caracterización de mutaciones del gen HOM3 que alteran la regulación de la síntesis de treonina y metionina en Saccharomyces cerevisiae. Ph.D. thesis. Seville, Spain: University of Seville; 1996. [Google Scholar]
- 2.Boeke J D, Garfinkel C A, Styles C A, Fink G R. Ty elements transpose through an RNA intermediate. Cell. 1985;40:491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- 3.Bram R J, Kornberg R D. Specific protein binding to far upstream activating sequences in polymerase II promoters. Proc Natl Acad Sci USA. 1985;82:43–47. doi: 10.1073/pnas.82.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craig E A. The heat-shock response of Saccharomyces cerevisiae. In: Jones E W, Pringle J R, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 501–537. [Google Scholar]
- 5.Delgado M A, Guerrero J, Conde J. Genetic and biochemical study of threonine-overproducing mutants of Saccharomyces cerevisiae. Mol Cell Biol. 1982;2:731–736. doi: 10.1128/mcb.2.7.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 7.Farfán M J, Martín-Rendón E, Calderón I L. Effect of gene amplification on threonine production by yeast. Biotechnol Bioeng. 1996;49:667–674. doi: 10.1002/(SICI)1097-0290(19960320)49:6<667::AID-BIT8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 8.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 9.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:142. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Hama M, Sumita Y, Kakutani Y, Tsuda M, Tsuchiya T. Target of serine inhibition in Escherichia coli. Biochem Biophys Res Commun. 1990;168:1211–1216. doi: 10.1016/0006-291x(90)91157-n. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan D. Techniques for transformation of E. coli. In: Glover D M, editor. DNA cloning. A practical approach. Oxford, United Kingdom: IRL Press; 1985. pp. 109–114. [Google Scholar]
- 11.Hwang P K, Tugendreich S, Fletterick R J. Molecular analysis of GPH1, the gene encoding glycogen phosphorylase in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:1659–1666. doi: 10.1128/mcb.9.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnston M, Flick J S, Pexton T. Multiple mechanisms provide rapid and stringent glucose repression of GAL gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3834–3841. doi: 10.1128/mcb.14.6.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnston S A, Hopper J E. Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Proc Natl Acad Sci USA. 1982;79:6971–6975. doi: 10.1073/pnas.79.22.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleemann A, Leuchtenberger W, Hoppe B, Tanner H. Amino acids. In: Bartholomé E, Biekert E, Hellmann H, editors. Ullmann’s encyclopedia of industrial chemistry. A2. Weinheim, Germany: VCH; 1985. pp. 57–97. [Google Scholar]
- 15.Köhrer K, Domdey H. Preparation of high molecular weight RNA. Methods Enzymol. 1991;194:398–405. doi: 10.1016/0076-6879(91)94030-g. [DOI] [PubMed] [Google Scholar]
- 16.Kramer B, Kramer W, Fritz H J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984;38:879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- 17.Leuchtenberger W. Amino acids. Technical production and use. In: Roehr M, editor. Biotechnology. Products of primary metabolism. Weinheim, Germany: VCH; 1996. pp. 465–502. [Google Scholar]
- 18.Mager W H, Moradas Ferreira P. Stress response of yeast. Biochem J. 1993;290:1–13. doi: 10.1042/bj2900001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchler G, Schüller C, Adam G, Ruis H. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 1993;12:1997–2003. doi: 10.1002/j.1460-2075.1993.tb05849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martínez-Force E, Benítez T. Selection of amino-acid overproducer yeast mutants. Curr Genet. 1992;21:191–196. [Google Scholar]
- 21.Martín-Rendón E, Farfán M J, Ramos C, Calderón I L. Isolation of a mutant allele that deregulates the threonine biosynthesis in Saccharomyces cerevisiae. Curr Genet. 1993;24:465–471. doi: 10.1007/BF00351707. [DOI] [PubMed] [Google Scholar]
- 22.Mountain H A, Byström A S, Larsen J T, Korch C. Four major transcriptional responses in the methionine/threonine biosynthetic pathway of Saccharomyces cerevisiae. Yeast. 1991;7:781–803. doi: 10.1002/yea.320070804. [DOI] [PubMed] [Google Scholar]
- 23.Petersen J G L, Kielland-Brandt M C, Nilsson-Tillgren T, Bornæs C, Holmberg S. Molecular genetics of serine and threonine catabolism in Saccharomyces cerevisiae. Genetics. 1988;119:527–534. doi: 10.1093/genetics/119.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramos C, Calderón I L. Overproduction of threonine by yeast mutants resistant to hydroxynorvaline. Appl Environ Microbiol. 1992;58:1677–1682. doi: 10.1128/aem.58.5.1677-1682.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Ramos F, Wiame J M. Occurrence of a catabolic l-serine (l-threonine) deaminase in Saccharomyces cerevisiae. Eur J Biochem. 1982;123:571–576. doi: 10.1111/j.1432-1033.1982.tb06570.x. [DOI] [PubMed] [Google Scholar]
- 25.Reinscheid D J, Kronemeyer W, Eggeling L, Eikmanns B J, Sahm H. Stable expression of hom-1–thrB in Corynebacterium glutamicum and its effect on the carbon flux to threonine and related amino acids. Appl Environ Microbiol. 1994;60:126–132. doi: 10.1128/aem.60.1.126-132.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rose M D, Novick P, Thomas J H, Botstein D, Fink G R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 28.Seibold M, Nill K, Poralla K. Homoserine and threonine pools of borrelidin resistant Saccharomyces cerevisiae mutants with altered aspartokinase. Arch Microbiol. 1981;129:368–370. doi: 10.1007/BF00406464. [DOI] [PubMed] [Google Scholar]
- 29.Sherman F, Fink G R, Hicks J B. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 30.Slater M R, Craig E A. Transcriptional regulation of an hsp70 heat shock gene in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:1906–1916. doi: 10.1128/mcb.7.5.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sulser H, DePizzol J, Büchi W. A probable flavoring principle in vegetable-protein hydrolysates. J Food Sci. 1967;32:611–615. [Google Scholar]