Abstract
Buchnera aphidicola is an obligate intracellular symbiont of aphids. One of its proposed functions is the synthesis of essential amino acids, nutrients required by aphids but deficient in their diet of plant phloem sap. The genetic organization of the tryptophan pathway in Buchnera from proliferous aphids of the family Aphididae has previously been shown to reflect a capacity to overproduce this essential amino acid (C.-Y. Lai, L. Baumann, and P. Baumann, Proc. Natl. Acad. Sci. USA 91:3819–3823, 1994). This involved amplification of the genes for the first enzyme in the pathway, anthranilate synthase (TrpEG), on a low-copy-number plasmid. Here we report on the finding and molecular characterization of TrpEG-encoding plasmids in Buchnera from aphids of the distantly related family Pemphigidae. Buchnera from Tetraneura caerulescens contained a 3.0-kb plasmid (pBTc2) that carried a single copy of trpEG and resembled trpEG plasmids of Buchnera from the Aphididae. The second plasmid (pBPs2), isolated from Buchnera of Pemphigus spyrothecae, contained a different replicon. It consisted of a putative origin of replication containing iterons and an open reading frame, designated repAC, which showed a high similarity to the gene encoding the replication initiation protein RepA of the RepA/C replicon from the broad-host-range IncA/C group of plasmids. The plasmid population was heterogeneous with respect to the number of tandem repeats of a 1.8-kb unit carrying repAC1, trpG, and remnants of trpE. The two principal forms consisted of either five or six copies of this repeat and a single-copy region carrying repAC2, the putative origin of replication, and trpE. The unexpected finding of elements of the RepA/C replicon in previously characterized trpEG plasmids from Buchnera of the Aphididae suggests that a replacement of replicons has occurred during the evolution of these plasmids, which may point to a common ancestry for all Buchnera trpEG amplifications.
Aphids are dependent on an intracellular symbiont (Buchnera aphidicola, Proteobacteria) for normal growth and reproduction (7, 19, 45). The bacteria reside in specialized cells in the aphid hemocele and are transmitted maternally through infection of eggs or embryos (11, 26). Phylogenetic studies have revealed two major characteristics of the evolutionary history of the association (37, 39); (i) the symbiosis had a single origin, dated about 150 million to 250 million years ago; and (ii) host and symbiont lineages have since diverged strictly in parallel. The association, like other symbioses in insects feeding on restricted and unbalanced diets, is thought to have a nutritional basis (5–7, 20). Aphids feed on plant phloem sap, a diet rich in carbohydrates but deficient in nitrogenous compounds, including most essential amino acids (16, 18, 27, 41). Buchnera has been proposed as the source of essential amino acids for the aphid (14), which has been supported by evidence from nutritional and physiological studies (17, 20–22, 45) and, more recently, by the finding of genetic modifications in the tryptophan and leucine biosynthetic pathways in Buchnera from several aphid species. In both cases, genes encoding key enzymes in the respective pathways were found to be amplified and relocated to plasmids (10, 30).
Lai et al. (30) found that the genes for the two subunits of anthranilate synthase (trpE and trpG), the first enzyme in the pathway leading to tryptophan, are contained on a low-copy-number plasmid in Buchnera from the aphid Schizaphis graminum (Aphididae). The plasmid consisted of four identical tandem repeats of a 3.6-kb trpEG-containing unit. trpEG was amplified about 16-fold over the remaining genes of the pathway, which reside in a single locus [trpDC(F)BA] on the Buchnera chromosome (38). trpEG-encoding plasmids have since been found in Buchnera from various species of the Aphididae (4, 32, 42, 43), and their overall similarity suggests that the amplification is ancestral to this lineage (32). The Aphididae is the largest and evolutionarily most successful family of aphids. Many of its species have high growth and reproductive rates, and it includes a number of major agricultural pests (8).
In contrast, Buchnera from the aphid Schlechtendalia chinensis, a member of the distantly related family Pemphigidae, was found to carry all the genes of the tryptophan pathway on the chromosome, organized into two single-copy linkage groups [trpEG and trpDC(F)BA] (31). This difference in organization, which is assumed to reflect a difference in the capacity to overproduce tryptophan, has been linked to potentially varying requirements for the amino acid by aphid hosts. S. chinensis has a long development time and a low reproductive rate, and its demand for tryptophan may therefore be lower than in the highly prolific aphids of the Aphididae (5–7, 9, 31).
Here we report on the finding and molecular characterization of trpEG-containing plasmids in Buchnera from the aphids Tetraneura caerulescens and Pemphigus spyrothecae, both belonging to the Pemphigidae. We propose a scenario for the evolution of trp in Buchnera in which there was a single ancestral transfer of trpEG to a RepA/C-like replicon followed by independent events of replicon replacement and back-transfer of trpEG to the chromosome in different lineages.
MATERIALS AND METHODS
Aphid material and B. aphidicola.
Leaves carrying galls produced by the aphid P. spyrothecae were collected from poplar trees (La Yesa, Spain). T. caerulescens was collected from galls on elm trees (Bugarra, Spain). The galls were kept at 4°C until needed for further manipulation of the animals.
General methods.
Buchnera plasmid and genomic DNA was isolated as previously described (49). DNA was manipulated by standard methods (2, 44). Southern hybridizations were done with either digoxigenin-labeled probes (Boehringer Mannheim) or the enhanced chemiluminescence (ECL) system (Amersham), as specified by the manufacturers.
Cloning and sequencing.
Cloning of B. aphidicola (T. caerulescens) plasmid DNA has been described previously (49). One clone with an EcoRI insert of 3.0 kb was completely sequenced. Populations of aphids from the Pemphigidae are often small, and only a limited number of P. spyrothecae animals could be collected. Consequently, the total amount of plasmid DNA available for experiments was reduced and a shotgun-cloning approach was taken to maximize cloning efficiency. Approximately 30 ng of B. aphidicola (P. spyrothecae) plasmid DNA was digested with XbaI and ligated to XbaI-digested, phosphatase-treated pBluescript II SK (Stratagene). After transformation of Escherichia coli DH5α with the ligation and screening of recombinant colonies, clones carrying inserts of 2.1 and 1.8 kb were analyzed by restriction enzyme digestion and end sequencing (see Fig. 4A). All the clones were identical, and one clone of each size was completely sequenced. Sequence information obtained from these clones was used to design oligonucleotide primers complementary to repAC and trpE that would allow the PCR amplification of a region spanning a 1.5-kb XbaI fragment that had remained uncloned during shotgun cloning (repACPs1,5′-AGA GCA ATG AAA AAC GCT TCT CG-3′; and trpEPs1,5′-TCA GGT GAC GCT CCA AAT AAG G-3′). Cycling was performed with the GeneAmp 2400 System (Perkin Elmer) and consisted of 5 cycles of 94°C for 30 s, 62°C for 1 min, and 72°C for 1 min followed by 30 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s, with a final 10-min extension at 72°C. The PCR yielded fragments of 1.53 and 1.70 kb, which were ligated into a T vector (36) and transformed into E. coli XL1Blue (Stratagene). Two clones with inserts corresponding to the sizes of the two PCR fragments were completely sequenced (see Fig. 4A). Nested deletions of all selected cloned fragments were generated in both directions by using the nested deletions kit (Pharmacia). Nucleotide sequencing was performed with the AmpliTaqF Dye Deoxy Terminator cycle-sequencing kit (Perkin Elmer) and the 373 automated DNA sequencer (Perkin-Elmer) as recommended by the manufacturer.
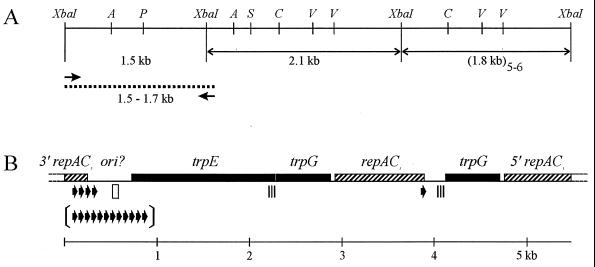
FIG. 4.
Physical map of cloned and sequenced portion of pBPs2 from B. aphidicola (P. spyrothecae). (A) Restriction site and clone map. Double-headed arrows, cloned XbaI restriction fragments; arrows, oligonucleotide primers used for PCR; dotted line, cloned PCR product. Restriction enzyme sites: A, AccI; P, PvuII; S, SacI; C, ClaI; V, EcoRV. (B) Genetic map. All genes are transcribed in the rightward direction. The region denoted ori? contains the putative origin of replication. Arrows, 19-bp iteron unique to pBPs2; arrows between brackets illustrate the copy number of the same iteron in the 1.70-kb PCR fragment; open rectangle, 19-bp element differing in one nucleotide from consensus sequence of RepA/C iterons; triple strip, 129-bp sequence identical to the 3′ end of trpE.
Quantitative hybridization.
The ratio between the copy number of the 1.8- and 2.1-kb XbaI fragments per plasmid was determined by densitometric scans of Southern blots containing eight XbaI digests of B. aphidicola (P. spyrothecae) total DNA and hybridized with a probe containing trpG and repAC1. Scanning was performed with a Gelstation (TDI) image analyzer, and images were analyzed with the Intelligent Quantifier Bioimage software (Bio Systems Corp.).
Serial partial digestion of B. aphidicola (P. spyrothecae) total DNA.
B. aphidicola (P. spyrothecae) total DNA was first digested to completion with AccI and SacI to remove most of the single-copy region from the plasmid that contained an XbaI site. Partial digestions with serial dilutions of XbaI were then performed by the method of Ausubel et al. (2), starting with 5 U of enzyme and using approximately 200 ng of DNA per digest. Controls included DNAs digested to completion with XbaI, AccI, SacI, and AccI plus SacI. Electrophoresis was performed overnight on 0.5% agarose gels in 0.5 × Tris-borate-EDTA at 1.2 V/cm and was followed by Southern blotting. Hybridization was done with the cloned 1.8-kb XbaI fragment as a probe. We used 1-kb ladder DNA (Gibco-BRL) and HindIII-digested λ DNA as molecular weight markers.
Computer analysis of the DNA sequences.
DNA sequence data were assembled and analyzed with the Genetics Computer Group (GCG) program package 8.0 for the VAX/VMS (15). BLASTP searches (1) were done at the network server of the National Center for Biotechnology Information. Phylogenetic analysis of amino acid sequences was performed with the PROTDIST (Dayhoff PAM distances) and the NEIGHBOR (neighbor-joining method) programs as implemented in PHYLIP version 3.5 (23).
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper have been deposited in the GenEMBL databases under accession no. AJ012333 and AJ012334 (PBTc2 and PBPs2, respectively).
RESULTS
Detection of plasmids.
B. aphidicola (T. caerulescens) has previously been shown to carry a small, cryptic plasmid (pBTc1, 1.74 kb) that contained the repA1 replicon (related to IncFII plasmids) found in leucine plasmids (49). Restriction enzyme analysis of plasmid DNA preparations from this species indicated the presence of a second plasmid of about 3 kb. Similar observations were made with plasmid DNA isolated from B. aphidicola (P. spyrothecae) (48). Plasmid DNA preparations from both species were here further analyzed by Southern hybridization, cloning, and nucleotide sequencing. The two natural plasmids described below are designated pBTc2 and pBPs2, after their source B. aphidicola (T. caerulescens) and B. aphidicola (P. spyrothecae, respectively.
Characterization of pBTc2.
An EcoRI shotgun cloning of B. aphidicola (T. caerulescens) plasmid DNA yielded four recombinants with an insert of 3.0 kb. Restriction fragment analysis showed these to be identical, and one clone was completely sequenced. A physical map of plasmid pBTc2 is presented in Fig. 1A. The G+C content of the 3.0-kb fragment is 20.74 mol%, which is similar to the 21.2 mol% of plasmid pBTc1 from this species (49) and is consistent with the low G+C content (28 mol%) of Buchnera genomic DNA (28). Two open reading frames (ORFs) were found, which correspond to trpE and trpG. The deduced amino acid sequences are 52 and 53% identical to TrpE and TrpG, respectively, from Buchnera of the phylogenetically treated aphid S. chinensis (Pemphigidae). Potential regulatory elements for gene expression (33) could not be identified.
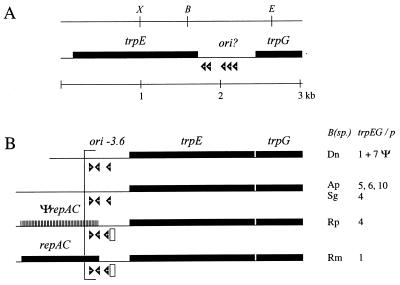
FIG. 1.
Linearized physical maps of B. aphidicola trpEG plasmids. All genes are transcribed in the rightward direction. Open arrowheads indicate the approximate position and orientation of DnaA boxes. (A) Restriction site and genetic map of pBTc2 from B. aphidicola (T. caerulescens). The region denoted ori? contains the putative origin of replication. Restriction enzyme sites: X, XbaI; B, BglII; E, EcoRI. (B) Genetic maps of the repeated units of B. aphidicola (Aphididae) trpEG plasmids (adapted from reference 42 with permission from the publisher). B(sp.), species names (see below); trpEG/p, number of repeat units per plasmid (42); ori-3.6, putative origin of replication; Ψ, repeat units containing trpEG pseudogenes (32); striped box and ΨrepAC, location of a putative repAC pseudogene; open rectangle, 19-bp element similar to consensus sequence of RepA/C iterons. Species abbreviations: Dn, Diuraphis noxia; Ap, Acyrthosiphon piseum; Sg, Schizaphis graminum; Rp, Rhopalosiphum padi; Rm, Rhopalosiphum maidis.
Most trpEG-containing plasmids of Buchnera from aphids of the family Aphididae share a conserved 500- to 600-nucleotide (nt) sequence immediately upstream of trpE which has features characteristic of an origin of replication (29). The region has been designated ori-3.6 and contains three DnaA boxes (consensus 5′-TTATCCACA-3′) and regions of low G+C content (Fig. 1B) (43). A similar region, 717 nt in length, is present downstream of trpE on pBTc2 and may therefore contain the origin of replication of the plasmid. It has a very low G+C content (11 mol%) and contains five DnaA boxes in direct orientation, only one of which deviates by a single nucleotide from the consensus sequence. Beyond a high A+T content, the region has no further sequence similarity to ori-3.6.
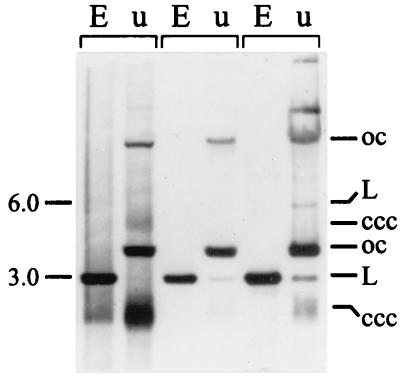
B. aphidicola (Aphididae) trpEG plasmids are typically composed of tandemly repeated, identical 3.2- to 3.6-kb units, each carrying a copy of trpEG and ori-3.6 (42, 43). To determine the composition of pBTc2, a Southern hybridization analysis was performed of various EcoRI-digested and undigested B. aphidicola (T. caerulescens) DNA preparations, using cloned pBTc2 as a probe. For each preparation, two strongly hybridizing bands were detected in lanes with undigested DNA and a single strong band was detected in lanes with EcoRI-digested DNA (Fig. 2). These three bands correspond to, respectively, the open-circular, covalently closed supercoiled, and linear forms of a plasmid of 3.0 kb. In addition, three larger, more weakly hybridizing bands were detected in undigested DNAs. Their sizes correspond to the different forms of a plasmid of 6.0 kb. EcoRI digestion converted all the different forms into a single linear fragment of 3.0 kb. These findings suggest that pBTc2 consists mainly of one copy of the sequenced 3.0-kb unit but that a fraction of the plasmid population is present as a dimer. In one preparation, molecules of an even higher molecular weight were detected (Fig. 2, right lane), indicating that further multimerization of the plasmid occurs.
FIG. 2.
Southern blot analysis of EcoRI-digested (E) and undigested (u) B. aphidicola (T. caerulescens) DNA preparations. Cloned pBTc2 was used as a hybridization probe. The different forms of the putative plasmids are marked: open circular (oc), linear (L), and covalently closed circular (ccc).
Identification of pBPs2.
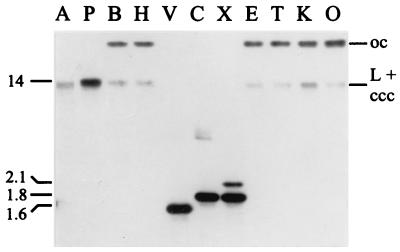
Restriction enzyme analysis of a plasmid DNA preparation from B. aphidicola (P. spyrothecae) indicated the presence of at least two plasmids. One of these (pBPs2) was larger than 12 kb as estimated from the migration of bands of undigested plasmid DNA in ethidium bromide-stained agarose gels. Analogous to trpEG plasmids of B. aphidicola (Aphididae), it seemed to be composed of repeated elements, because summation of three restriction fragments generated with XbaI (2.1, 1.8, and 1.5 kb) added up to only 5.4 kb. Moreover, the 1.8-kb band stained more intensely than the other two fragments. Southern hybridization with a trpG-containing probe revealed that the plasmid carried sequences homologous to trpG and reinforced the assumption that it was composed of repeated elements (Fig. 3). AccI and PvuII each yielded a single, slightly blurred band of 13 to 15 kb, while EcoRI, PstI, ScaI, and XhoI yielded two bands, one of which also migrated at about 13 kb and the second of which migrated at more than 23 kb. Digests with EcoRV, ClaI, and XbaI yielded either one or two bands migrating at 1.5 to 2.1 kb. These hybridization patterns were at first explained in terms of the 13- to 15-kb AccI and PvuII bands representing the linear form of the putative plasmid. The fact that the plasmid could be linearized, in contrast to most previously characterized trpEG plasmids (42), implied that it contained a single-copy region in addition to repeated elements. Recognition sites for EcoRI, PstI, ScaI, and XhoI are absent from the plasmid, and hence the two detected bands correspond to the open-circular (at 23 kb) and the comigrating linear and closed-circular forms of a 13- to 15-kb plasmid. Finally, recognition sites for EcoRV, ClaI, and XbaI are present in the repeated part of the plasmid and hence convert it into the constituent repeat units.
FIG. 3.
Southern blot analysis of B. aphidicola (P. spyrothecae) plasmid DNA. Hybridizations were performed with a probe complementary to trpG. Restriction enzymes: A, AccI; P, PvuII; B, BamHI; H, HindIII; V, EcoRV; C, ClaI; X, XbaI; E, EcoRI; T, PstI; K, ScaI; O, XhoI. oc, open circular; L, linear; ccc, covalently closed circular.
Cloning and sequencing.
B. aphidicola (P. spyrothecae) plasmid DNA was shotgun cloned with XbaI. Screening of 108 recombinant colonies showed that 10% of plasmids had an insert of 2.1 kb and 20% had an insert of 1.8 kb, corresponding in size to the two XbaI fragments detected in the Southern hybridization (Fig. 3). The 1.5-kb XbaI fragment, initially detected in agarose gels, was not found. Restriction enzyme analysis and end sequencing of the recombinant plasmids suggested that all clones of the same size were identical and one of each group was completely sequenced (Fig. 4B).
The 2.1-kb fragment contained three ORFs, the first two of which corresponded to the 3′ half of trpE and a complete copy of trpG, with the stop and start codons of the two genes overlapping. BLASTP homology searches with the putative product of the third ORF yielded a high probability score (i.e., 893) with the N-terminal part of replication initiation protein RepA encoded by E. coli plasmid RA1, which belongs to the broad-host-range IncA/C incompatibility group (34). The amino acid sequences were 69% identical over the first 243 positions. The B. aphidicola (P. spyrothecae) gene is designated repAC1. The 1.8-kb fragment also contained three ORFs, the first of which was similar to the gene encoding the C-terminal part of the same RepA protein that was lacking from the 2.1-kb fragment. Because the coding sequences are in frame, the two fragments are most probably contiguous on the plasmid. The second ORF corresponded to a complete copy of the trpG gene, and the third ORF, again, corresponds to the gene encoding the N-terminal part of RepA. Both sequences were completely identical to the homologous sequences in the 2.1-kb fragment.
The 2.1-kb XbaI fragment contained single AccI and SacI restriction sites (Fig. 4A). Since the former enzyme was at first assumed to linearize the plasmid, this implied that the fragment contained part of a single-copy region of the plasmid. However, neither of the two sequenced fragments contained a restriction site for PvuII, suggesting that this site had to be present in the uncloned 1.5-kb XbaI fragment and, hence, that the 1.8-kb fragment represented the repeated unit of the plasmid. In addition, the uncloned 1.5-kb XbaI fragment was expected to contain the missing 5′ end of the trpE gene and possibly the 3′ end of a repAC gene. Oligonucleotide primers complementary to trpE and repAC1 that would allow the amplification of this fragment were designed (Fig. 4A).
PCR yielded two products, which were 1.53 and 1.70 kb in size. After cloning, one recombinant plasmid from each size group was completely sequenced on both strands (Fig. 4B). Both fragments contained the 3′ end of a repAC gene and the missing 5′ part of trpE, as well as a single PvuII site. However, the 3′ end of the repAC sequences differed slightly from the homologous sequence of the 1.8-kb XbaI fragment. The latter contained only one copy of a 19-bp element (5′-CAACAAACTATAAAAAAAA-3′), located 30 nt upstream of the stop codon TAA (Fig. 4B). In the smaller of the two PCR products, this element occurred at exactly the same position within the coding sequence but was directly followed by three additional copies, the second of which contained the stop codon (TAA) of repAC. The 1.7-kb PCR product contained 12 copies of the element. Since the third copy again included the stop codon, the repAC coding sequences of the two PCR products were identical, and this variant of the gene is designated repAC2. The presence of a repeated 19-bp element resembles the iterons found in the origin of replication of the RepA/C replicon of plasmid RA1 (34). The difference of eight copies of this element in the region downstream of repAC2 explains the size difference of about 160 bp between the two PCR products. This length variation most probably reflects a heterogeneous plasmid population within our DNA preparation (see below).
The total length of the three assembled contiguous fragments, including the sequence derived from the smaller, 1.53-kb PCR product, is 5,477 bp (Fig. 4B). The sequence has a G+C content of 25.6 mol% and 85% is occupied by putative ORFs. In contrast to pBTc2, putative −35 (TTGAC) and −10 (AGTTAA) promoter elements could be identified, 125 bp upstream of trpE. The elements are separated by 16 bp and resemble previously identified Buchnera promoters (32). A second possible −10 box (TAACTA), which is identical to the E. coli trp −10 sequences (33), was found overlapping 3 bp with the first. The intergenic region between repAC1 and trpG contained a 129-nt sequence that was identical to the 3′ end of trpE. Similar “remnants” of trpE have been described for the trpEG plasmid of B. aphidicola (Uroleucon sonchi) (4). The intergenic region between repAC2 and trpE was highly AT rich (85 mol%) and contained one copy of a 19-bp element (5′-TATATGGGAATGTTGCACA-3′) that differs in only one position from the consensus sequence (5′–-AT-TGGG---G-TGCACG–3′) of a 13-copy iteron described for the origin of replication of E. coli plasmid RA1 (Fig. 4B) (34, 35). In addition, ori of this replicon contains one copy of a DnaA box. This element was absent from our sequence, but it may not be required for replication (40). The high A+T content and similarity with respect to the presence of iterons suggest that the repAC2-trpE intergenic region contains the origin of replication of pBPs2.
Structure of pBPs2.
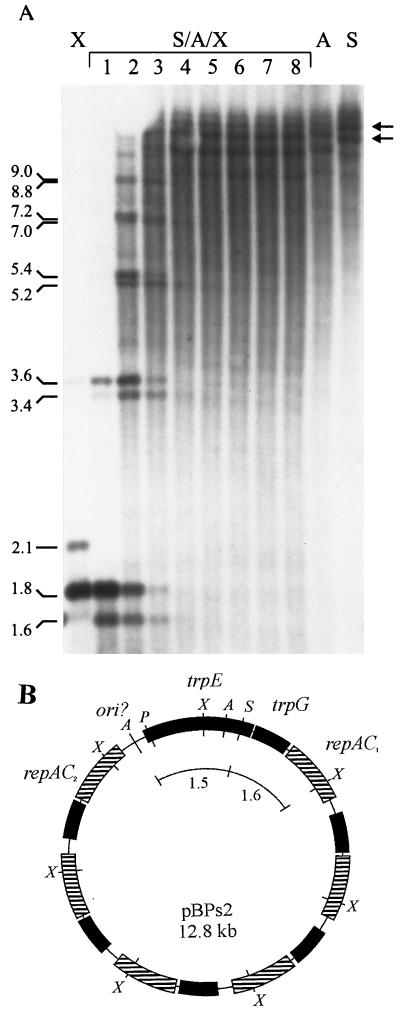
To establish the overall structure of the plasmid, we performed the following experiments. First, quantitative hybridizzations were carried out to determine the relative abundance of the two XbaI fragments detected in the initial hybridizations (Fig. 3). Densitometric assays of Southern blots containing XbaI digested plasmid DNA indicated that the 1.8-kb fragment was 5.19 ± 0.97 times more abundant than the 2.1-kb fragment (data not shown). A copy number of 5 or 6 of the 1.8-kb fragment relative to one copy each of the 2.1- and 1.5-kb fragments adds up to a plasmid size of either 12.8 or 14.6 kb, which was in good agreement with the initial estimations. Second, partial XbaI digestions of plasmid DNA predigested with AccI plus SacI resulted in a ladder of bands in which each step consisted of two bands differing 200 bp in size (Fig. 5A). The size difference between the double steps corresponded exactly to multiples of 1.8 kb plus 1.6 kb and multiples of 1.8 kb only. These findings were in agreement with the expectation based on the restriction site map (Fig. 4A), the 1.6-kb fragment resulting from the SacI site in the 2.1-kb XbaI fragment.
FIG. 5.
(A) Southern blot analysis of partial digestion of AccI-SacI-digested B. aphidicola (P. spyrothecae) DNA by using serial dilutions of XbaI. Lanes: 1 to 8, digestions with 5.0, 2.5, 1.24, 0.63, 0.31, 0.16, 0.08, and 0.04 U of XbaI, respectively, for 15 min. X, A, S, control digestions performed to completion (X, XbaI; S, SacI; A, AccI. Arrows indicate the two principal forms of the plasmid. (B) Proposed structure of pBPs2 from B. aphidicola (P. spyrothecae) containing five tandem repeats of a 1.8-kb unit. Boxes with a similar pattern represent identical genes. All genes are transcribed in a clockwise direction. The region denoted ori? contains the putative origin of replication. Arcs correspond to restriction fragments removed from molecules with AccI and SacI before partial digestion experiment with XbaI. The restriction enzyme sites shown are as in Fig. 4A.
Control digestions with AccI and SacI revealed that the plasmid population was in fact heterogeneous and was composed of molecules of different sizes. A single digestion with SacI, which linearizes the plasmid, yielded a short ladder of bands, differing in size by steps of 1.8 kb (Fig. 5A). The two most strongly hybridizing bands were 12.8 and 14.6 kb, corresponding to molecules containing either five or six copies of the 1.8-kb XbaI repeat, respectively. Four more weakly hybridizing bands measured approximately 18.2, 16.4, 11.0, and 9.2 kb, corresponding to molecules containing 8, 7, 4, and 3 copies of the same unit. A similar ladder of bands was observed for AccI, although all the bands were 1.3 kb smaller, which is consistent with the presence of two sites, 1.3 kb apart, in the single-copy region of the plasmid (Fig. 4A). Superimposed on the length variation due to differences in repeat number is the length variation accounted for by the difference in copy number of the 19-bp direct repeat downstream of repAC2. The inferred overall structure of a plasmid containing five tandem repeats of the 1.8-kb XbaI unit is presented in Fig. 5B.
Similarity between pBPs2 repAC and B. aphidicola (Rhopalosiphum) trpEG plasmids.
The single-copy region between repAC2 and trpE of pBPs2 was found to share similarities with the origin of replication of plasmid RA1, but no significant similarities to the 500- to 700-nt putative ori of pBTc2 or the B. aphidicola (Aphididae) trpEG plasmids were observed. However, a comparison of the repAC sequence to the latter group of plasmids revealed a striking similarity (73% identical positions in a 955-bp overlap) to the region immediately upstream of ori-3.6 of the B. aphidicola (R. maidis) plasmid (Fig. 1B). Analysis of the recent update of this sequence (provided by P. Baumann) showed that this plasmid in fact carries a potentially intact repAC gene, whose deduced amino acid sequence was 71% identical to the two pBPs2 RepAC sequences and 64% identical to the plasmid RA1 homologue (Fig. 6). In addition, the 19-bp element (5′-TATATGGGAATGTTGCACA-3′) present in a single copy in the putative ori of pBPs2 and in 13 copies in the ori of RA1 (34, 35) also occurs in the ori of B. aphidicola (R. maidis) plasmid, with only a 1-bp difference from the pBPs2 sequence.
FIG. 6.
Multiple alignment of translations of repAC-like sequences. Black background indicates stop codons (x). The C-terminal residues differing between RepAC1 and RepAC2 are shown in boldface type. Ps_RepAC, repAC of pBPs2; Rm_RepAC, repAC of trpEG plasmid of B(R. maidis); RA1_RepAC, repAC of E. coli plasmid RA1.
A subsequent comparison of B. aphidicola (R. maidis) repAC to the region upstream of ori-3.6 from the closely related B. aphidicola (R. paid) trpEG plasmid yielded a similarity of 71% in an 843-bp overlap. The latter sequence, however, did not contain an uninterrupted reading frame. A putative remnant of the 19-bp element (5′-TATACGAGAATAGTGCACA-3′) was also found in exactly the same position as in the B. aphidicola (R. maidis) sequence. The similarity of repAC to the upstream region of ori in trpEG plasmids from other species was much lower (44 to 59%), and the 19-bp element or remnants thereof could not be identified. Nevertheless, the high sequence similarity in B. aphidicola (Rhopalosiphum) suggests that at least the B. aphidicola (R. padi) sequence contains an repAC pseudogene. This mode of gene silencing has a well-documented precedent in the trpEG plasmids of Buchnera from the aphids Diruaphis noxia (32) and Uroleucon sonchi (4). Both plasmids carry only one functional copy of trpEG, in addition to a variable number of repeat units containing trpEG or trpE pseudogenes.
The multiple alignment further illustrates differences among the termini of the RepA sequences (Fig. 6). RepA from plasmid RA1 has an additional 33 residues at its N-terminal end, while the B. aphidicola (P. spyrothecae) sequences are longer at their C-terminal end. Remarkably, however, a high similarity to the RA1 sequence is retained at the nucleotide level after the stop codon, in the second reading frame of the RA1 sequence. The similarity drops immediately after the stop codon of the pBPs2 repAC sequences. Because it is unlikely that Buchnera would have retained such a high similarity in a noncoding sequence, it is possible that the published RA1 sequence contains a sequencing error. A reestimation of the size of RepA in the sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of Fig. 2 of Llanes et al. (35) yielded a molecular mass of 42 kDa, in contrast to the 35-kDa estimate reported by the authors. Our estimate corresponds exactly to a RepA protein of 366 residues (Fig. 6).
TrpEG phylogenetic relationships.
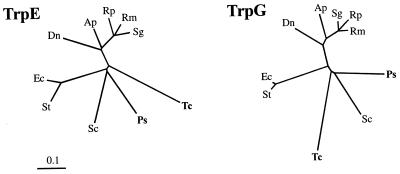
A phylogenetic analysis of Buchnera TrpE and TrpG sequences was conducted to assess the relationships of the newly sequenced genes. In accordance with previous 16S rDNA phylogenies (39, 49), the TrpG sequences from the B. aphidicola (Pemphigidae) form a cluster (Fig. 7). In the TrpE analysis, the outgroup sequences (E. coli and Salmonella typhimurium) rooted the tree within the B. aphidicola (Pemphigidae). The branch lengths of the groupings around the root were relatively short, particularly in the TrpE phylogeny, while the longer branches leading to the B. aphidicola (T. caerulescens) sequences indicate that the substitution rate in this lineage has been higher than in any of the other lineages.
FIG. 7.
Phylogenetic trees constructed by the neighbor-joining method (23) from estimated distances between TrpE and TrpG sequences. Species designations: Ec, Escherichia coli; St, Salmonella typhimurium; Ps, B. aphidicola (Pemphigus spyrothecae); Tc, B. aphidicola (Tetraneura caerulescens; Sc, B. aphidicola (Schlechtendalia chinensis); Dn, B. aphidicola (Diuraphis noxia); Ap, B. aphidicola (Acyrthosiphon pisum); Sg, B. aphidicola (Shizaphis graminum); Rp, B. aphidicola (Rhopalosiphum padi); Rm, B. aphidicola (Rhopalosiphum maidis).
DISCUSSION
Recent genetic studies have substantiated the importance of essential amino acid biosynthesis by B. aphidicola in its symbiotic association with aphids (7, 10, 30, 49). To date, rearrangements in the pathways leading to tryptophan and leucine remain the sole genetic modifications found in Buchnera that are indicative of a specific, enhanced biosynthetic capability (5–7, 12). However, amplification of the genes (trpEG) encoding anthranilate synthase has until now only been reported for Buchnera from the highly proliferous species of the Aphididae. Here we present the first report on Buchnera trpEG-encoding plasmids from aphids not belonging to the Aphididae.
Properties of B. aphidicola (Pemphigidae) trpEG plasmids.
B. aphidicola (T. caerulescens) carried a 3.0-kb plasmid (pBTc2) which resembled the trpEG plasmids found in Buchnera from the Aphididae. With the exception of the B. aphidicola (R. maidis) plasmid (see below), none of these plasmids encodes proteins potentially involved in replication. The distinctive feature of their putative origin of replication is the presence of multiple DnaA boxes. B. aphidicola (P. spyrothecae) contained trpEG-encoding plasmids (pBPs2) with a gene content and putative replicon different from those described above, including a putative replication gene that was highly similar to the replication initiation gene repA of the RepA/C replicon from E. coli plasmid RA1 (34) (Fig. 6). The origin of replication of the RepA/C replicon is located downstream of repA and contains iterons, which serve as binding sites for RepA and are required for replication. pBPs2 contains 4 or 12 copies of a 19-bp iteron that overlap the 3′ end of the repAC2 gene and the downstream putative origin of replication (Fig. 4B). The sequence of this iteron was unique to pBPs2, but it also carries a single copy of the 19-bp element that constitutes the iteron of RepA/C.
Besides carrying multiple copies of this replication gene, a noticeable feature of pBPs2 is the amplification of trpG over trpE. This only further occurs in the trpEG plasmid of B. aphidicola (U. sonchi) (4). Similar to this plasmid, trpG in the repeat unit of pBPs2 is preceded by a remnant of trpE (Fig. 4B). This suggests either that a region containing only the 3′ end of trpE was initially duplicated or that after the initial duplication of an entire repAC-trpEG unit, trpE was silenced through deletion. In either case, we suspect that the amplification is merely a by-product of recombination. A functional explanation would have to invoke an involvement of TrpG in a pathway other than tryptophan biosynthesis. In some bacteria, including Pseudomonas acidovorans, Bacillus subtilis, and Acinetobacter calcoaceticus (13), TrpG participates in both anthranilate synthase and p-aminobenzoate synthase activities. The latter enzyme synthesizes p-aminobenzoate, which is a component of the vitamin folic acid (50). In most other bacteria, however, including the closest known relative of Buchnera, E. coli (47), this activity is provided by a different though significantly similar protein (PabA) (13).
Structural variation of trpEG plasmids.
The most conspicuous characteristic of Buchnera trpEG plasmids is their structural fluidity. B. aphidicola (Aphididae) contains plasmids of a concatenate structure, consisting of 4 to 10 tandem duplications of a basic, 3.6-kb trpEG-containing unit (42). Exceptions are B. aphidicola (R. maidis), which contains a plasmid consisting of only one 3.6-kb unit (42), and B. aphidicola (D. noxia) and B. aphidicola (U. sonchi), which carry plasmids consisting of one 3.2-kb unit encoding a functional copy of TrpEG and a variable number of repeat units carrying trpEG pseudogenes (4, 32). The plasmid population in B. aphidicola (T. caerulescens) was composed mainly of molecules containing a single 3.0-kb trpEG unit, but Southern hybridization indicated the presence of multimers (Fig. 2).
The molecular basis of concatenation in these plasmids is unknown, but it is likely to be initiated by multimerization, which involves homologous recombination between plasmid monomers (25, 46). An indication that recombination (and mismatch repair) occurs in the trpEG plasmids comes from the observation by Lai et al. of gene conversion among trpEG pseudogenes (32). Multimerization of low-copy-number plasmids increases the chance of their loss at cell division (46). Several maintenance systems are known to counteract this risk, including multimer resolution and partitioning systems (46, 51). There is no evidence for the presence of such systems on the trpEG plasmids. However, stable inheritance may not be essential if selection for trpEG copy number does not act primarily on individual cells. In its intracellular environment, the slowing dividing Buchnera (3) may be viable with a reduced number of trpEG plasmids or even with none. This could shift the need for plasmid stability in individual cells to the need for partitioning bacteria with an adequate genetic composition to the next generation of aphids. We speculate that selection acting at this higher level underlies the observed structural variability of Buchnera trpEG plasmids.
Phylogenetic analysis of TrpEG.
The fact that pBTc2 was more similar to the B. aphidicola (Aphididae) trpEG plasmids than to pBPs2 raised the possibility that B. aphidicola (T. caerulescens) acquired its plasmid through horizontal transfer. Phylogenetic analysis, however, showed that the plasmid-borne sequences of B. aphidicola (T. caerulescens) and B. aphidicola (P. spyrothecae) are most closely related to the chromosomal TrpEG sequence of B. aphidicola (S. chinensis) (Fig. 7). This refutes horizontal acquisition of pBTc2 by B. aphidicola (T. caerulescens). The slight incongruence at the root between the TrpE and TrpG topologies can be attributed to an accelerated substitution rate in B. aphidicola (T. caerulescens), causing its long branch to be “artifactually attracted” (24) by the B. aphidicola (Aphididae) sequences in the case of the TrpE phylogeny.
Replicons in Buchnera trpEG plasmids.
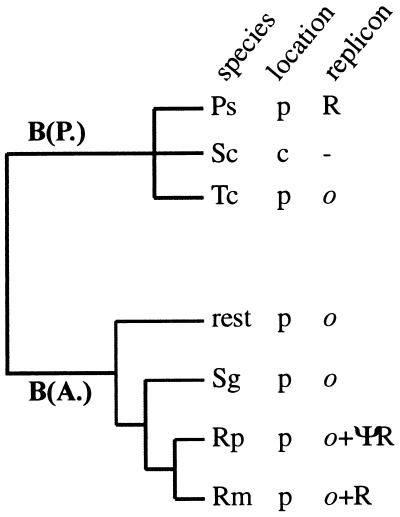
Two replicons are possibly linked to the amplification of trpEG: (i) a RepA/C-like replicon, in which replication is most probably initiated by plasmid-encoded RepAC, and (ii) an ori-3.6-like replicon, which is characterized by the presence of multiple DnaA boxes in the ori and for which DnaA is thought to be involved in initiation of replication (30, 42, 43). After the discovery of RepA/C in B. aphidicola (P. spyrothecae), computer analysis of previously characterized trpEG plasmids (42) revealed the presence of a putative intact repAC gene in B. aphidicola (R. maidis) and a repAC pseudogene in B. aphidicola (R. padi) (Fig. 1B and 6). In addition, ori-3.6 of both plasmids contained a single copy of the 19-bp iteron of the RepA/C replicon. None of these elements could be identified unequivocally in any of the other B. aphidicola (Aphididae) plasmids or in pBTc2. The phylogenetic distribution of the two replicons is summarized in Fig. 8.
FIG. 8.
Phylogenetic distribution of trpEG-associated replicons in Buchnera from aphids of the families Pemphigidae [B(P.)] nd Aphididae [B(A.)]. Species designations are as in Fig. 7. Location: p, plasmid; c, chromosomal. Replicon designation: R, RepA/C; ΨR, RepAC pseudogene; o; ori-3.6 like.
The presence of conserved elements of both replicons in the B. aphidicola (R. maidis) plasmid and a repAC pseudogene in its closest relative B. aphidicola (R. padi) demonstrates that repAC can be silenced in the presence of an ori-3.6-like replicon. The silenced repAC gene of B. aphidicola (R. padi) apparently diverged rapidly from the B. aphidicola (R. maidis) sequence through the accumulation of substitutions (only 71% similarity at the nucleotide level). This observation, together with the fact that the noncoding regions between trpG and ori-3.6 are often unusually long [up to 1 kb in B. aphidicola (A. pisum) (Fig. 1)], raises the possibility that a RepA/C-like replicon was ancestral to all B. aphidicola (Aphididae) plasmids but that the respective repAC genes have been silenced in parallel lineages and have diverged beyond recognition.
Evolution of trp in Buchnera.
Lai et al. (32) have previously proposed a scenario for the evolution of trp in Buchnera. In brief, they hypothesized that the ancestor of all present-day Buchnera species had the trp genes arranged in two linkage groups [trpEG and trpDC(F)BA] on its chromosome, an organization still found in B. aphidicola (S. chinensis) (31). trpEG was relocated to a plasmid in the ancestor of the B. aphidicola (Aphididae), and the resulting enhanced capacity of tryptophan production promoted increased growth and reproductive rates of the insect host. This scenario must be amended in the light of two significant findings made in the present study. Firstly, trpEG plasmids are found in Buchnera species from the two most divergent lineages of aphids, i.e., the Aphididae and Pemphigidae. This suggests that selection for amplification of trpEG in Buchnera may be as ancient as the symbiosis itself. Second, similar replicons are found in both lineages (Fig. 8). On the basis of these findings, we hypothesized that the transfer of trpEG to a plasmid carrying a RepA/C-like replicon occurred in the ancestor of all present-day Buchnera species. Like the related E. coli RA1 plasmid, the origin of replication may have contained a single DnaA box. An ori-3.6 like origin of replication evolved de novo on this plasmid through amplification of the resident DnaA box. (Note that, in contrast to the RepA/C-like replicon and the repA1 replicon linked to leucine plasmids [49], there are no reports on bacterial replicons related to the ori-3.6-like replicon.) De novo evolution of ori-3.6 must have occurred independently in the lineages leading to B. aphidicola (T. caerulescens) and B. aphidicola (Aphididae) but not in B. aphidicola (P. spyrothecae). In the presence of ori-3.6 and its hypothesized involvement in a regulatory mechanism of initiation of chromosome replication (32), control over replication of the trpEG plasmid was more advantageously exerted by the DnaA protein than by RepAC. This led to silencing of the ancestral RepA/C replicon in most lineages. Although it has yet to be established whether repAC is functional in B. aphidicola (R. maidis), it remains difficult to explain why the elements of RepA/C are much more highly conserved in this species than in any of the other B. aphidicola (Aphididae) strains.
Our scenario implies that trpEG in the lineage leading to B. aphidicola (S. chinensis) must have been transferred back into the chromosome (Fig. 8) rather than that its organization represents the ancestral state (31). In E. coli, the closest-known relative of Buchnera (47), all the genes of the trp pathway are organized into a single operon (13). If a similar organization was present in the presymbiotic ancestor of Buchnera, then the proposed reversal in B. aphidicola (S. chinensis) provides an explanation for the organization of its trp genes into two separate linkage groups.
Finally, our scenario would predict that (i) trpEG plasmids, containing elements of either RepA/C or ori-3.6 like replicons (or both), are likely to be found in Buchnera strains from other families of aphids and (ii) in lineages where trpEG is chromosomally encoded, its location is expected to be different from the one found in B. aphidicola (S. chinensis).
ACKNOWLEDGMENTS
We are indebted to Fernando González-Candelas for support and to the Servei de Bioinformàtica and the S.C.S.I.E. (Universitat de València) for providing computing and sequencing facilities, respectively. We gratefully thank J. M. Michelena for identification of the aphid species.
This work was supported by an EU HCM research grant (CHRX-CT94-0660) to R.V. and grant PB96-0793 C04-01 from DGES to A.M.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Green Publishing; 1989. [Google Scholar]
- 3.Baumann L, Baumann P. Growth kinetics of the endosymbiont Buchnera aphidicola in the aphid Schizaphis graminium. Appl Environ Microbiol. 1994;60:3440–3443. doi: 10.1128/aem.60.9.3440-3443.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumann L, Clark M A, Rouhbakhsh D, Baumann P, Moran N A, Voegtlin DJ. Endosymbionts (Buchnera) of the aphid Uroleucon sonchi contain plasmids with trpEG and remnants of trpE pseudogenes. Curr Microbiol. 1997;35:18–21. [Google Scholar]
- 5.Baumann P, Moran N A. Non-cultivable microorganisms from symbiotic associations of insects and other hosts. Antonie Leeuwenhoek. 1997;72:39–48. doi: 10.1023/a:1000239108771. [DOI] [PubMed] [Google Scholar]
- 6.Baumann P, Moran N A, Baumann L. The evolution and genetics of aphid endosymbionts. BioScience. 1997;47:12–20. [Google Scholar]
- 7.Baumann P, Baumann L, Lai C Y, Rouhbakhsh D, Moran N A, Clark M A. Genetics, physiology, and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu Rev Microbiol. 1995;49:55–94. doi: 10.1146/annurev.mi.49.100195.000415. [DOI] [PubMed] [Google Scholar]
- 8.Blackman R L, Eastop V F. Aphids on the world’s crops. Chichester, United Kingdom: John Wiley & Sons, Ltd.; 1984. [Google Scholar]
- 9.Blackman R L, Eastop V F. Aphids on the world’s crops. Wallingford, United Kingdom: CAB International; 1994. [Google Scholar]
- 10.Bracho A M, Martinez-Torres D, Moya A, Latorre A. Discovery and molecular characterization of a plasmid localized in Buchnera sp. bacterial endosymbiont of the aphid Rhopalosiphum padi. J Mol Evol. 1995;41:67–73. doi: 10.1007/BF00174042. [DOI] [PubMed] [Google Scholar]
- 11.Brough C N, Dixon A F G. Ultrastructural features of egg development in oviparae of the vetch aphid, Megoura viciae Buckton. Tissue Cell. 1990;22:51–63. doi: 10.1016/0040-8166(90)90089-r. [DOI] [PubMed] [Google Scholar]
- 12.Clark M A, Baumann L, Baumann P. Sequence analysis of a 34.7-kb DNA segment from the genome of Buchnera aphidicola (endosymbiont of aphids) containing groEL, dnaA, the atp operon, gidA, and rho. Curr Microbiol. 1998;36:158–163. doi: 10.1007/pl00006760. [DOI] [PubMed] [Google Scholar]
- 13.Crawford I P. Evolution of a biosynthetic pathway: the tryptophan paradigm. Annu Rev Microbiol. 1989;43:567–600. doi: 10.1146/annurev.mi.43.100189.003031. [DOI] [PubMed] [Google Scholar]
- 14.Dadd R H. Nutrition: organisms. In: Kerkut G A, Gilbert LI, editors. Comprehensive insect physiology, biochemistry and pharmacology. Vol. 4. Elmsford, N.Y: Pergamon Press; 1985. pp. 315–319. [Google Scholar]
- 15.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Douglas A E. Mycetocyte symbiosis in insects. Biol Rev. 1989;69:409–434. doi: 10.1111/j.1469-185x.1989.tb00682.x. [DOI] [PubMed] [Google Scholar]
- 17.Douglas A E. Nutritional interactions between Myzus persicae and its symbionts. In: Campbell R K, Eikenbary R D, editors. Aphid-plant genotype interactions. Amsterdam, The Netherlands: Elsevier Biomedical Press; 1990. pp. 319–327. [Google Scholar]
- 18.Douglas A E. The nutritional quality of phloem sap utilized by natural aphid populations. Ecol Entomol. 1993;18:31–38. [Google Scholar]
- 19.Douglas A E. Reproductive failure and the amino acid pools in pea aphids (Acyrthosiphon pisum) lacking symbiotic bacteria. J Insect Physiol. 1996;42:247–255. [Google Scholar]
- 20.Douglas A E. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol. 1998;43:17–37. doi: 10.1146/annurev.ento.43.1.17. [DOI] [PubMed] [Google Scholar]
- 21.Douglas A E, Prosser W A. Synthesis of the essential amino acid tryptophan in pea aphid (Acyrthosiphon pisum symbiosis. J Insect Physiol. 1992;38:565–568. [Google Scholar]
- 22.Febvay G, Liadouze I, Guillaud J, Bonnet G. Analysis of energetic amino acid metabolism in Acrythosiphon pisum—a multidimensional approach to amino acid metabolism in aphids. Arch Insect Biochem Physiol. 1995;29:45–69. [Google Scholar]
- 23.Felsenstein J. Phylogenetic Inference Package (PHILIP) version 3.5. Seattle: University of Washington; 1993. [Google Scholar]
- 24.Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 1998;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- 25.Guhathakurtta A, Viney I, Summers D. Accessory proteins impose site selectivity during ColE1 dimer resolution. Mol Microbiol. 1996;20:613–620. doi: 10.1046/j.1365-2958.1996.5471072.x. [DOI] [PubMed] [Google Scholar]
- 26.Hinde R. Maintenance of aphid cells and the intracellular symbiotes of aphids in vitro. J Invertebr Pathol. 1971;17:333–338. [Google Scholar]
- 27.Houk E J, Griffiths G W. Intracellular symbiotes of the Homoptera. Annu Rev Entomol. 1980;24:161–187. [Google Scholar]
- 28.Ishikawa H. Nucleotide composition and kinetic complexity of the genomic DNA of an intracellular symbiont of the pea aphid Acyrthosiphon pisum. J Mol Evol. 1987;24:205–211. [Google Scholar]
- 29.Kornberg A, Baker T A. DNA replication. 2nd ed. New York, N.Y: W. H. Freemann & Co.; 1992. [Google Scholar]
- 30.Lai C-Y, Baumann L, Baumann P. Amplification of trpEG: adaptation of Buchnera aphidicola to an endosymbiotic association with aphids. Proc Natl Acad Sci USA. 1994;91:3819–3823. doi: 10.1073/pnas.91.9.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai C-Y, Baumann P, Moran N A. Genetics of the tryptophan biosynthetic pathway of the prokaryotic endosymbiont (Buchnera) of the aphid Schlechtendalia chinensis. Insect Mol Biol. 1995;4:47–59. doi: 10.1111/j.1365-2583.1995.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 32.Lai C-Y, Baumann P, Moran N A. The endosymbiont (Buchnera sp.) of the aphid Diuraphis noxia contains plasmids consisting of trpEG and tandem repeats of trpEG pseudogenes. Appl Environ Microbiol. 1996;62:332–339. doi: 10.1128/aem.62.2.332-339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lisser S, Margalit H. Compilation of E. coli mRNA promoter sequences. Nucleic Acids Res. 1993;21:1507–1516. doi: 10.1093/nar/21.7.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Llanes C, Gabant P, Couturier M, Michel-Briand Y. Cloning and characterization of the Inc A/C plasmid RA1 replicon. J Bacteriol. 1994;176:3403–3407. doi: 10.1128/jb.176.11.3403-3407.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Llanes C, Gabant P, Couturier M, Bayer L, Plesiat P. Molecular analysis of the replication elements of the broad-host-range RepA/C replicon. Plasmid. 1996;36:26–35. doi: 10.1006/plas.1996.0028. [DOI] [PubMed] [Google Scholar]
- 36.Marchuk D, Drumm M, Saulino A, Collins F S. Construction of T-vectors, a rapid general system for direct cloning of unmodified PCR products. Nucleic Acids Res. 1992;19:1154. doi: 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moran N A, Munson M A, Baumann P, Ishikawa H. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc R Soc London Ser B. 1993;243:167–171. [Google Scholar]
- 38.Munson M A, Baumann P. Molecular cloning and nucleotide sequence analysis of a putative trpDC(F)BA operon in Buchnera aphidicola (endosymbiont of the aphid Schizaphis graminum) J Bacteriol. 1993;173:6321–6324. doi: 10.1128/jb.175.20.6426-6432.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munson M A, Baumann P, Clark M A, Baumann L, Moran N A. Evidence for the establishment of aphid-eubacterium endosymbiosis in an ancestor of four aphid families. J Bacteriol. 1991;173:6321–6324. doi: 10.1128/jb.173.20.6321-6324.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nieto C, Giraldo R, Fernandez-Tresguerres E, Diaz R. Genetic and functional analysis of the basic replicon of pPS10, a plasmid specific for Pseudomonas isolated from Pseudomonas synrigae pathovar savastanoi. J Mol Biol. 1992;223:415–426. doi: 10.1016/0022-2836(92)90661-3. [DOI] [PubMed] [Google Scholar]
- 41.Raven J A. Phytophages of xylem and phloem: a comparison of plant sap-feeders. Adv Ecol Res. 1983;13:136–234. [Google Scholar]
- 42.Rouhbakhsh D, Lai C-Y, von Dohlen C, Clark M A, Baumann L, Baumann P, Moran N A, Voegtlin D J. the tryptophan biosynthetic pathway of aphid endosymbionts (Buchnera): genetics and evolution of plasmid-associated anthranilate synthase (trpEG) within the Aphididae. J Mol Evol. 1996;42:414–421. doi: 10.1007/BF02498635. [DOI] [PubMed] [Google Scholar]
- 43.Rouhbakhsh D, Clark M A, Baumann L, Moran N A, Baumann P. Evolution of the tryptophan biosynthetic pathway in Buchnera (aphid endosymbionts): studies of plasmid-associated trpEG within the genus Uroleucon. Mol Phylogenet Evol. 1997;8:167–176. doi: 10.1006/mpev.1997.0419. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 45.Sasaki T, Hayashi H, Ishikawa H. Growth and reproduction of symbiotic and aposymbiotic pea aphids, Acyrtosiphon pisum, maintained on artificial diets. J Insect Physiol. 1991;37:85–92. [Google Scholar]
- 46.Summers D K. The kinetics of plasmid loss. Trends Biotechnol. 1991;9:273–278. doi: 10.1016/0167-7799(91)90089-z. [DOI] [PubMed] [Google Scholar]
- 47.Untermann B M, Baumann P, McLean D L. Pea aphid symbiont relationships established by analysis of 16S rRNAs. J Bacteriol. 1989;171:2970–2974. doi: 10.1128/jb.171.6.2970-2974.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Ham, R. C. H. J., and A. Latorre. Unpublished data.
- 49.van Ham R C H J, Moya A, Latorre A. Putative evolutionary origin of plasmids carrying the genes involved in leucine biosynthesis in Buchnera aphidicola (endosymbiont of aphids) J Bacteriol. 1997;179:4768–4777. doi: 10.1128/jb.179.15.4768-4777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viswanathan V K, Green J M, Nichols B P. Kinetic characterization of 4-amino-4-deoxychorismate synthase from Escherichia coli. J Bacteriol. 1995;177:5918–5923. doi: 10.1128/jb.177.20.5918-5923.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Williams D R, Thomas C M. Active partitioning of bacterial plasmids. J Gen Microbiol. 1992;138:1–16. doi: 10.1099/00221287-138-1-1. [DOI] [PubMed] [Google Scholar]