Abstract
Five‐fluorouracil (5‐FU) is a chemotherapeutic agent that is mainly metabolized by the rate‐limiting enzyme dihydropyrimidine dehydrogenase (DPD). The DPD enzyme activity deficiency involves a wide range of severities. Previous studies have demonstrated the effect of a DPYD single nucleotide polymorphism on 5‐FU efficacy and highlighted the importance of studying such genes for cancer treatment. Common polymorphisms of DPYD in European ancestry populations are less frequently present in Koreans. DPD is also responsible for the conversion of endogenous uracil (U) into dihydrouracil (DHU). We quantified U and DHU in plasma samples of healthy male Korean subjects, and samples were classified into two groups based on DHU/U ratio. The calculated DHU/U ratios ranged from 0.52 to 7.12, and the two groups were classified into the 10th percentile and 90th percentile for untargeted metabolomics analysis using liquid chromatography‐quantitative time‐of‐flight‐mass spectrometry. A total of 4440 compounds were detected and filtered out based on a coefficient of variation below 30%. Our results revealed that six metabolites differed significantly between the high activity group and low activity group (false discovery rate q‐value < 0.05). Uridine was significantly higher in the low DPD activity group and is a precursor of U involved in pyrimidine metabolism; therefore, we speculated that DPD deficiency can influence uridine levels in plasma. Furthermore, the cutoff values for detecting DPD deficient patients from previous studies were unsuitable for Koreans. Our metabolomics approach is the first study that reported the DHU/U ratio distribution in healthy Korean subjects and identified a new biomarker of DPD deficiency.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Dihydropyrimidine dehydrogenase (DPD) deficiency can cause severe 5‐fluorouracil toxicity. To predict DPD activity, DPYD genotyping and DPD phenotyping using dihydrouracil and uracil ratio were used.
WHAT QUESTION DID THIS STUDY ADDRESS?
This study aimed to identify the distribution of DPD activity in a Korean population and explore new biomarkers of DPD activity using untargeted metabolomics.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
This study recognized different distributions of DPD activity between a Korean population and populations of other ethnicities. The cutoff values for activity deficiency based on other ethnic groups could not be considered for East Asian populations, including Korean populations.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
This study indicated that a new cutoff value should be further validated to overcome the variability in DPD phenotyping in East Asian populations. The new biomarker for DPD deficiency uridine has the potential for more feasible and convenient prediction analysis.
INTRODUCTION
Dihydropyrimidine dehydrogenase (DPD) is the rate‐limiting enzyme that catabolizes 5‐fluorouracil (5‐FU). 1 DPD plays a pivotal role in that over 80% of 5‐FU is degraded by DPD in the liver, and a deficiency in DPD activity causes severe toxicity. 2 DPYD is a DPD encoding gene, and its variant alleles are related to DPD activity. 2 Previous studies have suggested DPYD single nucleotide polymorphisms to predict drug toxicities prior to the administration of drugs to patients who were heterozygous or homozygous for a mutant DPYD allele. 2 In addition, the Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines have updated the guided dosing of fluoropyrimidine. 3 The distinguished DPYD variant c.190511G>A (*2A) is recognized to have the strongest impact on DPD activity. 4 DPYD*2A genotype‐guided dosing has improved the safety and cost savings of fluoropyrimidine therapy. 5 However, common polymorphisms of DPYD, such as DPYD*2A and DPYD*13 in European ancestry populations and other populations, are less frequently present in East Asian populations. 6 , 7 Considering that common polymorphisms of DPYD*2A and DPYD*13 hardly exist in East Asian populations, two novel DPYD genetic variants, −832 G>A and −131 C>A, were reported in a previous study and have been associated with enzyme activity in a Korean population. 7 Individuals with higher DPD enzyme activity possessed genetic variant, –832 G>A and DPD enzyme activity was reduced in individuals with genetic variant, −131 C>A. 7
Over the years, diverse strategies for DPD phenotyping methods have been studied and discussed for predicting 5‐FU‐induced toxicity. 8 One of the methods to determine DPD deficiency is measuring DPD enzyme activity in peripheral blood mononuclear cells (PBMCs). 9 A previous study showed DPD activity in PBMCs is highly correlated with liver DPD activity in patients. 10 Besides, it has high sensitivity to distinguish enzyme activity; however, DPD activity in PBMCs highly fluctuates in both healthy volunteers and patients. 8 , 11 The DPD enzyme also catabolizes endogenous uracil (U) and thymine (T) to dihydrouracil (DHU) and dihydrothymine (DHT). 12 Several studies have been reported based on the knowledge that DPD deficiency perhaps decreases the metabolic ratio of U, which results in a higher concentration of U in patients with severe toxicity of 5‐FU. 13 , 14 , 15 , 16 To predict the efficacy and toxicity of 5‐FU, DPD phenotyping using the metabolic ratio of U is useful to determine patients with DPD deficiency. 14 The endogenous metabolic ratio of U in plasma showed a correlation with the 5‐FU plasma concentration and clearance. 15
Global metabolomics profiling is an omics technology that provides information to understand individual variability in various physiological conditions and snapshots of entire physiology associated with the outcome phenotype. 17 More, metabolomics offers feasibility of biomarker discovery that metabolites act an important role in biological system. 18 Considering the low frequency of common DPYD polymorphism in the Korean population, the metabolic ratio of U might be different between Korean and other ethnicities, and a new biomarker would yield the more feasible and practical application of DPD deficiency screening. Therefore, the aim of this study was to evaluate the metabolic ratio of U as a predictive marker of DPD enzyme activity in a Korean population and explore a new biomarker of DPD activity deficiency using an untargeted metabolomics tool. To the best of our knowledge, this is the first study analyzing the metabolic ratio of U in a Korean population and provides a new marker of DPD deficiency using untargeted metabolomics.
METHODS
Study design and sample collection
Healthy subject plasma was provided by the Blood Bank in Seoul National University Hospital and stored at −70°C until use. The study protocol was approved by the Institutional Review Board of the Seoul National University Hospital (study identification number: H‐1608‐170‐789). Subjects were enrolled from 2015 to 2016 and gave written informed consent. The average age of healthy subjects was 30 ± 2.3 years. A total of 511 samples collected in a fasted state from healthy Korean male subjects were included for this targeted analysis. For untargeted analysis, the 90th percentile and 10th percentile groups consisted of 51 subjects according to their metabolic ratio of U and classified into low and high DPD activity groups. To clarify the results, quantitation of a significant marker was performed using 507 out of 511 plasma samples. The workflow of this study was summarized in Figure 1.
FIGURE 1.
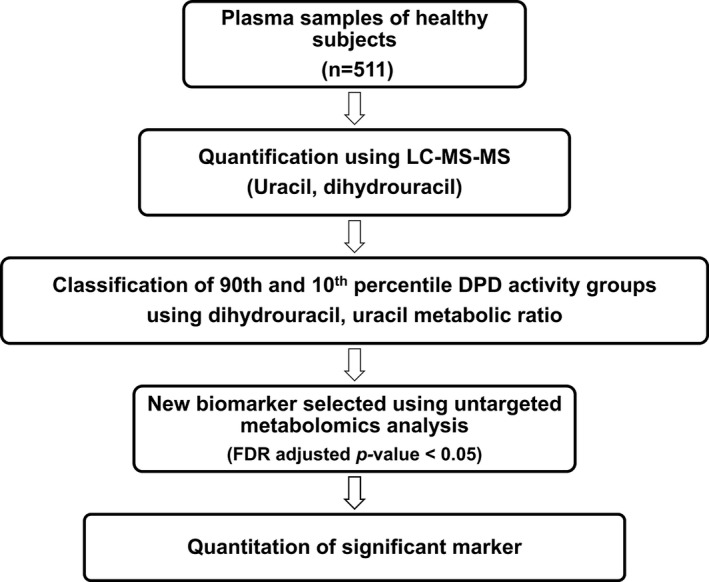
Study work flow. DPD, dihydropyrimidine dehydrogenase; FDR, false discovery rate; LC‐MS‐MS, liquid chromatography tandem mass spectrometry
Targeted metabolomics analysis for U and T
Sample preparation for U, DHU, T, and DHT quantitation
Standard stock solutions of U, DHU, T, and DHT, internal standard (IS), and quality controls (QCs) were prepared separately and diluted with 50% methanol. Thymine‐d4 was used as the IS. The IS (50 ng/ml) was added to 300 µl of plasma sample. Plasma proteins were precipitated with 1 ml of ethyl acetate. The mixture was vortexed for 1 min and centrifuged at 12,000 g for 10 min. Then, the supernatant was evaporated under nitrogen gas. Dried samples were reconstituted using 100 µl of 50% methanol. Calibration curve concentrations ranged from 2 to 650 ng/ml for U and from 1 to 650 ng/ml for T. For DHU and DHT, the calibration concentrations ranged from 2 to 6500 ng/ml.
Sample preparation for uridine quantitation
Standard stock solutions of uridine, IS, and QCs were prepared separately and diluted with 50% methanol. Uridine‐2‐13C‐1,3‐15N2 was used as the IS. The IS (1 µg/ml) was added to 50 µl of plasma sample. Plasma proteins were precipitated with 500 µl of acetonitrile. The mixture was vortexed for 1 min and centrifuged at 14,000 g for 10 min. Then, the supernatant was transferred to an autosampler vial. Calibration curve concentrations ranged from 100 to 10,000 ng/ml for uridine.
Liquid chromatography and mass spectrometry
Mass spectrometric analyses were performed using an AB SCIEX QTRAP 5500 instrument (AB SCIEX, Foster City, CA, USA) coupled with an ultrahigh‐performance liquid chromatography (UPLC) instrument (Waters, Milford, MA, USA). For U, T, DHU, and DHT quantification, one microliter of reconstituted sample was injected directly onto Synergi 4 µm Polar‐RP 80 A LC column (150 × 4.6 mm; Phenomenex, Torrance, CA, USA). The isocratic mobile phase consisted of high‐performance liquid chromatography (HPLC) grade water (solvent A) and acetonitrile (solvent B) at a flow rate of 0.6 ml/min. For uridine quantification, one microliter of supernatant was injected directly onto 2.1 mm × 100 mm ACQUITY 1.7 μm UPLC BEH C18 Column (Waters, USA). The isocratic mobile phase consisted of HPLC grade water with 0.1% formic acid (solvent A) and methanol with 0.1% formic acid (solvent B) at a flow rate of 0.3 ml/min. Mass spectrometric detection was conducted in negative electrospray ionization mode. Calibration curves were generated using linear regression after plotting the peak areas against the analyte concentrations within a dynamic range.
Untargeted metabolomics analysis
Sample preparation
The 70 µl of plasma sample was mixed with 280 µl of pre‐chilled 1:1 acetonitrile/methanol (v/v) and vortexed briefly. The mixture was centrifuged at 14,000 g for 10 min, and then 300 µl of supernatant dried under nitrogen gas. Dried samples were reconstituted using 400 µl of 80% acetonitrile.
Liquid chromatography and mass spectrometry
Untargeted metabolomics was performed by a Waters SYNAPT G2‐S time of flight platform (Waters, USA). A 4 µl aliquot of each reconstituted sample was injected into a Waters UPLC system with a 2.1 mm × 100 mm ACQUITY 1.7 μm BEH HILIC column. The mobile phase was composed of 0.1% formic acid (solvent A) and methanol containing 0.1% formic acid (solvent B). The gradient was as follows: 0–1.5 min, 99% B; 1.50–11.50 min, 50% B; and the equilibrium time was 6.5 min. Both positive and negative ionization modes were operated. Pooled QC replicates were analyzed with samples for consistency and stability.
Data processing
Metabolomic data were imported into EZinfo 2.0 software (Umetrics, Umea, Sweden) for multivariate analysis. Progenesis QI software (version 2.3; Nonlinear Dynamics, Newcastle, UK) was used for peak alignment, peak picking, deconvolution, normalization, and statistical analysis. Peak alignment and peak picking was processed by pooled QC replicates. Most suitable reference was selected to normalize all detected ions. A quantitative abundance ratio between the run and the reference run was calculated to normalize compounds. Metabolic features were filtered with a coefficient of variation (CV) of abundance greater than 30% in the QCs. Significantly different features were selected with a false discovery rate adjusted p value (q‐value) less than 0.05. The volcano plot was conducted with log2 transformed data using GraphPad Prism software version 7.0 (GraphPad Software Inc., San Diego, CA, USA).
RESULTS
U and DHU quantification and distribution
Plasma samples from a total of 511 healthy subjects were used for quantitation of U and DHU. The median and mean DHU/U ratios were 2.14 and 2.26, respectively. The median and mean DHT/T were 61.20 and 63.37, respectively (Figure S1). Our current data showed a weak correlation between the metabolic ratios of U and T, coinciding with those from a previous study in healthy controls and patients with cancer (Figure S2). 12 Samples were classified into high and low DPD activity groups according to the distribution of the metabolic ratio, corresponding to the 90th and 10th percentiles of metabolic ratio of U, respectively (Figure 2). The 90th percentile metabolic ratio of U ranged from 3.11 to 7.12. The 10th percentile metabolic ratio of U ranged from 0.52 to 1.571 (Figure 2). Both groups were subjected to untargeted metabolomics analysis using liquid chromatography‐quantitative time‐of‐flight‐mass spectrometry.
FIGURE 2.
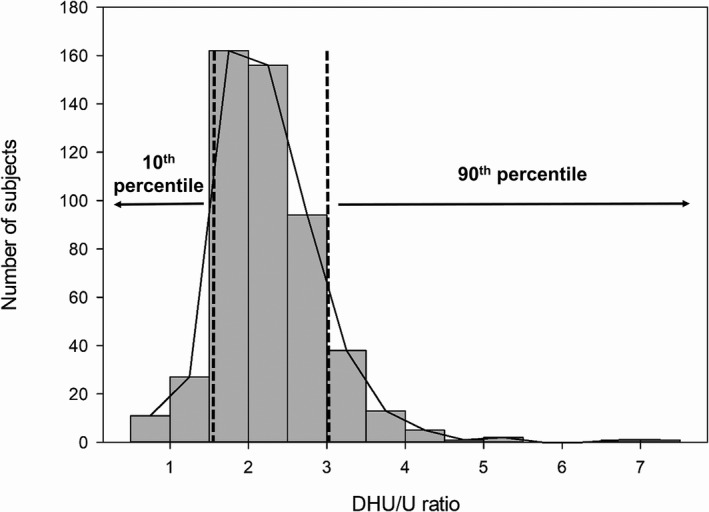
Distribution of DHU/U ratio in 511 healthy subjects. The 90th percentile and 10th percentile groups consisted of 51 subjects. The dashed line represents the threshold of each 90th percentile and 10th percentile. DHU/U, dihydrouracil/uracil
Plasma metabolic profiling using untargeted metabolomics
Untargeted global profiling of plasma was performed for samples in the 90th and 10th percentiles of the metabolic ratio of U. In total, 4440 compounds were detected and filtered out based on a CV below 30%. Principal component analysis displayed no clear separation between the high DPD activity group and the low DPD activity group (Figure 3a). Ions with a q‐value less than 0.05 between the two groups were selected. The volcano plot highlighted six metabolites that differed significantly between the high DPD activity group and the low DPD activity group (q‐value < 0.05; Figure 3b). One distinguishable marker was identified as uridine using authentic compound (Figure S3). However, the other five ions (m/z 419.3554, 249.9205, 855.5117, 864.5028, and 311.2943) were not identified.
FIGURE 3.
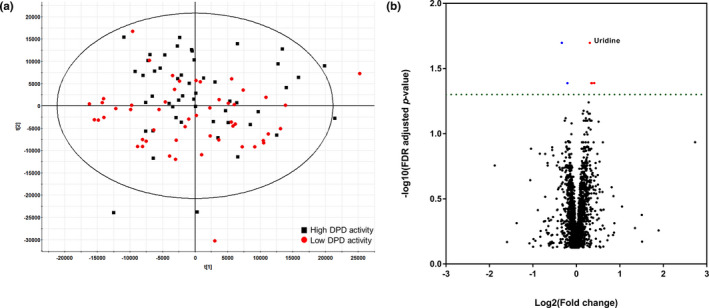
Multivariate analysis of plasma metabolomic profiles. (a) Principal component analysis (PCA) unsupervised clustering plots for the high DPD activity group (black square) and low DPD activity group (red circle). (b) The volcano plot shows t‐test results of samples from the high DPD activity group and low DPD activity group. Red dots are upregulated metabolites and blue dots are downregulated metabolites. The dashed horizontal line reflects the filtering area (FDR adjusted p value < 0.05). DPD, dihydropyrimidine dehydrogenase; FDR, false discovery rate
Depending on the metabolic ratio of U, uridine levels showed changes in plasma from healthy subjects
The uridine was significantly higher in the low DPD activity group than in the high activity group (Figure 4). The concentration of uridine showed a weak correlation with the calculated metabolic ratio of U and U concentrations (Figure S4). The plasma level of uridine showed alterations between the 90th and 10th percentiles of the metabolic ratio of U. Uridine is a precursor of U, and both metabolites are involved in pyrimidine metabolism. 19
FIGURE 4.
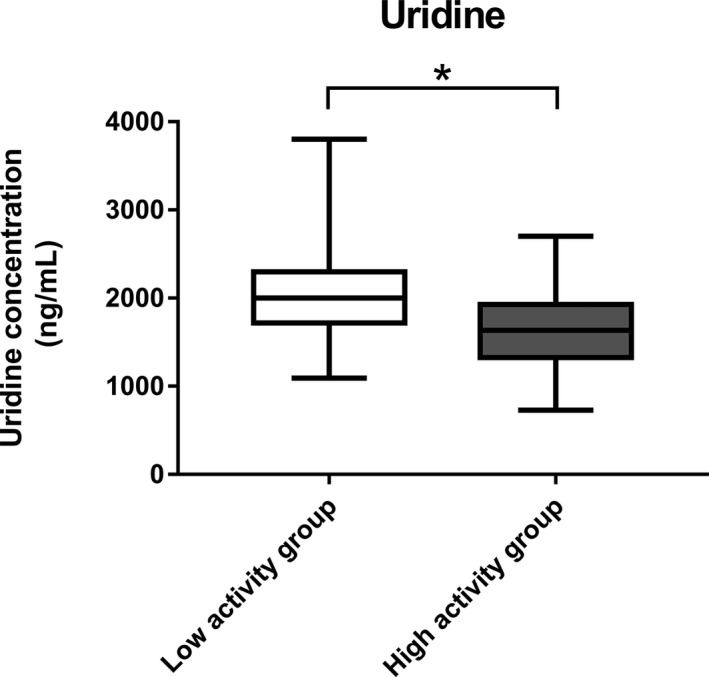
Box plot of uridine. This plot represents the concentration of uridine. A t‐test was used for statistical evaluation. * p value <0.0001
DISCUSSION
To our knowledge, this is the first study to evaluate the metabolic ratio of U in a Korean population. Although some studies reported an association between the metabolic ratio of U and DPD activity, 13 , 14 this study is the first to address a new biomarker of DPD activity deficiency using untargeted metabolomics. Our results indicated the following: (i) in the Korean population, the metabolic ratio of U indicating the prediction of 5‐FU toxicity shows dissimilar distribution with other populations; and (ii) our untargeted approach identified a new biomarker of DPD deficiency, uridine, in plasma.
To date, quantifying U and DHU is one of the DPD phenotyping methods for evaluating its activity. 8 Several studies have suggested the metabolic ratio of U measurement as a screening method for DPD deficiency. 13 , 14 , 16 , 20 Based on those studies, we categorized the low and high activity groups of DPD according to the percentile of the metabolic ratio of U (Figure 1). In healthy Korean male subjects, the distribution of the metabolic ratio of U ranged from 0.52 to 7.12; the median was 2.14. This range is lower than a previously reported range of 0.002 to 17.3, and a median was of 7.3 in 252 French White patients with colorectal carcinoma. 21 In addition, the metabolic ratio of U in healthy volunteers was previously reported to be 2.3 to 28.9, and the median was 11.2. 11 An interethnic difference in the metabolic ratio of U may be present in various populations.
In previous studies, cutoff values were assumed for a plasma ratio of 4.0 or 6.0 to predict if patients have severe 5‐FU toxicity in other ethnic groups. 14 , 16 , 20 A total of 501 of 511 (98%) samples in our Korean population had a DPD deficiency using the cutoff value of 4.0 and 509 of 511 (99.6%) Korean population had DPD deficiency using the cutoff value of 6.0. However, East Asians had lower rates of toxicity induced by fluoropyrimidine containing regimens than other populations. 22 , 23 Our data indicated that the cutoff value in a previous study with other races may be inappropriate to predict whether Korean patients will present severe toxicity. New cutoff values should be further explored and validated in East Asian populations.
We speculated that DPD activity deficiency can lead to the accumulation of uridine in pyrimidine metabolism (Figure 5). Uridine homeostasis is regulated uridine phosphorylase, an enzyme that converts uridine to U. 24 The uridine level was elevated in the low activity group, which showed that DPD deficiency affected the uridine catabolism pathway. Uridine homeostasis is linked to pyrimidine metabolism. 25 In a previous study, uridine levels showed an association with the metabolic ratio of U in plasma level and similar patterns with U. 26 This result is consistent with our results, which showed a weak correlation between uridine and U and the metabolic ratio of U (Figure S4). Although many studies have shown the effectiveness of the metabolic ratio of U, 13 , 14 , 20 from analytical perspectives, single compound analysis is simpler and more feasible than analysis of multiple compounds. Hence, uridine may be a new predictive marker of DPD deficiency.
FIGURE 5.
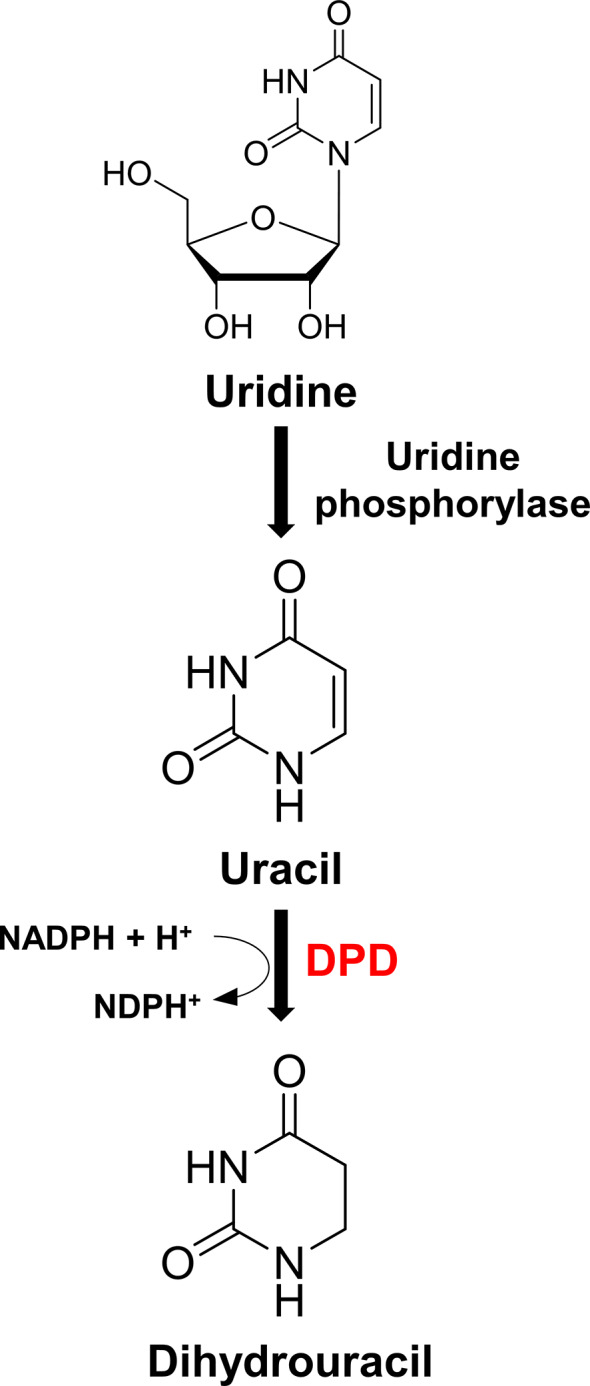
Uridine, uracil, and dihydrouracil in pyrimidine metabolism. Metabolomic analysis indicated that DPD activity deficiency can lead to the accumulation of uridine in pyrimidine metabolism. DPD, dihydropyrimidine dehydrogenase; NADPH, nicotinamide adenine dinucleotide phosphate
The main limitation of our study is the lack of evaluation of patient’s clinical outcomes and consideration of the DPYD genotype. Most studies have investigated the metabolic ratio of U in patients with cancer to predict toxicity related to a fluoropyrimidine or 5‐FU. 13 , 14 , 20 We recognized the distribution of the metabolic ratio of U in healthy Korean male subjects and divided low and high DPD activity groups using the metabolic ratio of U based on published studies. Additionally, there was the lack of female subjects in our study. However, a previous study showed no significant correlation between sex and DPD activity; the distribution of healthy Korean male subjects is well‐represented. 27 Nevertheless, this result is not the gold standard to detect DPD enzyme activity, and further studies should be conducted in patients who struggle with severe toxicity related to 5‐FU to confirm these findings.
In conclusion, this is the first large sample size study to show different distributions of DPD activity using quantification of U, DHU, T, and DHT in a Korean population. Beyond providing a range of metabolic ratios of U in a Korean population to support further investigation of new cutoff values, our study also explored a new biomarker, uridine, and, consequently, measuring pretreatment uridine levels may provide a feasible strategy for predicting DPD deficiency.
CONFLICT OF INTEREST
The authors declared no competing interests for this work.
AUTHOR CONTRIBUTIONS
J.K., S.H.L., and J.Y.C. wrote the manuscript. A.H.K., I.J.J., S.H.L., and J.Y.C. designed the research. J.K., I.J., and J.O. performed the research and analyzed the data.
Supporting information
Figure S1‐S4
ACKNOWLEDGEMENTS
Jihyun Kang received a scholarship from the BK21FOUR education program.
Kang J, Kim AH, Jeon I, et al. Endogenous metabolic markers for predicting the activity of dihydropyrimidine dehydrogenase. Clin Transl Sci. 2022;15:1104‐1111. doi: 10.1111/cts.13203
SeungHwan Lee and Joo‐Youn Cho have equally contributed to this work and are co‐corresponding authors.
Funding information
No funding was received for this work.
Contributor Information
SeungHwan Lee, Email: leejh413@snu.ac.kr.
Joo‐Youn Cho, Email: joocho@snu.ac.kr.
REFERENCES
- 1. van Kuilenburg AB, Haasjes J, Richel DJ, et al. Clinical implications of dihydropyrimidine dehydrogenase (DPD) deficiency in patients with severe 5‐fluorouracil‐associated toxicity: identification of new mutations in the DPD gene. Clin Cancer Res. 2000;6:4705‐4712. [PubMed] [Google Scholar]
- 2. Omura K. Clinical implications of dihydropyrimidine dehydrogenase (DPD) activity in 5‐FU‐based chemotherapy: mutations in the DPD gene, and DPD inhibitory fluoropyrimidines. Int J Clin Oncol. 2003;8:132‐138. [DOI] [PubMed] [Google Scholar]
- 3. Amstutz U, Henricks LM, Offer SM, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for dihydropyrimidine dehydrogenase genotype and fluoropyrimidine dosing: 2017 update. Clin Pharmacol Ther. 2018;103:210‐216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nie Q, Shrestha S, Tapper EE, et al. Quantitative contribution of rs75017182 to dihydropyrimidine dehydrogenase mRNA splicing and enzyme activity. Clin Pharmacol Ther. 2017;102:662‐670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deenen MJ, Meulendijks D, Cats A, et al. Upfront genotyping of DPYD*2A to individualize fluoropyrimidine therapy: a safety and cost analysis. J Clin Oncol. 2016;34:227‐234. [DOI] [PubMed] [Google Scholar]
- 6. Khushman M, Patel GK, Hosein PJ, et al. Germline pharmacogenomics of DPYD*9A (c.85T>C) variant in patients with gastrointestinal malignancies treated with fluoropyrimidines. J Gastrointest Oncol. 2018;9:416‐424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shin J‐G, Kang TS, Cheong HS, et al. Determination of DPYD enzyme activity in Korean population. Ther Drug Monit. 2015;37:147‐151. [DOI] [PubMed] [Google Scholar]
- 8. Knikman JE, Gelderblom H, Beijnen JH, Cats A, Guchelaar H‐J, Henricks LM. Individualized dosing of fluoropyrimidine‐based chemotherapy to prevent severe fluoropyrimidine‐related toxicity: what are the options? Clin Pharmacol Ther. 2021;109:591‐604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chazal M, Etienne MC, Renée N, et al. Link between dihydropyrimidine dehydrogenase activity in peripheral blood mononuclear cells and liver. Clin Cancer Res. 1996;2:507‐510. [PubMed] [Google Scholar]
- 10. Lu Z, Zhang R, Diasio RB. Dihydropyrimidine dehydrogenase activity in human peripheral blood mononuclear cells and liver: population characteristics, newly identified deficient patients, and clinical implication in 5‐fluorouracil chemotherapy. Cancer Res. 1993;53:5433‐5438. [PubMed] [Google Scholar]
- 11. Sistonen J, Büchel B, Froehlich TK, et al. Predicting 5‐fluorouracil toxicity: DPD genotype and 5,6‐dihydrouracil:uracil ratio. Pharmacogenomics. 2014;15:1653‐1666. [DOI] [PubMed] [Google Scholar]
- 12. Kuilenburg ABP, Meijer J, Tanck MWT, et al. Phenotypic and clinical implications of variants in the dihydropyrimidine dehydrogenase gene. Biochim Biophys Acta. 2016;1862:754‐762. [DOI] [PubMed] [Google Scholar]
- 13. Neto OV, Raymundo S, Franzoi MA, et al. DPD functional tests in plasma, fresh saliva and dried saliva samples as predictors of 5‐fluorouracil exposure and occurrence of drug‐related severe toxicity. Clin Biochem. 2018;56:18‐25. [DOI] [PubMed] [Google Scholar]
- 14. Launay M, Dahan L, Duval M, et al. Beating the odds: efficacy and toxicity of dihydropyrimidine dehydrogenase‐driven adaptive dosing of 5‐FU in patients with digestive cancer. Br J Clin Pharmacol. 2016;81:124‐130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gamelin E, Boisdron‐Celle M, Guérin‐Meyer V, et al. Correlation between uracil and dihydrouracil plasma ratio, fluorouracil (5‐FU) pharmacokinetic parameters, and tolerance in patients with advanced colorectal cancer: A potential interest for predicting 5‐FU toxicity and determining optimal 5‐FU dosage. J Clin Oncol. 1999;17:1105. [DOI] [PubMed] [Google Scholar]
- 16. Thomas F, Hennebelle I, Delmas C, et al. Genotyping of a family with a novel deleterious DPYD mutation supports the pretherapeutic screening of DPD deficiency with dihydrouracil/uracil ratio. Clin Pharmacol Ther. 2016;99:235‐242. [DOI] [PubMed] [Google Scholar]
- 17. Clish CB. Metabolomics: an emerging but powerful tool for precision medicine. Cold Spring Harb Mol Case Stud. 2015;1: a000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Monteiro MS, Carvalho M, Bastos ML, Guedes de Pinho P. Metabolomics analysis for biomarker discovery: advances and challenges. Curr Med Chem. 2013;20:257‐271. [DOI] [PubMed] [Google Scholar]
- 19. Olivares J, Dubus I, Barrieux A, Samuel JL, Rappaport L, Rossi A. Pyrimidine nucleotide synthesis is preferentially supplied by exogenous cytidine in adult rat cultured cardiomyocytes. J Mol Cell Cardiol. 1992;24:1349‐1359. [DOI] [PubMed] [Google Scholar]
- 20. Kristensen MH, Pedersen P, Mejer J. The value of dihydrouracil/uracil plasma ratios in predicting 5‐fluorouracil‐related toxicity in colorectal cancer patients. J Int Med Res. 2010;38:1313‐1323. [DOI] [PubMed] [Google Scholar]
- 21. Boisdron‐Celle M, Remaud G, Traore S, et al. 5‐Fluorouracil‐related severe toxicity: a comparison of different methods for the pretherapeutic detection of dihydropyrimidine dehydrogenase deficiency. Cancer Lett. 2007;249:271‐282. [DOI] [PubMed] [Google Scholar]
- 22. Haller DG, Cassidy J, Clarke SJ, et al. Potential regional differences for the tolerability profiles of fluoropyrimidines. J Clin Oncol. 2008;26:2118‐2123. [DOI] [PubMed] [Google Scholar]
- 23. Loh M, Chua D, Yao Y, et al. Can population differences in chemotherapy outcomes be inferred from differences in pharmacogenetic frequencies? Pharmacogenomics J. 2013;13:423‐429. [DOI] [PubMed] [Google Scholar]
- 24. Cao D, Pizzorno G. Uridine phosophorylase: an important enzyme in pyrimidine metabolism and fluoropyrimidine activation. Drugs Today. 2004;40:431‐443. [DOI] [PubMed] [Google Scholar]
- 25. Le TT, Ziemba A, Urasaki Y, et al. Disruption of uridine homeostasis links liver pyrimidine metabolism to lipid accumulation. J Lipid Res. 2013;54:1044‐1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Henricks LM, Jacobs BAW, Meulendijks D, et al. Food‐effect study on uracil and dihydrouracil plasma levels as marker for dihydropyrimidine dehydrogenase activity in human volunteers. Br J Clin Pharmacol. 2018;84:2761‐2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Coenen MJH, Paulussen ADC, Breuer M, et al. Evolution of dihydropyrimidine dehydrogenase diagnostic testing in a single center during an 8‐year period of time. Curr Ther Res Clin Exp. 2019;90:1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1‐S4


