Abstract
The roles that natural killer (NK) cells play in liver disease and transplantation remain ill‐defined. Reports on the matter are often contradictory, and the mechanisms elucidated are complex and dependent on the context of the model tested. Moreover, NK cell attributes, such as receptor protein expression and function differ among species, make study of primate or rodent transplant models challenging. Recent insights into NK function and NK‐mediated therapy in the context of cancer therapy may prove applicable to transplantation. Of specific interest are immune checkpoint molecules and the mechanisms by which they modulate NK cells in the tumor micro‐environment. In this review, we summarize NK cell populations in the peripheral blood and liver, and we explore the data regarding the expression and function of immune checkpoint molecules on NK cells. We also hypothesize about the roles they could play in liver transplantation and discuss how they might be harnessed therapeutically in transplant sciences.
INTRODUCTION
Natural killer (NK) cells play a significant, yet often overlooked role in promoting graft tolerance during liver transplantation. Donor NK cells that reside within graft tissue mediate tolerance through the direct killing of recipient alloreactive T cells, which might otherwise promote graft rejection. The potential role that enhanced donor NK cells could have on promoting graft tolerance is largely unexplored, due to most established transplant therapies inhibiting immune function rather than enhancing it. Immune checkpoint inhibitors are an emerging therapy developed in the context of cancer. If these same drugs were instead administered to graft NK cells, they might promote graft tolerance through heightened NK‐mediated killing of alloreactive T cells.
INTRODUCTION TO NATURAL KILLER CELL BIOLOGY
NK cells are cytotoxic innate immune cells of the lymphoid lineage. 1 , 2 NK cells play a central role in the recognition and killing of virus‐infected cells 3 , 4 and tumor cells. 5 , 6 , 7 NK cells mediate their cytotoxic function through the secretion of cytotoxic granules, which contain perforin, granzymes, and other cytotoxic proteins. 8 In contrast to the lymphoid B and T cells, NK cells express germline‐encoded receptors to mediate their cytotoxic function. NK cell cytotoxic function is governed by the net of activating and inhibitory signals it receives through various receptors expressed on the cell surface. 9 , 10
In human peripheral blood, NK cells have traditionally been divided into two major subpopulations marked by the expression level of the CD56 and CD16 surface receptors. High expression of the CD56 surface receptor (CD56brightCD16−) display strong cytokine secretion, but weak cytotoxic activity with lower amounts of intracellular perforin and granzyme A and B. 11 , 12 , 13 This subset is enriched in lymph nodes, and is less frequent in peripheral blood. 14 Conversely, CD56dimCD16+ NK cells have stronger cytotoxic activity and are the most abundant NK cell detected in the peripheral blood. 15 Although differentially expressed, the precise molecular functions of CD56 on NK cells remains elusive. It is not thought to be play a significant role in any major NK‐mediated effector functions, cytotoxic functions, or tolerance mechanisms. CD16 is a marker of NK maturity and cytotoxic function; it is the fragment crystallizable (Fc) gamma receptor and mediates the NK cell’s antibody‐dependent cellular cytotoxicity. Recent investigations have revealed the presence of other subpopulations of NK cells, including tissue‐resident NK cells (trNK) present in the uterus, 16 lungs, 17 and liver. 13 Human NK cells have also been observed to differentiate into adaptive/memory‐like phenotypes in response to various stimuli, which have been described and reviewed elsewhere. 18 , 19 , 20
NK CELLS MEDIATE ACTIVATING AND INHIBITORY FUNCTION THROUGH GERMLINE‐ENCODED SURFACE RECEPTORS
The interplay among activating and inhibitory NK cell surface receptors is diverse and still being defined. 21 Many receptors belong to either the immunoglobulin‐like superfamily of proteins or the C‐type lectin superfamily. The killer immunoglobulin‐like receptor (KIR) genes are in the leukocyte receptor complex (LRC) on chromosome 19, 22 whereas the C‐type lectin superfamily is located in the natural killer gene complex (NKC) on chromosome 12. 23 These member receptors mediate activating or inhibitory phenotypes through conserved motifs known as immunoreceptor tyrosine‐based inhibitory motifs (ITIMs) or immunoreceptor tyrosine‐based activating motifs (ITAMs). 21 The ITAMs are typically found within constitutively expressed transmembrane adaptor proteins, such as DAP‐12, the common Fc receptor gamma chain (FcRγ), and the CD3‐ζ chain. 21 These are recruited to charged lysine residues within the transmembrane portion of activating receptors upon ligand binding. Examples of these activating receptors include natural cytotoxicity receptors (NCRs), most Fc receptors, natural killer group 2D (NKG2D), and activating KIRs (aKIRs). 21 On the other hand, ITIMs are typically found within the cytoplasmic tails of the inhibitory receptors. Examples include inhibitory killer cell immunoglobulin‐like proteins, NKG2A, and programmed cell death protein 1 (PD‐1). 24
As stated above, most KIRs are members of the immunoglobulin superfamily on the LRC locus. 25 These receptors mediate both activating and inhibitory function. The inhibitory receptors within this family bind to major histocompatibility complex (MHC) class I molecules (such as human leukocyte antigens [HLA]‐A, ‐B and ‐C) and thus provide the machinery required for NK education of “self” versus “non‐self” recognition. 26 , 27 Activating KIRs possess a much lower binding affinity for HLA ligands and are expressed at a lower frequency compared to their inhibitory counterparts. 28 Some activating KIRs have been suggested to play a role in protection against chronic viral infection. 29 , 30
The C‐type lectin superfamily of NK cell receptors includes the killer‐cell lectin‐like receptors (KLR). 31 The KLR are both activating and inhibitory receptors and are located at the NKC locus on chromosome 12. KLRs can be further divided into separate subfamilies based on ligand interaction. 31 KLRs that bind ligands with an MHC class I‐like fold include the CD94/NKG2 family of receptors. 32 The activating receptor NKG2D is the most prominent member of this subfamily, due to its central role in infection and tumor surveillance and clearing. 33 Unlike NKG2D, natural killer group 2A (NKG2A) and natural killer group 2C (NKG2C) dimerize with CD94 to form either the inhibitory CD94/NKG2A heterodimer or the activating CD94/NKG2C heterodimer, both of which bind major histocompatibility complex, class I‐E (HLA‐E). 34 , 35 Not unlike their KIR counterparts, NKG2A contains an ITIM in its cytoplasmic tail, and NKG2C contains a charged residue in its tail that recruits DAP‐12 36 (Figure 1).
FIGURE 1.
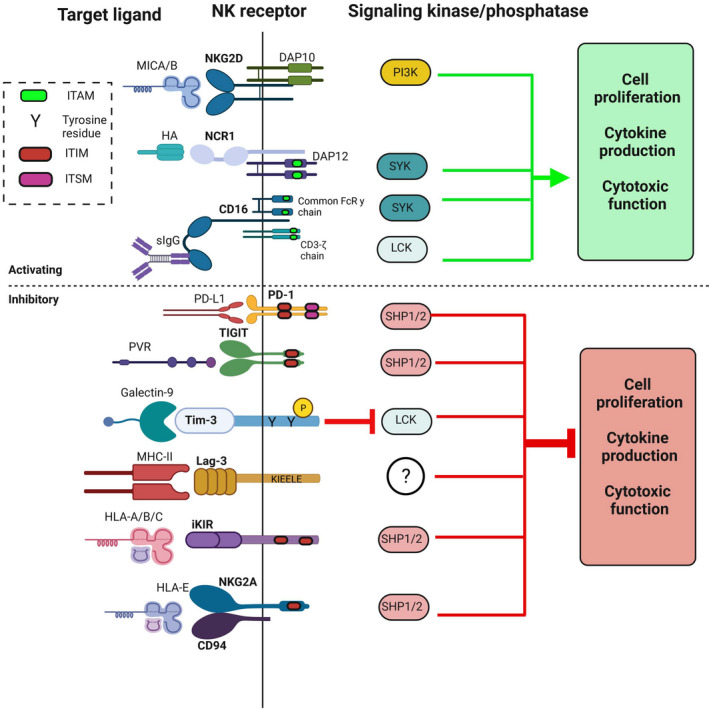
A summary of critical NK cell receptors and checkpoint inhibitors, their cognate ligands, and intracellular signaling cascade. In general, activating receptors will increase cell proliferation and cytokine production and function
HEPATIC TISSUE‐RESIDENT NK CELLS ARE PHENOTYPICALLY AND FUNCTIONALLY DISTINCT FROM PERIPHERAL NK CELLS
NK cells make up ~10% of peripheral blood lymphocytes, but account for 30–50% of intrahepatic lymphocytes. 37 , 38 Hence, great effort has been put forth to understand their role in liver disease and transplantation. Although NK cells are the predominant hepatic lymphoid cell, two separate and phenotypically distinct NK cell populations within the liver have been identified. These include conventional NK cells traveling through the liver, and the hepatic trNK cells whose role in the liver microenvironment have been the topic of numerous investigations. 13
Conventional NK cells, which may coexist alongside trNK cells in the liver, are phenotypically similar peripheral circulating NK cells. Hepatic conventional NK cells are CD56dimCD16+, and functionally behave similar to other peripheral CD56dim NK populations with increased production of interferon‐gamma and increased cytotoxicity. 39 Originally, NK cells were thought to develop from hematopoietic stem cells in the bone marrow. However, later developmental stages are known to occur in secondary lymphoid tissues and other organs, 40 suggesting local development of certain NK populations and giving rise to the distinctiveness of the hepatic NK cells.
Peng and colleagues first described specialized hepatic trNK cells in the murine liver. 41 In mouse models, conventional peripheral NK cells express the murine‐specific marker DX5, but do not express CD49a− (a marker for a subunit of integrin alpha). In contrast, Peng and colleagues identified unique CD49a+DX5− cells which reside in the hepatic sinusoids, but not in the efferent or afferent hepatic blood supply. 41 These CD49a+DX5− liver resident NK cells are functionally different from their conventional counterpart as they exhibit unique memory‐like effects against antigens. CD49a+DX5− NK cells, which had previously been sensitized to antigen had a much more robust antigen contact hypersensitivity response when challenged as compared to the CD49a−DX5+ conventional NK cells. 42 The authors suggested that this memory response may reflect NK cell priming in the liver.
Detection of this unique subset of trNK cells in murine models led to investigation for comparable human liver trNK cells. Marquardt and colleagues identified CD3−CD49a+CD56+ intrahepatic NK cells that could be a human counterpart of the previously identified mouse trNK cells. 43 This human liver trNK cell subpopulation is CD56bright and lacks CD16 expression. However, the CD49+ trNK also express high levels of mostly inhibitory KIR and the activating receptor NKG2C, an expression pattern seen in conventional CD56dim NK cells and after viral infection. 44 Interestingly, high‐resolution KIR phenotyping of the trNK showed an oligoclonal expression pattern, suggesting a clonal‐like expansion of NK cells in this subset. Upon stimulation, these cells produced higher levels of proinflammatory cytokines, but lower levels of perforin with subsequent poor degranulation and cytotoxicity compared to conventional CD56dim NK cells. 43 This functional analysis is consistent with the murine hepatic trNK cells previously described and may reflect a population of human NK cells with adaptive or memory‐like capabilities.
Additional investigations have reported other phenotypic and functional descriptions of these human liver trNK cells. Hudspeth and colleagues note the CD49a+ trNK represents a fraction of the total trNK cell population. 45 CD49a−CD56brightCD16− intrahepatic NK cells have also been described that have high expression of surface C‐X‐C motif chemokine receptor 6 (CXCR6), a protein that interacts with C‐C motif chemokine ligand 16 (CCL16) expressed in liver sinusoids. 38 , 45 , 46 These CD49a−CD56bright NK cells are a population of hepatic trNK cells retained within hepatic sinusoids. In addition to CXCR6, this population also expresses the chemokine receptor C‐C motif chemokine receptor 5 (CCR5) and the tissue activation marker CD69, surface markers not expressed on conventional CD56dim NK cells. 45 This subset also displays enhanced degranulation and efficient IFN‐γ and tumor necrosis factor‐alpha (TNF‐α) production, and they could be key to inflammatory responses. However, they also express high levels of TNF‐related apoptosis‐inducing ligand (TRAIL) and can mediate the elimination of activated T cells, perhaps contributing to the more tolerant liver environment. 38 , 46
NATURAL KILLER CELLS PLAY VARIABLE ROLES IN LIVER TRANSPLANT TOLERANCE AND REJECTION
The overall role of NK cells in liver transplantation remains poorly understood. With reperfusion of the donor graft upon transplantation, donor hepatic CD56brightCD16− trNK and donor conventional NK cells are transferred to the recipient. These cells are notable for the increased expression of the T‐box transcription factor eomesodermin (Eomes) in nearly half of the hepatic trNK cells, separating them from the uniformly Eomeslo peripheral blood NK counterparts. 47 More than 95% of trNK expressed the tissue activation marker CD69, whereas samples of donor peripheral blood showed only 4–7% of all NK cell populations expressed CD69. 48 Of interest, donor Eomeshi hepatic trNK seem to be long‐lived, and can persist in an allograft for 13 years. Donor hepatic Eomeslo NK cells can enter the recipient’s circulation and are nearly undetectable more than a few years after liver transplantation. 47 These data would suggest that CD56brightCD16−Eomeshi NK cells may be the most “liver resident” NK cell population.
EomeshiCD56bright hepatic trNK cells express lower levels of KIR, whereas CD94 (part of the inhibitory coreceptor complex with NKG2A) is expressed at high levels. Additionally, levels of perforin and granzyme B are reduced, suggesting decreased cytotoxicity of this cell population. 47 On the other hand, the whole population of hepatic donor CD56bright NK cells when studied by Moroso and colleagues was more cytotoxic compared with their peripheral counterparts. 48 Together, this might suggest that the initial passenger CD56bright liver donor NK cells could attack the recipient’s infiltrating lymphocytes and prevent early graft rejection, but that longer‐lived, Eomeshi trNK from the donor would eventually contribute to a more tolerogenic milieu. 48
This more tolerogenic environment is well‐recognized within transplant medicine. An estimated 20% of liver transplant recipients wean off immunosuppressive medications completely, developing “operational tolerance” without development of graft rejection. 49 The cellular mechanisms which lead to operational tolerance are not well understood; however, NK cells may contribute to its development. 50 Notably, de la Garza and colleagues found a larger percentage of NK cells in peripheral blood samples in patients who developed operational tolerance as compared to those who went on to develop acute rejection following immunosuppression withdrawal. 51 Relatedly, Li and colleagues identified 13 genes which were highly expressed in operationally tolerant children and adults. All 13 genes were enriched in NK cell (CD56+) populations, suggesting NK cells contributions to tolerance. 52
Adding to these ideas, Pagano and colleagues found that NK cells (based on a CD3−CD56+ cell population) made up approximately one third of lymphocytes in the liver perfusate of deceased donors. The majority of these NK cells expressed activating markers, including NKG2D. 53 Of 46 donor liver perfusates analyzed, 11 recipients experienced an episode of acute cellular rejection. The patients who experienced acute cellular rejection showed a significantly lower percentage of NK cells in the liver perfusate (35% NK cells for non‐rejectors and 28% for rejectors). Having a smaller donor liver NK cell population could translate as decreased capability of donor NK cells to prevent recipient immune response against the graft, thus increasing the likelihood of recipient rejection of the allograft. 53
When considering NK cell activity in promoting graft tolerance versus rejection, one must consider the role of circulating recipient NK cells. Following graft transplant and restoration of blood flow through the graft, recipient NK cells are detected in the graft within hours. 54 T cell mediated rejection is well accepted as the primary form of liver graft rejection. 55 The innate immune system—specifically NK cells—may also participate in allograft rejection. Obara and colleagues identified the rapid recruitment of recipient NK cells to liver allograft in a murine model within 12 h of transplantation. 56 Upon graft infiltration, recipient NK cells produced pro‐inflammatory IFN‐γ and helped recruit T cells to the graft. When peripheral blood NK cells were depleted, allografts had statistically significant decreases in intrahepatic IFN‐γ expression and prolonged survival. 56
Understanding the role of circulating NK cells and graft infiltration in humans has been more challenging with conflicting reports between studies. Jamil and colleagues found an increase in peripheral CD56bright NK cells after transplant, but it is unclear if this increase was recipient or donor derived. 57 The CD56bright NK cells had decreased expression of activating receptors NKp30 and NKp46. This downregulation resulted in decreased NK functional capacities with impaired degranulation and IFN‐γ production. 57 If this same hypofunctional NK cell population trafficked from the recipient into the donor graft, then they could play a role in promoting graft tolerance. Alternatively, hypofunctional donor NK cells could perhaps allow increased activity against the allograft by recipient immune cells.
Distinct from the findings of Jamil and colleagues, Pham et al. noted a statistically significant decrease in the number total NK cells and also the proportion of conventional CD56dim NK cells circulating in peripheral blood post‐transplantation. 58 This decrease was transient and may reflect the effects of immunosuppressive medications versus trafficking of NK cells to the graft. In addition, whereas the total number of NK cells circulating peripherally decreased in the immediate post‐transplant period, those that did remain in the periphery showed higher levels of activating receptor NKp30 in both the CD56bright and CD56dim populations, contrasting with the findings described by Jamil and colleagues. 57
Further in vivo studies delineating NK cell phenotypic changes in response to the physiologic stress of transplantation, immunosuppressive medications, and the role of NK cells as a bridge between the innate and adaptive immune systems at the time of transplant and during acute cellular rejection are necessary. This will allow increased understanding of how NK cells might be harnessed to modulate the allograft immune response (Figure 2).
FIGURE 2.

Donor (a) and recipient (b) NK cell activity in liver transplantation tolerance and rejection. In theory, targeted inhibition of the PD‐1 pathway in donor NK cells could enhance the killing of alloreactive recipient immune cells and limit rejection (a). However, inhibiting immune checkpoints in recipient NK cells and other immune cell populations could augment the anti‐graft response (b)
IMMUNE CHECKPOINT MOLECULAR PATHWAYS ARE IMPORTANT TO TRANSPLANT TOLERANCE
Many immunosuppressive medications were developed within the context of the “two signal” model of T cell activation. Signal one corresponds to T cell recognition (TCR/HLA axis), and signal 2 to co‐stimulatory pathways (including the prototypical CD28‐CD80/86 T cell stimulatory signaling pathway, among others). 59 , 60 , 61 Overall, clinical immunosuppressants used to treat liver and other solid organ transplant recipients typically target signal one. 62
However, important second signal pathways also include co‐inhibitory pathways, meant to modulate unchecked immune activity from activated immune cells. Co‐inhibitory pathways that abrogate anti‐graft immune activity include cytotoxic T lymphocyte‐associated protein‐4 (CTLA‐4; also known as CD152), PD‐1 (also known as CD279), and its ligands PD‐L1 (B7‐H1; CD274) and PD‐L2 (B7‐DC; CD273). 63 , 64 In the setting of persistent activation of the T cell, CTLA‐4 and PD‐1 interactions with their ligands serve as inhibitory signals to regulate activation and prevent disordered immune activity, including autoimmune disease and rejection.
The PD‐1 “checkpoint” is key to maintaining peripheral tolerance. 65 On the other hand, inhibiting this pathway has become an exploitable target for increasing immune activity. 66 Such a strategy is useful in the typically tolerant tumor microenvironment where inhibition can augment antitumor immune responses. In T‐cells, PD‐1 inhibitory function is primarily mediated through the phosphorylated immunoreceptor tyrosine‐based switch motif (ITSM), which recruits Src homology region 2 domain‐containing protein tyrosine phopsphatase‐2 (SHP‐2). Although the exact mechanism with T‐cells remains elusive, triggering of PD‐1 shows inhibition of phosphatidylinositol‐3‐kinase NF‐kβ and Ras/MEK/Erk pathways, resulting in impaired interleukin‐2 (IL‐2) production upon TCR/CD3 stimulation and cell cycle arrest. 67 PD‐1 signaling is also present in NK cells. PD‐1 surface expression is negligible in healthy CD56dim NK cells, however, may be induced. PD‐1+ NK cells are most commonly found in cytomegalovirus (CMV)‐infected individuals, as well as in several types of cancers. 68 It is unclear to what degree PD‐1 is upregulated, or what downstream targets it impacts, during the various mechanisms of NK cell activation. Because T and NK cells share many signaling molecules and coreceptors involved in cytotoxic activation (the CD‐3ζ chain present in both the TCR and CD16 activation complexes, for example), it stands to reason that downstream targets of PD‐1 activation overlap in NK and T cells (Figure 3).
FIGURE 3.

PD‐1 signaling cascade in T cells. The PD‐1 pathway is an important regulator of T cell activation, acting as a “brake” to modulate the T cell response. PD‐1 activation inhibits signaling through both the nuclear factor kappa‐light‐chain‐enhancer of activated B cells (NF‐κB) and extracellular signal‐regulated kinase (ERK) pathways. This in turn leads to reduced cell proliferation and metabolism
IMMUNE CHECKPOINT INHIBITORS MAY PROMOTE TRANSPLANT REJECTION
Inhibitors of the immune checkpoint have revolutionized cancer immunotherapy by promoting an effector immune cell environment, whereas the individuals own immune system can become increasingly activated and destroy the tumor. Therapeutic strategies known as immune checkpoint blockade (ICB) to target and block CTLA‐4, PD‐1 and PD‐L1 have been successful in multiple tumor types, such as melanoma, non‐small cell lung cancer, breast cancer, and cervical cancer. 69 , 70 Unfortunately, their utilization for a patient with a solid organ transplant is often limited due to concerns of potentiating transplant organ rejection. For example, PD‐1 activation plays a role in maintaining graft tolerance after transplantation in part by preventing T cell infiltration to the graft. 71 Morita and colleagues found that blocking the inhibitory PD‐1 pathway in a murine transplanted liver led to severe acute rejection with organ necrosis due to profound T cell infiltration of the graft. 64
Of interest, ICB is increasingly used in patients with hepatocellular carcinoma (HCC). Patients with HCC can be prime candidates for liver transplant, but graft and patient survival is affected by tumor recurrence. Because immune checkpoint inhibition is thought to promote graft rejection, these drugs are not usually prescribed in the context of liver graft recipients. Despite this, a small number of post‐transplant patients with HCC have received ICB to treat tumor recurrence, typically as a last resort. Out of 19 cases of liver transplant recipients with advanced HCC who received ICB, 37% saw graft rejection. 72 Another recent study has documented a single successful treatment of disseminated HCC post‐liver transplant with nivolumab, a monoclonal antibody targeting PD‐1. 73
Clinical studies of checkpoint molecule inhibitors in solid organ transplant recipients are largely limited to case series reports and have shown mixed results. Fisher and colleagues completed a systematic review of 36 articles (2 retrospective studies and 34 case reports/series) with a total of 57 solid organ transplant recipients. 74 In total, 37% of patients experienced graft rejection and 14% died from rejection when treated with a PD‐1 or CTLA‐4 inhibitor. 74 When considering immune checkpoint inhibitor therapy in the setting of liver transplant specifically, Munker and De Toni reported that four out of 14 cases of liver transplant recipients who had received immune checkpoint inhibitors rejected the graft, with 75% mortality rate in those who experienced rejection. 75
EXPLOITATION OF NK IMMUNE CHECKPOINT MOLECULES TO PROMOTE TOLERANCE IN TRANSPLANTATION
PD‐1/PDL‐1 blockade has garnered increased attention as a target in cancer therapies, 76 , 77 , 78 but little is known about exploiting activation of this pathway for therapeutic utility in organ transplant. PD‐1 expression on hepatic T cells has been well‐documented in viral hepatitis, where, as a marker of exhaustion, PD‐1 leads to poor T cell adaptive immune response and poor virus elimination. 79 , 80 CD49a+ hepatic trNK cells may also be characterized by high expression of regulatory surface markers that include the PD‐1 ligand, PD‐L1. 81 These hepatic trNK cells were shown to influence the adaptive immune response to viral infection by inhibiting the antiviral T cell response via the PD‐1/PD‐L1 pathway, thus inhibiting viral clearance and contributing to the more tolerant microenvironment of the liver. 81
Hepatitis C viral infection has served as a model in better understanding the role of NK cells and immune checkpoints in viral clearance as well as progressive liver disease post‐transplant. Inhibition of viral clearance secondary to reduced NK cell activity associated with increased PD‐1 expression has been shown to contribute to the development of chronic hepatitis C viral infection. 82 Collister and colleagues further defined this role in noting that high hepatitis C viral loads correlated with higher expression of PD‐1 on NK cells, finding that hepatitis C proteins were able to induce NK cell exhaustion via the PD‐1 pathway. 83 Direct acting antiviral agents have revolutionized hepatitis C viral therapy by inhibiting viral replication, but the mechanisms of immune modulation by these new antiviral agents may be nuanced. Szereday and colleagues demonstrated that treatment with direct acting antiviral agents resulted in decreased expression of immune checkpoint ligands, allowing for restoration of the previously exhausted immune response. 84
Beyond viral infections, the presence of CD49a+ trNK cells in human HCC was associated with deteriorating disease conditions—including tumor thrombus and lack of a tumor capsule—in addition to shorter overall and disease‐free survival. 85 The presence of CD49+ cells was also associated with increased NK cell expression of the inhibitory receptors NKG2A and PD‐1, suggesting a tolerogenic NK cell presence within liver tumor. 85
These studies show that in the setting of viral infection and cancer, activation of the PD‐1/PD‐L1 axis for T cells and NK cells limits the adaptive immune response, the primary regulator of graft rejection. In the setting of liver transplant, Shi and colleagues demonstrated that PD‐L1 is expressed on liver graft hepatocytes. 86 During rejection, PD‐L1 is upregulated on lobular hepatocytes and sinusoids and portal cholangiocytes. In addition, graft infiltrating T cells were shown to have high expression of PD‐1. Blockade of the PD‐1/PD‐L1 pathway led to increased intragraft T cell proliferation and further activation of the immune system. 86 Little is known about the role of hepatic NK cells and the PD‐1/PD‐L1 axis in transplant; however, the current literature suggests that hepatic NK cells are associated with increased graft tolerance. If these same liver resident NK cells express PD‐1 and PD‐L1, and blockade of the PD‐1/PD‐L1 axis increases T cell trafficking to the graft, then this suggests that intrahepatic PD‐1 potentiation could limit T cell trafficking, increasing self‐tolerance and liver graft tolerance.
OTHER NK IMMUNE CHECKPOINT MOLECULES, AND POTENTIAL ROLES IN LIVER TRANSPLANT TOLERANCE
In comparison to PD1/PDL1, much less is known about the individual role other NK immune checkpoints play during organ transplant. Recent investigation supports the hypothesis that these inhibitory receptors may promote NK self‐tolerance in the setting of infection or tumor. 87 , 88
T cell immunoreceptor with Ig and ITIM domains
The T cell immunoreceptor with Ig and ITIM domains (TIGIT)/nectin‐like (Necl)/DNAX accessory molecule‐1 (DNAM‐1) axis plays a central role in NK cell maturation, education, and tumor clearing. 89 DNAM‐1 is an activating receptor expressed on NK cells and T cells. Upon binding nectin/Necl, the epithelial cell adhesion molecules poliovirus receptor (PVR/CD155), and nectin‐2 (CD112), DNAM‐1 triggers NK cell cytotoxic function, and facilitates the adhesion of NK cells to target cells bearing these adhesion molecules. 89 TIGIT binds PVR and nectin‐2, and inhibits NK cytotoxic function 90 and cytokine secretion 91 through an inhibitory signaling cascade mediated by the ITIM domain in its cytoplasmic tail.
Expression of TIGIT in healthy human NK cells varies, with one study suggesting that TIGIT expression is inversely correlated with NK cytokine production and cytotoxic potential, and that cytokine stimulation does not significantly impact TIGIT expression level. 92 The authors of this study suggest that human NK cells naturally express high levels of TIGIT, which contrasts with results obtained from mouse studies. 88 Other recent studies have shown that TIGIT might contribute to NK education in an MHC‐independent manner 93 and inhibit cytokine production (namely IFN‐γ) in mice. 91 Studies in mice have also revealed that blockade of TIGIT enhances NK effector function in infection and cancer models. 94 , 95 In the context of immunotherapy, Roche’s anti‐TIGIT tiragolumab has recently been granted a US Food and Drug Administration (FDA) Breakthrough Therapy designation when combined with atezolizumab (PD‐L1 monoclonal antibody) in treating non‐small cell lung cancer. A clinical trial evaluating the safety and therapeutic potential of combining PD‐1, PVRIG, and TIGIT inhibition in treating solid advanced tumors is currently underway (NCT04570839). Another trial underway will compare therapeutic potential of the already established elotuzumab (anti‐SLAMF7 antibody)/lenalidomide (thalidomide derivate that inhibits tumor angiogenesis)/dexamethasone (corticoid steroid used to reduce inflammation) multiple myeloma therapy versus TIGIT blockade/lenalidomide/dexamethasone or lymphocyte activating 3 (LAG3) blockade/lenalidomide/dexamethasone in patients with relapsed multiple myeloma (NCT04150965).
A study from 2014 suggests that during liver regeneration in mice, NK cells selectively upregulate TIGIT along with PVR expression on hepatocytes. 88 Using a murine model, Bi and colleagues identified liver NK cells that upregulate TIGIT in response to adenovirus infection. Subsequently, TIGIT blockade resulted in increased NK cell activation and liver injury, suggesting that TIGIT expression by NK cells plays a key role in controlling immune response to active infection and limiting NK mediated cellular destruction. 88 The implications that this might have for hepatic trNK cells in the transplanted graft remain unclear, but the increased expression or stimulation of TIGIT in recipient infiltrating rtNK could be an effective immune modulator.
T‐cell immunoglobulin and mucin domain‐containing protein 3
T‐cell immunoglobulin and mucin domain‐containing protein 3 (TIM‐3) was first described as limiting IFN‐γ secretion in cytotoxic and helper T‐cells. 96 Since then, it has been reported in many other immune cells, including NK cells. 97 Ligands that have been identified for TIM‐3 include the soluble ligands galectin‐9 and high mobility group box 1 (HMGB1), as well as the cell surface ligand ceacam‐1. 98 TIM‐3 expression in human NK cells is a marker of NK maturity that suppresses NK cytotoxic function when cross‐linked. 97 Although named and described as a T cell protein, TIM‐3 is most highly transcribed in NK cells compared to other lymphocytes. TIM‐3 is expressed in resting and activated NK cells; expression may be enhanced via cytokine stimulation or through CD16/Fc interactions. 22 Induction of TIM‐3 with its cognate ligand galectin‐9 enhances IFN‐γ production in vitro. 97 Under specific culture conditions, TIM‐3 on NK cells may become downregulated in response to cancer. 97 TIM‐3 expression on NK cells has been associated with poor prognosis in various solid cancers 99 , 100 and decreased expression was shown to correlate with better prognosis in patients with severe autoimmune aplastic anemia. 101 Clinical trials evaluating therapeutic potential of TIM3 blockade on solid cancers alone or in combination with LAG3 and PD‐1 blockade are currently underway (NCT02817633 and NCT03739710).
To better understand the role of immune checkpoint molecules in the development of HCC, Tan and colleagues bridged the gap between murine and human models by identifying TIM‐3+ NK cells in both species. 87 CD49a+ murine liver trNK cells and CD49a− conventional NK cells and the equivalent human CXCR6+ and CXCR6− NK cells showed higher TIM‐3 expression in tumor‐infiltrated cells as compared to normal tissue. 87 This upregulation resulted in suppressed cytokine secretion and cytotoxic activity. Given that donor hepatic NK cells mediate tolerance through cytokine secretion and cytotoxicity, one could speculate that potentiation of the inhibitory checkpoint pathway with upregulation of TIM‐3 may result in decreased cytotoxicity/cytokine secretion of graft‐infiltrating recipient NK cells could also work to promote tolerance.
Lymphocyte activating 3 expression
Lymphocyte activating 3 (LAG3) expression may be induced on a number of lymphocyte populations, including NK cells. Liver and lymph node sinusoidal endothelial cell C‐type lectin (LSECtin) serves as a ligand for LAG3 and is most prominently expressed in sinusoidal endothelial cells in the liver and lymph nodes. In T‐cells, LAG3 serves as a marker of exhaustion in the context of cancer. 102 Preliminary knockout studies in mice suggested that LAG3 might promote NK cytotoxic function, 103 however, this has not been observed in any human in vitro models. 104 The NK subgroups that express LAG3 in response to stimulation tend to be mature, cytokine secreting NK cells that have higher glycolytic activity when compared to LAG3‐ NK cells. 105 A clinical trial evaluating safety and immunotherapeutic potential of LAG3 blockade alone or with PD‐1 blockade (NCT01968109) in treating solid tumors is currently underway.
FUTURE DIRECTIONS
Incorporating artificial intelligence and precision therapeutics into NK cell‐based treatment strategies in transplantation will be paramount. As an example, performing KIR‐ligand mismatching prior to hematopoietic stem cell transplantation has already become increasingly common, especially in treating acute myeloid leukemia. 106 In this application of precision medicine, NK‐mediated graft‐versus‐recipient phenotype is correlated with improved survivability and reduced risk of acute myeloblastic leukemia (AML) relapse. The NK cells derived from stem cell transplant are thought to display superior killing against the mismatched HLA ligands on the surface of cancer cells. 107 , 108 Importantly, KIR‐ligand mismatch does not seem to have any significant impact on the onset of pathological graft‐versus‐host disease or graft rejection. The impact that KIR‐ligand mismatch has during solid organ transplant has not been investigated extensively, however, mismatched donor NK cells could potentially display superior killing against host alloreactive T cells that might otherwise mediate allograft rejection. In the context of liver transplant during liver cancer, donor NK cells could display enhanced killing of tumor cells, however, the persistence and potency of donor hepatic NK cells within the graft environment have not been fully characterized. Machine learning may aid in future efforts to improve precision medicine techniques during liver transplantation. Indeed, genetic predictive models of tolerance during solid organ transplant have been established via machine learning in kidney 109 and pancreatic islet cell transplant. 110 Potentially, these methodologies could be used to decipher the KIR‐HLA axis and find other positive phenotype matches for liver transplantation.
INCREASING TRANSPLANT ALLOGRAFT ACCEPTANCE BY INCORPORATING CHECKPOINT PATHWAY TARGETS
In summary, recent insights have highlighted two distinct liver NK cells populations—the conventional NK cell population which phenotypically and functionally are similar to circulating NK cells and the liver resident NK cells. Liver transplantation results in a unique interface of these two cell populations with transfer of donor liver trNK cells to recipient and infiltration of recipient circulating NK cells into the graft within hours of transplantation. Following transplantation, donor liver resident NK cells are found in an activated state with increased cytolytic and cytotoxic activity, which helps mitigate infiltration of recipient lymphocytes to the graft and thus allograft rejection. In contrast, recipient NK cells have been implicated in acute graft rejection, although these mechanisms remain unclear.
Immune checkpoint pathways, such as PD‐1/PDL‐1, act to inhibit immune dysregulation and have been implicated as key mediators of preventing excess lymphocyte infiltration and acute cellular rejection of liver grafts. Just as inhibition of the pathway has led to graft rejection, exploitation through PD‐1 promotors which increase checkpoint molecule expression on recipient NK cells may further reduce graft infiltration and thus improve graft tolerance. In contrast, inhibition of PD‐1 and other checkpoint molecules leads to unmetered T cell activation with resultant hepatocellular damage. Although the current checkpoint inhibition immunotherapies that act broadly against cancer cells, T cells, and NK cells often result in intrinsic liver damage, a targeted checkpoint inhibition for donor liver resident NK cells could result in increased killing of recipient immune cells, decreased graft infiltration, and ultimately improved graft survival.
The identification of tissue resident, phenotypically and functionally distinct NK cells provides a framework for understanding the role of NK cells in organ transplant; however, much is still unknown with regard to how NK cells promote tolerance versus induce rejection in liver transplantation. Future studies are needed to better understand how phenotypic and functional changes of NK cells affect graft outcomes. Immune checkpoint molecules are present on tissue resident NK cells and have been implicated in promoting graft tolerance. Additional studies should look to delineate the mechanisms by which these inhibitory pathways regulate NK cell activation. Of particular interest would be further development of immunotherapy with honing of the checkpoint inhibitor pathway to the specific NK cell phenotype to promote graft tolerance and expansion of machine learning to advance our understanding of these complex cellular interactions.
CONFLICT OF INTEREST
The authors declared no competing interests for this work.
Halma J, Pierce S, McLennan R, Bradley T, Fischer R. Natural killer cells in liver transplantation: Can we harness the power of the immune checkpoint to promote tolerance? Clin Transl Sci. 2022;15:1091‐1103. doi: 10.1111/cts.13208
Funding information
No funding was received for this work.
REFERENCES
- 1. Kondo M, Akashi K, Domen J, Sugamura K, Weissman IL. Bcl‐2 rescues T lymphopoiesis, but not B or NK cell development, in common gamma chain‐deficient mice. Immunity. 1997;7(1):155‐162. [DOI] [PubMed] [Google Scholar]
- 2. Abel AM, Yang C, Thakar MS, Malarkannan S. Natural killer cells: development, maturation, and clinical utilization. Front Immunol. 2018;9:1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Orange JS, Ramesh N, Remold‐O'Donnell E, et al. Wiskott‐Aldrich syndrome protein is required for NK cell cytotoxicity and colocalizes with actin to NK cell‐activating immunologic synapses. Proc Natl Acad Sci USA. 2002;99(17):11351‐11356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brandstadter JD, Yang Y. Natural killer cell responses to viral infection. J Innate Immun. 2011;3(3):274‐279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rosenau W, Moon HD. Lysis of homologous cells by sensitized lymphocytes in tissue culture. J Natl Cancer Inst. 1961;27:471‐483. [PubMed] [Google Scholar]
- 6. Herberman RB, Nunn ME, Holden HT, Lavrin DH. Natural cytotoxic reactivity of mouse lymphoid cells against syngeneic and allogeneic tumors. II. Characterization of effector cells. Int J Cancer. 1975;16(2):230‐239. [DOI] [PubMed] [Google Scholar]
- 7. Yang Q, Goding S, Hagenaars M, et al. Morphological appearance, content of extracellular matrix and vascular density of lung metastases predicts permissiveness to infiltration by adoptively transferred natural killer and T cells. Cancer Immunol Immunother. 2006;55(6):699‐707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krzewski K, Coligan JE. Human NK cell lytic granules and regulation of their exocytosis. Front Immunol. 2012;3:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tassi I, Klesney‐Tait J, Colonna M. Dissecting natural killer cell activation pathways through analysis of genetic mutations in human and mouse. Immunol Rev. 2006;214:92‐105. [DOI] [PubMed] [Google Scholar]
- 10. Bryceson YT, Chiang SC, Darmanin S, et al. Molecular mechanisms of natural killer cell activation. J Innate Immun. 2011;3(3):216‐226. [DOI] [PubMed] [Google Scholar]
- 11. Cooper MA, Fehniger TA, Turner SC, et al. Human natural killer cells: a unique innate immunoregulatory role for the CD56(bright) subset. Blood. 2001;97(10):3146‐3151. [DOI] [PubMed] [Google Scholar]
- 12. Poli A, Michel T, Theresine M, Andres E, Hentges F, Zimmer J. CD56 bright natural killer (NK) cells: an important NK cell subset. Immunology. 2009;126(4):458‐465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mikulak J, Bruni E, Oriolo F, Di Vito C, Mavilio D. Hepatic natural killer cells: organ‐specific sentinels of liver immune homeostasis and physiopathology. Front Immunol. 2019;10:946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Melsen JE, Lugthart G, Lankester AC, Schilham MW. Human circulating and tissue‐resident CD56(bright) natural killer cell populations. Front Immunol. 2016;7:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moretta L, Locatelli F, Pende D, Marcenaro E, Mingari MC, Moretta A. Killer Ig‐like receptor‐mediated control of natural killer cell alloreactivity in haploidentical hematopoietic stem cell transplantation. Blood. 2011;117(3):764‐771. [DOI] [PubMed] [Google Scholar]
- 16. Sojka DK, Yang L, Yokoyama WM. Uterine natural killer cells. Front Immunol. 2019;10:960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cong J, Wei H. Natural killer cells in the lungs. Front Immunol. 2019;10:1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. O'Leary JG, Goodarzi M, Drayton DL, von Andrian UH. T cell‐ and B cell‐independent adaptive immunity mediated by natural killer cells. Nat Immunol. 2006;7(5):507‐516. [DOI] [PubMed] [Google Scholar]
- 19. Reeves RK, Li H, Jost S, et al. Antigen‐specific NK cell memory in rhesus macaques. Nat Immunol. 2015;16(9):927‐932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pierce S, Geanes ES, Bradley T. Targeting natural killer cells for improved immunity and control of the adaptive immune response. Front Cell Infect Microbiol. 2020;10:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9(5):495‐502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barrow AD, Colonna M. Exploiting NK cell surveillance pathways for cancer therapy. Cancers (Basel). 2019;11(1):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huysamen C, Brown GD. The fungal pattern recognition receptor, Dectin‐1, and the associated cluster of C‐type lectin‐like receptors. FEMS Microbiol Lett. 2009;290(2):121‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van Dam LS, de Zwart VM, Meyer‐Wentrup FAG. The role of programmed cell death‐1 (PD‐1) and its ligands in pediatric cancer. Pediatr Blood Cancer. 2015;62(2):190‐197. [DOI] [PubMed] [Google Scholar]
- 25. Wende H, Colonna M, Ziegler A, Volz A. Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mamm Genome. 1999;10(2):154‐160. [DOI] [PubMed] [Google Scholar]
- 26. Campbell KS, Purdy AK. Structure/function of human killer cell immunoglobulin‐like receptors: lessons from polymorphisms, evolution, crystal structures and mutations. Immunology. 2011;132(3):315‐325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boudreau JE, Hsu KC. Natural killer cell education in human health and disease. Curr Opin Immunol. 2018;50:102‐111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pende D, Falco M, Vitale M, et al. Killer Ig‐like receptors (KIRs): their role in NK cell modulation and developments leading to their clinical exploitation. Front Immunol. 2019;10:1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Körner C, Altfeld M. Role of KIR3DS1 in human diseases. Front Immunol. 2012;3:326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martin MP, Nelson G, Lee JH, et al. Cutting edge: susceptibility to psoriatic arthritis: influence of activating killer Ig‐like receptor genes in the absence of specific HLA‐C alleles. J Immunol. 2002;169(6):2818‐2822. [DOI] [PubMed] [Google Scholar]
- 31. Weis WI, Taylor ME, Drickamer K. The C‐type lectin superfamily in the immune system. Immunol Rev. 1998;163:19‐34. [DOI] [PubMed] [Google Scholar]
- 32. Gunturi A, Berg RE, Forman J. The role of CD94/NKG2 in innate and adaptive immunity. Immunol Res. 2004;30(1):29‐34. [DOI] [PubMed] [Google Scholar]
- 33. Smyth MJ, Wallace ME, Nutt SL, Yagita H, Godfrey DI, Hayakawa Y. Sequential activation of NKT cells and NK cells provides effective innate immunotherapy of cancer. J Exp Med. 2005;201(12):1973‐1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Borrego F, Masilamani M, Kabat J, Sanni TB, Coligan JE. The cell biology of the human natural killer cell CD94/NKG2A inhibitory receptor. Mol Immunol. 2005;42(4):485‐488. [DOI] [PubMed] [Google Scholar]
- 35. LaBonte ML, Choi EI, Letvin NL. Molecular determinants regulating the pairing of NKG2 molecules with CD94 for cell surface heterodimer expression. J Immunol. 2004;172(11):6902‐6912. [DOI] [PubMed] [Google Scholar]
- 36. Bartel Y, Bauer B, Steinle A. Modulation of NK cell function by genetically coupled C‐type lectin‐like receptor/ligand pairs encoded in the human natural killer gene complex. Front Immunol. 2013;4:362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lysakova‐Devine T, O'Farrelly C. Tissue‐specific NK cell populations and their origin. J Leukoc Biol. 2014;96(6):981‐990. [DOI] [PubMed] [Google Scholar]
- 38. Harmon C, Robinson MW, Fahey R, et al. Tissue‐resident Eomes(hi) T‐bet(lo) CD56(bright) NK cells with reduced proinflammatory potential are enriched in the adult human liver. Eur J Immunol. 2016;46(9):2111‐2120. [DOI] [PubMed] [Google Scholar]
- 39. Ishiyama K, Ohdan H, Ohira M, Mitsuta H, Arihiro K, Asahara T. Difference in cytotoxicity against hepatocellular carcinoma between liver and periphery natural killer cells in humans. Hepatology. 2006;43(2):362‐372. [DOI] [PubMed] [Google Scholar]
- 40. Freud AG, Becknell B, Roychowdhury S, et al. A human CD34(+) subset resides in lymph nodes and differentiates into CD56bright natural killer cells. Immunity. 2005;22(3):295‐304. [DOI] [PubMed] [Google Scholar]
- 41. Peng H, Sun R, Tang L, Wei H, Tian Z. CD62L is critical for maturation and accumulation of murine hepatic NK cells in response to viral infection. J Immunol. 2013;190(8):4255‐4262. [DOI] [PubMed] [Google Scholar]
- 42. Peng H, Jiang X, Chen Y, et al. Liver‐resident NK cells confer adaptive immunity in skin‐contact inflammation. J Clin Investig. 2013;123(4):1444‐1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marquardt N, Beziat V, Nystrom S, et al. Cutting edge: identification and characterization of human intrahepatic CD49a+ NK cells. J Immunol. 2015;194(6):2467‐2471. [DOI] [PubMed] [Google Scholar]
- 44. Björkström NK, Lindgren T, Stoltz M, et al. Rapid expansion and long‐term persistence of elevated NK cell numbers in humans infected with hantavirus. J Exp Med. 2011;208(1):13‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hudspeth K, Donadon M, Cimino M, et al. Human liver‐resident CD56(bright)/CD16(neg) NK cells are retained within hepatic sinusoids via the engagement of CCR5 and CXCR6 pathways. J Autoimmun. 2016;66:40‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stegmann KA, Robertson F, Hansi N, et al. CXCR6 marks a novel subset of T‐bet(lo)Eomes(hi) natural killer cells residing in human liver. Sci Rep. 2016;6:26157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cuff AO, Robertson FP, Stegmann KA, et al. Eomeshi NK cells in human liver are long‐lived and do not recirculate but can be replenished from the circulation. J Immunol. 2016;197(11):4283‐4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Moroso V, Metselaar HJ, Mancham S, et al. Liver grafts contain a unique subset of natural killer cells that are transferred into the recipient after liver transplantation. Liver Transpl. 2010;16(7):895‐908. [DOI] [PubMed] [Google Scholar]
- 49. Lerut J, Sanchez‐Fueyo A. An appraisal of tolerance in liver transplantation. Am J Transplant. 2006;6(8):1774‐1780. [DOI] [PubMed] [Google Scholar]
- 50. Harmon C, Sanchez‐Fueyo A, O'Farrelly C, Houlihan DD. Natural killer cells and liver transplantation: orchestrators of rejection or tolerance? Am J Transplant. 2016;16(3):751‐757. [DOI] [PubMed] [Google Scholar]
- 51. Garcia de la Garza R, Sarobe P, Merino J, et al. Immune monitoring of immunosuppression withdrawal of liver transplant recipients. Transpl Immunol. 2015;33(2):110‐116. [DOI] [PubMed] [Google Scholar]
- 52. Li L, Wozniak LJ, Rodder S, et al. A common peripheral blood gene set for diagnosis of operational tolerance in pediatric and adult liver transplantation. Am J Transplant. 2012;12(5):1218‐1228. [DOI] [PubMed] [Google Scholar]
- 53. Pagano D, Badami E, Conaldi PG, et al. Liver perfusate natural killer cells from deceased brain donors and association with acute cellular rejection after liver transplantation: a time‐to‐rejection analysis. Transplantation. 2019;103(2):371‐380. [DOI] [PubMed] [Google Scholar]
- 54. Heerwagen C, Schuster M, Bornscheurer A, et al. Rapid exchange of large numbers of donor‐ and host leukocytes after human liver transplantation. Transpl Int. 2001;14(4):240‐247. [DOI] [PubMed] [Google Scholar]
- 55. Ronca V, Wootton G, Milani C, Cain O. The immunological basis of liver allograft rejection. Front Immunol. 2020;11:2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Obara H, Nagasaki K, Hsieh CL, et al. IFN‐gamma, produced by NK cells that infiltrate liver allografts early after transplantation, links the innate and adaptive immune responses. Am J Transplant. 2005;5(9):2094‐2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jamil KM, Hydes TJ, Cheent KS, et al. STAT4‐associated natural killer cell tolerance following liver transplantation. Gut. 2017;66(2):352‐361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pham B, Piard‐Ruster K, Silva R, et al. Changes in natural killer cell subsets in pediatric liver transplant recipients. Pediatr Transplant. 2012;16(2):176‐182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ford ML. T Cell cosignaling molecules in transplantation. Immunity. 2016;44(5):1020‐1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen L, Flies DB. Molecular mechanisms of T cell co‐stimulation and co‐inhibition. Nat Rev Immunol. 2013;13(4):227‐242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mueller DL, Jenkins MK, Schwartz RH. Clonal expansion versus functional clonal inactivation: a costimulatory signalling pathway determines the outcome of T cell antigen receptor occupancy. Annu Rev Immunol. 1989;7:445‐480. [DOI] [PubMed] [Google Scholar]
- 62. Conti F, Morelon E, Calmus Y. Immunosuppressive therapy in liver transplantation. J Hepatol. 2003;39(5):664‐678. [DOI] [PubMed] [Google Scholar]
- 63. Francisco LM, Sage PT, Sharpe AH. The PD‐1 pathway in tolerance and autoimmunity. Immunol Rev. 2010;236:219‐242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Morita M, Fujino M, Jiang G, et al. PD‐1/B7‐H1 interaction contribute to the spontaneous acceptance of mouse liver allograft. Am J Transplant. 2010;10(1):40‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Riella LV, Paterson AM, Sharpe AH, Chandraker A. Role of the PD‐1 pathway in the immune response. Am J Transplant 2012;12(10):2575‐2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pan C, Liu H, Robins E, et al. Next‐generation immuno‐oncology agents: current momentum shifts in cancer immunotherapy. J Hematol Oncol. 2020;13(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Boussiotis VA, Chatterjee P, Li L. Biochemical signaling of PD‐1 on T cells and its functional implications. Cancer J. 2014;20(4):265‐271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pesce S, Greppi M, Tabellini G, et al. Identification of a subset of human natural killer cells expressing high levels of programmed death 1: A phenotypic and functional characterization. J Allergy Clin Immunol. 2017;139(1):335‐346 e333. [DOI] [PubMed] [Google Scholar]
- 69. Petitprez F, Meylan M, de Reyniès A, Sautès‐Fridman C, Fridman WH. The tumor microenvironment in the response to immune checkpoint blockade therapies. Front Immunol. 2020;11:784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schnell A, Bod L, Madi A, Kuchroo VK. The yin and yang of co‐inhibitory receptors: toward anti‐tumor immunity without autoimmunity. Cell Res. 2020;30(4):285‐299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Brunner‐Weinzierl MC, Rudd CE. CTLA‐4 and PD‐1 Control of T‐Cell Motility and Migration: Implications for Tumor Immunotherapy. Front Immunol. 2018;9:2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. De Bruyn P, Van Gestel D, Ost P, et al. Immune checkpoint blockade for organ transplant patients with advanced cancer: how far can we go? Curr Opin Oncol. 2019;31(2):54‐64. [DOI] [PubMed] [Google Scholar]
- 73. Amjad W, Kotiah S, Gupta A, Morris M, Liu L, Thuluvath PJ. Successful treatment of disseminated hepatocellular carcinoma after liver transplantation with nivolumab. J Clin Exp Hepatol. 2020;10(2):185‐187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fisher J, Zeitouni N, Fan W, Samie FH. Immune checkpoint inhibitor therapy in solid organ transplant recipients: A patient‐centered systematic review. J Am Acad Dermatol. 2020;82(6):1490‐1500. [DOI] [PubMed] [Google Scholar]
- 75. Munker S, De Toni EN. Use of checkpoint inhibitors in liver transplant recipients. United European Gastroenterol J. 2018;6(7):970‐973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Guo Y, Yu P, Liu Z, et al. Prognostic and clinicopathological value of programmed death ligand‐1 in breast cancer: a meta‐analysis. PLoS One. 2016;11(5):e0156323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Beldi‐Ferchiou A, Lambert M, Dogniaux S, et al. PD‐1 mediates functional exhaustion of activated NK cells in patients with Kaposi sarcoma. Oncotarget. 2016;7(45):72961‐72977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Benson DM Jr, Bakan CE, Mishra A, et al. The PD‐1/PD‐L1 axis modulates the natural killer cell versus multiple myeloma effect: a therapeutic target for CT‐011, a novel monoclonal anti‐PD‐1 antibody. Blood. 2010;116(13):2286‐2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Radziewicz H, Ibegbu CC, Fernandez ML, et al. Liver‐infiltrating lymphocytes in chronic human hepatitis C virus infection display an exhausted phenotype with high levels of PD‐1 and low levels of CD127 expression. J Virol. 2007;81(6):2545‐2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Fisicaro P, Valdatta C, Massari M, et al. Antiviral intrahepatic T‐cell responses can be restored by blocking programmed death‐1 pathway in chronic hepatitis B. Gastroenterology. 2010;138(2):682‐693, 693 e681–684. [DOI] [PubMed] [Google Scholar]
- 81. Zhou J, Peng H, Li K, et al. Liver‐resident NK cells control antiviral activity of hepatic T cells via the PD‐1‐PD‐L1 Axis. Immunity. 2019;50(2):403‐417 e404. [DOI] [PubMed] [Google Scholar]
- 82. Golden‐Mason L, Klarquist J, Wahed AS, Rosen HR. Cutting edge: programmed death‐1 expression is increased on immunocytes in chronic hepatitis C virus and predicts failure of response to antiviral therapy: race‐dependent differences. J Immunol. 2008;180(6):3637‐3641. [DOI] [PubMed] [Google Scholar]
- 83. Collister M, Ellison C, Li Q, Minuk GY, Rempel JD, Kung SK. The Influence of hepatitis C viral loads on natural killer cell function. Gastroenterology Res. 2019;12(1):8‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Szereday L, Meggyes M, Berki T, et al. Direct‐acting antiviral treatment downregulates immune checkpoint inhibitor expression in patients with chronic hepatitis C. Clin Exp Med. 2020;20(2):219‐230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Sun H, Liu L, Huang Q, et al. Accumulation of tumor‐infiltrating CD49a(+) NK cells correlates with poor prognosis for human hepatocellular carcinoma. Cancer Immunol Res. 2019;7(9):1535‐1546. [DOI] [PubMed] [Google Scholar]
- 86. Shi XL, Mancham S, Hansen BE, et al. Counter‐regulation of rejection activity against human liver grafts by donor PD‐L1 and recipient PD‐1 interaction. J Hepatol. 2016;64(6):1274‐1282. [DOI] [PubMed] [Google Scholar]
- 87. Tan S, Xu Y, Wang Z, et al. Tim‐3 hampers tumor surveillance of liver‐resident and conventional NK cells by disrupting PI3K signaling. Cancer Res. 2020;80(5):1130‐1142. [DOI] [PubMed] [Google Scholar]
- 88. Bi J, Zheng X, Chen Y, Wei H, Sun R, Tian Z. TIGIT safeguards liver regeneration through regulating natural killer cell‐hepatocyte crosstalk. Hepatology. 2014;60(4):1389‐1398. [DOI] [PubMed] [Google Scholar]
- 89. Martinet L, Ferrari De Andrade L, Guillerey C, et al. DNAM‐1 expression marks an alternative program of NK cell maturation. Cell Rep. 2015;11(1):85‐97. [DOI] [PubMed] [Google Scholar]
- 90. Stanietsky N, Simic H, Arapovic J, et al. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc Natl Acad Sci USA. 2009;106(42):17858‐17863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Li M, Xia P, Du Y, et al. T‐cell immunoglobulin and ITIM domain (TIGIT) receptor/poliovirus receptor (PVR) ligand engagement suppresses interferon‐gamma production of natural killer cells via beta‐arrestin 2‐mediated negative signaling. J Biol Chem. 2014;289(25):17647‐17657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wang F, Hou H, Wu S, et al. TIGIT expression levels on human NK cells correlate with functional heterogeneity among healthy individuals. Eur J Immunol. 2015;45(10):2886‐2897. [DOI] [PubMed] [Google Scholar]
- 93. He Y, Peng H, Sun R, et al. Contribution of inhibitory receptor TIGIT to NK cell education. J Autoimmun. 2017;81:1‐12. [DOI] [PubMed] [Google Scholar]
- 94. Morvan MG, Lanier LL. NK cells and cancer: you can teach innate cells new tricks. Nat Rev Cancer. 2016;16(1):7‐19. [DOI] [PubMed] [Google Scholar]
- 95. Zhang Q, Bi J, Zheng X, et al. Blockade of the checkpoint receptor TIGIT prevents NK cell exhaustion and elicits potent anti‐tumor immunity. Nat Immunol. 2018;19(7):723‐732. [DOI] [PubMed] [Google Scholar]
- 96. Gao X, Zhu Y, Li G, et al. TIM‐3 expression characterizes regulatory T cells in tumor tissues and is associated with lung cancer progression. PLoS One. 2012;7(2):e30676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ndhlovu LC, Lopez‐Vergès S, Barbour JD, et al. Tim‐3 marks human natural killer cell maturation and suppresses cell‐mediated cytotoxicity. Blood. 2012;119(16):3734‐3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wolf Y, Anderson AC, Kuchroo VK. TIM3 comes of age as an inhibitory receptor. Nat Rev Immunol. 2020;20(3):173‐185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zheng Y, Li Y, Lian J, et al. TNF‐alpha‐induced Tim‐3 expression marks the dysfunction of infiltrating natural killer cells in human esophageal cancer. J Transl Med. 2019;17(1):165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Dao TN, Utturkar S, Atallah Lanman N, Matosevic S. TIM‐3 expression is downregulated on human NK cells in response to cancer targets in synergy with activation. Cancers. 2020;12(9):2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Zhang X, Zhang H, Chen L, Feng Z, Gao L, Li Q. TIGIT expression is upregulated in T cells and causes T cell dysfunction independent of PD‐1 and Tim‐3 in adult B lineage acute lymphoblastic leukemia. Cell Immunol. 2019;344:103958. [DOI] [PubMed] [Google Scholar]
- 102. Yang ZZ, Kim HJ, Villasboas JC, et al. Expression of LAG‐3 defines exhaustion of intratumoral PD‐1(+) T cells and correlates with poor outcome in follicular lymphoma. Oncotarget. 2017;8(37):61425‐61439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Miyazaki T, Dierich A, Benoist C, Mathis D. LAG‐3 is not responsible for selecting T helper cells in CD4‐deficient mice. Int Immunol. 1996;8(5):725‐729. [DOI] [PubMed] [Google Scholar]
- 104. Huard B, Tournier M, Triebel F. LAG‐3 does not define a specific mode of natural killing in human. Immunol Lett. 1998;61(2–3):109‐112. [DOI] [PubMed] [Google Scholar]
- 105. Merino A, Zhang B, Dougherty P, et al. Chronic stimulation drives human NK cell dysfunction and epigenetic reprograming. J Clin Investig. 2019;129(9):3770‐3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Stringaris K, Marin D, Barrett AJ, et al. KIR gene haplotype: an independent predictor of clinical outcome in MDS patients. Blood. 2016;128(24):2819‐2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Valiante NM, Parham P. Natural killer cells, HLA class I molecules, and marrow transplantation. Biol Blood Marrow Transplant. 1997;3(5):229‐235. [PubMed] [Google Scholar]
- 108. Ruggeri L, Mancusi A, Capanni M, et al. Donor natural killer cell allorecognition of missing self in haploidentical hematopoietic transplantation for acute myeloid leukemia: challenging its predictive value. Blood. 2007;110(1):433‐440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Fu Q, Agarwal D, Deng K, et al. An unbiased machine learning exploration reveals gene sets predictive of allograft tolerance after kidney transplantation. Front Immunol. 2021;12: 695806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Ceballos GA, Hernandez LF, Paredes D, Betancourt LR, Abdulreda MH. A machine learning approach to predict pancreatic islet grafts rejection versus tolerance. PLoS One. 2020;15(11):e0241925. [DOI] [PMC free article] [PubMed] [Google Scholar]


