Version Changes
Revised. Amendments from Version 1
The manuscript was revised and refined by a language style corrector. Some sentences in the methods and results section were improved giving more detail and explanations in accord to revisor suggestions.
Abstract
Background: In autoimmune vasculitis, autoantibodies to Human Proteinase 3 (PR3), a human serine protease, seems to have a role on the inception of c-ANCA associated vasculitis. The origin of this autoreactive response remains unclear. However, for several autoreactive responses, molecular mimicry between environmental antigens and human proteins is key to trigger autoantibodies and finally autoimmunity manifestations. Considering that PR3 is a serine protease and house dust mite (HDM) group 3 allergens share this biochemical activity, the aim of this study was to identify cross-reactive epitopes between serine proteases from human and mites using an in silico approach.
Methods: Multi alignment among amino acid sequences of PR3 and HDM group 3 allergens was performed to explore identity and structural homology. ElliPro and BepiPred in silico tools were used to predict B and T cell epitopes. Consurf tool was used to conduct identification of conserved regions in serine proteases family.
Results: PR3 and HDM group 3 allergens shared moderate identity and structural homology (root mean square deviation < 1). One B cell cross reactive epitope among serine proteases was identified (29I, 30V, 31G, 32G, 34E, 36K, 37A, 38L, 39A and 54C) and two T cell epitopes.
Conclusions: PR3 have structural homology and share cross reactive epitopes with HDM group 3 allergens.
Keywords: Serine proteases, human proteinase 3, house dust mites group 3 allergens, ANCA associated vasculitis, sequence homology, T and B cell epitopes, cross-reactivity, epitope modelling
Introduction
Anti-neutrophil cytoplasmic antibody (ANCA)-associated vasculitis (AAV) is a life-threatening autoimmune disease affecting small vessels, compromising the respiratory mucosa, skin, lung, and the kidney 1 . This group of small vessel vasculitis includes various diseases: granulomatosis with polyangiitis, microscopic polyangiitis, kidney-limited vasculitis and Eosinophilic granulomatosis with polyangiitis, all of them having in common some degree of autoimmune response to the Human Proteinase 3 protein (PR3). Previous studies have shown that autoantibody binding to PR3 expressed on the neutrophil surface may activate its degranulation, eliciting tissue damage in small vessels and their irrigated organs. Also, while proinflammatory effector T cells have been implicated in vasculitis pathogenesis 2 , a specific PR3 T cell epitope has not been reported in AAV patients 3 . PR3 is a serine protease physiologically expressed in human neutrophils. Due to its enzymatic activity, it degrades various intercellular gap-junction proteins and collagen and may play a role in neutrophil transendothelial migration. In addition, this protein is an important autoantigen in AAV, and sera from patients with severe and relapsing forms of the disease can bind it in IgG ELISA assays 4– 6 . Further, although a cause-effect relationship between PR3-autoantibodies and vasculitis is not clearly defined, animal models support a pathogenic role 7, 8 , revealing that they may be involved in disease inception, progression and severity 1 .
Environmental exposures, specially to microbial components mimicking self-antigens have been proposed as triggers of autoimmunity 9, 10 . Also, in AAV, it has been proposed that an endogenous immune response to a complementary protein to PR3 autoantigen could be implicated in disease inception, and this antisense protein harbors homology to various bacterial peptides 11 . PR3 crystal structure has been elucidated, and various epitopes are recognized by patients suffering AAV; however, its cross-reactivity with environmental antigens is poorly studied 12– 14 .
Previous studies have shown that specific IgE to some self-proteins have been identified in autoimmune and allergic diseases like lupus, urticaria, dermatitis, allergic pulmonary aspergillosis and have a strong association with disease activity 15– 18 . Some allergens can cross-react with human proteins and participates in autoimmunity inception in pemphigus vulgaris by a “hit-and-run” mechanism, opening the theoretical possibility for a similar mechanism to occur in another autoimmune disease such as AAV 19– 22 .
In the tropics, house dust mites (HDM) are important ubiquitous allergen sources and exposure is perennial, increasing the possibilities of exposure in the general population 23 , and IgE sensitization to their components 24, 25 . Sensitization to HDM group 3 allergens is common 26 , as they harbor serine protease activity and conserved structural homology 27 , making them potential PR3 cross reactive antigens; this has not been explored before. Here, we show in silico data suggesting cross-reactivity and epitope sharing between PR3 and HDM group 3 allergens.
Methods
Searching homologous with BLAST (Basic Local Alignment Search Tool)
The amino acid sequence from the human PR3 (Uniprot accession: P24158) was used as query to perform a search for serine protease homologous reported in allergenic sources: Dermatophagoides pteronyssinus (Der p 3: Accession number P39675), Blomia tropicalis (Blo t 3: A1KXI1), Glycyphagus domesticus (Gly d 3: Q1M2M8), Lepidoglyphus destructor (Led p 3: Q1M2L7) and Tyrophagus putrescentiae (Tyr p 3: C6ZDB5) with the PSI-BLAST tool. Parameters were set as default.
Multiple alignment
Identity among all allergenic sequences homologous to PR3 was analyzed using the Jalview tool2.11.0 28 . First, all allergens and human PR3 codes were used as inputs in the Jalview tool. Second, the T coffee tool was chosen to assess alignment. Third, alignment was displayed as an identity percentage.
Construction of 3D model
The 3D model of Der p 3, a serine protease of Dermatophagoides pteronyssinus was generated by homology in the SWISS-MODEL server using the zymogen catalytic region of human MASP-2 (PDB: 1zjk f) as a template. The 3D model of Der p 3 was loaded into the ProSA-web server 29 , which was used to analyze its quality.
The model was refined in DeepView v4.1 (energy minimization and rotamer replacements). Its quality was evaluated by several tools, including Ramachandran graphs, WHATIF, QMEAN4 index, and energy values (GROMOS96 force field). For the validation of the Der p 3 structure we used the Minimize Structure option in the UCSF Chimera software, a procedure that adjust the energy and reduce the entropy of the model 30 .
Three-dimensional structure (PDB: 1FUJ) of the human PR3 serine protease was retrieved from the Protein Data Bank. A cartoon model was created using Pymol software v2.4. Root median square deviation (RMSD) value between Der p 3 and PR3 was calculated using Chimera software v1.0 30 .
B and T cell epitope prediction
ElliPro v3.0 and BepiPred v2.0 tools were used to predict B and T cell epitopes on Der p 3 31, 32 . With ElliPro, the 3D structure of Der p 3 was used to predict epitopes. Minimum score and maximum distance (Angstrom) were set to 0.5 and 6, respectively. Epitopes with high conserved rates were visualized in the 3D model. For prediction using BepiPred, an amino acid sequence of Der p 3 was used as input.
Conservation analysis
The 3D structure of Der p 3 was submitted to the ConSurf server to generate evolutionarily related conservation scores to help to identify functional regions in the proteins. HMMER algorithm, 1 iteration, E-value cutoff (0.0001) and UNIREF-90 database was set as default to generate multiple alignment, prior to evolutive analysis. All amino acid sequences in FASTA format were used.
Results
Human PR3 and HDM group 3 allergens exhibited identity and features of the serine protease family
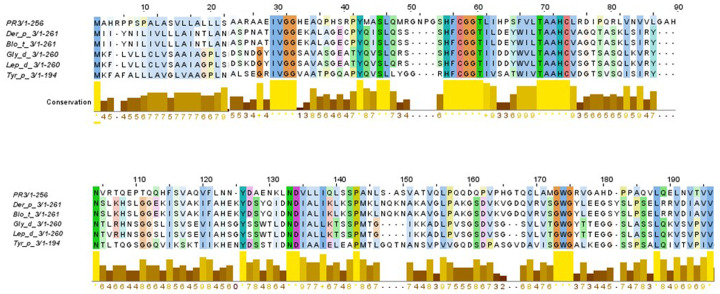
BLAST search identified various serine protease family members from HDM as homologous. The multiple sequence alignment analysis showed that Der p 3, Blo t 3, Gly d 3, Led p 3 and Tyr p 3 allergens shared 45% of identity in their aminoacid sequences with PR3. The most conserved region is located between residues 53 to 75, indicating the existence of molecular mimicry ( Figure 1). Among the members of HDM group 3 allergens, an identity until 41% was reported ( Table 1), and a highly conserved region between residues 40 to 90 was found. When identity between PR3 and each allergen used in study was analyzed, a moderate level of identity was found (30%) ( Table 1).
Figure 1. Multiple alignment among PR3 and the HDM allergens belonging to group 3.
An identity of 45% in their amino acid sequences was found.
Table 1. Identity matrix among serine proteases used in study.
All comparisons of PR3 with HDM group 3 allergens showed a moderate identity.
| PR3 | Der p 3 | Blo t 3 | Gly d 3 | Lep d 3 | Tyr p 3 | |
| PR3 | 100 | 33 | 27 | 30 | 30 | 27 |
| Der p 3 | 33 | 100 | 48 | 52 | 53 | 43 |
| Blo t 3 | 27 | 48 | 100 | 58 | 58 | 47 |
| Gly d 3 | 30 | 52 | 58 | 100 | 99 | 41 |
| Lep d 3 | 30 | 53 | 58 | 99 | 100 | 41 |
| Tyr p 3 | 27 | 43 | 47 | 41 | 41 | 100 |
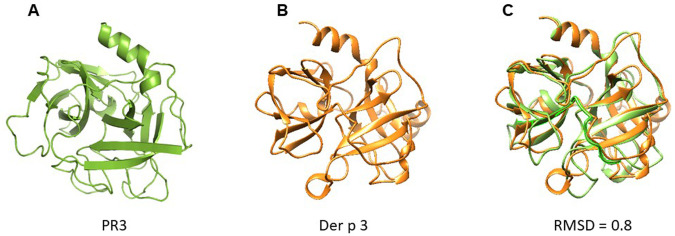
A structural model of Der p 3 was obtained by homology modelling using the 3D structure of PR3 reported in the PDB database. According to modelling, the Der p 3 tertiary structure exhibited a typical fold of serine protease family, conformed by four α-helixes and fifteen β-strands with structural homology with PR3 (RMSD = 0.8) ( Figure 2).
Figure 2. Cartoon models of PR3 and Der p 3.
( A) PR3; ( B) Der p 3. According to modelling by homology, Der p 3 exhibited a typical fold of serine protease family. ( C) Overlapping of 3D model of PR3 and Der p 3. RMSD value of 0.8 is reported, suggesting structural homology.
T and B cell cross-reactive epitopes were predicted between HDM group 3 allergens and PR3
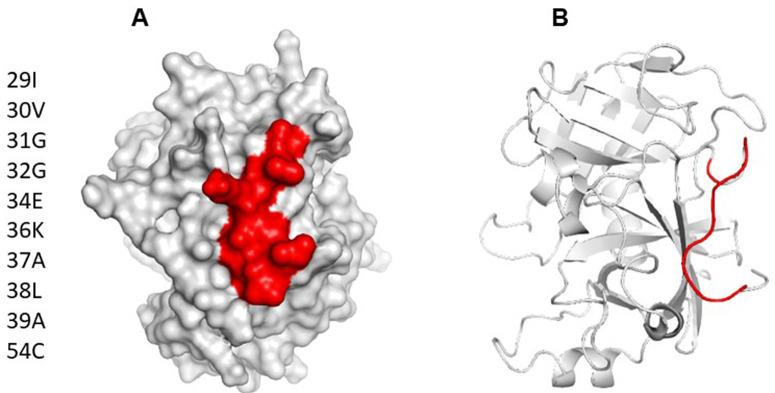
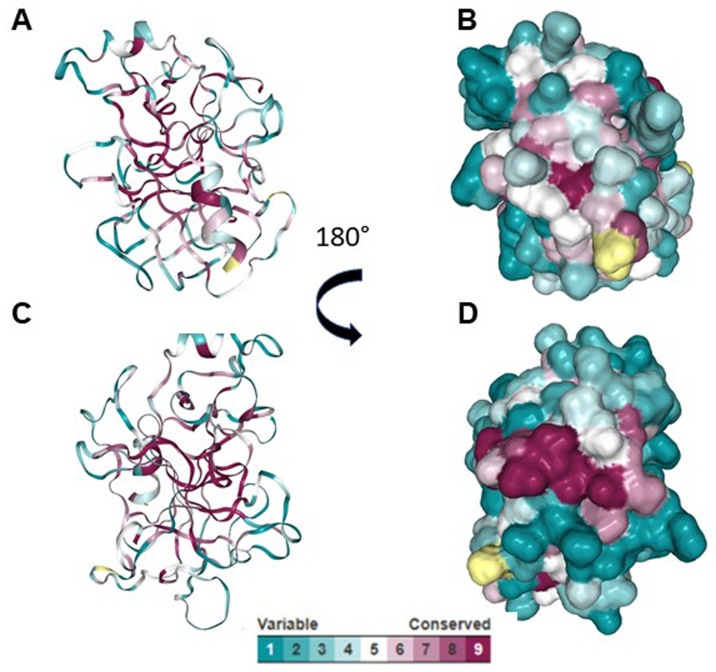
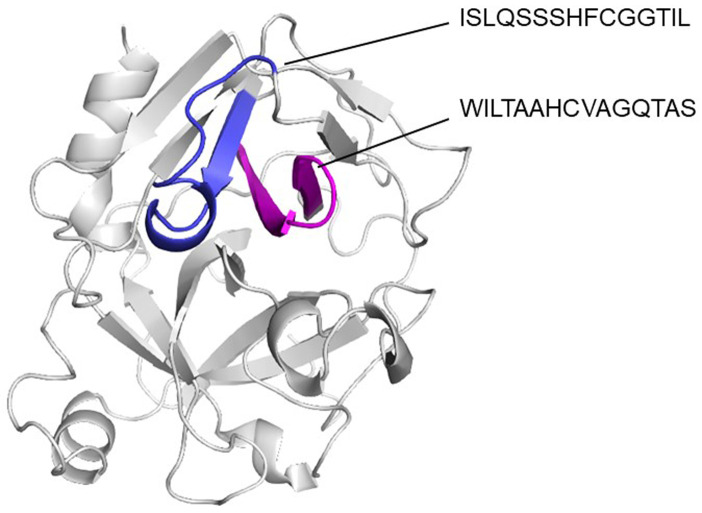
Using ElliPro and BepiPred servers, a cross reactive B cell epitope was predicted on all serine protease used in this study. This epitope is formed by ten residues and is on the N-terminal region, spanning amino acids 29 and 39 with a surface area of 470 Å, not forming part of any domain within the protein. Conservative analysis indicated that the antigenic region predicted was highly conserved in the serine proteases ( Figure 3). According to ConSurf analysis, the region covering the cross-reactive epitope is conserved among the serine protease family ( Figure 4). T cell epitope prediction identified at least two epitopes with potential cross-reactivity among all sequences analyzed. Both epitopes are located on the first and second β strands: the first epitope spans the 45 to 59 region (ISLQSSSHFCGGTIL); and the second, the 63 to 77 region (WILTAAHCVAGQTAS) ( Figure 5; Table 2).
Figure 3.
( A) Surface model of Der p 3, showing area occupied by B cell epitope predicted as cross reactive. ( B) Cartoon model showing location of epitope on tridimensional structure. It can be appreciated that the predicted epitope is on a loop spanning residues 29 to 39.
Figure 4. Phylogenetic analysis of the serine proteases using Consurf.
( A and C) Cartoon models showing the conserved region among serine proteases. ( B and D) Surface models showing the conserved region among serine proteases.
Figure 5. Cartoon model showing the location of T cell epitopes in their tridimensional structure.
It can be appreciated that predicted epitopes are in a continuous β strands (blue and magenta).
Table 2. T cell epitopes predicted in silico.
Both are conserved among PR3 and HDM group 3 allergens.
| Allele | Start | End | Sequence |
|---|---|---|---|
| HLA-DRB1*01:01 | 45 | 59 | ISLQSSSHFCGGTIL |
| HLA-DRB1*01:01 | 63 | 77 | WILTAAHCVAGQTAS |
Discussion
In this study we found that PR3 and HDM group 3 serine protease allergens have conserved identity and homology. Also, for the first time, we predicted various T and B cell cross reactive epitopes between them through an in silico approach. PR3 is an important autoantigen in small vessel vasculitis and it seems to participate in disease inception, progression, and severity 1 . Our results have potential implications for the understanding of autoreactive response in AAV and open the possibility for a new environmental trigger of the autoreactive response in AAV.
In AAV, it has been proposed that autoantibodies directed to a complementary protein to PR3 autoantigen could be implicated in disease inception, and this antisense protein harbors homology to various bacterial peptides 11 – a theory named autoantigen complementarity 33 . However, in epidemiological studies, autoantigen complementarity hypothesis testing has showed conflicting results, since sera from some patients suffering from AAV do not recognize complementary PR3, while others do 34– 36 . Also, molecular mimicry of PR3 protein by infectious microorganism components have been proposed as a possible environmental trigger of the disease based on the reports of infections preceding the manifestations of vasculitis 10, 37– 39 , although a cross reactive antigen have not been reported yet.
In their seminal publication, Pendergraft et al. run a BLAST query to find homologues of PR3 protein in microbial or fungal microorganisms, and do not find matching sequences at that time 11 . However, they do not include Arachnida or other environmental sources of cross-reactivity. In our analysis we find matching PR3 protein sequences with various HDM group 3 serine protease allergens, and at least theoretically this finding could have many implications for the understanding of inception and even diagnosis of autoreactive response in AAV. Recently, Qian et al. have shown that some allergens can cross-react with human proteins 19 and participate in autoimmunity inception in pemphigus vulgaris by a “hit-and-run” mechanism 22 , opening the theoretical possibility for a similar mechanism to occur in another autoimmune disease such as AAV. Similarly, in atopic dermatitis, Valenta and collaborators observe that some patients with severe complications from the disease, had IgE directed to the profilin of the Betula verrucosa, but also to the human homologue 40 .
In the tropics, HDM are important ubiquitous sources of protease allergens. Exposure is perennial, increasing the possibilities of exposure and IgE sensitization to their components in the general population 23– 25 . Sensitization to HDM group 3 allergens is common 26 , and they harbor serine protease activity 27 , a characteristic that make them highly allergenic. Moreover, their conserved structural homology makes them highly immunogenic 41, 42 and suitable for epitope spreading 43 . In this context, “hit-and-run” and epitope spreading establish framework mechanisms for environmental allergens with homology to autoantigens to potentially participate in the development of autoimmunity. We speculate that HDM group 3 allergens harbor two characteristics that make them suitable candidates for environmental triggering of AAV: their proteolytic activity that, as other protease allergens, set a tissue damaging microenvironment during antigen recognition 41 ; and molecular homology-epitope sharing with human PR3, that would elicit B cell autoantibody production and autoreactive T cell receptor generation. In conclusion, we observe that PR3 and HDM group 3 serine protease allergens have conserved identity, and for the first time we predict cross-reactive epitopes between them through an in silico approach.
Data availability
UniProtKB: PRTN3_HUMAN, Accession number P24158: https://www.uniprot.org/uniprot/P24158
Protein Data Bank: PR3 (MYELOBLASTIN), Accession number 1FUJ: https://www.rcsb.org/structure/1FUJ
UniProtKB: Mite allergen Der p 3, Accession number P39675: https://www.uniprot.org/uniprot/P39675
UniProtKB: Trypsin Blo t 3, Accession number A1KXI1: https://www.uniprot.org/uniprot/A1KXI1
UniProtKB: Gly d 3, Accession number Q1M2M8: https://www.uniprot.org/uniprot/Q1M2M8
UniProtKB: Allergen Lep d 3, Accession number Q1M2L7: https://www.uniprot.org/uniprot/Q1M2L7
UniProtKB: Trypsin Tyr p 3.0101, Accession number C6ZDB5: https://www.uniprot.org/uniprot/C6ZDB5
Funding Statement
Buendía, E. received a postdoctoral grant (Fondo Nacional de Financiamiento para la Ciencia, la Tecnología y la Innovación "FRANCISCO JOSÉ DE CALDAS") from Ministerio de Ciencia, Tecnología e Innovación, Colombia (grant number 80740-249-2020).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 2; peer review: 1 approved
References
- 1. Jennette JC, Falk RJ: Pathogenesis of antineutrophil cytoplasmic autoantibody-mediated disease. Nat Rev Rheumatol. 2014;10(8):463–73. 10.1038/nrrheum.2014.103 [DOI] [PubMed] [Google Scholar]
- 2. Wilde B, Thewissen M, Damoiseaux J, et al. : T cells in ANCA-associated vasculitis: what can we learn from lesional versus circulating T cells? Arthritis Res Ther. 2010;12(1):204. 10.1186/ar2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van der Geld YM, Huitema MG, Franssen CF, et al. : In Vitro T lymphocyte responses to proteinase 3 (PR3) and linear peptides of PR3 in patients with Wegener's granulomatosis (WG). Clin Exp Immunol. 2000;122(3):504–13. 10.1046/j.1365-2249.2000.01415.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hogan SL, Falk RJ, Chin H, et al. : Predictors of relapse and treatment resistance in antineutrophil cytoplasmic antibody-associated small-vessel vasculitis. Ann Intern Med. 2005;143(9):621–31. 10.7326/0003-4819-143-9-200511010-00005 [DOI] [PubMed] [Google Scholar]
- 5. Müller A, Voswinkel J, Gottschlich S, et al. : Human proteinase 3 (PR3) and its binding molecules: implications for inflammatory and PR3-related autoimmune responses. Ann N Y Acad Sci. 2007;1109:84–92. 10.1196/annals.1398.010 [DOI] [PubMed] [Google Scholar]
- 6. Specks U: What you should know about PR3-ANCA. Conformational requirements of proteinase 3 (PR3) for enzymatic activity and recognition by PR3-ANCA. Arthritis Res. 2000;2(4):263–7. 10.1186/ar99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Falk RJ, Terrell RS, Charles LA, et al. : Anti-neutrophil cytoplasmic autoantibodies induce neutrophils to degranulate and produce oxygen radicals In Vitro. Proc Natl Acad Sci U S A. 1990;87(11):4115–9. 10.1073/pnas.87.11.4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xiao H, Heeringa P, Hu P, et al. : Antineutrophil cytoplasmic autoantibodies specific for myeloperoxidase cause glomerulonephritis and vasculitis in mice. J Clin Invest. 2002;110(7):955–63. 10.1172/JCI15918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Benoist C, Mathis D: Autoimmunity. The pathogen connection. Nature. 1998;394(6690):227–8. 10.1038/28282 [DOI] [PubMed] [Google Scholar]
- 10. Rojas M, Restrepo-Jiménez P, Monsalve DM, et al. : Molecular mimicry and autoimmunity. J Autoimmun. 2018;95:100–23. 10.1016/j.jaut.2018.10.012 [DOI] [PubMed] [Google Scholar]
- 11. Pendergraft WF, 3rd, Preston GA, Shah RR, et al. : Autoimmunity is triggered by cPR-3(105-201), a protein complementary to human autoantigen proteinase-3. Nat Med. 2004;10(1):72–9. 10.1038/nm968 [DOI] [PubMed] [Google Scholar]
- 12. Fujinaga M, Chernaia MM, Halenbeck R, et al. : The Crystal Structure of PR3, a Neutrophil Serine Proteinase Antigen of Wegener's Granulomatosis Antibodies. J Mol Biol. 1996;261(2):267–78. 10.1006/jmbi.1996.0458 [DOI] [PubMed] [Google Scholar]
- 13. Bruner BF, Vista ES, Wynn DM, et al. : Anti-neutrophil cytoplasmic antibodies target sequential functional proteinase 3 epitopes in the sera of patients with Wegener’s granulomatosis. Clin Exp Immunol. 2010;162(2):262–70. 10.1111/j.1365-2249.2010.04251.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuhl A, Korkmaz B, Utecht B, et al. : Mapping of conformational epitopes on human proteinase 3, the autoantigen of Wegener's granulomatosis. J Immunol. 2010;185(1):387–99. 10.4049/jimmunol.0903887 [DOI] [PubMed] [Google Scholar]
- 15. Hasni S, Gupta S, Davis M, et al. : Safety and Tolerability of Omalizumab: A Randomized Clinical Trial of Humanized Anti-IgE Monoclonal Antibody in Systemic Lupus Erythematosus. Arthritis Rheumatol. 2019;71(7):1135–40. 10.1002/art.40828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Glaser AG, Menz G, Kirsch AI, et al. : Auto- and cross-reactivity to thioredoxin allergens in allergic bronchopulmonary aspergillosis. Allergy. 2008;63(12):1617–23. 10.1111/j.1398-9995.2008.01777.x [DOI] [PubMed] [Google Scholar]
- 17. Roesner LM, Heratizadeh A, Wieschowski S, et al. : α-NAC-Specific Autoreactive CD8 + T Cells in Atopic Dermatitis Are of an Effector Memory Type and Secrete IL-4 and IFN-γ. J Immunol. 2016;196(8):3245–52. 10.4049/jimmunol.1500351 [DOI] [PubMed] [Google Scholar]
- 18. Sánchez J, Sánchez A, Cardona R: Causal Relationship Between Anti-TPO IgE and Chronic Urticaria by In Vitro and In Vivo Tests. Allergy Asthma Immunol Res. 2019;11(1):29–42. 10.4168/aair.2019.11.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qian Y, Jeong JS, Maldonado M, et al. : Cutting Edge: Brazilian pemphigus foliaceus anti-desmoglein 1 autoantibodies cross-react with sand fly salivary LJM11 antigen. J Immunol. 2012;189(4):1535–9. 10.4049/jimmunol.1200842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roesner LM, Ernst M, Chen W, et al. : Human thioredoxin, a damage-associated molecular pattern and Malassezia-crossreactive autoallergen, modulates immune responses via the C-type lectin receptors Dectin-1 and Dectin-2. Sci Rep. 2019;9(1):11210. 10.1038/s41598-019-47769-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Balaji H, Heratizadeh A, Wichmann K, et al. : Malassezia sympodialis thioredoxin-specific T cells are highly cross-reactive to human thioredoxin in atopic dermatitis. J Allergy Clin Immunol. 2011;128(1):92–9.e4. 10.1016/j.jaci.2011.02.043 [DOI] [PubMed] [Google Scholar]
- 22. Lin L, Moran TP, Peng B, et al. : Walnut antigens can trigger autoantibody development in patients with pemphigus vulgaris through a "hit-and-run" mechanism. J Allergy Clin Immunol. 2019;144(3):720–8.e4. 10.1016/j.jaci.2019.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Acevedo N, Zakzuk J, Caraballo L: House Dust Mite Allergy Under Changing Environments. Allergy Asthma Immunol Res. 2019;11(4):450–69. 10.4168/aair.2019.11.4.450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zakzuk J, Mercado D, Bornacelly A, et al. : Hygienic conditions influence sensitization to Blomia tropicalis allergenic components: Results from the FRAAT birth cohort. Pediatr Allergy Immunol. 2019;30(2):172–8. 10.1111/pai.13004 [DOI] [PubMed] [Google Scholar]
- 25. Acevedo N, Sánchez J, Zakzuk J, et al. : Particular characteristics of allergic symptoms in tropical environments: follow up to 24 months in the FRAAT birth cohort study. BMC Pulm Med. 2012;12:13. 10.1186/1471-2466-12-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sánchez-Borges M, Fernandez-Caldas E, Thomas WR, et al. : International consensus (ICON) on: clinical consequences of mite hypersensitivity, a global problem. World Allergy Organ J. 2017;10(1):14. 10.1186/s40413-017-0145-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stewart GA, Ward LD, Simpson RJ, et al. : The group III allergen from the house dust mite Dermatophagoides pteronyssinus is a trypsin-like enzyme. Immunology. 1992;75(1):29–35. [PMC free article] [PubMed] [Google Scholar]
- 28. Clamp M, Cuff J, Searle SM, et al. : The Jalview Java alignment editor. Bioinformatics. 2004;20(3):426–7. 10.1093/bioinformatics/btg430 [DOI] [PubMed] [Google Scholar]
- 29. Wiederstein M, Sippl MJ: ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(Web Server issue):W407–10. 10.1093/nar/gkm290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pettersen EF, Goddard TD, Huang CC, et al. : UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–12. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 31. Ponomarenko J, Bui HH, Li W, et al. : ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics. 2008;9:514. 10.1186/1471-2105-9-514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jespersen MC, Peters B, Nielsen M, et al. : BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45(W1):W24–w9. 10.1093/nar/gkx346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pendergraft WF, 3rd, Pressler BM, Jennette JC, et al. : Autoantigen complementarity: a new theory implicating complementary proteins as initiators of autoimmune disease. J Mol Med (Berl). 2005;83(1):12–25. 10.1007/s00109-004-0615-3 [DOI] [PubMed] [Google Scholar]
- 34. Tadema H, Kallenberg CG, Stegeman CA, et al. : Reactivity against complementary proteinase-3 is not increased in patients with PR3-ANCA-associated vasculitis. PLoS One. 2011;6(3):e17972. 10.1371/journal.pone.0017972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hewins P, Belmonte F, Charles Jennette J, et al. : Longitudinal studies of patients with ANCA vasculitis demonstrate concurrent reactivity to complementary PR3 protein segments cPR3m and cPR3C and with no reactivity to cPR3N. Autoimmunity. 2011;44(2):98–106. 10.3109/08916934.2010.491843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang J, Bautz DJ, Lionaki S, et al. : ANCA patients have T cells responsive to complementary PR-3 antigen. Kidney Int. 2008;74(9):1159–69. 10.1038/ki.2008.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mezzano S, Valderrama G, Olavarria F, et al. : Antineutrophil-cytoplasmic-autoantibodies in poststreptococcal nephritis. Adv Exp Med Biol. 1993;336:449–53. 10.1007/978-1-4757-9182-2_80 [DOI] [PubMed] [Google Scholar]
- 38. Hellmich B, Ehren M, Lindstaedt M, et al. : Anti-MPO-ANCA-positive microscopic polyangiitis following subacute bacterial endocarditis. Clin Rheumatol. 2001;20(6):441–3. 10.1007/pl00011214 [DOI] [PubMed] [Google Scholar]
- 39. Pudifin DJ, Duursma J, Gathiram V, et al. : Invasive amoebiasis is associated with the development of anti-neutrophil cytoplasmic antibody. Clin Exp Immunol. 1994;97(1):48–51. 10.1111/j.1365-2249.1994.tb06578.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Valenta R, Duchêne M, Pettenburger K, et al. : Identification of profilin as a novel pollen allergen; IgE autoreactivity in sensitized individuals. Science. 1991;253(5019):557–60. 10.1126/science.1857985 [DOI] [PubMed] [Google Scholar]
- 41. Cayrol C, Duval A, Schmitt P, et al. : Environmental allergens induce allergic inflammation through proteolytic maturation of IL-33. Nat Immunol. 2018;19(4):375–85. 10.1038/s41590-018-0067-5 [DOI] [PubMed] [Google Scholar]
- 42. Westernberg L, Schulten V, Greenbaum JA, et al. : T-cell epitope conservation across allergen species is a major determinant of immunogenicity. J Allergy Clin Immunol. 2016;138(2):571–8.e7. 10.1016/j.jaci.2015.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Burastero SE: T-cell receptor-mediated cross-reactivity to different allergens is driven by recognition of homologous, phylogenetically conserved epitopes. J Allergy Clin Immunol. 2016;138(4):1237. 10.1016/j.jaci.2016.04.055 [DOI] [PubMed] [Google Scholar]