CONSPECTUS
The modern healthcare system faces an unrelenting threat from microorganisms, evidenced by global outbreaks of new viral diseases, emerging antimicrobial resistance, and the rising incidence of healthcare-associated infections (HAIs). An effective response to these threats requires rapid and accurate diagnostic tests that can identify causative pathogens at the point-of-care (POC). Such tests could eliminate diagnostic uncertainties, facilitating patient triaging, minimizing empiric use of antimicrobial drugs, and enabling targeted treatments. Current standard methods, however, often fail to meet the needs of rapid diagnosis in POC settings. Culture-based assays entail long processing times and require specialized laboratory infrastructure; nucleic-acid (NA) tests are often limited to centralized hospitals due to assay complexity and high costs.
Here we discuss two new POC tests developed in our groups to enable rapid diagnosis of infection. The first is nanoPCR that takes advantages of core-shell magnetoplasmonic nanoparticles (MPNs): i) Au shell significantly accelerates thermocycling via volumetric, plasmonic light-to-heat conversion, ii) magnetic core enables sensitive in-situ fluorescent detection via magnetic clearing. By adopting a Ferris wheel module, the system expedites multi-samples in parallel with minimal setup. When applying to COVID-19 diagnosis, nanoPCR detected SARS-CoV-2 RNA down to 3.2 copy/μL within 17 min. In particular, nanoPCR diagnostics accurately identified COVID-19 cases in clinical samples (n = 150), validating its clinical applicability.
The second is a polarization anisotropy diagnostic (PAD) system that exploits the principle of fluorescence polarization (FP) as a detection modality. Fluorescent probes were designed to alter their molecular weight upon recognizing target NAs. This event modulates probes’ tumbling rate (Brownian motion), which leads to changes in FP. The approach is robust against environmental noise and benefits from the ratiometric nature of the signal readout. We applied PAD to detect clinically relevant HAI bacteria (Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Staphylococcus aureus). The PAD assay demonstrated detection sensitivity down to the single bacterium level and determined both drug resistance and virulence status.
In summary, these new tests have a potential to become powerful tools for rapid diagnosis in the infectious disease space. They do not require highly skilled personnel or labor-intensive analyses, and the assays are quick and cost effective. These attributes will make nanoPCR and PAD well-aligned with a POC workflow to aid physicians to initiate prompt and informed patient treatment.
Graphical Abstract
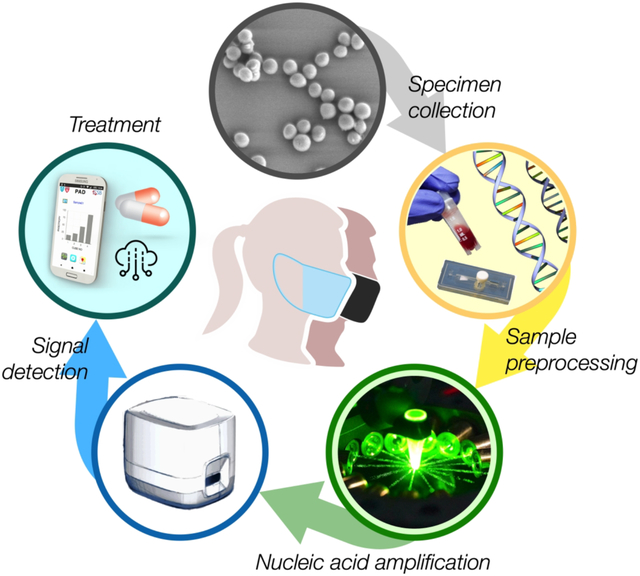
1. INTRODUCTION
The threat of infectious diseases is globally omnipresent.5–9 The ongoing COVID-19 pandemic has claimed more than 2.6 million lives within the first 15 months of its outbreak; the global economy reportedly has lost over $7 trillion dollars in 2020 gross domestic product.10 Emerging antimicrobial-resistance (AMR) is another lurking threat. In the US alone, close to three million people are infected with antimicrobial-resistant bacteria, and more than 30,000 people die as a result of such infections.11–13 The economic burden is also substantial. The annual cost from healthcare-associated infections (HAIs) is over $30 billion dollars in the US.14,15
The most effective way to combat infectious diseases is to implement rapid, accurate diagnostic tests at the point-of-care (POC).16–18 Such tests would eliminate diagnostic delays and uncertainties, enabling the timely initiation of the most appropriate therapies and minimizing emergence of resistance through indiscriminate use of antibiotics. Unfortunately, current diagnostic methods often fail to meet these goals, because the assays are either too lengthy or not readily available at the POC settings. For example, culture-based tests for bacterial detection and AMR monitoring, albeit the gold standard, have drawbacks for on-site use, including long processing time (several days), personnel cost, and needs for specialized equipment and species-specific protocols.19 As an alternative, nucleic-acid (NA) assays have been increasingly adopted for clinical diagnostics.20–23 These tests can produce comprehensive information for pathogens; the target sequence library is rapidly expanding with advances in whole-genome sequencing.24 Technical constraints, however, limit the use of NA tests to centralized hospital laboratories: most tests currently cost > $100 in order to recoup the large capital expenditure on equipment ($10,000 – $150,000). Furthermore, NA-based systems are bulky and/or house sophisticated optics which require routine calibration.
The workflow of POC testing consists of i) sample preprocessing to extract NA from biological samples (i.e. blood, swab, saliva, urine), ii) NA assay to amplify the signal, iii) signal detection, and iv) decision making. The key challenge is to streamline the entire process so that the test can be completed while patients are waiting. Aiming at this goal, we have been developing new biosensing modalities whose functions range from sample preprocessing,1,25 rapid NA amplification,2 to signal detection.3,4,26–34 Our recent efforts have focused on exploring new, integrated technologies that improve signal transduction and/or assay speed; this led to the development of i) a nanoPCR approach that significantly sped up conventional polymerase chain reaction (PCR) through plasmonic heating,2 and ii) a fluorescence polarization (FP) method that yielded robust results compared to direct fluorescence.3,4,28 Here, we will review the development of these assay systems, covering the detection principles, devices for sample preprocessing and signal detection, clinical applications, and future directions.
2. Disposable fluidic cartridges for sample processing
Purifying or conditioning native clinical samples is a key step that significantly influences the accuracy of downstream analyses. Various commercial kits are available for such sample pre-processing, but most of them require benchtop equipment (e.g., centrifuge) and involve multiple fluidic transfers. To streamline liquid handling while minimizing external setups, we adopted microfluidics and developed self-contained devices. Figure 1A shows a fluidic chip designed for sample purification. We sandwiched a fluidic chip between two magnetic arrays, capturing magnetically labeled cells in flow.1 This system, in conjunction with target-specific magnetic nanoparticles, was used to enrich pathogens (positive selection) or remove other blood cells (negative selection); these processes effectively concentrated pathogen targets and thereby improved assay sensitivity.
Figure 1. Fluidic cartridges for sample processing.
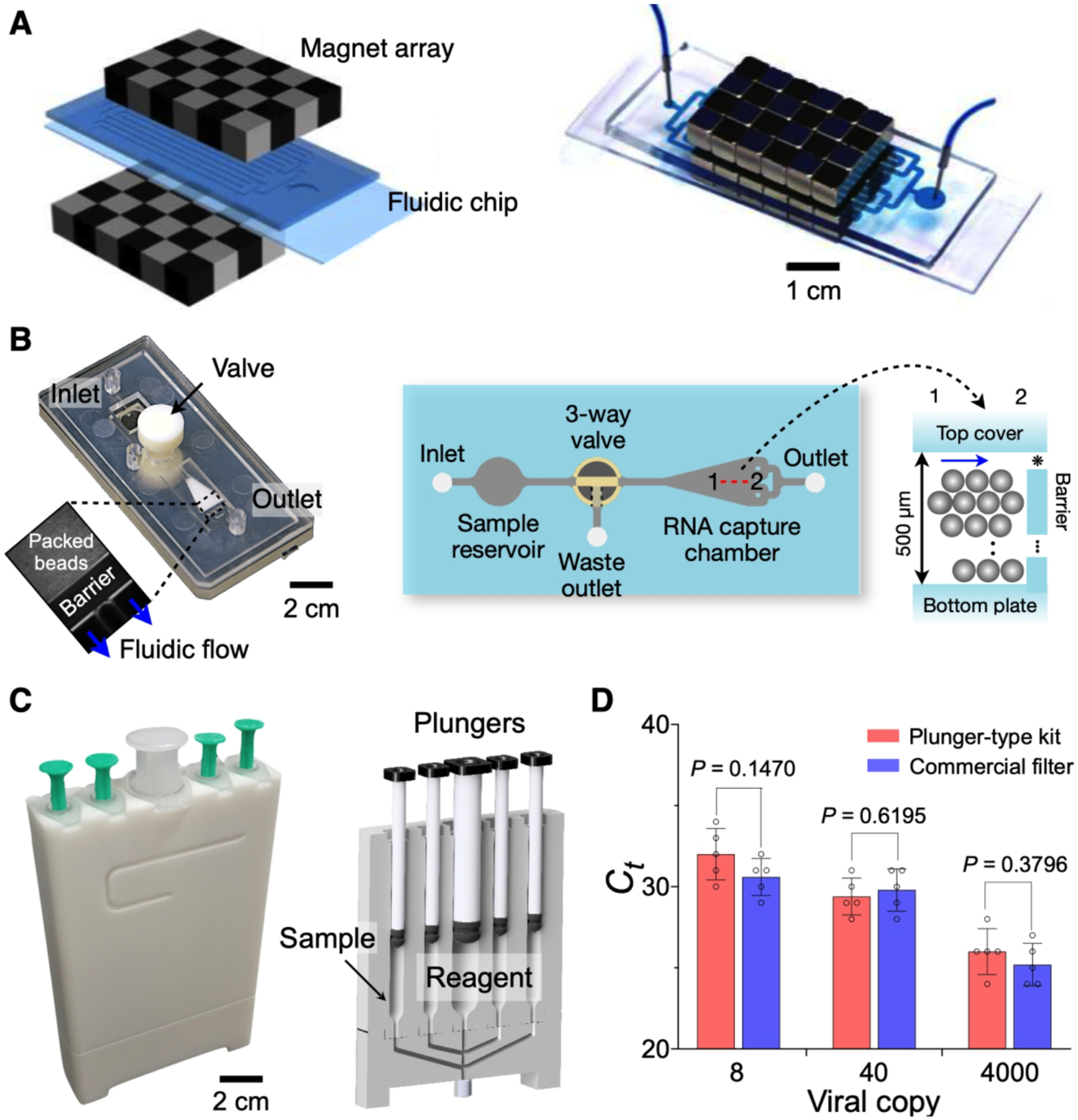
(A) Magnetic separator to capture magnetically labeled cells. The device was constructed by placing a fluidic device between checkerboard-type magnetic arrays. Through immunomagnetic selection, either pathogenic cells are captured or host cells are removed. (B) Photograph (left) and top view (right) of the fluidic NA extraction chip. Glass beads are filled in the capture chamber and retained in the barrier, allowing for fluidic flow (inset). Gap (*), 10 μm. The 3-way valve with push-button separates wastes to the waste outlet. (C) Photograph (left) and cross-sectional configuration (right) of a plunger-type RNA extraction kit. (D) Comparison of RNA extraction yield between the plunger-type kit and a commercial filter using RT-qPCR. The results were comparable to each other (t-test). (A), adapted with permission from ref 1. Copyright © 2017, American Chemical Society. (B), adapted with permission from ref 3. Copyright 2016 © The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. (C, D) adapted with permission from ref 2. Copyright © 2020, The Authors, under exclusive license to Springer Nature Limited.
For on-site NA detection, we adapted the principle of solid-phase NA extraction into fluidic devices. The extraction used silica as substrates; negatively charged NAs would bind to silica surface in the presence of chaotropic salts. Following wash steps, adsorbed NAs are eluted with low salt solution. We used either pure silica beads,2,3 or plastic microbeads coated with silica.25 Figure 1B shows a fluidic chip implemented in poly(methylmethacrylate) via injection molding.3 The chip had a fluidic chamber containing glass beads (30 μm in diameter). Samples and other reagents were sequentially injected for NA extraction. When compared to a commercial column filter, the fluidic chip showed comparable performance in RNA quality.3 More recently, we have designed a plunger-type kit (Figure 1C) that eliminated the need for external fluidic connection.2 The kit had multiple chambers preloaded with NA extraction reagents and a silica filter. After initial sample loading, we sequentially pushed plungers to perform pathogen lysis, RNA capture, washing, and elution. RNA extraction yields, measured by RT-qPCR, were comparable between the plunger-type kit and commercial filters (Figure 1D), but the operation was equipment-free and fast (3 min) with the plunger kit.
3. NanoPCR for rapid thermocycling and detection
Thermoplasmonics, which exploits light-to-heat conversion mediated by plasmonic substrates,35 is an emerging technique to replace slow conductive heating process. We specifically explored the use of hybrid magnetoplasmonic nanoparticles (MPNs) that consisted of a magnetic core and a gold (Au) shell (Figure 2A). The choice was motivated by the observation that most plasmonic effect is confined near the surface of metallic nanoparticles. We reason that MPNs would enable i) efficient plasmonic heating through the Au shell and ii) rapid clearing of particles through the magnetic interaction between the core and external magnetic fields.
Figure 2. Characterization of MPNs.
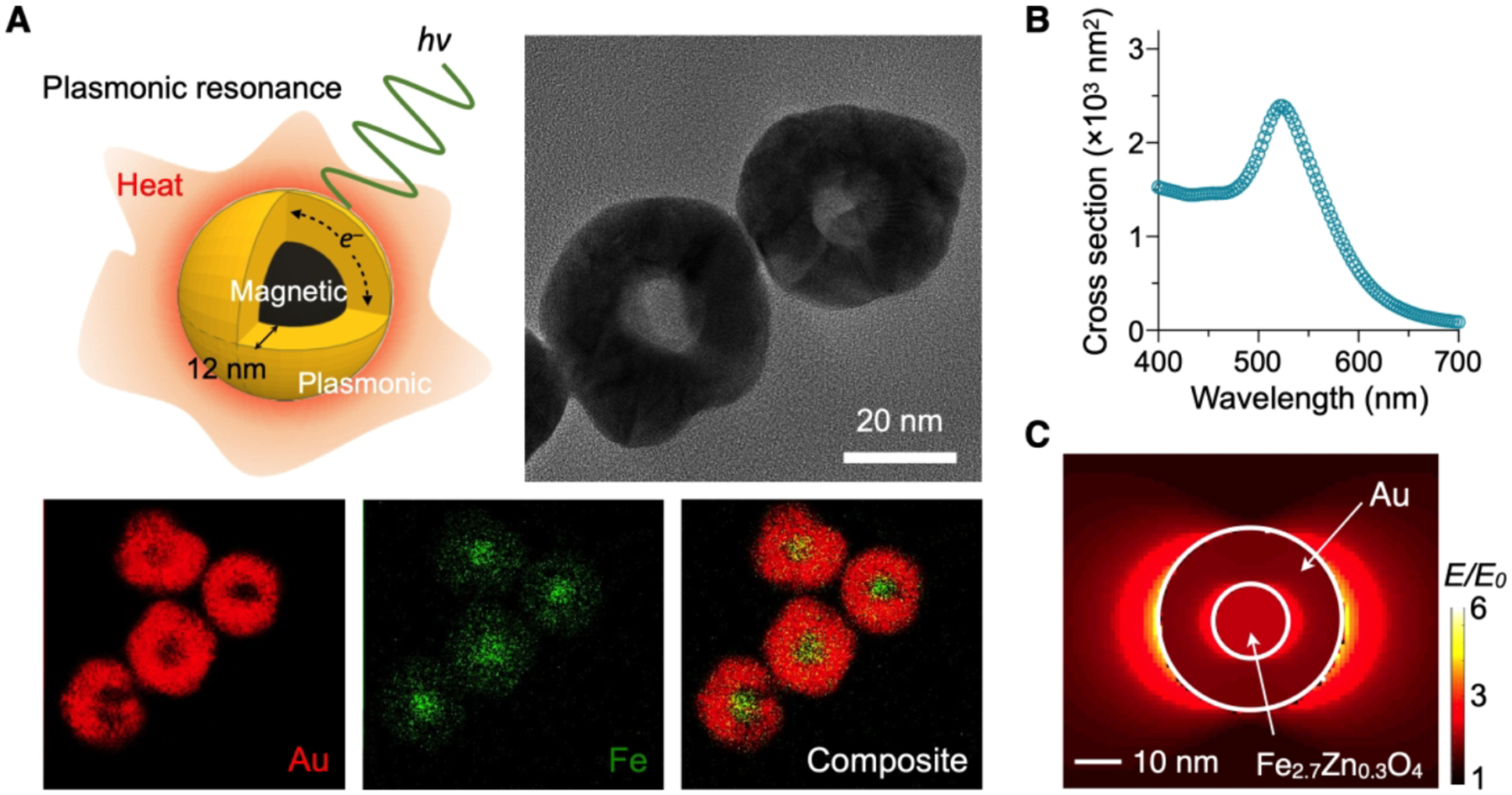
(A) Schematic and TEM images of MPNs (top) and elemental mapping for Au and Fe (bottom). A magnetic core was coated with a 12-nm thick Au shell. (B) Cross-sectional absorption spectrum of MPNs. For 12-nm thick Au shell, the peak absorbance was at 535 nm. (C) Electric field simulation at the surface plasmon resonance condition. Electric field of MPN was mainly confined to the inner and outer surfaces and its field enhancement factor (E/E0 = 5.8) at the outer one was almost the same as that of 40 nm AuNP (E/E0 = 6.0). Adapted with permission from ref 2. Copyright © 2020, The Authors, under exclusive licence to Springer Nature Limited.
3.1. Synthesis and characterization of MPNs.
MNPs had a 16-nm magnetic core (Zn0.4Fe2.6O4) and a 12 nm-thick Au shell (Figure 2A).2,36 We further coated particles with phosphine-sulfonate ligands to impart negative surface charges for colloidal stability. MPNs showed plasmon resonance at a wavelength of λ = 535 nm (Figure 2B). Numerical simulation showed that the electrical field enhancement would be confined to the inner and the outer surfaces of Au shell under plasmonic resonance condition (Figure 2C). The field enhancement and the absorption cross-section of a MPN would be similar to those of a 40 nm AuNP.2
MPNs in solution quickly heated the entire sample upon light illumination (Figure 3A). The heating was effective when the incident wavelength matched with the MPNs’ plasmonic resonance, confirming the underlying thermoplasmonic mechanism (Figure 3B).35,37,38 Using MPNs as a heating source, we could achieve rapid thermocycling required for PCR: i) the cycling time from 58 to 90 and back to 58 °C was 8.91 sec and ii) the coefficient of variations were < 2% (Figure 3C). Sample cooling (~6.5 s from 90 to 58 °C) was based on a convective heat transfer to air.
Figure 3. MPN-mediated thermocycling.
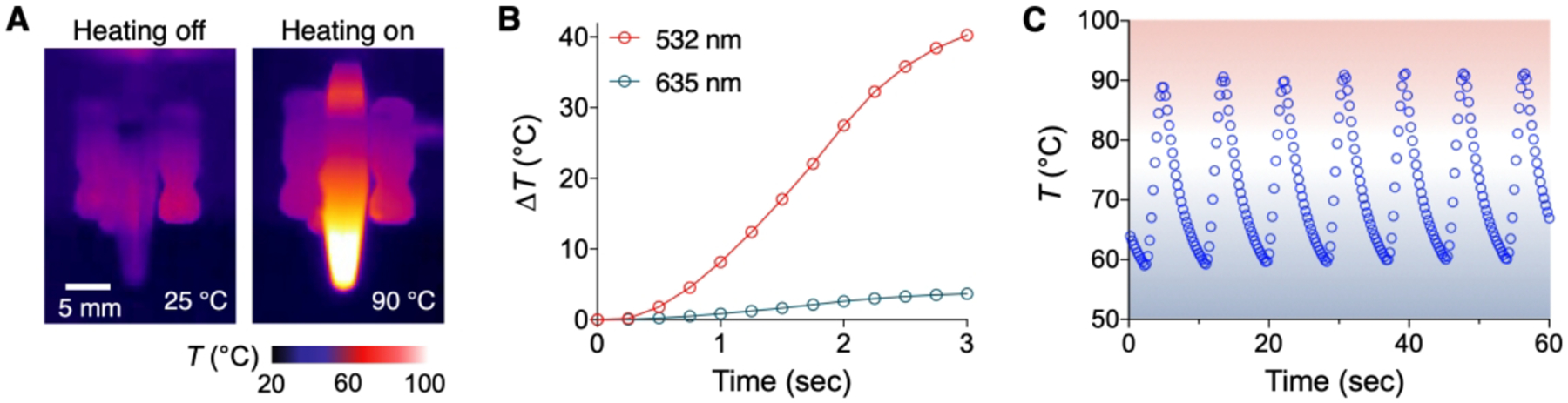
(A) Thermal images of MPN solutions. Upon illumination, the solution temperature increased from 25 to 90 °C via plasmonic heating (light source = 1 W at 532 nm, solution volume = 10 μL, [MPN] = 2.6 × 1011 particles/mL). (B) Temperature profile of MPN solution at different illumination wavelengths. Temperature increased rapidly when the illumination wavelength matched with the peak absorption wavelength of MPNs (535 nm). (C) Plasmonic thermocycling profile of MPN solution. Seven cycles (58 ↔ 90 °C) were completed within a minute. Adapted with permission from ref 2. Copyright © 2020, The Authors, under exclusive licence to Springer Nature Limited.
3.2. NanoPCR instrument
To facilitate its POC operation, we have built a compact nanoPCR system (15×15×18.5 cm3, 3 kg; Figure 4A). The system integrated a light source for plasmonic heating, a rotating sample holder (i.e., ‘Ferris wheel’), a movable magnet assembly, and fluorescent detection optics. As a light source, we used a circular array of low-power laser diodes (80 mW) rather than a single high-power laser (Figure 4A, right); such an arrangement promoted uniform heating and eliminated the need to cool the light source. For multi-sample processing, we employed a Ferris wheel scheme (Figure 4B). The rotating wheel sequentially placed a sample under focused light while others being cooled. By syncing wheel rotation with an illumination timing (2.43 s under light; 6.48 s with air cooling), we were able to perform PCR reactions in multiple samples using a single light source (Figure 4C). A microcontroller coordinated all the system operation, including thermocycling, MPN collection, fluorescent measurement, and data display.
Figure 4. nanoPCR system.
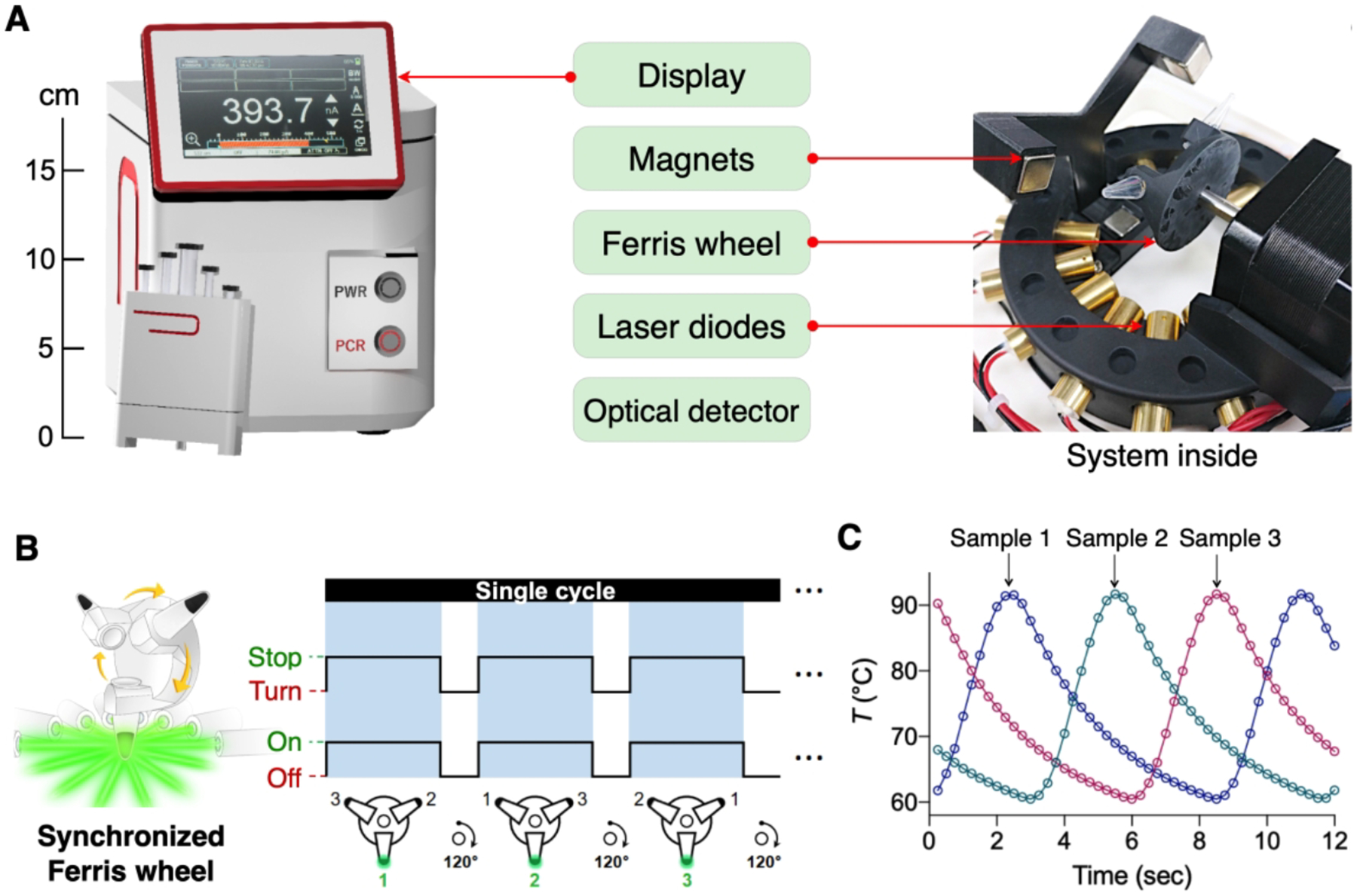
(A) The system housed a light source for plasmonic heating, a Ferris wheel for multi-sample processing, a magnet assembly, optics for fluorescent detection, and a screen for prompt result display. The system was automated to conduct the entire procedures with a single button push. (B) A circular array of the low-powered laser diodes heated samples on the Ferris wheel. Syncing laser illumination with Ferris wheel rotation enabled the processing of multiple samples without compromising the total assay time. (C) Temperature profile of each sample on the wheel. As the system was designed to heat one sample while others cooled, individual profiles were interleaved. Adapted with permission from ref 2. Copyright © 2020, The Authors, under exclusive licence to Springer Nature Limited.
3.3. COVID-19 diagnosis
We set up the overall nanoPCR protocol for rapid COVID-19 diagnoses (Figure 5A). Because SARS-CoV-2 is an RNA virus, an initial reverse transcription (RT) step was necessary to convert RNA into complementary DNA using reverse transcriptase. To achieve the constant temperate (42 °C) required for reverse transcriptase activity, we modulated the on/off duty cycle of the light source (Figure 5A, inset). Following the RT (5 min) step, the nanoPCR system carried out fast thermocycling (6 min) on three targets: nucleocapsid genes (N1 and N2) of SARS-CoV-2 and human ribonuclease P30 subunit gene (RPP30).17 The nanoPCR (11 min) and a conventional benchtop (2 hr) systems showed comparable performances; the gel electrophoresis on PCR products showed matching bands in size and intensity (Figure 5B). For in-situ fluorescence detection after thermal cycling, the nanoPCR system moved the magnet assembly to sample tubes, which sedimented MPNs to the tube bottom. This step minimized the optical interference from MPNs whose absorbance (λpeak = 535 nm) overlaps with fluorescent emission (λpeak = 517 nm) of amplicons. Indeed, the fluorescent signal was barely detectable after thermocycling and only to appear after magnetic pulling (Figure 5C). We set the pulling time to 3 min, which recovered ≥ 50% of the signal from its saturation level. The limit of detection (LOD) was determined to be 3.2 copy/μL from the dilution series of N1 samples (Figure 5D), which was comparable to that of conventional RT-qPCR (2.0 copy/μL). In addition, the designed N1 and N2 probes had negligible cross-reactivity with other zoonotonic coronaviruses, SARS-CoV and MERS-CoV (Figure 5E).
Figure 5. NanoPCR for SARS-CoV-2 RNA detection.
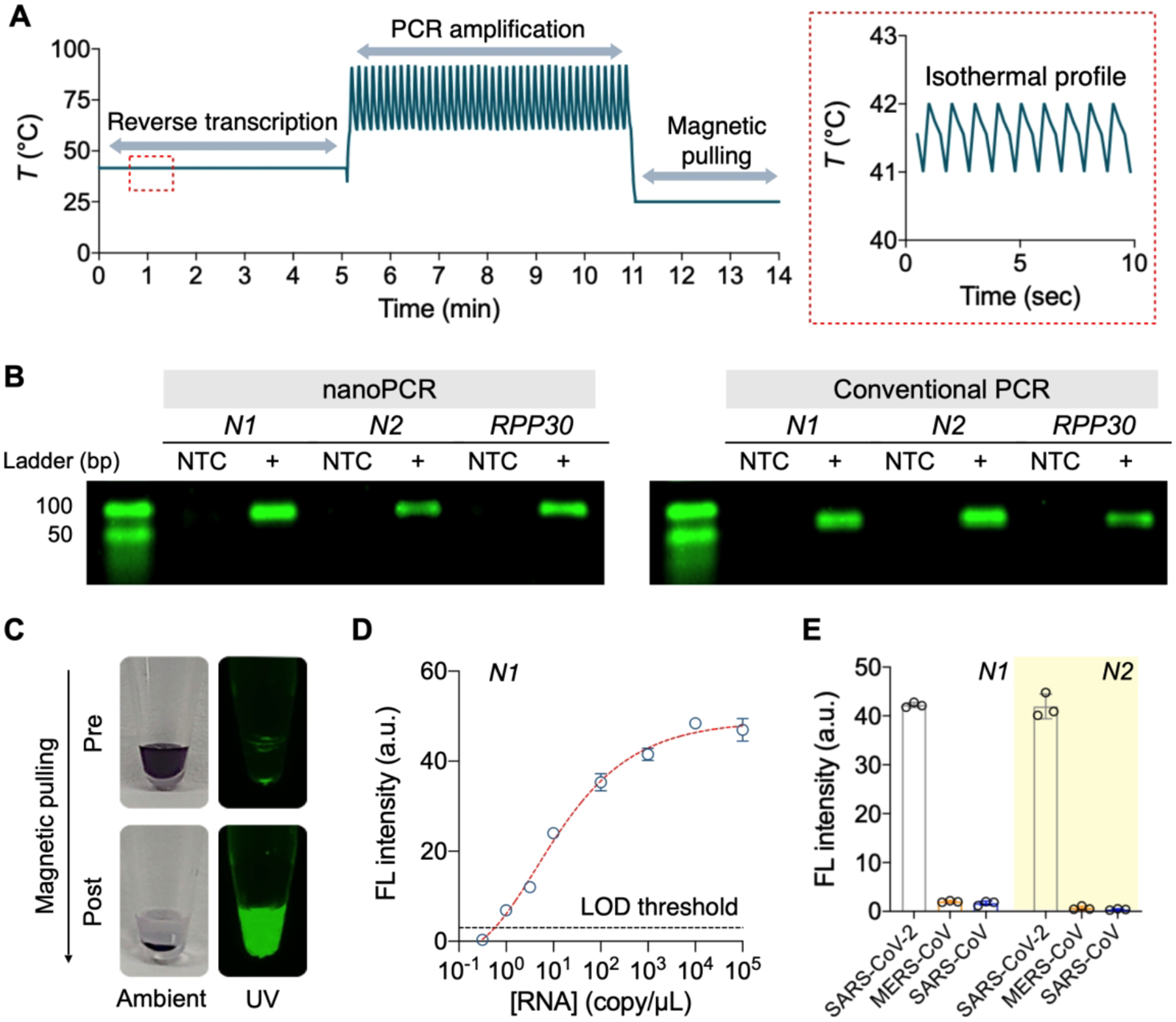
(A) Temperature profile of nanoPCR for SARS-CoV-2 RNA detection. After RT step at 42 °C for 5 min (enlarged in a red box), PCR was performed for 6 min (40 cycles), followed by signal detection (3 min). (B) Gel electrophoresis images of PCR products (N1, N2, and RPP30 genes). NanoPCR produced almost identical bands to those of conventional benchtop thermocycler. NTC: non-target control. (C) Photographs of an amplified sample before and after magnetic pulling. The fluorescence signal was recovered as MPNs were pulled down. (D) Quantitative analysis of N1 RNA by nanoPCR. The estimated limit of detection (LOD) was 3.2 copy/μL. (E) Specific detection of target genes of SARS-CoV-2 RNA. NanoPCR assay distinguished SARS-CoV-2 from other zoonotonic coronaviruses. All data are mean ± s.d. (triplicates unless otherwise indicated). Adapted with permission from ref 2. Copyright © 2020, The Authors, under exclusive licence to Springer Nature Limited.
3.4. Clinical application of nanoPCR
As a pilot clinical application, we used nanoPCR to diagnose COVID-19 in clinical specimens (i.e., nasopharyngeal/oropharyngeal swabs and sputum). We tested 100 samples as a discovery cohort (50 COVID-19 patients and 50 controls) and another 50 samples (25 COVID-19 patients and 25 controls) as a validation cohort. Due to the safety concern, we used RNA extracts from clinical specimens. Each RNA sample was divided into two aliquots; one was processed by nanoPCR and the other by conventional RT-qPCR (Figure 6A). We compared nanoPCR’s fluorescence intensity, I with −Log2Ct, where Ct is the cycle cutoff of RT-qPCR. These two quantities would be proportional to target gene concentrations, and indeed showed a good concordance (Figure 6B). We next evaluated the diagnostic accuracy of nanoPCR. As an analytical metrics, we defined FN1 and FN2 for N1 and N2 genes, respectively, by normalizing target-induced fluorescent intensities with that of the positive control (RPP30). Both FN1 and FN2 values were significantly higher (P < 0.0001, two-sided t-test) in COVID-19 patients than non-COVID-19 controls (Figure 6C).
Figure 6. Clinical application of nanoPCR to COVID-19 diagnosis.
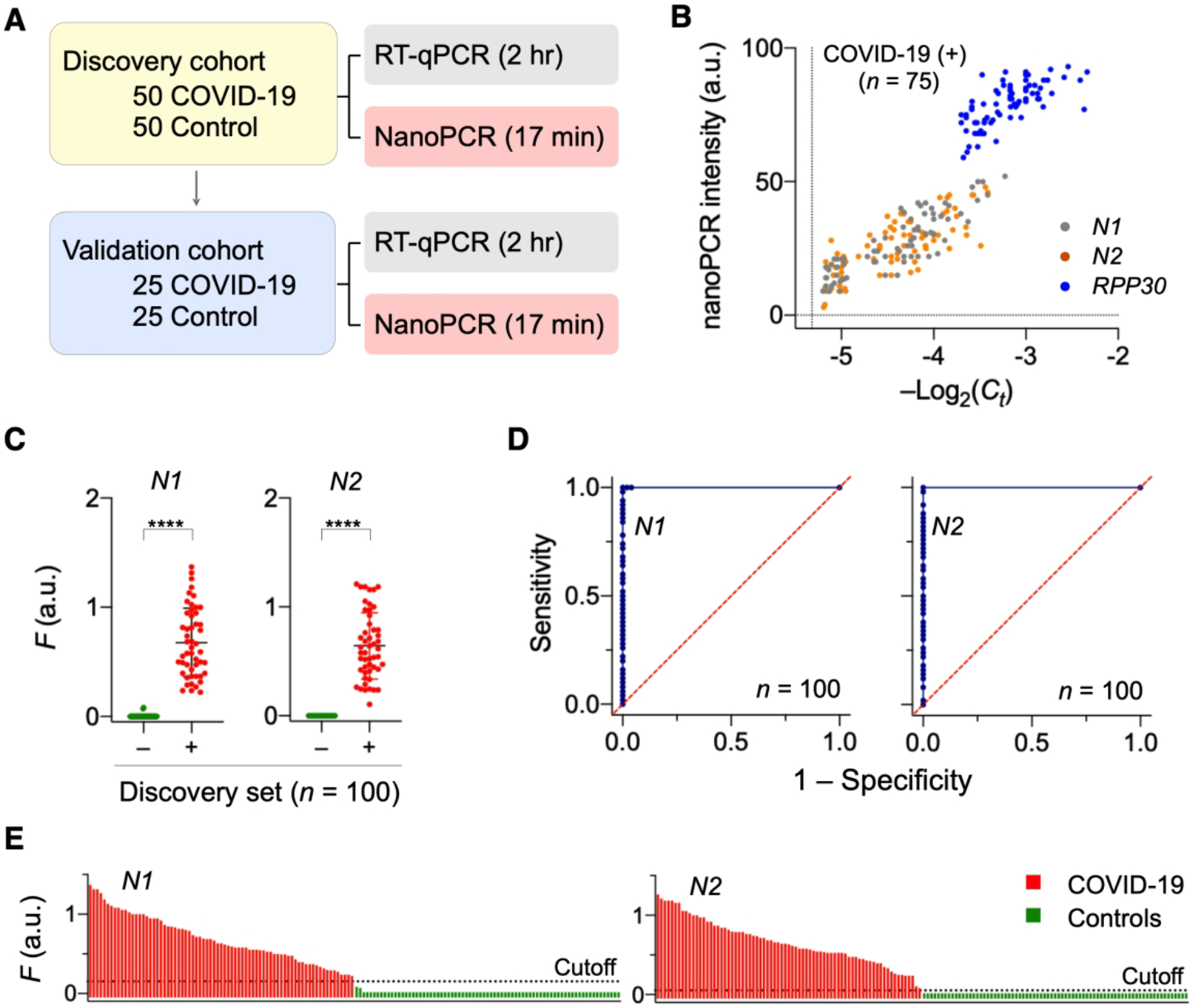
(A) Clinical study design (n = 150). First 100 samples were used as a discovery cohort while the other 50 samples a validation set. Each sample was aliquoted for nanoPCR and conventional RT-qPCR. (B) Analytical concordance between nanoPCR and RT-qPCR. The results for target genes were positively correlated (Pearson’s r values: rN1 = 0.87, rN2 = 0.78, rRPP30 = 0.70). (C) Analysis of the discovery cohort. Normalized signals (FN1 and FN2) from COVID-19 patients (+) were significantly higher than those from controls (−) (****P < 0.0001; two-sided t-test; n = 100). (D) Receiver operation characteristic (ROC) curves for the discovery cohort. The cut-off F values for N1 and N2 were determined from ROC curves. (E) Waterfall plots of FN1 and FN2 of the all samples (n = 150). Adapted with permission from ref 2. Copyright © 2020, The Authors, under exclusive licence to Springer Nature Limited.
We further constructed receiver operating characteristic (ROC) curves for FN1 and FN2, graphically plotting the true positive rate (sensitivity) against the false positive rate (1 – specificity) at varying FN1 and FN2 values. ROC curves characterize the diagnostic capability of a binary classifier system and can be used to determine the optimal cutoff values for diagnostic decision making. The diagnostic accuracy was excellent with an area under the curve of 1 in ROC curve analyses (Figure 6D). The cutoff values were determined to be 0.15 for FN1 and 0.05 for FN2, that maximized the sum of sensitivity and specificity. When these cutoffs were applied to the validation set, nanoPCR maintained its high diagnostic power. Overall, nanoPCR correctly classified all clinical samples (n = 150; Figure 6E).
4. Fluorescence polarization system
FP measurement relies on differential rotational motion of fluorescent molecules according to their molecular weight (Figure 7A).39 Large molecules tumble slowly in solution so their emission will have a polarization angle that is very similar to that of the excitation light. In contrast, small molecules will rotate quickly, thereby their fluorescent emission effectively depolarized. FP’s analytical metric (r) is the ratio of light intensities, r = (I∥ – I⊥) / (I∥ + 2·I⊥), where I∥ (I⊥) is the parallel (perpendicular) component of fluorescence intensities relative to the exciting light. Small fluorescent molecules will have negligible r as I∥ is close to I⊥. Conversely, large molecules (or molecules bound to a stationary target) will show high r values. FP readout is advantageous over measuring raw fluorescence. The stability requirement for the excitation light source can be less stringent as FP is independent of the incident light intensity. FP detection is also robust to environmental noise (e.g., electrical and optical signal fluctuations). Dual optical detectors, each capturing an orthogonal polarization component, can compensate one another in the event of common noise. These detectors can also be monitored to reject single-channel noise that causes the signal variance to increase asymmetrically between channels (Figure 7B).
Figure 7. Overview of FP technique.
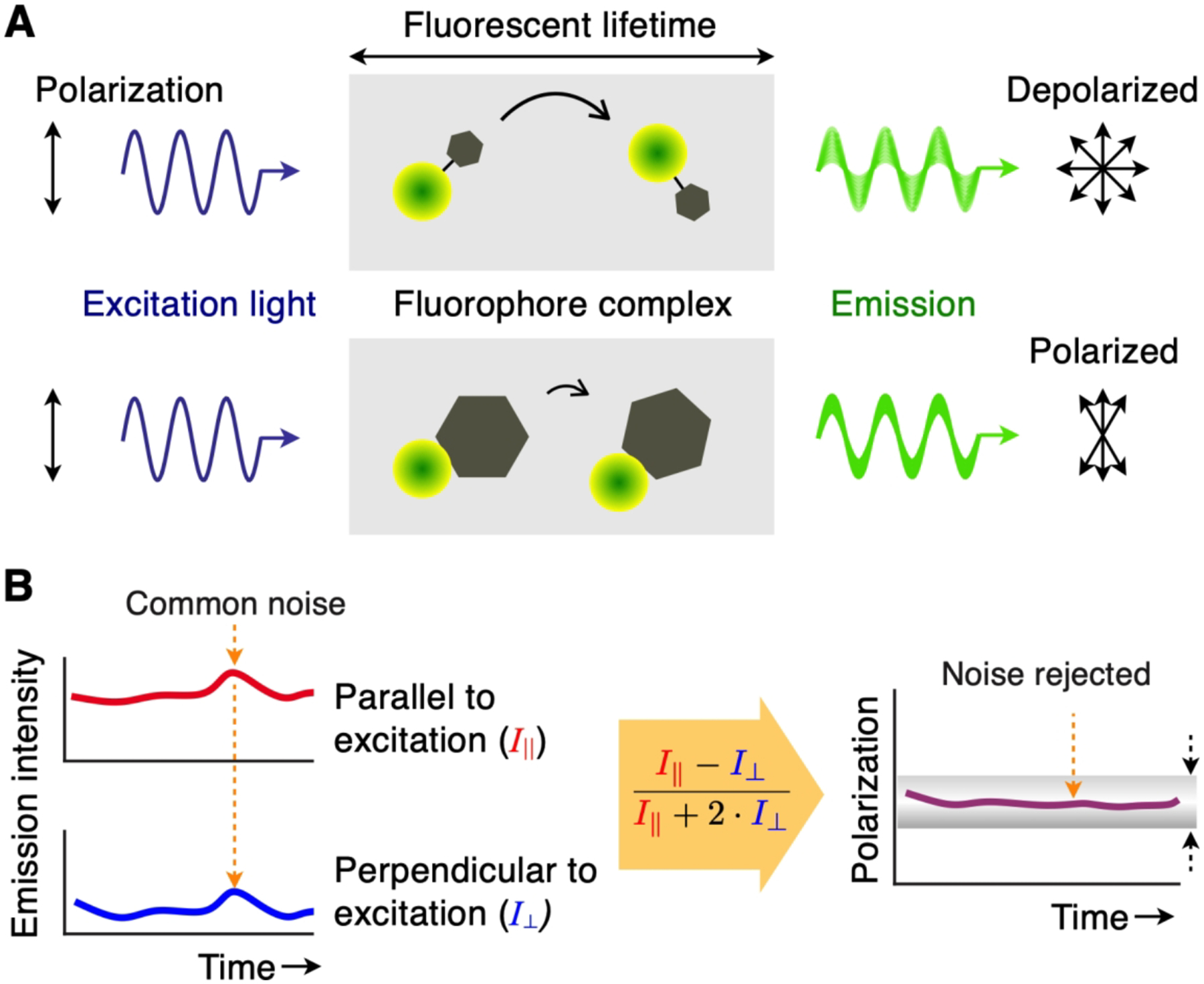
(A) Upon absorbing light (blue sinusoid), a fluorophore will emit longer-wavelength photons (green sinusoid) with identical polarization angles. If the fluorophore is small, it tumbles quickly during the fluorescence lifetime and the observed emission is depolarized (top). If the fluorophore binds a larger particle, the complex would rotate slowly and the emission light largely maintains the same polarization as the excitation light (bottom). (B) FP signal acquisition. Intensity readings from dual optical detectors are captured continuously, and a FP value is calculated in real-time. Common noise (dotted orange arrows) present on both channels is rejected due to the ratiometric nature of the measurement (i.e. the subtraction of parallel and perpendicular signals). In addition, the measured FP value can be monitored to ensure that its variance is within a threshold (dotted black arrows), which guards against temporal or momentary noise.
We adopted FP as a signal transduction mechanism in NA detection. Assays were designed to change the molecular weight of fluorescent probes when target NAs were present. One approach was to detach fluorescent dyes from fluorescent DNA probes, wherein freed dyes led to low r values. This method was used in our initial work on bacterial profiling,3 which will be discussed in the following sections. More recently, we have combined the clustered-regularly-interspaced-short-palindromic-repeats (CRISPR) technique with FP measurements. When CRISPR-associated protein and guide-RNA complexes recognized target NA, they became active and indiscriminately cut down fluorescent DNA probes to fragments (lower r values). This alternative method enabled SARS-CoV-2 RNA detection within 20 min and with an LOD of 3 copy/μL. Details on this assay can be found in our recent report.4
4.1. Miniaturized FP detection system
To provide FP assays in the POC settings, we have advanced portable detection systems (Figures 8A, B).3,4 An important design factor was to adopt the lock-in method to achieve high signal-to-noise. During the fluorescent detection, we controlled the excitation light source to generate an on-off or a sinusoidal intensity patterns. The emission light would follow the same intensity profile of the excitation, and could be extracted from the noisy background by referencing (“locking-in”) the excitation light. Figure 8C shows an example of a miniaturized FP setup. We used a light-emitting diode (LED) as an affordable light source. The illumination light was linearly polarized and focused onto a sample to excite its fluorophores. The emission light was then measured by a pair of photodiodes. We modulated the intensity of the excitation light at a carrier frequency of 1 kHz. The emission signals were then frequency-locked in the detection circuit.
Figure 8. FP detection system.
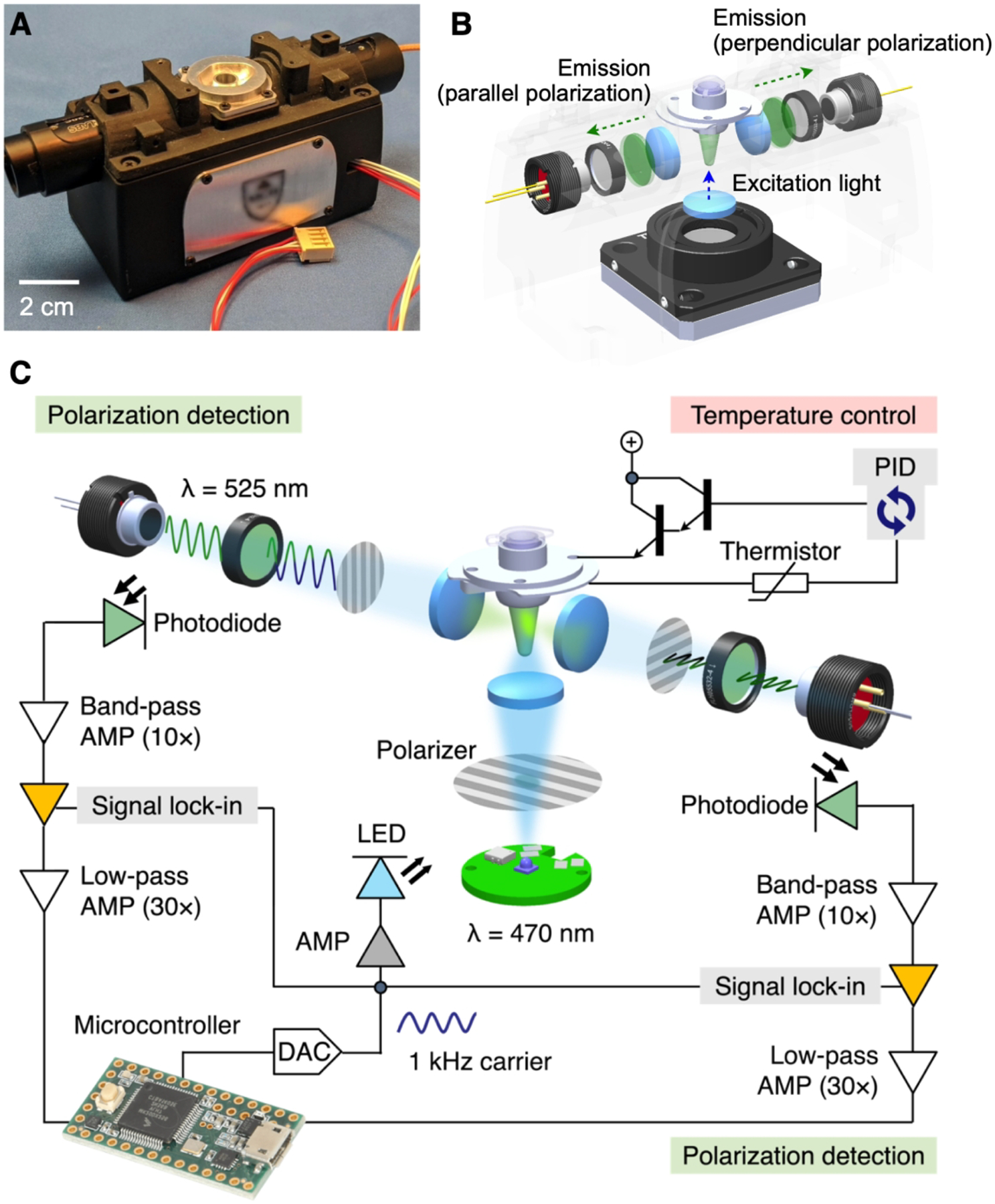
(A) Photograph of the portable device for onsite FP detection. (B) FP optics. A linearly polarized light illuminates a sample from its bottom side. Two photodetectors measure orthogonal polarization of fluorescent emission from the sample. (C) To enhance the signal-to-noise ratio (SNR), the system uses the optical lock-in detection. The sample is illuminated with linearly polarized light oscillating at 1 kHz. Fluorescence is measured by two photodetectors, each consisting of a photodiode, a 525 nm bandpass filter and a linear polarizer. The signal is processed by a sequence of integrated filtering/amplification steps: 10 × band pass, lock-in, and 30 × lowpass. Sample’s temperature can be controlled through a feedback loop control. AMP, amplifier; DAC, digital-to-analog converter; PID, proportional–integral–derivative. Adapted with permission from ref 4. Copyright © 2021 Elsevier B.V. All rights reserved.
4.2. FP assays for bacteria detection
We have adopted the FP principle to identify major pathogens that cause HAIs. After bacterial NA extraction, target sequences of 16S ribosomal RNA (16S rRNA) or mRNA were pre-amplified with asymmetric RT-PCR. The assay, termed PAD (polarization anisotropy diagnostics),3 detected amplified NAs with a dual-probe set composed of a detection probe and a reporter DNA. The detection probe (detection key) had a DNA polymerase-specific aptamer adjoined with an overhang sequence complementary to target NAs. Upon hybridization with target NA, the detection key became stable to bind to DNA polymerase, suppressing polymerase activity.28,40 The reporter DNA then remained intact, maintaining high fluorescence anisotropy (r) due to slow rotational motion. Conversely, in the absence of target NA, the uninhibited DNA polymerase catalyzed primer extension reactions on the reporter DNA so that fluorophores were cut out from reporter probes. The free fluorophores rotated rapidly, leading to low r value (Figure 9A).
Figure 9. Bacterial detection with PAD.
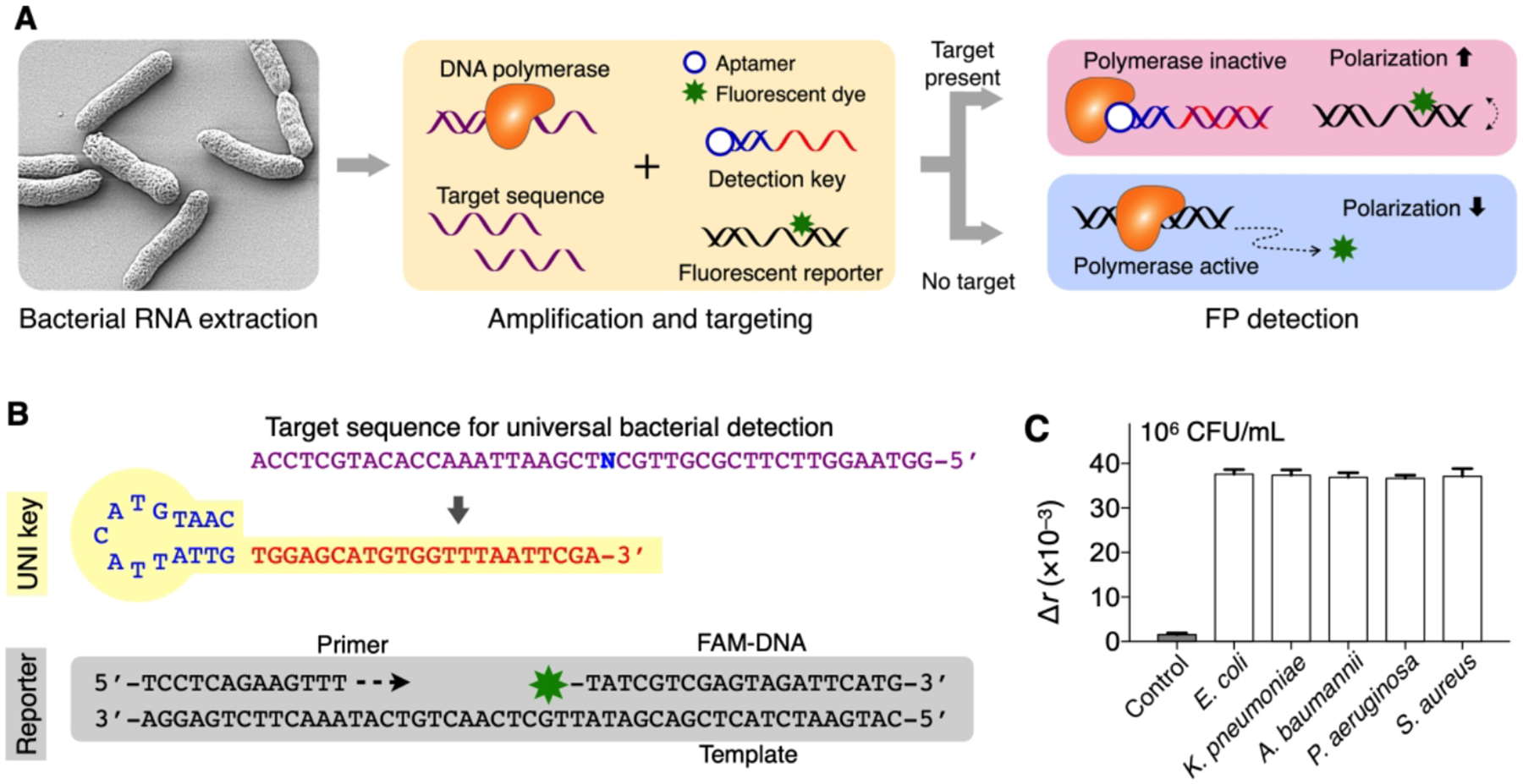
(A) Assay principle. Following the extraction and amplification of target NAs, the sample is mixed with a dual probe set. In the presence of target NA, the detection key is stabilized through hybridization and locks into DNA polymerase, which deactivates polymerase activity. The fluorescent reporter DNA then retains its structure and assumes high fluorescence anisotropy (r). In the absence of target NA, unlocked DNA polymerase cleaves the reporter’s fluorophore during the extension reaction, which leads to low r values. (B) Universal bacteria detection with UNI key. A conserved sequence was targeted (N = A for Escherichia, Klebsiella, Acinetobacter; N = T for Pseudomonas, Staphylococcus). (C) Five different HAI pathogens (106 CFU/mL) were detected. The signals were statistically identical (P = 0.886, one-way ANOVA) among concentration-matched samples. The bar graphs display mean ± s.d. (triplicates). Adapted with permission from ref 3. Copyright 2016 © The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science.
We have designed three types of PAD probes. Each type was specialized in analyzing either bacterial infection, bacterial species, or antibiotic-resistance and virulence status. To detect bacterial infection, we designed a single, universal probe (UNI key) that targets a conserved region of 16S rRNA in different bacterial species (Figure 9B). Applying the designed assay (UNI-PAD), we observed consistent signal values in concentration-matched samples regardless of bacterial species (E. coli, K. pneumoniae, A. baumannii, P. aeruginosa, S. aureus; Figure 9C); this result supported the use of UNI-PAD in estimating total bacterial load.
To differentiate pathogens, we designed a set of detection probes (HAI keys). Each key targeted the hypervariable region of 16S rRNA in different bacterial species. The sequence homology among genus types was kept < 50% to minimize cross-reactivity. The HAI keys assumed high specificity. For example, Escherichia key (Figure 10A) showed high PAD signal only with its intended target, whereas off-target signals were negligible even in high biological background (Figure 10B). Similarly, other HAI keys displayed excellent specificity with minimal crosstalk (Figure 10C). We further made probes for antimicrobial resistance (AMR keys). These keys targeted bacterial genes that make pathogens antibiotic-resistant or highly virulent. As a model system, we profiled samples for mecA (key factor conferring methicillin-resistance on S. aureus); PVL, nuc, and femB (virulence factors contributing to the pathogenicity of S. aureus). We used two representative MRSA strains: healthcare–associated MRSA (HA-MRSA; mecA+, PVL−) and community-acquired MRSA (CA-MRSA; mecA+, PVL+). Control samples were MSSA (mecA−), E. coli and P. aeruginosa (mecA−, PVL−, nuc−, femB−). The AMR-PAD correctly genotyped bacteria, agreeing with RT-qPCR and culture results (Figure 10D).
Figure 10. Bacteria typing.
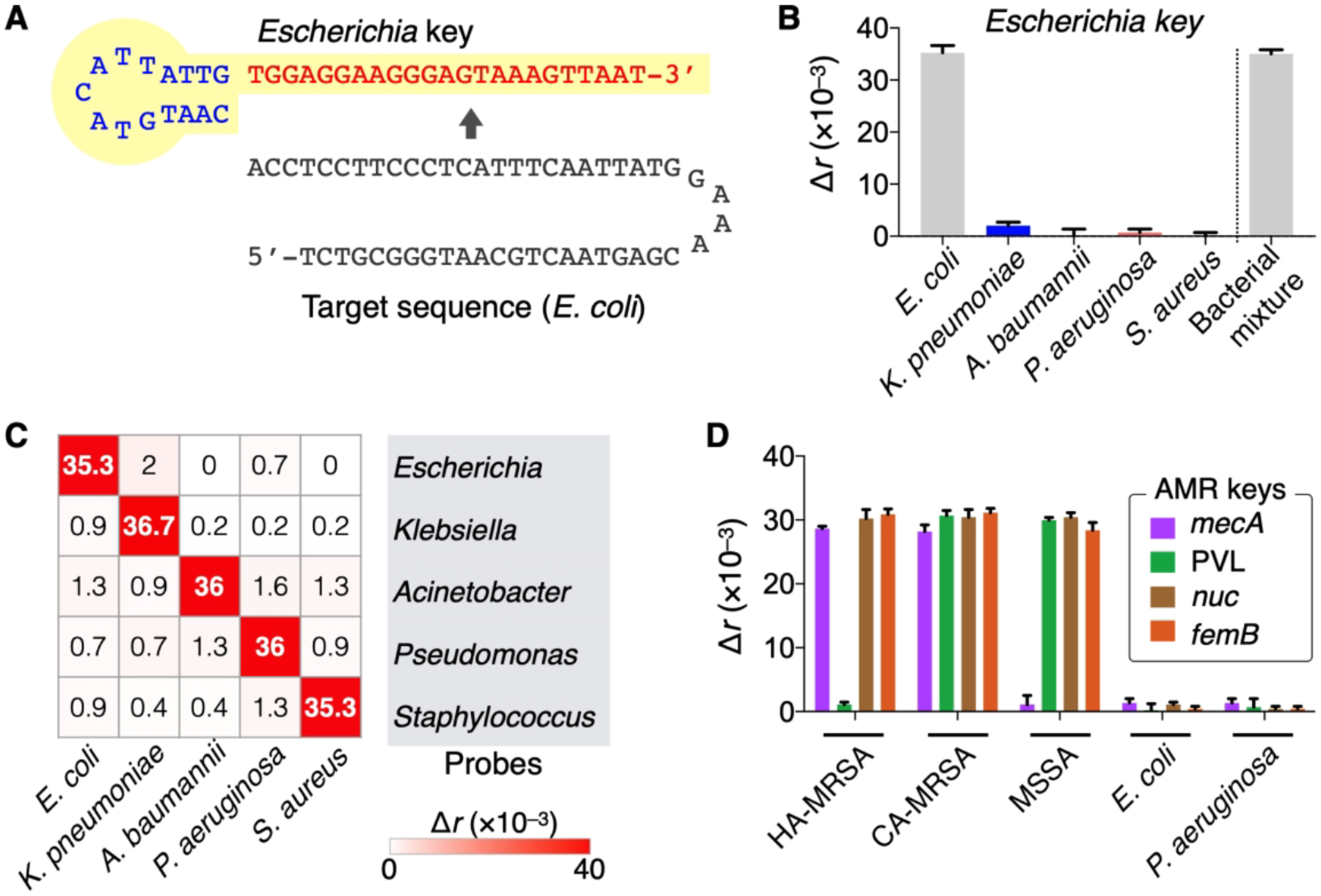
(A) Pathogen-specific detection keys (HAI keys) were prepared, targeting variable 16S rRNA region. Escherichia probe is shown as an example. (B) Specificity of HAI keys. The signal was high only in the presence of the target species ([E. coli] = 106 CFU/mL) even in the mixture of other bacterial species (106 CFU/mL per each). (C) Heat-map (Δr) showing the detection of five pathogens. (D) AMR keys identified HA-MRSA and CA-MRSA by targeting the specific regions in mecA and PVL genes. The heat-map and bar graphs display mean and mean ± s.d. (triplicates), respectively. Adapted with permission from ref 3. Copyright 2016 © The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science.
4.3. Proof-of-principle clinical study
Using the first PAD prototype, we performed a pilot clinical study to detect pathogens in human samples.3 The PAD test (2 hr) results showed an excellent match with the culture (3–5 days) (Figure 11). The UNI-PAD correctly detected infection; samples negative with UNI-PAD were also negative with HAI-PADs, suggesting the potential use of universal detection for sample triaging. Among six UNI-PAD–positive samples, HAI-PAD detected HAI pathogens in five samples, and the differentiation results matched the bacterial culture readouts. One patient (No. 6) was positive with bacterial load but was negative with HAI keys; the patient was later found to be infected with Providencia rettgeri (non-targeted in this system). For the S. aureus-infected sample (No. 2), AMR-PAD identified no mecA, which matched the MRSA-negative pathology.
Figure 11. Clinical application of PAD.
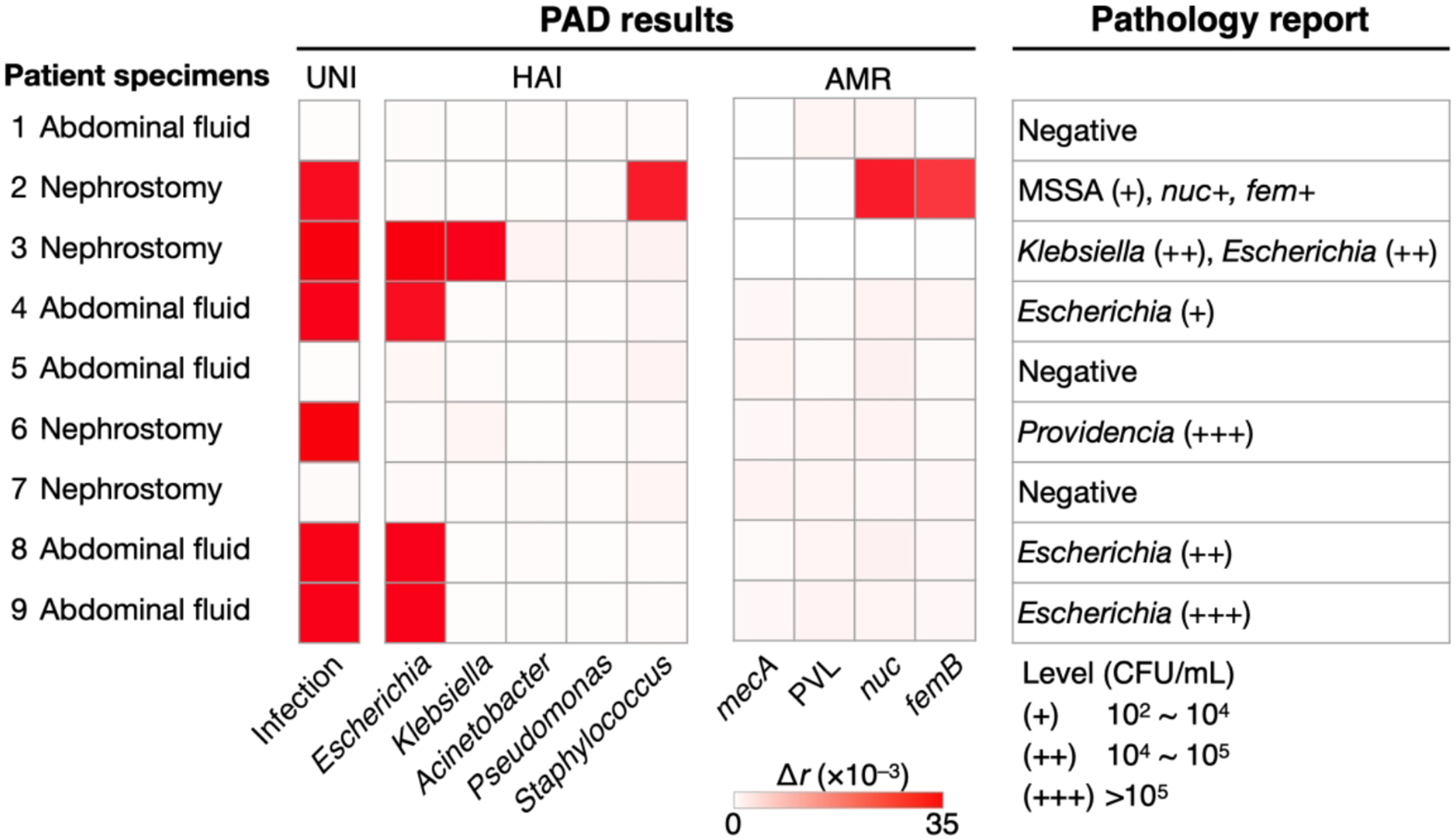
Patient samples were processed by the PAD for bacterial load (UNI), presence of the HAI species (HAI), and resistance/virulence status (AMR). The PAD and pathology reports (i.e., culture and RT-qPCR) agreed with each other. Adapted with permission from ref 3. Copyright 2016 © The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science.
5. CONCLUSIONS AND OUTLOOK
We have discussed two complementary components in POC NA tests, namely nanoPCR for rapid NA amplification and FP system for reliable signal detection. The key merit of nanoPCR is its fast assay speed achieved through the use of hybrid MPNs. Volumetric, plasmonic heating through Au shells in MPNs enables fast thermocycling; magnetic cores allow for in-situ fluorescent detection via magnetic pulling. As such, nanoPCR is faster than conventional RT-PCR instruments as well as other commercial POC system (e.g., Accula SARS-CoV-2 Test, GeneXpert, Talis One COVID-19 assay, HiberGene’s HG COVID-19 test) that are based on RT-PCR or isothermal amplification.41–44 Furthermore, we have engineered a compact nanoPCR device for POC operations, integrating a ferris wheel module and detection optics. The resulting system automatically processed multiple samples with a single button push. These technical advances along with a pilot clinical testing highlight nanoPCR as a promising, clinical POC diagnostics.
FP is a powerful analytical approach that has been broadly used in chemistry,45 drug development,46,47 and now infectious disease diagnostics. The sensing is inherently robust against environmental noise; incorporating the optical lock-in method further enhances such robustness. These merits in turn make FP detection systems relatively simple and cheap to design. Second, the assay is cost-effective. It uses a dual-probe scheme wherein target NAs are recognized by a detection probe but the signal is generated by a common reporter. This method lowers the reagent cost ($2 per assay), while allowing the assay to achieve detection accuracy comparable to bacterial culture, the clinical gold standard. Third, the assay is simple as reactions can take place in a single tube without any washing steps. These advantages make the FP platform well-suited to the POC workflow, facilitating rapid infection care in hospitals and ambulatory clinics. Our pilot studies proved the concept that PAD system can comprehensively interrogate bacterial HAI.
Further improvements will bring these technologies closer to clinical translation. i) We need to improve sample preprocessing devices to enable continuous workflow. This will reduce the likelihood of errors from sample contamination and user intervention. ii) The assay throughput should be increased to detect different targets at once. A straightforward approach would be to integrate multiple sensors, leveraging on system’s small size and low cost. A more innovative solution might be to engineer both detection optics and probes compatible with multiple fluorescent wavelengths, which would allow for the detection of a plurality of targets in a convenient one-pot assay format. iii) A probe library should be expanded to incorporate additional causative pathogens, antibiotic-resistance, and host responses. A highly specific diagnosis based on a comprehensive panel of probes will allow clinicians to effectively triage patients having similar symptoms to respond quickly and decisively to emerging outbreaks.
ACKNOWLEDGEMENTS
This work was supported in part by the US National Institutes of Health (R01CA229777, R21DA049577, U01CA233360, R01CA239078, R01CA237500, R01CA239078, R01CA237500); the US Department of Defense (W81XWH1910199, W81XWH1910194); MGH Scholar Fund; and Institute for Basic Science (IBS-R026-D1).
Biographies
Chang Yeol Lee is a postdoctoral researcher in the Center for Systems Biology (CSB), Massachusetts General Hospital (MGH) and Institute for Basic Science, Center for NanoMedicine (IBS-CNM). He received his Ph.D. in the Department of Chemical and Biomolecular Engineering from KAIST. His research focuses on nucleic acid-based biosensors for infection diagnosis.
Ismail Degani is a Ph.D. candidate in the Department of Electrical Engineering and Computer Science at the Massachusetts Institute of Technology (MIT). He received an M.S. in Electrical Engineering and Computer Science from MIT. His research interests include signal processing, deep learning, and diagnostic platform development.
Jiyong Cheong is Ph.D. candidate in Nano-BioMedical Engineering (nanoBME), Advanced Science Institute (ASI), Yonsei University. He received his B.S. in Biotechnology and Life Science from Yonsei University. His research interests are in creating a platform for nanomaterial-based molecular diagnostics.
Ralph Weissleder is the Thrall Professor of Radiology and Professor of Systems Biology at Harvard Medical School (HMS), Director of CSB-MGH, and attending clinician (Interventional Radiology) at MGH. His research interests include the development of novel molecular diagnostics, systems analyses, and clinical translation.
Jae-Hyun Lee is Associate Professor at ASI, Yonsei University. He graduated from Yonsei University with his Ph.D. in chemistry in 2012. His current research interests are the fabrication of nanoscale devices and nano-bio interfaces at single cellular level.
Jinwoo Cheon is the Horace G. Underwood Professor of Chemistry at Yonsei University and the Director of IBS-CNM. His research interests include inorganic nanocrystal chemistry and nanomedicine.
Hakho Lee is Associate Professor in Radiology (HMS), Director of the Biomedical Engineering Program at the CSB, MGH, and Hostetter MGH Research Scholar. His research focuses on developing novel biomedical sensors for clinical applications.
REFERENCES
- (1).Park KS; Kim H; Kim S; Lee K; Park S; Song J; Min C; Khanam F; Rashu R; Bhuiyan TR; Ryan ET; Qadri F; Weissleder R; Cheon J; Charles RC; Lee H Nanomagnetic System for Rapid Diagnosis of Acute Infection. ACS Nano 2017, 11, 11425–11432. [DOI] [PMC free article] [PubMed] [Google Scholar]; A miniaturized magnetic fluidic device was constructed for effective capturing of pathogenic cells.
- (2).Cheong J; Yu H; Lee CY; Lee JU; Choi HJ; Lee JH; Lee H; Cheon J Fast Detection of SARS-CoV-2 RNA Via the Integration of Plasmonic Thermocycling and Fluorescence Detection in a Portable Device. Nat. Biomed. Eng 2020, 4, 1159–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]; A nanoPCR system was developed by combining fast thermocycling via plasmonic heating and in-situ fluorescence detection with magnetoplasmonic nanoparticles; this system detected SARS-CoV-2 RNA in 17 min.
- (3).Park KS; Huang CH; Lee K; Yoo YE; Castro CM; Weissleder R; Lee H Rapid Identification of Health Care-Associated Infections With an Integrated Fluorescence Anisotropy System. Sci. Adv 2016, 2, e1600300. [DOI] [PMC free article] [PubMed] [Google Scholar]; A new detection scheme, based on fluorescence polarization (FP), was implemented as a clinical tool for infection diagnostics, which can quantify bacterial load, identify its strains, and detect antibiotic resistance.
- (4).Lee CY; Degani I; Cheong J; Lee JH; Choi HJ; Cheon J; Lee H Fluorescence Polarization System for Rapid COVID-19 Diagnosis. Biosens. Bioelectron 2021, 178, 113049. [DOI] [PMC free article] [PubMed] [Google Scholar]; A one-pot assay based on the CRISPR technology was developed for virus detection in 20 min.
- (5).Baud D; Gubler DJ; Schaub B; Lanteri MC; Musso D An Update on Zika Virus Infection. Lancet 2017, 390, 2099–2109. [DOI] [PubMed] [Google Scholar]
- (6).Boucher HW; Talbot GH; Bradley JS; Edwards JE; Gilbert D; Rice LB; Scheld M; Spellberg B; Bartlett J Bad Bugs, No Drugs: No Eskape! An Update From the Infectious Diseases Society of America. Clin. Infect. Dis 2009, 48, 1–12. [DOI] [PubMed] [Google Scholar]
- (7).Fauci AS Infectious Diseases: Considerations for the 21st Century. Clin. Infect. Dis 2001, 32, 675–685. [DOI] [PubMed] [Google Scholar]
- (8).Fauci AS; Touchette NA; Folkers GK Emerging Infectious Diseases: A 10-Year Perspective From the National Institute of Allergy and Infectious Diseases. Emerg. Infect. Dis 2005, 11, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Morens DM; Folkers GK; Fauci AS The Challenge of Emerging and Re-Emerging Infectious Diseases. Nature 2004, 430, 242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Cutler DM; Summers LH The COVID-19 Pandemic and the $16 Trillion Virus. JAMA 2020, 324, 1495–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Blake KS; Choi J; Dantas G Approaches for Characterizing and Tracking Hospital-Associated Multidrug-Resistant Bacteria. Cell. Mol. Life. Sci 2021, 78, 2585–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Antibiotic Resistance Threats in the United States, 2019; US Department of Health and Human Services, CDC: USA, 2019. [Google Scholar]
- (13).Frieri M; Kumar K; Boutin A Antibiotic Resistance. J. Infect. Public Health 2017, 10, 369–378. [DOI] [PubMed] [Google Scholar]
- (14).Magill SS; Edwards JR; Bamberg W; Beldavs ZG; Dumyati G; Kainer MA; Lynfield R; Maloney M; McAllister-Hollod L; Nadle J; Ray SM; Thompson DL; Wilson LE; Fridkin SK Multistate Point-Prevalence Survey of Health Care-Associated Infections. N. Engl. J. Med 2014, 370, 1198–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Haque M; Sartelli M; McKimm J; Abu Bakar M Health Care-Associated Infections - an Overview. Infect. Drug Resist 2018, 11, 2321–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Weissleder R; Lee H; Ko J; Pittet MJ COVID-19 Diagnostics in Context. Sci. Transl. Med 2020, 12, eabc1931. [DOI] [PubMed] [Google Scholar]
- (17).Kilic T; Weissleder R; Lee H Molecular and Immunological Diagnostic Tests of COVID-19: Current Status and Challenges. iScience 2020, 23, 101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Allegranzi B; Bagheri Nejad S; Combescure C; Graafmans W; Attar H; Donaldson L; Pittet D Burden of Endemic Health-Care-associated Infection in Developing Countries: Systematic Review and Meta-Analysis. Lancet 2011, 377, 228–241. [DOI] [PubMed] [Google Scholar]
- (19).Ahmad A; Iram S; Hussain S; Yusuf NW Diagnosis of Paediatric Sepsis By Automated Blood Culture System and Conventional Blood Culture. J. Pak. Med. Assoc 2017, 67, 192–195. [PubMed] [Google Scholar]
- (20).Salipante SJ; Kawashima T; Rosenthal C; Hoogestraat DR; Cummings LA; Sengupta DJ; Harkins TT; Cookson BT; Hoffman NG Performance Comparison of Illumina and Ion Torrent Next-Generation Sequencing Platforms for 16s rRNA-Based Bacterial Community Profiling. Appl. Environ. Microbiol 2014, 80, 7583–7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Dark PM; Dean P; Warhurst G Bench-to-bedside Review: The Promise of Rapid Infection Diagnosis During Sepsis Using Polymerase Chain Reaction-Based Pathogen Detection. Crit. Care 2009, 13, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Pechorsky A; Nitzan Y; Lazarovitch T Identification of Pathogenic Bacteria in Blood Cultures: Comparison Between Conventional and PCR Methods. J. Microbiol. Methods 2009, 78, 325–330. [DOI] [PubMed] [Google Scholar]
- (23).Maurer JJ Rapid Detection and Limitations of Molecular Techniques. Annu. Rev. Food Sci. Technol 2011, 2, 259–279. [DOI] [PubMed] [Google Scholar]
- (24).Loman NJ; Constantinidou C; Chan JZ; Halachev M; Sergeant M; Penn CW; Robinson ER; Pallen MJ High-Throughput Bacterial Genome Sequencing: An Embarrassment of Choice, a World of Opportunity. Nat. Rev. Microbiol 2012, 10, 599–606. [DOI] [PubMed] [Google Scholar]
- (25).Hong S; Park KS; Weissleder R; Castro CM; Lee H Facile Silicification of Plastic Surface for Bioassays. Chem. Commun 2017, 53, 2134–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Huang CH; Park YI; Lin HY; Pathania D; Park KS; Avila-Wallace M; Castro CM; Weissleder R; Lee H Compact and Filter-Free Luminescence Biosensor for Mobile in Vitro Diagnoses. ACS Nano 2019, 13, 11698–11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Park KS; Chung HJ; Khanam F; Lee H; Rashu R; Bhuiyan MT; Berger A; Harris JB; Calderwood SB; Ryan ET; Qadri F; Weissleder R; Charles RC A Magneto-DNA Nanoparticle System for the Rapid and Sensitive Diagnosis of Enteric Fever. Sci. Rep 2016, 6, 32878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Park KS; Charles RC; Ryan ET; Weissleder R; Lee H Fluorescence Polarization Based Nucleic Acid Testing for Rapid and Cost-Effective Diagnosis of Infectious Disease. Chem. Eur. J 2015, 21, 16359–16363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Chung HJ; Castro CM; Im H; Lee H; Weissleder R A Magneto-DNA Nanoparticle System for Rapid Detection and Phenotyping of Bacteria. Nat. Nanotechnol 2013, 8, 369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Lee H; Sun E; Ham D; Weissleder R Chip-NMR Biosensor for Detection and Molecular Analysis of Cells. Nat. Med 2008, 14, 869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Liong M; Hoang AN; Chung J; Gural N; Ford CB; Min C; Shah RR; Ahmad R; Fernandez-Suarez M; Fortune SM; Toner M; Lee H; Weissleder R Magnetic Barcode Assay for Genetic Detection of Pathogens. Nat. Commun 2013, 4, 1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Chung HJ; Reiner T; Budin G; Min C; Liong M; Issadore D; Lee H; Weissleder R Ubiquitous Detection of Gram-Positive Bacteria With Bioorthogonal Magnetofluorescent Nanoparticles. ACS Nano 2011, 5, 8834–8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Liong M; Fernandez-Suarez M; Issadore D; Min C; Tassa C; Reiner T; Fortune SM; Toner M; Lee H; Weissleder R Specific Pathogen Detection Using Bioorthogonal Chemistry and Diagnostic Magnetic Resonance. Bioconjug. Chem 2011, 22, 2390–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Issadore D; Min C; Liong M; Chung J; Weissleder R; Lee H Miniature Magnetic Resonance System for Point-of-care Diagnostics. Lab Chip 2011, 11, 2282–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Brongersma ML; Halas NJ; Nordlander P Plasmon-Induced Hot Carrier Science and Technology. Nat. Nanotechnol 2015, 10, 25–34. [DOI] [PubMed] [Google Scholar]
- (36).Kim JW; Seo D; Lee JU; Southard KM; Lim Y; Kim D; Gartner ZJ; Jun YW; Cheon J Single-Cell Mechanogenetics Using Monovalent Magnetoplasmonic Nanoparticles. Nat. Protoc 2017, 12, 1871–1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).You M; Li Z; Feng S; Gao B; Yao C; Hu J; Xu F Ultrafast Photonic PCR Based on Photothermal Nanomaterials. Trends Biotechnol. 2020, 38, 637–649. [DOI] [PubMed] [Google Scholar]
- (38).Ndukaife JC; Shalaev VM; Boltasseva A Plasmonics—turning Loss Into Gain. Science 2016, 351, 334–335. [DOI] [PubMed] [Google Scholar]
- (39).Ameloot M; vandeVen M; Acuña AU; Valeur B Fluorescence Anisotropy Measurements in Solution: Methods and Reference Materials (IUPAC Technical Report). Pure Appl. Chem 2013, 85, 589–608. [Google Scholar]
- (40).Park KS; Lee CY; Park HG Target DNA Induced Switches of DNA Polymerase Activity. Chem. Commun 2015, 51, 9942–9945. [DOI] [PubMed] [Google Scholar]
- (41).Hogan CA; Garamani N; Lee AS; Tung JK; Sahoo MK; Huang C; Stevens B; Zehnder J; Pinsky BA Comparison of the Accula SARS-CoV-2 Test With a Laboratory-Developed Assay for Detection of SARS-CoV-2 RNA in Clinical Nasopharyngeal Specimens. J. Clin. Microbiol 2020, 58, e01072–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Goldenberger D; Leuzinger K; Sogaard KK; Gosert R; Roloff T; Naegele K; Cuénod A; Mari A; Seth-Smith H; Rentsch K; Hinić V; Hirsch HH; Egli A Brief Validation of the Novel GeneXpert Xpress SARS-CoV-2 PCR Assay. J. Virol. Methods 2020, 284, 113925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Loeffelholz MJ; Tang YW Laboratory Diagnosis of Emerging Human Coronavirus Infections - the State of the Art. Emerg. Microbes. Infect 2020, 9, 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Nath P; Kabir A; Khoubafarin Doust S; Kreais ZJ; Ray A Detection of Bacterial and Viral Pathogens Using Photonic Point-of-care Devices. Diagnostics (Basel) 2020, 10, 841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Jameson DM; Ross JA Fluorescence Polarization/anisotropy in Diagnostics and Imaging. Chem. Rev 2010, 110, 2685–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Vinegoni C; Fumene Feruglio P; Brand C; Lee S; Nibbs AE; Stapleton S; Shah S; Gryczynski I; Reiner T; Mazitschek R; Weissleder R Measurement of Drug-Target Engagement in Live Cells By Two-Photon Fluorescence Anisotropy Imaging. Nat. Protoc 2017, 12, 1472–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Vinegoni C; Feruglio PF; Gryczynski I; Mazitschek R; Weissleder R Fluorescence Anisotropy Imaging in Drug Discovery. Adv. Drug. Deliv. Rev 2019, 151–152, 262–288. [DOI] [PMC free article] [PubMed] [Google Scholar]


