Abstract
Heterochromatin is characterized by dimethylated or trimethylated histone H3 Lys9 (H3K9me2 or H3K9me3, respectively) and is found at transposable elements, satellite repeats and genes, where it ensures their transcriptional silencing. The histone methyltransferases (HMTs) that methylate H3K9 — in mammals Suppressor of variegation 3–9 homologue 1 (SUV39H1), SUV39H2, SET domain bifurcated 1 (SETDB1), SETDB2, G9A and G9A-like protein (GLP) — and the ‘readers’ of H3K9me2 or H3K9me3 are highly conserved and show considerable redundancy. Despite their redundancy, genetic ablation or mistargeting of an individual H3K9 methyltransferase can correlate with impaired cell differentiation, loss of tissue identity, premature aging and/or cancer. In this Review, we discuss recent advances in understanding the roles of the known H3K9-specific HMTs in ensuring transcriptional homeostasis during tissue differentiation in mammals. We examine the effects of H3K9-methylation-dependent gene repression in haematopoiesis, muscle differentiation and neurogenesis in mammals, and compare them with mechanistic insights obtained from the study of model organisms, notably Caenorhabditis elegans and Drosophila melanogaster. In all these organisms, H3K9-specific HMTs have both unique and redundant roles that ensure the maintenance of tissue integrity by restricting the binding of transcription factors to lineage-specific promoters and enhancer elements.
Subject terms: Gene silencing, Histone post-translational modifications, Nuclear envelope
Histone H3 Lys9 (H3K9)-methylated heterochromatin ensures transcriptional silencing of repetitive elements and genes, and its deregulation leads to impaired cell and tissue identity, premature aging and cancer. Recent studies in mammals clarified the roles H3K9-specific histone methyltransferases in ensuring transcriptional homeostasis during tissue differentiation.
Introduction
More than half of the vertebrate genome is packaged into condensed, transcriptionally repressed heterochromatin. The epigenetic hallmark of heterochromatin is dimethylated or trimethylated histone H3 Lys9 (H3K9me2 or H3K9me3, respectively). The DNA classically attributed to methylated H3K9 (H3K9me) heterochromatin comprises repetitive non-coding sequences such as pericentric satellites and subtelomeric repeats, which are silenced and clustered together, forming constitutive heterochromatin. However, recent work shows that this well-conserved histone modification serves a second, more dynamic function in the repression of tissue-specific genes. The six mammalian histone methyltransferases (HMTs) that target H3K9 have distinct modes of action and a complex interplay as they establish facultative heterochromatin, which controls lineage-specific gene repression during development in a tissue-specific manner.
Early data suggesting that H3K9 methylation had a role in repressing genes and not only repetitive elements were based on the study of individual loci (for example, IFNB1 (ref.1), Magea genes2 and Bmi1 (ref.3)). H3K9me2 catalysed by the mammalian HMT G9A was especially shown to regulate transcription in these pioneering studies. Obtaining a genome-wide view was hindered by the difficulty of mapping H3K9 methylation accurately4 and by the fact that the six HMTs that methylate H3K9 in mice and humans showed partial redundancy. The redundancy issue was resolved in part by studying H3K9 methylation in simpler organisms, such as Drosophila melanogaster, which has three enzymes, and Caenorhabditis elegans, which has only two. Remarkably, the deletion of both H3K9 methyltransferases in worms, which eliminated all detectable H3K9 methylation, only slightly impaired embryo to adult development5–7.
In this Review, we focus on the best characterized mammalian H3K9 methyltransferases — Suppressor of variegation 3–9 homologue 1 (SUV39H1), SUV39H2, SET domain bifurcated 1 (SETDB1), SETDB2, G9A (also known as EHMT2) and G9A-like protein (GLP; also known as EHMT1)8 — and on their related enzymes in D. melanogaster and C. elegans (Fig. 1a). We discuss their modes of action and roles in lineage-specific gene expression. Elegant work in fission yeast detailing the role of H3K9 methylation and its methyltransferase Clr4, which sets up self-propagating domains of heterochromatin at centromeres, telomeres and the mating type locus, has been covered elsewhere9,10.
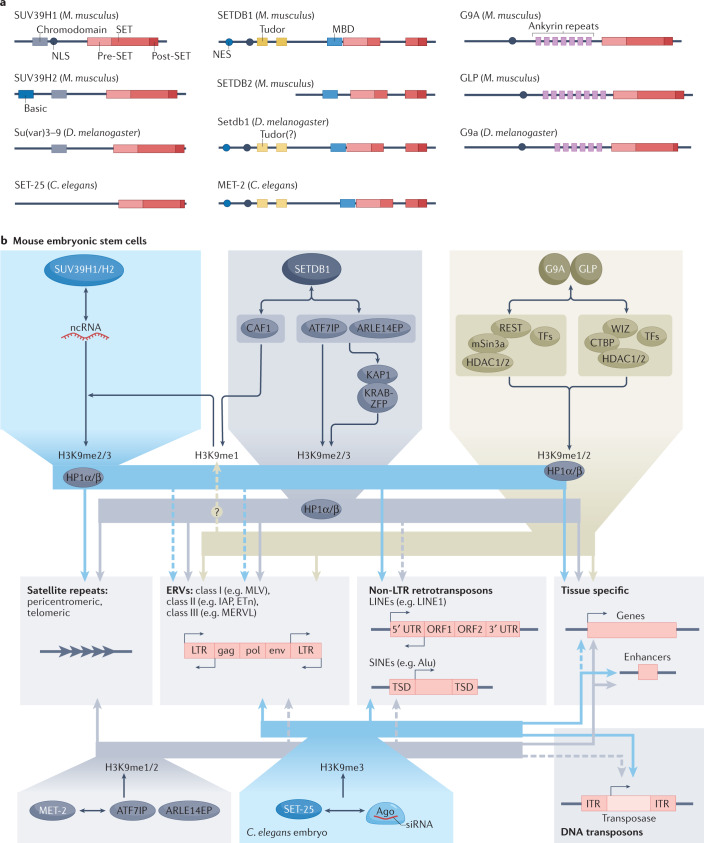
Fig. 1. The major H3K9 methyltransferases, cofactors and targets.
a | The structural domains of the major histone H3 Lys9 (H3K9) methyltransferases in Mus musculus, Drosophila melanogaster and Caenorhabditis elegans. The SET domain contains the catalytic site. The enzymes are grouped by structural relatedness. One outlier is C. elegans SET-25, which has a SUV39-like SET and pre-SET domain, but does not have the methylated-lysine-binding chromodomain like other SUV39 enzymes. The amino-terminal basic domain in SUV39H2 is found uniquely in the mouse, and not the human, enzyme. SET domain bifurcated 1 (SETDB1), Setdb1 and MET-2 are structurally closely related and also share conserved essential cofactors. Tudor domains, ankyrin repeats and methyl-CpG-binding domains (MBDs) have roles in targeting the histone methyltransferases (HMTs) to specific sites of action. b | The three families of H3K9 methyltransferases are shown with cofactors and interacting proteins that enable site-specific HMT recruitment. Suppressor of variegation 3–9 homologue 1 (SUV39H1) and SUV39H2 are often recruited to chromatin by non-coding RNA (ncRNA), or in worms by endogenous siRNA bound to Argonaut (Ago) proteins. SUV39 enzymes bind both to heterochromatin protein 1 (HP1) proteins and to dimethylated or trimethylated H3K9 (H3K9me2/3), which help to further recruit the HMT. SETDB1 class enzymes form a complex with activating transcription factor 7-interacting protein 1 (ATF7IP) (LIN-65 in worms) and ARLE14EP (adenosine 5′-diphosphate-ribosylation factor-like 14 (ARLE-14) in worms) or with chromatin assembly factor 1 (CAF1). With CAF1, SETDB1 monomethylates H3K9, whereas with ATF7IP, it binds either Krüppel-associated box (KRAB)-interacting protein 1 (KAP1) together with KRAB domain-containing zinc-finger proteins (KRAB-ZFPs) at endogenous retroviruses (ERVs) or is directly recruited by HP1 proteins to produce H3K9me2/3. In worms, MET-2 binds LIN-65 and ARLE-14. The enzymes G9A and G9A-like protein (GLP) bind a variety of transcription factors (TFs), together with histone deacetylases (HDACs), mSin3a and other chromatin modifiers. They also bind monomethylated H3K9 (H3K9me1) or H3K9me2 with their ankyrin repeats. The repeat elements targeted by these HMTs belong to three classes: centromeric or telomeric satellites (simple repeats), ERVs, which are long terminal repeat (LTR)-containing retrotransposons, or non-LTR-containing transposons such as long interspersed nuclear elements (LINEs) and short interspersed nuclear elements (SINEs). All HMTs are also recruited to genes and enhancer elements. CTBP, C-terminal-binding protein 1; ETn, early transposon; IAP, intracisternal A-particle; MERVL, murine endogenous retrovirus type L; MLV, murine leukaemia virus; NES, nuclear export signal; NLS, nuclear localization signal; ORF, open reading frame; TSD, target site duplication; UTR, untranslated region; WIZ, widely interspaced zinc-finger-containing protein.
Features and functions of H3K9 methyltransferases
The six mammalian H3K9 methyltransferases fall into three main families, with two variants in each. The fact that each family has homologues in simpler multicellular organisms, on the basis of homology within and beyond the catalytic SET domain, argues for conserved modes of recruitment and secondary activities. Genetic approaches that ablate H3K9 methyltransferases confirm that the mechanisms that regulate each HMT family are distinct, although the HMTs silence partially overlapping sets of target loci.
The ‘Suppressor of variegation’ HMT family: SUV39H1, SUV39H2, Su(var)3–9 and SET-25
One of the best studied H3K9 methyltransferases is the fruitfly enzyme Suppressor of variegation 3–9 (Su(var)3–9), which was identified genetically as a suppressor of position effect variegation (PEV)11. PEV is a genetic assay used in screens, which provided much of our basic insight into individual heterochromatin proteins and how they function12 (Box 1). Following up on genetic assays that implicated Su(var)3–9 in the establishment and spreading of centromeric heterochromatin in flies, it was shown that its two mouse homologues, SUV39H1 and SUV39H2 (SUV39H1/H2)13, as well as Su(var)3–9 itself, function as H3K9-specific HMTs11.
Su(var)3–9 is a founding member of the SET domain-containing superfamily of methyltransferases (Fig. 1a). Su(var)3–9 and its homologues are characterized by a central SET domain flanked by conserved pre-SET and post-SET domains. These Cys-rich modules bind the three zinc ions necessary for the catalytic activity of the central SET domain, which was originally named for a larger conserved region present in the D. melanogaster proteins Su(var)3–9, Enhancer of zeste and Trithorax14.
Although SUV39H1 can methylate unmodified H3K9 in vitro13, it was shown to bind preferentially to H3K9me1, which may serve as its recruitment signal at centromeric repeats15. In vivo, although SUV39H1/H2 are together responsible for most H3K9me2 and H3K9me3 at pericentric heterochromatin and interspersed satellite repeats, they are not essential for H3K9me1 (refs16–19) (Fig. 1b). Thus, it is widely assumed, albeit not proven, that in organisms H3K9me1 is a prerequisite for SUV39H1/H2 activity. Interestingly, the catalytic activities of SUV39H1/H2 are reinforced by their binding to H3K9me2 and H3K9me3 (ref.20) through a chromodomain located in their amino termini21,22, suggesting a role for these HMTs in the propagation of existing H3K9 methylation. The chromodomain is conserved in the D. melanogaster Su(var)3–9 and the fission yeast Clr4, but is missing in a related C. elegans homologue, SET-25 (Fig. 1a). Activity of both Clr4 and SUV39H2 also appears to be regulated by automethylation within an inhibitory loop downstream of the SET domain23.
Besides binding H3K9me2 and H3K9me3 directly, SUV39H1/H2 interact with members of the heterochromatin protein 1 (HP1) family, which are characterized by an amino-terminal chromodomain specific for H3K9me2 and H3K9me3, and a dimer-promoting chromoshadow domain in their carboxyl terminus. These domains are linked by a flexible hinge region that is thought to be involved in binding RNA24. The HP1 proteins function as adaptors that recruit other proteins to heterochromatin and mediate repression. In mammals the binding of HP1α or HP1β to existing H3K9me2 and H3K9me3 helps recruit SUV39H1/H2, providing a mechanism that promotes the spread of H3K9 methylation from sites of initial HP1 recruitment25,26, reminiscent of mechanisms described in fission yeast9,10 (Fig. 1b).
Neither the direct binding nor the indirect binding of an HMT to existing H3K9me2 and H3K9me3 explains, however, the de novo establishment of heterochromatin at target sequences, a capacity clearly demonstrated for the C. elegans HMT SET-25 (ref.27). SET-25 appears to be recruited to its target regions at least in part through Argonaute proteins and endogenous siRNAs to nucleate repression28–30 (Fig. 1b). Interestingly, recent work revealed that, in addition to binding H3K9me2 and H3K9me3, the chromodomain of SUV39H1 recognizes RNA transcribed from major satellite repeats31,32. A similar activity was assigned to the first 81 amino acids of mouse SUV39H2, a domain that binds single-stranded major satellite repeat RNA, ensuring retention of the enzyme on mitotic chromosomes33. The RNA-mediated recruitment of SUV39H1/H2 provides an elegant mechanism for nucleation of heterochromatin precisely at the sites where repetitive sequences are aberrantly transcribed. Moreover, in two-cell-stage mouse embryos, a burst of transcription from the major satellite repeats is required for the organization of heterochromatin into chromocentres34–36 (Box 2; Fig. 2).
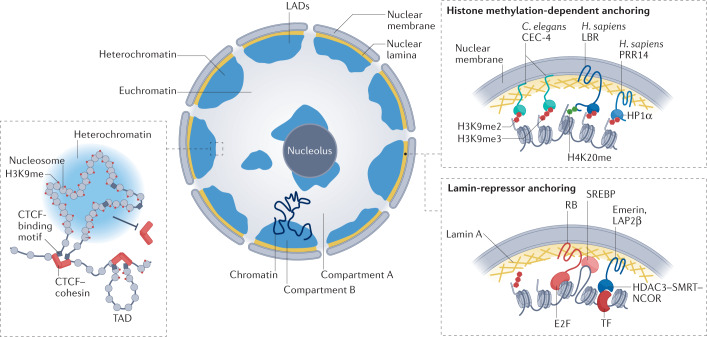
Fig. 2. Roles of H3K9 methylation in long-range chromatin interactions and nuclear organization.
An interphase nucleus in a differentiated vertebrate tissue is shown with heterochromatin (blue) sequestered against the nucleolus and against the nuclear lamina.The separation of active and inactive chromatin domains can manifest itself as lamin-associated domains (LADs)207 or as A and B compartments208 (Box 2). B compartments correspond to gene-inactive chromatin (heterochromatin) and largely include LADs. A compartments correspond to gene-active chromatin, which is further organized into topologically associating domains (TADs; left), which enable enhancer–promoter interaction that stimulates gene expression (not shown). In differentiated tissues, methylated histone H3 Lys9 (H3K9me) blocks TAD formation by preventing the binding of CCCTC-binding factor (CTCF) and cohesin complexes, which delineate TAD borders. On the right are shown several mechanisms for heterochromatin anchorage at the nuclear periphery. In Caenorhabditis elegans, the nuclear envelope-associated protein CEC-4 recognizes all forms of H3K9me (monomethylated H3K9 (H3K9me1), dimethylated H3K9 (H3K9me2) and trimethylated H3K9 (H3K9me3)) through its chromodomain, with affinity roughly equal to that of heterochromatin protein 1 (HP1)209. CEC-4 contributes directly to heterochromatin anchoring in worm embryos209–211, but upon tissue differentiation, as in mammals, additional anchoring pathways are induced, which in worms are independent of H3K9me212. Anchoring mechanisms present in differentiated mammalian cells include a histone methylation-dependent pathway with at least two potential anchors: proline-rich protein 14 (PRR14; potentially a functional homologue of CEC-4) is a non-transmembrane protein that anchors heterochromatin through H3K9me and HP1, and lamin B receptor (LBR), which is a transmembrane factor that binds both HP1α and dimethylated histone H4 Lys20 (H4K20me2) through its Tudor domain213–216. Another mechanism is lamin A dependent214 and likely involves its interaction with RB and/or with transcription factors (TFs), such as SREBP. Emerin and lamina-associated polypeptide 2, isoform-β (LAP2β) are additional lamin-associated factors that bind chromatin through the histone deacetylase 3 (HDAC3)–nuclear receptor co-repressor 1 (NCOR)–silencing mediator of retinoic acid and thyroid hormone receptor (SMRT; also known as NCOR2) complex and potentially through tissue-specific transcription factors215,216 (Box 2). H. sapiens, Homo sapiens.
In addition to RNA transcribed from major satellite repeats, the long non-coding RNA Oct4P4, which is transcribed from an Oct4 pseudogene, has been shown to form a complex with SUV39H1 and to direct H3K9me3 to the promoter of the functional Oct4 gene, ensuring its transcriptional silencing37. This important example demonstrates that RNA-mediated SUV39H1 recruitment can occur in trans. Similarly, the long non-coding RNA telomeric repeat-containing RNA (TERRA) associates with SUV39H1 to promote an accumulation of H3K9me3 at damaged telomeres and prevent end-to-end chromosomal fusions38, potentially acting in trans. The molecular determinants of specificity in the interaction of SUV39H1/H2 with RNA are unclear, yet these results argue that RNA-mediated recruitment of SUV39H1/H2 may be a general mechanism for context-dependent establishment of an H3K9me-marked domain (see ref.10 for siRNA-mediated heterochromatin establishment in Schizosaccharomyces pombe).
Determinants of specificity of HMT recruitment are of particular interest given the diverse set of targets that depend on SUV39H1/H2 for repression18,19,39. Namely, in addition to pericentric heterochromatin, SUV39H1/H2 are recruited to class II endogenous retroviruses (ERVs), particularly intracisternal A-particles (IAPs; such as IAPEz and IAPEy) and early transposons (especially MMETn and ETnERV), and to intact long interspersed nuclear elements (LINEs) such as LINE1 in mouse embryonic stem cells (ESCs)19 (Fig. 1b). In mouse ESCs, the loss of SUV39H1/H2 led to a loss H3K9me3 and upregulation of transcripts from intact LINE1 elements18,19. By contrast, IAP elements responded to the deletion of both Suv39h1 and Suv39h2 by losing their flanking H3K9me3, yet they retained H3K9me3 over the repeat element itself19 and remained silent, likely owing to the recruitment and activity of another H3K9 methyltransferase, for example SETDB1 (ref.19). Similar redundancy among H3K9 methyltransferases was observed at telomeres: although the deletion of both Suv39h1 and Suv39h2 resulted in abnormally long telomeres with lower levels of H3K9me2 and H3K9me3, the levels of H3K9me1 increased39, as did the recruitment of SETDB1, thereby ensuring that H3K9me3 was not completely lost40. Such redundancy, found both within and across the H3K9 methyltransferase families, makes it difficult to use loss-of-function genetic screens to identify recruitment factors.
Whereas satellite repeats and LINE1 transposons are the best studied targets of SUV39H1/H2-mediated repression, these HMTs silence other important targets. For example, SUV39H1 and its homologues are essential for the repression of non-transcribed ribosomal DNA (rDNA) repeats in flies41, worms7 and mice42. In flies, the loss of Su(var)3–9 results not only in elevated levels of ribosomal RNA transcripts but also in chromosomal instability at the rDNA repeat loci, a phenotype also observed in ago2 mutants41, which lose Argonaute-2. In contrast to the constitutive silencing of satellite repeats, mouse SUV39H1 at rDNA repeats is regulated by the energy status of the cell, potentially by the NAD+/NADH ratio and the histone deacetylase (HDAC) SIRT1 (ref.42). SIRT1 is important for the deacetylation of histones H3 and H4, to allow their subsequent methylation42, but it also directly regulates SUV39H1 through the deacetylation of Lys266 in its SET domain. Lys266 deacetylation increases SUV39H1 activity43 and prevents its polyubiquitylation on Lys87 and subsequent degradation, which occur under oxidative stress44. SUV39H1 activity is further regulated by the HDAC SIRT6, which promotes monoubiquitylation of Cys222, Cys226 and Cys232 in the pre-SET domain by the E3 ubiquitin ligase SKP2 (ref.45). These mechanisms help regulate SUV39H1/H2 activity at repeat sequences, yet they may also be relevant in the repression of tissue-specific genes, as discussed later.
Box 1 Position effect variegation.
The classical position effect variegation (PEV) assay is based on an inversion in the Drosophila melanogaster X chromosome, which brings the white gene within ~25 kb of a domain of pericentric heterochromatin, shifting the border between heterochromatin and euchromatin towards the telomere. The inversion leads to the silencing of white and loss of the red eye colour in flies (fly genes are named for the mutant phenotypes)12. With use of the PEV assay, it was observed that sequences organized into heterochromatin had a tendency to be silenced in a semi-stable manner, generating phenotypic variegation. Similar variegated phenotypes arise when genes are inserted into subtelomeric chromatin in yeast221. In the case of the translocated white gene, PEV manifests itself as a random pattern of eye facets coloured white, instead of red. Various research groups have used PEV-reporter strains to screen for mutant genes that encode proteins that either modulate or are essential for heterochromatic gene repression, with the histone H3 Lys9 (H3K9) methyltransferase gene Su(var)3–9 being one of the most prominent hits, along with Su(var)2–5, which encodes heterochromatin protein 1 (Hp1), a protein that binds methylated H3K9 (ref.12).
Box 2 H3K9 methyltransferases and the spatial organization of chromatin.
Euchromatin and heterochromatin are spatially segregated in the interphase nucleus. The identification of A and B compartments within the genome stems from sequence interaction data generated by Hi-C208 or ARC-C222, and reflects the observation that interactions between loci on a megabase scale are favoured within each compartment rather than between them. The A compartment comprises largely open chromatin, and the B compartment is generally closed (Fig. 2). The former is further subdivided into smaller, looped topologically associating domains (TADs)222–224, within which sequences also preferentially interact to regulate transcription. TAD looping and insulation is mediated primarily by cohesin complexes225 and the insulator protein CTCF in mammals226,227. Finally, juxtaposed to the nuclear envelope, lamin-associated domains (LADs) can be identified by Dam-ID207. LADs are enriched in heterochromatin in mammals and worms228–231, and dimethylated histone H3 Lys9 (H3K9me2) loss compromises their lamina anchoring in worm embryos6.
H3K9me2 is enriched at LADs, but is not exclusively found in LADs229, which significantly overlap with the B compartment222,232. H3K9me2 levels are regulated by different histone methyltransferases (HMTs) in A and B compartments: in embryonic stem cells (ESCs) and differentiated mammalian cells, A-compartment H3K9me2 levels depend on SET domain bifurcated 1 (SETDB1) and especially on G9A and/or G9A-like protein (GLP), whereas in B compartments, they depend on all five main HMTs (G9A, GLP, SETDB1, Suppressor of variegation 3–9 homologue 1 (SUV39H1) and SUV39H2)233,234. The separation of A and B compartments is less pronounced in undifferentiated ESCs, and SETDB1 ablation in ESCs resulted in changes in H3K9 methylation in both compartments233,235.
G9A-deficient and SETDB1-deficient cells show aberrant CTCF and cohesin binding, resulting in the formation of additional chromatin contacts and diminished TAD insulation107,236. The loss of widely interspaced zinc-finger-containing protein (WIZ), a G9A cofactor, decreased CTCF and cohesin colocalization and chromatin looping and destabilized G9A237, implicating HMTs and H3K9me2 in establishment and maintenance of the inactive compartment.
Connecting heterochromatin to the nuclear envelope is well characterized in Caenorhabditis elegans, where a nuclear envelope-associated chromodomain protein, CEC-4, binds equally to monomethylated H3K9, H3K9me2 and trimethylated H3K9 (H3K9me3) peptides209–211,216 (Fig. 2). CEC-4 chromodomain integrity is required for the correct positioning of methylated H3K9 (H3K9me)-marked domains in worm embryos, but not for transcription repression, which depends instead on ‘readers’, namely heterochromatin protein 1 (HP1) homologues and the protein LIN-61 (refs82,209). H3K9me, but not perinuclear anchoring, was shown to contribute to megabase-scale TAD organization on the worm X chromosome by the condensin dosage compensation complex (DCC)238. In met-2 (SETDB1 homologue) set-25 (Su(var)3–9 homologue) double mutants, DCC-dependent TAD boundaries were compromised, while DCC-independent boundaries were not238, showing that multiple pathways segregate active and inactive chromatin domains212.
Whereas ablation of the H3K9me2 HMT MET-2 in worm embryos largely abolished heterochromatin anchoring6,60,210, loss of the mouse HMT G9A had a more nuanced effect on nuclear organization. Similarly, in G9A-deficient mouse ESCs or G9A inhibitor-treated hepatocytes, only a subset of nuclear envelope-bound genes were derepressed234,239, likely reflecting redundancy between G9A, SETDB1, SUV39H1 and SUV39H2 (ref.240) and redundancy between anchors212,214 (Fig. 2). In mammals, multiple factors contribute to heterochromatin anchoring. First, PRR14 is a bifunctional heterochromatin anchor with separable HP1 and lamin interaction domains241,242. It is phosphorylated and dissociates from chromatin at mitosis and reassociates in early G1 phase to anchor H3K9-methylated chromatin243. Second, lamin itself contributes to the spatial segregation of active TADs from inactive LADs in mammalian cells244, Drosophila melanogaster S2 cells245 and C. elegans246,247. Third, vertebrate lamin A and the lamin B receptor contribute with various degrees to heterochromatin tethering in differentiated-cell nuclei214,215, along with other tissue-specific pathways found in many species214. In cultured D. melanogaster cells, lamin ablation led to chromatin repositioning from the nuclear envelope, an accumulation of active TADs (A type) and TAD decompaction245. This phenotype also occurred upon lamin disruption in mammals244. Finally, lamin point mutations can both disrupt and enhance perinuclear sequestration of genes in differentiated tissues181,214,248,249.
Although H3K9me has a direct role in nuclear positioning of chromatin and in gene repression, positioning at the nuclear periphery is not sufficient to repress transcription6,83,250–252. Nonetheless, in a promoter-dependent manner, perinuclear localization can dampen expression215. For instance, histone deacetylase 3 (HDAC3) associates with the nuclear lamina and with the SMRT–NCOR1 complex, which interacts with various tissue-specific transcription factors215,253. These interactions appear to antagonize H3K27 acetylation, which drives both transcription activation and relocation of nuclear envelope genes in differentiated cells212,214.
Importantly, the association of HP1 with H3K9me is not sufficient for nuclear envelope anchoring: heterochromatin centromeric satellite repeats are found clustered in nucleus-internal chromocentres254 in both mouse embryonic fibroblasts and D. melanogaster cells. Although deletion of both SUV39H1 and SUV39H2 resulted in H3K9me3 loss from chromocentres, these compartments remained structurally intact and the chromatin remained compacted17,255. The integrity of chromocentres was partially lost in SUV39H1–SUV39H2–SETDB1 triple mutants and was extensively lost upon the ablation of all six mammalian HMTs8, underscoring their functional redundancy.
SETDB1 and MET-2
SETDB1, like SUV39H1, is an abundant SET domain-containing H3K9 methyltransferase. SETDB1 is characterized by an unusual bifurcated SET domain, which contains a 347 amino acid insertion in the middle of an otherwise well conserved and catalytically active SET domain as well as characteristic pre-SET and post-SET domains46,47 (Fig. 1a). In contrast to SUV39H1/H2, however, SETDB1 can methylate unmodified H3K9 to generate H3K9me1, H3K9me2 and H3K9me3 both in vitro and in vivo48. Its catalytic activity is regulated by several modifications at or near the SET domain49–52. These modifications include monoubiquitylation of Lys867 in the SET domain insertion, which has been shown to increase SETDB1 processivity in mammals49,50 and to enhance protein stability and nuclear localization in D. melanogaster52. In addition, sumoylation of the insertion sequence may regulate SETDB1 recruitment to chromatin51. Su(var)2-10, the fly homologue of the SUMO E3 ligase PIAS, is essential to mediate SETDB1-dependent and H3K9me3-dependent repression by PIWI-interacting RNAs in the female germ line53. Collectively, these observations suggest that the regulation of SETDB1 function through ubiquitylation and sumoylation is conserved. Interestingly, the H3K9me reader HP1 and the transcription regulator Krüppel-associated box (KRAB)-interacting protein 1 (KAP1; also known as TIF1β and TRIM28), which recruits SETDB1, are also regulated by sumoylation54,55.
Monoubiquitylation of SETDB1 has been suggested to be promoted by the conserved cofactor of SETDB1, activating transcription factor 7-interacting protein 1 (ATF7IP)56,57. Consistently, mammalian ATF7IP and its fly homologue (Windei) and worm homologue (LIN-65) are essential for robust SETDB1-mediated H3K9 methylation58. As well as regulating SETDB1 activity, ATF7IP (like Windei and LIN-65) is necessary for nuclear localization and chromatin association of SETDB1 (refs52,56,57,59,60). Loss of ATF7IP further coincides with enhanced, but incomplete, degradation of SETDB1, a phenomenon also observed for the C. elegans homologue MET-2 upon loss of LIN-65 (refs56,57,60). It is likely that these various roles are interdependent, as ATF7IP and LIN-65 binding also correlates with the stable presence of SETDB1 and MET-2, respectively, in heterochromatin foci or condensates60.
An integral element of SETDB1 control appears to be its nuclear depletion, which has been observed in differentiating muscle cells in mice61, upon heat stress in C. elegans60 and when the protein is overexpressed in fusion with GFP (compare ref.6 with ref.60). Indeed, the SETDB1 amino terminus contains both nuclear localization and nuclear export signals62, and the ATF7IP-binding site52,57,59 (Fig. 1a). The fluctuation of nuclear levels of SETDB1 in response to environmental stress suggests that active nuclear uptake or export may be a physiological regulator of SETDB1 function60,61,63. Indeed, MET-2 shifts from cytoplasm to the nucleus during early development, coincident with the bulk accumulation of H3K9me2 (ref.59).
In addition to the SET domain, SETDB1 contains a double Tudor domain64 that has been implicated in its recruitment to histones that are bivalent for H3K9me1 and H3K4 acetylation65. Although the bivalency is proposed to recruit SETDB1 to specific loci, it is unclear whether or where the bivalency is relevant in vivo. SETDB1 further contains a putative methyl-CpG-binding domain47,48 that is conserved across mammals, flies and C. elegans (Fig. 1a). The methyl-CpG-binding domain is proposed to help target SETDB1 to regions of DNA methylation, yet it likely serves other functions given that D. melanogaster and C. elegans lack DNA methylation (Box 3). In worms, adenosine 5′-diphosphate-ribosylation factor-like 14 (ARLE-14) potentially helps to recruit MET-2 to gene promoters59,60,63 (Fig. 1b).
SETDB1 has been extensively studied in mouse ESCs, where its loss affects both cell viability and differentiation potential66. In these cells the best characterized targets of SETDB1 are class I and class II long terminal repeat (LTR)-containing retroviruses, such as murine leukaemia virus elements and IAPs67–75 (Fig. 1b). Both of these ERV families are evolutionarily young with intact repeat copies that were shown to have the potential to be active in mice76. SETDB1 is recruited to these elements through its interaction with KAP1 (ref.48), which functions as a scaffold that bridges between KRAB domain-containing zinc-finger proteins (KRAB-ZFPs) and various heterochromatin proteins. KRAB-ZFPs bind to ERV LTRs to ensure the sequence-specific establishment of H3K9me3 domains77. The co-evolution of genetically unstable ERVs and their cognate KRAB-ZFPs has been proposed to drive the enormous expansion of the latter in mammalian genomes78.
Although ERV silencing is the best described function of SETDB1, several other repetitive elements, as well as genes, are SETDB1 targets (Fig. 1b). At telomeric repeats, SETDB1-mediated H3K9me3 drives recombination-based alternative telomere lengthening, whereas SUV39H1/H2 appear to suppress recombination40. At pericentric heterochromatin, SETDB1 is part of the machinery involved in silencing both major and minor satellite repeats79, possibly by providing H3K9me1 for SUV39-mediated trimethylation8,80. This mode of action, in which two HMTs sequentially methylate H3K9, was also shown in C. elegans for MET-2 and SET-25. MET-2 deposits H3K9me2, which is converted to H3K9me3 by SET-25 at full-length transposons and tissue-specific genes7,81,82. In worms, the ablation of set-25 did not derepress many genes or satellite repeats, and derepressed only a subset of transposons, because MET-2-mediated H3K9 dimethylation was sufficient (and necessary) to maintain transcriptional repression27,81,83, likely in cooperation with a range of other heterochromatin proteins82.
Partial redundancy between SUV39H1 and/or SUV39H2 and SETDB1 was also observed in the silencing of the mammalian LINE1 retrotransposons. Recruitment of SETDB1 to these non-LTR transposons dependent not on KAP1 (ref.84) but rather on its interaction with the human silencing hub (HUSH) complex, which is composed of transgene activation suppressor protein (TASOR), M-phase phosphoprotein 8 (MPP8) and periphilin85. SETDB1 recruitment appears to require interaction between MPP8 and methylated ATF7IP86, while TASOR is the central adaptor connecting MPP8 with periphilin 1 and MORC2, which is an ATPase involved in heterochromatin compaction87,88. In addition, TASOR interacts with RNA-processing proteins and/or regulators of RNA polymerase II activity89.
There are two mechanisms that ensure the recruitment of HUSH and SETDB1 to their target sites. MPP8 can bind H3K9me3 through its chromodomain90, which would in principle promote SETDB1 spreading through a reader–writer mechanism, much like HP1 and SUV39H1/H2 (refs25,26,91). This, however, has been contested by another study that argues that HUSH–SETDB1 cannot promote the spread of heterochromatin on its own, even though it can methylate nucleosomes at LINE1 elements19. A non-exclusive alternative would be that SETDB1 recruitment is mediated by the ability of periphilin 1 to bind large, intronless RNA derived from endogenous retroelements (for example, LINE1) and pseudogenes, and exogenous RNA transcribed from plasmids or viruses92. This capacity would allow a transcription-dependent, but sequence-independent, establishment of H3K9me3 domains92.
During DNA replication, SETDB1 is also found in complex with other chromatin-targeting factors, namely the histone chaperone chromatin assembly factor 1 (CAF1) and HP1α80,93 (Fig. 1b). Interestingly, formation of the CAF1–HP1α–SETDB1 complex is mutually exclusive with formation of the ATF7IP–SETDB1 complex80, and whereas CAF1–HP1α–SETDB1 mediates H3K9 monomethylation at pericentric heterochromatin, ATF7IP–SETDB1 drives most SETDB1-mediated H3K9 trimethylation. SETDB1 is ubiquitously expressed throughout mammalian development, yet most work aimed at understanding SETDB1 function and recruitment is performed in HeLa cells or undifferentiated ESCss. These are inadequate models for explaining the diverse roles of SETDB1 in cell type-specific gene repression during tissue differentiation, especially when compared to C. elegans.
Box 3 The relationship between H3K9 methyltransferases and DNA methylation.
In mammals, but not in Drosophila melanogaster, Caenorhabditis elegans or fission yeast, repeat elements and genes can be repressed through DNA methylation of cytosine to 5-methylcytosine (reviewed in ref.256). DNA methylation has been shown to depend on individual histone H3 Lys9 (H3K9) methyltransferases to various degrees. Loss of G9A in mouse embryonic stem cells leads to reduced levels of DNA methylation at retrotransposons, satellite repeats, CpG-rich promoters and imprinting control regions110,257,258. However, H3K9 methyltransferase redundancy ensures that most repetitive elements retain H3K9 trimethylation and remain silent, despite the loss of DNA methylation110. Loss of SET domain bifurcated 1 (SETDB1) or its cofactor Krüppel-associated box (KRAB)-interacting protein 1 (KAP1) results in loss of de novo DNA methylation at long terminal repeat retrotransposons and telomeres in early embryos40,259, and deletion of both Suppressor of variegation 3–9 homologue 1 (SUV39H1) and SUV39H2 leads to displacement of part of the DNA methyltransferase machinery from pericentric heterochromatin158,260. In mouse embryonic stem cells that lack G9A and G9A-like protein (GLP), DNA methylation is lost from imprinting control regions257, including from the Prader–Willi syndrome imprinting centre258, thereby generating a parent-of-origin disease. The general consensus is that H3K9 methylation precedes DNA methylation.
SETDB2
SETDB2 is a closely related paralogue of SETDB1 that also has H3K9 methylation activity (Fig. 1a). It is one of the six mouse HMTs that must be ablated to generate cells lacking all detectable H3K9me8, yet its catalytic activity appears to be restricted to the generation of H3K9me3 from H3K9me1 or H3K9me2 (ref.94). In zebrafish, the loss of Setdb2 specifically affects H3K9me3 and disrupts left–right asymmetry during development95. In mammals, several recent studies have linked SETDB2 to the silencing of pro-inflammatory chemokine and cytokine genes during influenza virus96 or severe acute respiratory syndrome coronavirus 2 (ref.97) infections. Other studies found low SETDB2 levels in individuals with diabetes with pro-inflammatory phenotypes98. In general, however, SETDB2 is an understudied H3K9 methyltransferase.
G9A, GLP and SET-25
The two SET domain-containing HMTs G9A and GLP define a third class of H3K9-specific HMT complexes (Fig. 1). The two enzymes are both ubiquitously expressed and share 44.5% sequence identity (76.5% similarity). Whereas each can form functional homodimers and heterodimers in vitro, only the G9A–GLP heterodimer has been successfully isolated from mouse ESCs99, and loss or inhibition of either HMT results in major loss of H3K9me2 in mouse ESCs99,100. Nonetheless, G9A and GLP appear to have non-overlapping functions in muscle development and terminal differentiation101.
Like SUV39H1 and SETDB1, G9A and GLP also contain characteristic pre-SET and post-SET domains, even though their SET domains can catalyse only H3K9 monomethylation and dimethylation. This is because the hydroxyl group of Tyr1067 in the G9A SET domain (Tyr1124 in GLP) restricts the orientation of the dimethylamine of H3K9me2, thereby preventing transfer of a third methyl group102,103. To date it is unclear whether H3K9me1 and H3K9me2 catalysed by G9A and GLP serve as substrates for other HMTs, although most genomes do contain large domains bearing H3K9me2 only. These domains could arise from unique recruitment of G9A and GLP, from the activity of specific H3K9me1 and/or H3K9me2 readers that block subsequent trimethylation by other HMTs or by the localized action of specific lysine demethylases104,105 that prevent H3K9me3 accumulation.
On the basis of early immunofluorescence analysis of H3K9me2 and/or H3K9me3 in G9A and GLP mutants, both HMTs were initially considered to be ‘euchromatic H3K9 methyltransferases’ (that is, specifically involved in the regulation of genes)2,99,106. However, we now know that also class III ERVs, such as murine endogenous retrovirus type L (specifically MT2), are marked by H3K9me2 and repressed in a G9A-dependent manner107,108. In addition, G9A was uniquely required for de novo establishment, but not for maintenance, of transcriptional silencing and DNA methylation of integrated vectors of Moloney murine leukaemia virus109. Interestingly, in mouse ESCs lacking G9A, a number of IAP and class I MusD retroelements lose H3K9me2, but maintain H3K9me3, suggesting redundancy between G9A and SETDB1 (Fig. 1b). By contrast, a C. elegans set-25 null mutant exhibited loss of H3K9me3, but a gain of MET-2-mediated H3K9me2. Such compensation illustrates the importance of analysing both dimethylation and trimethylation H3K9 states both in the wild type and in the relevant mutants81,110,111.
Further evidence for a broad function of G9A and GLP in transposon silencing comes from analyses in mouse testis, where G9A is both necessary and sufficient to silence LINE1a and IAP elements in the absence of a functional PIWI-interacting RNA pathway (Mili−/−; also known as Piwil2−/−) 112. Confusingly, IAP silencing is also dependent on SETDB1 in testis113, suggesting either that individual copies of these transposons are silenced by different HMTs in some tissues or that the two HMTs are interdependent in testis, but not in mouse ESCs108. Finally, similarly to SUV39H1 mutants, the inhibition of G9A resulted in the derepression of rDNA repeats, in nucleolar fragmentation and in accumulation of nucleolar R-loops114.
Like SUV39H1/H2, G9A and GLP can directly bind H3K9me, albeit not through a chromodomain, but through seven or eight ankyrin repeats, respectively, that bind H3K9me1 and H3K9me2 (ref.115). In addition, G9A and GLP bind the HP1 proteins99,116 through an interaction regulated by automethylation of G9A and GLP at Lys165 (ref.116). This automethylation falls within a motif (ARKT) that resembles the histone H3K9 context (ARKS). Interestingly, the same motif is found methylated in the SETDB1 cofactor ATF7IP86, suggesting that regulation through methylation may be conserved among multiple HMTs.
In addition to interactions that recruit G9A to pre-existing H3K9me1 and H3K9me2, G9A was shown to bind a range of sequence-specific transcription factors in different cell types. In CD8+ T cells and U2OS cells, G9A binds the transcription factor PRDM1 (refs1,117), whose multiple roles include repression of the interferon-β gene response to regulate T cell development following viral infection117. In HeLa cells, G9A and GLP are part of a complex containing chromodomain Y-like protein, HDAC1 or HDAC2 and the transcriptional repressor REST116,118, whereas in liver cells, G9A represses important developmental genes (for example, FGF21 and CYP7A1) through interactions with the transcription factors E4BP4 (also known as NFIL3)119,120 and small heterodimer partner (SHP)121,122. SHP is an unusual orphan nuclear receptor that lacks a DNA-binding domain, which is involved in the repression of nuclear-receptor targets by recruiting G9A and the chromatin remodeller mSin3a complex121,122 (Fig. 1b). The interaction of G9A and GLP with HDACs or with known HDAC adapter proteins such as mSin3a is found in multiple tissues. Indeed, the best known cofactor of G9A and GLP is the transcription factor widely interspaced zinc-finger-containing protein (WIZ), which bridges between the HMTs and the co-repressor CTBP123. CTBP associates with HDAC1, HDAC2 and HDAC3 (ref.124), which, in addition to deacetylating histones, may have a role in the anchoring of inactive genes to the nuclear lamina (Box 2; Fig. 2).
Overall, the mechanisms through which H3K9 methyltransferases work together to establish heterochromatin are only partly understood. Nor is it clear in which context they establish H3K9 methylation de novo as opposed to the maintenance of an existing methylation state that antagonizes acetylation. The spatial separation of euchromatin from heterochromatin may itself have a role in the maintenance of silent states (Box 2; Fig. 2). Clearly, understanding both specific HMT targeting and domain segregation remains an important goal for deciphering how H3K9me-mediated silencing influences organismal development.
HMTs during and after differentiation
Whereas much of the cellular H3K9me2 and H3K9me3 marks repetitive or ‘constitutive’ heterochromatin, detailed analyses of tissue-specific H3K9 methylation patterns and transcriptomics in both wild type and HMT mutant organisms implicate this modification in the silencing of tissue-specific genes. Among the different physiological roles proposed for H3K9me2 and H3K9me3, dominant is the maintenance of cell identity by preventing inappropriate expression of tissue-specific genes125. Compelling support for this role came from the discovery that H3K9me itself is a barrier to the reprogramming of differentiated cells into so-called induced pluripotent stem cells, presumably by partially blocking the binding to DNA of the pluripotency transcription factors OCT4, SOX2, Krüppel-like factor 4 (KLF4) and MYC126,127. Similarly, genomic regions resistant to gene-expression reprogramming following somatic nuclear transfer are marked by H3K9me3, and both the depletion of SUV39H1/H2 and the overexpression of H3K9me demethylases enhance the efficiency of somatic cell nuclear reprogramming128,129. Conversely, ectopic expression of the oncogenic mutation of H3 Lys9 to Met (H3K9M), which inhibits Lys9 methylation even in wild type histone H3, impairs ESC differentiation by allowing the continued expression of OCT4, SOX2 and another pluripotency factor, NANOG130. Despite these deficiencies, H3K9M-expressing mice survived for up to 1 year, although they experienced expansion of multipotent progenitors, aberrant lymphopoiesis and thrombocytosis, and, in some animals, aggressive T cell leukaemia or lymphomas130. Collectively, these results argue strongly for a role for H3K9 methylation in stabilizing differentiated cell type identity and countering oncogenic transformation131.
In C. elegans, where H3K9me is not essential for mitosis, the complete loss of H3K9 methylation (that is, in the met-2 set-25 double mutant) did not prevent development from embryos to adults, despite widespread misexpression of both repetitive elements and tissue-specific genes5–7,82. Nonetheless, the H3K9me-deficient worms displayed stochastic developmental delay and reduced fertility7,132 and became highly dependent on factors involved in DNA repair and small-RNA pathways for survival27,81,82,133. In contrast to worms, mice with knockout of G9a (also known as Ehmt2) or Glp (also known as Ehmt1) were early embryonic lethal (approximately embryonic day 9.5 to embryonic day 12.5) and showed gross morphological defects across all developing cell lineages2. Maternally contributed G9A was able to silence a subset of four cell stage-specific genes134, and ensure proper chromosome segregation135, but was presumably insufficient to allow prolonged development134. Deletion of Setdb1 resulted in similar embryonic lethal phenotypes at 3.5–5.5 days after coitus, blastocysts in vivo showed sub-Mendelian survival66 and ESC survival in vitro was compromised66. By contrast, knockout of Suv39h1 or Suv39h2 showed no superficial developmental defects, although double-null mice were also born at sub-Mendelian ratios and were characterized by prenatal lethality linked to genome instability16.
The developmental defects that stem from the loss of H3K9 methylation in vertebrates can be aggravated by the mis-segregation of chromosomes, which arises from altered pericentromeric heterochromatin16,136. In addition, H3K9 methyltransferases are known to have a few non-histone targets (for example, G9A methylates oestrogen receptor-α137 and ligase 1 (ref.138), SETDB1 and SETDB2 methylate the HIV-1 protein Tat139, and SUV39H2 methylates RAG2 and the lysine methyltransferases SET8 and DOT1L140). Loss of methylation of these non-histone targets could compromise viability; thus, phenotypes driven by the loss of HMTs may not exclusively reflect the absence of H3K9 methylation.
Given their ubiquitous expression in tissues141–143, including in testis22,141,142, oocytes and zygotes144, it is not surprising that vertebrate organisms lacking H3K9 methyltransferases are embryonic lethal. To study the roles of individual HMTs in differentiated cells, use of conditional tissue-specific knockouts or conditional degradation through tissue-specific degrons is required. Given the complexity of producing organisms with HMT-deficient tissues, work in tissues was mainly based on loss of SUV39H1 alone, despite evidence for its redundancy with SUV39H2 (ref.111) and the role of the latter in keratinocyte gene repression145. Similarly, largely on the basis of the interdependency between G9A and GLP in mouse ESCs, primarily G9A mutants have been analysed in tissues, despite evidence that the two HMTs have non-overlapping target genes in differentiating myoblasts101. Nonetheless, studies of haematopoiesis, muscle development and neuronal development have provided ample evidence that specific H3K9me methyltransferases have key roles in the formation and maintenance of differentiated tissues. We discuss these studies in the following sections, and include C. elegans and D. melanogaster results for additional mechanistic insights.
Haematopoiesis
Haematopoietic cells, and especially the lymphoid cell lineages, are among the best characterized models of lineage-dependent differentiation (reviewed in refs146–148). Haematopoiesis is thus an interesting model for the analysis of mutants with pleiotropic effects, such as deletions of epigenetic modifiers.
The adult lymphoid system originates from haematopoietic stem cells (HSCs) located in the bone marrow146 (Fig. 3). In HSCs, SETDB1 is required for stem cell maintenanace149 by repressing genes associated with other, non-haematopoietic cell lineages150. Loss of H3K9me3 owing to deletion of Setdb1 in the mouse, however, was restricted to a fraction of repressed genes and transposable elements150. Among the SETDB1-modified genes, only a subset was actually transcribed in the Setdb1−/− mice. Transposable elements in SETDB1-deficient HSCs also showed only a marginal loss of H3K9me3, even at the class I and class II retrotransposons that in mouse ESCs require SETDB1 for repression. Reductions in H3K9me3 levels were insufficient to trigger loss of DNA methylation at transposons150. Nevertheless, mice with HSC-specific Setdb1 deficiency rapidly lost HSCs and progenitor cells from the bone marrow150.
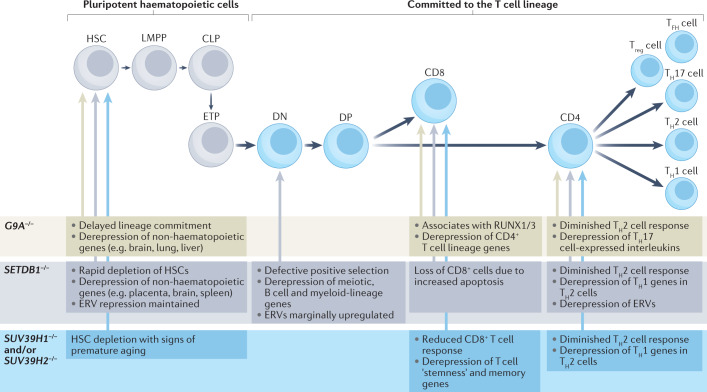
Fig. 3. Roles of H3K9 methyltransferases in T cell lineage determination.
The well-characterized differentiation steps from haematopoietic stem cells (HSCs) into terminally differentiated T helper cells (T helper 1 (TH1) cells, TH2 cells, T follicular helper (TFH) cells and TH17 cells) are shown. The effects of deletion of SUV39H1 and/or SUV39H2, SETDB1 or G9A are listed. Although not shown, histone H3 Lys9 (H3K9) methyltransferases also have a role in the maturation of B cell progenitors (pro-B cells). In this case, the loss of SETDB1 leads to derepression of both specific endogenous retroviruses (ERVs)69,217 and non-lineage-specific genes, including genes specific to innate immunity69. The selection of genes and repeats that are derepressed likely depends on the variable presence of lineage-specific transcription factors, as described in Caenorhabditis elegans83 (Box 4). CLP, common lymphoid progenitor; DN, double negative (CD4−CD8−); DP, double positive (CD4+CD8+); ETP, early T cell progenitor; LMPP, lymphoid primed multipotent progenitor; Treg cell, regulatory T cell.
Knockout of both Suv39h1 and Suv39h2, but not of either of them, resulted in reduction of the HSC pool and a nuanced loss of differentiation. Surviving HSCs were characterized by an increase in the number of nucleoli, disrupted nuclear lamina and the appearance of phosphorylated histone H2AX (γ-H2AX) foci, which mark DNA damage151. There was also a loss of 4′,6-diamidino-2-phenylindole (chromatin)-dense regions overlapping with HP1α151. Interestingly, these phenotypes are similarly associated with HSCs isolated from older individuals150, and appear during cellular aging152. Indeed, HSCs isolated from humans older than 70 years showed an ∼50% reduction in SUV39H1 expression compared with humans younger than 35 years153. A similar reduction in the levels of both SUV39H1 and H3K9me3 was observed in HSCs from old mice151,153. Thus, in the haematopoietic lineage, the loss of SUV39H1/SUV39H2 generates aging-related phenotypes. By contrast, the deletion of G9A in HSCs more closely resembled the loss of SETDB1, leading to the transcription of genes from a variety of non-haematopoietic tissues, such as lung, liver, saliva and brain154. It is unclear whether the similarity in the sets of genes derepressed upon loss of G9A or SETDB1 represents an interdependency among the enzymes, or the fact that they have overlapping targets (Fig. 3).
During both B cell differentiation and T cell differentiation, the ablation of SUV39H1, SETDB1 or G9A individually did not block differentiation per se, but each HMT was required to stabilize well-defined differentiation steps, as illustrated in Fig. 3 for T cells. In wild type mice, naive CD4+ T helper (TH) cells differentiate into two main cell types, TH1 and TH2 cells, and the ratio between the two cell types is an important determinant of the immune response155. However, isolated naive CD4+ T cells deficient in Suv39h1 or lacking the H3K9me2 and H3K9me3 reader HP1α were unable to stably differentiate into CD4+ TH2 cells. These unstable Suv39h1-deficient TH2 cells were characterized by derepression of TH1-specific genes, which correlated with reduced H3K9me3 levels156. Consequently, TH2 cell to TH1 cell transdifferentiation increased and caused a skewed TH1 cell response in a mouse model of TH2 cell-driven allergic asthma, thereby decreasing the associated lung pathology156. Interestingly, Suv39h1-deficient cytotoxic CD8+ T lymphocytes also showed a reduced antigen-specific CD8+ T cell response due to defects in the silencing of T cell ‘stemness’ memory genes157. Thus, even in closely related cell lineages, the loss of H3K9me2 and/or H3K9me3 affects different target genes, and the aberrantly expressed cohort of genes is strongly cell type specific.
Given the phenotypes of SUV39H1 deficiency in late differentiation, it was somewhat surprising to find that SETDB1 was also essential for the stable, terminal differentiation of CD4+ T cells into TH1/TH2 CD4+ T cells70. SETDB1 deletion in naive CD4+ T cells resulted in hyperactivated TH1 cells and an unstable commitment to the TH2 cell lineage (Fig. 3). In contrast to SUV39H1, SETDB1 is not thought to directly control promoter activity of a majority of the misexpressed TH1-specific genes. Instead, gene derepression correlated with the loss of silencing of nearby ERV elements, which then functioned as aberrant enhancers or enhancer control elements for at least some of the induced genes70. This mechanism raised two interesting questions. First, why are TH1-specific genes so sensitive to the loss of H3K9me despite the likely redundancy between H3K9 methyltransferases? Second, are ERV-linked control elements part of the normal gene regulation process during wild type haematopoiesis?
Deletion of SETDB1 at an earlier stage (that is, in CD4−CD8− thymocytes) resulted in a partial block of the production of both CD4+ T cells and CD8+ T cells. The residual naive CD4+ SETDB1−/− T cells showed misexpression of genes typically expressed during meiosis or in other haematopoietic cell lineages, and this was independent of ERV misexpression72. Thus, depending on the cell type, SETDB1 may also mediate gene repression directly, not through transposable-element repression.
Mice with a HSC-specific G9a deletion had no discernible defects in the generation of T cells158,159. However, similarly to Suv39h1-knockout and Setdb1-knockout mice, these mice failed to develop a protective TH2 cell response following T cell activation160. Instead, these activated G9a−/− T cells lost H3K9me2 and gained the expression of genes specific to TH17 cells158,160, a TH cell lineage with important functions in modulating the immune response161. Interestingly, the presence of H3K9me2 and/or H3K9me3 at the interleukin-17A (IL-17A) promoter is triggered by the transcription factor OX40 (ref.160), which is a T cell co-stimulatory molecule in the tumour necrosis factor receptor superfamily and which is highly expressed in activated T cells162. This example illustrates how a specific transcription factor can regulate the heterochromatin state of cell type-specific genes without affecting H3K9me levels overall. However, given the large diversity of transcription factors that interact with HMTs, the rules that govern the specificity of their interaction with HMTs remain unknown.
One of the early phenotypes in haematopoiesis found in Suv39h1-knockout mice was the increased risk of late-onset B cell lymphomas, which resemble non-Hodgkin lymphoma in humans16. The exact molecular mechanism driving tumorigenesis in SUV39H1 mutants is not understood, yet one of the central B cell fate-determining steps — V(D)J recombination of antibody light and heavy chains — is controlled by H3K9 methylation. In non-B cells, the complete immunoglobulin heavy chain variable region (Vh) locus is marked by H3K9me31, and the forced methylation of H3K9 by artificially targeting G9A in cultured B cells is sufficient to prevent V(D)J recombination163. Interestingly, the transcription factor PAX5, which is a master regulator of B cell development, is required for the removal of H3K9me2 during B cell differentiation31. Thus, not only are H3K9me2 and H3K9me3 domains dynamic during development but chromatin modifiers are able to trigger localized loss or formation of heterochromatin. Whereas individual knockout of Suv39h1, G9a or Setdb1 was not sufficient to compromise H3K9me at the Vh locus, Suv39h1 deficiency in B cells nonetheless reduced isotype-specific IgA switching in mice69,164,165.
Bulk H3K9me2 levels increase during the course of haematopoiesis154, even though KDM3B, the dual-purpose arginine demethylase (targeting dimethylated histone H4 Arg3) and lysine demethylase (targeting H3K9me2), is also needed for proper gene activation during the maturation and differentiation of HSCs166,167. This finding suggests that H3K9me2 and/or H3K9me3 is important both for keeping genes silent until a cell-fate decision occurs and for de novo repression of genes that were active in earlier stages. Similarly, studies in C. elegans and mice confirmed that H3K9me2 and H3K9me3 are gained and lost at different subsets of tissue-specific genes during development83,111. Analysis of C. elegans muscles following loss of MET-2, which deposits H3K9me2, showed that the resulting cell type-specific pattern of gene derepression was driven by a specific set of transcription factors that have developmental stage-specific and lineage-specific expression patterns83 (Box 4). Importantly, without transcription factor expression, the drop in H3K9 methylation level was not sufficient for gene derepression in either muscle or hypoderm83 (Fig. 4).
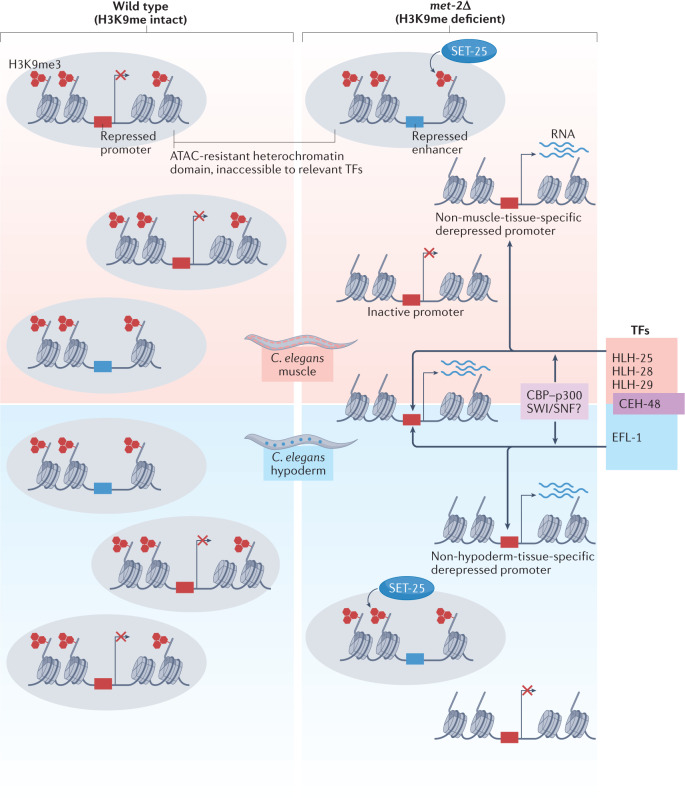
Fig. 4. Transcription factors control tissue-specific transcription derepression upon H3K9me loss.
A major question is whether histone H3 Lys9 (H3K9) methylation contributes to gene repression in differentiated tissues by blocking transcription factor (TF) binding to chromatin218. A recent study addressed this question in Caenorhabditis elegans83 by comparing the effects of loss of the SET domain bifurcated 1 (SETDB1)-like histone methyltransferase MET-2 on gene expression in embryos and in isolated muscle and hypoderm tissues. As depicted schematically, the genes upregulated by MET-2 loss constitute only a fraction (12% in embryos and 13% in muscle) of the genes marked by trimethylated H3K9 (H3K9me3), and the subset of genes affected is both tissue specific and developmental stage specific. In muscle and hypoderm, H3K9 methylation by MET-2 represses tissue-specific genes expressed in other cell lineages, including, but not restricted to, germline-specific genes83,219,220. In addition, MET-2 and SET-25 redundantly repress a large number of enhancer elements in muscle. Methylated H3K9 (H3K9me) heterochromatin ensures transcription inactivity by restricting the binding of a defined set of transcription factors at promoters and enhancers, which are either tissue specific — for example, HLH25, HLH-28 and HLH-29 in muscle and EFL-1 in hypoderm — or more widely expressed, for example CEH-48 (ref.83). As measured by assay for transposase-accessible chromatin with high-throughput sequencing (ATAC–seq), increased accessibility upon loss of H3K9me is neither sufficient nor necessary to drive transcription, but a subset of β-helix–turn–helix and bZip-containing factors that can recruit chromatin remodelling complexes such as SWI/SNF or the acetyltransferase complex CBP–p300 drive transcription at the genes that become both accessible and upregulated83. These results suggest that H3K9 methylation confers tissue-specific gene expression by restricting transcription factor access. The met-2 mutation compromises both muscle ultrastructure and worm mobility.
The cell-type specificity of gene derepression upon loss of an H3K9 methyltransferase in mammals is not lymphoid specific. The loss of SETDB1 triggers a hyperactive pro-inflammatory response in macrophages at the promoter of pro-inflammatory cytokines, such as IL-6 (ref.168). In these cells, H3K9me3 normally restricts the recruitment of NF-κB, a central regulator of the inflammatory response in the immune system168, which also plays critical roles in the development, survival and activation of B cells169. While it is unclear why the effect of H3K9me3 on the NF-κB pathway is specific to the macrophage lineage, such observations suggest that stress-induced signalling pathways regulate H3K9me in a cell type-specific manner.
Box 4 Mechanisms of H3K9me-mediated silencing in differentiated tissues.
Histone H3 Lys9 (H3K9) dimethylation and trimethylation anticorrelate with transcription and chromatin accessibility17,125,238,255,261,262. However, how H3K9 methylation prevents transcription is not well understood. There are theoretically three ways in which methylated H3K9 (H3K9me) and chromatin compaction could suppress transcription: first, they could limit access of RNA polymerase complexes; second, they could restrict transcript elongation or the stability of mRNAs; or third, they could block the access or function of activating transcription factors. The first two scenarios predict that in the absence of dimethylated H3K9 (H3K9me2) and/or trimethylated H3K9 (H3K9me3), all heterochromatin sequences become broadly transcribed. If true, one would have to attribute the derepression of cell type-specific gene sets in individual H3K9 methyltransferase mutants to target-gene redundancy among them. However, a recent study analysing the transcriptomes of different tissues of Caenorhabditis elegans lacking detectable H3K9me2 and H3K9me3 argues that only a small subset of heterochromatin genes become aberrantly transcribed (upregulated) upon loss of H3K9me, even though a very large number of loci become accessible according to the assay for transposase-accessible chromatin with high-throughput sequencing (ATAC–seq) analysis83. The subset of genes upregulated by H3K9me loss is highly cell type specific83.
A comparison of the derepressed genes showed that a specific set of transcription factors is required for gene derepression in the absence of H3K9me2 and H3K9me3 (ref.83) (Fig. 4). In wild-type cells, heterochromatin restricts their binding, thereby ensuring the maintenance of tissue identity, potentially through heterochromatin protein 1 (HP1)-like readers263. Similarly, the loss of a large number of heterochromatin-associated proteins in human fibroblasts had only a limited effect on gene and repeat derepression206. By contrast, overexpression of transcription factors linked to cell type reprogramming resulted in more widespread and more pronounced transcription activation when heterochromatin was challenged206. Interestingly, the most pronounced effect on gene expression was observed not following depletion of individual H3K9 methyltransferases but following depletion of Enhancer of rudimentary homologue (ERH), a factor that appears to regulate global levels of H3K9me3 in the cells studied. The exact mechanism by which ERH enhances H3K9me3 levels is unclear, yet once again it seems to involve a complex redundancy among H3K9 methyltransferases.
Taken together, these studies illustrate that H3K9me2 and/or H3K9me3 prevents transcription by restricting transcription factor accessibility. The loss of H3K9me creates the potential for gene activation, which ultimately depends on the expression pattern of specific sets of transcription factors83,264. The need for sequence-specific transcription factors has also been observed for the reactivation of retroelements during early human embryogenesis265 and in human fibroblasts206. Although these observations explain why the upregulation of H3K9me-modified genes is tissue specific, it does not explain why this particular set of transcription factors are able to activate genes upon loss of H3K9me, whereas others cannot.
Muscle differentiation and maintenance
The differentiation of muscle cells is a multistep process, driven initially by basic helix–loop–helix transcription factors such as myoblast determination protein 1 (MYOD1) in mammals170,171. Differentiation into muscle begins when myoblast precursors exit the cell cycle, rendering them unresponsive to mitogenic signals172. During this transition, S-phase genes acquire H3K9me3 and become permanently silenced172,173. Evidence that H3K9me might be an important epigenetic regulator of cell cycle exit was provided by the depletion of SUV39H1/H2, which resulted in aberrant transcription of a subset of S-phase-specific genes in differentiating (postmitotic) cells, but not in cycling muscle precursor cells173. The derepressed S-phase genes were largely regulated by the RB–E2F pathway, the central regulator of G1/S phase transition and cell cycle exit.
Whereas the E2F-type transcription factors function as activators, when E2F is bound by RB, the complex mediates transcriptional repression174. Silencing through RB has been attributed to interactions with HDAC1 (refs175,176) and with SUV39H1 (refs177–179). Interestingly, the deposition of H3K9me3 and silencing of a subset of E2F targets correlates with their spatial clustering with pericentric heterochromatin in muscle cells180. Related to these results is a study of a C. elegans model of Emery–Dreyfuss muscular dystrophy, which arises from the introduction of a laminopathic mutant in otherwise wild-type worms (lmn-1Y59C or LMNAY45C in mammals). In this gain-of-function mutant, a large number of loci were aberrantly associated with the nuclear lamina in differentiated muscle in C. elegans, and the relevant promoters were statistically enriched for E2F binding sites. As expected, they became hyper-repressed181. Moreover, the association of these loci with the nuclear periphery was sensitive to loss of the H3K9me reader CEC-4 (ref.181), confirming that the lmn-1Y59C mutation led to a gain in H3K9me. Surprisingly, the loss of H3K9me2 by ablation of MET-2 in otherwise wild-type C. elegans muscle upregulated a range of genes not necessarily enriched for E2F binding sites83.
In contrast to the role of SUV39H1, G9A promotes myoblast proliferation and inhibits cell cycle exit during myogenic differentiation182. Loss of G9a resulted in the upregulation of genes that were either expressed in non-muscle lineages (for example, genes of immune cells) or regulated cell cycle exit. Interestingly, these genes were not E2F targets, but instead were regulated by MYOD1182. Thus, although SUV39H1 and G9A both methylate H3K9, HMT loss can have opposite effects in differentiating cells. This divergence in phenotype contrasts with the highly similar effects that the same HMT mutants have on late T cell differentiation. Obviously, before any clinical application of H3K9 methylation inhibition, the specific pathways and genes targeted by a given HMT in the relevant tissue must be understood.
A direct comparison between SUV39H1, G9A and SETDB1 in myogenesis is complicated by the intriguing role of SETDB1 in adult muscle regeneration. In adult mice, SETDB1 (ref.61), but not G9A183, is required for the expansion of activated muscle stem cells (satellite cells), which are needed for muscle regeneration61. Again, contrasting with the role of SETDB1 in haematopoiesis, depletion of SETDB1 in adult muscle stem cells resulted in the premature expression of genes that would normally be expressed in terminally differentiated muscle and not the misexpression of non-muscle genes. Many of the upregulated loci were direct targets of SETDB1, as SETDB1 and H3K9me3 were both found at the relevant promoters.
An analysis of SETDB1 and SUV39H1 function in wild type muscle development showed that upon muscle stem cell activation, WNT3A signalling triggers the export of SETDB1 from the nucleus by exportin 1 (ref.61). The cytosolic accumulation of SETDB1 correlated with loss of chromatin-bound SETDB1 and partial activation of its target genes61. This phenotype resembles the loss of the SETDB1 cofactor ATF7IP57 or the response of MET-2, the SETDB1 homologue in C. elegans, following exposure to heat stress60,63. In both cases, the HMT was depleted from the nucleus and there was a global reduction in the level of H3K9me, leading to selective gene upregulation. By contrast, ischaemic or oxidative stress in rat myocytes was shown to trigger a rapid transcriptional upregulation of SUV39H1, and a coordinated downregulation of the acetylated H3K9 deacetylase SIRT6 (ref.184). Thus, in addition to the control of differentiation-induced patterns of gene expression by H3K9me, stress can illicit global responses by altering the level of H3K9me.
The effects of stress on H3K9me levels may be relevant in rat neonatal ventricular myocytes, where an exposure to oxidative stress resulted in an increased likelihood of myocardial infarction184. Suv39h1-knockout mice and Suv39h2-knockout mice were both protected from myocardial infarction and showed a reduced accumulation of reactive oxygen species184. Currently it is unclear how stress-induced overexpression of SUV39H1 affects gene transcription and whether this transient state leads to lasting changes in heterochromatin. Nonetheless, it is an intriguing possibility that specific tissues undergo a long-term adaptation to stress through changes in their epigenetic landscape44,60,63,185,186.
Neurogenesis
Neurogenesis is a complex process that results in an enormous diversity of cell types, ranging from oligodendrocytes to different types of astrocytes, and a vast number of specialized neurons. In addition to the process of differentiation, the nervous system provides interesting models for postmitotic adaptation, otherwise known as neuronal plasticity. Various studies have shown that H3K9me-mediated silencing contributes to neuronal plasticity, and to early stages of neurogenesis.
As demonstrated in other tissues, all the major H3K9 methyltransferases are required to prevent non-lineage gene expression during neuronal development. Loss of SETDB1 in the neuronal lineage impairs differentiation of neurons and increases the chance of neuronal precursor cells developing into astrocytes187. In C. elegans the SETDB1 homologue MET-2 is required to restrict neuronal subtype-specific gene expression during terminal neuronal differentiation188. Setdb1-deficient mouse neurons are characterized by the upregulation of genes specific to glial cells and of germline-specific genes. Similarly to what has been observed in SETDB1-deficient CD4+ T cells, derepression in a minority of cases was mediated by a chimeric transcript originating at a nearby derepressed ERV187.
Similarly to SETDB1, G9A and its catalytic activity are required for neuronal differentiation in cell culture models189 and for the integrity of differentiated neurons in the postnatal brain. Postnatal depletion of GLP or G9A in neurons led to the derepression of non-neuron and neuronal progenitor genes, and ultimately resulted in behavioural phenotypes190. The most striking evidence for a role for H3K9me2, H3K9me3 and G9A in the maintenance of cell-fate choice, however, comes from the olfactory system. Olfactory sensory neurons are characterized by the monogenic and monoallelic expression of a specific olfactory receptor in each neuron, chosen from more than 1,000 available receptor genes in the genome191. The transcriptionally silent receptor genes are marked by H3K9me2 in olfactory epithelial stem cells and acquire H3K9me3 upon differentiation192. Interestingly, loss of G9a, which itself performs only H3K9 dimethylation, was sufficient to prevent H3K9 trimethylation at the olfactory receptor gene cluster, thereby interfering with normal monogenic receptor expression193. Although the enzyme or enzymes responsible for H3K9me3 at these genes are not known, the stage-specific, sequential methylation suggests an interplay between G9A and either SUV39H1/H2 or SETDB1/2 (ref.193).
Evidence for an important role of SUV39H2 in neurogenesis, and/or neuron function, comes from a recent description of a rare loss-of-function mutation (A211S) in the pre-SET domain of SUV39H2 in an individual with autism spectrum disorder (ASD)194. There is also a general reduction of SUV39H1/H2 levels in neuronal regions of individuals with ASD194. Subsequent molecular analysis showed that SUV39H2 is essential for the repression of a cluster of protocadherin-β genes in the developing brain of mice, and its deletion triggered ASD-relevant symptoms194.
In contrast to other tissues, the expression of the main H3K9 methyltransferases in neurons is subject to extensive regulation. SETDB1 is highly expressed in neuronal precursor cells, but its expression decreases as cells exit the cell cycle and progress through terminal differentiation. SETDB1 overexpression in differentiated neurons has been shown to alter several aspects of neuronal physiology, causing both cognitive impairment195 and antidepressant-like effects196. SETDB1 overexpression can be observed in humans with Huntington disease, and the pharmacological inhibition of SETDB1 ameliorated symptoms in a mouse model of the disease197. Non-pathogenic huntingtin protein has been proposed to sequester and thus inactivate the ATF7IP–SETDB1 complex198. Accordingly, depletion of huntingtin in human ESCs or expression of mutant huntingtin in induced pluripotent stem cells derived from individuals with Huntington disease led to an increase in H3K9me3 levels, which suggests that a feedback loop from huntingtin to SETDB1 may control genes involved in neuronal differentiation198.
In the mouse brain, G9A is expressed during the embryonic and synaptogenic periods, but its expression is reduced in later development199. Interestingly, during neuronal differentiation, G9A undergoes alternative splicing to include its E10 exon, which leads to increased nuclear localization of G9A and elevated H3K9me2 levels189, which promotes neuronal polarization200. In addition, G9A expression and H3K9me2 levels are increased following nerve injury in the dorsal root ganglion. The increase in promoter H3K9me2 levels correlates with a downregulation of the central K+ channel and hypersensitivity to pain201,202, which can be suppressed by G9A inhibition201,202. A similar increase in G9A expression and H3K9me2 levels has been observed during neonatal ethanol-induced neurodegeneration, with G9A upregulation contributing to neuronal apoptosis199. Conversely, the downregulation of G9A and a reduction of H3K9me2 levels has been observed in the brain of mice repeatedly exposed to cocaine203. Experimental downregulation of G9A correlates with increased neuronal plasticity and expression of genes that regulate dendritic plasticity203,204. Interestingly, haploinsufficiency of GLP, but not of G9a, is linked to Kleefstra syndrome, which is a rare genetic disease characterized by intellectual disability, autistic-like features, childhood hypotonia and facial dysmorphisms205. Considering this wide array of HMT mutant effects, it is likely that the modulation of HMT levels will be therapeutically helpful for neuronal diseases.
Conclusions and future directions
Taken together, the studies discussed herein demonstrate that H3K9me2-mediated and H3K9me3-mediated silencing is an important epigenetic regulator of gene expression throughout development, affecting cell type-specific genes. Surprisingly, this silencing function does not seem to be essential for cell or organismal viability, but rather it ensures the robustness of cell-fate decisions and the integrity of differentiated tissues. The limited and highly cell type-specific transcriptional changes observed in H3K9 methyltransferase mutants suggest that the loss of this heterochromatin mark is often not sufficient to induce transcription. Indeed, recent experiments in C. elegans show that most of the repressed genes remain transcriptionally silent in differentiated cells that have lost all H3K9me, and that a specific cell type-dependent set of transcription factors is necessary to drive transcription activation of genes that had been embedded in heterochromatin83 (Box 4; Fig. 4). Similarly, depletion of a large number of heterochromatin-associated proteins in human cell culture permits the aberrant binding of transcription factors to repressed non-lineage genes, but not an extensive, nonspecific activation of genes and repeats embedded in heterochromatin206. Understanding the relevant molecular features of these transcription factors and what additional changes to chromatin allow active transcription of previously H3K9me-repressed genes will be essential if we are to understand the role of H3K9 methylation in development. The partial redundancy observed among the HMTs in mammals makes it necessary to compare single and double or even triple mutants of H3K9 methyltransferases directly to reveal the full picture. This complexity poses major challenges for experimental design, and encourages further study of heterochromatin in model organisms that have fewer H3K9-methylating enzymes. The involvement of H3K9 methyltransferases in various stress responses, their misexpression and aberrant recruitment in a range of human cancers, and the overall loss of H3K9me during aging highlight the medical relevance of deeply understanding the role of heterochromatin in differentiated cells.
Acknowledgements
The authors apologize to all those whose articles could not be cited due to space limitations, and they recognize that there are many studies on histone methyltransferases and mammalian cell differentiation beyond those cited here. The authors thank the Friedrich Miescher Institute for Biomedical Research for an excellent research infrastructure and all members of the epigenetics and quantitative biology sections for interaction and discussion. S.M.G. thanks the European Research Council for support through the European Union’s Horizon 2020 research and innovation programme (Epiherigans — agreement no. 743312). J.P. and S.P.M. were both supported by EMBO long-term fellowships in the Gasser laboratory.
Glossary
- Constitutive heterochromatin
Traditional designation of dimethylated or trimethylated H3 Lys9 chromatin that is generally transcriptionally inactive in a manner that persists throughout the cell cycle and throughout development. This chromatin type is neither cell type-specific nor regulated during development.
- Histone methyltransferases
(HMTs). Class of enzymes that transfer methyl groups from S-adenosyl-l-methionine to Lys and Arg amino acids, predominantly in histones.
- Facultative heterochromatin
Transcriptionally silent chromatin with dimethylated or trimethylated H3 Lys9, which is repressed in a cell type-specific manner that is regulated during development.
- Short interspersed nuclear elements
(SINEs). Abundant non-autonomous non-long-terminal-repeat retrotransposons that hijack long interspersed nuclear element proteins to mobilize.
- Argonaute
A class of small single-stranded RNA-binding proteins implicated in small RNA-mediated gene silencing.
- Chromocentres
In some cells, clustering of centromeric satellite DNA in the nucleus, representing centromeres of multiple chromosomes.
- Endogenous retroviruses
(ERVs). Proviral remnants of ancient retroviral infections. Full-length ERVs encode viral proteins such as envelope proteins (env), polymerases (pol) and structural proteins (gag) that can mediate ERV amplification and in some cases encode full viral particles.
- Intracisternal A-particles
(IAPs). A rodent-specific class of transposable elements that retain the ability to undergo transposition. Among the most mutagenic long terminal repeat retrotransposons in mice.
- Long interspersed nuclear elements
(LINEs). Autonomous non-long-terminal-repeat retrotransposons that constitute the largest transposable element class in human and mice.
- PIWI-interacting RNAs
Germline-specific small RNAs that mediate homology-dependent silencing through their interaction with PIWI-clade Argonaute proteins.
- Long terminal repeat
(LTR). A direct repeat flanking the coding sequences of LTR retrotransposons, containing regulatory elements and polyadenylation signals. They promote transcription of the retrotransposons and potentially of neighbouring genes.
- Imprinting control regions
Genomic regions that epigenetically regulate parental-specific or maternal-specific allelic expression of genes established during germline development.
- R-loops
RNA–DNA hybrids formed through hybridization of one of the DNA strands with a complementary RNA, forming a potentially mutagenic structure.
- Assay for transposase-accessible chromatin with high-throughput sequencing
(ATAC–seq). A method for monitoring accessibility of chromatin in intact cells. The hyperactive transposase Tn5 is expressed endogenously, and high-throughput sequencing is used to detect sites of transposase cleavage and thus assess levels of accessibility.
Author contributions
The authors contributed equally to all aspects of the article.
Peer review
Peer review information
Nature Reviews Molecular Cell Biology thanks Yoichi Shinkai and the other, anonymous, reviewers for their contribution to the peer review of this work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Győry I, Wu J, Fejér G, Seto E, Wright KL. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat. Immunol. 2004;5:299–308. doi: 10.1038/ni1046. [DOI] [PubMed] [Google Scholar]
- 2.Tachibana M, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kubicek S, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol. Cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 4.Becker JS, et al. Genomic and proteomic resolution of heterochromatin and its restriction of alternate fate genes. Mol. Cell. 2017;68:1023–1037.e15. doi: 10.1016/j.molcel.2017.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garrigues JM, Sidoli S, Garcia BA, Strome S. Defining heterochromatin in C. elegans through genome-wide analysis of the heterochromatin protein 1 homolog HPL-2. Genome Res. 2015;25:76–88. doi: 10.1101/gr.180489.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Towbin BD, et al. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- 7.Zeller P, et al. Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat. Genet. 2016;48:1385–1395. doi: 10.1038/ng.3672. [DOI] [PubMed] [Google Scholar]
- 8.Montavon T, et al. Complete loss of H3K9 methylation dissolves mouse heterochromatin organization. Nat. Commun. 2021;12:4359. doi: 10.1038/s41467-021-24532-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allshire RC, Madhani HD. Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 2018;19:229–244. doi: 10.1038/nrm.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schotta G, et al. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elgin SC, Reuter G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb. Perspect. Biol. 2013;5:a017780. doi: 10.1101/cshperspect.a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rea S, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 14.Jenuwein T, Laible G, Dorn R, Reuter G. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell Mol. Life Sci. 1998;54:80–93. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell. 2006;24:309–316. doi: 10.1016/j.molcel.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 16.Peters AH, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/S0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 17.Peters AH, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell. 2003;12:1577–1589. doi: 10.1016/S1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 18.Martens JHA, et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulut-Karslioglu A, et al. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol. Cell. 2014;55:277–290. doi: 10.1016/j.molcel.2014.05.029. [DOI] [PubMed] [Google Scholar]
- 20.Wang T, et al. Crystal structure of the human SUV39H1 chromodomain and its recognition of histone H3K9me2/3. PLoS ONE. 2012;7:e52977. doi: 10.1371/journal.pone.0052977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melcher M, et al. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol. Cell Biol. 2000;20:3728–3741. doi: 10.1128/MCB.20.10.3728-3741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Carroll D, et al. Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol. Cell Biol. 2000;20:9423–9433. doi: 10.1128/MCB.20.24.9423-9433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iglesias N, et al. Automethylation-induced conformational switch in Clr4 (Suv39h) maintains epigenetic stability. Nature. 2018;560:504–508. doi: 10.1038/s41586-018-0398-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Canzio D, Larson A, Narlikar GJ. Mechanisms of functional promiscuity by HP1 proteins. Trends Cell Biol. 2014;24:377–386. doi: 10.1016/j.tcb.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 26.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 27.Padeken J, et al. Argonaute NRDE-3 and MBT domain protein LIN-61 redundantly recruit an H3K9me3 HMT to prevent embryonic lethality and transposon expression. Genes Dev. 2021;35:82–101. doi: 10.1101/gad.344234.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guang S, et al. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465:1097–1101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gushchanskaia ES, Esse R, Ma Q, Lau NC, Grishok A. Interplay between small RNA pathways shapes chromatin landscapes in C. elegans. Nucleic Acids Res. 2019;47:5603–5616. doi: 10.1093/nar/gkz275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lev I, Gingold H, Rechavi O. H3K9me3 is required for inheritance of small RNAs that target a unique subset of newly evolved genes. eLife. 2019;8:e40448. doi: 10.7554/eLife.40448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson WL, et al. RNA-dependent stabilization of SUV39H1 at constitutive heterochromatin. eLife. 2017;6:e25299. doi: 10.7554/eLife.25299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shirai A, et al. Impact of nucleic acid and methylated H3K9 binding activities of Suv39h1 on its heterochromatin assembly. eLife. 2017;6:e25317. doi: 10.7554/eLife.25317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Velazquez Camacho O, et al. Major satellite repeat RNA stabilize heterochromatin retention of Suv39h enzymes by RNA-nucleosome association and RNA:DNA hybrid formation. eLife. 2017;6:e25293. doi: 10.7554/eLife.25293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Probst AV, et al. A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev. Cell. 2010;19:625–638. doi: 10.1016/j.devcel.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 35.Casanova M, et al. Heterochromatin reorganization during early mouse development requires a single-stranded noncoding transcript. Cell Rep. 2013;4:1156–1167. doi: 10.1016/j.celrep.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 36.Burton A, Torres-Padilla ME. Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:723–734. doi: 10.1038/nrm3885. [DOI] [PubMed] [Google Scholar]
- 37.Scarola M, et al. Epigenetic silencing of Oct4 by a complex containing SUV39H1 and Oct4 pseudogene lncRNA. Nat. Commun. 2015;6:7631. doi: 10.1038/ncomms8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Porro A, et al. Functional characterization of the TERRA transcriptome at damaged telomeres. Nat. Commun. 2014;5:5379. doi: 10.1038/ncomms6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.García-Cao M, O’Sullivan R, Peters AH, Jenuwein T, Blasco MA. Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- 40.Gauchier M, et al. SETDB1-dependent heterochromatin stimulates alternative lengthening of telomeres. Sci. Adv. 2019;5:eaav3673. doi: 10.1126/sciadv.aav3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat. Cell Biol. 2007;9:25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murayama A, et al. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008;133:627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 43.Vaquero A, et al. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 44.Bosch-Presegué L, et al. Stabilization of Suv39H1 by SirT1 is part of oxidative stress response and ensures genome protection. Mol. Cell. 2011;42:210–223. doi: 10.1016/j.molcel.2011.02.034. [DOI] [PubMed] [Google Scholar]
- 45.Santos-Barriopedro I, et al. SIRT6-dependent cysteine monoubiquitination in the PRE-SET domain of Suv39h1 regulates the NF-κB pathway. Nat. Commun. 2018;9:101. doi: 10.1038/s41467-017-02586-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harte PJ, Wu W, Carrasquillo MM, Matera AG. Assignment of a novel bifurcated SET domain gene, SETDB1, to human chromosome band 1q21 by in situ hybridization and radiation hybrids. Cytogenet. Cell Genet. 1999;84:83–86. doi: 10.1159/000015220. [DOI] [PubMed] [Google Scholar]
- 47.Yang L, et al. Molecular cloning of ESET, a novel histone H3-specific methyltransferase that interacts with ERG transcription factor. Oncogene. 2002;21:148–152. doi: 10.1038/sj.onc.1204998. [DOI] [PubMed] [Google Scholar]
- 48.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun L, Fang J. E3-independent constitutive monoubiquitination complements histone methyltransferase activity of SETDB1. Mol. Cell. 2016;62:958–966. doi: 10.1016/j.molcel.2016.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishimoto K, et al. Ubiquitination of lysine 867 of the human SETDB1 protein upregulates its histone H3 lysine 9 (H3K9) methyltransferase activity. PLoS ONE. 2016;11:e0165766. doi: 10.1371/journal.pone.0165766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng Q, et al. Senp2 regulates adipose lipid storage by de-SUMOylation of Setdb1. J. Mol. Cell Biol. 2018;10:258–266. doi: 10.1093/jmcb/mjx055. [DOI] [PubMed] [Google Scholar]
- 52.Osumi K, Sato K, Murano K, Siomi H, Siomi MC. Essential roles of Windei and nuclear monoubiquitination of Eggless/SETDB1 in transposon silencing. EMBO Rep. 2019;20:e48296. doi: 10.15252/embr.201948296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ninova M, et al. Su(var)2-10 and the SUMO pathway link piRNA-guided target recognition to chromatin silencing. Mol. Cell. 2020;77:556–556–570.e6. doi: 10.1016/j.molcel.2019.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ivanov AV, et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol. Cell. 2007;28:823–837. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee YK, Thomas SN, Yang AJ, Ann DK. Doxorubicin down-regulates Kruppel-associated box domain-associated protein 1 sumoylation that relieves its transcription repression on p21WAF1/CIP1 in breast cancer MCF-7 cells. J. Biol. Chem. 2007;282:1595–1606. doi: 10.1074/jbc.M606306200. [DOI] [PubMed] [Google Scholar]
- 56.Timms RT, Tchasovnikarova IA, Antrobus R, Dougan G, Lehner PJ. ATF7IP-mediated stabilization of the histone methyltransferase SETDB1 is essential for heterochromatin formation by the HUSH complex. Cell Rep. 2016;17:653–659. doi: 10.1016/j.celrep.2016.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsusaka T, Shimura C, Shinkai Y. ATF7IP regulates SETDB1 nuclear localization and increases its ubiquitination. EMBO Rep. 2019;20:e48297. doi: 10.15252/embr.201948297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang H, et al. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol. Cell. 2003;12:475–487. doi: 10.1016/j.molcel.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 59.Mutlu B, et al. Regulated nuclear accumulation of a histone methyltransferase times the onset of heterochromatin formation in C. elegans embryos. Sci. Adv. 2018;4:eaat6224. doi: 10.1126/sciadv.aat6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Delaney CE, et al. Heterochromatic foci and transcriptional repression by an unstructured MET-2/SETDB1 co-factor LIN-65. J. Cell Biol. 2019;218:820–838. doi: 10.1083/jcb.201811038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Beyer S, et al. Canonical Wnt signalling regulates nuclear export of Setdb1 during skeletal muscle terminal differentiation. Cell Discov. 2016;2:16037. doi: 10.1038/celldisc.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cho S, Park JS, Kang YK. Regulated nuclear entry of over-expressed Setdb1. Genes Cells. 2013;18:694–703. doi: 10.1111/gtc.12068. [DOI] [PubMed] [Google Scholar]
- 63.Delaney CE, et al. SETDB1-like MET-2 promotes transcriptional silencing and development independently of its H3K9me-associated catalytic activity. Nat. Struct. Mol. Biol. 2022;29:85–96. doi: 10.1038/s41594-021-00712-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pek JW, Anand A, Kai T. Tudor domain proteins in development. Development. 2012;139:2255–2266. doi: 10.1242/dev.073304. [DOI] [PubMed] [Google Scholar]
- 65.Jurkowska RZ, et al. H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1. Nat. Commun. 2017;8:2057. doi: 10.1038/s41467-017-02259-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dodge JE, Kang YK, Beppu H, Lei H, Li E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol. Cell Biol. 2004;24:2478–2486. doi: 10.1128/MCB.24.6.2478-2486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karimi MM, et al. DNA Methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell. 2011;8:676–687. doi: 10.1016/j.stem.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matsui T, et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 69.Collins PL, Kyle KE, Egawa T, Shinkai Y, Oltz EM. The histone methyltransferase SETDB1 represses endogenous and exogenous retroviruses in B lymphocytes. Proc. Natl Acad. Sci. USA. 2015;112:8367–8372. doi: 10.1073/pnas.1422187112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Adoue V, et al. The histone methyltransferase SETDB1 controls T helper cell lineage integrity by repressing endogenous retroviruses. Immunity. 2019;50:629–644.e8. doi: 10.1016/j.immuni.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 71.Kato M, Takemoto K, Shinkai Y. A somatic role for the histone methyltransferase Setdb1 in endogenous retrovirus silencing. Nat. Commun. 2018;9:1683. doi: 10.1038/s41467-018-04132-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takikita S, et al. A histone methyltransferase ESET is critical for T cell development. J. Immunol. 2016;197:2269–2279. doi: 10.4049/jimmunol.1502486. [DOI] [PubMed] [Google Scholar]
- 73.Fasching L, et al. TRIM28 represses transcription of endogenous retroviruses in neural progenitor cells. Cell Rep. 2015;10:20–28. doi: 10.1016/j.celrep.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Južnić L, et al. SETDB1 is required for intestinal epithelial differentiation and the prevention of intestinal inflammation. Gut. 2021;70:485–498. doi: 10.1136/gutjnl-2020-321339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang R, et al. Gut stem cell necroptosis by genome instability triggers bowel inflammation. Nature. 2020;580:386–390. doi: 10.1038/s41586-020-2127-x. [DOI] [PubMed] [Google Scholar]
- 76.Stocking C, Kozak CA. Endogenous retroviruses. Cell. Mol. Life Sci. 2008;65:3383–3398. doi: 10.1007/s00018-008-8497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huntley S, et al. A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 2006;16:669–677. doi: 10.1101/gr.4842106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Helleboid P-Y, et al. The interactome of KRAB zinc finger proteins reveals the evolutionary history of their functional diversification. EMBO J. 2019;38:e101220. doi: 10.15252/embj.2018101220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cruz-Tapias P, Robin P, Pontis J, Maestro LD, Ait-Si-Ali S. The H3K9 methylation writer SETDB1 and its Reader MPP8 cooperate to silence satellite DNA repeats in mouse embryonic stem cells. Genes. 2019;10:750. doi: 10.3390/genes10100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Loyola A, et al. The HP1alpha-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 2009;10:769–775. doi: 10.1038/embor.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Padeken J, et al. Synergistic lethality between BRCA1 and H3K9me2 loss reflects satellite derepression. Genes Dev. 2019;33:436–451. doi: 10.1101/gad.322495.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McMurchy AN, et al. A team of heterochromatin factors collaborates with small RNA pathways to combat repetitive elements and germline stress. eLife. 2017;6:e21666. doi: 10.7554/eLife.21666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Methot SP, et al. H3K9me selectively blocks transcription factor activity and ensures differentiated tissue integrity. Nat. Cell Biol. 2021;23:1163–1175. doi: 10.1038/s41556-021-00776-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robbez-Masson L, et al. The HUSH complex cooperates with TRIM28 to repress young retrotransposons and new genes. Genome Res. 2018;28:836–845. doi: 10.1101/gr.228171.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tchasovnikarova IA, et al. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science. 2015;348:1481–1485. doi: 10.1126/science.aaa7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tsusaka T, et al. Tri-methylation of ATF7IP by G9a/GLP recruits the chromodomain protein MPP8. Epigenetics Chromatin. 2018;11:56. doi: 10.1186/s13072-018-0231-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tchasovnikarova IA, et al. Hyperactivation of HUSH complex function by Charcot-Marie-Tooth disease mutation in MORC2. Nat. Genet. 2017;49:1035–1044. doi: 10.1038/ng.3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Douse CH, et al. Neuropathic MORC2 mutations perturb GHKL ATPase dimerization dynamics and epigenetic silencing by multiple structural mechanisms. Nat. Commun. 2018;9:651. doi: 10.1038/s41467-018-03045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Douse CH, et al. TASOR is a pseudo-PARP that directs HUSH complex assembly and epigenetic transposon control. Nat. Commun. 2020;11:4940. doi: 10.1038/s41467-020-18761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kokura K, Sun L, Bedford MT, Fang J. Methyl-H3K9-binding protein MPP8 mediates E-cadherin gene silencing and promotes tumour cell motility and invasion. EMBO J. 2010;29:3673–3687. doi: 10.1038/emboj.2010.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Timms RT, Tchasovnikarova IA, Lehner PJ. Position-effect variegation revisited: HUSHing up heterochromatin in human cells. Bioessays. 2016;38:333–343. doi: 10.1002/bies.201500184. [DOI] [PubMed] [Google Scholar]
- 92.Seczynska M, Bloor S, Cuesta SM, Lehner PJ. Genome surveillance by HUSH-mediated silencing of intronless mobile elements. Nature. 2022;601:440–445. doi: 10.1038/s41586-021-04228-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kourmouli N, Sun Y-M, van der Sar S, Singh PB, Brown JP. Epigenetic regulation of mammalian pericentric heterochromatin in vivo by HP1. Biochem. Biophys. Res. Commun. 2005;337:901–907. doi: 10.1016/j.bbrc.2005.09.132. [DOI] [PubMed] [Google Scholar]
- 94.Falandry C, et al. CLLD8/KMT1F is a lysine methyltransferase that is important for chromosome segregation. J. Biol. Chem. 2010;285:20234–20241. doi: 10.1074/jbc.M109.052399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu PF, et al. Setdb2 restricts dorsal organizer territory and regulates left-right asymmetry through suppressing fgf8 activity. Proc. Natl Acad. Sci. USA. 2010;107:2521–2526. doi: 10.1073/pnas.0914396107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schliehe C, et al. The methyltransferase Setdb2 mediates virus-induced susceptibility to bacterial superinfection. Nat. Immunol. 2015;16:67–74. doi: 10.1038/ni.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Melvin WJ, et al. Coronavirus induces diabetic macrophage-mediated inflammation via SETDB2. Proc. Natl Acad. Sci. USA. 2021;118:e2101071118. doi: 10.1073/pnas.2101071118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kimball AS, et al. The histone methyltransferase Setdb2 modulates macrophage phenotype and uric acid production in diabetic wound repair. Immunity. 2019;51:258–271.e5. doi: 10.1016/j.immuni.2019.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tachibana M, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- 101.Battisti V, et al. Unexpected distinct roles of the related histone H3 lysine 9 methyltransferases G9a and G9a-like protein in myoblasts. J. Mol. Biol. 2016;428:2329–2343. doi: 10.1016/j.jmb.2016.03.029. [DOI] [PubMed] [Google Scholar]
- 102.Collins RE, et al. In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J. Biol. Chem. 2005;280:5563–5570. doi: 10.1074/jbc.M410483200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wu H, et al. Structural biology of human H3K9 methyltransferases. PLoS ONE. 2010;5:e8570. doi: 10.1371/journal.pone.0008570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Whetstine JR, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 105.Shen H, Xu W, Lan F. Histone lysine demethylases in mammalian embryonic development. Exp. Mol. Med. 2017;49:e325. doi: 10.1038/emm.2017.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 107.Jiang Q, et al. G9a plays distinct roles in maintaining DNA methylation, retrotransposon silencing, and chromatin looping. Cell Rep. 2020;33:108315. doi: 10.1016/j.celrep.2020.108315. [DOI] [PubMed] [Google Scholar]
- 108.Maksakova IA, et al. Distinct roles of KAP1, HP1 and G9a/GLP in silencing of the two-cell-specific retrotransposon MERVL in mouse ES cells. Epigenetics Chromatin. 2013;6:15. doi: 10.1186/1756-8935-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Leung DC, et al. Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. Proc. Natl Acad. Sci. USA. 2011;108:5718–5723. doi: 10.1073/pnas.1014660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dong KB, et al. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J. 2008;27:2691–2701. doi: 10.1038/emboj.2008.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nicetto D, et al. H3K9me3-heterochromatin loss at protein-coding genes enables developmental lineage specification. Science. 2019;363:294–297. doi: 10.1126/science.aau0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Di Giacomo M, Comazzetto S, Sampath SC, Sampath SC, O’Carroll D. G9a co-suppresses LINE1 elements in spermatogonia. Epigenetics Chromatin. 2014;7:24. doi: 10.1186/1756-8935-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu S, et al. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev. 2014;28:2041–2055. doi: 10.1101/gad.244848.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhou H, et al. H3K9 Demethylation-Induced R-loop accumulation is linked to disorganized nucleoli. Front. Genet. 2020;11:43. doi: 10.3389/fgene.2020.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Collins RE, et al. The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat. Struct. Mol. Biol. 2008;15:245–250. doi: 10.1038/nsmb.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sampath SC, et al. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol. Cell. 2007;27:596–608. doi: 10.1016/j.molcel.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 117.Shin HM, et al. Epigenetic modifications induced by blimp-1 regulate CD8+ T cell memory progression during acute virus infection. Immunity. 2013;39:661–675. doi: 10.1016/j.immuni.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shi Y, et al. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- 119.Tong X, et al. Transcriptional repressor E4-binding protein 4 (E4BP4) regulates metabolic hormone fibroblast growth factor 21 (FGF21) during circadian cycles and feeding. J. Biol. Chem. 2010;285:36401–36409. doi: 10.1074/jbc.M110.172866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tong X, et al. Recruitment of histone methyltransferase G9a mediates transcriptional repression of Fgf21 gene by E4BP4 protein. J. Biol. Chem. 2013;288:5417–5425. doi: 10.1074/jbc.M112.433482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fang S, et al. Coordinated recruitment of histone methyltransferase G9a and other chromatin-modifying enzymes in SHP-mediated regulation of hepatic bile acid metabolism. Mol. Cell. Biol. 2007;27:1407–1424. doi: 10.1128/MCB.00944-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kemper JK, Kim H, Miao J, Bhalla S, Bae Y. Role of an mSin3A-Swi/Snf chromatin remodeling complex in the feedback repression of bile acid biosynthesis by SHP. Mol. Cell Biol. 2004;24:7707–7719. doi: 10.1128/MCB.24.17.7707-7719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ueda J, Tachibana M, Ikura T, Shinkai Y. Zinc finger protein Wiz links G9a/GLP histone methyltransferases to the co-repressor molecule CtBP. J. Biol. Chem. 2006;281:20120–20128. doi: 10.1074/jbc.M603087200. [DOI] [PubMed] [Google Scholar]
- 124.Subramanian T, Chinnadurai G. Association of class I histone deacetylases with transcriptional corepressor CtBP. FEBS Lett. 2003;540:255–258. doi: 10.1016/S0014-5793(03)00275-8. [DOI] [PubMed] [Google Scholar]
- 125.Nicetto D, Zaret KS. Role of H3K9me3 heterochromatin in cell identity establishment and maintenance. Curr. Opin. Genet. Dev. 2019;55:1–10. doi: 10.1016/j.gde.2019.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chen J, et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat. Genet. 2013;45:34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- 128.Matoba S, et al. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell. 2014;159:884–895. doi: 10.1016/j.cell.2014.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu X, et al. H3K9 demethylase KDM4E is an epigenetic regulator for bovine embryonic development and a defective factor for nuclear reprogramming. Development. 2018;145:dev158261. doi: 10.1242/dev.158261. [DOI] [PubMed] [Google Scholar]
- 130.Brumbaugh J, et al. Inducible histone K-to-M mutations are dynamic tools to probe the physiological role of site-specific histone methylation in vitro and in vivo. Nat. Cell Biol. 2019;21:1449–1461. doi: 10.1038/s41556-019-0403-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Griffin GK, et al. Epigenetic silencing by SETDB1 suppresses tumour intrinsic immunogenicity. Nature. 2021;595:309–314. doi: 10.1038/s41586-021-03520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Checchi PM, Engebrecht J. Caenorhabditis elegans histone methyltransferase MET-2 shields the male X chromosome from checkpoint machinery and mediates meiotic sex chromosome inactivation. PLoS Genet. 2011;7:e1002267. doi: 10.1371/journal.pgen.1002267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lev I, et al. MET-2-dependent H3K9 methylation suppresses transgenerational small RNA inheritance. Curr. Biol. 2017;27:1138–1147. doi: 10.1016/j.cub.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 134.Zylicz JJ, et al. G9a regulates temporal preimplantation developmental program and lineage segregation in blastocyst. eLife. 2018;7:e33361. doi: 10.7554/eLife.33361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Au Yeung WK, et al. Histone H3K9 methyltransferase G9a in oocytes is essential for preimplantation development but dispensable for CG methylation protection. Cell Rep. 2019;27:282–293.e4. doi: 10.1016/j.celrep.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 136.Chang C-R, Wu C-S, Hom Y, Gartenberg MR. Targeting of cohesin by transcriptionally silent chromatin. Genes Dev. 2005;19:3031–3042. doi: 10.1101/gad.1356305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang X, et al. G9a-mediated methylation of ERα links the PHF20/MOF histone acetyltransferase complex to hormonal gene expression. Nat. Commun. 2016;7:10810. doi: 10.1038/ncomms10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ferry L, et al. Methylation of DNA ligase 1 by G9a/GLP recruits UHRF1 to replicating DNA and regulates DNA methylation. Mol. Cell. 2017;67:550–565.e5. doi: 10.1016/j.molcel.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 139.Van Duyne R, et al. Lysine methylation of HIV-1 Tat regulates transcriptional activity of the viral LTR. Retrovirology. 2008;5:40. doi: 10.1186/1742-4690-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kudithipudi S, Schuhmacher MK, Kebede AF, Jeltsch A. The SUV39H1 protein lysine methyltransferase methylates chromatin proteins involved in heterochromatin formation and VDJ recombination. ACS Chem. Biol. 2017;12:958–968. doi: 10.1021/acschembio.6b01076. [DOI] [PubMed] [Google Scholar]
- 141.Thul PJ, et al. A subcellular map of the human proteome. Science. 2017;356:eaal3321. doi: 10.1126/science.aal3321. [DOI] [PubMed] [Google Scholar]
- 142.Uhlén M, et al. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 143.Smith CM, et al. The mouse gene expression database (GXD): 2019 update. Nucleic Acids Res. 2018;47:D774–D779. doi: 10.1093/nar/gky922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Burton A, et al. Heterochromatin establishment during early mammalian development is regulated by pericentromeric RNA and characterized by non-repressive H3K9me3. Nat. Cell Biol. 2020;22:767–778. doi: 10.1038/s41556-020-0536-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Balmer P, et al. SUV39H2 epigenetic silencing controls fate conversion of epidermal stem and progenitor cells. J. Cell Biol. 2021;220:e201908178. doi: 10.1083/jcb.201908178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pucella JN, Upadhaya S, Reizis B. The source and dynamics of adult hematopoiesis: insights from lineage tracing. Annu. Rev. Cell Dev. Biol. 2020;36:529–550. doi: 10.1146/annurev-cellbio-020520-114601. [DOI] [PubMed] [Google Scholar]
- 147.Hosokawa H, Rothenberg EV. How transcription factors drive choice of the T cell fate. Nat. Rev. Immunol. 2021;21:162–176. doi: 10.1038/s41577-020-00426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nagasawa T. Microenvironmental niches in the bone marrow required for B-cell development. Nat. Rev. Immunol. 2006;6:107–116. doi: 10.1038/nri1780. [DOI] [PubMed] [Google Scholar]
- 149.Pasquarella A, Nuber A, Schotta G. Deletion of the histone methyltransferase Setdb1 during hematopoiesis results in hematopoietic stem cell failure and abrogates B cell development. Exp. Hematol. 2013;41:S19. doi: 10.1016/j.exphem.2013.05.072. [DOI] [Google Scholar]
- 150.Koide S, et al. Setdb1 maintains hematopoietic stem and progenitor cells by restricting the ectopic activation of nonhematopoietic genes. Blood. 2016;128:638–649. doi: 10.1182/blood-2016-01-694810. [DOI] [PubMed] [Google Scholar]
- 151.Keenan CR, et al. Extreme disruption of heterochromatin is required for accelerated hematopoietic aging. Blood. 2020;135:2049–2058. doi: 10.1182/blood.2019002990. [DOI] [PubMed] [Google Scholar]
- 152.Ermolaeva M, Neri F, Ori A, Rudolph KL. Cellular and epigenetic drivers of stem cell ageing. Nat. Rev. Mol. Cell Biol. 2018;19:594–610. doi: 10.1038/s41580-018-0020-3. [DOI] [PubMed] [Google Scholar]
- 153.Djeghloul D, et al. Age-associated decrease of the histone methyltransferase SUV39H1 in HSC perturbs heterochromatin and B lymphoid differentiation. Stem Cell Rep. 2016;6:970–984. doi: 10.1016/j.stemcr.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen X, et al. G9a/GLP-dependent histone H3K9me2 patterning during human hematopoietic stem cell lineage commitment. Genes Dev. 2012;26:2499–2511. doi: 10.1101/gad.200329.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Luckheeram RV, Zhou R, Verma AD, Xia B. CD4+ T cells: differentiation and functions. Clin. Dev. Immunol. 2012;2012:925135. doi: 10.1155/2012/925135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Allan RS, et al. An epigenetic silencing pathway controlling T helper 2 cell lineage commitment. Nature. 2012;487:249–253. doi: 10.1038/nature11173. [DOI] [PubMed] [Google Scholar]
- 157.Pace L, et al. The epigenetic control of stemness in CD8+ T cell fate commitment. Science. 2018;359:177–186. doi: 10.1126/science.aah6499. [DOI] [PubMed] [Google Scholar]
- 158.Lehnertz B, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–1200. doi: 10.1016/S0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 159.Thomas LR, et al. Functional analysis of histone methyltransferase g9a in B and T lymphocytes. J. Immunol. 2008;181:485–493. doi: 10.4049/jimmunol.181.1.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Xiao X, et al. The costimulatory receptor OX40 inhibits interleukin-17 expression through activation of repressive chromatin remodeling pathways. Immunity. 2016;44:1271–1283. doi: 10.1016/j.immuni.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Maddur MS, Miossec P, Kaveri SV, Bayry J. Th17 cells: biology, pathogenesis of autoimmune and inflammatory diseases, and therapeutic strategies. Am. J. Pathol. 2012;181:8–18. doi: 10.1016/j.ajpath.2012.03.044. [DOI] [PubMed] [Google Scholar]
- 162.Sugamura K, Ishii N, Weinberg AD. Therapeutic targeting of the effector T-cell co-stimulatory molecule OX40. Nat. Rev. Immunol. 2004;4:420–431. doi: 10.1038/nri1371. [DOI] [PubMed] [Google Scholar]
- 163.Osipovich O, et al. Targeted inhibition of V(D)J recombination by a histone methyltransferase. Nat. Immunol. 2004;5:309–316. doi: 10.1038/ni1042. [DOI] [PubMed] [Google Scholar]
- 164.Bradley SP, Kaminski DA, Peters AH, Jenuwein T, Stavnezer J. The histone methyltransferase Suv39h1 increases class switch recombination specifically to IgA. J. Immunol. 2006;177:1179–1188. doi: 10.4049/jimmunol.177.2.1179. [DOI] [PubMed] [Google Scholar]
- 165.George-Alexander L-E, Kania A, Mi T, Scharer CD, Boss JM. H3K9 dimethyltransferase G9a deficiency modulates B-cell response to LPS. J. Immunol. 2021;206:63.08–63.08. [Google Scholar]
- 166.Li S, et al. JMJD1B demethylates H4R3me2s and H3K9me2 to facilitate gene expression for development of hematopoietic stem and progenitor cells. Cell Rep. 2018;23:389–403. doi: 10.1016/j.celrep.2018.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kim JY, et al. KDM3B is the H3K9 demethylase involved in transcriptional activation of lmo2 in leukemia. Mol. Cell Biol. 2012;32:2917–2933. doi: 10.1128/MCB.00133-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hachiya R, et al. The H3K9 methyltransferase Setdb1 regulates TLR4-mediated inflammatory responses in macrophages. Sci. Rep. 2016;6:28845. doi: 10.1038/srep28845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sasaki Y, Iwai K. Roles of the NF-κB pathway in B-lymphocyte biology. Curr. Top. Microbiol. Immunol. 2016;393:177–209. doi: 10.1007/82_2015_479. [DOI] [PubMed] [Google Scholar]
- 170.Buckingham M. Skeletal muscle formation in vertebrates. Curr. Opin. Genet. Dev. 2001;11:440–448. doi: 10.1016/S0959-437X(00)00215-X. [DOI] [PubMed] [Google Scholar]
- 171.Chen L, Krause M, Sepanski M, Fire A. The Caenorhabditis elegans MYOD homologue HLH-1 is essential for proper muscle function and complete morphogenesis. Development. 1994;120:1631–1641. doi: 10.1242/dev.120.6.1631. [DOI] [PubMed] [Google Scholar]
- 172.Walsh K, Perlman H. Cell cycle exit upon myogenic differentiation. Curr. Opin. Genet. Dev. 1997;7:597–602. doi: 10.1016/S0959-437X(97)80005-6. [DOI] [PubMed] [Google Scholar]
- 173.Ait-Si-Ali S, et al. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 2004;23:605–615. doi: 10.1038/sj.emboj.7600074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Matthews HK, Bertoli C, de Bruin RAM. Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 2021;23:74–88. doi: 10.1038/s41580-021-00404-3. [DOI] [PubMed] [Google Scholar]
- 175.Luo RX, Postigo AA, Dean DC. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/S0092-8674(00)80940-X. [DOI] [PubMed] [Google Scholar]
- 176.Brehm A, et al. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 177.Vandel L, et al. Transcriptional repression by the retinoblastoma protein through the recruitment of a histone methyltransferase. Mol. Cell Biol. 2001;21:6484–6494. doi: 10.1128/MCB.21.19.6484-6494.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Vaute O, Nicolas E, Vandel L, Trouche D. Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases. Nucleic Acids Res. 2002;30:475–481. doi: 10.1093/nar/30.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nielsen SJ, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- 180.Guasconi V, et al. Preferential association of irreversibly silenced E2F-target genes with pericentromeric heterochromatin in differentiated muscle cells. Epigenetics. 2010;5:704–709. doi: 10.4161/epi.5.8.13025. [DOI] [PubMed] [Google Scholar]
- 181.Harr JC, et al. Loss of an H3K9me anchor rescues laminopathy-linked changes in nuclear organization and muscle function in an Emery-Dreifuss muscular dystrophy model. Genes Dev. 2020;34:560–579. doi: 10.1101/gad.332213.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Rao VK, et al. G9a promotes proliferation and inhibits cell cycle exit during myogenic differentiation. Nucleic Acids Res. 2016;44:8129–8143. doi: 10.1093/nar/gkw483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zhang R-H, Judson RN, Liu DY, Kast J, Rossi FMV. The lysine methyltransferase Ehmt2/G9a is dispensable for skeletal muscle development and regeneration. Skelet. Muscle. 2016;6:22. doi: 10.1186/s13395-016-0093-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yang G, et al. The histone H3K9 methyltransferase SUV39H links SIRT1 repression to myocardial infarction. Nat. Commun. 2017;8:14941. doi: 10.1038/ncomms14941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Riahi H, et al. The histone methyltransferase G9a regulates tolerance to oxidative stress–induced energy consumption. PLoS Biol. 2019;17:e2006146. doi: 10.1371/journal.pbio.2006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Klosin A, Casas E, Hidalgo-Carcedo C, Vavouri T, Lehner B. Transgenerational transmission of environmental information in C. elegans. Science. 2017;356:320–323. doi: 10.1126/science.aah6412. [DOI] [PubMed] [Google Scholar]
- 187.Tan SL, et al. Essential roles of the histone methyltransferase ESET in the epigenetic control of neural progenitor cells during development. Development. 2012;139:3806–3816. doi: 10.1242/dev.082198. [DOI] [PubMed] [Google Scholar]
- 188.Zheng C, Karimzadegan S, Chiang V, Chalfie M. Histone methylation restrains the expression of subtype-specific genes during terminal neuronal differentiation in Caenorhabditis elegans. PLoS Genet. 2013;9:e1004017. doi: 10.1371/journal.pgen.1004017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Fiszbein A, et al. Alternative splicing of G9a regulates neuronal differentiation. Cell Rep. 2016;14:2797–2808. doi: 10.1016/j.celrep.2016.02.063. [DOI] [PubMed] [Google Scholar]
- 190.Schaefer A, et al. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron. 2009;64:678–691. doi: 10.1016/j.neuron.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/S0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 192.Magklara A, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145:555–570. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lyons DB, et al. Heterochromatin-mediated gene silencing facilitates the diversification of olfactory neurons. Cell Rep. 2014;9:884–892. doi: 10.1016/j.celrep.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Balan S, et al. A loss-of-function variant in SUV39H2 identified in autism-spectrum disorder causes altered H3K9 trimethylation and dysregulation of protocadherin β-cluster genes in the developing brain. Mol. Psychiatry. 2021;26:7550–7559. doi: 10.1038/s41380-021-01199-7. [DOI] [PubMed] [Google Scholar]
- 195.Bharadwaj R, et al. Conserved higher-order chromatin regulates NMDA receptor gene expression and cognition. Neuron. 2014;84:997–1008. doi: 10.1016/j.neuron.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Jiang Y, et al. Setdb1 histone methyltransferase regulates mood-related behaviors and expression of the NMDA receptor subunit NR2B. J. Neurosci. 2010;30:7152–7167. doi: 10.1523/JNEUROSCI.1314-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ryu H, et al. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington’s disease. Proc. Natl Acad. Sci. USA. 2006;103:19176–19181. doi: 10.1073/pnas.0606373103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Irmak D, et al. Mechanism suppressing H3K9 trimethylation in pluripotent stem cells and its demise by polyQ-expanded huntingtin mutations. Hum. Mol. Genet. 2018;27:4117–4134. doi: 10.1093/hmg/ddy304. [DOI] [PubMed] [Google Scholar]
- 199.Subbanna S, et al. G9a-mediated histone methylation regulates ethanol-induced neurodegeneration in the neonatal mouse brain. Neurobiol. Dis. 2013;54:475–485. doi: 10.1016/j.nbd.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wilson C, et al. The histone methyltransferase G9a controls axon growth by targeting the RhoA signaling pathway. Cell Rep. 2020;31:107639. doi: 10.1016/j.celrep.2020.107639. [DOI] [PubMed] [Google Scholar]
- 201.Laumet G, et al. G9a is essential for epigenetic silencing of K+ channel genes in acute-to-chronic pain transition. Nat. Neurosci. 2015;18:1746–1755. doi: 10.1038/nn.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Liang L, et al. G9a participates in nerve injury-induced Kcna2 downregulation in primary sensory neurons. Sci. Rep. 2016;6:37704. doi: 10.1038/srep37704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Maze I, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Maze I, et al. G9a influences neuronal subtype specification in striatum. Nat. Neurosci. 2014;17:533–539. doi: 10.1038/nn.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kleefstra T, et al. Further clinical and molecular delineation of the 9q subtelomeric deletion syndrome supports a major contribution of EHMT1 haploinsufficiency to the core phenotype. J. Med. Genet. 2009;46:598–606. doi: 10.1136/jmg.2008.062950. [DOI] [PubMed] [Google Scholar]
- 206.McCarthy RL, et al. Diverse heterochromatin-associated proteins repress distinct classes of genes and repetitive elements. Nat. Cell Biol. 2021;23:905–914. doi: 10.1038/s41556-021-00725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Pickersgill H, et al. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- 208.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Gonzalez-Sandoval A, et al. Perinuclear anchoring of H3K9-methylated chromatin stabilizes induced cell fate C. elegans embryos. Cell. 2015;163:1333–1347. doi: 10.1016/j.cell.2015.10.066. [DOI] [PubMed] [Google Scholar]
- 210.Sawh AN, et al. Lamina-dependent stretching and unconventional chromosome compartments in early C. elegans embryos. Mol. Cell. 2020;78:96–111.e6. doi: 10.1016/j.molcel.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Snyder MJ, et al. Anchoring of heterochromatin to the nuclear lamina reinforces dosage compensation-mediated gene repression. PLoS Genet. 2016;12:e1006341. doi: 10.1371/journal.pgen.1006341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Cabianca DS, et al. Active chromatin marks drive spatial sequestration of heterochromatin in C. elegans nuclei. Nature. 2019;569:734–739. doi: 10.1038/s41586-019-1243-y. [DOI] [PubMed] [Google Scholar]
- 213.Hirano Y, et al. Lamin B receptor recognizes specific modifications of histone H4 in heterochromatin formation. J. Biol. Chem. 2012;287:42654–42663. doi: 10.1074/jbc.M112.397950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Solovei I, et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell. 2013;152:584–598. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 215.Hoskins VE, Smith K, Reddy KL. The shifting shape of genomes: dynamics of heterochromatin interactions at the nuclear lamina. Curr. Opin. Genet. Dev. 2021;67:163–173. doi: 10.1016/j.gde.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Manzo SG, Dauban L, van Steensel B. Lamina-associated domains: tethers and looseners. Curr. Opin. Cell Biol. 2022;74:80–87. doi: 10.1016/j.ceb.2022.01.004. [DOI] [PubMed] [Google Scholar]
- 217.Pasquarella A, et al. Retrotransposon derepression leads to activation of the unfolded protein response and apoptosis in pro-B cells. Development. 2016;143:1788–1799. doi: 10.1242/dev.130203. [DOI] [PubMed] [Google Scholar]
- 218.Fisher AG. Cellular identity and lineage choice. Nat. Rev. Immunol. 2002;2:977–982. doi: 10.1038/nri958. [DOI] [PubMed] [Google Scholar]
- 219.Rechtsteiner A, et al. Repression of germline genes in Caenorhabditis elegans somatic tissues by H3K9 dimethylation of their promoters. Genetics. 2019;212:125–140. doi: 10.1534/genetics.118.301878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Carpenter BS, et al. Caenorhabditis elegans establishes germline versus soma by balancing inherited histone methylation. Development. 2021;148:dev196600. doi: 10.1242/dev.196600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-Z. [DOI] [PubMed] [Google Scholar]
- 222.Huang N, et al. Accessible region conformation capture (ARC-C) gives high-resolution insights into genome architecture and regulation. Genome Res. 2022;32:357–366. doi: 10.1101/gr.275669.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Rao SSP, et al. Cohesin loss eliminates all loop domains. Cell. 2017;171:305–320.e24. doi: 10.1016/j.cell.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-G. [DOI] [PubMed] [Google Scholar]
- 227.Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/S0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 228.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 229.Kind J, et al. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 230.Wu R, Terry AV, Singh PB, Gilbert DM. Differential subnuclear localization and replication timing of histone H3 lysine 9 methylation states. Mol. Biol. Cell. 2005;16:2872–2881. doi: 10.1091/mbc.e04-11-0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.See K, et al. Histone methyltransferase activity programs nuclear peripheral genome positioning. Dev. Biol. 2020;466:90–98. doi: 10.1016/j.ydbio.2020.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Kind J, et al. Genome-wide maps of nuclear lamina interactions in single human cells. Cell. 2015;163:134–147. doi: 10.1016/j.cell.2015.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Fukuda K, et al. Regulation of mammalian 3D genome organization and histone H3K9 dimethylation by H3K9 methyltransferases. Commun. Biol. 2021;4:571. doi: 10.1038/s42003-021-02089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Yan Z, et al. G9a/GLP-sensitivity of H3K9me2 demarcates two types of genomic compartments. Genomics Proteomics Bioinformatics. 2020;18:359–370. doi: 10.1016/j.gpb.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Mattout A, et al. Heterochromatin protein 1β (HP1β) has distinct functions and distinct nuclear distribution in pluripotent versus differentiated cells. Genome Biol. 2015;16:213. doi: 10.1186/s13059-015-0760-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Jiang Y, et al. The methyltransferase SETDB1 regulates a large neuron-specific topological chromatin domain. Nat. Genet. 2017;49:1239–1250. doi: 10.1038/ng.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Justice M, Carico ZM, Stefan HC, Dowen JM. A WIZ/cohesin/CTCF complex anchors DNA loops to define gene expression and cell identity. Cell Rep. 2020;31:107503. doi: 10.1016/j.celrep.2020.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Bian Q, Anderson EC, Yang Q, Meyer BJ. Histone H3K9 methylation promotes formation of genome compartments in Caenorhabditis elegans via chromosome compaction and perinuclear anchoring. Proc. Natl Acad. Sci. USA. 2020;117:11459–11470. doi: 10.1073/pnas.2002068117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Escamilla-Del-Arenal M, et al. Cdyl, a new partner of the inactive X chromosome and potential reader of H3K27me3 and H3K9me2. Mol. Cell Biol. 2013;33:5005–5020. doi: 10.1128/MCB.00866-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Yokochi T, et al. G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proc. Natl Acad. Sci. USA. 2009;106:19363–19368. doi: 10.1073/pnas.0906142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Poleshko A, et al. The human protein PRR14 tethers heterochromatin to the nuclear lamina during interphase and mitotic exit. Cell Rep. 2013;5:292–301. doi: 10.1016/j.celrep.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Dunlevy KL, et al. The PRR14 heterochromatin tether encodes modular domains that mediate and regulate nuclear lamina targeting. J. Cell Sci. 2020;133:jcs240416. doi: 10.1242/jcs.240416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Poleshko A, et al. H3K9me2 orchestrates inheritance of spatial positioning of peripheral heterochromatin through mitosis. eLife. 2019;8:e49278. doi: 10.7554/eLife.49278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Zheng X, et al. Lamins organize the global three-dimensional genome from the nuclear periphery. Mol. Cell. 2018;71:802–815.e7. doi: 10.1016/j.molcel.2018.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Ulianov SV, et al. Nuclear lamina integrity is required for proper spatial organization of chromatin in Drosophila. Nat. Commun. 2019;10:1176. doi: 10.1038/s41467-019-09185-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Towbin BD, Meister P, Pike BL, Gasser SM. Repetitive transgenes in C. elegans accumulate heterochromatic marks and are sequestered at the nuclear envelope in a copy-number- and lamin-dependent manner. Cold Spring Harb. Symp. Quant. Biol. 2010;75:555–565. doi: 10.1101/sqb.2010.75.041. [DOI] [PubMed] [Google Scholar]
- 247.Rzepecki R, Gruenbaum Y. Invertebrate models of lamin diseases. Nucleus. 2018;9:227–234. doi: 10.1080/19491034.2018.1454166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Salvarani N, et al. The K219T-lamin mutation induces conduction defects through epigenetic inhibition of SCN5A in human cardiac laminopathy. Nat. Commun. 2019;10:2267. doi: 10.1038/s41467-019-09929-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Briand N, Collas P. Laminopathy-causing lamin A mutations reconfigure lamina-associated domains and local spatial chromatin conformation. Nucleus. 2018;9:216–226. doi: 10.1080/19491034.2018.1449498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J. Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Finlan LE, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 253.Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat. Rev. Genet. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- 254.Padeken J, Heun P. Nucleolus and nuclear periphery: velcro for heterochromatin. Curr. Opin. Cell Biol. 2014;28:54–60. doi: 10.1016/j.ceb.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 255.Pinheiro I, et al. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell. 2012;150:948–960. doi: 10.1016/j.cell.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 256.Schübeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 257.Zhang T, et al. G9a/GLP complex maintains imprinted DNA methylation in embryonic stem cells. Cell Rep. 2016;15:77–85. doi: 10.1016/j.celrep.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Xin Z, et al. Role of histone methyltransferase G9a in CpG methylation of the Prader-Willi syndrome imprinting center. J. Biol. Chem. 2003;278:14996–15000. doi: 10.1074/jbc.M211753200. [DOI] [PubMed] [Google Scholar]
- 259.Rowe HM, et al. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development. 2013;140:519–529. doi: 10.1242/dev.087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Saksouk N, et al. Redundant mechanisms to form silent chromatin at pericentromeric regions rely on BEND3 and DNA methylation. Mol. Cell. 2014;56:580–594. doi: 10.1016/j.molcel.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 261.Wallrath LL, Elgin SC. Position effect variegation in Drosophilais associated with an altered chromatin structure. Genes Dev. 1995;9:1263–1277. doi: 10.1101/gad.9.10.1263. [DOI] [PubMed] [Google Scholar]
- 262.Costello ME, Petrella LN. C. elegans synMuv B proteins regulate spatial and temporal chromatin compaction during development. Development. 2019;146:dev174383. doi: 10.1242/dev.174383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Patel T, Hobert O. Coordinated control of terminal differentiation and restriction of cellular plasticity. eLife. 2017;6:e24100. doi: 10.7554/eLife.24100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Burton A, Torres-Padilla M-E. Deconfining heterochromatin for expression. Nat. Cell Biol. 2021;23:814–816. doi: 10.1038/s41556-021-00726-6. [DOI] [PubMed] [Google Scholar]
- 265.Grow EJ, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]