Abstract
Sites of Listeria monocytogenes contamination in a cold-smoked rainbow trout (Oncorhynchus mykiss) processing plant were detected by sampling the production line, environment, and fish at different production stages. Two lots were monitored. The frequency of raw fish samples containing L. monocytogenes was low. During processing, the frequency of fish contaminated with L. monocytogenes clearly rose after brining, and the most contaminated sites of the processing plant were the brining and postbrining areas. A total of 303 isolates from the raw fish, product, and the environment were characterized by pulsed-field gel electrophoresis (PFGE). PFGE yielded nine pulsotypes, which formed four clusters. The predominating L. monocytogenes pulsotypes of the final product were associated with brining and slicing, whereas contaminants of raw fish were not detected in the final product. Air-mediated contamination in the plant could not be proved. In accordance with these results, an L. monocytogenes eradication program was planned. The use of hot steam, hot air, and hot water seemed to be useful in eliminating L. monocytogenes. None of the control samples taken in the 5 months after the eradication program was implemented contained L. monocytogenes.
Food-borne Listeria monocytogenes has been the causative agent of several outbreaks of human listeriosis (9). Implicated foods have been milk and dairy products, vegetables, salads, and meat products. Although L. monocytogenes has been found in a variety of fishery products (1) and these products have been considered potential sources of listeriosis, only recently have they been linked with human listeriosis (8, 16). In 1997 Ericsson et al. (8) reported an outbreak of listeriosis which they suspected had been due to consumption of vacuum-packaged rainbow trout. Thus, the prevention of L. monocytogenes contamination in fishery products, especially in ready-to-eat products like vacuum-packaged cold-smoked fish, is of major importance.
The processing of cold-smoked rainbow trout does not inactivate L. monocytogenes. The products are mainly vacuum packaged, which ensures a long shelf life and potentially enables L. monocytogenes to grow (10). Furthermore, the products are consumed without further heating. However, the sources of L. monocytogenes and the routes and sites of contamination in cold-smoked fish processing plants are still poorly known. Rørvik et al. (20) showed by multilocus enzyme electrophoresis that contamination of smoked salmon might occur during production. To prevent contamination caused by L. monocytogenes in cold-smoked rainbow trout, it is very important to detect the contamination routes of L. monocytogenes in the processing plant.
Strain characterization by pulsed-field gel electrophoresis (PFGE) has been successfully applied to typing of L. monocytogenes in epidemiological and contamination investigations (4, 6, 8, 11, 14). PFGE typing has good discriminatory power and reproducibility for L. monocytogenes (3, 4, 6). In this study, PFGE typing was applied to an in-plant L. monocytogenes contamination analysis of cold-smoked rainbow trout. The contamination sites were detected by sampling of the production line and fish at different production steps for L. monocytogenes and by typing of isolates by PFGE.
MATERIALS AND METHODS
Product manufacture and the processing plant.
The processing plant used ready-filleted rainbow trout as raw material for production of cold-smoked rainbow trout. In the processing plant, the skin was removed and the fillets were brined with a commercial injection machine. Brined fish was cold smoked (16 h at 22°C), sliced, and vacuum packaged. The ambient temperature in the plant was 10°C. After each day’s production, a cleanup crew carried out complete cleaning and sanitation procedures.
Sampling.
A total of 587 samples were collected. For contamination analyses 493 samples were examined, and for evaluating the success of the eradication program 94 samples were examined. The samples were transported to the laboratory together with plastic-encased ice packs in insulated boxes and were analyzed within 14 h of arrival.
Sixty samples were taken from the heads of fish, including skin and slime, in the rainbow trout slaughterhouse. Fish was examined as 25-g samples. In the processing plant fish was sampled after every processing step (see Table 1). Two production lots were monitored. From filleted rainbow trout, skin and a 0.5-cm-deep layer of flesh from immediately beneath the skin were cut. Skinned fillets were examined by taking a 0.5-cm-deep layer of surface flesh. Skin samples were taken after skinning, and each sample contained five randomly selected skins. The flesh of fish was examined after brining, after smoking, and after slicing. The final product was examined by enrichment and direct plating on the production day and on the manufacturer’s recommended sell-by date after storage for 28 days at 3°C.
TABLE 1.
Incidence of L. monocytogenes in samples obtained at different production stages and restriction patterns of the isolates identified by PFGE
| Sampling site and sample source | No. of positive samples (total no. of samples) | No. of isolates | Restriction pattern
|
Pulsotype | No. of isolates | |
|---|---|---|---|---|---|---|
| AscI | SmaI | |||||
| Slaughterhouse | ||||||
| Raw fish | 1 (60) | 4 | H | E | VIII | 4 |
| Processing plant | ||||||
| Arrival area | ||||||
| Ice packs | 1 (7) | 2 | C | C | III | 2 |
| Fish | 0 (35) | |||||
| Environmenta | 0 (6) | |||||
| Airb | 0 (9) | |||||
| Skinning, brining, and smoking areas | ||||||
| Environment | 4 (48) | 8 | A | A | I | 4 |
| B | B | II | 2 | |||
| I | F | IX | 2 | |||
| Machines | 15 (62) | 30 | A | A | I | 16 |
| B | B | II | 10 | |||
| C | C | III | 4 | |||
| Brine solution | 4 (6) | 8 | A | A | I | 8 |
| Employeesc | 4 (9) | 8 | A | A | I | 4 |
| C | C | III | 2 | |||
| G | D | VII | 2 | |||
| Fish after skinning | 0 (10) | |||||
| Skins | 0 (4) | |||||
| Fish after brining | 7 (10) | 28 | A | A | I | 15 |
| B | B | II | 13 | |||
| Fish after smoking | 8 (10) | 29 | A | A | I | 19 |
| B | B | II | 3 | |||
| D | D | IV | 6 | |||
| E | D | V | 1 | |||
| Air | 0 (66) | |||||
| Slicing and packaging area | ||||||
| Environment | 5 (19) | 10 | A | A | I | 6 |
| C | C | III | 4 | |||
| Machines | 5 (22) | 10 | A | A | I | 4 |
| C | C | III | 6 | |||
| Employees | 2 (10) | 4 | A | A | I | 3 |
| C | C | III | 1 | |||
| Fish after slicing | 7 (10) | 25 | A | A | I | 17 |
| C | C | III | 7 | |||
| F | D | VI | 1 | |||
| Air | 0 (42) | |||||
| Cleaning aread | 2 (12) | 4 | B | B | II | 2 |
| C | C | III | 2 | |||
| Departure area | ||||||
| Environment | 0 (6) | |||||
| Air | 0 (8) | |||||
| Product, enrichment | 22 (22) | 74 | A | A | I | 64 |
| B | B | II | 4 | |||
| C | C | III | 2 | |||
| D | D | IV | 3 | |||
| E | D | V | 1 | |||
| Product, direct plating | 9 (22) | 59 | A | A | I | 41 |
| B | B | II | 3 | |||
| C | C | III | 1 | |||
| D | D | IV | 10 | |||
| F | D | VI | 4 | |||
Samples were taken from different plant surfaces and from drains.
Air samples in each sampling site were collected with four sampling methods (see Materials and Methods).
Samples were taken from aprons and gloves of employees.
The cleaning area was situated between the brining and slicing areas and was used for cleaning smoking tracks.
Samples from the processing environment (surfaces and drains), machines, and employees (aprons and gloves) were taken before the start of the daily production, when the plant had been completely washed and disinfected. Samples were also taken during production, when the lots were handled in the area. The processing environment, machines, and equipment were examined by swabbing with 7.5- cm by 7.5-cm sterile gauze pads or with sterile swabs premoistened in sterile 0.1% peptone water. Seven pooled samples, each containing samples from five ice packs, were taken from the ice packs in the transport boxes for the raw fish. The ice packs were used to avoid direct contact between fish and ice. Floor drains were examined by taking 25 ml of water from each of the nine drains. Samples from the gloves of the fish handlers were taken by washing the gloves with 100 ml of 0.1% peptone water, ensuring that only the outer surfaces of the gloves contacted the liquid. Liquid samples (minimum volume, 500 ml) were taken from the brine solution, rinsing water, and tap water. Wooden splinters used for the smoke oven and salt and sugar (25 g for each sample) were also collected. Liquid samples and dissolved brine ingredients (salt and sugar) were filtered through a 0.45-μm-pore-size membrane which was placed in the enrichment broth.
Air samples were taken during processing and during the washing of the plant after production. Each sampling site was examined by the following four sampling methods: by sedimentation, by liquid impingement (AGI-30 impinger; Ace Glass Inc., Vineland, N.J.), with a portable high-volume surface air system impactor sampler (Air Sampler SAS-SUPER90; International, Milan, Italy), and a Reuter centrifugal air sampler (Biotest AG, Dreieich, Germany). For each of 30 sedimentation samples, 10 ml of LEB (primary Listeria enrichment broth without supplement; Oxoid, Basingstoke, United Kingdom) was poured into a petri dish, which was left open for 10 min. In addition, five samples were taken within an 8-h sampling time. The AGI-30 impinger, containing 20 ml of LEB (Oxoid), was operated at a flow rate of 12.5 liters/min for 10 min. In the surface air system impactor and Reuter centrifugal air samplers, Oxford agar (Oxoid)-containing plates and strips were used, and these samplers were operated at flow rates of 180 liters/min for 1 min and 40 liters/min for 5 min, respectively.
Eradication program.
Based on the results of the contamination analyses, an eradication program for decreasing the level of L. monocytogenes contamination was planned. Skinning, slicing, and brining machines were disassembled and thoroughly cleaned and disinfected, and the pieces were either placed in hot water (80°C), heated in an oven (80°C), or treated with gas flame. The production line, floors, and walls were treated thoroughly with hot steam.
Bacteriological analysis.
Examination for L. monocytogenes was carried out according to the recommendations of the Nordic Committee on Food Analysis (19) by a two-step enrichment method. Each sample was incubated in LEB1 (primary Listeria enrichment broth; Oxoid) at 30°C for 24 h. Volumes (0.1 ml each) of LEB1 were then transferred to LEB2 (secondary Listeria enrichment broth; Oxoid) and incubated at 30°C for 24 h. LEB2 was plated on Oxford agar (Oxoid) and incubated at 37°C for 48 h. Ten suspected colonies from Oxford agar were streaked on blood agar. The beta-hemolytic colonies were identified by Gram staining, catalase reaction, and motility at 25°C and further identified by using API Listeria kit (BioMérieux SA, Marcy l’Etoile, France). Four and two isolates from each positive fish sample and environmental sample, respectively, were preserved frozen at −70°C. Sedimentation and impingement air samples were preincubated at 30°C for 6 to 8 h before primary enrichment. Direct plating was performed on Oxford plates as described by Loncarevic et al. (17).
In situ DNA isolation and PFGE.
Brain heart infusion (Difco, Detroit, Mich.) broth was inoculated with a single colony grown on blood agar. Cells were harvested from 2 ml of brain heart infusion broth after overnight incubation. DNA isolation was performed as described by Björkroth et al. (2), with the following modifications. The plugs were lysed for only 3 h. Only a single 2-h wash with ESP at 50°C was used. SmaI and AscI (New England Biolabs, Beverly, Mass.) were used for restriction endonuclease digestion. The samples were electrophoresed through 1.0% (wt/vol) agarose gel (SeaKem Gold; FMC Bioproducts, Rockland, Maine) in 0.5× Tris-borate-EDTA (45 mM Tris, 4.5 mM boric acid [pH 8.3], and 1 mM sodium EDTA) at 200 V and 14°C in a Gene Navigator system with a hexagonal electrode (Pharmacia, Uppsala, Sweden). The pulse times ramped from 1 to 15 s for 18 h and from 1 to 35 s for 18 h for SmaI and AscI, respectively. The gels were stained with ethidium bromide and photographed under UV transillumination. Lambda ladder PFG marker (New England Biolabs) was used for fragment size determination.
PFGE pattern analysis.
AscI macrorestriction patterns (MRP) were analyzed with the GelCompar software (version 4.0; Applied Maths, Kortrijk, Belgium). The similarities between MRPs were expressed by Dice coefficient correlation, and clustering by the unweighted pair group method with arithmetic averages was used for the construction of a dendrogram. The genotypes resulting from MRP analyses were clustered at a similarity level of 75%.
Serotyping.
One strain from each pulsotype was serotyped with commercial Listeria O antisera (Denka Seiken, Tokyo, Japan) as described by the manufacturer.
RESULTS
Only one sample (2%) among the total of 60 fish from the slaughterhouse contained L. monocytogenes (Table 1). None of the 49 samples of filleted fish, skinned fish, and pooled skin was positive for L. monocytogenes. The number of fish samples positive for L. monocytogenes clearly increased after the brining stage.
Of the environmental samples, 13% (16 of 122 samples) of those taken from the washed and disinfected environment before production and 30% (19 of 65 samples) of those taken during production were positive for L. monocytogenes. The most contaminated sites were the areas for brining, slicing, and packaging (Table 2). L. monocytogenes was detected in the drains in the slicing and packaging area both before and during processing, whereas in the skinning and brining area the drains were contaminated with L. monocytogenes only during processing. No L. monocytogenes was detected in samples taken from the arrival and departure rooms. One of the seven pooled samples taken from the ice packs of the transport boxes for the raw fish fillet contained L. monocytogenes. Fresh brine solution and its ingredients (tap water, salt, and sugar) were not contaminated with L. monocytogenes, nor were the wooden splinters used for the smoke oven. By contrast, recirculated brine solution was found to contain L. monocytogenes both during and at the end of production of both lots (Table 1). None of the samples taken from workers’ gloves before brining contained L. monocytogenes, but two of five samples taken from gloves of workers postbrining were contaminated. Three of eight samples taken from the aprons of the workers were positive for L. monocytogenes. No air samples were positive for L. monocytogenes.
TABLE 2.
L. monocytogenes-positive samples of nonfish origin and division of the isolates by PFGE according to sampling time
| Source | Before production
|
During production
|
||
|---|---|---|---|---|
| No. of positive samples (total no. of samples) | Pulsotype | No. of positive samples (total no. of samples) | Pulsotype | |
| Processing plant | ||||
| Arrival area | ||||
| Ice packs | 1 (7) | III | ||
| Skinning, brining, and smoking areas | ||||
| Environmenta | 1 (20) | I | 3 (15) | I, II, IX |
| Machines | 8 (47) | I, II, III | 7 (16) | I, II |
| Brine solution | 4 (4) | I | ||
| Employeesb | 2 (6) | III, VII | 2 (3) | I |
| Slicing and packaging area | ||||
| Environment | 3 (7) | I, III | 2 (6) | I, III |
| Machines | 3 (10) | I, III | 2 (12) | I, III |
| Employees | 2 (4) | I, III | ||
| Cleaning areac | 2 (6) | II, III | ||
Samples were taken from different plant surfaces and from drains.
Samples were taken from aprons and gloves of employees.
Cleaning area was situated between brining and slicing areas and was used for cleaning smoking tracks.
After the eradication program 94 samples were taken from critical contamination points determined during the sampling for contamination analysis, of which none was positive for L. monocytogenes (Table 3).
TABLE 3.
Sampling at fish processing plant after eradication program for L. monocytogenes
| Source of sample | No. of samples | No. of positive samples |
|---|---|---|
| Machines | 72 | 0 |
| Brine solutiona | 2 | 0 |
| Final productb | 20 | 0 |
| Total | 94 | 0 |
Samples were taken from recirculated brine solution at the end of production.
Final product samples were examined on sell-by date.
PFGE with SmaI yielded seven restriction patterns, and PFGE with AscI yielded nine restriction patterns, for the isolated L. monocytogenes strains, dividing the isolates into nine different pulsotypes. The pulsotypes of the isolates from different sources are shown in Table 1, and the pulsotypes of isolates of nonfish origin are shown according to the sampling time in Table 2. Pulsotype I was the predominating pulsotype in the plant, whereas pulsotype II was associated with the brining and pulsotype III was associated with the slicing and packaging area. Twenty-three samples were contaminated with two or more different pulsotypes. Seven of those samples were environmental, and 16 were fish samples. Four pulsotypes, at most, could be detected in a single sample.
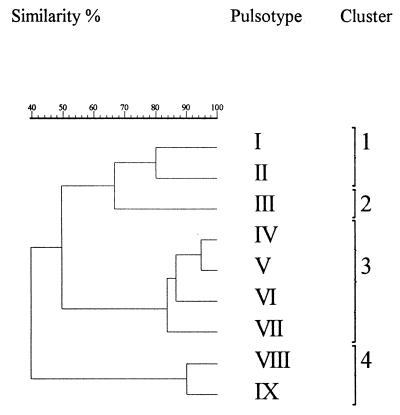
Figure 1 shows a dendrogram of the isolates based upon AscI MRPs. The isolates formed four clusters at a similarity level of 75%. The isolates from final products belonged to clusters 1, 2, and 3. The environmental isolates in cluster 1 were associated mostly with brining, and those in cluster 2 were associated mostly with slicing. The isolates from raw fish and the isolates of possible raw fish origin were in cluster 4. All strains belonged to serogroup 1/2.
FIG. 1.
Dendrogram of Listeria monocytogenes isolates based on AscI macrorestriction profiles. Similarity analysis was performed with the Dice coefficient and clustering by unweighted pair group method with arithmetic averages.
DISCUSSION
The frequency of fish samples containing L. monocytogenes clearly rose after brining, and the most-contaminated sites of the processing plant were the brining and postbrining areas. In addition, the brine solution became contaminated with L. monocytogenes during production of both lots. Of the pulsotypes identified by PFGE typing, two pulsotypes (I and II) were associated with brining. Pulsotype I, the major contaminant of fish and the processing environment, was most frequently found in the skinning, brining, and smoking areas. In addition, pulsotype I was the only pulsotype detected in the recirculated brine-solution samples. Pulsotype II was detected in the brining machine, the fish sampled during processing, and the final product, whereas it was not detected in any samples taken from the slicing and packaging area. Moreover, fish sampled after brining contained only pulsotypes I and II. Thus, it seemed that the fish were contaminated during brining with these two pulsotypes, which form cluster 1. In addition, the gloves of employees working on the production line after brining were positive for L. monocytogenes, whereas no L. monocytogenes was detected on the gloves of employees working prebrining. These findings indicate that brining was the most critical point of L. monocytogenes contamination. The fillets were brined by using a commercial injection machine. The recirculated brine solution contained fat and protein from brined fillets, and during circulation this material probably got attached to the inside of the brining machine, offering a culture and adhesion medium for L. monocytogenes and inhibiting the efficiency of cleaning.
The contaminants of the slicing and packaging area were pulsotypes I and III, and in five instances both pulsotypes could be detected in a single sample. The fish, already containing pulsotype I, had probably contaminated the slicing machine with pulsotype I, and thereafter the slicing machine continued to spread pulsotype I. Pulsotype III was associated with the slicing and packaging area, especially with the slicing machine, and was found in fish only after slicing. These findings indicate that the slicing machine had been contaminated with pulsotype III. Pulsotype III was also found in two places in earlier production steps: in one skinning machine and in one sample from the ice packs of the fillet transporting boxes, which indicated that raw fish was a source of that pulsotype. However, the skinning machine was movable and had been used in the slicing and packaging room, which may explain the presence of pulsotype III in that machine. The other skinning machine was used only in the skinning and brining area and was contaminated with pulsotype I, which was the most frequent contaminant in that area. Employees removed the ice packs from the boxes before sampling. Pulsotype III was also recovered before the shift started from the apron of an employee working in the area that day. That may explain the finding of pulsotype III in the ice pack sample.
Our results indicate that contamination of fish with L. monocytogenes occurred during processing. Raw fish seemed not to be the most important factor in the contamination of cold-smoked rainbow trout. Firstly, the frequency of L. monocytogenes in raw fish was low, and secondly, the pulsotype of L. monocytogenes in raw fish, pulsotype VIII, was found neither in any samples taken from the processing plant nor in the final products. Pulsotype IX, which formed cluster 4 with pulsotype VIII, was detected only in a floor drain sampled at the early stages of the processing on the line, when the lot was handled in the area. Eklund et al. (7) suggested that raw fish is the primary source of plant contamination. In the absence of molecular typing, these conclusions were based on the incidence of L. monocytogenes in raw salmon. Using multilocus enzyme electrophoresis, Rørvik et al. (20) found that raw fish were not the essential source of contamination. Although the numbers of L. monocytogenes cells in raw fish, both in the present study and in the study by Rørvik et al. (20), were small, raw fish as a primary source of L. monocytogenes contamination cannot be ruled out. Therefore, more research is needed to establish the role of raw fish in cold-smoked fish product contamination with L. monocytogenes.
Strains from clusters 1 and 2 were the predominating strains in this plant, whereas some strains were detected only sporadically, and pulsotype VIII, the contaminant of raw fish, was not found in any samples taken from the processing plant. These results indicate that some pulsotypes seem to predominate in the plant whereas some pulsotypes can be found only sporadically. Predominating pulsotypes were detected in the processing environment even before daily production began (Table 2), whereas the sporadic pulsotype IX was detected only during production. The cleaning methods seemed to be insufficient to prevent the colonization by dominant L. monocytogenes strains in the production environment. Endemic strains have been documented to persist for several years in food plants (15, 18, 24). It is not clear why certain L. monocytogenes clones seem to be more able to contaminate plants than others and even to contaminate certain areas within a plant. These strains may have characteristics promoting survival, colonization, and multiplication under certain industrial conditions.
The role of employees in the spreading of L. monocytogenes contamination is not clear. It is most likely that the gloves were contaminated when the workers handled the fish, rather than that the gloves themselves were the vehicle of contamination. Destro et al. (6) made the same observation in a shrimp processing plant. In this study, aprons were often contaminated with L. monocytogenes even before the shift had started, which was obviously due to a failure in the cleaning and disinfection of the aprons. The pulsotypes detected on aprons were I, III, and VII. Pulsotype VII was detected only on one apron and was not found at any other sampling site. Pulsotype VII belongs to cluster 3; other pulsotypes in this cluster were detected only in fish and only at the postsmoking stages. However, in instances where L. monocytogenes contamination levels are low, the role of employees in the spreading of contamination should not be underestimated. In particular, the rotation of assigned duties between departments has been described as the strongest expressed risk factor for L. monocytogenes contamination of smoked salmon (21).
None of the 125 air samples were positive for L. monocytogenes. This is in agreement with the results of Jacquet et al. (12) and Salvat et al. (22), who did not find any Listeria in samples of air from food processing plants, although they examined a very limited number of samples. On the other hand, Spurlock and Zottola (23) in their experimental study showed that large populations of L. monocytogenes cells survive in aerosol suspensions for hours and suggested that practices encouraging aerosol formation should not be used in a food processing environment. In our study air samples were taken by impaction sampling at two different flow rates, by liquid impingement, and by sedimentation, both during the peak of activity and at the beginning of cleaning, when a large number of aerosols should have been in the air. The level of L. monocytogenes contamination in the processing environment was also high; however, no Listeria was found. These findings indicate that air-mediated contamination with L. monocytogenes may not be of major importance.
In 23 of the samples, more than one pulsotype could be detected in a single sample. This result is in agreement with the findings reported by Danielsson-Tham et al. (5) and Loncarevic et al. (17), who identified different L. monocytogenes clones in a single sample by the direct plating method. Therefore, several isolates from a single sample should be typed in epidemiological and contamination studies. The finding of several pulsotypes in a single product sample may be due to contamination at many different sites.
To conclude, this cold-smoked rainbow trout processing plant had two major contamination sites: those associated with brining and with slicing. In order to decrease the level of L. monocytogenes contamination an eradication program was planned. The mechanical systems, e.g., brining and slicing machines, used in food processing are often complex and difficult to clean. Therefore, skinning, brining, and slicing machines were disassembled and thoroughly cleaned and disinfected, and the pieces were either placed in hot water (80°C), heated in an oven (80°C), or treated with gas flame. The production line, floors, and walls were treated thoroughly with hot steam. The hot steam, hot air, and hot water seemed to be useful tools in cleaning to eliminate L. monocytogenes from the fish processing plant. In the 5 months immediately after eradication none of 94 samples taken from critical contamination points, the recirculated brine solution, and products on the sell-by date were positive for L. monocytogenes (Table 3). Even though L. monocytogenes could not be detected in any of the samples taken after eradication, the total elimination of L. monocytogenes may be impossible. As proved in this study and several other studies (7, 13, 20), incoming fish are contaminated by L. monocytogenes to a certain level. Strict attention must be paid to cleaning and disinfection to control the level of L. monocytogenes and to avoid in-plant colonization by L. monocytogenes.
ACKNOWLEDGMENTS
We are grateful to Sirkku Ekström and Raija Keijama for their excellent technical assistance.
This work was supported by the Technology Development Center (TEKES) and the Walter Ehrström Foundation.
REFERENCES
- 1.Ben Embarek P K. Presence, detection and growth of Listeria monocytogenes in seafoods: a review. Int J Food Microbiol. 1994;23:17–34. doi: 10.1016/0168-1605(94)90219-4. [DOI] [PubMed] [Google Scholar]
- 2.Björkroth J, Ridell J, Korkeala H. Characterization of Lactobacillus sake strains associating with production of ropy slime by randomly amplified polymorphic DNA (RAPD) and pulsed-field gel electrophoresis (PFGE) patterns. Int J Food Microbiol. 1996;31:59–68. doi: 10.1016/0168-1605(96)00964-6. [DOI] [PubMed] [Google Scholar]
- 3.Brosch R, Buchrieser C, Rocourt J. Subtyping of Listeria monocytogenes serovar 4b by use of low-frequency-cleavage restriction endonucleases and pulsed-field gel electrophoresis. Res Microbiol. 1991;142:667–675. doi: 10.1016/0923-2508(91)90080-t. [DOI] [PubMed] [Google Scholar]
- 4.Buchrieser C, Brosch R, Catimel B, Rocourt J. Pulsed-field gel electrophoresis applied for comparing Listeria monocytogenes strains involved in outbreaks. Can J Microbiol. 1993;39:395–401. doi: 10.1139/m93-058. [DOI] [PubMed] [Google Scholar]
- 5.Danielsson-Tham M L, Bille J, Brosch R, Buchrieser C, Persson K, Rocourt J, Schwarzkopf A, Tham W, Ursing J. Characterization of Listeria strains isolated from soft cheese. Int J Food Microbiol. 1993;18:161–166. doi: 10.1016/0168-1605(93)90220-b. [DOI] [PubMed] [Google Scholar]
- 6.Destro M T, Leitao M F, Farber J M. Use of molecular typing methods to trace the dissemination of Listeria monocytogenes in a shrimp processing plant. Appl Environ Microbiol. 1996;62:705–711. doi: 10.1128/aem.62.2.705-711.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eklund M W, Poysky F T, Paranjpye R N, Lashbrook L C, Peterson M E, Pelroy G A. Incidence and sources of Listeria monocytogenes in cold-smoked fishery products and processing plants. J Food Prot. 1995;58:502–508. doi: 10.4315/0362-028X-58.5.502. [DOI] [PubMed] [Google Scholar]
- 8.Ericsson H, Eklow A, Danielsson-Tham M L, Loncarevic S, Mentzing L O, Persson I, Unnerstad H, Tham W. An outbreak of listeriosis suspected to have been caused by rainbow trout. J Clin Microbiol. 1997;35:2904–2907. doi: 10.1128/jcm.35.11.2904-2907.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farber J M, Peterkin P I. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev. 1991;55:476–511. doi: 10.1128/mr.55.3.476-511.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guyer S, Jemmi T. Behavior of Listeria monocytogenes during fabrication and storage of experimentally contaminated smoked salmon. Appl Environ Microbiol. 1991;57:1523–1527. doi: 10.1128/aem.57.5.1523-1527.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacquet C, Catimel B, Brosch R, Buchrieser C, Dehaumont P, Goulet V, Lepoutre A, Veit P, Rocourt J. Investigations related to the epidemic strain involved in the French listeriosis outbreak in 1992. Appl Environ Microbiol. 1995;61:2242–2246. doi: 10.1128/aem.61.6.2242-2246.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacquet C, Rocourt J, Reynaud A. Study of Listeria monocytogenes contamination in a dairy plant and characterization of the strains isolated. Int J Food Microbiol. 1993;20:13–22. doi: 10.1016/0168-1605(93)90056-m. [DOI] [PubMed] [Google Scholar]
- 13.Jemmi T, Keusch A. Occurrence of Listeria monocytogenes in freshwater fish farms and fish-smoking plants. Food Microbiol. 1994;11:309–316. [Google Scholar]
- 14.La Scola B, Fournier P E, Musso D, Tissot-Dupont H. Pseudo-outbreak of listeriosis elucidated by pulsed-field gel electrophoresis. Eur J Clin Microbiol Infect Dis. 1997;16:756–760. doi: 10.1007/BF01709260. [DOI] [PubMed] [Google Scholar]
- 15.Lawrence L M, Gilmour A. Characterization of Listeria monocytogenes isolated from poultry products and from the poultry-processing environment by random amplification of polymorphic DNA and multilocus enzyme electrophoresis. Appl Environ Microbiol. 1995;61:2139–2144. doi: 10.1128/aem.61.6.2139-2144.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loncarevic S, Danielsson-Tham M L, Gerner-Smidt P, Sahlström L, Tham W. Potential sources of human listeriosis in Sweden. Food Microbiol. 1998;15:65–69. [Google Scholar]
- 17.Loncarevic S, Tham W, Danielsson-Tham M L. The clones of Listeria monocytogenes detected in food depend on the method used. Lett Appl Microbiol. 1996;22:381–384. doi: 10.1111/j.1472-765x.1996.tb01184.x. [DOI] [PubMed] [Google Scholar]
- 18.Nesbakken T, Kapperud G, Caugant D A. Pathways of Listeria monocytogenes contamination in the meat processing industry. Int J Food Microbiol. 1996;31:161–171. doi: 10.1016/0168-1605(96)00978-6. [DOI] [PubMed] [Google Scholar]
- 19.Nordic Committee on Food Analysis. Listeria monocytogenes. Detection in foods. Espoo, Finland: Nordic Committee on Food Analysis; 1990. [Google Scholar]
- 20.Rørvik L M, Caugant D A, Yndestad M. Contamination pattern of Listeria monocytogenes and other Listeria spp. in a salmon slaughterhouse and smoked salmon processing plant. Int J Food Microbiol. 1995;25:19–27. doi: 10.1016/0168-1605(94)00080-p. [DOI] [PubMed] [Google Scholar]
- 21.Rørvik L M, Skjerve E, Knudsen B R, Yndestad M. Risk factors for contamination of smoked salmon with Listeria monocytogenes during processing. Int J Food Microbiol. 1997;37:215–219. doi: 10.1016/s0168-1605(97)00057-3. [DOI] [PubMed] [Google Scholar]
- 22.Salvat G, Toquin M T, Michel Y, Colin P. Control of Listeria monocytogenes in the delicatessen industries: the lessons of a listeriosis outbreak in France. Int J Food Microbiol. 1995;25:75–81. doi: 10.1016/0168-1605(94)00087-m. [DOI] [PubMed] [Google Scholar]
- 23.Spurlock A T, Zottola E A. The survival of Listeria monocytogenes in aerosols. J Food Prot. 1991;54:910–912. doi: 10.4315/0362-028X-54.12.910. [DOI] [PubMed] [Google Scholar]
- 24.Unnerstad H, Bannerman E, Bille J, Danielsson-Tham M L, Waak E, Tham W. Prolonged contamination of a dairy with Listeria monocytogenes. Neth Milk Dairy J. 1996;50:493–499. [Google Scholar]