Abstract
Four strains of filamentous bacteria were isolated from slimes collected in different paper mill factories. Morphological and physiological characterization of the isolates indicated an affiliation with the genus Sphaerotilus. However, while the physiological properties of the isolates were almost identical, pronounced physiological differences between the isolates and Sphaerotilus natans DSM 6575T, DSM 565, and DSM 566 with respect to their ability to metabolize complex polysaccharides, sugars, polyalcohols, or organic acids as carbon sources were detected. In contrast to the analyzed culture collection strains of S. natans, all paper mill isolates were able to grow at elevated temperatures of up to 40°C. Comparative sequence analysis of nearly complete 16S ribosomal DNA (rDNA) sequences from the four new isolates demonstrated that the retrieved sequences were highly similar to each other (99.6 to 99.8% similarity) and to previously published partial 16S rDNA sequences of S. natans DSM 6575T and ATCC 15291. Polyphasic characterization of the isolated Sphaerotilus strains revealed interesting adaptations of the strains to the environmental paper mill conditions with regard to temperature tolerance and utilization of cellulose and starch.
Sphaerotilus natans, a member of the beta-1 subdivision of the class Proteobacteria (5), occurs mainly in flowing water, sewage, and activated sludge (10, 18, 28, 31). This filamentous bacterium is characterized by a tubular sheath enclosing the rod-shaped cells. It is known to cause technological problems, such as pipe clogging and bulking of activated sludge in wastewater treatment (8, 9, 22). These problems are partly due to the ability of Sphaerotilus to settle on a solid surface by entanglement or by means of an adhesive basal element at one end of the filament (20). Moreover, the relatively good growth of Sphaerotilus under conditions of low oxygen concentrations (7) and its capability to utilize a great variety of organic carbon compounds cause its relative dominance in biological deposits (22, 28), such as paper mill slimes (30).
In paper mill plants, unique environmental conditions (available carbohydrates, high moisture levels, moderate temperatures, and the recycling of the process water) favor the development of microorganisms. Sphaerotilus enters the system primarily via fresh water containing low levels of chlorine and is implicated in the appearance of biological slimes (1, 2, 13). Tenacious microbiological deposits are formed when suspended matter, such as particles, cellulose fibers, or microorganisms, are trapped in the filamentous Sphaerotilus structures. In small quantities, Sphaerotilus natans also contributes to the stability of colonies of other organisms like Klebsiella sp. and Pseudomonas sp. (23). Biofilm development induces technical problems in the paper machines (2) and reduces the hygienic and technical quality of the paper that is produced. Preventive measures against slime entail expansive deposit control programs with the use of chemical biocides.
Most Sphaerotilus strains studied so far were isolated from rivers, sewage, or activated sludge. In spite of the well-described importance of S. natans in paper mill plants, Sphaerotilus strains originating from paper mill slimes have not been studied yet. Investigation of such strains might be particularly interesting since the environmental conditions present in paper factories differ significantly from those found in other habitats of S. natans and might therefore select for strains with unusual physiological properties. Understanding the ecology of paper mill Sphaerotilus strains is a prerequisite for the development of more efficient control strategies against slime formation.
In the present investigation, we used a polyphasic approach to characterize four Sphaerotilus-like isolates of filamentous bacteria obtained from paper mill slimes. Morphological and physiological properties as well as 16S ribosomal DNA (rDNA) sequences were determined for the new isolates and compared to culture collection strains of S. natans.
MATERIALS AND METHODS
Materials and chemicals.
All bacteriological products were purchased from Biokar (Biokar Diagnostics, Beauvais, France). All common chemicals and carbohydrates were purchased from Sigma (Sigma-Aldrich Chimie, Saint Quentin Fallavier, France).
Bacterial strains and preservation.
The reference strains were S. natans DSM 6575T, DSM 565, and DSM 566 and Leptothrix discophora ATCC 43182. Isolates from paper mill slimes were named IF4, IF5, IF9, and IF14. They were collected in the different paper mills listed in Table 1. Pure cultures were maintained at −80°C as follows. Cell suspensions (10 ml) from a 1-day-old culture in FIL broth (19) with orbital shaking (150 rpm) at 30°C (25°C for DSM 565) were harvested by centrifugation (10 min, 5,000 × g). The cell pellet was transferred to 1 ml of FIL broth containing 10% dimethylsulfoxide and thoroughly mixed before transferring it to 2-ml vials for storage at −80°C in a Nalgene Cryo Freezing Container with isoamyl alcohol as cooling rate controller.
TABLE 1.
Characteristics of Sphaerotilus strains
| Strain (paper mill of origin [location]) | Size of hydrated cells (mm)a
|
Type of colony formation on FIL agar mediumb | Type of growth in FIL brothc | Abundance of single cells (in FIL broth) | |
|---|---|---|---|---|---|
| Width | Length | ||||
| IF4 (du Rhin [Mulhouse]) | 1.5 | 5 | R | Pellicular | + |
| IF5 (de l’Aa [Saint-Omer]) | 2 | 4 | R | Pellicular | + |
| IF9 (de Veuze [Angoulême]) | 2 | 6 | R | Pellicular | + |
| IF14 (Chapelle Darblay) | 1.5 | 6 | R/S | Pellicular | ++ |
| DSM 565 | 2 | 6 | R/S | Pellicular | ++ |
| DSM 566 | 1.5 | 6 | S | Flocculent | +++ |
| DSM 6575T | 1 | 4 | S | Homogeneous | +++ |
Average measurements obtained from 20 samples.
R, rough; S, smooth.
On broth surface.
+, rare; ++, observed in each microscope field; +++, abundant.
Sampling and isolation procedure.
Slime samples were collected in sterile flasks from the “wet end” steel surfaces of machines (i.e., those situated before the press section), transported, and stored at room temperature. Filamentous strains were isolated from slimes as follows. The deposits (approx. 0.5 g) were rinsed fivefold in 5 ml of sterile distilled water in order to remove contaminant cells. The washed pieces were placed on a surface of FIL medium solidified with 10 g of agar liter−1 containing cycloheximide (0.005 g liter−1). Incubation was performed for 72 h at 30°C. Agar plates were examined with an inverted microscope (Leica DMIL, 090-131.001) at a magnification of ×200. Surface colonies of Sphaerotilus were tentatively identified by their characteristic filamentous appearance. When rough colonies (the typical type of Sphaerotilus colonies) were observed, they were removed with a piece of agar and transferred to a sterile agar plate to separate Sphaerotilus from the rest of the slime flora. This procedure was repeated until pure cultures were obtained.
Strain characterization.
Isolates were characterized by using the method previously described by Farquhar and Boyle (11). A dilute crystal violet staining solution was used to highlight the sheath of the Sphaerotilus-like bacteria. The ability of sheathed microorganisms to oxidize ferrous iron and deposit it within their sheaths was detected by the Prussian blue reaction: filaments of paper mill isolates were incubated for 60 min with gentle agitation in 5 ml of FIL broth containing 0.7 mg of FeSO4 · 7H2O. A fixed dried smear of filament was covered with 2% potassium ferrocyanide (15 min) and then with 2% HCl (10 sec). Sheaths containing iron are stained blue. The ability of microorganisms to oxidize and deposit manganese was detected as follows. Each strain was streaked onto FIL agar plates containing 2% MnCO3. Manganese oxide covering bacterial sheaths colors the organisms in dark brown. A dilute Sudan black staining solution was used in demonstrating the presence of poly-β-hydroxybutyrate intracellular granules. All determinations were made in triplicate.
Biochemical tests.
Oxidase activities were carried out using an oxidase test (bioMérieux, Marcy-l’Etoile, France). The presence of β-galactosidase activity was tested by using ONPG (o-nitrophenyl-β-d-galactopyranoside) disk tests (Sanofi Diagnostics Pasteur, Marne-la-Coquette, France). Other enzyme activities were studied according to the method described by Marchal et al. (17).
Cultivation conditions.
Incubations in Erlenmeyer flasks were performed on a rotary shaker at 150 rpm. Growth and biochemical tests were carried out at 30°C (the average temperature in paper factories) except for tests with S. natans DSM 565, which was cultivated at 25°C. Growth rate determination was performed in 1,000-ml Erlenmeyer flasks containing 400 ml of FIL broth. Precultures were obtained with 48 h of incubation in 5 ml of FIL broth. The ability of the Sphaerotilus strains to use sugars or polyalcohols was analyzed in 5 ml of modified FIL broth containing the tested compound (2 g liter−1), pancreatic digest of meat peptone (2 g liter−1), basal salts components (19) and phenol red (0.05 g liter−1) as pH indicator. Tested substrates were sterilized separately by filtration (pore size, 0.22 μm; Millipore, Saint Quentin Yvelines, France) and added to the medium after autoclaving. Cultures were observed during a 7-day incubation period for yellow color formation indicating medium acidification and the capacity to use the tested carbon sources. The ability to metabolize polysaccharides and organic acids was analyzed in 250-ml Erlenmeyer flasks containing 75 ml of FIL broth and the tested component (2 g liter−1). Precultures were obtained by 24 h of orbital shaking incubation in 5 ml of FIL broth. One milliliter of preculture was added to the medium, and the measured biomass obtained during growth was compared to that of a reference sample.
pH limits for growth.
pH tests were performed in a modified FIL medium from which phosphate was omitted. pH levels were established with 0.2 M phosphate buffer (final molarity, 0.02 M). When necessary, media were adjusted to various pHs with HCl (1 M) and NaOH (1 M). Observations were made after a 4-day incubation period. Presence of growth, visually determined, indicated the ability of Sphaerotilus strain to support the tested pH.
Biomass measurement.
To measure biomass, 5 to 20 ml of culture broth was harvested by centrifugation (10 min, 5,000 × g), rinsed twofold with 1 ml of Tris-HCl buffer (0.05 M, pH 7), and centrifuged (10 min, 5,000 × g). The pellet was suspended in 1 ml of Tris-HCl buffer and disintegrated over a 3-min period with an ultrasonic disintegrator (Sonics Materials, model no. VC 50 240U). This disintegration time was required to disrupt most of the cells as determined microscopically. The lysate was centrifuged (10 min, 8,000 × g) to remove cellular remains, and the concentration of proteins in the supernatant was measured using the Protein Assay Kit (Bio-Rad, Yvry sur Seine, France) according to the manufacturer’s instructions. The deduced amount of bacterial protein concentration of original broth culture was considered as a biomass measurement.
Scanning electron microscopy.
Pure culture samples were fixed on microscope slides at room temperature with 2% (vol/vol) glutaraldehyde (TAAB; Saint-Germain en Laye, France) in 0.1 M phosphate buffer (pH 7.35). Chemical fixation for 30 min was followed by dehydration in a graded acetone series (50, 70, and 90%) ending with absolute acetone. The samples were critical-point dried in liquid carbon dioxide with a Balzers Union CPD 030 and coated with gold in a Balzers Union FL 9496. Samples were examined with a JEOL JMC 35C scanning electron microscope at 15 kV.
PCR amplification, cloning, and sequencing of the 16S rDNA.
Isolates were cultivated overnight in FIL medium. Cells were harvested from 2 ml of medium by centrifugation, washed with ddH2O, and resuspended in 100 μl of ddH2O. Cells were lysed by heating for 20 min at 94°C. Cell debris was pelleted by centrifugation, and the supernatant was transferred to a new tube. Nucleic acid concentration in the supernatant was estimated by spectrophotometric measurement of optical density at 260 nm. Oligonucleotide primers targeting 16S rDNA signature regions, which are highly conserved within the domain Bacteria, were used for PCR with a thermal capillary cycler (Idaho Technology, Idaho Falls, Idaho). The nucleotide sequences of the primers were 5′-AGAGTTTGATYMTGGCTCAG-3′ (Escherichia coli 16S rDNA positions 8 to 27 [3]) and 5′-CAKAAAGGAGGTGATCC-3′ (E. coli 16S rDNA positions 1529 to 1545 [3]). Amplification was performed using 1 μl of the “boiled-cell” supernatant (containing approx. 100 ng/μl) and 10 pmol of each primer according to the manufacturer’s recommendations in a total volume of 50 μl by using the 20 mM MgCl2 reaction buffer. After initial heating to 94°C for 3 min, 30 cycles consisting of denaturation (94°C, 30 sec), annealing (48°C, 20 sec), and extension (72°C, 30 sec) were performed. Cycling was completed by a final elongation step (72°C, 60 sec). Positive controls containing purified DNA from E. coli were included in all sets of amplifications along with negative controls (no DNA added). The presence and size of the amplification products were determined by agarose (1%) gel electrophoresis of the reaction product. For isolates IF4 and IF5, 1 to 2 μl of the PCR product was directly ligated into the cloning vector pCR 2.1 (TA Cloning Kit; Invitrogen Corp., San Diego, Calif.) and transformed in E. coli. Nucleotide sequences of the cloned DNA fragments were determined by the dideoxynucleotide method (4) by cycle sequencing of purified plasmid preparations (Qiagen, Hilden, Germany) with a Thermo Sequenase Cycle Sequencing kit (Amersham Life Science, Little Chalfont, England) and an infrared automated DNA sequencer (MWG-Biotech, Ebersberg, Germany). Dye-labeled vector-specific sequencing primers (M13pucV, M13pucR, and primer 610V [5′-GTGCCAAGCAGCCGCGGT-3′, E. coli 16S rDNA positions 515 to 531]) (MWG-Biotech) were used. For isolates IF9 and IF14, the amplified DNA fragments were sequenced directly (24) by using 1 to 3 μl of the PCR product after purification (QIAgen 28106; Qiagen) as template and dye-labeled primers (5′-CATGCAAGTCGAACG-3′, E. coli 16S rDNA positions 53 to 86; 5′-GCTACCTTGTTACGACTT-3′, E. coli 16S rDNA positions 1491 to 1509) for cycle sequencing as described above.
Phylogenetic analysis.
The obtained 16S rDNA sequences were added to the 16S rRNA sequence database of the Technischen Universität München (encompassing about 10,000 published and unpublished homologous small-subunit rRNA primary structures (6, 16) by use of the program package ARB (29). Alignment of the new 16S rDNA sequences was performed by using the ARB automated alignment tool. The alignments were refined by visual inspection and by secondary structure analysis. Phylogenetic analyses were performed by applying the ARB parsimony tool, distance matrix, and maximum likelihood methods to different data sets. To determine the robustness of the phylogenetic trees, phylogenetic analyses were performed on the one hand on the original data set and on the other hand on a data set from which highly variable positions were removed by use of a 50% conservation filter (15) for the members of the beta subclass of Proteobacteria.
Nucleotide sequence accession numbers.
The 16S rDNA sequences obtained in this study have been deposited in GenBank under accession no. AF0772914, AF072915, AF072916, and AF072917 for isolates IF4, IF5, IF9, and IF14, respectively.
RESULTS AND DISCUSSION
Morphological and physiological characterization of filamentous paper mill isolates.
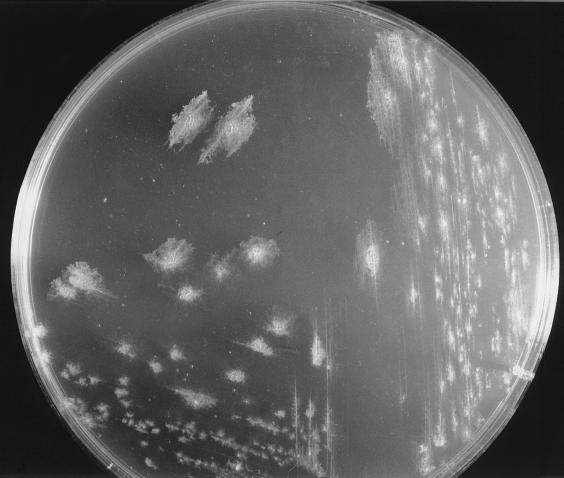
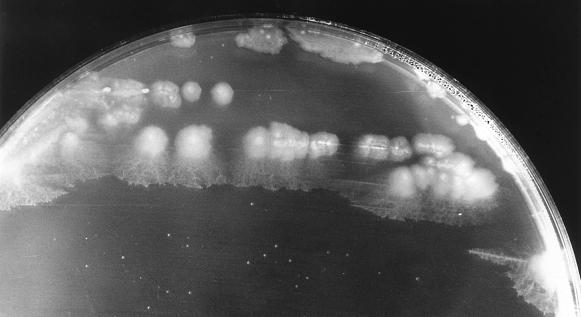
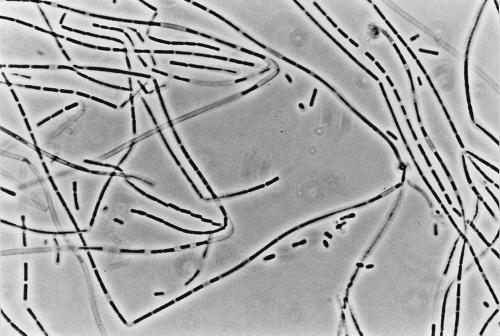
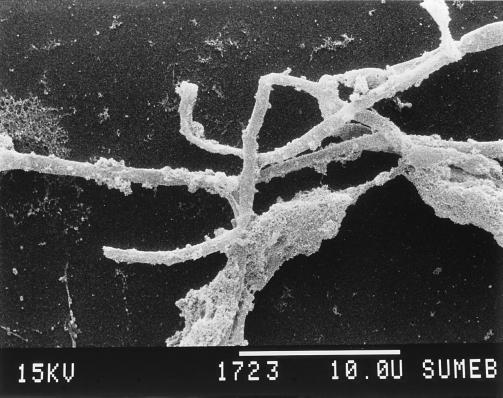
Four filamentous bacteria (IF4, IF5, IF9, and IF14) isolated from slimes collected in different paper mills were studied. Strains IF4, IF5, and IF9 developed rough colonies on FIL agar media (Fig. 1; Table 1). IF14 produced a colony type characterized by a smooth central region and a “short-haired” periphery (Fig. 2). After 36 h of stationary broth culture, isolates developed a pellicule on the broth surface, mainly composed of long, unbranched, and ensheathed filaments (Fig. 3) which occasionally exceeded 500 μm in length. Like those observed by Gaudy and Wolfe (12), these filaments were embedded in an abundant slime layer (Fig. 4) which may constitute the paper mill slime body (30). These morphological characteristics (summarized in Table 1) of the filamentous bacteria isolated from the paper mill slimes indicated an affiliation with the Sphaerotilus-Leptothrix group. The capability of all isolates to oxidize iron and to produce holdfasts on solid surfaces and their inability to oxidize manganous ions (11) strongly supported the idea that all isolates are related to the genus Sphaerotilus (19, 20, 31) (Table 2). While previous investigators noticed that the composition of the slime biota varies notably according to season (14) and to the age of the slime (8), the successful isolation of Sphaerotilus sp. from slime samples collected during winter (IF4 and IF5), spring (IF14), and summer (IF9) indicates that Sphaerotilus sp. is a common year-round inhabitant of paper mill slimes.
FIG. 1.
S. natans IF4 producing rough colonies on FIL agar medium (magnification, ×1.5).
FIG. 2.
S. natans IF14 developing rough-smooth colonies on FIL agar medium (magnification, ×2).
FIG. 3.
S. natans IF4 producing long unbranched and ensheathed filaments (magnification, ×284).
FIG. 4.
Scanning electron micrograph showing slimy matrix covering the sheath of strain IF4 (magnification, ×3,154).
TABLE 2.
Characteristics of Sphaerotilus and L. discophora
| Characteristic | Result for strain:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| S. natans DSM 565 | S. natans DSM 566 | S. natans DSM 6575T | L. discophora ATCC 43182 | IF4 | IF5 | IF9 | IF14 | |
| Gram staining | − | − | − | − | − | − | − | − |
| Sheath | + | + | − | + | + | + | + | + |
| Mobility of single cellsa | + | + | + | + | + | + | + | + |
| Oxidation of iron | + | + | + | + | + | + | + | + |
| Oxidation of manganous ions | − | − | − | + | − | − | − | − |
| Deposition of PHBb | + | + | + | + | + | + | + | + |
| Holds fast to surface | + | + | + | − | + | + | + | + |
| Genera | SPHc | SPH | SPH | LEPd | SPH | SPH | SPH | SPH |
Stain Flagella (Difco).
PHB, poly-β-hydroxybutyrate.
SPH, Sphaerotilus natans.
LEP, Leptothrix discophora.
Enzymatic activities and detailed carbon source utilization patterns (various sugars, polysaccharides, polyalcohols, and organic acids) were determined for the filamentous paper mill isolates and compared to respective parameters of the S. natans reference strains (Tables 3 and 4). Characterization of enzymatic activities demonstrated a remarkable homogeneity between all four isolates but differences with S. natans reference strains. In regard to its enzymatic activities the type strain of S. natans DSM 6575T was most similar to the paper mill isolates differing only in the absence of arginine dihydrolase activity. Concerning their ability to use sugars, polyalcohols, and organic acids as carbon sources, all Sphaerotilus-like isolates and the S. natans reference strains showed quite homogeneous behavior that accorded with the observations of Stokes (28). While all Sphaerotilus-like isolates and two of the three analyzed S. natans strains were ONPG positive (Table 3), only isolate IF4 and S. natans DSM 565 were able to use lactose as carbon source (Table 4) and consequently were β-galactosidase positive. For the other ONPG-positive strains (IF5, IF9, IF14, and DSM 6575T), the inability to use lactose despite a positive ONPG character was due to either the presence of an enzymatic activity different from the β-galactosidase activity or the absence of a lactose permease. Except for S. natans DSM 565 and DSM 6575T, which were both unable to use acetate as carbon source, all other strains by their acetate-positive character possessed a functional glyoxylate cycle. Interestingly, isolates IF4 and IF14 were, in contrast to previously analyzed Sphaerotilus strains (31), able to utilize starch and cellulose as carbon source, which provides these strains with a significant nutritional advantage in their paper mill habitat.
TABLE 3.
Enzymatic activities of Sphaerotilus isolates and S. natans strains
| Enzyme or process | Result for strain:
|
||||||
|---|---|---|---|---|---|---|---|
| IF4 | IF5 | IF9 | IF14 | DSM 565 | DSM 566 | DSM 6575T | |
| Catalase | + | + | + | + | + | + | + |
| Oxidase | + | + | + | + | − | − | + |
| Lysine decarboxylase | − | − | − | − | − | − | − |
| Ornithine decarboxylase | − | − | − | − | − | − | − |
| Arginine dihydrolase | + | + | + | + | − | + | − |
| Nitrate reductase | + | + | + | + | + | − | + |
| Nitrite reductase | − | − | − | − | − | − | − |
| ONPG utilization | + | + | + | + | + | − | + |
TABLE 4.
Utilization of carbon compounds by Sphaerotilus strains
| Compound | Result for strain:
|
||||||
|---|---|---|---|---|---|---|---|
| IF4 | IF5 | IF9 | IF14 | DSM 565 | DSM 566 | DSM 6575T | |
| Fructose | + | + | + | + | + | + | + |
| Glucose | + | + | + | + | + | + | + |
| Sucrose | + | + | + | + | + | + | + |
| Maltose | + | + | + | + | + | + | + |
| Trehalose | + | + | + | + | + | + | + |
| Arabinose | − | − | − | − | − | − | − |
| Raffinose | − | − | + | − | − | − | − |
| Rhamnose | − | − | − | − | − | − | + |
| Lactose | + | − | − | − | + | − | − |
| Galactose | + | − | − | + | + | − | + |
| Mannose | + | + | − | + | + | + | − |
| Xylose | − | + | − | + | + | − | − |
| Starch | + | − | − | + | − | − | − |
| Cellulose | + | − | − | + | − | − | − |
| Arabic gum | − | − | − | − | − | − | − |
| Guar gum | + | − | − | − | − | − | − |
| Glycerol | + | + | + | + | + | + | + |
| myo-inositol | − | + | + | + | + | + | + |
| Mannitol | + | + | + | + | + | − | − |
| Sorbitol | − | + | + | + | + | − | + |
| Lactate | + | + | + | + | + | + | + |
| Pyruvate | + | + | + | + | + | + | + |
| Succinate | + | + | + | + | + | + | + |
| Acetate | + | + | + | + | − | + | − |
| Citrate | − | − | − | − | − | + | − |
| Tartrate | − | − | − | − | − | + | + |
Growth kinetics.
Since the slime samples were collected from paper machines operating in a temperature range from 25 to 55°C, we compared growth kinetics of the Sphaerotilus-like isolates and S. natans reference strains within this temperature range. Quantitative analysis of the growth of the Sphaerotilus-like isolates by spectrophotometric analysis appeared to be difficult due to their filamentous and flocculent growth. Consequently, a procedure determining total cell proteins to measure bacterial biomass was selected. Initially, we validated this method with S. natans DSM 6575T and DSM 566 by comparative spectrophotometric and cell protein analysis. Under the applied culture conditions, strain DSM 6575T grew mainly as single cells, and the resulting culture was homogeneous in regard to its turbidity. Strain DSM 566 was characterized by the occurrence of swarmer cells and short ensheathed filaments containing 5 to 10 cells. Therefore, for both strains, spectrophotometric analysis gave reproducible results well suited for obtaining a growth curve. Similar growth curves were derived from cell protein analysis (conducted in parallel) for strains DSM 6575T and DSM 566 (data not shown). Growth kinetics of paper mill isolates and reference strains at different temperatures are presented in Table 5. Interestingly, the four Sphaerotilus-like isolates appeared to be well adapted to the elevated temperatures of the paper mill environment since they showed shortest generation times at 35°C while maximum growth rates of S. natans reference strains required lower temperatures, which is consistent with previous findings of van Veen et al. (31). In addition, all paper mill Sphaerotilus-like isolates were able to grow at 40°C after 48 h of incubation, in contrast to all S. natans reference strains.
TABLE 5.
Growth kinetics of S. natans strains
| Temperature (°C) | Generation time (min) of straina:
|
||||||
|---|---|---|---|---|---|---|---|
| IF4 | IF5 | IF9 | IF14 | DSM 565 | DSM 566 | DSM 6575T | |
| 25 | 100 | 120 | 90 | 120 | 210 | 120 | 140 |
| 30 | 90 | 95 | 80 | 60 | NG | 100 | 60 |
| 35 | 60 | 60 | 50 | 50 | 210 | 78 | |
| 40 | 180 | 240 | 140 | 150 | NG | NG | |
| 45 | NG | NG | NG | NG | |||
NG, no growth observed after a 48-h incubation period.
pH limits for growth.
The paper machines were characterized by pH values varying from 5 to 8 (21) depending on the type and quality of paper produced. Regarding pH growth limitation, all but one of the Sphaerotilus strains did not grow at a pH lower than 5.4 and above 9 and showed reduced growth in the pH ranges 5.4 to 6.3 and 8.4 to 9 (data not shown). Strain DSM 565 was even more sensitive to basic pH and tolerated only a pH range around neutrality for growth (5.8 to 7.8). These results match well with those of a previous study demonstrating that Sphaerotilus sp. is not able to tolerate very acidic or alkaline conditions and grows best at neutral pH (31).
Phylogenetic inference.
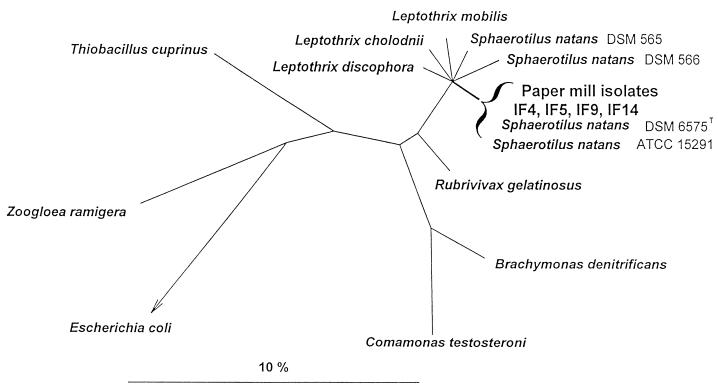
Nearly complete 16S rDNA sequences were amplified, cloned, and sequenced from the Sphaerotilus-like paper mill isolates. Comparative sequence analysis revealed that all four 16S rDNA sequences were highly similar to each other (99.6 to 99.8% similarity) (Table 6) and to two identical previously published partial 16S rDNA sequences of S. natans DSM 6575T and ATCC 15291 (99.0 to 100% similarity [25]). Since the 16S rDNA similarities to the validly described S. natans DSM 6575T and ATCC 15291 of the four isolates appear to be significantly greater than 97%, DNA-DNA reassociation studies will have to be performed to clarify the species affiliation of the isolates (27). Phylogenetic analysis demonstrated that the retrieved isolate sequences clustered together with S. natans DSM 6575T and ATCC 15291 within the beta-1 subdivision of the class Proteobacteria. The tree given in Fig. 5 is based on the results of a maximum likelihood analysis of all available 16S rDNA sequences from representatives of the beta subclass of Proteobacteria, for which more than 1,000 nucleotide positions have been determined, and of a selection of members of the major lines of descent among the Bacteria. Only sequence positions which share the same nucleotides in at least 50% of all available sequences from the beta subclass of Proteobacteria were included, to reduce potential treeing artifacts which may result from multiple base changes (15). To enhance clarity, several phylogenetic groups within the beta subclass and the outgroup organisms were subsequently removed from the tree without changing its topology. Short partial 16S rDNA sequences of S. natans DSM 6575T, DSM 566, and ATCC 15291 (25) were subsequently added to the tree without changing its topology. The topology of the tree was evaluated by maximum parsimony and distance matrix analyses of a variety of data sets differing with respect to the inclusion of sequence positions and outgroup reference sequences. The different treeing methods consistently supported clustering of the isolate sequences with S. natans DSM 6575T and ATCC 15291 but, consistent with previously published phylogenetic analyses (26), an unambiguous pattern of the respective branches within the Sphaerotilus-Leptothrix group could not be determined on the basis of the available data set.
TABLE 6.
Overall 16S rDNA sequence similarities for paper mill isolates and selected reference organisms of the beta subclass of Proteobacteriaa
| Organism | Similarity (%) to organism:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| 1. Zoogloea ramigera | ||||||||||||
| 2. Brachymonas denitrificans | 88.0 | |||||||||||
| 3. Comamonas testosteroni | 88.0 | 94.8 | ||||||||||
| 4. Rubrivivax gelatinosus | 89.5 | 92.8 | 91.2 | |||||||||
| 5. Thiobacillus cuprinus | 89.2 | 89.5 | 88.8 | 91.7 | ||||||||
| 6. Leptothrix cholodnii | 88.7 | 92.3 | 91.0 | 96.9 | 92.1 | |||||||
| 7. Leptothrix discophora | 89.1 | 92.8 | 91.2 | 97.0 | 92.3 | 96.8 | ||||||
| 8. Leptothrix mobilis | 89.5 | 92.0 | 90.6 | 97.2 | 92.2 | 97.7 | 97.1 | |||||
| 9. Sphaerotilus natans DSM 565 | 88.8 | 91.2 | 90.7 | 95.5 | 92.2 | 96.7 | 96.8 | 96.5 | ||||
| 10. IF5 (paper mill isolate) | 88.9 | 91.5 | 90.6 | 95.8 | 91.8 | 96.3 | 96.3 | 96.5 | 97.2 | |||
| 11. IF4 (paper mill isolate) | 89.0 | 91.8 | 91.1 | 96.2 | 92.2 | 96.5 | 96.6 | 96.9 | 97.4 | 99.7 | ||
| 12. IF14 (paper mill isolate) | 89.4 | 92.1 | 90.9 | 96.5 | 92.0 | 96.3 | 96.5 | 96.7 | 97.3 | 99.6 | 99.8 | |
| 13. IF9 (paper mill isolate) | 89.6 | 92.3 | 91.1 | 96.5 | 92.0 | 96.4 | 96.6 | 96.8 | 97.4 | 99.6 | 99.8 | 99.6 |
Only alignment positions for which nucleotides were determined unambiguously were included in the calculations.
FIG. 5.
Phylogenetic tree reflecting the relationships of the Sphaerotilus sp. isolates obtained from paper mill slime within the beta subclass of Proteobacteria. The tree is based on the results of maximum likelihood analysis. The multifurcation connects branches for which a relative order could not be unambiguously determined. The bar indicates 10% estimated sequence divergence.
In conclusion, our results demonstrated the year-round presence of Sphaerotilus spp. within paper mill slimes. Polyphasic characterization of the isolated Sphaerotilus strains revealed interesting adaptations of the strains to the environmental paper mill conditions in regard to temperature tolerance and utilization of cellulose and starch. Future studies will focus on the design and application of rRNA-targeted oligonucleotide probes for studying in situ abundance and spatial organization of Sphaerotilus spp. in paper mill slimes. Improved understanding of the ecology of paper mill slimes will be key to their control.
ACKNOWLEDGMENTS
This work was supported by Rhodia/Texel. M.W. and S.J. were supported by a grant from the EU (MACOBS-PL970349).
Jiri Snaidr is acknowledged for helpful discussions.
REFERENCES
- 1.Bennett C. The control of microbiological problems in the paper industry. Int Biodeterior. 1988;24:381–386. [Google Scholar]
- 2.Blanco M A, Negro C, Gaspar I, Tijero J. Slime problems in the paper and board industry. Appl Microbiol Biotechnol. 1996;46:203–208. [Google Scholar]
- 3.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 4.Chen E Y, Seeburg P H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985;4:165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- 5.Corstjens P, Muyzer G. Phylogenetic analysis of the metal-oxidizing bacteria Leptothrix discophora and Sphaerotilus natans using 16S rDNA sequencing data. Syst Appl Microbiol. 1993;16:219–223. [Google Scholar]
- 6.De Rijk P, Van de Peer Y, De Wachter R. Database on the structure of large ribosomal subunit RNA. Nucleic Acids Res. 1996;24:92–97. doi: 10.1093/nar/24.1.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dias F F, Dondero N C, Finstein M S. Attached growth of Sphaerotilus and mixed population in continuous-flow apparatus. Appl Microbiol. 1968;16:1191–1199. doi: 10.1128/am.16.8.1191-1199.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dondero N C. Sphaerotilus, its nature and economic significance. Adv Appl Microbiol. 1961;3:77–107. doi: 10.1016/s0065-2164(08)70507-0. [DOI] [PubMed] [Google Scholar]
- 9.Dondero N C. The Sphaerotilus-Leptothrix group. Annu Rev Microbiol. 1975;29:407–428. doi: 10.1146/annurev.mi.29.100175.002203. [DOI] [PubMed] [Google Scholar]
- 10.Eikelboom D H. Filamentous organisms observed in activated sludge. Water Res. 1975;9:365–388. [Google Scholar]
- 11.Farquhar G J, Boyle W C. Occurrence of filamentous microorganisms in activated sludge. J Water Pollut Control Fed. 1971;43:604–622. [PubMed] [Google Scholar]
- 12.Gaudy E, Wolfe R S. Composition of an extracellular polysaccharide produced by Sphaerotilus natans. Appl Microbiol. 1962;10:200–205. doi: 10.1128/am.10.3.200-205.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harju-Jeanty P, Väätänen P. Detrimental micro-organisms in paper and cardboard mills. Pap Puu. 1984;3:245–259. [Google Scholar]
- 14.Hugues-van Kregten H C. Slime flora of New Zealand paper mills. Appita J. 1988;41:470–474. [Google Scholar]
- 15.Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weissenegger M, Neumeier J, Bachleitner M, Scleifer K-H. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis. 1998;19:554–568. doi: 10.1002/elps.1150190416. [DOI] [PubMed] [Google Scholar]
- 16.Maidak B L, Larsen N, McCaughey M J, Overbeek R, Olsen G J, Fogel K, Blandy J, Woese C R. The ribosomal database project. Nucleic Acids Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchal N, Bourdon J L, Richard D. Les milieux de culture pour l’isolement et l’identification biochimique des bactéries. 4th ed. Paris, France: Doin Editeurs; 1991. Milieux et techniques d’identification d’application générale; pp. 47–160. [Google Scholar]
- 18.Mulder E G. Iron bacteria, particularly those of the Sphaerotilus-Leptothrix group, and industrial problems. J Appl Bacteriol. 1964;27:151–173. [Google Scholar]
- 19.Mulder E G. Genus Sphaerotilus Kützing 1833, 386AL. In: Holt J G, et al., editors. Bergey’s manual of determinative bacteriology. 8th ed. Baltimore, Md: The Williams & Wilkins Co.; 1989. pp. 1994–1998. [Google Scholar]
- 20.Mulder E G, Deinema M H. The sheathed bacteria. In: Balows A, Trüpper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. Berlin, Germany: Springer-Verlag; 1992. pp. 2612–2624. [Google Scholar]
- 21.Purvis M R, Tomlin J L. Microbiological growth and control in the papermaking process. TAPPI Chem Processing Aids. 1991;1991:69–77. [Google Scholar]
- 22.Richard M, Hao O, Jenkins D. Growth kinetics of Sphaerotilus species and their significance in activated sludge bulking. J Water Pollut Control Fed. 1985;57:68–81. [Google Scholar]
- 23.Safade T L. Tackling the slime problem in a paper-mill. Paper Technol Ind. 1988;1988:280–285. [Google Scholar]
- 24.Sanger J, Nicklen S, Coulsen A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;7:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siering P L, Ghiorse W C. Phylogeny of the Sphaerotilus-Leptothrix group inferred from morphological comparison, genomic fingerprinting, and 16S ribosomal DNA sequence analyses. Int J Syst Bacteriol. 1996;46:173–182. doi: 10.1099/00207713-46-1-173. [DOI] [PubMed] [Google Scholar]
- 26.Spring S, Kämpfer P, Ludwig W, Schleifer K-H. Polyphasic characterization of the genus Leptothrix: new descriptions of Leptothrix mobilis sp. Nov. and Leptothrix discophora sp. Nov. nom. Rev. And emended description of Leptothrix cholodnii emend. Syst Appl Microbiol. 1996;19:634–643. [Google Scholar]
- 27.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 28.Stokes J L. Studies on the filamentous sheathed iron bacterium Sphaerotilus natans. J Bacteriol. 1954;67:278–291. doi: 10.1128/jb.67.3.278-291.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strunk O, Ludwig W. ARB software package. 1997. http://www.biol.chemie.tu-muenchen.de/pub/ARB/ http://www.biol.chemie.tu-muenchen.de/pub/ARB/. . [Google Scholar]
- 30.Väisänen O M, Nurmiaho-Lassila E-L, Marmo S A, Salkinoja-Salonen M S. Structure and composition of biological slimes on paper and board machines. Appl Environ Microbiol. 1994;60:641–653. doi: 10.1128/aem.60.2.641-653.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Veen W L, Mulder E G, Deinema M H. The Sphaerotilus-Leptothrix group of bacteria. Microbiol Rev. 1978;42:329–356. doi: 10.1128/mr.42.2.329-356.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]