Abstract
The BRCA1/2 germline and/or somatic pathogenic variants (PVs) are key players in the hereditary predisposition and therapeutic response for breast, ovarian and, more recently, pancreatic and prostate cancers. Aberrations in other genes involved in homologous recombination and DNA damage response (DDR) pathways are being investigated as promising targets in ongoing clinical trials. However, DDR genes are not routinely tested worldwide. Due to heterogeneity in cohort selection and dissimilar sequencing approaches across studies, neither the burden of PVs in DDR genes nor the prevalence of PVs in genes in common among pancreatic and prostate cancer can be easily quantified. We aim to contextualize these genes, altered in both pancreatic and prostate cancers, in the DDR process, to summarize their hereditary and somatic burden in different studies and harness their deficiency for cancer treatments in the context of currently ongoing clinical trials. We conclude that the inclusion of DDR genes, other than BRCA1/2, shared by both cancers considerably increases the detection rate of potentially actionable variants, which are triplicated in pancreatic and almost doubled in prostate cancer. Thus, DDR alterations are suitable targets for drug development and to improve the outcome in both pancreatic and prostate cancer patients. Importantly, this will increase the detection of germline pathogenic variants, thereby patient referral to genetic counseling.
Keywords: DNA damage response, BRCA, mismatch repair, homologous recombination, genetics, PARP inhibitors, pancreatic cancer, prostate cancer
1. Introduction
At each cell division, there is a risk of errors occurring in the DNA replication machinery. DNA replication errors occur more frequently in the presence of DNA damaging agents, both endogenous and exogenous. For instance, reactive oxygen species (ROS) generated during metabolism and the inflammatory process can alter the biochemical structure of nucleic acids. Exogenous factors influencing the likelihood of replication errors include UV rays, which alter the structure of nucleic acids and can lead to the formation of pyrimidine dimers. Moreover, ionizing radiation, such as X-rays and γ-rays, can cause both single-strand and double-strand DNA breaks, whereas exposure to alkylating agents can lead to the formation of DNA adducts and DNA crosslinks [1].
To correctly maintain the fidelity of the genetic code, cells have developed sophisticated methods to sense and repair DNA replication errors. When those errors cannot be repaired, mechanisms are put in place to force the cell to undergo senescence and/or eliminate the cell through apoptosis. However, the disruption of the DNA damage response (DDR), resulting in the escape of death/senescence or uncontrolled cell proliferation with DNA replication errors, leads to genomic instability, one of the hallmarks of cancer [2].
2. DNA Damage Response (DDR)
More than 400 proteins and multiple pathways are involved in the regulatory machinery that constitutes the DDR [3]. The main DDR pathways are: base excision repair (BER), nucleotide excision repair (NER), mismatch repair (MMR), homologous recombination (HR), and non-homologous end joining (NHEJ). Although a subset of genes operates within a single pathway, the different DDR pathways are intertwined, and several genes are involved in the correct functioning of multiple pathways [4].
2.1. Base Damage and DNA Single-Strand Breaks
Alterations that change or remove a single base, such as UV-induced cytosine deamination, are addressed by DNA glycosylases belonging to the BER pathway [5]. After the altered base is removed by DNA glycosylases, specific endonucleases, such as Ape1, introduce DNA single-strand (ssDNA) breaks, which are then repaired by DNA polymerase β (POLβ) and XRCC1-DNA ligase IIIa, recruited to the ssDNA break site by the poly(ADP-ribose) polymerase inhibitor (PARP) 1 (PARP1) [6].
Another major DNA excision repair is NER, which removes a broad spectrum of single-strand lesions that impair correct DNA coiling [7]. Unlike BER, NER consists of the removal of an oligonucleotide, followed by the repair of the excision using the opposite DNA strand as a template [8].
DNA base–base mismatches and insertion–deletion loops (IDL) can be generated during DNA replication. These types of DNA errors are identified and repaired by the MMR pathway [9]. The main MMR genes frequently altered in cancer are: MLH1, MSH2, MSH6, and PMS2 genes. The MMR is identified and initiated by the MSH2/MSH6 heterodimer (mutSα), and then completed by another heterodimer (mutLα) formed by MLH1 and PMS2 [10].
In addition to the editing of mismatched base pairs, MMR genes can also regulate the HR pathway, in order to maintain the correct functioning of DNA double-strand (dsDNA) break repair [11].
2.2. DNA Double-Strand Breaks
dsDNA breaks are the most severe form of DNA damage, resulting in DNA replication arrest if not repaired [12]. HR is a high-fidelity pathway involved in the restoration of dsDNA breaks [13]. In HR, DNA damage is sensed by ATM, which activates several proteins, including BRCA1 and BRCA2, after which DNA ends are resected from 5′ to 3′ by the MRN complex (formed by the proteins RAD50, MRE11, and NBS) [14]. An array of other molecules, including the RAD51 paralogs, invade with the stranded filaments the sister chromatid, which is then used as a template by DNA polymerases to elongate the stranded filaments [15].
An alternative dsDNA repair is carried out through the NHEJ pathway, which does not require an intact template [16]. Briefly, through the interaction of the kinase proteins Ku70 and Ku80, DNA-PKcs, and ATM, and the MRN complex, the stranded ends are cleaved to a lesser extent than that of HR and are then ligated together by specific ligases. Although this mechanism repairs dsDNA lesions, it is more error-prone, as it results in deletions with the consequent loss of genetic information [17].
3. Harnessing DDR Deficiency for Cancer Treatment
With the advent of targeted therapies, DDR genes, frequently altered in cancer, have been studied to implement and personalize cancer medical treatment [18].
The rationale of using DDR-targeting agents is to induce cell death through synthetic lethality by blocking a complementary pathway in cells lacking one DDR pathway [19]. Specifically, poly(ADP-ribose) polymerase 1 and 2 (PARP1 and PARP2) enzymes are essential for the normal functioning of BES and its blockade results in the lack of repair of single-strand DNA breaks, with the consequent increased number of errors leading to DNA double-strand breaks that, in the absence of the HR pathway in BRCA1/2 deficient cells, can only be repaired by error-prone mechanisms, such as NHEJ [20]. Moreover, several PARP-inhibitors (PARP-i) also cause an entrapment of PARP at the replication fork, which becomes stalled, and cannot be restarted unless the HR pathway is functioning. There is a growing amount of evidence that this latter mechanism plays a major role in cell death by PARP-i molecules [21,22]
Starting from BRCA1/2 studies in breast and ovarian cancer [23,24], DDR-targeting drugs are being studied in other neoplasms with a deficiency of BRCA1/2 or other HR genes, especially pancreatic cancer [25] and castration-resistant prostate cancer [26]. Indeed, clinical data on olaparib, the first PARP-i approved, in pancreatic and prostate cancer were published for the first time in 2014, and even though the size of the study cohort was small (23 pancreatic and 8 prostate), these data led to further clinical investigations [27]. Following the success of PARP-i, novel molecules targeting other DDR genes and pathways are being studied.
4. Pancreatic Cancer and Prostate Cancer
Exocrine pancreatic cancer is one of the most lethal malignancies, being the fourth cause of death by cancer considering both sexes together, and predicted to be the second by 2030, with a survival rate at five years from diagnosis lower than 10% [28]. Pancreatic adenocarcinoma, in particular ductal adenocarcinoma, is the most frequent form of pancreatic cancer, constituting more than 85% of all pancreatic cancer cases [29].
Traditional chemotherapy and radiotherapy regimens can hardly overcome the aggressiveness of this disease, and do not guarantee the same response in different treated patients. Therefore, research is ongoing to identify potentially actionable genes and pathways involved in the genesis and progression of this disease, to improve and personalize pancreatic cancer medical treatment [30].
A subset of 5 to 15% of individuals who develop pancreatic cancer are either younger than expected for this malignancy or have a positive family history of pancreatic cancer and/or multi-tumor syndromes [31]. A germline pathogenic variant in a known pancreatic cancer predisposition gene can be found in less than 20% of these patients, depending on selection criteria and genes tested.
The recent literature shows that genes associated with breast and ovarian cancer risk are also the most strongly associated with pancreatic cancer risk, with the exception of CDKN2A in some populations [32,33,34]. For example, the DDR genes BRCA1, BRCA2 and ATM, or PALB2, each one usually found mutated in no more than 3.5% of cases, increase pancreatic cancer risk when altered at the germline level [35]. Based on the available literature, it is estimated that 17 to 25% of pancreatic cancer harbor somatic PVs in one of the genes involved in DDR, mainly those implicated in homologous recombination DNA damage response and repair (HR), such as BRCA1, BRCA2, ATM, PALB2, ATRX, and RAD51 [36,37,38,39,40,41,42,43,44].
Prostate cancer is the second most frequent malignancy in males worldwide (the first in western countries and in most African countries), representing 14.3% of all new cancers in males 2020, and the fifth cause of death by cancer in this population [28]. Although the majority of prostate cancers are low risk and/or diagnosed at an early stage, a subset of them displays an aggressive behavior. The initial medical approach to prostate cancer is based on the use of androgen-blocking agents, but a high proportion of metastatic prostate cancers tend to rapidly develop resistance to androgen-blocking agents. Metastatic castration-resistant prostate cancer (mCRPC) patients have a dismal prognosis, as median survival does not exceed two years [45].
The majority of mCRPC samples harbor clinically actionable molecular alterations. With regard to DDR genes, somatic mutations are found in around 23% of mCRPC, and up to 10% of individuals diagnosed with metastatic prostate cancer harbor a germline mutation [46,47]. Of the latter, more than half show loss of heterozygosity in the tumor [48].
In this review, we provide an overview on the DDR genes altered in both pancreatic and prostate cancers. Considering that the mutation rates of DDR genes vary considerably across different studies, and that differences in size cohorts and DNA sequencing methods are likely to be among the reasons of these discrepancies, we only considered original papers, reviews, and systematic reviews/meta-analyses involving at least 200 cases analyzed through multi-gene panel, exome, or genome sequencing, and we set a cut-off of at least 0.2% for reporting mutation rates.
In addition, we summarize the clinical implications of targeting these genes in the context of currently ongoing clinical trials.
5. Potentially Actionable DDR Genes in Common between Pancreatic Adenocarcinoma and Castration-Resistant Prostate Cancer (mCRPC)
5.1. BRCA1 and BRCA2
Identified in 1994 and 1995 by positional cloning [47], BRCA1 and BRCA2 are two of the main genes that control chromosomal stability. Indeed, upon phosphorylation by protein kinases, such as ATM, ATR, and CHK2, BRCA1 and BRCA2 become part of the macromolecular complexes necessary to repair DNA double-strand breaks through HR [49]. BRCA1- and BRCA2-deficient cells, lacking both copies of either of the two genes, show a high rate of mutations in multiple genes, including gatekeeper genes, such as CDKN2A, a phenomenon that can lead to neoplastic degeneration.
Germline biallelic pathogenic variants (PVs) in BRCA1 or BRCA2 result in different forms of Fanconi Anemia (FA), a syndrome characterized by short stature, multi-organ malformations, neurodevelopmental disorders, and cancer susceptibility [50]. On the other hand, the inheritance of a single allele with a PV predispose to several types of cancers following a second hit, including pancreatic cancer [51]. BRCA1 and BRCA2 PVs can be found in up to nearly 5% of the primary tumors of pancreatic cancer, with the highest frequencies in cohorts enriched for high-risk pancreatic cancer cohorts enriched for familiar cases [52].
In prostate cancer, BRCA1/2 mutations represent around 13% of the of DDR genes alterations in tumor samples and 51% of all germline variants found in individuals affected by prostate cancer, with BRCA2 harboring the majority of variants [46].
Germline BRCA1 and BRCA2 PVs increase the developing prostate cancer with a higher likelihood of aggressive disease for BRCA2 PV carriers [53,54,55,56]. Overall, BRCA1/2 PVs can be found in both germline and somatic samples from pancreatic cancer patients at similar rates. Conversely, BRCA1/2 PVs in prostate cancer occur more frequently as a somatic hit.
5.2. ATM
Ataxia–Telangiectasia mutated (ATM) is a large (351KD) PI3/Pi4 kinase with pleiotropic functions. In addition to playing a key role in HR by activating BRCA1 and BRCA2, ATM is involved in DNA double-strand break repair via NHEJ. Moreover, ATM is essential for the correct maturation of lymphocytes [57] and the central nervous system [58]. Indeed, the carriers of biallelic ATM PVs are affected by ataxia–telangiectasia, a rare syndrome characterized by progressive cerebellar ataxia, skin telangiectasias, an increased susceptibility to hematologic and solid tumors, and immunodeficiency. Heterozygous carriers of ATM PVs are at increased risk of several types of cancer, including pancreatic cancer, and it is estimated that up to 3% of high-risk individuals who develop pancreatic cancer harbor an ATM PV [25,35,59,60].
ATM has also been proposed as a prostate cancer predisposition gene and has been found altered at the germline level in both prostate cancer patients with suspected familial cancer syndromes and in apparently sporadic prostate cancer patients [48,61,62]
Somatic ATM loss rate can also happen in sporadic pancreatic cancer and prostate cancer [25,36,37,61,63,64]. Indeed, results from a large pan-cancer WGS study include ATM among the top 26 driver genes in these cancers, being found altered in 9 and 7% of pancreatic cancer and prostate cancer samples, respectively [63].
5.3. ATR
The Ataxia–Telangiectasia and Rad3-Related Protein (ATR) encodes for a serine-threonine kinase that acts as a DNA stress sensor. Specifically, in the presence of DNA ionizing and UV radiation and other genotoxic stressors, as well as in the case of the stalling of the replication fork, ATR activates checkpoint inhibitors, such as CHK1, to arrest cell cycle. Moreover, in cells lacking ATR, the inhibition of ATM or proteins in the ATM pathway is synthetically lethal [65]. The rates of ATR PVs, higher in at least 0.2% of the analyzed samples, have been found only as a germline event in pancreatic cancer and prostate cancer patients [35,48].
5.4. BRIP1
BRCA1-interacting protein (BRIP1) is a helicase that interacts with BRCA1 and promotes its DNA repair activity. It is also known as FANCJ, as it is part of the FA complex J [66]. Similar to BRCA1/2, biallelic germline PVs in the BRIP1 gene are found in children with FA syndrome. Germline monoallelic PVs increase the risk of cancer and are found in up to 1% of pancreatic cancer patients. [35,60]. A similar rate can also be found in sporadic pancreatic cancer samples [36,37]. In prostate cancer, BRIP1 PVs are less frequent, and rates higher that 0.2% are exclusively of germline origin [61,62].
5.5. CHEK1 and CHEK2
Checkpoint Kinase 1 (CHEK1) and Checkpoint Kinase 2 (CHEK2) encode for two serine-threonine kinases (CHK1 and CHK2, respectively) that are effector kinases acting downstream of ATM and ATR and are involved in cell cycle arrest following DNA damage [67].
ssDNA breaks as well as ssDNA generated following DSB resection in dsDNA break repair or during stalled replication fork ultimately result in the activation of the ATR/ATRIP complex, with subsequent CHK1 phosphorylation [68]. CHK2, on the other hand, is activated through phosphorylation by ATM following dsDNA breaks [67]. Both proteins activate signaling networks, leading to cell cycle arrest [67].
Germline PVs in CHEK2 increase the risk of breast cancer, albeit the penetrance has not yet been defined [69]. Moreover, germline PVs in both genes have been found in several other cancers, but their role those cancers is still under investigation [70,71,72,73].
PVs in one or both of those genes are more frequent at the germline level, both in pancreatic cancer and prostate cancer. In the latter, germline CHEK1/CHEK2 PVs have been found in up to 4.1% of affected individuals [48,61].
5.6. FANCA
The FA Complementation Group A (FANCA) gene is the main causative gene of FA, being altered at the germline levels in at least 60% of the affected children [74]. FANCA is part of the FA core complex, a macromolecular structure with ubiquitin-ligase functions belonging to the FA pathway, involved in DNA crosslinks repair and signaling upon replication stress, and acts in close interaction with BRCA1/2 and RAD51 to protect the replication fork from stalling [75,76].
FANCA has been associated with pancreatic cancer, being among those genes with germline and PV rates higher than 1% [35]. At the somatic level, FANCA has been found mutated in both cancers, with a higher prevalence of PVs in prostate cancer.
5.7. Mismatch Repair (MMR) Genes
The mismatch genes are MLH1, MSH2, MSH6, and PMS2. Germline PVs in MMR genes predispose to Lynch syndrome, which is characterized by a higher risk of developing non-polyposis-associated colorectal cancer (HNPCC) as well as extracolonic neoplasms, including pancreatic cancer and prostate cancer [77], whereas microsatellite instability (MSI) due to somatic impairment of MMR can be found in sporadic colorectal cancer. Recently, MMR genes have been implicated in the development of pancreatic and prostate cancers. Indeed, individuals with Lynch syndrome have a higher risk of developing both pancreatic and prostate cancers [78,79].
Germline PVs in MMR genes have been described in both pancreatic cancer and mCRPC unselected for family history [35,60]. MMR PVs are also present in 0.8% of pancreatic cancer samples and in up to 3% of prostate cancer samples [36,61].
5.8. NBN
The Nijmegen Breakage Syndrome 1 gene, also called Nibrin (NBN), is part of the NBN-MRE11-RAD50 complex. Upon activation by dsDNA breaks, the MRN complex participates in both HR and NHEJ [80]. The biallelic absence of NBN characterizes the Nijmegen Breakage Syndrome, a recessive disorder that includes intrauterine growth restriction, microcephaly, increased susceptibility to upper and lower airway infections, and several types of cancer [81,82].
NBN germline PVs range from 0.21% in unselected pancreatic cancer patients to 0.59% in familial pancreatic cancer patients [35], as well as in 2% of prostate cancer patients unselected for family history [48]. Conversely, NBN somatic variants are not found at rates higher than 2% in either of the two cancers.
5.9. PALB2
Partner and Localizer of BRCA2 (PALB2), also known as FANCN, is a moderate-risk breast cancer susceptibility gene involved in both to the FA pathway. Moreover, PALB2 binds BRCA1 and BRCA2, forming a complex necessary for HR [83]. Biallelic loss-of-function PALB2 variants cause a form of Fanconi anemia. PALB2 somatic PV occur in both pancreatic cancer and prostate cancer samples, whereas germline PV have been described in pancreatic cancer patients, at a slightly higher rate in high-risk compared to apparently sporadic patients (0.97% and 0.1–0.65%, respectively) [35,59].
5.10. RAD51 Paralogs
The RAD51 paralogs are: RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3. Each of these proteins work in an intertwined way with the others, forming macromolecule complexes essential for HR.
Considering that cells with null RAD51 paralog genes show deficient HR, tumors that lack at least one of those genes could be considered targets for therapy with PARP-I and other drugs directed at HR. Indeed, human cell lines deficient for RAD51 paralogs, with the exception of RAD51B, show marked genomic instability and sensitivity to both mytomicin C and olaparib [84].
Of all RAD51 paralogs, the germline RAD51C and RAD51D variants can be found in both pancreatic cancer and prostate cancer, at a rate lower than 0.5%. Only in prostate cancer, however, RAD51D is altered in up to 4% of somatic samples.
5.11. The Burden of DDR Deficiency in Pancreatic and Prostate Adenocarcinomas
Overall, genomic aberrations in 11 non-BRCA DDR genes are shared by pancreatic cancer and prostate cancer. PVs in those genes are found in up to 16% of germline pancreatic cancer samples. The addition of non-BRCA DDR genes triplicates the mutational burden given by BRCA1 and BRCA2 alone. When looking at somatic PVs, the scenario is comparable, as the highest rate of PVs found in the literature increases from 4.8% to 18.5% when adding other DDR genes to BRCA1/2. As for prostate cancer, the addition of the genes included in this review to BRCA1 and BRCA2 more than doubles the mutational burden in both germline and in somatic sample PVs (8.6% to 20.7% and 15.2% to 36.9%).
Overall, PVs in both BRCA1/2 and in non-BRCA genes grouped together are slightly more frequent at the somatic level in pancreatic cancer, whereas in prostate cancer, somatic PVs are twice as many as germline PVs.
In fact, there is a striking difference between the two types of tumors in what concerns the rate of somatic PVs, which are almost doubled in prostate cancer compared to pancreatic cancer (36.94% vs. 18.5%). A detailed overview of the mutation rates of the above-mentioned 11 genes is shown in Table 1, and their role in DDR pathways is summarized in Figure 1.
Table 1.
Frequency of the pathogenic variants in DDR genes shared by pancreatic and prostate cancers.
| Pancreatic Cancer | Prostate Cancer | |||||||
|---|---|---|---|---|---|---|---|---|
| Germline [25,35,59,60] | Somatic [25,36,37,63] | Germline [48,61,62] | Somatic [61,63,64] | |||||
| Gene | Range | Max | Range | Max | Range | Max | Range 1 | Max |
| BRCA1/2 | 0.9–5 | 5 | 0.9–4.8 | 4.80 | 0.3–8.6 | 8.60 | 0.6–15.2 | 15.2 |
| ATM | 2–3.09 | 2.2–9 | 1.59–2.3 | 1.9–7.3 | ||||
| ATR | 0.2 | 0.29 | ||||||
| BRIP1 | 0.22–1 | 0.48–1 | 0.28–0.45 | |||||
| CHEK1-2 | 0.3–2.2 | 0.2–0.6 | 1.87–4.1 | 0.9–1.9 | ||||
| FANCA | 1.04 | 0.6 | 3 | |||||
| MMR | 0.39–1.2 | 0.8 | 0.14–1.7 | 3–6 | ||||
| NBN | 0.2–0.59 | 0.29–2 | ||||||
| PALB2 | 0.1–0.97 | 0.2–1.2 | 0.45–0.56 | 0.4–2 | ||||
| RAD50 | 0.3–0.36 | 0.5 | 1 | |||||
| RAD51 | 0.35 2 | 0.14–0.57 2,3 | 0.54 2,3 | |||||
| Non-BRCA1/2 | 11 | 13.7 | 12.07 | 21.74 | ||||
| TOTAL | 16 | 18.5 | 20.67 | 36.94 | ||||
Frequencies are reported as percentages. 1 mCRPC, 2 RAD51C, 3 RAD51D.
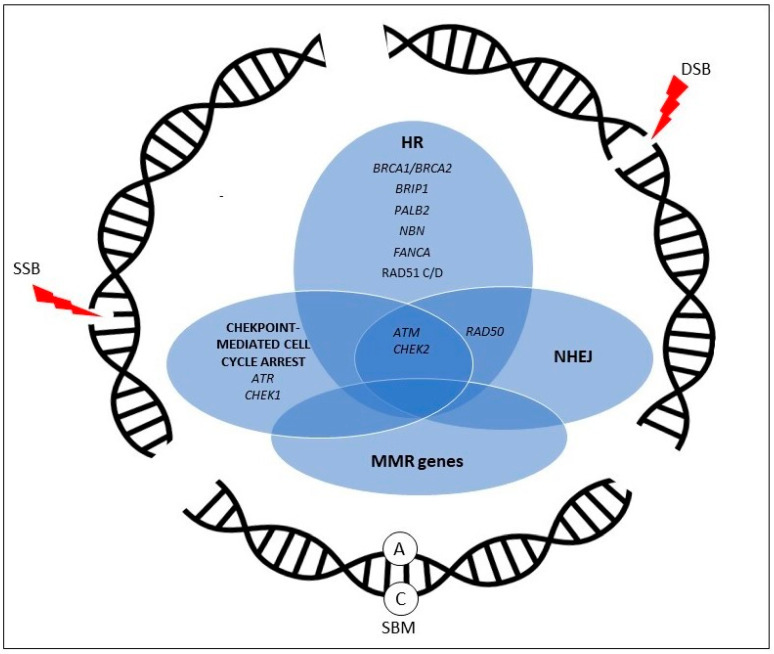
Figure 1.
Overview of DNA damage response pathways with genes altered in both pancreatic and prostate cancer. HR = homologous recombination, NHEJ = nonhomologous end-joining, MMR = mismatch repair, SBM = single-base mismatch.
6. DDR and Cancer Treatment in Pancreatic Adenocarcinomas
6.1. DDR Pathogenic Variants in Pancreatic Cancer
DDR gene alterations are correlated with increased overall survival (OS) (17.9 versus 9.6 months, p = 0.03) compared to patients without a DDR gene alteration [85]. Indeed, in a recent article, the median overall survival (mOS) in patients with ATM alterations was 40.2 months compared with 15.5 months in the control population (hazard ratio (HR) = 0.14, 95% confidence interval (CI) = 0.04 to 0.47, 2-sided p = 0.001). These findings suggest that pathogenic ATM alterations may be prognostic for improved outcomes in patients with pancreatic cancer [86]. Interestingly, DDR mutations seem to correlate with a significantly longer OS in patients treated with 5-Fluorouracile, irinotecan, and oxaliplatin (FOLFIRINOX) compared to patients without mutations in DDR genes [87]. The success of novel target therapies with PARP1-i in pancreatic cancer has prompted researchers to explore to a greater extent the role of DDR in pancreatic cancer genesis and progression, in order to broaden the set of patients who could benefit from those therapies and also identify potential targets for the development of other targeted therapies focused on DDR deficiency.
In this review, we report the different mutations in DDR pathways currently investigated in pancreatic cancer patients as promising targetable alterations.
6.2. Clinical Trials Results in Pancreatic Cancer
Ongoing clinical trials in pancreatic cancer are reported in Table 2.
Table 2.
Ongoing clinical trials on PARP-i in pancreatic cancer.
| PARP-i | Clinical Trial | Phase | Patient Population | Somatic Mutations/Germline PVs | Treatment Arm(s) |
|---|---|---|---|---|---|
| Olaparib | NCT04548752 | 2 | Pancreatic cancer | BRCA1/2 | Olaparib + pembrolizumab |
| NCT04005690 | 1 | Pancreatic cancer | nd | Olaprib + Cobimetinib | |
| NCT02498613 | 2 | Advanced Solid Tumors | nd | Olaparib + cediranib | |
| NCT03162627 | 1 | Solid Tumors | nd | Olaparib + selumetinib | |
| NCT03842228 | 1 | Advanced Solid Tumors | ARID1A, ATM, ATRX, BARD1, BRCA1, BRCA2, BRIP1, CDK12, CHEK1, CHEK2, FANCA, FANCL, MRE11A, MSH2, PALB2, PARP1, POLD1, PP2R2A, RAD51B, RAD51C, RAD51D, RAD54L, XRCC2, PTEN, PIK3CA | Olaparib + durvalumab + Copalinsib | |
| NCT02511223 | 2 | Pancreatic cancer | BRCAness | Olaparib alone | |
| NCT02677038 | 2 | Pancreatic cancer | Somatic BRCA mutation, Fanconi anemia genes, ATM or RAD51 mutations | Olaparib alone | |
| NCT02576444 | 2 | Solid tumors | ATM, CHK2, MRN (MRE11/NBS1/RAD50), CDKN2A/B, APOBEC, IDH1/IDH2, TP52, KRAS, PTEN, PIK3CA, AKT, or ARID1A | Olaparib + AZD1775 OR AZD5363 OR AZD6738 | |
| Rucaparib | NCT03140670 | 2 | Pancreatic cancer | BRCA 1/2 or PALB2 | Maintenance after platino-based chemo |
| NCT03337087 | 1–2 | Pancreatic, colorectal, gastroesophageal, or biliary cancer | BRCA 1/2 or PALB2 | Liposomal Irinotecan, Fluorouracil, Leucovorin Calcium, and Rucaparib | |
| NCT04171700 | 2 | Solid Tumors | BRCA1, BRCA2, PALB2, RAD51C, RAD51D, BARD1, BRIP1, FANCA, NBN, RAD51 or RAD51B. | Rucaparib alone | |
| Veliparib | NCT02890355 | 2 | Pancreatic cancer | nd | FOLFIRI or mFOLFIRI + Veliparib as II Line |
| NCT01585805 | 2 | Pancreatic cancer | BRCA1/2 or PALB2 Germline PV | Gemcitabine + Cisplatin with or without veliparib or veliparib wlone | |
| NCT02723864 | 1 | Solid tumors | nd | M6620 (ATR inhibitor) + veliparib + cisplatin | |
| Niraparib | NCT03601923 | 2 | Pancreatic cancer | Germline PVs or somatic mutation of one of these: BRCA1/2, PALB2, CHECK2 or ATM | Niraparib alone |
| NCT04409002 | 2 | Pancreatic cancer | nd | Niraparib + Dostarlimab + RT | |
| NCT03553004 | 2 | Pancreatic cancer | DDR family mutation | Niraparib alone | |
| NCT04493060 | 2 | Pancreatic cancer | BRCA1/2 or PALB2 | Niraparib + Dostarlimab | |
| NCT03404960 | 1–2 | Pancreatic cancer | nd | Niraparib + Nivolumab or Ipilimumab after platinum-based chemotherapy | |
| NCT03207347 | 2 | Solid tumors | ARID1A, ATM, ATR, BACH1 (BRIP1), BAP1, BARD1, BLM, CHEK1, CHEK2, CDK2, CDK4, ERCC, FAM175A, FEN1, IDH1, IDH2, MRE11A, NBN (NBS1), PALB2, POLD1, PRKDC (DNA-PK) PTEN, RAD50, RAD51, RAD52, RAD54, RPA1, SLX4, WRN, or XRCC | Niraparib | |
| NCT03209401 | 1 | Solid tumors | ARID1A, ATM, ATRX, MRE11A, NBN, PTEN, RAD50/51/51B, BARD1, BLM, BRCA1, BRCA2, BRIP1, FANCA/C/D2/E/F/G/L, PALB2, WRN, CHEK2, CHEK1, BAP1, FAM175A, SLX4, MLL2 or XRCC | Niraparib + carboplatin | |
| Talazoparib | NCT02286687 | 2 | Solid tumors | Somatic BRCA1 or BRCA2; germline BRCA, ATM, PALB2, Fanconi Anemia genes, ARID1A, MER11, RAD50, NBS1, ATR; amplification of EMSY | Talazoparib |
| NCT03565991 | 2 | Solid tumors | ATM or BRCA | Avelumab and talazoparib | |
| Fluzoparib | NCT04300114 | 3 | Pancreatic cancer | Germline BRCA/PALB2 | Maintenance after platinum |
| Prexasertib | NCT02873975 | 2 | Solid tumors | MYC amplification, Rb loss, FBXW7 mutation, BRCA1, BRCA2, PALB2, RAD51C, RAD51D, ATR, ATM, CHK2, the Fanconi anemia pathway genes, CCNE1 amplification of 6-fold or greater, or other genomic or somatic mutation in a known HR gene | Prexasertib (CHK inhibitor) |
| NCT03057145 | 1 | Solid tumors | nd | Prexasertib + olaparib | |
| BTT-114 | NCT02950064 | 1 | Pancreatic, breast, ovarian, or prostate cancer | BRCA or other DNA repair mutations, such as ATM, CHEK2, PALB2, and RAD51D | BTT-114, a novel platino product |
| ABT-144 | NCT01489865 | 1 | Pancreatic cancer | BRCA1/2 or PALB2 or FANC mutation or family history | ABT-144 + mFOLFOX6 |
| AZD0156 | NCT02588105 | 1 | Solid tumors | nd | (ATM/ATR inhibitor) Alone or in combination |
| M6620 (VX-970) | NCT02595931 | 1 | Solid tumors | nd | M6620 + irinotecan |
| AZD6738 | NCT03682289 | 2 | Renal, urothelial or pancreatic cancer | ATM or ARID1A | AZD6738 (ATR inhibitor) +/− olaparib |
| NCT02223923 | 1 | Solid tumor | nd | AZD6738 (ATR inhibitor) + radiotherapy | |
| NCT02630199 | 1 | Solid tumors | nd | AZD6738 (ATR inhibitor) + paclitaxel | |
| NCT03669601 | 1 | Solid tumor | nd | AZD6738 (ATR inhibitor) + Gemcitabine | |
| Ceralasertib | NCT02264678 | 1–2 | Solid tumors | ATM and BRCA evaluation | Ceralasertib +/− other drugs |
| BAY1895344 | NCT03188965 | 1 | Solid tumors | ATM or other DDR defects | BAY1895344 (ATR inhibitor) |
| NCT04514497 | 1 | Solid tumors | ATM and other DDR defects | BAY1895344 (ATR inhibitor) + irinotecan |
The POLO study is a phase III clinical trial conducted on BRCA1/2 mutated metastatic pancreatic cancer patients treated with olaparib vs. placebo as a maintenance therapy after a platinum-based chemotherapy. In this study, 3315 patients were screened, 154 underwent a 3:2 ratio randomization, and 92 received olaparib. The progression free survival (PFS) was longer in the olaparib group (7.4 vs. 3.8 months; HR = 0.53) [88]. Results from the POLO trial led to olaparib being approved by the US Food and Drug Administration (FDA) and the European Medicines Agency (EMA) in pancreatic cancer with germline BRCA1/2 PV, as maintenance after a platinum-based chemotherapy. Updated results in 2021 showed that mOS was similar between olaparib and placebo, although this is probably due to 29% of the crossover of patients from placebo to PARP inhibitor upon progression. More importantly, the OS rate at 36 months was 33.9% for olaparib and 17.8% for placebo, which are impressive results considering the poor prognosis of pancreatic cancer patients [89].
Another PARP-i that has been investigated in pancreatic cancer patients harboring DDR gene mutations is Veliparib. In a phase I trial, Veliparib, in association with gemcitabine and cisplatin as first-line treatment in both BRCA1/2 germline mutated and wild-type patients, showed a good safety profile, although the clinical response was reported exclusively in the mutated group [90]. On the other hand, a phase II trial involving BRCA1/2 or PALB2 mutated pancreatic cancer patients did not demonstrate a benefit in response rate adding veliparib to Cisplatin + Gemcitabine treatment [91]. However, this trial established cisplatin + gemcitabine as a possible new standard treatment in patients harboring BRCA1/2 or PALB2 mutation [91].
Moreover, a phase II trial tested Veliparib in previously treated BRCA1/2 mutated pancreatic cancer patients. The trial did not show an objective response rate (ORR), although 25% of patients were stable for 4 months [92]
An ongoing clinical phase II clinical trial (NCT02890355) is testing 5-fluorouracile + irinotecan (FOLFIRI) + veliparib as second-line treatment in pancreatic cancer patients with or without BRCA1/2 mutation. Preliminary results showed no difference in OS between the FOLFIRI group and FOLFIRI + veliparib [93].
In a phase I/II trial, veliparib in association with 5-fluorouracile and oxaliplatin showed promising results in terms of ORR in patients harboring a DDR mutation with a good safety profile [94]. Moreover, veliparib in combination with gemcitabine and radiotherapy demonstrated high tolerability and better mOSin in patients with a DDR pathway alteration compared to patients without DDR mutations [95].
Talazoparib is a new promising PARP-i. In vitro experiments demonstrated that talazoparib selectively targets tumor cells with BRCA1/2 or PTEN mutations with 20- to more than 200-fold greater efficacy than the old generation of PARP-i [96]. A phase I clinical trial of talazoparib in different BRCA1/2-mutated tumors showed a good safety profile and promising antitumor activity [97]. Phase II clinical trials in solid tumors are currently ongoing (see Table 2).
The phase II RUCAPANC trial enrolled pancreatic cancer patients with a BRCA1/2 PV (either germline or somatic) to receive rucaparib. The disease control rate was 31.6% (6 out of 19 patients); hence, the insufficient response rate prompted the closure of the study [98].
Recently, Pishivain et al. published data on 1028 pancreatic cancer patients, 189 of whom harbored an actionable mutation. Of these, 46 (24%) received a molecularly matched therapy [99]. The most common pathway mutated was the DDR (94 of 189 patients). In a subgroup analysis on patients harboring a DDR mutation, 27 received a matched therapy and 67 received an unmatched one. In the subgroup treated with target therapy (PARP-i or ATR inhibitor), the mOS was significantly longer. Similarly, the mOS in patients with an actionable non-DDR mutation was longer in the group treated with the matched therapy [99]. This is a key study to understand the clinical role of using matched therapy in mutated pancreatic cancer patients. According to this study, harboring an actionable alteration and receiving a molecularly matched therapy can predict treatment response and improve mOS compared to receiving a non-matched therapy.
An interesting aspect is the use of HRDness inducers, which can create artificial vulnerabilities allowing the use of PARP-i in patients without BRCA mutation, thus improving the number of patients who can benefit from this therapy [100].
Several ongoing trials are exploring the use of PARP inhibitors in PDAC patients, both as a monotherapy and in combination with other treatments [101,102].
6.3. Beyond BRCA
Two phase II ongoing trials are evaluating olaparib in patients with a negative BRCA germline mutation, a tumor with a BRCAness phenotype, and a family history of BRCA-related cancers, as a second or further line of therapy. Both studies (NCT02677038, NCT02511223) have shown encouraging results, although caution must be taken due to the small sample size (21 and 11 patients); hence, further studies are required [103].
The ataxia–telangiectasia mutated (ATM) gene plays an important role in the DDR. Preclinical data in ATM-deficient mouse model showed efficacy of PARP-i and ATM-inhibitors (ATM-i) [104]. Different ATM-i have been developed in recent years: KU55933 and KU60019 have shown to be potent radiosensitizers, while AZ31 improved the efficacy of irinotecan therapy [105]. The aToM study is a phase I trial evaluating the safety and efficacy of the ATM-i AZD0156 at increasing doses alone or in combination with other anti-cancer treatments (olaparib or FOLFIRI schedule) in patients with advanced cancers, including pancreatic cancer. Preclinical data demonstrate that the combination of ATM-i with PARP-i enhances the activity of the latter improving DNA DSB and then cell apoptosis [106].
In two phase 2 nonrandomized clinical trials, olaparib was well tolerated and showed limited antitumor activity in patients with advanced, platinum-sensitive pancreatic cancer with alterations in DDR genes, including ATM [107].
Preclinical data showed the high activity of ATR-inhibitors (ATR-i) in tumors with a somatic mutation of the ATM pathway, since the ATM-deficient cells rely on the ATR pathway for survival [108,109]. Currently, there are no clinical data available on ATR-i, but several clinical trials are ongoing (see Table 2).
As reported in the previous chapters, checkpoint kinase 1 and 2 (CHK1/CHK2) are activated by ATR and ATM in response to DNA damage or stress [110]. A preclinical study on a CHK1 inhibitor in association with chemotherapy (gemcitabine) and radiotherapy demonstrated a synergic role in killing pancreatic tumor cells [111]. Furthermore, a CHK2 inhibitor has been tested in association with gemcitabine, demonstrating an increased apoptosis of pancreatic tumor cells [111]. No clinical data are available to date, yet CHK inhibitors are being investigated in some clinical trials (see Table 2).
PALB2 has a key role in orchestrating DNA repair, and it is strongly linked to the BRCA1/2, ATM, and ATR pathways [112]. Currently, some trials are evaluating the prognostic role of PALB2 in PARPi-treated patients with mutations in this gene (see Table 2). Nowadays, pancreatic cancer remains a tumor with a poor prognosis and the search for actionable mutations, in order to improve the outcome, is a very hot topic in both preclinical and clinical research.
However, although encouraging results are available in the preclinical setting, clinical data are needed to confirm and validate them.
In the past decade, several treatment strategies have been approved for mCRPC patients and, recently, a better understanding of the underlying biology of prostate cancer allowed researchers to identify and investigate novel therapeutic agents.
The DDR pathways are one of the main actionable molecular alterations of mCRPC, and the investigation of PARP-i has opened a new prospective in the advanced setting of prostate cancer [113].
7. DDR and Cancer Treatment in Prostate Adenocarcinomas
Prostate cancer is strongly driven by androgen receptors (ARs) at the beginning of the tumor natural history (“castration sensitive”), while the “castration-resistant” phase of prostate cancer is characterized by tumor heterogeneity caused by the onset of genomic and transcriptomic alterations [113].
HR genes (mainly BRCA1, BRCA2, and ATM), are present in up to 20% of mCRPC patients, at either germline or somatic levels [114].
Testing DDR gene mutations in prostate cancer is clinically relevant, due to their prognostic and predictive values [115]. In prostate cancer patients, harboring a DDR gene mutation, especially a BRCA2 mutation, is associated with a worse prognosis and a higher Gleason score and stage at diagnosis, as well as with an increased risk of developing distant metastases [116,117].
Moreover, DDR mutations were shown to be positive predictive markers of sensitivity to the platinum-based chemotherapy regimen and PARP-i response in different tumors, including breast, ovarian, and prostate cancers [118].
Due to the promising results of the use of PARP-i in ovarian or breast cancer patients harboring BRCA1/2 mutations, several studies have investigated the efficacy of PARP-i in DDR-mutated mCRPC patients, leading to FDA approval for some of these molecules (Table 3) [119,120].
Table 3.
Main clinical trials and results of PARP-i in mCRPC patients with a HR mutation.
| PARP-i | Clinical Trial | Phase and Study Type |
Patient Population | Treatment Arm | N pts | Results | Status in February 2022 |
|---|---|---|---|---|---|---|---|
| Olaparib | PROFOUND (NCT02987543) |
Phase 3, randomized | Progression to ≥1 novel HT 1 Cohort A: BRCA1m, BRCA2m, ATMm. Cohort B: other HR. |
Olaparib vs. Enzalutamide or Abiraterone acetate + prednisone |
Cohort A: 245 Cohort B: 142 |
Cohort A: Olaparib > Hormonal therapy in PFS = 7.4 vs. 3.6 mo, HR 0.34; p < 0.0001 OS = 18.5 vs. 15.1 mo, HR 0.64; p = 0.02 ORR = 33% vs. 2% Cohort A + B: Olaparib > Hormonal therapy in PFS = 5.8 vs. 3.5 mo, HR 0.49; p < 0.0001 OS = 17.5 vs. 14.3 mo, HR 0.67 ORR = 22% vs. 4% |
FDA-approved in May 2020 Active, not recruiting |
| KEYNOTE-365 (NCT02861573) | 1b-2, single arm |
mCRPC (molecularly unselected, docetaxel-pretreated) |
Pembrolizumab + Olaparib (Cohort A) |
102 | BRCA+ vs. BRCA - PSA response: 50% vs. 14% ORR: 33% vs. 6% HR+ vs. HR- PSA response: 22% vs. 13% ORR: 8% vs. 3% |
Active, recruiting |
|
| PROpel (NCT03732820) | Phase 3, randomized | mCRPC 1 L treatment after failure of ADT |
Olaparib + Abiraterone Acetate | 796 | rPFS: 24.8 vs. 16.6 months, HR 0.66, p < 0.0001 OS: HR 0.86 ORR: 58.4% vs. 48.1% |
Active, not recruiting |
|
| Rucaparib | TRITON2 (NCT02952534) |
Phase 2, single arm | Progression to 1–2 novel HT 1 AND 1 taxane-based CT |
Rucaparib | 115 BRCAm |
ORR IRR = 43.5% ORR IA = 50.8% PSA RR = 54.8% m-rPFS IRR = 9.0 mo m-rPFS IA = 8.5 mo 12-mo OS = 73.0% |
FDA-approved in May 2020. Completed |
| Talazoparib | TALAPRO-1 (NCT03148795) |
Phase 2, single arm | Progression to ≥1 novel HT AND 1–2 CT regimens (≥1 taxane-based CT) |
Talazoparib | 86 overall population 46 BRCA1/2m 4 PALB2m 18 ATMm |
ORR overall population = 28% ORR BRCA1/2m = 43.9% ORR PALB2m = 33.3% ORR ATMm = 11.8% m-rPFS BRCA1/2m = 9.3 mo m-rPFS PALB2m = 7.4 mo m-rPFS ATMm = 5.5 mo |
Active, not recruiting |
| Niraparib | GALAHAD (NCT02854436) |
Phase 2, single arm | Progression to ≥1 novel HT 1 AND ≥1 taxane-based CT |
Niraparib | 46 BRCA 1/2m 35 non-BRCAm |
BRCA1/2m vs. non-BRCAm ORR = 41% vs. 9% PSA RR = 50% vs. 3% m-rPFS = 8.2 vs. 5.3 mo mOS = 12.6 vs. 14 mo |
Active, not recruiting |
| MAGNITUDE (NCT03748641) | Phase 3, randomized | mCRPC 1 L treatment after failure of ADT |
Niraparib + Abiraterone Acetate | 423 HR patients |
BRCA1/2m vs. non-BRCAm rPFS: 16.6 vs. 10.9 mo, HR 0.53 ORR: 52% vs. 31% HR+ vs. HR- rPFS: 16.5 vs. 13.7 mo, HR 0.73 ORR: 60% vs. 28% |
Active, not recruiting |
HT: hormonal therapy; N: number; pts: patients; BRCA1m: BRCA1 mutation; BRCA2m: BRCA2 mutation; ATMm: ATM mutation; HR: homologous recombination DNA damage response and repair; CT: chemotherapy; PALB2m: PALB2 mutation; PFS: progression-free survival; mo: month; HR: hazard ratio; p: p value; OS: overall survival; ORR: overall response rate; IRR: independent radiology review; IA: investigator assessment; PSA RR: prostate-specific antigen response rate; m-rPFS: median radiological progression-free survival; 12-mo OS: overall survival at 12 months; FDA: Food and Drug Administration. 1 Novel hormonal therapy, e.g.; abiraterone acetate and/or enzalutamide.
In this review, we report the main clinical trials on the use of PARP-i in mCRPC.
7.1. Clinical Trials Results in Prostate Cancer
Several phase 2/3 trials have investigated the efficacy and safety of PARP-i in mCRPC patients (Table 3).
The PROFOUND phase 3 trial assessed olaparib compared with AR-directed therapy (enzalutamide or abiraterone acetate) in mCRPC patients with multiple loss-of-function DDR alterations who progressed to the new hormonal agents [121]. Cohort A included 245 patients with at least one mutation in BRCA1, BRCA2, or ATM, while cohort B included 142 patients who had a mutation in any of other 12 DDR genes. The somatic mutation status was evaluated with a tissue gene panel analysis. The primary endpoint was radiological PFS (rPFS) in cohort A and secondary endpoints included PFS in cohort B and ORR and OS in cohort A.
The analysis revealed that in cohort A olaparib significantly improved rPFS (7.4 vs. 3.6 months, hazard ratio of 0.34; 95% CI: 0.25–0.47, p < 0.0001), the OS of the interim analysis (18.5 vs. 15.1 months, hazard ratio of 0.64; 95% CI: 0.43–0.97, p = 0.02), and the objective response rate (ORR) (33% vs. 2%) compared to the hormonal therapy arm. Moreover, olaparib also improved rPFS, OS, and ORR in the overall population (cohorts A and B). This study confirmed the survival improvement and the clinical benefits of olaparib in mCRPC patients harboring DDR alterations opening the path for a new promising class. On the basis of these results, in May 2020, olaparib was approved by the FDA for germline or somatic HR gene-mutated mCRPC patients who progressed to enzalutamide or abiraterone.
The results of the final OS analysis have been subsequently reported [122], confirming the significantly longer OS in patients treated with olaparib in cohort A (19.1 vs. 14.7 months, hazard ratio of 0.69; 95% CI: 0.50–0.97; p = 0.02) than in cohort B (14.1 vs. 11.5 months) and overall population (17.3 vs. 14.0 months), despite the substantial crossover from the control therapy to olaparib (66%). Moreover, a sensitivity analysis adjusted for crossover to olaparib showed hazard ratios for death of 0.42 in cohort A, 0.83 in cohort B, and 0.55 in the overall population.
The phase 2 single-arm TRITON2 trial investigated rucaparib 600 mg twice daily in patients with mCRPC with germline or somatic HR gene alterations, detected on blood and/or tumor biopsy. Patients had to be progressed after one or two lines of next-generation AR-directed therapy and one taxane-based chemotherapy. The primary endpoint was ORR and PSA response rate (decrease ≥ 50% from baseline) and secondary endpoints included rPFS and OS.
The study demonstrated the efficacy of rucaparib in 115 mCRPC patients with a BRCA alteration, reporting confirmed ORRs per independent radiology review (IRR) and investigator assessment (IA) of 43.5% and 50.8%, respectively, and a PSA response rate of 54.8% [123]. No difference in ORR was seen in patients with a germline versus somatic BRCA alteration and in patients with a BRCA1 versus BRCA2 mutation, while a higher PSA response rate was observed BRCA2 mutation. (59.8% vs. 15.4%). Promising survival results were also reported: the median rPFS was 9.0 months per IRR assessment and 8.5 months per investigator assessment. Additionally, although OS data were not mature at the time of the analysis, the 12-month OS reported was 73.0%.
A subgroup analysis revealed that non-BRCA DDR gene alterations, including ATM, CDK12, or CHEK2 mutations, were associated with limited radiographic/PSA responses to rucaparib, while promising responses were observed with mutations in genes that directly interact with the BRCA complex (e.g., PALB2, BRIP1, FANCA, and RAD51B) [124].
On the basis of this study, in May 2020, rucaparib was approved by the FDA for germline or somatic BRCA-mutated mCRPC patients treated with AR-directed therapy and a taxane-based chemotherapy.
In addition to olaparib and rucaparib, other PARP-i have shown promising efficacy results and could be the next FDA-approved PARP-i.
The phase 2 TALAPRO-1 evaluated the efficacy and safety of talazoparib in 104 mCRCP patients with DRR gene defects and measurable disease who received one or two taxane-based chemotherapy regimens for metastatic disease and AR-directed therapy (enzalutamide or/and abiraterone), for metastatic castration-resistant prostate cancers [125]. The study showed that talazoparib was associated with an ORR (primary endpoint) of 30% in the overall population, with a greater antitumor activity in BRCA1/2-mutated patients than in those with PALB2 and ATM mutations (46% vs. 25% vs. 12%, respectively). Additionally, the rPFS was higher in BRCA1/2-mutated patients compared to the other DDR alterations (overall population: 5.6 months; BRCA1/2-mutated patients: 11.2 months; PALB2-mutated patients: 5.6 months; ATM-mutated patients: 3.5 months). The composite response rate (CRR) (ORR and/or ≥50% PSA decline and/or circulating tumor cell conversion) was 51% in the overall population and was similar in BRCA1/2-mutated and PALPB2-mutated patients, but lower in ATM-mutated patients (72%, 75%, and 24%, respectively).
These results showed that talazoparib has encouraging antitumor activity in heavily pretreated mCRPC patients with DDR–HRR gene alterations, especially those with BRCA1/2 mutations.
The phase 2 GALAHAD trial investigated the efficacy and safety of niraparib (300 mg daily) in mCRPC patients with DDR defects who progressed on AR-directed therapy and taxane-based chemotherapy [126]. The primary endpoint was ORR and secondary endpoints included PSA response, CRR, rPFS, and OS. In the final study analysis, 289 patients were included in the overall efficacy analysis population showing an ORR in the measurable BRCA cohort (n = 76) (primary endpoint) of 34.2%, a median rPFS of 8.08 months, and a mOS of 13.01 months. These results in the BRCA1/2-mutated patients were better than those observed in the measurable non-BRCA cohort (ORR 10.6%, median rPFS 3.71 months and mOS 9.63 months). Additionally, this study concluded that niraparib has encouraging antitumor activity in heavily pretreated mCRPC patients with DDR–HRR gene alterations, especially those with BRCA1/2 mutations.
At the ESMO Congress 2021, the biomarker analysis of cohort A of the phase 1b/2 KEYNOTE-365 trial on the combination of pembrolizumab + olaparib in molecularly unselected, docetaxel-pretreated mCRPC patients was presented [127]. The primary endpoints (PSA response and ORR) were higher in BRCA-mutated patients compared with patients without a BRCA mutation (50% vs. 14% and 33% vs. 6%, respectively) and in patients with an HR mutation compared with those without an HR mutation (22% vs. 13% and 8% vs. 3%, respectively). According to these results, the combination of pembrolizumab + olaparib have shown promising activity results, regardless of HR mutation status, even though higher response rates were observed in mutated patients.
Recently, two randomized, double-blind phase 3 studies on PARP-i (the PROpel trial and the MAGNITUDE trial) reported the first analyses at the ASCO Genitourinary Cancers Symposium of February 2022 [128,129]
The PROpel trial investigated the combination of abiraterone acetate plus olaparib versus abiraterone acetate + placebo as a first-line therapy of mCRPC patients [129]. The treatment with abiraterone acetate + olaparib significantly prolonged rPFS (primary endpoint) irrespective of the HRR status (24.8 vs. 16.6 months; hazard ratio of 0.66, 95% CI: 0.54–0.81; p < 0.0001). OS is currently immature, but a trend in OS favoring abiraterone acetate plus olaparib was observed (HR 0.86). The secondary endpoints of ORR (58.4% vs. 48.1%), time to first subsequent treatment (hazard ratio of 0.74), and time to second PFS (hazard ratio of 0.69) were supportive of activity and long-term benefits.
The MAGNITUDE trial analyzed the combination of abiraterone acetate plus niraparib versus abiraterone acetate plus placebo as a first-line therapy in mCRPC patients with and without HR [128]. Niraparib + abiraterone acetate did not show any benefit in terms of rPFS or biochemical PFS, and for this reason the accrual in this cohort of patients was interrupted. The combination of niraparib + abiraterone acetate significantly improved rPFS (primary endpoint) in the BRCA1/2 subgroup (hazard ratio of 0.53, 16.6 vs. 10.9 months) and in all HR+ patients (hazard ratio of 0.73, 16.5 vs. 13.7 months). The first interim analysis of OS is immature, but a trend in OS favoring abiraterone acetate + niraparib was observed (hazard ratio of 0.77). The advantage was also observed in terms of ORR, time to subsequent chemotherapy, and time to symptomatic and biochemical progression, in both BRCA1/2-mutated and HR+ patients.
7.2. Ongoing Clinical Trials
PARP-i represents a novel treatment option for mCRPC patients harboring HR mutations who progressed to chemotherapy and/or next generation AR-targeted therapy.
However, primary resistance to PARP-i may be present upfront and acquired resistance to them may occur, mainly related to the restoration of the HR mechanism. For this reason, ongoing clinical trials are currently investigating PARP-i in different treatment settings and in combination with different types of oncological therapies, including AR-direct therapy, immunotherapy, and tyrosine kinase inhibitor (TKI) [114,130] (Table 4).
Table 4.
Active clinical trials on PARP-i in prostate cancer.
| PARP-i | Clinical Trial | Phase | Patient Population | Somatic Mutations/Germline PVs | Treatment Arm(s) |
|---|---|---|---|---|---|
| Olaparib |
NCT03434158 (IMANOL) |
2 | mCRPC | BRCA1, BRCA2, ATM, FANC, CHEK2, MLH1, MSH2, MSH6, PMS2, PALB2, RAD51C, MRE11 | Olaparib |
| NCT03012321 | 2 | mCRPC |
BRCA1, BRCA2, ATM Other DDR mutations |
Abiraterone alone OR Olaparib alone OR Abiraterone + Olaparib |
|
| NCT03516812 | 2 | CRPC | MMR deficiency, HR deficiency |
Olaparib + Testosterone | |
| NCT03317392 | 1–2 | mCRPC | Not required | Olaparib + Radium 233 | |
|
NCT03834519 (KEYLYNK-010) |
3 | mCRPC | Not available | Pembrolizumab + Olaparib Vs Abiraterone OR Enzalutamide |
|
| NCT03432897 | 2 | Locally advanced Prostate cancer |
BRCA1, BRCA 2, ATM, CHEK1, CHEK2, FANCONI ANEMIA (FANCL), HDAC2, PALB2, BARD1, BRIP1, CDK12, PPP2R2A, RAD51B, RAD51C, RAD51D, or RAD54L |
Olaparib alone | |
| NCT03810105 | 2 | Castration sensitive AND biochemically recurrent prostate cancer | BRCA1, BRCA2, ATM, CHEK2, FANCA, RAD51C, RAD51D, PALB2, BRIP1, BARD1, or CDK12 | Olaparib + Durvalumab | |
| Niraparib | NCT02854436 | 2 | mCRPC | BRCA1, BRCA2, or other DDR alteration | Niraparib alone |
| NCT04037254 | 2 | High-risk prostate cancer | Not required | Niraparib + ADT | |
| NCT04030559 | 2 | High-risk localized prostate cancer | DDR deficiency | Neoadjuvant Niraparib | |
| Rucaparib | NCT03442556 | 2 | mCRPC | DDR deficiency | Rucaparib |
|
NCT02975934 (TRITON3) |
3 | mCRPC | BRCA1, BRCA2 or ATM | Rucaparib OR Docetaxel OR Abiraterone OR Enzalutamide |
|
|
NCT03413995 (TRIUMPH) |
2 | mHSPC | BRCA1, BRCA2, ATM, CHEK2, NBN, RAD50, RAD51C, RAD51D, PALB2, MRE11, FANCA, FANCB, FANCC, FANCD2, FANCE, FANCF, FANCG, FANCI, FANCL, FANCM | Rucaparib alone | |
| NCT03533946 | 2 | Non-metastatic prostate cancer | ATM, ATR, BARD1, BRCA1, BRCA2, BRIP1, CDK12, CHEK1, CHEK2, ERCC3, FAM175A, FANCA, FANCL, GEN1, HDAC2, MLH1, MRE11, NBN, PALB2, PPP2R2A, RAD51, RAD54L | Rucaparib alone | |
| Talazoparib |
NCT03395197 (TALAPRO-2) |
3 | mCRPC | DDR assessment required | Talazoparib + Enzalutamide |
| NCT03330405 | 2 | mCRPC | ATM, BRCA1 and BRCA2 | Talazoparib + Avelumab | |
| NCT04332744 | 2 | mHSPC | Not required | Talazoparib + Enzalutamide |
mCRPC: metastatic castration-resistant prostate cancer; mHSPC: metastatic hormone-sensitive prostate cancer; Prostate cancer: prostate cancer; MMR: mismatch repair; HR: homologous recombination; DDR: DNA damage repair; ADT: androgen-deprivation therapy.
Moreover, some clinical trials included as inclusion criteria the presence or absence of specific HR mutations.
7.2.1. PARP-i in Different Treatment Settings
PARP-i are currently investigated in the early stages of prostate cancer as, e.g., neoadjuvant treatment for locally advanced prostate cancer (BrUOG 337 trial—NCT03432897) or high-risk localized prostate cancer (NCT04030559) and in the metastatic hormone sensitive prostate cancer (mHSPC) (TRYUMPH trial—NCT03413995).
7.2.2. PARP-i plus AR-Direct Therapy
Preclinical data have shown synergy between olaparib and drugs targeting the AR-pathway [131,132]. The exact mechanisms are still not completely known, but it seems that novel hormonal agents can induce an HR phenotype in prostate cancer cells, the signaling of AR pathway may be involved in DNA repair, and the PARP1 gene may be activated in AR transcriptional activity [133].
Therefore, a phase 2 randomized trial was conducted by Clarke et al. to assess the efficacy and tolerability of olaparib in combination with abiraterone compared with placebo plus abiraterone in mCRPC patients previously received docetaxel, irrespective of their HR mutation status [134]. Olaparib in combination with abiraterone provided an additional clinical efficacy benefit compared with abiraterone alone, even though with an increase in serious adverse events.
Upon on the same rationale, other ongoing phase 3 randomized trials are comparing PARP-i versus placebo in addition to new hormonal agents (e.g., TALAPRO-2 study—NCT03395197 [135]).
7.2.3. PARP-i plus Immunotherapy
PARP-i have shown to have an immunomodulatory effect modulating the tumor immune microenvironment by a wide range of molecular and cellular mechanisms, such as increasing genomic instability, activating immune pathways and increasing PD-L1 expression on cancer cells, which might promote responsiveness to immune checkpoint inhibitors (ICIs) [136].
According to this biological rationale, several studies are investigating the combination of PARP-i and ICIs [137].
The phase I/II study NCT02484404 reported that the combination of olaparib and durvalumab demonstrates efficacy in terms of PSA response (reduction ≥50%) in mCRPC patients who have received prior enzalutamide and/or abiraterone. Patients with DDR mutations exhibited greater survival (in terms of PFS) benefit compared with those without known alterations [138]. Another phase II study is also assessing the efficacy of the combination olaparib plus durvalumab in castration-sensitive biochemically recurrent non-metastatic prostate cancer harboring at least one DDR deleterious mutation (NCT03810105).
The phase 1–2 KEYNOTE-365 study (NCT02861573) investigated the association of pembrolizumab plus olaparib showing antitumor activity in docetaxel-pretreated and molecularly unselected mCRPC patients who previously received less than two second-generation hormone treatments [139].
A phase 3 study (KEYLYNK-010, NCT03834519) is currently ongoing assessing the efficacy and safety of this combination in molecularly unselected mCRPC patients who progressed to taxane chemotherapy and at least one novel hormonal therapy.
Other phase 2 are currently investigating the combination of different PARP-i with ICIs in mCRPC patients (e.g., rucaparib plus nivolumab in the CheckMate 9 KD—NCT03338790).
7.2.4. PARP-i Associated with TKI
Tumor cell growth is influenced by several factors, including the mechanisms of cell repair, which are contrasted by PARP-i, and the signaling of growth factors, which are contrasted by TKI [130]. The double targeting of these patterns may help to treat patients with CRPC.
TKIs include inhibitors of angiogenesis (e.g., cediranib in NCT02893917) or of the AKT pathway (e.g., ipatasertib in NCT03840200].
7.2.5. PARP-i Associated with Other Treatments
Other combination strategies with PARP-i include therapies that induce DNA damage/replication stress enhancing the activity of PARP-i [130]. Other agents target DDR (ATR-i AZD6738 in NCT03787680), radionuclides (Radium-223 dichloride in NCT03076203 and NCT03317392; Lutetium 177 dotatate in NCT03874884), radiotherapy (NCT04037254), and bipolar androgen therapy (alternating between castration and supraphysiologic testosterone in NCT03516812).
7.3. Beyond BRCA in Prostate Cancer
As the main clinical trials’ results on PARP-i in mCRPC were obtained in patients with BRCA1/2 mutations and data for other DDR genes come from subgroup analyses, the efficacy of PARP-i remains unclear in patients harboring non-BRCA DDR mutations. The “BRCAness” status is defined as a HR alteration not due to BRCA1/2 mutations, but to other DDR genes, such as PALB2, ATM, ATR, CDK12, CHEK1, FANC, and RAD51/54 [140].
7.3.1. PARP-i
The phase 3 PROfound trial reported that olaparib was less effective in terms of ORR and PFS in the cohort B of patients harboring non-BRCA DDR alterations compared to cohort A of BRCA-mutated patients [141]. These data were also observed in two phase II trials on olaparib (TOPARP-B) and rucaparib (TRITON2) [120,124].
The phase II TOPARP-B study assessed the association between olaparib and non-BRCA DDR mutations with the aim to extend the validation of olaparib in DDR-mutated mCRPC [120].
In this study, the antitumor activity of olaparib was higher in BRCA1/2 patients, but it was also observed in other DDR gene mutations, especially in the PALB2 and ATM subgroups [120].
The ad hoc analysis of the TRITON2 trial on patients with a non-BRCA DDR gene alteration, especially ATM, CDK2, or CHECK2 mutations, showed a limited ORR and PSA response rate. On the other hand, responses were observed in patients with PALB2, FANCA, BRIP1, and RAD5B alterations [124].
These results suggest that PARP-i might have a role as monotherapy or in combination with other therapies also in BRCAness mCRPC patients, although with a lower magnitude of benefit compared to BRCA-mutated patients. Further, ad hoc studies are needed to assess the therapeutic role of PARP-i in BRCAness mCRPC patients.
7.3.2. Other DDR Gene Inhibitors
ATM and ATR genes are involved in complex DDR pathways [114]. Some evidence has suggested that targeting ATM/ATR mutated cancer with both PARP-i and ATR-i/ATM-i may be more efficacious than using PARP-i alone [142]. Several ATR-i and ATM-i are currently being investigated in early clinical trials as monotherapy or in association with PARP-i, as a double blockade of the DDR pathway, immunotherapy, and hormonotherapy chemotherapy [114].
Other promising preclinical data were obtained with compounds targeting other DDR alterations, including CHK1, WEE1, CDK12, and DNA-PKcs inhibitors, which were tested for activity in preclinical prostate cancer models and are currently under investigation in phase 1/2 trials in different solid tumors, including prostate cancer patients, or specifically in prostate cancer patients [114].
8. Conclusions
Pancreatic and prostate cancer were the first cancers after breast and ovarian cancer for which the efficacy of PARP inhibitors was evaluated in the presence of BRCA1/2 mutations. Since then, the presence and actionability of DDR deficiency in these tumors has been investigated. However, due to differences in cohort selection and DNA sequencing approaches, the burden of PVs in non-BRCA1/2 DDR genes shared by pancreatic and prostate cancers is not completely defined.
The picture that emerges from the examined literature shows that the addition of other DDR genes to BRCA1/2 markedly increases the burden of actionable variants, even when looking only at point mutations. We conclude that the inclusion of DDR genes other than BRCA1/2 shared by both cancers considerably increases the detection rate of potentially actionable variants, which are triplicated in pancreatic and almost doubled in prostate cancer. For prostate cancer, this is particularly relevant at the somatic level, where DDR mutations are almost doubled compared to those found in germline samples, and a germline testing-based approach would miss a large amount of DDR-deficient tumors. Considering the growing applications of DDR-targeting agents in cancer therapy, and the importance of timely genetic testing for patient access to treatment, multi-gene panels for pancreatic cancer and prostate cancer that include these genes should be routinely used in the clinical setting for both cancers.
Overall, DDR alterations are suitable targets for drug development and to improve the outcome in both pancreatic and prostate cancer patients. Importantly, this will increase the detection of germline pathogenic variants, thereby patient referral to genetic counseling.
Acknowledgments
S.E.R. and G.F. would like to thank the Italian Ministry of Health (Ricerca Corrente 2018–2021 grants) that financially supports their current research focused on the identification of prognostic and predictive markers for patients with genitourinary tumors. P.G. would like to thank AR3 onlus and patients’ families support for pancreatic cancer research studies.
Author Contributions
Conceptualization, methodology and writing—original draft preparation: B.D., A.P., S.E.R. and P.G.; writing—review and editing and supervision: F.C., R.B., M.L.I., W.B., G.F., S.S. and P.G. All authors have made substantial contributions to this review. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from Lega Italiana per la Lotta contro i Tumori (LILT) 5 × 1000 IG 2019 to P.G., Italian Ministry of Health (Ospedale Policlinico San Martino Ricerca Corrente and 5 × 1000 funds 2018–2021) to P.G., Associazione Ricerca Tumori Rari ed Ereditari (AR3) onlus to P.G., and Italian Ministry of Health (Ricerca Corrente 2018–2021) grants to S.E.R. and G.F.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest for this work.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chatterjee N., Walker G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017;58:235–263. doi: 10.1002/em.22087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D., Weinberg R.A. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Pearl L.H., Schierz A.C., Ward S.E., Al-Lazikani B., Pearl F.M. Therapeutic opportunities within the DNA damage response. Nat. Rev. Cancer. 2015;15:166–180. doi: 10.1038/nrc3891. [DOI] [PubMed] [Google Scholar]
- 4.Jackson S.P., Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.David S.S., O’Shea V.L., Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giglia-Mari G., Zotter A., Vermeulen W. DNA damage response. Cold Spring Harb. Perspect. Biol. 2011;3:a000745. doi: 10.1101/cshperspect.a000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoeijmakers J.H. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 8.Ogi T., Limsirichaikul S., Overmeer R.M., Volker M., Takenaka K., Cloney R., Nakazawa Y., Niimi A., Miki Y., Jaspers N.G., et al. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol. Cell. 2010;37:714–727. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 9.Li G.M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 10.Hsieh P., Zhang Y. The Devil is in the details for DNA mismatch repair. Proc. Natl. Acad. Sci. USA. 2017;114:3552–3554. doi: 10.1073/pnas.1702747114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spies M., Fishel R. Mismatch repair during homologous and homeologous recombination. Cold Spring Harb. Perspect. Biol. 2015;7:a022657. doi: 10.1101/cshperspect.a022657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cannan W.J., Pederson D.S. Mechanisms and Consequences of Double-Strand DNA Break Formation in Chromatin. J. Cell Physiol. 2016;231:3–14. doi: 10.1002/jcp.25048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X., Heyer W.D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powell S.N., Kachnic L.A. Roles of BRCA1 and BRCA2 in homologous recombination, DNA replication fidelity and the cellular response to ionizing radiation. Oncogene. 2003;22:5784–5791. doi: 10.1038/sj.onc.1206678. [DOI] [PubMed] [Google Scholar]
- 15.Prakash R., Zhang Y., Feng W., Jasin M. Homologous recombination and human health: The roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol. 2015;7:a016600. doi: 10.1101/cshperspect.a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang H.H.Y., Pannunzio N.R., Adachi N., Lieber M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017;18:495–506. doi: 10.1038/nrm.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shrivastav M., De Haro L.P., Nickoloff J.A. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 18.Gourley C., Balmana J., Ledermann J.A., Serra V., Dent R., Loibl S. Moving From Poly (ADP-Ribose) Polymerase Inhibition to Targeting DNA Repair and DNA Damage Response in Cancer Therapy. J. Clin. Oncol. 2019;37:2257–2269. doi: 10.1200/JCO.18.02050. [DOI] [PubMed] [Google Scholar]
- 19.Lord C.J., Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017;355:1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashworth A., Lord C.J. Synthetic lethal therapies for cancer: What’s next after PARP inhibitors? Nat. Rev. Clin. Oncol. 2018;15:564–576. doi: 10.1038/s41571-018-0055-6. [DOI] [PubMed] [Google Scholar]
- 21.Murai J., Huang S.Y., Das B.B., Renaud A., Zhang Y., Doroshow J.H., Ji J., Takeda S., Pommier Y. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012;72:5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hopkins T.A., Shi Y., Rodriguez L.E., Solomon L.R., Donawho C.K., DiGiammarino E.L., Panchal S.C., Wilsbacher J.L., Gao W., Olson A.M., et al. Mechanistic Dissection of PARP1 Trapping and the Impact on In Vivo Tolerability and Efficacy of PARP Inhibitors. Mol. Cancer Res. 2015;13:1465–1477. doi: 10.1158/1541-7786.MCR-15-0191-T. [DOI] [PubMed] [Google Scholar]
- 23.Fong P.C., Boss D.S., Yap T.A., Tutt A., Wu P., Mergui-Roelvink M., Mortimer P., Swaisland H., Lau A., O’Connor M.J., et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 24.Turk A.A., Wisinski K.B. PARP inhibitors in breast cancer: Bringing synthetic lethality to the bedside. Cancer. 2018;124:2498–2506. doi: 10.1002/cncr.31307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casolino R., Paiella S., Azzolina D., Beer P.A., Corbo V., Lorenzoni G., Gregori D., Golan T., Braconi C., Froeling F.E.M., et al. Homologous Recombination Deficiency in Pancreatic Cancer: A Systematic Review and Prevalence Meta-Analysis. J. Clin. Oncol. 2021;39:2617–2631. doi: 10.1200/JCO.20.03238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sztupinszki Z., Diossy M., Krzystanek M., Borcsok J., Pomerantz M.M., Tisza V., Spisak S., Rusz O., Csabai I., Freedman M.L., et al. Detection of Molecular Signatures of Homologous Recombination Deficiency in Prostate Cancer with or without BRCA1/2 Mutations. Clin. Cancer Res. 2020;26:2673–2680. doi: 10.1158/1078-0432.CCR-19-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaufman B., Shapira-Frommer R., Schmutzler R.K., Audeh M.W., Friedlander M., Balmana J., Mitchell G., Fried G., Stemmer S.M., Hubert A., et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J. Clin. Oncol. 2015;33:244–250. doi: 10.1200/JCO.2014.56.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 29.Hackeng W.M., Hruban R.H., Offerhaus G.J., Brosens L.A. Surgical and molecular pathology of pancreatic neoplasms. Diagn. Pathol. 2016;11:47. doi: 10.1186/s13000-016-0497-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leroux C., Konstantinidou G. Targeted Therapies for Pancreatic Cancer: Overview of Current Treatments and New Opportunities for Personalized Oncology. Cancers. 2021;13:799. doi: 10.3390/cancers13040799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts N.J., Norris A.L., Petersen G.M., Bondy M.L., Brand R., Gallinger S., Kurtz R.C., Olson S.H., Rustgi A.K., Schwartz A.G., et al. Whole Genome Sequencing Defines the Genetic Heterogeneity of Familial Pancreatic Cancer. Cancer Discov. 2016;6:166–175. doi: 10.1158/2159-8290.CD-15-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghiorzo P. Genetic predisposition to pancreatic cancer. World J. Gastroenterol. 2014;20:10778–10789. doi: 10.3748/wjg.v20.i31.10778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghiorzo P., Fornarini G., Sciallero S., Battistuzzi L., Belli F., Bernard L., Bonelli L., Borgonovo G., Bruno W., De Cian F., et al. CDKN2A is the main susceptibility gene in Italian pancreatic cancer families. J. Med. Genet. 2012;49:164–170. doi: 10.1136/jmedgenet-2011-100281. [DOI] [PubMed] [Google Scholar]
- 34.Harinck F., Kluijt I., van der Stoep N., Oldenburg R.A., Wagner A., Aalfs C.M., Sijmons R.H., Poley J.W., Kuipers E.J., Fockens P., et al. Indication for CDKN2A-mutation analysis in familial pancreatic cancer families without melanomas. J. Med. Genet. 2012;49:362–365. doi: 10.1136/jmedgenet-2011-100563. [DOI] [PubMed] [Google Scholar]
- 35.Astiazaran-Symonds E., Goldstein A.M. A systematic review of the prevalence of germline pathogenic variants in patients with pancreatic cancer. J. Gastroenterol. 2021;56:713–721. doi: 10.1007/s00535-021-01806-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pishvaian M.J., Bender R.J., Halverson D., Rahib L., Hendifar A.E., Mikhail S., Chung V., Picozzi V.J., Sohal D., Blais E.M., et al. Molecular Profiling of Patients with Pancreatic Cancer: Initial Results from the Know Your Tumor Initiative. Clin. Cancer Res. 2018;24:5018–5027. doi: 10.1158/1078-0432.CCR-18-0531. [DOI] [PubMed] [Google Scholar]
- 37.Heeke A.L., Pishvaian M.J., Lynce F., Xiu J., Brody J.R., Chen W.J., Baker T.M., Marshall J.L., Isaacs C. Prevalence of Homologous Recombination-Related Gene Mutations across Multiple Cancer Types. [(accessed on 15 March 2022)];JCO Precis. Oncol. 2018 2018 doi: 10.1200/PO.17.00286. Available online: https://pubmed.ncbi.nlm.nih.gov/30234181/ [DOI] [Google Scholar]
- 38.Aguirre A.J., Nowak J.A., Camarda N.D., Moffitt R.A., Ghazani A.A., Hazar-Rethinam M., Raghavan S., Kim J., Brais L.K., Ragon D., et al. Real-time Genomic Characterization of Advanced Pancreatic Cancer to Enable Precision Medicine. Cancer Discov. 2018;8:1096–1111. doi: 10.1158/2159-8290.CD-18-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Witkiewicz A.K., McMillan E.A., Balaji U., Baek G., Lin W.C., Mansour J., Mollaee M., Wagner K.U., Koduru P., Yopp A., et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat. Commun. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lowery M.A., Jordan E.J., Basturk O., Ptashkin R.N., Zehir A., Berger M.F., Leach T., Herbst B., Askan G., Maynard H., et al. Real-Time Genomic Profiling of Pancreatic Ductal Adenocarcinoma: Potential Actionability and Correlation with Clinical Phenotype. Clin. Cancer Res. 2017;23:6094–6100. doi: 10.1158/1078-0432.CCR-17-0899. [DOI] [PubMed] [Google Scholar]
- 41.Waddell N., Pajic M., Patch A.M., Chang D.K., Kassahn K.S., Bailey P., Johns A.L., Miller D., Nones K., Quek K., et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bailey P., Chang D.K., Nones K., Johns A.L., Patch A.M., Gingras M.C., Miller D.K., Christ A.N., Bruxner T.J., Quinn M.C., et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 43.Biankin A.V., Waddell N., Kassahn K.S., Gingras M.C., Muthuswamy L.B., Johns A.L., Miller D.K., Wilson P.J., Patch A.M., Wu J., et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collisson E.A., Sadanandam A., Olson P., Gibb W.J., Truitt M., Gu S., Cooc J., Weinkle J., Kim G.E., Jakkula L., et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011;17:500–503. doi: 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mehtala J., Zong J., Vassilev Z., Brobert G., Gabarro M.S., Stattin P., Khanfir H. Overall survival and second primary malignancies in men with metastatic prostate cancer. PLoS ONE. 2020;15:e0227552. doi: 10.1371/journal.pone.0227552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson D., Van Allen E.M., Wu Y.M., Schultz N., Lonigro R.J., Mosquera J.M., Montgomery B., Taplin M.E., Pritchard C.C., Attard G., et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mateo J., Boysen G., Barbieri C.E., Bryant H.E., Castro E., Nelson P.S., Olmos D., Pritchard C.C., Rubin M.A., de Bono J.S. DNA Repair in Prostate Cancer: Biology and Clinical Implications. Eur. Urol. 2017;71:417–425. doi: 10.1016/j.eururo.2016.08.037. [DOI] [PubMed] [Google Scholar]
- 48.Pritchard C.C., Mateo J., Walsh M.F., De Sarkar N., Abida W., Beltran H., Garofalo A., Gulati R., Carreira S., Eeles R., et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N. Engl. J. Med. 2016;375:443–453. doi: 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Venkitaraman A.R. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/S0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 50.Sawyer S.L., Tian L., Kahkonen M., Schwartzentruber J., Kircher M., University of Washington Centre for Mendelian Genomics. FORGE Canada Consortium. Majewski J., Dyment D.A., Innes A.M., et al. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov. 2015;5:135–142. doi: 10.1158/2159-8290.CD-14-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pilarski R. The Role of BRCA Testing in Hereditary Pancreatic and Prostate Cancer Families. Am. Soc. Clin. Oncol. Educ. Book. 2019;39:79–86. doi: 10.1200/EDBK_238977. [DOI] [PubMed] [Google Scholar]
- 52.Petersen G.M. Familial pancreatic cancer. Semin. Oncol. 2016;43:548–553. doi: 10.1053/j.seminoncol.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lecarpentier J., Silvestri V., Kuchenbaecker K.B., Barrowdale D., Dennis J., McGuffog L., Soucy P., Leslie G., Rizzolo P., Navazio A.S., et al. Prediction of Breast and Prostate Cancer Risks in Male BRCA1 and BRCA2 Mutation Carriers Using Polygenic Risk Scores. J. Clin. Oncol. 2017;35:2240–2250. doi: 10.1200/JCO.2016.69.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Castro E., Goh C., Leongamornlert D., Saunders E., Tymrakiewicz M., Dadaev T., Govindasami K., Guy M., Ellis S., Frost D., et al. Effect of BRCA Mutations on Metastatic Relapse and Cause-specific Survival After Radical Treatment for Localised Prostate Cancer. Eur. Urol. 2015;68:186–193. doi: 10.1016/j.eururo.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 55.Castro E., Goh C., Olmos D., Saunders E., Leongamornlert D., Tymrakiewicz M., Mahmud N., Dadaev T., Govindasami K., Guy M., et al. Germline BRCA mutations are associated with higher risk of nodal involvement, distant metastasis, and poor survival outcomes in prostate cancer. J. Clin. Oncol. 2013;31:1748–1757. doi: 10.1200/JCO.2012.43.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carter H.B., Helfand B., Mamawala M., Wu Y., Landis P., Yu H., Wiley K., Na R., Shi Z., Petkewicz J., et al. Germline Mutations in ATM and BRCA1/2 Are Associated with Grade Reclassification in Men on Active Surveillance for Prostate Cancer. Eur. Urol. 2019;75:743–749. doi: 10.1016/j.eururo.2018.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alt F.W., Zhang Y., Meng F.L., Guo C., Schwer B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 2013;152:417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Woodbine L., Neal J.A., Sasi N.K., Shimada M., Deem K., Coleman H., Dobyns W.B., Ogi T., Meek K., Davies E.G., et al. PRKDC mutations in a SCID patient with profound neurological abnormalities. J. Clin. Investig. 2013;123:2969–2980. doi: 10.1172/JCI67349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lowery M.A., Wong W., Jordan E.J., Lee J.W., Kemel Y., Vijai J., Mandelker D., Zehir A., Capanu M., Salo-Mullen E., et al. Prospective Evaluation of Germline Alterations in Patients With Exocrine Pancreatic Neoplasms. J. Natl. Cancer Inst. 2018;110:1067–1074. doi: 10.1093/jnci/djy024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yurgelun M.B., Chittenden A.B., Morales-Oyarvide V., Rubinson D.A., Dunne R.F., Kozak M.M., Qian Z.R., Welch M.W., Brais L.K., Da Silva A., et al. Germline cancer susceptibility gene variants, somatic second hits, and survival outcomes in patients with resected pancreatic cancer. Genet. Med. 2019;21:213–223. doi: 10.1038/s41436-018-0009-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abida W., Armenia J., Gopalan A., Brennan R., Walsh M., Barron D., Danila D., Rathkopf D., Morris M., Slovin S., et al. Prospective Genomic Profiling of Prostate Cancer Across Disease States Reveals Germline and Somatic Alterations That May Affect Clinical Decision Making. [(accessed on 15 March 2022)];JCO Precis. Oncol. 2017 2017 doi: 10.1200/PO.17.00029. Available online: https://pubmed.ncbi.nlm.nih.gov/28825054/ [DOI] [Google Scholar]
- 62.Nicolosi P., Ledet E., Yang S., Michalski S., Freschi B., O’Leary E., Esplin E.D., Nussbaum R.L., Sartor O. Prevalence of Germline Variants in Prostate Cancer and Implications for Current Genetic Testing Guidelines. JAMA Oncol. 2019;5:523–528. doi: 10.1001/jamaoncol.2018.6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Consortium ITP-CAoWG Pan-cancer analysis of whole genomes. Nature. 2020;578:82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lozano R., Castro E., Aragon I.M., Cendon Y., Cattrini C., Lopez-Casas P.P., Olmos D. Genetic aberrations in DNA repair pathways: A cornerstone of precision oncology in prostate cancer. Br. J. Cancer. 2021;124:552–563. doi: 10.1038/s41416-020-01114-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kantidze O.L., Velichko A.K., Luzhin A.V., Petrova N.V., Razin S.V. Synthetically Lethal Interactions of ATM, ATR, and DNA-PKcs. Trends Cancer. 2018;4:755–768. doi: 10.1016/j.trecan.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 66.Cantor S.B., Guillemette S. Hereditary breast cancer and the BRCA1-associated FANCJ/BACH1/BRIP1. Future Oncol. 2011;7:253–261. doi: 10.2217/fon.10.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blackford A.N., Jackson S.P. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell. 2017;66:801–817. doi: 10.1016/j.molcel.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 68.Dai Y., Grant S. New insights into checkpoint kinase 1 in the DNA damage response signaling network. Clin. Cancer Res. 2010;16:376–383. doi: 10.1158/1078-0432.CCR-09-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kleiblova P., Stolarova L., Krizova K., Lhota F., Hojny J., Zemankova P., Havranek O., Vocka M., Cerna M., Lhotova K., et al. Identification of deleterious germline CHEK2 mutations and their association with breast and ovarian cancer. Int. J. Cancer. 2019;145:1782–1797. doi: 10.1002/ijc.32385. [DOI] [PubMed] [Google Scholar]
- 70.Lopes J.L., Chaudhry S., Lopes G.S., Levin N.K., Tainsky M.A. FANCM, RAD1, CHEK1 and TP53I3 act as BRCA-like tumor suppressors and are mutated in hereditary ovarian cancer. Cancer Genet. 2019;235–236:57–64. doi: 10.1016/j.cancergen.2019.04.061. [DOI] [Google Scholar]
- 71.Tlemsani C., Takahashi N., Pongor L., Rajapakse V.N., Tyagi M., Wen X., Fasaye G.A., Schmidt K.T., Desai P., Kim C., et al. Whole-exome sequencing reveals germline-mutated small cell lung cancer subtype with favorable response to DNA repair-targeted therapies. Sci. Transl. Med. 2021;13:eabc7488. doi: 10.1126/scitranslmed.abc7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kumari N., Singh R.K., Mishra S.K., Raghvendra L., Mohindra S., Krishnani N. Prevalence and spectrum of pathogenic germline variants in intestinal and pancreatobiliary type of ampullary cancer. Pathol. Res. Pract. 2021;217:153309. doi: 10.1016/j.prp.2020.153309. [DOI] [PubMed] [Google Scholar]
- 73.Vahteristo P., Tamminen A., Karvinen P., Eerola H., Eklund C., Aaltonen L.A., Blomqvist C., Aittomaki K., Nevanlinna H. p53, CHK2, and CHK1 genes in Finnish families with Li-Fraumeni syndrome: Further evidence of CHK2 in inherited cancer predisposition. Cancer Res. 2001;61:5718–5722. [PubMed] [Google Scholar]
- 74.Shimamura A., Alter B.P. Pathophysiology and management of inherited bone marrow failure syndromes. Blood Rev. 2010;24:101–122. doi: 10.1016/j.blre.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Crossan G.P., Patel K.J. The Fanconi anaemia pathway orchestrates incisions at sites of crosslinked DNA. J. Pathol. 2012;226:326–337. doi: 10.1002/path.3002. [DOI] [PubMed] [Google Scholar]
- 76.Schlacher K., Wu H., Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Idos G., Valle L. Lynch Syndrome. University of Washington; Seattle, WA, USA: 2021. [(accessed on 15 March 2022)]. GeneReviews® [Internet] Available online: https://www.ncbi.nlm.nih.gov/books/NBK1211/ [Google Scholar]
- 78.Raymond V.M., Mukherjee B., Wang F., Huang S.C., Stoffel E.M., Kastrinos F., Syngal S., Cooney K.A., Gruber S.B. Elevated risk of prostate cancer among men with Lynch syndrome. J. Clin. Oncol. 2013;31:1713–1718. doi: 10.1200/JCO.2012.44.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haraldsdottir S., Hampel H., Wei L., Wu C., Frankel W., Bekaii-Saab T., de la Chapelle A., Goldberg R.M. Prostate cancer incidence in males with Lynch syndrome. Genet. Med. 2014;16:553–557. doi: 10.1038/gim.2013.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lamarche B.J., Orazio N.I., Weitzman M.D. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 2010;584:3682–3695. doi: 10.1016/j.febslet.2010.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chrzanowska K.H., Gregorek H., Dembowska-Baginska B., Kalina M.A., Digweed M. Nijmegen breakage syndrome (NBS) Orphanet J. Rare Dis. 2012;7:13. doi: 10.1186/1750-1172-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Varon R., Demuth I., Chrzanowska K.H. Nijmegen Breakage Syndrome. University of Washington; Seattle, WA, USA: 2021. [(accessed on 15 March 2022)]. GeneReviews® [Internet] Available online: https://www.ncbi.nlm.nih.gov/books/NBK1176/ [Google Scholar]
- 83.Sy S.M., Huen M.S., Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc. Natl. Acad. Sci. USA. 2009;106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Garcin E.B., Gon S., Sullivan M.R., Brunette G.J., Cian A., Concordet J.P., Giovannangeli C., Dirks W.G., Eberth S., Bernstein K.A., et al. Differential Requirements for the RAD51 Paralogs in Genome Repair and Maintenance in Human Cells. PLoS Genet. 2019;15:e1008355. doi: 10.1371/journal.pgen.1008355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goldstein J.B., Zhao L., Wang X., Ghelman Y., Overman M.J., Javle M.M., Shroff R.T., Varadhachary G.R., Wolff R.A., McAllister F., et al. Germline DNA Sequencing Reveals Novel Mutations Predictive of Overall Survival in a Cohort of Patients with Pancreatic Cancer. Clin. Cancer Res. 2020;26:1385–1394. doi: 10.1158/1078-0432.CCR-19-0224. [DOI] [PubMed] [Google Scholar]
- 86.Hannan Z., Yu S., Domchek S., Mamtani R., Reiss K.A. Clinical Characteristics of Patients With Pancreatic Cancer and Pathogenic ATM Alterations. JNCI Cancer Spectr. 2021;5:pkaa121. doi: 10.1093/jncics/pkaa121. [DOI] [Google Scholar]
- 87.Sehdev A., Gbolahan O., Hancock B.A., Stanley M., Shahda S., Wan J., Wu H.H., Radovich M., O’Neil B.H. Germline and Somatic DNA Damage Repair Gene Mutations and Overall Survival in Metastatic Pancreatic Adenocarcinoma Patients Treated with FOLFIRINOX. Clin. Cancer Res. 2018;24:6204–6211. doi: 10.1158/1078-0432.CCR-18-1472. [DOI] [PubMed] [Google Scholar]
- 88.Golan T., Hammel P., Reni M., Van Cutsem E., Macarulla T., Hall M.J., Park J.O., Hochhauser D., Arnold D., Oh D.Y., et al. Maintenance Olaparib for Germline BRCA-Mutated Metastatic Pancreatic Cancer. N. Engl. J. Med. 2019;381:317–327. doi: 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Golan T., Hammel P., Reni M., Cutsem E.V., Macarulla T., Hall M.J., Park J.O., Hochhauser D., Arnold D., Oh D.-Y., et al. Overall survival from the phase 3 POLO trial: Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. J. Clin. Oncol. 2021;39:378. doi: 10.1200/JCO.2021.39.3_suppl.378. [DOI] [Google Scholar]
- 90.O’Reilly E.M., Lee J.W., Lowery M.A., Capanu M., Stadler Z.K., Moore M.J., Dhani N., Kindler H.L., Estrella H., Maynard H., et al. Phase 1 trial evaluating cisplatin, gemcitabine, and veliparib in 2 patient cohorts: Germline BRCA mutation carriers and wild-type BRCA pancreatic ductal adenocarcinoma. Cancer. 2018;124:1374–1382. doi: 10.1002/cncr.31218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.O’Reilly E.M., Lee J.W., Zalupski M., Capanu M., Park J., Golan T., Tahover E., Lowery M.A., Chou J.F., Sahai V., et al. Randomized, Multicenter, Phase II Trial of Gemcitabine and Cisplatin With or Without Veliparib in Patients With Pancreas Adenocarcinoma and a Germline BRCA/PALB2 Mutation. J. Clin. Oncol. 2020;38:1378–1388. doi: 10.1200/JCO.19.02931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lowery M.A., Kelsen D.P., Capanu M., Smith S.C., Lee J.W., Stadler Z.K., Moore M.J., Kindler H.L., Golan T., Segal A., et al. Phase II trial of veliparib in patients with previously treated BRCA-mutated pancreas ductal adenocarcinoma. Eur. J. Cancer. 2018;89:19–26. doi: 10.1016/j.ejca.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chiorean E.G., McDonough S., Philip P.A., Swisher E.M., Pishvaian M.J., Guthrie K., Lowy A.M., Hochster H.S. Randomized phase II study of 2nd-line FOLFIRI versus modified FOLFIRI with PARP inhibitor ABT-888 (veliparib) (NSC-737664) in metastatic pancreatic cancer (mPC): SWOG S1513. J. Clin. Oncol. 2017;35:TPS4147. doi: 10.1200/JCO.2017.35.15_suppl.TPS4147. [DOI] [Google Scholar]
- 94.Pishvaian M.J., Wang H., Parenti S., He A.R., Hwang J.J., Ley L., Difebo H., Smaglo B.G., Kim S.S., Weinberg B.A., et al. Final report of a phase I/II study of veliparib (Vel) in combination with 5-FU and oxaliplatin (FOLFOX) in patients (pts) with metastatic pancreatic cancer (mPDAC) J. Clin. Oncol. 2019;37:4015. doi: 10.1200/JCO.2019.37.15_suppl.4015. [DOI] [Google Scholar]
- 95.Tuli R., Shiao S.L., Nissen N., Tighiouart M., Kim S., Osipov A., Bryant M., Ristow L., Placencio-Hickok V., Hoffman D., et al. A phase 1 study of veliparib, a PARP-1/2 inhibitor, with gemcitabine and radiotherapy in locally advanced pancreatic cancer. EBioMedicine. 2019;40:375–381. doi: 10.1016/j.ebiom.2018.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shen Y., Rehman F.L., Feng Y., Boshuizen J., Bajrami I., Elliott R., Wang B., Lord C.J., Post L.E., Ashworth A. BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin. Cancer Res. 2013;19:5003–5015. doi: 10.1158/1078-0432.CCR-13-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.de Bono J., Ramanathan R.K., Mina L., Chugh R., Glaspy J., Rafii S., Kaye S., Sachdev J., Heymach J., Smith D.C., et al. Phase I, Dose-Escalation, Two-Part Trial of the PARP Inhibitor Talazoparib in Patients with Advanced Germline BRCA1/2 Mutations and Selected Sporadic Cancers. Cancer Discov. 2017;7:620–629. doi: 10.1158/2159-8290.CD-16-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shroff R.T., Hendifar A., McWilliams R.R., Geva R., Epelbaum R., Rolfe L., Goble S., Lin K.K., Biankin A.V., Giordano H., et al. Rucaparib Monotherapy in Patients With Pancreatic Cancer and a Known Deleterious BRCA Mutation. [(accessed on 15 March 2022)];JCO Precis. Oncol. 2018 2018 doi: 10.1200/PO.17.00316. Available online: https://pubmed.ncbi.nlm.nih.gov/30051098/ [DOI] [Google Scholar]
- 99.Pishvaian M.J., Blais E.M., Brody J.R., Lyons E., DeArbeloa P., Hendifar A., Mikhail S., Chung V., Sahai V., Sohal D.P.S., et al. Overall survival in patients with pancreatic cancer receiving matched therapies following molecular profiling: A retrospective analysis of the Know Your Tumor registry trial. Lancet Oncol. 2020;21:508–518. doi: 10.1016/S1470-2045(20)30074-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Roberti M., Schipani F., Bagnolini G., Milano D., Giacomini E., Falchi F., Balboni A., Manerba M., Farabegoli F., De Franco F., et al. Rad51/BRCA2 disruptors inhibit homologous recombination and synergize with olaparib in pancreatic cancer cells. Eur. J. Med. Chem. 2019;165:80–92. doi: 10.1016/j.ejmech.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 101.Principe D.R. Precision Medicine for BRCA/PALB2-Mutated Pancreatic Cancer and Emerging Strategies to Improve Therapeutic Responses to PARP Inhibition. Cancers. 2022;14:897. doi: 10.3390/cancers14040897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Catalano F., Borea R., Puglisi S., Boutros A., Gandini A., Cremante M., Martelli V., Sciallero S., Puccini A. Targeting the DNA Damage Response Pathway as a Novel Therapeutic Strategy in Colorectal Cancer. Cancers. 2022;14:1388. doi: 10.3390/cancers14061388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Golan T., Varadhachary G.R., Sela T., Fogelman D.R., Halperin N., Shroff R.T., Halparin S., Xiao L., Aderka D., Maitra A., et al. Phase II study of olaparib for BRCAness phenotype in pancreatic cancer. J. Clin. Oncol. 2018;36:297. doi: 10.1200/JCO.2018.36.4_suppl.297. [DOI] [Google Scholar]
- 104.Perkhofer L., Schmitt A., Romero Carrasco M.C., Ihle M., Hampp S., Ruess D.A., Hessmann E., Russell R., Lechel A., Azoitei N., et al. ATM Deficiency Generating Genomic Instability Sensitizes Pancreatic Ductal Adenocarcinoma Cells to Therapy-Induced DNA Damage. Cancer Res. 2017;77:5576–5590. doi: 10.1158/0008-5472.CAN-17-0634. [DOI] [PubMed] [Google Scholar]
- 105.Armstrong S.A., Schultz C.W., Azimi-Sadjadi A., Brody J.R., Pishvaian M.J. ATM Dysfunction in Pancreatic Adenocarcinoma and Associated Therapeutic Implications. Mol. Cancer Ther. 2019;18:1899–1908. doi: 10.1158/1535-7163.MCT-19-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Riches L.C., Trinidad A.G., Hughes G., Jones G.N., Hughes A.M., Thomason A.G., Gavine P., Cui A., Ling S., Stott J., et al. Pharmacology of the ATM Inhibitor AZD0156: Potentiation of Irradiation and Olaparib Responses Preclinically. Mol. Cancer Ther. 2020;19:13–25. doi: 10.1158/1535-7163.MCT-18-1394. [DOI] [PubMed] [Google Scholar]
- 107.Javle M., Shacham-Shmueli E., Xiao L., Varadhachary G., Halpern N., Fogelman D., Boursi B., Uruba S., Margalit O., Wolff R.A., et al. Olaparib Monotherapy for Previously Treated Pancreatic Cancer With DNA Damage Repair Genetic Alterations Other Than Germline BRCA Variants: Findings From 2 Phase 2 Nonrandomized Clinical Trials. JAMA Oncol. 2021;7:693–699. doi: 10.1001/jamaoncol.2021.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Reaper P.M., Griffiths M.R., Long J.M., Charrier J.D., Maccormick S., Charlton P.A., Golec J.M., Pollard J.R. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat. Chem. Biol. 2011;7:428–430. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 109.Dunlop C.R., Wallez Y., Johnson T.I., Bernaldo de Quiros Fernandez S., Durant S.T., Cadogan E.B., Lau A., Richards F.M., Jodrell D.I. Complete loss of ATM function augments replication catastrophe induced by ATR inhibition and gemcitabine in pancreatic cancer models. Br. J. Cancer. 2020;123:1424–1436. doi: 10.1038/s41416-020-1016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Huang R.X., Zhou P.K. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct. Target. Ther. 2020;5:60. doi: 10.1038/s41392-020-0150-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Engelke C.G., Parsels L.A., Qian Y., Zhang Q., Karnak D., Robertson J.R., Tanska D.M., Wei D., Davis M.A., Parsels J.D., et al. Sensitization of pancreatic cancer to chemoradiation by the Chk1 inhibitor MK8776. Clin. Cancer Res. 2013;19:4412–4421. doi: 10.1158/1078-0432.CCR-12-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nepomuceno T.C., De Gregoriis G., de Oliveira F.M.B., Suarez-Kurtz G., Monteiro A.N., Carvalho M.A. The Role of PALB2 in the DNA Damage Response and Cancer Predisposition. Int. J. Mol. Sci. 2017;18:1886. doi: 10.3390/ijms18091886. [DOI] [Google Scholar]
- 113.Devlies W., Eckstein M., Cimadamore A., Devos G., Moris L., Van den Broeck T., Montironi R., Joniau S., Claessens F., Gevaert T. Clinical Actionability of the Genomic Landscape of Metastatic Castration Resistant Prostate Cancer. Cells. 2020;9:2494. doi: 10.3390/cells9112494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wengner A.M., Scholz A., Haendler B. Targeting DNA Damage Response in Prostate and Breast Cancer. Int. J. Mol. Sci. 2020;21:8273. doi: 10.3390/ijms21218273. [DOI] [Google Scholar]
- 115.Messina C., Cattrini C., Soldato D., Vallome G., Caffo O., Castro E., Olmos D., Boccardo F., Zanardi E. BRCA Mutations in Prostate Cancer: Prognostic and Predictive Implications. J. Oncol. 2020;2020:4986365. doi: 10.1155/2020/4986365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Castro E., Romero-Laorden N., Del Pozo A., Lozano R., Medina A., Puente J., Piulats J.M., Lorente D., Saez M.I., Morales-Barrera R., et al. PROREPAIR-B: A Prospective Cohort Study of the Impact of Germline DNA Repair Mutations on the Outcomes of Patients With Metastatic Castration-Resistant Prostate Cancer. J. Clin. Oncol. 2019;37:490–503. doi: 10.1200/JCO.18.00358. [DOI] [PubMed] [Google Scholar]
- 117.Giri V.N., Knudsen K.E., Kelly W.K., Cheng H.H., Cooney K.A., Cookson M.S., Dahut W., Weissman S., Soule H.R., Petrylak D.P., et al. Implementation of Germline Testing for Prostate Cancer: Philadelphia Prostate Cancer Consensus Conference 2019. J. Clin. Oncol. 2020;38:2798–2811. doi: 10.1200/JCO.20.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mota J.M., Barnett E., Nauseef J.T., Nguyen B., Stopsack K.H., Wibmer A., Flynn J.R., Heller G., Danila D.C., Rathkopf D., et al. Platinum-Based Chemotherapy in Metastatic Prostate Cancer With DNA Repair Gene Alterations. JCO Precis. Oncol. 2020;4:355–366. doi: 10.1200/PO.19.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mateo J., Carreira S., Sandhu S., Miranda S., Mossop H., Perez-Lopez R., Nava Rodrigues D., Robinson D., Omlin A., Tunariu N., et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N. Engl. J. Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mateo J., Porta N., Bianchini D., McGovern U., Elliott T., Jones R., Syndikus I., Ralph C., Jain S., Varughese M., et al. Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARP-B): A multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 2020;21:162–174. doi: 10.1016/S1470-2045(19)30684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.de Bono J., Kang J., Hussain M. Olaparib for Metastatic Castration-Resistant Prostate Cancer. Reply. N. Engl. J. Med. 2020;383:891. doi: 10.1056/NEJMc2023199. [DOI] [PubMed] [Google Scholar]
- 122.Hussain M., Mateo J., Fizazi K., Saad F., Shore N., Sandhu S., Chi K.N., Sartor O., Agarwal N., Olmos D., et al. Survival with Olaparib in Metastatic Castration-Resistant Prostate Cancer. N. Engl. J. Med. 2020;383:2345–2357. doi: 10.1056/NEJMoa2022485. [DOI] [PubMed] [Google Scholar]
- 123.Abida W., Patnaik A., Campbell D., Shapiro J., Bryce A.H., McDermott R., Sautois B., Vogelzang N.J., Bambury R.M., Voog E., et al. Rucaparib in Men With Metastatic Castration-Resistant Prostate Cancer Harboring a BRCA1 or BRCA2 Gene Alteration. J. Clin. Oncol. 2020;38:3763–3772. doi: 10.1200/JCO.20.01035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Abida W., Campbell D., Patnaik A., Shapiro J.D., Sautois B., Vogelzang N.J., Voog E.G., Bryce A.H., McDermott R., Ricci F., et al. Non-BRCA DNA Damage Repair Gene Alterations and Response to the PARP Inhibitor Rucaparib in Metastatic Castration-Resistant Prostate Cancer: Analysis From the Phase II TRITON2 Study. Clin. Cancer Res. 2020;26:2487–2496. doi: 10.1158/1078-0432.CCR-20-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.de Bono J.S., Mehra N., Scagliotti G.V., Castro E., Dorff T., Stirling A., Stenzl A., Fleming M.T., Higano C.S., Saad F., et al. Talazoparib monotherapy in metastatic castration-resistant prostate cancer with DNA repair alterations (TALAPRO-1): An open-label, phase 2 trial. Lancet Oncol. 2021;22:1250–1264. doi: 10.1016/S1470-2045(21)00376-4. [DOI] [PubMed] [Google Scholar]
- 126.Smith M.R., Scher H.I., Sandhu S., Efstathiou E., Lara P.N., Jr., Yu E.Y., George D.J., Chi K.N., Saad F., Stahl O., et al. Niraparib in patients with metastatic castration-resistant prostate cancer and DNA repair gene defects (GALAHAD): A multicentre, open-label, phase 2 trial. Lancet Oncol. 2022;23:362–373. doi: 10.1016/S1470-2045(21)00757-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yu E., Piulats J.M., Gravis G., Fong P., Todenhöfer T., Laguerre B., Arranz J., Oudard S., Massard C., Stoeckle M., et al. 73P Association between homologous recombination repair mutations and response to pembrolizumab (pembro) plus olaparib (ola) in metastatic castration-resistant prostate cancer (mCRPC): KEYNOTE-365 Cohort A biomarker analysis. Ann. Oncol. 2021;32:S387. doi: 10.1016/j.annonc.2021.08.353. [DOI] [Google Scholar]
- 128.Chi K.N., Rathkopf D.E., Smith M.R., Efstathiou E., Attard G., Olmos D., Lee J.Y., Small E.J., Gomes A.J., Roubaud G., et al. Phase 3 MAGNITUDE study: First results of niraparib (NIRA) with abiraterone acetate and prednisone (AAP) as first-line therapy in patients (pts) with metastatic castration-resistant prostate cancer (mCRPC) with and without homologous recombination repair (HRR) gene alterations. J. Clin. Oncol. 2022;40:12. doi: 10.1200/JCO.21.01891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Saad F., Armstrong A.J., Thiery-Vuillemin A., Oya M., Loredo E., Procopio G., Janoski de Menezes J., Girotto G.C., Arslan C., Mehra N., et al. PROpel: Phase III trial of olaparib (ola) and abiraterone (abi) versus placebo (pbo) and abi as first-line (1L) therapy for patients (pts) with metastatic castration-resistant prostate cancer (mCRPC) J. Clin. Oncol. 2022;40:11. doi: 10.1200/JCO.2022.40.6_suppl.011. [DOI] [Google Scholar]
- 130.Pezaro C. PARP inhibitor combinations in prostate cancer. Ther. Adv. Med. Oncol. 2020;12:1758835919897537. doi: 10.1177/1758835919897537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Asim M., Tarish F., Zecchini H.I., Sanjiv K., Gelali E., Massie C.E., Baridi A., Warren A.Y., Zhao W., Ogris C., et al. Synthetic lethality between androgen receptor signalling and the PARP pathway in prostate cancer. Nat. Commun. 2017;8:374. doi: 10.1038/s41467-017-00393-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schiewer M.J., Goodwin J.F., Han S., Brenner J.C., Augello M.A., Dean J.L., Liu F., Planck J.L., Ravindranathan P., Chinnaiyan A.M., et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov. 2012;2:1134–1149. doi: 10.1158/2159-8290.CD-12-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Antonarakis E.S., Gomella L.G., Petrylak D.P. When and How to Use PARP Inhibitors in Prostate Cancer: A Systematic Review of the Literature with an Update on On-Going Trials. Eur. Urol. Oncol. 2020;3:594–611. doi: 10.1016/j.euo.2020.07.005. [DOI] [PubMed] [Google Scholar]
- 134.Clarke N., Wiechno P., Alekseev B., Sala N., Jones R., Kocak I., Chiuri V.E., Jassem J., Flechon A., Redfern C., et al. Olaparib combined with abiraterone in patients with metastatic castration-resistant prostate cancer: A randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol. 2018;19:975–986. doi: 10.1016/S1470-2045(18)30365-6. [DOI] [PubMed] [Google Scholar]
- 135.Agarwal N., Azad A., Shore N.D., Carles J., Fay A.P., Dunshee C., Karsh L.I., Paccagnella M.L., Santo N.D., Elmeliegy M., et al. Talazoparib plus enzalutamide in metastatic castration-resistant prostate cancer: TALAPRO-2 phase III study design. Future Oncol. 2022;18:425–436. doi: 10.2217/fon-2021-0811. [DOI] [PubMed] [Google Scholar]
- 136.Peyraud F., Italiano A. Combined PARP Inhibition and Immune Checkpoint Therapy in Solid Tumors. Cancers. 2020;12:1502. doi: 10.3390/cancers12061502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chabanon R.M., Muirhead G., Krastev D.B., Adam J., Morel D., Garrido M., Lamb A., Henon C., Dorvault N., Rouanne M., et al. PARP inhibition enhances tumor cell-intrinsic immunity in ERCC1-deficient non-small cell lung cancer. J. Clin. Invest. 2019;129:1211–1228. doi: 10.1172/JCI123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Karzai F., VanderWeele D., Madan R.A., Owens H., Cordes L.M., Hankin A., Couvillon A., Nichols E., Bilusic M., Beshiri M.L., et al. Activity of durvalumab plus olaparib in metastatic castration-resistant prostate cancer in men with and without DNA damage repair mutations. J. Immunother. Cancer. 2018;6:141. doi: 10.1186/s40425-018-0463-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yu E.Y., Piulats J.M., Gravis G., Laguerre B., Arija J.A.A., Oudard S., Fong P.C.C., Kolinsky M.P., Augustin M., Feyerabend S., et al. KEYNOTE-365 cohort A updated results: Pembrolizumab (pembro) plus olaparib in docetaxel-pretreated patients (pts) with metastatic castration-resistant prostate cancer (mCRPC) J. Clin. Oncol. 2020;38:100. doi: 10.1200/JCO.2020.38.6_suppl.100. [DOI] [PubMed] [Google Scholar]
- 140.Dhawan M., Ryan C.J. BRCAness and prostate cancer: Diagnostic and therapeutic considerations. Prostate Cancer Prostatic Dis. 2018;21:488–498. doi: 10.1038/s41391-018-0069-2. [DOI] [PubMed] [Google Scholar]
- 141.Stopsack K.H. Efficacy of PARP Inhibition in Metastatic Castration-resistant Prostate Cancer is Very Different with Non-BRCA DNA Repair Alterations: Reconstructing Prespecified Endpoints for Cohort B from the Phase 3 PROfound Trial of Olaparib. Eur. Urol. 2021;79:442–445. doi: 10.1016/j.eururo.2020.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jette N.R., Kumar M., Radhamani S., Arthur G., Goutam S., Yip S., Kolinsky M., Williams G.J., Bose P., Lees-Miller S.P. ATM-Deficient Cancers Provide New Opportunities for Precision Oncology. Cancers. 2020;12:687. doi: 10.3390/cancers12030687. [DOI] [Google Scholar]