Abstract
Predation by bacteriophages is thought to control bacterial numbers and facilitate gene transfer among bacteria in the biosphere. A thorough understanding of phage population dynamics is therefore necessary if their significance in natural environments is to be fully appreciated. Here we describe the in situ population dynamics of three separate phage populations predating on separate bacterial species, living on the surface of field-grown sugar beet (Beta vulgaris var. Amethyst), as recorded over a 9-month period. The distributions of the three phage populations were different and fluctuated temporally in 1996 (peak density, ∼103 PFU g−1). One of these populations, predating on the indigenous phytosphere bacterium Serratia liquefaciens CP6, consisted of six genetically distinct DNA phages that varied in relative abundance to the extent that an apparent temporal succession was observed between the two most abundant phages, ΦCP6-1 and ΦCP6-4.
The population dynamics of phages have been successfully monitored in chemostats, and the resulting mathematical models have served to provide important insights into the ecology and coevolutionary biology of not only phages and their host bacteria but also predator-prey relationships generally (5, 10, 13, 14). However, comparable in situ data from natural environments is largely lacking. Currently, information is restricted to aquatic habitats, where the total number of viruslike particles has been monitored (3, 4), but methodological limitations have prevented monitoring changes of specific viruses.
Thus far, terrestrial studies have concentrated on specific phage-host interactions within otherwise sterile microcosms. These have been mainly simple soil systems (6, 9, 19, 25), although Kidambi et al. (12) did monitor a phage and its hosts for 11 days on the leaves of aseptically grown bean seedlings during their transduction experiments.
To learn more of the population ecology of specific phages in a natural environment, we investigated the microflora living on sugar beet. Plant surfaces (i.e., phytosphere) are good sites to look for phages, because they are nutritionally richer environments than bulk soil and support many metabolically active bacterial species (22). To date, phage studies on phytosphere bacteria have been limited to sugar beet (18, 20) and bean (12) seedlings grown in sterile soil. To learn more of the predator-prey relationship between phage and host in situ, we investigated field-grown sugar beet over two growing seasons.
MATERIALS AND METHODS
Bacterial strains.
The bacteria used in this study (Table 1) were stored in 50% glycerol at −80°C and maintained on tryptone soya broth agar (TSBA) (15) supplemented with the appropriate antibiotics (rifampin, 100 μg ml−1; kanamycin, 100 μg ml−1).
TABLE 1.
Bacterial strains used in this study
| Strain | Descriptiona | Additional information | Source or reference |
|---|---|---|---|
| P. fluorescens | |||
| SBW25 | Wild type | Sugar beet phytosphere isolate | 2 |
| SBW25EeZY6KX | Kmr, lacZY xylE | Genetically engineered variant of SBW25 | 2 |
| SBW25EeZY6KX(pQBR103) | Kmr, lacZY xylE, Hgr | Previous strain containing a plasmid encoding Hgr | 16 |
| SBW25Rif | Rifr | Spontaneous rifampin-resistant mutant of SBW25 | This study |
| MCP1 | Wild type | Sugar beet phytosphere isolate | This study |
| S. liquefaciens | |||
| CP6 | Wild type | Sugar beet phytosphere isolate | This study |
Kmr, kanamycin resistant; lacZY, β-galactosidase and lactose permease activity; xylE, catechol-2,3-dioxygenase activity; Hgr, mercury resistant; Rifr, rifampin resistant.
Field site.
The field work took place at Oxford University Farm, Wytham, Oxford, United Kingdom. Sugar beet had been continuously grown at the site since 1990, before which the site had been undisturbed pasture land. The present study was carried out between 1994 and 1997.
Preliminary field experiment.
Starting on 20 April 1994 (day 0), untreated sugar beet and beet inoculated with Pseudomonas fluorescens SBW25EeZY6KX with or without pQBR103 were grown at the field site. Full details of this field trial are recorded elsewhere (15, 16). The experiment involved growing the sugar beet in nine 5-m2 plots. On days 28, 41, 78, 102, 148, 167, and 202 after sowing, three plants were taken from each plot (i.e., nine plants per treatment). Homogenates were prepared from the leaves, roots, and leaf buds of each plant and plated on, among other media, Pseudomonas selective agar (PSA) (Oxoid CM559 plus C-F-C supplement SR103), amended with 0.01% (wt/vol) 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and 100 μg of kanamycin ml−1, to select for the released SBW25 strains (15, 16). For the present study, these plates were then assayed for the presence of phages antagonistic towards SBW25. Bacteria from each plate were resuspended in 10 ml of sterile quarter-strength Ringer’s (QSR; Oxoid BR52). These suspensions were centrifuged (for 5 min at 14,000 × g), and the resulting supernatants were checked for phages by the overlay agar method of Adams (1) with SBW25 as the host.
Second field experiment.
A second release of P. fluorescens SBW25 (this time SBW25Rif) took place in 1996. Retaining the field design used the previous season, three of the 5-m2 plots (labelled 1 to 3) were used. Plot 1 had previously supported uninoculated sugar beet, while plots 2 and 3 had held plants inoculated with SBW2525EeZY6KX and SBW2525EeZY6KX(pQBR103), respectively. In February 1995, each plot was cleared of sugar beet from the preceding season. The field was then left fallow until April 1996, when, approximately 1 month prior to sowing, the plots were weeded and the soil was turned over. Three weeks later, the plots were fertilized and treated with herbicides and pesticides as described previously (15, 23). On 7 May 1996 (day 0), immediately prior to sowing, one soil sample from each quarter of the three plots was collected for analysis (n = 12).
Sugar beet seeds (EB3 pellets; Germains UK Ltd., Kings Lynn, United Kingdom) were inoculated with P. fluorescens SBW25Rif as described previously (15, 16). They were then sown into half of each of the three 5-m2 plots, with two seeds being planted at 23-cm intervals in a 5-by-10 matrix. Appropriately treated uninoculated seeds (15, 16) were similarly sown in the remaining half of each plot and as three rows of guard plants around each plot. Unused seeds were retained to assess the mean inoculum density by homogenizing in QSR and drop-plating onto selective media after dilution.
On days 29, 48, 78, 99, 119, 141, 156, 176, 197, 216, and 282, at least two plants were randomly sampled from both the treated and untreated sections of each plot. On days 29, 48, 176, 197, and 282, plants were taken from all three plots. On all other occasions, due to unexpected poor growth in plot 2, only plots 1 and 3 were sampled. Thus, in total, 20 plants were sampled on days 29 and 48, 12 were sampled on days 78 to 156, inclusive, 14 were sampled on days 176 and 197, 10 were sampled on day 216, and 11 were sampled on day 282. Eleven soil samples were also collected on day 282.
Rhizosphere, leaf, and leaf bud homogenates were prepared from each plant (15). Soil homogenates (0.5 g in 10 ml of sterile QSR) were similarly produced. Homogenates were plated onto TSBA (total viable count), PSA (total viable pseudomonad count), PSA plus rifampin (SBW25Rif count), and PSA plus kanamycin and X-Gal (previously released genetically engineered microorganisms) and screened for phages antagonistic toward SBW25 by using the overlay agar method (1). Phage screening was done the following day. Where no phages were detected, aliquots of the homogenates were enriched with nutrient broth, spiked with SBW25, and reassayed. Aliquots of each homogenate were stored at −80°C in 50% glycerol; the remainder was stored at 4°C.
On days 141 and 176 after sowing, two further phytosphere bacteria (designated CP6 and MCP1, respectively) were isolated. These strains were identified from their fatty acid methyl ester profiles, after comparison with the Microbial Identification Software aerobe (TSBA) library version 3.70 (MIS, Newark, Del.), as Serratia liquefaciens CP6 (similarity index, 0.774) and P. fluorescens MCP1 (similarity index, 0.834). Both proved useful host strains in phage assays and so were used, on all subsequent sampling occasions, in parallel with P. fluorescens SBW25. In addition, the homogenates stored at 4°C on previous dates were rescreened for phages antagonistic toward these two strains. On several sampling occasions, sugar beet growing in other parts of the field site (unassociated with the present experiment) were also sampled and assayed for the presence of phages for these strains.
Characterization of bacteriophages.
Plaque morphologies were examined, and differences were noted. Phage lysates were prepared from representative plaques from every positive overlay plate. DNA was extracted from these lysates (7) and cut with EcoRI or ClaI, as described by the manufacturer (Promega). Restriction profiles were compared by running on 0.7% agarose gels at 0.13 to 0.32 V cm−2 along with HindIII-cut lambda DNA (D-9780; Sigma).
Putative lysogens were isolated from plaque centers and, after purification, assessed for phage production to confirm their lysogen status.
Statistics.
Where possible, data sets were compared by analysis of variance (8). Log10 transformation was used for bacterial counts and phage titers, while arcsine transformation was used for percentages. Group means were then compared by using the Tukey-Kramer method for calculating minimum significant difference at P = 0.05 (8). When the assumptions of analysis of variance could not be met, group medians were compared by the Kruskal-Wallis test (8). Confidence interval notches (95%) for plotted medians were calculated by the method of Velleman and Hoaglin (24). Smoothed lines were calculated by using the 4253H twice algorithm (24).
RESULTS
Preliminary field experiment.
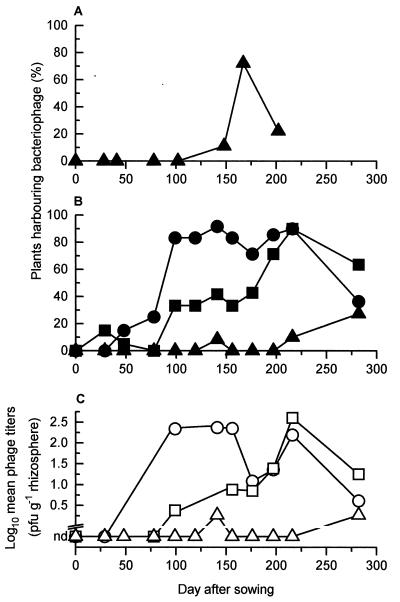
By assaying for phages present in the harvests of plates selective for P. fluorescens SBW25EeZY6KX or SBW25EeZY6KX(pQBR103), we determined the percentage of plants harboring SBW25 phages during 1994 (Fig. 1A). SBW25 phages were detected from day 148 until the end of the experiment on day 202 and were found throughout the phytosphere (42.1% of root samples, 57.9% of leaf samples, and 47.4% of leaf bud samples harbored phages).
FIG. 1.
Temporal variation in occurrence of phages isolated from sugar beet during 1994 (A) or 1996 to 1997 (B and C) and antagonistic toward P. fluorescens SBW25 (triangles), P. fluorescens MCP1 (squares), or S. liquefaciens CP6 (circles).
Second field experiment. (i) Presowing analysis of plots.
Soil taken from the plots immediately before sowing produced mean total counts of viable bacteria of 2.01 × 107 CFU g of soil−1 and mean total counts of viable pseudomonads of 4.48 × 105 CFU g of soil−1. No significant variation in total viable counts (P = 0.102) or total pseudomonad counts (P = 0.442) was detected among the plots. SBW25EeZY6KX, released in 1994, was not detected, and no pseudomonad sharing the rifampin-resistant phenotype of SBW25Rif was detected (limit of detection, 40 CFU g of soil−1). No phages antagonistic toward either SBW25, MCP1, or CP6 could be detected (limit of detection after nutrient enriching and spiking with host bacteria, 2 PFU g of soil−1).
(ii) Monitoring of the introduced inoculum.
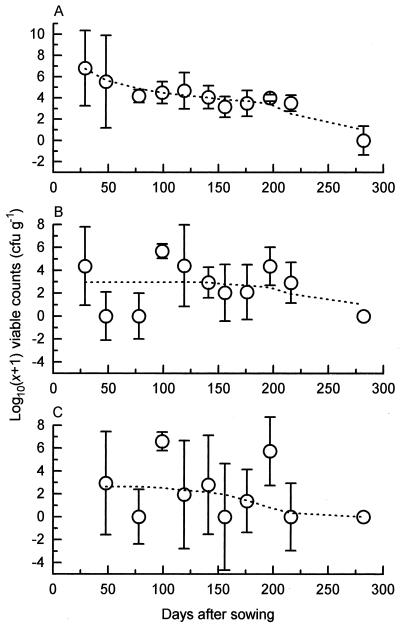
P. fluorescens SBW25Rif was successfully introduced onto the surface of the inoculated sugar beet seeds (mean count, 1.10 × 107 CFU seed−1, n = 30) and subsequently maintained a substantial population throughout the phytosphere of the resulting plants (Fig. 2). P. fluorescens SBW25EeZY6KX was not found.
FIG. 2.
Temporal variation during 1996 to 1997 of viable counts of P. fluorescens SBW25 established in the phytosphere microflora of sugar beet rhizosphere (A), leaf (B), and leaf bud (C) samples. Group medians for each sampling occasion are plotted (circles), along with their respective boxplot notches (bars [27]), enabling a pairwise comparison of group medians at the 95% level; overlapping bars indicate medians that are not significantly different. Dotted lines represent the same data after smoothing with the 4253H twice algorithm (27).
(iii) Monitoring of SBW25 phages.
From days 0 to 119, inclusive, no SBW25 phages were isolated from the samples, even after nutrient enrichment. On day 141, phages were again not detected; however after the homogenates were left on the bench overnight, phages could be isolated from the root and leaf bud of one plant (Fig. 1B and C). No further phages were found until days 216 and 282, when, again after overnight incubation, phages were detected (Fig. 1B and C). The mean limits of detection for phages isolated from root, leaf, and leaf bud samples without recourse to nutrient enrichment were 1.9 × 102, 2.0 × 102, and 5.6 × 102 PFU g (wet weight) of plant material−1, respectively. The limits of detection after enrichment were 10-fold lower. All plants testing positive for phages came from the treated sectors of plots 1 and 3.
(iv) Monitoring of non-SBW25 phages.
On day 141, an indigenous phytosphere bacterium, S. liquefaciens CP6, was isolated. When CP6 was used as an alternative to SBW25 in overlay agar assays, phages antagonistic toward this strain were successfully isolated from 11 of the 12 plants sampled on that day. Similarly, on day 176, another indigenous strain, P. fluorescens MCP1, was isolated which also proved successful in isolating phages (6 of the 14 plants sampled on day 176). The fatty acid methyl ester profile of MCP1 was remarkably similar to that of SBW25 (similarity index, 0.854), suggesting that they were closely related. In spite of this, MCP1 was completely insensitive to all the SBW25 phages we had isolated. Equally, all subsequently isolated MCP1 phages failed to lyse SBW25. MCP1 also lacked any of the markers associated with either SBW25Rif or SBW25EeZY6KX.
By retrospectively scoring stored homogenates for the presence of phages antagonistic toward CP6 or MCP1 and using both strains in phage assays during subsequent sampling occasions, phages for both these strains were found on a large number of sugar beet plants throughout the experiment but most commonly on plants at least 3 months old (Fig. 1B).
CP6 and MCP1 phage titers were also recorded for the most of these homogenates. Control experiments indicated that homogenate titers increased by a median factor of 42 (n = 17) with storage but that no further significant increase occurred after 24 h for at least up to 3 months (P = 0.145). Thus, with this correction factor in mind, mean phage titers for both CP6 and MCP1 could also be tentatively plotted (Fig. 1C). The similarity between Fig. 1B and C reflects the constancy of titers obtained from homogenates harboring phages. Indeed, a comparison of the most reliable titer data, collected after the discovery of CP6 and MCP1 (i.e., days 141 and 176 onward), showed that there was no significant variation in titer among sampling occasions when phages were present (P = 0.153). Over this period, CP6 phage titers ranged from 2.1 × 100 to 5.3 × 103 (mean, 1.1 × 103) PFU g−1 and MCP1 phage titers ranged from 1.8 × 100 to 4.2 × 104 (mean, 4.0 × 103) PFU g−1.
In total, 87 of the 149 plants sampled over the 9-month experiment were found to harbor detectable levels of CP6 phage whereas 53 plants harbored MCP1 phage. Both CP6 and MCP1 phages were found in all plot sectors, with no significant difference detected (P = 0.867). Sugar beet from other regions of the field site, outside of our three plots and thus unassociated with the present study, were also assayed and found to harbor CP6 and MCP1 phages.
As with SBW25 phages, MCP1 phages were found in all the phytosphere regions examined (86.8, 43.4, and 32.1% of root, leaf, and leaf bud samples, respectively, of harbored phages). In contrast, CP6 phages were isolated from the root samples of every plant found to harbor CP6 phage, while only 9.2% of these had CP6 phages on their leaves. No CP6 phages were isolated from leaf buds.
Soil samples taken at both the beginning and the end of the field experiment were also assayed for phages. No phages were isolated from the 12 day 0 samples. However, on day 282, MCP1 phages were isolated from 4 of the 11 soil samples taken, with phage counts of 4.00 × 102 to 6.2 × 103 PFU g of soil−1. CP6 phages were isolated from one soil sample at 4.00 × 103 PFU g−1.
(v) Characterization of bacteriophages.
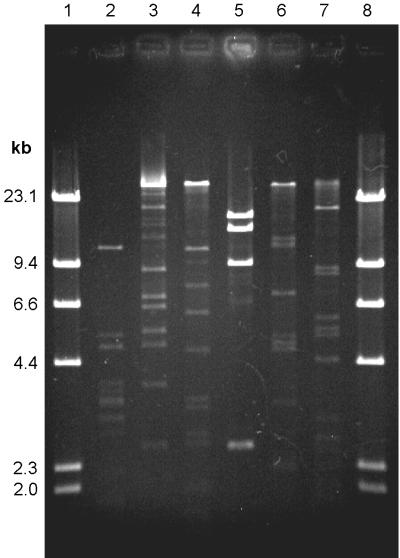
Variation in plaques produced on overlay plates indicated that all three host bacteria were parasitized by more than one phage species. The plaques varied in both turbidity and size. This variability was confirmed when phage DNA was extracted and restriction digests were compared. In this way, two MCP1 phages (labelled ΦMCP1-1 and ΦMCP1-2), at least three SBW25 phages, and six CP6 phages (ΦCP6-1 to ΦCP6-6) (Fig. 3) were identified.
FIG. 3.
ClaI restriction digests, run on a 0.7% agarose gel, illustrating the six different phages antagonistic toward S. liquefaciens CP6, isolated from sugar beet during 1996 to 1997. Lanes 1 and 8, HindIII-cut lambda DNA ladder; lanes 2 to 7, phages ΦCP6-1 to ΦCP6-6, respectively.
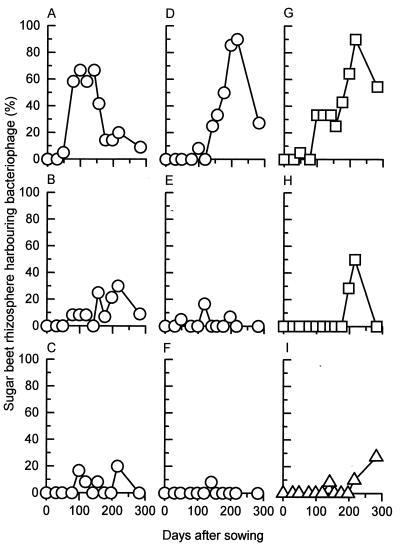
No significant difference (P > 0.05) in distribution of the individual phages could be detected between the two plots with the most complete data sets (plots 1 and 3), although significant variations in abundances of the various phage types were identified, with phages ΦCP6-1, ΦCP6-4, and ΦMCP1-1 being the most abundant (Fig. 4). The restriction profiles of these abundant phages did not vary either between plots or over time.
FIG. 4.
Temporal variation in occurrence of individual phage species in sugar beet rhizosphere samples over the 1996 to 1997 experimental period. Phages ΦCP6-1 to ΦCP6-6 (A to F, respectively) were antagonistic toward S. liquefaciens CP6, while phages ΦMCP1-1 and ΦMCP1-2 (G and H) infected P. fluorescens MCP1. Phages antagonistic toward P. fluorescens SBW25 are also represented (I). Twenty sugar beet plants were sampled on days 29 and 48, 12 were sampled on days 78 to 156 inclusive, 14 were sampled on days 176 and 197, 10 were sampled on day 216, and 11 were sampled on day 282.
(vi) Temporal variation by individual phages.
When plotted against time, a distinct variation in the occurrence of these genetically distinct phages was observed for CP6 and MCP1 (and to a lesser extent SBW25) in the rhizosphere (Fig. 4). For the MCP1 phages, this variation was a straightforward increase in occurrence late in the season, which subsequently declined by the end of the experiment (Fig. 4G and H). However, with CP6 phages the situation was more complex (Fig. 4A to F). Most striking was a clear temporal difference between phages ΦCP6-1 (Fig. 4A) and ΦCP6-4 (Fig. 4D), with the former predominating within the experimental plot early in the season, to be succeeded by the latter as the experiment progressed. Besides having different restriction profiles, these two phages differed dramatically in their plaque morphology. On bacterial lawns grown overnight at 15°C, phage ΦCP6-1 produced 1-mm-diameter turbid plaques whereas ΦCP6-4 formed equivalent-sized clear plaques. Further incubation failed to elicit any major change in ΦCP6-1 plaque morphology; however, with ΦCP6-4, several concentric rings developed around the initial clear plaque, so that after 24 h these plaques had increased to three times their original diameter. The presence of the turbid plaques suggested that ΦCP6-1 was a temperate phage. This hypothesis was confirmed when lysogens were successfully isolated. In contrast, ΦCP6-4 failed to produce any lysogens, strongly suggesting that it was virulent.
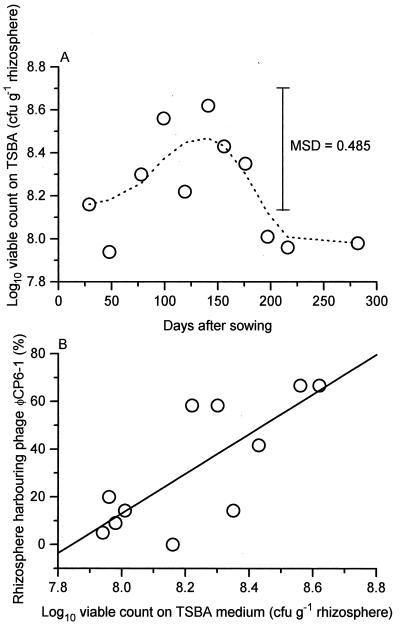
The change in distribution of phage ΦCP6-1 also coincided with a significant increase and subsequent decrease in overall bacterial numbers on the surface of the sugar beet roots, as determined on TSBA (P < 0.05) (Fig. 5A). When plotted against ΦCP6-1, a positive correlation was observed (correlation coefficient of 0.769, P < 0.05) (Fig. 5B), indicating a significant linear association between the increased distribution of that phage and overall bacterial numbers.
FIG. 5.
Temporal variation in total numbers of viable bacteria, isolated on TSBA medium, within the sugar beet rhizosphere during 1996 to 1997. Group means (n = 10 to 20, depending on the sampling day) are plotted against time (A) and phage ΦCP6-1 abundance (B). Smoothed data (––––) was calculated from the means by using the 4253H twice algorithm (27). Least-squares linear regression (9) was used to calculate the regression line (——). Phage abundance (%) = −652.2 + 83.2 × viable count. MSD, minimum significant difference.
DISCUSSION
P. fluorescens SBW25, P. fluorescens MCP1, and S. liquefaciens CP6 are natural components of the sugar beet microflora. During this study, populations of phages, antagonistic toward these bacteria, were also shown to be part of this microflora. All three phage populations fluctuated over time and to differing degrees. The phages were most readily found on sugar beet from roughly 3 months after sowing onwards (i.e., after day 78), coinciding with the period of most rapid sugar beet growth (60 to 120 days).
The three phage populations also had different distributions on the sugar beet plants, presumably reflecting variations in the spatial distribution of their respective hosts. Thus, phages antagonistic toward SBW25 or MCP1 were found throughout the phytosphere, while CP6 phages predominated in the rhizosphere.
Each phage population consisted of various phage species, as indicated by differences in plaque morphology and confirmed by restriction analysis. The CP6 phage population was particularly varied, with six genetically distinct phages being identified. This led to the observation that not only did overall phage populations change with time but also the occurrence of individual phages making up these populations fluctuated, so much so that a distinct temporal variation was observed between phages ΦCP6-1 and ΦCP6-4, the two most dominant phages predating on CP6. These dominant phages occurred in all the plots sampled (as well as on sugar beet unassociated with the experiment), with no detectable variation in restriction profile. Their restriction profiles did not change in any detectable way with time, suggesting the presence of stable genomes.
In 1994, we detected a bloom in SBW25 phages late in the season, which coincided with sugar beet maturation and succeeded a bloom in conjugative-gene transfer events noted between phytosphere bacteria occurring the same year at the same site (15, 16). These results suggested a specific period during the growing season of sugar beet during which phage infection, in concert with conjugative-gene transfer, was more likely. However, in 1996, no such bloom was observed, although small numbers of phages were found on day 141 and at the end of the experiment on days 216 and 282. These results illustrate the inherent variability between years, which is not surprising given the number of environmental factors likely to affect bacterial populations and thus phage numbers.
In contrast to SBW25, CP6 and MCP1 phage populations were monitored without prior inoculation of the sugar beet with these strains. Both CP6 and MCP1 were isolated and identified as useful indicator strains during the 1996 field trial. These were purely serendipitous events occurring when indigenous bacteria, carried over from the homogenates onto the overlay agar plates during SBW25 phage assays, were left to develop faint but discernible bacterial lawns in their own right, superimposed over the intended SBW25 lawn. Within these lawns, apparent large plaques were observed. Subsequent isolation and purification of both the contaminating bacteria and phage from these plaques produced not only indigenous bacteria but also their associated phages from the same sample. That CP6 and MCP1 were isolated when they were (days 141 and 176, respectively) was probably because only then did the large-plaque-forming ΦCP6-4 and ΦMCP1-2 phages start to become more abundant and thus noticeable.
This relatively late discovery of CP6 and MCP1 precluded us from accurately monitoring changes in in situ phage numbers over time for these two bacteria. Control experiments demonstrated that homogenate titers increased with storage, albeit by a constant factor, which did enable a tentative estimation of phage numbers to be made (Fig. 1C). Nevertheless, by recording the presence or absence of phages antagonistic toward these two strains in each homogenate collected, we had a method that was insensitive to such storage problems. Therefore, we were able to reliably monitor changes in phage abundance, in terms of their level of distribution within the field site, over time (Fig. 1B), and it is worth noting that these fluctuations closely correlate with our tentative estimation of mean phage titers over the same period (Fig. 1C).
By this approach, we showed that CP6 and MCP1 phage populations experienced dramatic increases as the crop developed (in stark contrast to SBW25 phages). Where recorded, their in situ titers were also typically an order of magnitude higher than that estimated for SBW25. Such results highlight the importance of choosing not only the correct indicator host for study during any particular year but also the most appropriate strain of that host. After all, P. fluorescens MCP1 and SBW25 had remarkably similar fatty acid profiles and yet were predated on by completely separate phage populations with dramatically different abundances. Our experience with SBW25 during two separate years also cautions us against placing too great a predictive value on our single year of CP6 and MCP1 data, highlighting the need, for data of this sort, to be amassed over a number of years to properly assess trends.
Nevertheless, our results are notable in two respects. They demonstrate, for the first time, temporal changes in several phage populations antagonistic toward different bacteria occurring in a terrestrial environment. Previous in situ studies have concentrated on aquatic environments, and even then, these studies involved tracing overall phage morphology types observed by transmission electron microscopy rather than viable phage populations for specific hosts (3).
Also, our results are unique in that they describe changing relative distributions of the specific phages making up these populations. Figure 4A to F demonstrate that over about 6 months, the composition of the population of phages predating on CP6 changed from one dominated by phage ΦCP6-1 to one dominated by phage ΦCP6-4. In addition, less dramatic changes in the other CP6 phages occurred.
The domination of phages ΦCP6-1 and ΦCP6-4 at different times suggests they are adapted to two quite different temporal niches in the rhizosphere. Being obligate parasites, phages are largely dependent on the availability and physiological status of their host. Thus, the temporal variation observed may well reflect changes in host physiology and numbers, which are likely to have been due to physiological changes in the sugar beet, environmental conditions, or both, as the year progressed.
Clearly, phages ΦCP6-1 and ΦCP6-4 are physiologically different. The temperate nature of ΦCP6-1, suggested by its turbid plaques and confirmed when lysogens were produced, contrasts with ΦCP6-4, which appeared to be entirely virulent. The fact that ΦCP6-4 plaques continued to grow long after ΦCP6-1 plaques had stopped growing also suggests a difference in the capacities of the two phages to infect the bacterial host as it undergoes the inevitable physiological changes associated with an ageing bacterial lawn.
What these differences might say about the respective niches of these two phages is unclear, due, in the main, to our current lack of knowledge of the changing population dynamics of the host. It has been postulated that temperate phages are more likely to predominate over virulent phages when relatively small numbers of physiologically suitable host bacteria are present (21). If true, this would suggest that numbers of metabolically active CP6 increased later in the growing season when the occurrence of ΦCP6-1 decreased and that of ΦCP6-4 increased.
However, the prolonged lysis of ΦCP6-4 on CP6 lawns suggests that this phage might have a selective advantage over ΦCP6-1 when the host is not so metabolically active. Thus, a decrease in numbers of metabolically active CP6 late in the season may be a more realistic scenario. Certainly, a small but significant drop in overall bacterial counts was recorded, but whether this reflected an equivalent change in CP6 numbers is, of course, unknown.
Both Pseudomonas and Serratia spp. are regularly occurring members of the sugar beet phytosphere (22) and are known to interact with the “host” plant (11, 17). The observation that phage population dynamics of the sort described in this paper occurred for two natural isolates strongly suggests that such changes are common among phytosphere bacteria. From a broader viewpoint, our observations highlight the need for a greater understanding of phage ecology in all natural environments, especially in habitats where a thorough understanding of microbial community dynamics is required.
ACKNOWLEDGMENTS
This work was supported by Ministry of Agriculture Fisheries and Food grant RG0112. A.K.L. was in receipt of a NERC personal fellowship.
We thank Vicki Chesters, Silké Hagen, and Susan Norris for technical assistance.
REFERENCES
- 1.Adams M H. Bacteriophages. New York, N.Y: Interscience Publishers; 1959. [Google Scholar]
- 2.Bailey M J, Lilley A K, Thompson I P, Rainey P B, Ellis R J. Site directed chromosomal marking of a fluorescent pseudomonad isolated from the phytosphere of sugar beet; stability and potential for marker gene transfer. Mol Ecol. 1995;4:755–763. doi: 10.1111/j.1365-294x.1995.tb00276.x. [DOI] [PubMed] [Google Scholar]
- 3.Bergh Ø, Børsheim K Y, Bratbak G, Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–468. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- 4.Bratbak G, Heldal M, Norland S, Thingstad F. Viruses as partners in spring bloom microbial trophodynamics. Appl Environ Microbiol. 1990;56:1400–1405. doi: 10.1128/aem.56.5.1400-1405.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chao L, Levin B R, Stewart F M. A complex community in a simple habitat: an experimental study with bacteria and phage. Ecology. 1977;58:369–378. [Google Scholar]
- 6.Cresswell N, Herron P R, Saunders V A, Wellington E M H. The fate of introduced streptomycetes, plasmid and phage populations in a dynamic soil system. J Gen Microbiol. 1992;138:659–666. [Google Scholar]
- 7.Day M J, Marchesi J R. Transduction in the aquatic environment. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1996. pp. 1–21. [Google Scholar]
- 8.Fry J C. One-way analysis of variance. In: Fry J C, editor. Biological data analysis. Oxford, United Kingdom: IRL Press; 1992. pp. 1–39. [Google Scholar]
- 9.Germida J J. Population dynamics of Azospirillum brasilense and its bacteriophage in soil. Plant Soil. 1986;90:117–128. [Google Scholar]
- 10.Horne M T. Coevolution of Escherichia coli and bacteriophages in chemostat culture. Science. 1970;168:992–993. doi: 10.1126/science.168.3934.992-a. [DOI] [PubMed] [Google Scholar]
- 11.Kalbe C, Marten P, Berg G. Strains of the genus Serratia as beneficial rhizobacteria of oilseed rape with antifungal properties. Microbiol Res. 1996;151:433–439. doi: 10.1016/S0944-5013(96)80014-0. [DOI] [PubMed] [Google Scholar]
- 12.Kidambi S P, Ripp S, Miller R V. Evidence for phage-mediated gene transfer among Pseudomonas aeruginosa strains on the phylloplane. Appl Environ Microbiol. 1994;60:496–500. doi: 10.1128/aem.60.2.496-500.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lenski R E, Levin B. Constraints on the coevolution of bacteria and virulent phage: a model, some experiments, and predictions for natural communities. Am Nat. 1985;125:585–602. [Google Scholar]
- 14.Levin B R, Stewart F M, Chao L. Resource-limited growth, competition, and predation: a model and experimental studies with bacteria and bacteriophage. Am Nat. 1977;111:3–24. [Google Scholar]
- 15.Lilley A K, Bailey M J. The acquisition of indigenous plasmids by a genetically marked pseudomonad population colonizing the sugar beet phytosphere is related to local environmental conditions. Appl Environ Microbiol. 1997;63:1577–1583. doi: 10.1128/aem.63.4.1577-1583.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lilley A K, Bailey M J. Impact of plasmid pQBR103 acquisition and carriage on the phytosphere fitness of Pseudomonas fluorescens SBW25: burden and benefit. Appl Environ Microbiol. 1997;63:1584–1587. doi: 10.1128/aem.63.4.1584-1587.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Sullivan D J, O’Gara F. Traits of fluorescent Pseudomonas spp. involved in suppression of plant root pathogens. Microbiol Rev. 1992;56:662–676. doi: 10.1128/mr.56.4.662-676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Sullivan M, Stephens P M, O’Gara F. Interactions between the soil-borne bacteriophage Fo-1 and Pseudomonas spp. on sugarbeet roots. FEMS Microbiol Lett. 1990;68:329–334. [Google Scholar]
- 19.Pantastico-Caldas M, Duncan K E, Istock C A. Population dynamics of bacteriophage and Bacillus subtilis in soil. Ecology. 1992;73:1888–1902. [Google Scholar]
- 20.Stephens P M, O’Sullivan M, O’Gara F. Effect of bacteriophage on colonization of sugar beet roots by fluorescent Pseudomonas spp. Appl Environ Microbiol. 1987;53:1164–1167. doi: 10.1128/aem.53.5.1164-1167.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stewart F M, Levin B R. The population biology of bacterial viruses: why be temperate. Theor Popul Biol. 1984;26:93–117. doi: 10.1016/0040-5809(84)90026-1. [DOI] [PubMed] [Google Scholar]
- 22.Thompson I P, Bailey M J, Fenlon J S, Fermor T R, Lilley A K, Lynch J M, McCormack P J, McQuilken M P, Purdy K J, Rainey P B, Whipps J M. Quantitative and qualitative seasonal changes in the microbial community from the phyllosphere of sugar beet (Beta vulgaris) Plant Soil. 1993;150:177–191. [Google Scholar]
- 23.Thompson I P, Lilley A K, Ellis R J, Bramwell P A, Bailey M J. Survival, colonization and dispersal of genetically modified Pseudomonas fluorescens SBW25 in the phytosphere of field grown sugar beet. Bio/Technology. 1995;13:1493–1497. [Google Scholar]
- 24.Velleman P F, Hoaglin D C. Applications, basics, and computing of exploratory data analysis. Boston, Mass: Duxbury Press; 1981. [Google Scholar]
- 25.Williams S T, Mortimer A M, Manchester L. Ecology of soil bacteriophages. In: Goyal S H, Gerba C P, Bitton G, editors. Phage ecology. New York, N.Y: John Wiley & Sons, Inc.; 1987. pp. 157–179. [Google Scholar]