Abstract
Acanthamoebae are ubiquitous soil and water bactivores which may serve as amplification vehicles for a variety of pathogenic facultative bacteria and as hosts to other, presently uncultured bacterial endosymbionts. The spectrum of uncultured endosymbionts includes gram-negative rods and gram-variable cocci, the latter recently shown to be members of the Chlamydiales. We report here the isolation from corneal scrapings of two Acanthamoeba strains that harbor gram-negative rod endosymbionts that could not be cultured by standard techniques. These bacteria were phylogenetically characterized following amplification and sequencing of the near-full-length 16S rRNA gene. We used two fluorescently labelled oligonucleotide probes targeting signature regions within the retrieved sequences to detect these organisms in situ. Phylogenetic analyses demonstrated that they displayed 99.6% sequence similarity and formed an independent and well-separated lineage within the Rickettsiales branch of the alpha subdivision of the Proteobacteria. Nearest relatives included members of the genus Rickettsia, with sequence similarities of approximately 85 to 86%, suggesting that these symbionts are representatives of a new genus and, perhaps, family. Distance matrix, parsimony, and maximum-likelihood tree-generating methods all consistently supported deep branching of the 16S rDNA sequences within the Rickettsiales. The oligonucleotide probes displayed at least three mismatches to all other available 16S rDNA sequences, and they both readily permitted the unambiguous detection of rod-shaped bacteria within intact acanthamoebae by confocal laser-scanning microscopy. Considering the long-standing relationship of most Rickettsiales with arthropods, the finding of a related lineage of endosymbionts in protozoan hosts was unexpected and may have implications for the preadaptation and/or recruitment of rickettsia-like bacteria to metazoan hosts.
Members of the Rickettsiales comprise a diverse group of bacteria, most of which are small gram-negative rods that exist as either parasitic or mutualistic symbionts within eukaryotic cells. Beyond these common characteristics, uncertainty about their classification is mostly due to the difficulties of working with obligate intracellular bacteria. In addition to the usual genera (Rickettsia, Orientia, Neorickettsia, Ehrlichia, Wolbachia, Cowdria, and Bartonella, among others) historically included in the Rickettsiales on the basis of phenotypic and/or genotypic data (12, 36, 46, 48), a large number of “rickettsia-like” endosymbiotic bacteria that are associated with protozoa, insects and other invertebrates, and fungi are incompletely described (33).
Previously, we reported the occurrence of noncultured bacterial endosymbionts in both clinical and environmental isolates of Acanthamoeba spp. (16). To date, 17 (22%) of 78 axenically growing Acanthamoeba strains we maintain contain endosymbionts, including the presence of gram-negative rods (GNR) in 17% (13 amoebic isolates) and gram-negative cocci (GNC) in 5% (4 amoebic isolates) (15). Preliminary phylogenetic analyses of three of the GNC strains revealed that they were most closely related to but distinct from the genus Chlamydia, while one of the GNR was shown to display affinities to the Rickettsiales (19). This is consistent with other recent reports describing the recovery of Chlamydia-like endosymbionts in Acanthamoeba spp. (2, 8) and an Ehrlichia-like endosymbiont within an isolate of Saccamoeba (30). The finding of protozoal endosymbionts closely related to members of the Chlamydiales and Rickettsiales adds to the diversity of bacterial lineages that adapted themselves to intracellular survival within amoebae. While the life cycles of the Chlamydiales and Rickettsiales are typically dependent upon an intracellular habitat for survival and growth, a variety of facultatively growing bacteria, most notably members of the Legionellaceae, also survive and multiply within amoebic hosts. Such host-symbiont interactions are thought to be critical in the epidemiology of legionellosis (3, 6, 14, 37).
Acanthamoeba spp. are increasingly recognized as serious human pathogens responsible for keratitis, granulomatous encephalitis, and both focal and systemic disease in immunocompromised hosts, although the mechanisms of pathogenesis are poorly understood (20). Due to the recent observation of putative enhancement of cytopathogenicity of Acanthamoeba following acquisition of noncultured GNR and GNC bacterial endosymbionts (17) and the potential of GNC endosymbionts to directly produce human disease (8), a more detailed characterization of Acanthamoeba endosymbionts may be of clinical relevance. In this paper, we present details of the morphologic and phylogenetic analyses of two GNR endosymbionts infecting axenically maintained isolates of Acanthamoeba originally recovered from patients with amoebic keratitis. Because these bacterial isolates could not be cultivated by standard microbiological approaches, we undertook a comparative analysis of their 16S rRNA genes to determine their phylogenetic affiliations. Fluorescently labelled oligonucleotide probes targeting signature regions within the retrieved 16S rDNA sequences were subsequently designed for in situ hybridization to further assist with the characterization and intracellular localization of individual bacterial cells.
MATERIALS AND METHODS
Isolation and maintenance of Acanthamoeba strains.
The techniques used for recovery and maintenance of acanthamoebae from clinical and environmental sources are described elsewhere (16, 44). Briefly, primary isolation was performed from infected human corneal tissues by using 1.5% nonnutrient agar plates seeded with live Escherichia coli. Subsequent incubation was performed at ambient temperature (22 to 24°C) for up to 10 days. Upon evidence of growth, clonal cultures were established by transference of a single double-walled cyst to fresh medium. The use of heat-killed E. coli and/or incorporation of antibiotics (penicillin, 100 μg/ml; streptomycin, 10 μg/ml; and amphotericin B, 0.25 μg/ml) in subsequent subcultures resulted in axenic growth. Clones were then adapted to growth in sterile tryptic soy-yeast extract broth. Two isolates of Acanthamoeba (UWC8 and UWC36) known to be infected with intracellular, rod-shaped bacteria that are readily detected by Gram, Giemsa, and fluorochrome staining methods were included in this study. General phenotypic characteristics of both endosymbiont strains, including an electron micrograph of UWC8, have been described previously (16, 18).
DNA isolation, PCR amplification, cloning, and sequencing.
Amoebae and their endosymbionts were harvested from axenic cultures, washed twice with double-distilled water, and resuspended in 500 μl of an appropriate lysis buffer. UWC8 amoebae were lysed in STE buffer (2% sodium dodecyl sulfate [SDS], 10 mM EDTA, 50 mM Tris-HCl [pH 8.0]) containing 0.3 mg of proteinase K per ml by incubation at 37°C for 2 h, followed by 5 min of gentle inversion at room temperature (28); UWC36 amoebae were lysed in UNSET buffer (8 M urea, 2% SDS, 0.15 M NaCl, 0.001 M EDTA, 0.1 M Tris-HCl [pH 7.5]) by incubation at 60°C for 5 min (23). The lysates were extracted twice with phenol-chloroform, and DNA was precipitated with 2 volumes of absolute ethanol.
Oligonucleotide primers targeting highly conserved 16S rDNA signature regions within the domain Bacteria were used for PCR to obtain near-full-length bacterial 16S rRNA gene fragments. The nucleotide sequences of the forward and reverse primers used for amplification of UWC8 were, respectively, 5′-AGAGTTTGATCCTGGCTCAG-3′ and 5′-ACGGCTACCTTGTTACGACTT-3′ (47), while those used for UWC36 were, respectively, 5′-AGAGTTTGATYMTGGCTCAG-3′ (Escherichia coli 16S rDNA positions 8 to 27) and 5′-CAKAAAGGAGGTGATCC-3′ (E. coli 16S rDNA positions 1529 to 1546) (9). Amplification reactions for UWC8 were performed in a 100-μl reaction volume in a programmable thermal cycler (Perkin-Elmer, Foster City, Calif.) with the GeneAmp PCR reagent kit (Perkin-Elmer) as recommended by the manufacturer. Thermal cycling consisted of 35 cycles of denaturation at 94°C for 1.5 min, annealing at 42°C for 1 min, and elongation at 72°C for 4 min, with a final elongation step at 72°C for 20 min. Amplification reactions for UWC36 were performed in a reaction volume of 50 μl in a thermal capillary cycler with reaction mixtures, including a 20 mM MgCl2 reaction buffer, prepared as recommended by the manufacturer (Idaho Technology, Idaho Falls, Idaho) with Taq DNA polymerase (Promega, Madison, Wis.). Thermal cycling consisted of an initial denaturation step at 94°C for 30 s followed by 30 cycles of denaturation at 94°C for 20 s, annealing at 52°C for 15 s, and elongation at 72°C for 30 s, with a final elongation step at 72°C for 1 min. Positive controls containing purified DNA from E. coli were included along with negative controls (no DNA added). The presence and size of the amplification products were determined by 0.8% agarose gel electrophoresis and ethidium bromide staining of the reaction product.
Amplified DNA from UWC8 was purified by electrophoresis in low-melting-point agarose and ligated into the cloning vector Bluescript II (Stratagene, La Jolla, Calif.), while amplified DNA from UWC36 was directly ligated into the cloning vector pCR2.1 (Invitrogen, Carlsbad, Calif.), with subsequent transformation of E. coli by each vector. Nucleotide sequences of the cloned DNA fragments were determined by automated dideoxynucleotide methods with the Taq DyeDeoxy Terminator cycle-sequencing kit (Perkin-Elmer Applied Biosystems, Foster City, Calif.) for UWC8 and the Thermo Sequenase cycle-sequencing kit (Amersham Life Science, Little Chalfont, England) for UWC36.
Phylogenetic analysis.
16S rDNA sequences were added, with the program package ARB, to the 16S rRNA sequence database of the Technischen Universität München, which encompasses about 10,000 published and unpublished homologous small-subunit rRNA primary structures (11, 26, 42). Alignment of sequences was performed with the ARB automated alignment tool. Alignments were refined by visual inspection and by secondary-structure analysis. Phylogenetic analyses were performed by applying ARB parsimony, distance matrix, and maximum-likelihood methods to different data sets. To determine the robustness of phylogenetic trees, analyses were performed both on the original data set and on a data set from which highly variable positions were removed by use of a 50% conservation filter for the members of the Rickettsiales (24). A check for chimeric sequences was conducted by independently subjecting the first, second, and third 513-base positions (5′ to 3′) to independent phylogenetic analyses.
In situ identification and detection of Acanthamoeba endosymbionts.
Acanthamoeba cells were harvested from 3 ml of liquid broth culture by centrifugation (200 × g for 3 min), washed briefly with 1 ml of Page’s saline (44), and pretreated for in situ hybridization (39). The specific pretreatments included (i) resuspension of amoebic cells in a 1:3 ratio of Page’s saline and 4% paraformaldehyde for 12 h at 4°C, spotting of 30 μl of the cell suspension onto glass slides, and air drying; (ii) resuspension of cells in a 1:9 ratio of saturated mercuric chloride and Page’s saline for 12 h at 4°C, washing with 1 ml of Page’s saline, spotting of 30 μl onto slides, air drying, and dehydration by immersion in 80% ethanol for 5 to 10 s; (iii) resuspension of cells in 0.4% trichloroacetic acid in Page’s saline for 15 min at room temperature followed by processing as for (ii); (iv) resuspension of cells in Page’s saline followed by spotting of 50 μl of suspension on a glass slide, storing the slide in a moisture chamber for 2 h to permit natural cell attachment, immersion in 80% ethanol for 10 to 30 s, and air drying; and (v) resuspension of cells in a solution containing 0.05% (final concentration) agarose in Page’s saline, spotting of 20 μl onto a slide, air drying, and immersion in 80% ethanol for 5 to 10 s.
Oligonucleotide probes S-*-AcEnd-0090-a-A-18 (AcRic90) and S-*-AcEnd-1196-a-A-18 (AcRic1196), both specific for UWC8 and UWC36 endosymbionts, were designed by using the Probedesign/Probematch tools of ARB (42); the probes were designated according to the standard proposed by Alm et al. (1). Oligonucleotides were synthesized and directly labeled with the hydrophilic sulfoindocyanine fluorescent dye Cy3 or Cy5 (Interactiva, Ulm, Germany). Optimal hybridization conditions for the probes were determined by using the hybridization and wash buffers (with and without SDS) described by Manz et al. (27). Negative control in situ hybridization experiments were performed with Cy3- and Cy5-labelled derivatives of the oligonucleotide probe BET42a, specific for the beta subclass of Proteobacteria (27). The slides were examined with a confocal laser-scanning microscope (LSM 510; Carl Zeiss, Oberkochen, Germany) with two helium-neon lasers (543 and 633 nm). Image analysis processing was performed with the standard software package delivered with the instrument (version 1.5). Staining of endosymbiont-harboring amoebic cells with 4′,6-diamidino-2-phenylindole (DAPI) after in situ hybridization was performed by incubation with 1 μM aqueous DAPI solution for 4 min at room temperature.
Electron microscopy.
Amoebic strains in which symbioses were detected by conventional microscopy were further examined by electron microscopy, using a variation of published methods (21). Briefly, aliquots of amoebae in broth were fixed with 2% glutaraldehyde in 0.1 M cacodylate. The fixed amoebae were then pelleted in agar and embedded. Thin sections were stained with uranyl acetate and lead citrate and examined with a Philips CM-10 electron microscope.
Nucleotide sequence accession numbers.
Recovered 16S rDNA sequences were deposited in GenBank under accession no. AF069962 (endosymbiont of Acanthamoeba sp. strain UWC36) and AF069963 (endosymbiont of Acanthamoeba sp. strain UWC8).
RESULTS
Phylogenetic inference.
Two almost complete 16S rDNA sequences were amplified, cloned, and sequenced from two clinical Acanthamoeba isolates containing microscopically observable prokaryotic endosymbionts. Comparative sequence analysis revealed that the UWC8 and UWC36 endosymbiont 16S rDNA sequences were almost identical (99.6% sequence similarity) and clustered unequivocally with members of the alpha subclass of Proteobacteria (Table 1). Their closest neighbors included Rickettsia australis, R. sibirica, and R. typhi, with sequence similarities of approximately 85 to 86%.
TABLE 1.
Overall 16S rRNA sequence similarities for the UWC8 and UWC36 endosymbionts and affiliated bacteria
| Strain | % Similarity to:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| W. pipientis | Wolbachia sp. | E. canis | C. ruminantium | E. equi | N. helminthoeca | R. sibirica | R. australis | R. typhi | O. tsutsugamushi | C. caryophila | UWC36 endosymbiont | |
| Wolbachia pipientis | ||||||||||||
| Wolbachia sp. | 99.2 | |||||||||||
| Ehrlichia canis | 86.4 | 86.1 | ||||||||||
| Cowdria ruminantium | 86.8 | 86.5 | 96.3 | |||||||||
| Ehrlichia equi | 87.2 | 87.0 | 91.8 | 92.0 | ||||||||
| Neorickettsia helminthoeca | 84.8 | 84.2 | 84.9 | 85.2 | 85.5 | |||||||
| Rickettsia sibirica | 83.8 | 83.4 | 82.9 | 83.9 | 83.7 | 83.8 | ||||||
| Rickettsia australis | 83.7 | 83.1 | 82.6 | 83.6 | 83.4 | 83.1 | 98.8 | |||||
| Rickettsia typhi | 83.9 | 83.5 | 82.6 | 83.3 | 83.9 | 83.4 | 98.4 | 98.1 | ||||
| Orientia tsutsugamushi | 82.4 | 81.8 | 82.3 | 83.4 | 83.0 | 82.1 | 90.3 | 89.8 | 90.2 | |||
| Caedibacter caryophila | 82.2 | 81.4 | 81.2 | 82.6 | 82.3 | 81.6 | 84.5 | 84.8 | 84.4 | 84.3 | ||
| UWC36 endosymbiont | 83.4 | 82.8 | 82.7 | 84.7 | 83.7 | 81.7 | 85.5 | 85.6 | 85.5 | 83.4 | 84.5 | |
| UWC8 endosymbiont | 83.3 | 82.9 | 82.9 | 84.4 | 83.9 | 81.6 | 85.4 | 85.2 | 85.4 | 83.0 | 83.9 | 99.6 |
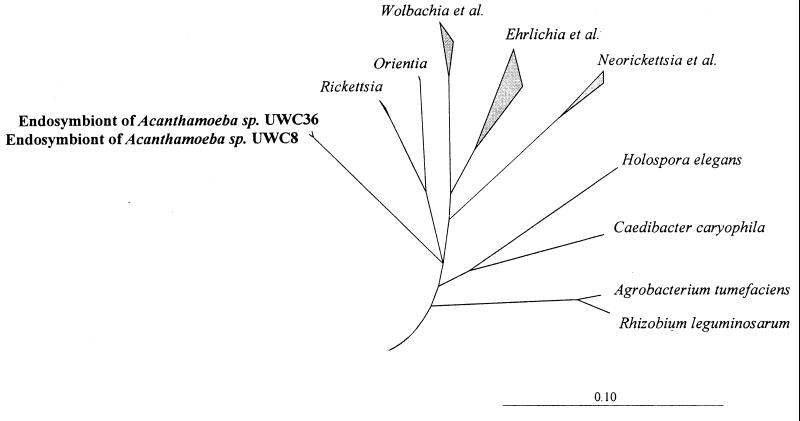
Phylogenetic analyses demonstrated that the retrieved sequences form an independent, well-separated lineage within the Rickettsiales (46). The neighbor-joining tree shown in Fig. 1 is based on the results of a distance matrix analysis of all available 16S rRNA sequences from representatives of the alpha subclass of Proteobacteria and a selection of members of the major lines of descent among the Bacteria. Only sequence positions that have the same nucleotides in at least 50% of all available sequences from the Rickettsiales were included, to reduce potential tree artifacts that may result from multiple base changes (24). To enhance clarity, several phylogenetic groups within the alpha subclass and the outgroup organisms were subsequently removed from the tree without changing its topology. The topology of the tree was further evaluated by parsimony and maximum-likelihood analyses of a variety of data sets differing with respect to the inclusion of sequence positions and outgroup reference sequences. Different tree-generating methods consistently supported deep branching of retrieved 16S rDNA sequences within the Rickettsiales, but an unambiguous pattern of the respective branch origins within the Rickettsiales could not be determined.
FIG. 1.
Neighbor-joining dendrogram showing relationships of endosymbionts of Acanthamoeba strains UWC8 and UWC36 to related members of the Rickettsiales and outgroups (the bar represents the estimated evolutionary distance). All tree-generating methods support deep branching of the retrieved 16S rDNA sequences, although an unambiguous pattern of the respective branch origins within the Rickettsiales could not be determined based upon the current data set, resulting in the presence of a multifurcation (24).
In situ analysis of endosymbionts by electron microscopy and in situ hybridization.
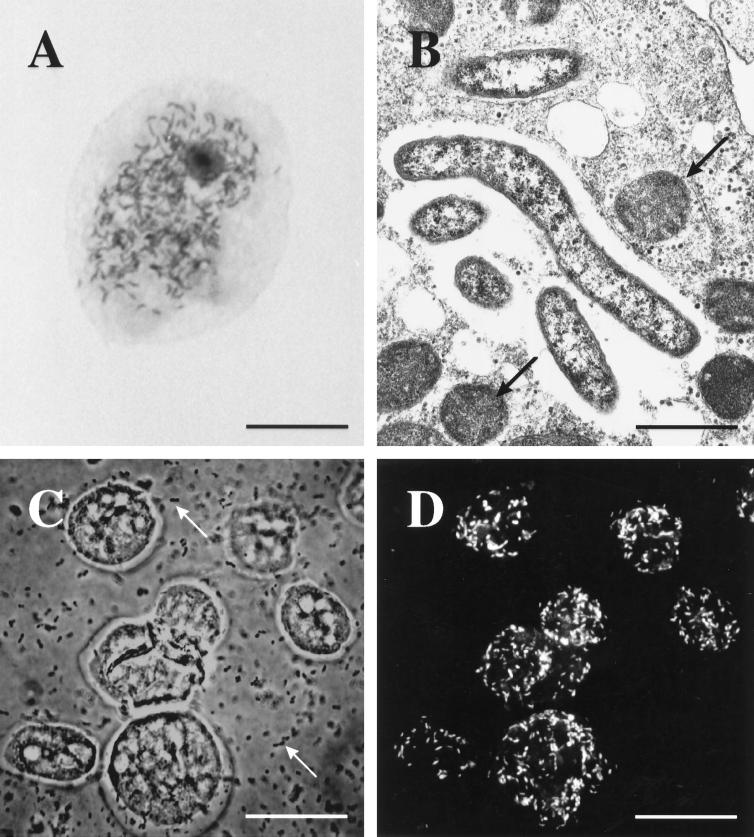
Light microscopic and ultrastructural analyses of UWC8 determined that the bacterial cells displayed a typical gram-negative cell wall, varied in shape from straight to curved, and were cytoplasmic. They often had an adjacent clear zone suggestive of capsules or slime layers (Fig. 2A and B). The symbionts varied considerably in size, being 0.3 to 0.5 μm wide by 0.8 to 2.3 μm long.
FIG. 2.
(A) Acanthamoeba trophozoites (UWC8 isolate) naturally infected with rod-shaped bacterial endosymbionts as seen with Hemacolor stain (bar, 7 μm). (B) Electron micrograph demonstrating intracellular bacterial symbionts (UWC8 isolate) and several mitochondria (arrows) in an Acanthamoeba trophozoite (bar, 1 μm). (C) Phase-contrast photomicrograph of fixed Acanthamoeba strain UWC8 trophozoites, with numerous E. coli food bacteria seen in the background (arrows) (bar, 15 μm). (D) Specific fluorescent in situ detection of the endosymbionts of Acanthamoeba strain UWC8 within the same field as seen in panel C; numerous rod-shaped intracellular bacteria are recognized by using probe AcRic1196 labelled with Cy3 (bar, 15 μm).
The oligonucleotide probes AcRic90 and AcRic1196 were designed complementary to specific target regions shared between both retrieved 16S rRNA sequences. Both probes had at least three mismatches with respect to all other available 16S rRNA sequences (Fig. 3). Use of these probes for in situ detection of the endosymbionts within their eukaryotic host cells by fixation with 4% paraformaldehyde and standard hybridization methods (27) was initially hampered by amoebic cell shrinkage accompanied by an increase in autofluorescence. Similar problems were observed after the use of HgCl2 and trichloroacetic acid-based fixation methods (35). Attempts to maintain amoebic cell morphology by capitalizing on their natural abilities to attach to a glass substrate, while effective, resulted in significant disruption of the host cells upon exposure to 80% ethanol. Consequently, we implemented an additional agarose-embedding step that successfully stabilized Acanthamoeba cell morphology despite treatment with 80% ethanol. Exclusion of SDS from the hybridization and washing buffers in subsequent in situ hybridization reactions further minimized the detrimental effects to the amoebic cells and allowed the unambiguous detection of probe-labeled, rod-shaped endosymbionts by confocal laser-scanning microscopy (Fig. 2C and D). Numbers of endosymbionts per host cell detected by in situ hybridization varied from 1 to approximately 100. Simultaneous application of endosymbiont-specific probes and DAPI staining verified that all DAPI-detectable endosymbiont cells were also visualized by probe-conferred fluorescence. A side effect of agarose embedding was the formation of large vacuoles within the Acanthamoeba cells (Fig. 2C).
FIG. 3.
Alignment of 16S rRNA target regions for endosymbionts of Acanthamoeba strains UWC8 and UWC36, along with other bacterial species displaying the smallest number of mismatches with respect to probes AcRic90 and AcRic1196.
The optimal hybridization stringency for both endosymbiont-targeted probes was determined by the addition of formamide to the hybridization buffer in 5% increments at a constant hybridization temperature of 46°C. Probe-conferred signals increased following the addition of formamide up to 10% for probe AcRic1196 and 25% for probe AcRic90, then decreased and eventually disappeared at 20 and 50%, respectively. An increase in probe sensitivity and specificity following the addition of formamide up to an optimal concentration was reported previously and may result from better access of the probe to its target site (i.e., denaturation) or from a direct effect on the probe, such as unfolding (4). Nonspecific binding of fluorescently labeled oligonucleotide probes to Acanthamoeba endosymbionts was ruled out by the application of Cy3- and Cy5-labelled derivatives of probe BET42a, specific for the beta subclass of Proteobacteria (data not shown). The specificity of the endosymbiont probes was further confirmed by the absence of detectable signals following in situ hybridization of paraformaldehyde-fixed municipal activated sludge with both probes (data not shown) (45). Consequently, positive hybridization reactions of the bacterial endosymbionts with the specific probes demonstrated that the retrieved Rickettsia-like 16S rDNA sequences did originate from the endosymbionts of Acanthamoeba strains UWC8 and UWC36.
DISCUSSION
In the past, the order Rickettsiales served as a convenient location for the taxonomic grouping of a large variety of gram-negative bacteria that have an obligate need to develop within eukaryotic cells. A reappraisal of this concept includes proposals to remove all Bartonella spp. (including those reclassified from the genus Rochalimaea), Afipia spp., and Coxiella burnetii from the order and to move R. tsutsugamushi to a new genus, Orientia. All Ehrlichia spp., most Wolbachia spp., Neorickettsia helminthoeca, Cowdria ruminatum, and Anaplasma marginale appear to form a series of related groups separate from the genus Rickettsia but clearly within the order and with a common ancestor (12, 36, 48). Within the genus Rickettsia, analysis of the 16S rRNA gene is proving to be less useful as a tool for evolutionary inference, with the analysis of other genes, including the one encoding citrate synthase, appearing to provide greater discriminatory potential (40). In the final analysis, strict intracellular location of gram-negative organisms can no longer be regarded as a definitive taxonomic marker only of the Rickettsiales (12).
We applied the techniques of small-subunit ribosomal gene sequence analysis to characterize two isolates of GNR endosymbionts stably infecting isolates of Acanthamoeba spp. recovered from keratitis specimens. It was surprising to find that these endosymbionts were related to the Rickettsiales, given that most members of the order are associated with arthropods. The finding is consistent, however, with previous studies on these and other endosymbionts that demonstrate their reliance upon intracellular growth, presence of a capsule or slime layer, and typical gram-negative cell wall (16, 21, 34). While indicating emergence from a common ancestor, the endosymbiont lineage does branch deeply from other members of the order in a pattern that is not clearly established by the tree-generating methods used (Fig. 1). Such ambiguity is displayed by the presence of a multifurcation, resolution of which will be forthcoming only through analysis of other meaningful data sets, such as additional gene sequences with phylogenetic potential or an increased data set of the various rickettsial lineages (24). Use of two fluorescently labelled oligonucleotide probes targeting signature regions at the 5′ and 3′ ends of the generated 16S rDNA sequence data did allow us to identify all individual bacterial cells with both probes within amoebic host cells and excluded a chimeric nature of the determined 16S rDNA sequences.
The finding of uncultured endosymbionts in Acanthamoeba spp. related to the Rickettsiales broadens the spectrum of bacteria known to interact with protozoa and may help to explain the appearance of host-symbiont specificity or cellular tropism known to exist with symbionts of ciliates (22, 33). Such specificity was previously demonstrated for the endosymbiont of Acanthamoeba sp. strain UWC8 and another uncharacterized GNR endosymbiont, which were shown to infect closely related strains of Acanthamoeba spp. but which failed to infect strains considered to be more distantly related, as determined by mitochondrial DNA restriction fragment length polymorphism analysis (18). Occasionally, the presence of a “killer” phenotype which appears to be dependent upon the degree of genetic relatedness of the originating and receiving hosts was observed in different groups of protozoa: contact between genetically matched pairs results in the creation of a stable symbiosis, whereas contact between mismatched though recognized pairs may result in host cell death (18, 22, 33). Theoretically, protozoal strains capable of maintaining stable symbiotic relationships may realize a substantial selective advantage from the ability to control competing, bactivorous populations of related protozoa, which succumb to the killer phenotype following acquisition of discharged symbionts. Michel et al. also demonstrated that an Ehrlichia-like organism found infecting an environmental isolate of Saccamoeba limax was able to infect certain other strains of Saccamoeba but was not able to develop within isolates representing nine other amoebic genera (30). Such cellular tropism, which is usually receptor mediated, is a characteristic presumably shared by all members of the Rickettsiales and is an important determinant of the particular disease presentations seen in higher mammals.
Many free-living soil and water protozoa mimic the role of professional phagocytes in their abilities to ingest and destroy large numbers of bacteria, and they undoubtedly serve as a natural testing ground for innumerable evolutionary experiments in intracellular survival (3, 41). The spectrum of pathogens able to survive and multiply to various degrees within acanthamoebae includes Legionella spp., Burkholderia pickettii, Listeria monocytogenes, Vibrio cholerae, Francisella tularensis, Mycobacterium avium, and Chlamydia pneumoniae (3, 5, 7, 10, 13, 14, 25, 29, 31, 32, 38, 40, 41, 43). For all of these organisms, acanthamoebae are potential reservoirs and vectors, due in part to their ubiquity in the environment, their resistant cyst stages, and their potential to grow in water supply, cooling, and humidification systems (7, 14, 37).
The recovery of rickettsia-like 16S rRNA gene sequences from endosymbionts of Acanthamoeba spp. is a novel finding that broadens the spectrum of the bacterium-host relationships documented among the Rickettsiales. This may reflect an evolutionary divergence of the protozoan endosymbiont lineage from the other recognized rickettsial lineages at a time before their acquisition by arthropods or may represent an earlier association with protozoa, which preadapted them to life in the intracellular environment, thus facilitating their ultimate recruitment to metazoan hosts.
ACKNOWLEDGMENTS
This study was supported by Deutsche Forschungsgemeinschaft grant WA 1027/2-1 to M.W. and K.-H.S. Stipend support to T.R.F. was provided by Public Health Service grant F06 TW02279-01 from the Fogarty International Center.
Technical assistance by Maria Marosvölgyi and Sibylle Schadhauser is gratefully acknowledged.
REFERENCES
- 1.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R, Springer N, Schönhuber W, Ludwig W, Schmid E N, Müller K, Michel R. Obligate intracellular bacterial parasites of Acanthamoebae related to Chlamydia spp. Appl Environ Microbiol. 1997;63:115–121. doi: 10.1128/aem.63.1.115-121.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker J, Brown M R W. Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology. 1994;140:1253–1259. doi: 10.1099/00221287-140-6-1253. [DOI] [PubMed] [Google Scholar]
- 4.Beimfohr C, Krause A, Amann R, Ludwig W, Schleifer K-H. In situ identification of lactococci, enterococci and streptococci. Syst Appl Microbiol. 1993;16:450–456. [Google Scholar]
- 5.Berdal B P, Mehl R, Meidell N K, Lorentzen-Styr A M, Scheel O. Field investigations of tularemia in Norway. FEMS Immunol Med Microbiol. 1996;13:191–195. doi: 10.1111/j.1574-695X.1996.tb00235.x. [DOI] [PubMed] [Google Scholar]
- 6.Berk S G, Ting R S, Turner G W, Ashburn R J. Production of respirable vesicles containing live Legionella pneumophila cells by two Acanthamoeba spp. Appl Environ Microbiol. 1998;64:279–286. doi: 10.1128/aem.64.1.279-286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birtles R J, Rowbotham T J, Raoult D, Harrison T G. Phylogenetic diversity of intra-amoebal legionellae as revealed by 16S rRNA gene sequence comparison. Microbiology. 1996;142:3525–3530. doi: 10.1099/13500872-142-12-3525. [DOI] [PubMed] [Google Scholar]
- 8.Birtles R J, Rowbotham T J, Storey C, Marrie T J, Raoult D. Chlamydia-like obligate parasite of free-living amoebae. Lancet. 1997;349:925–926. doi: 10.1016/s0140-6736(05)62701-8. [DOI] [PubMed] [Google Scholar]
- 9.Brosius J, Dull T L, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 10.Cirillo J D, Falkow S, Tompkins L S, Bermudez L E. Interaction of Mycobacterium avium with environmental amoebae enhances virulence. Infect Immun. 1997;65:3759–3767. doi: 10.1128/iai.65.9.3759-3767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Rijk P, Van de Peer Y, De Wachter R. Database on the structure of large ribosomal subunit RNA. Nucleic Acids Res. 1996;24:92–97. doi: 10.1093/nar/24.1.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drancourt M, Raoult D. Taxonomic position of the Rickettsiae: current knowledge. FEMS Microbiol Rev. 1994;13:13–24. doi: 10.1111/j.1574-6976.1994.tb00032.x. [DOI] [PubMed] [Google Scholar]
- 13.Essig A, Heinemann M, Simnacher U, Marre R. Infection of Acanthamoeba castellanii by Chlamydia pneumoniae. Appl Environ Microbiol. 1997;63:1396–1399. doi: 10.1128/aem.63.4.1396-1399.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fields B S. The molecular ecology of Legionella. Trends Microbiol. 1996;4:286–290. doi: 10.1016/0966-842x(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 15.Fritsche, T., and R. K. Gautom. Unpublished observations.
- 16.Fritsche T R, Gautom R K, Seyedirashti S, Bergeron D L, Lindquist T D. Occurrence of bacterial endosymbionts in Acanthamoeba spp. isolated from corneal and environmental specimens and contact lenses. J Clin Microbiol. 1993;31:1122–1126. doi: 10.1128/jcm.31.5.1122-1126.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fritsche T R, Sobek D, Gautom R K. Enhancement of in vitro cytopathogenicity of Acanthamoeba spp. following acquisition of bacterial endosymbionts. FEMS Microbiol Lett. 1998;166:231–236. doi: 10.1111/j.1574-6968.1998.tb13895.x. [DOI] [PubMed] [Google Scholar]
- 18.Gautom R, Fritsche T R. Transmissibility of bacterial endosymbionts between isolates of Acanthamoeba spp. J Eukaryot Microbiol. 1995;42:452–456. doi: 10.1111/j.1550-7408.1995.tb05890.x. [DOI] [PubMed] [Google Scholar]
- 19.Gautom R, Herwig R, Fritsche T R. Abstracts of the 96th General Meeting of the American Society for Microbiology 1996. Washington, D.C: American Society for Microbiology; 1996. Molecular phylogeny of bacterial endosymbionts of Acanthamoeba spp., abstr. R-29; p. 474. [Google Scholar]
- 20.Gautom R K, Fritsche T R, Lindquist T D. Acanthamoeba keratitis. In: Tasman W, Jaeger E A, editors. Duane’s foundations of clinical ophthalmology. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1997. pp. 1–15. [Google Scholar]
- 21.Hall J, Voelz H. Bacterial endosymbionts of Acanthamoeba sp. J Parasitol. 1985;71:89–95. [PubMed] [Google Scholar]
- 22.Heckmann K, Görtz H-D. Prokaryotic symbionts of ciliates. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. 2nd ed. Vol. 4. New York, N.Y: Springer-Verlag; 1992. pp. 3865–3890. [Google Scholar]
- 23.Hugo E R, Gast R J, Byers T J, Stewart V J. Purification of amoeba mtDNA using the UNSET procedure. In: Lee J J, Soldo A T, editors. Protocols in protozoology. Lawrence, Kans: Allen Press; 1992. pp. D7.1–D7.2. [Google Scholar]
- 24.Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, Neumaier J, Bachleitner M, Schleifer K-H. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis. 1998;19:554–568. doi: 10.1002/elps.1150190416. [DOI] [PubMed] [Google Scholar]
- 25.Ly T M, Muller H E. Ingested Listeria monocytogenes survive and multiply in protozoa. J Med Microbiol. 1990;33:51–54. doi: 10.1099/00222615-33-1-51. [DOI] [PubMed] [Google Scholar]
- 26.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The ribosomal database project. Nucleic Acids Res. 1996;24:82–85. doi: 10.1093/nar/24.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligonucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 28.McLaughlin G L, Brant F H, Visvesvara G S. Restriction fragment length polymorphism of the DNA of selected Naegleria and Acanthamoeba. J Clin Microbiol. 1988;26:1655–1658. doi: 10.1128/jcm.26.9.1655-1658.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michel R, Hauröder B. Isolation of an Acanthamoeba strain with intracellular Burkholderia pickettii infection. Zentbl Bakteriol. 1997;285:541–557. doi: 10.1016/s0934-8840(97)80116-8. [DOI] [PubMed] [Google Scholar]
- 30.Michel R, Müller K-D, Schmid E N. Ehrlichia-like organisms (KSL1) observed as obligate intracellular parasites of Saccamoeba species. Endocytobiosis Cell Res. 1995;11:69–80. [Google Scholar]
- 31.Michel R, Müller K-D, Amann R, Schmid E N. Legionella-like slender rods multiplying within a strain of Acanthamoeba sp. isolated from drinking water. Parasitol Res. 1998;60:84–88. doi: 10.1007/s004360050362. [DOI] [PubMed] [Google Scholar]
- 32.Newsome A L, Scott T M, Benson R F, Fields B F. Isolation of an amoeba naturally harboring a distinctive Legionella species. Appl Environ Microbiol. 1998;64:1688–1693. doi: 10.1128/aem.64.5.1688-1693.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Preer J R, Preer L B. Endosymbionts of protozoa. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 795–811. [Google Scholar]
- 34.Proca-Ciobanu M, Lupascu G H, Pertrovici A L, Ionescu M D. Electron microscopic studies of a pathogenic Acanthamoeba castellanii strain: the presence of bacterial endosymbionts. Int J Parasitol. 1975;5:49–56. doi: 10.1016/0020-7519(75)90097-1. [DOI] [PubMed] [Google Scholar]
- 35.Rice J, O’Connor C D, Sleigh M A, Burkill P H, Giles I G, Zubkov M V. Fluorescent oligonucleotide rDNA probes that specifically bind to a common nanoflagellate, Paraphysomonas vestita. Microbiology. 1997;143:1717–1727. doi: 10.1099/00221287-143-5-1717. [DOI] [PubMed] [Google Scholar]
- 36.Roux V, Rydkina E, Eremeeva M, Raoult D. Citrate synthase gene comparison, a new tool for phylogenetic analysis, and its application for the rickettsiae. Int J Syst Bacteriol. 1997;47:252–261. doi: 10.1099/00207713-47-2-252. [DOI] [PubMed] [Google Scholar]
- 37.Rowbotham T J. Current view on the relationship between amoebae, legionellae, and man. Isr J Med Sci. 1986;22:678–689. [PubMed] [Google Scholar]
- 38.Rowbotham T J. Legionella-like amoebal pathogens. In: Barbee J M, Breiman R F, Dufour A P, editors. Legionella: current status and emerging perspectives. Washington, D.C: American Society for Microbiology; 1993. pp. 137–140. [Google Scholar]
- 39.Springer N, Amann R, Ludwig W. The design and application of ribosomal RNA-targeted, fluorescent oligonucleotide probes for the identification of endosymbionts in protozoa. Methods Mol Biol. 1996;50:133–144. doi: 10.1385/0-89603-323-6:133. [DOI] [PubMed] [Google Scholar]
- 40.Springer N, Ludwig W, Drozanski W, Amann R, Schleifer K-H. The phylogenetic status of Sarcobium lyticum, an obligate intracellular bacterial parasite of small amoebae. FEMS Microbiol Lett. 1992;96:199–202. doi: 10.1016/0378-1097(92)90403-b. [DOI] [PubMed] [Google Scholar]
- 41.Steinert M, Birkness K, White E, Fields B, Quinn F. Mycobacterium avium bacilli grow saprophytically in coculture with Acanthmaoeba polyphaga and survive within cyst walls. Appl Environ Microbiol. 1998;64:2256–2261. doi: 10.1128/aem.64.6.2256-2261.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strunk O, Ludwig W. ARB: a software environment for sequence data. 1996. www.biol.chemie.tu-muenchen.de/pub/ARB/. Internet address: www.biol.chemie.tu-muenchen.de/pub/ARB/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thom S, Warhurst D, Drasar B S. Association of Vibrio cholerae with fresh water amoebae. J Med Microbiol. 1992;36:303–306. doi: 10.1099/00222615-36-5-303. [DOI] [PubMed] [Google Scholar]
- 44.Visvesvara G S. Pathogenic and opportunistic free-living amebae. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: American Society for Microbiology; 1995. pp. 1196–1203. [Google Scholar]
- 45.Wagner M, Assmuss B, Hartmann A, Hutzler P, Amann R. In situ analysis of microbial consortia in activated sludge using fluorescently labelled, rRNA-targeted oligonucleotide probes and confocal scanning laser microscopy. J Microsc. 1994;176:181–187. doi: 10.1111/j.1365-2818.1994.tb03513.x. [DOI] [PubMed] [Google Scholar]
- 46.Warren J H, Zhang W, Guo L R. Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc R Soc London Ser B. 1995;261:55–71. doi: 10.1098/rspb.1995.0117. [DOI] [PubMed] [Google Scholar]
- 47.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S Ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weisburg W G, Dobson M E, Samual J E, Dasch G A, Mallavia L P, Baca O, Mandelco L, Sechrest J E, Weiss E, Woese C R. Phylogenetic diversity of the rickettsiae. J Bacteriol. 1989;171:4202–4206. doi: 10.1128/jb.171.8.4202-4206.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]