Abstract
A multidisciplinary approach was used to study the effects of pollution from a marine fish farm on nitrification rates and on the community structure of ammonia-oxidizing bacteria in the underlying sediment. Organic content, ammonium concentrations, nitrification rates, and ammonia oxidizer most-probable-number counts were determined in samples of sediment collected from beneath a fish cage and on a transect at 20 and 40 m from the cage. The data suggest that nitrogen cycling was significantly disrupted directly beneath the fish cage, with inhibition of nitrification and denitrification. Although visual examination indicated some slight changes in sediment appearance at 20 m, all other measurements were similar to those obtained at 40 m, where the sediment was considered pristine. The community structures of proteobacterial β-subgroup ammonia-oxidizing bacteria at the sampling sites were compared by PCR amplification of 16S ribosomal DNA (rDNA), using primers which target this group. PCR products were analyzed by denaturing gradient gel electrophoresis (DGGE) and with oligonucleotide hybridization probes specific for different ammonia oxidizers. A DGGE doublet observed in PCR products from the highly polluted fish cage sediment sample was present at a lower intensity in the 20-m sample but was absent from the pristine 40-m sample station. Band migration, hybridization, and sequencing demonstrated that the doublet corresponded to a marine Nitrosomonas group which was originally observed in 16S rDNA clone libraries prepared from the same sediment samples but with different PCR primers. Our data suggest that this novel Nitrosomonas subgroup was selected for within polluted fish farm sediments and that the relative abundance of this group was influenced by the extent of pollution.
In recent years, the culture of Atlantic salmon (Salmo salar L.) has become of major industrial importance worldwide, leading to concern over the impact of fish farming on previously pristine marine environments. Intensive cage cultivation of fish leads to localized pollution of the adjacent seabed through accumulation of uneaten food and fecal material (14, 21). The impact of this organic enrichment on the underlying sediment is affected by several factors, such as water currents, turbulence, and depth and the duration, quality, and quantity of waste loading (41), and anaerobic conditions can develop, resulting in the outgassing of hydrogen sulfide, ammonia, and methane.
The major component of fish feed is protein, and the output from fish farms, therefore, has a high nitrogen content. This may lead to eutrophication, since nitrogen is the major nutrient limiting phytoplankton growth (9). Nitrification, the oxidation of ammonia to nitrate via nitrite, is central to the cycling of nitrogen in the environment and, when coupled with denitrification, alleviates the effects of eutrophication through removal of nitrogen to the atmosphere as nitrous oxide or dinitrogen gas (5). Nitrification, however, is sensitive to low oxygen tension, sulfur compounds, high ammonia and nitrite concentrations, and the presence of a broad range of organic compounds (3, 20, 22, 24). In situations of chronic pollution, nitrification and, consequently, the coupled process of denitrification are often completely suppressed, exacerbating the environmental impact of the aquaculture industry (21).
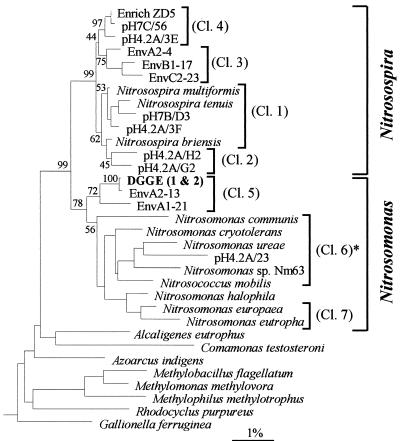
The ammonia-oxidizing bacteria carry out the first, rate-limiting stage of nitrification, the conversion of ammonia to nitrite. The intractability of these organisms to isolation and laboratory cultivation has, until recently, prevented meaningful study of their community structure and diversity in natural environments. Phylogenetic analysis of pure cultures, based on 16S rRNA sequence data, has demonstrated that the ammonia-oxidizing bacteria can be divided into two groups. The first contains Nitrosococcus oceanus and forms a deep branch within the γ subgroup of the class Proteobacteria (44). The second group, containing the majority of cultured strains, forms a tight cluster within the β-subgroup proteobacteria and can be subdivided into two clades, corresponding to Nitrosomonas spp. and Nitrosospira spp. (12, 43). Sequence analysis of partial 16S ribosomal DNA (rDNA) clones (1,099 bases) obtained from marine sediment and soil samples by using primers specific for the β-subgroup ammonia oxidizers (27) revealed that the environmental sequences which have been isolated so far from marine sediments and from soil form at least six clusters, four within the Nitrosospira clade and two within the Nitrosomonas clade (Fig. 1) (37). Stephen et al. (37) also presented the first evidence for the existence of marine representatives of the genus Nitrosospira. Analysis of alignments of shorter rDNAs, which included sequences from marine enrichment cultures, indicated that a seventh group, associated with Nitrosomonas europaea and Nitrosomonas eutropha, may also exist (37). Pure-culture representatives have now been isolated for all groups (40, 42) with the exception of Nitrosospira cluster 1 and Nitrosomonas cluster 5, although sequences from these two groups have been amplified from enrichment cultures of ammonia-oxidizing bacteria (37).
FIG. 1.
Neighbor-joining tree showing the relationship of the single sequence obtained from two excised DGGE gel bands, designated DGGE (1 & 2), to environmental sequences and reference β-subgroup proteobacteria based on analysis of 424 bases of aligned 16S rDNA sequences. The percentages of bootstrap support above 50% are shown for distance matrix analysis at nodes concerning major ammonia oxidizer relationships. Clusters (Cl.) 1 to 7 are indicated. Clone sequences obtained from sediment DNA libraries are prefixed Env. EnvA clones were obtained from polluted sediment beneath the fish cage, while EnvB and EnvC clones were obtained at distances of 20 and 40 m, respectively. Other environmental clone sequences (prefixed pH) and the enrichment sequence Enrich ZD5 were isolated from soil. For convenience, the tree has been pruned from a larger tree containing additional sequences from reference proteobacteria. The scale is 1% estimated change. For the design of the oligonucleotide probe, it was necessary to subdivide cluster 6, indicated by the asterisk, into two groups. Cluster 6a, represented by pH4.2A/23 on this tree, comprises a small cluster of three soil clones. All other known sequences belonging to cluster 6 are designated cluster 6b.
rRNA sequence analysis has been used to compare ammonia oxidizer populations within acid and neutral soils and in fish farm sediments with various degrees of organic pollution (37). A small group of novel Nitrosomonas sequences, designated cluster 5, was detected within polluted sediments but not within a pristine sediment, suggesting that nitrogen-rich organic pollution from fish farming may be selecting for or providing a source of particular ammonia oxidizers. Denaturing gradient gel electrophoresis (DGGE) (29) provides an alternative approach to the analysis of rDNA clone libraries and has recently been used successfully to demonstrate differences in ammonia oxidizer populations in sand dune sites (23). PCR amplification products are electrophoresed through a denaturing polyacrylamide gel, enabling investigation of population differences through the presence or absence of bands in environmental samples. Bands can then be identified by using a range of oligonucleotide probes with various specificities within the β-subgroup ammonia oxidizers (38). This approach has been used to confirm selection for particular ammonia oxidizers within acid- and neutral-pH agricultural soils (38).
The aim of this study was to investigate the effects of organic particulate material from a marine fish farm on the ammonia oxidizer community structure in the underlying sediment by using 16S rDNA-based techniques and to relate these effects to process measurements. Furthermore, because the same DNA extracts were used in this study and in that of Stephen et al. (37), it was possible to compare inferred population structures by using two different primer sets and different analytical methods.
MATERIALS AND METHODS
Fish farm location and cage history.
The fish farm used in this study consisted of two sets of six cages and a fallow site situated in Glenmore Bay, Loch Sunart, West Scotland, United Kingdom (Ordnance Survey Map coordinate 592 618) at a water depth of 16 m. Samples were collected from cages which had been stocked with Atlantic salmon (Salmo salar L.) for 11 months following a 4-month fallow period.
Sample collection.
Sediment samples (eight cores per sampling station) were collected in Perspex core tubes (59-mm inner diameter by 300-mm length) by divers at four stations. Station A was situated directly underneath a fish cage, while stations B and C were located on a transect leading from the cage, at distances of 20 and 40 m, respectively. An additional sample, from a transect at a distance of 10 m from the cage (station S), was also collected for chemical analysis, rate measurements, and cell counts only. Cores were transported to the laboratory in insulated boxes and stored at 8 to 10°C in the dark until processed (within 12 h).
After preliminary visual macroscopic inspection of the core appearance, the top 0.5 cm of each core was aseptically removed for analysis. Duplicate cores from stations A, B, and C were used in molecular analyses. After removal of subsamples of sediment for enrichment of marine ammonia-oxidizing bacteria (described in reference 37), samples (approximately 1 g each) were stored in sterile Eppendorf tubes at −70°C prior to DNA extraction. For nutrient determination, rate measurements, and most-probable-number (MPN) bacterial counts, two sets of three sediment cores (i.e., six cores) from stations A to C and station S were pooled to provide two replicates per sampling point.
Organic-matter content, porosity, and water content.
To determine the porosity and water content of sediments, six 5-ml subsamples were taken by using disposable syringes with the luer ends removed, and wet weight was determined in preweighed glass vials. Sediment samples were frozen and lyophilized to a constant weight to give sediment dry-weight values, and the water content and porosity of each of the sediments were calculated. The organic-matter content of sediment was determined as weight loss by ignition (500°C, 12 h) of freeze-dried samples (approximately 1 g each) (7).
Pore water nutrients.
Concentrations of ammonia and nitrate in pore water were determined for each station. The pore water was separated from sediment by centrifugation in a polypropylene tube at 2,000 × g for 10 min. Supernatants were removed, filtered through Whatman GF/F filters in an N2 atmosphere, and stored frozen until chemical analysis was performed.
Ammonia and nitrite oxidation rates.
Ammonia and nitrite oxidation rates for each station were determined by a modification of the method of Bianchi et al. (4). For each replicate sediment sample, three subsamples (5 ml each) were deposited into each of three glass vials containing 10 ml of filtered (Whatman GF/F) seawater. One vial from each set was used as a control (i.e., no additions). Allylthiourea and sodium chlorate (final concentrations, 10 mg liter−1 and 10 mM, respectively) were added to the second and third vials to inhibit ammonia and nitrite oxidation, respectively. Vials were placed in a shaking water bath at 4°C, and nitrite concentrations were determined for samples taken at 0, 4, 8, and 24 h.
Ammonia, nitrite, and nitrate analyses.
Samples were analyzed for ammonium and nitrate by the use of a Technicon II Automated Analyzer. Nitrite was measured by a scaled-down modification of the method of Parsons et al. (30). Samples (1 ml each) were pipetted into acid-washed 75- by 10-mm borosilicate glass test tubes, and 40 μl of sulfanilamide solution (0.01% [wt/vol] in 8.5 M hydrochloric acid) was added to each. The tube contents were vortexed and then incubated for 2 min, and 40 μl of n-(1-naphthyl)-ethylenediamine dihydrochloride (0.001% [wt/vol] in double-distilled water) was added to each. Tube contents were vortexed and allowed to stand for 30 min, and absorbance readings were obtained spectrophotometrically in 1-cm-light-path cells at 543 nm.
Denitrification rates.
Rates of denitrification were determined by a modification of the method of Raymond et al. (31). Subsamples (2 ml) of each replicate sediment core were transferred into four 10-ml bottles, each containing 2 ml of seawater. The bottles were flushed with chemically pure nitrogen (grade CP; BOC, Guildford, United Kingdom) and sealed with butyl rubber stoppers. Reduction of N2O to N2 was inhibited by addition of acetylene (15 kPa; BOC)-saturated seawater (0.4 ml) to each bottle with a hypodermic syringe, and the bottles were incubated at 20°C. Samples were taken at 0, 30, 60, and 120 min by vortexing vigorously for 1 min before removal of 2.7 ml of the headspace into a preevacuated Monovette tube (Sarstedt Ltd., Leicester, United Kingdom). N2O was detected with a Varian model 3400 gas chromatograph equipped with an electron capture detector. Measured N2O values were read against a standard curve prepared with N2O from Distillers MG Limited (Reigate, Surrey, United Kingdom).
Enumeration of nitrifying and nitrate-respiring bacteria.
Ammonia oxidizers, nitrite oxidizers, and nitrate-respiring bacteria within sediment samples were enumerated by the MPN technique in microtiter plates (32). This involved vigorously shaking a 1-ml sediment sample in 9 ml of sterile seawater containing approximately 10 sterile glass beads (3 mm in diameter) for 1 min. For each group of bacteria, 0.2 ml of the appropriate autoclaved medium was dispensed into each well of a 96-well microtiter plate, and eight replicate twofold serial dilutions of sediment slurry (0.2 ml) were performed. Following incubation, wells were assayed for the presence of each group of bacteria as described below, and numbers were calculated by using MPN tables (32).
Ammonia-oxidizing bacteria were enumerated in modified Watson’s medium (16) prepared in artificial seawater, with the pH of the medium being adjusted to 7.5 by addition of sterile 5% Na2CO3 after autoclaving (121°C for 15 min). The pH indicator phenol red was included in the medium to allow detection of a reduction in pH below 7. Inoculated MPN plates were incubated in the dark for 21 days at room temperature. Wells were scored as positive for ammonia oxidizers by acid production and by detection of NO2− and NO3− through development of a blue color after addition of 1 or 2 drops of 0.2% (wt/vol) diphenylamine in concentrated H2SO4.
Nitrite-oxidizing bacteria were enumerated in the medium of Alexander and Clark (2) prepared in artificial seawater. Wells were scored as positive for growth when nitrite was absent, as indicated by the lack of a color change upon addition of Griess Ilosvay reagents 1 and 2.
Nitrate respirers, i.e., those organisms capable of causing an increase in pH through reduction of nitrate to ammonia (dissimilatory nitrate reduction) or to nitrous oxide and nitrogen (denitrification), were enumerated in Alexander’s (1) medium containing bromophenol blue as a pH indicator, which changes in color from yellow to blue with an increase in pH. Inoculated microtiter plates were placed in an anaerobic jar, the jar was sealed and then flushed with oxygen-free nitrogen, and the plates were read after incubation in the dark for 7 days.
DGGE.
DNA was extracted directly from duplicate sediment samples from stations A, B, and C by a bead beating method (37). PCR amplification of a 498-bp PCR product and subsequent DGGE analysis were carried out as described previously (23), using the primers CTO178f-GC and CTO637r. This primer pair was designed for specific amplification of a 460-bp region of the 16S rDNA of ammonia-oxidizing bacteria belonging to the proteobacterial β-subgroup and incorporation of a 38-bp GC clamp (35). PCR products were visualized by standard agarose electrophoresis followed by ethidium bromide staining. Approximately 200 ng of each product, estimated by visual reference to molecular weight standards, was used in the subsequent DGGE analysis. PCR amplification and DGGE analysis were also performed with rDNA clones which represented each currently recognized cluster of marine β-subgroup ammonia oxidizers (37), as follows: EnvC1-17 (Nitrosomonas cluster 6b), EnvB2-11 (Nitrosospira cluster 1), and EnvA1-21 (Nitrosomonas cluster 5). Polyacrylamide gels were stained with ethidium bromide and washed twice for 10 min each in 0.5× TAE buffer (48.22 g of Tris base, 2.05 g of anhydrous sodium acetate, and 1.86 g of Na2EDTA · 2H2O [pH 8] in 1 liter of distilled water), and the UV gel image was then captured by using the Imager system (Ampligene, Illkirch, France). DNA was transferred to a nylon hybridization membrane, and the blot was hybridized with γ-32P-end-labelled oligonucleotide probes (listed in Table 1) by the procedure of Stephen et al. (38). The identities of bands for clusters 1 and 6b were confirmed in this way. Bands corresponding to cluster 5 sequences were identified by comparison of their migration profiles with those of control clones and by sequence analysis of excised bands (described below). This was achieved by densitometry. The autoradiograph obtained by probing with β-AO233 was scanned by using a model GS-700 imaging densitometer (Bio-Rad). For each lane on the gel, bands corresponding to each cluster were selected and the data were processed by using the volume analysis function of Molecular Analyst software (version 1.5; Bio-Rad) and correcting for background signal. The relative abundance of the three detected clusters in the environmental samples was calculated as the percentage contribution of bands from each cluster to the total hybridization signal within each lane. Data are expressed as the means of duplicate samples.
TABLE 1.
Oligonucleotide probes and hybridization temperatures used in analysis of DGGE banding patterns of PCR-amplified rDNA of β-subgroup ammonia-oxidizing bacteria from polluted and pristine marine sediments
| Probe | Probe sequence, 5′→3′ | Target group | Probe namea | Hybridization temp (°C) |
|---|---|---|---|---|
| 1 | AGCTAATCAGRCATCGG | All β-subgroup ammonia oxidizers | β-AO233 | 44 |
| 2 | CTTTTACCTTACCAACAA | Nitrosospira cluster 1 | NspCL1_249 | 42 |
| 3 | GGATCAGGCTTGCGCCC | Nitrosomonas cluster 6b | NmoCL6b_376 | 46 |
| 4 | GTAGGCCSTTACCCYACC | All Nitrosomonas spp. | Nmo254 | 43 |
| 5 | CTCTTTCTTTCCGACTAA | Nitrosomonas cluster 7 | NmoCL7_439 | 44 |
Recovery and sequence analysis of bands from the DGGE gel.
The putative cluster 5 bands, which could not be identified by specific oligonucleotide hybridization, were excised from the DGGE gel, reamplified, and sequenced (23). Sequences were aligned against representative prokaryote 16S rRNA sequences from the Ribosomal Database Project (26) and ammonia oxidizer sequences (27, 37). Sequence data were manipulated by using Genetic Data Environment (GDE) software, version 2.2, distributed by the Ribosomal Database Project. The GDE mask function was used to exclude from this analysis missing data or positions which could not be unambiguously aligned. Distance matrix analysis was performed by the Jukes and Cantor (19) correction and the neighbor-joining method (33) implemented in PHYLIP 3.5 (10) operated through GDE software.
Nucleotide sequence accession numbers.
The sequences of the upper and lower cluster 5 DGGE bands have been deposited in GenBank under accession no. AF006666 and AF006667.
RESULTS
Macroscopic appearance of sediment samples.
Core samples collected beneath the fish cage (station A) were extremely flocculant to a depth of 300 mm and consisted mostly of decomposing fish feed and feces. Sulfide-oxidizing bacteria formed a confluent white mat over the sediment surface, while the subsurface sediment was black due to the formation of ferrous sulfide. At station S, the sediment was coarser in texture but was otherwise similar to that at station A. At 20 m (station B), some signs of impaction were still apparent, although the ferrous sulfide layer penetrated to a depth of only 10 mm. It should be noted, however, that the region of the cores analyzed in this study (i.e., the top 0.5 cm) did not extend into the unaffected sediment layers. At station C, 40 m from the fish cage, there was no visual evidence of organic enrichment.
Organic content, water content, and sediment porosity.
Organic content was highest directly beneath the fish cage, reflecting the high sedimentation rate of organic material, and was significantly reduced (P < 0.05) at all other stations (Table 2). Porosity and water content values of polluted sediments (0 and 10 m) were high, as indicated by their more viscous nature, particularly at station A (0.88). All three parameters decreased with distance from the fish cage, and readings at the 40-m site were indicative of a tightly packed, pristine sediment, in contrast to the highly disturbed, liquefied surface of cores collected from station A.
TABLE 2.
Water content, porosity, and organic matter content of sediments sampled directly beneath and on a transect extending from an Atlantic salmon farm cage
| Station | Distance (m) | Water content | Porosity | Organic content (%) |
|---|---|---|---|---|
| A | 0 | 0.881 | 0.770 | 48.3 |
| S | 10 | 0.567 | 0.817 | 9.94 |
| B | 20 | 0.517 | 0.690 | 5.12 |
| C | 40 | 0.358 | 0.512 | 3.86 |
Ammonium, nitrate, and nitrous oxide concentrations.
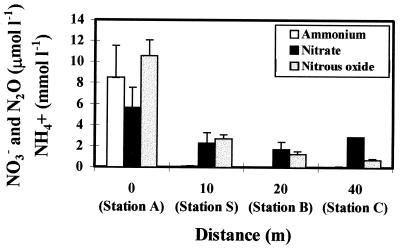
Ammonium levels directly beneath the fish cage were high (8 mmol liter−1) (Fig. 2), reflecting the nitrogen content of sedimenting detritus, and decreased dramatically with increasing distance from the cage. A similar profile was observed for sediment-bound ammonium (data not shown). Pore water nitrite concentrations, although considerably lower than ammonium levels, were elevated directly beneath the cage while showing little variation over the remainder of the transect (Fig. 2). Nitrous oxide (determined during time zero sampling of denitrification assays) was also detected, with concentrations decreasing with distance from the cage site (Fig. 2).
FIG. 2.
Changes in pore water ammonium (in millimoles per liter), nitrate, and nitrous oxide (both in micromoles per liter) concentrations in the 0- to 5-mm-depth-horizon sediment samples with increasing distance from the fish cage. Error bars represent standard errors for duplicate measurements.
Nitrification and denitrification rates.
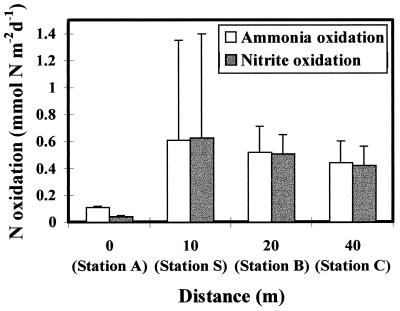
Ammonia and nitrite oxidation rates at increasing distances from the fish cage are shown in Fig. 3. At station A, directly beneath the fish cage, rates for both ammonia and nitrite oxidation were low (0.11 and 0.04 mmol of N m−2 day−1, respectively). At 10 m, ammonia and nitrite oxidation rates had increased considerably, with similar values being observed at stations B and C. Denitrification, measured as accumulation of N2O in the presence of acetylene, was not detected at any of the sampling points (data not shown).
FIG. 3.
Ammonia and nitrite oxidation rates (in millimoles per square meter per day) in the 0- to 5-mm-depth-horizon sediment samples with increasing distance from the fish cage. Error bars represent standard errors for duplicate measurements.
Enumeration of ammonia- and nitrite-oxidizing and nitrate-respiring bacteria.
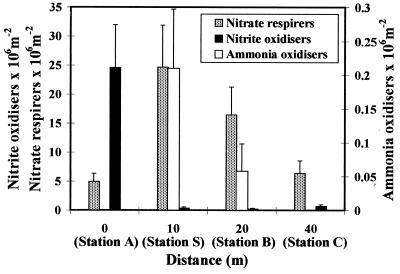
Numbers of ammonia-oxidizing bacteria were low at all stations and were generally only slightly above the lower detection limit (0.07 × 106 bacteria m−2) (Fig. 4). Despite the high concentration of ammonium at station A, there was no increase in the number of ammonia oxidizers, consistent with the low rates of ammonia oxidation measured at this station. A significant increase in numbers at station S was followed by a decrease over the remainder of the transect. A substantial population of nitrite-oxidizing bacteria was detected at station A, directly beneath the fish cage. The numbers decreased to detection level by 10 m and did not vary greatly over the remainder of the transect. MPN counts of nitrate-respiring bacteria were highest at station S, decreasing with distance from the cage.
FIG. 4.
MPN counts of ammonia and nitrite oxidizers and denitrifying bacteria in the 0- to 5-mm-depth-horizon sediment samples with increasing distance from the fish cage. Error bars represent standard errors for duplicate measurements.
DGGE analysis and oligonucleotide probing.
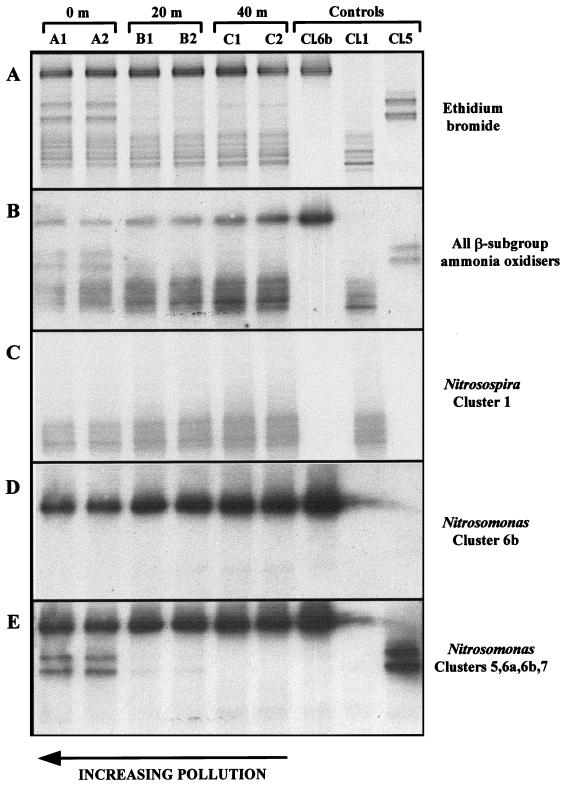
PCR amplification products, with an incorporated GC clamp for DGGE analysis, were obtained for all six samples. Figure 5 illustrates the ethidium bromide-stained polyacrylamide gel and autoradiographs obtained by the use of oligonucleotide probes specific for β-subgroup ammonia oxidizers (Table 1).
FIG. 5.
DGGE analysis and Southern oligonucleotide hybridization following PCR amplification of β-subgroup ammonia-oxidizing bacteria from 0- to 5-mm-depth-horizon sediment samples obtained directly beneath the fish cage (0 m) and at distances of 20 and 40 m. Lanes 1 to 6, duplicate samples from stations A (lanes 1 and 2), B (lanes 3 and 4), and C (lanes 5 and 6); lane 7, EnvC1-17 (Nitrosomonas cluster 6b); lane 8, EnvB2-11 (Nitrosospira cluster 1); lane 9, EnvA1-21 (Nitrosomonas cluster 5). (A) An ethidium bromide-stained DGGE gel. (B to E) Oligonucleotide hybridizations. (B) Probe 1 (all β-subgroup ammonia oxidizers); (C) probe 2 (Nitrosospira cluster 1); (D) probe 3 (Nitrosomonas cluster 6b); (E) probe 4 (Nitrosomonas clusters 5, 6a, 6b, and 7).
Southern hybridization with probe β-AMO233 (Fig. 5B), designed for detection of all proteobacterial β-subgroup ammonia oxidizers, resulted in binding to all bands present on the ethidium bromide-stained gel (Fig. 5A). The blots in Fig. 5C and D, probed with a cluster 1 marine Nitrosospira-specific oligonucleotide and a probe for marine Nitrosomonas spp. (cluster 6b only), respectively, demonstrated that these organisms were present in all sediment samples. In the blot shown in Fig. 5E, which illustrates the result of hybridization with a probe specific for all Nitrosomonas clusters (clusters 5 to 7), an additional band doublet was detected, with an intense signal in those lanes corresponding to highly polluted sediment (lanes 1 and 2), a less-intense signal at the 20-m station (lanes 3 and 4), and no signal at 40 m (lanes 5 and 6). While the intensity of this doublet decreased with decreasing pollutant levels, there appeared to be a corresponding increase in band intensity of the lower cluster 1 Nitrosospira sequences at the 20- and 40-m stations. This was confirmed by quantification of band densities (Fig. 6), which illustrates the relative signal contribution of bands from each subgroup to the total hybridization signal, estimated with the β-AO233 probe, within DGGE products obtained at each sampling station. The band doublets did not hybridize with either cluster 6b (Fig. 5D)- or cluster 7 (data not shown)-specific oligonucleotide probes but comigrated with the control clone for the novel, marine Nitrosomonas clones. On the basis of current sequence information, it is not possible to design cluster 5-specific oligonucleotide probes; hence, the identity was confirmed by sequence analysis of upper and lower bands. Both bands produced identical 422-bp sequences, and, as predicted, it appears that the degeneracy of the reverse PCR primer is responsible for the observed doublet effect (23). A similar explanation may apply to the multiple banding observed for the cluster 1 control. The cluster 5 DGGE sequences show >99% identity (1 bp mismatch) with the partial clone sequence EnvA2-13, obtained from the same sampling site by using different PCR primers. Phylogenetic analysis illustrates clearly that these DGGE band sequences cluster within Nitrosomonas cluster 5 (100% of bootstrapped replicates), as shown in Fig. 1.
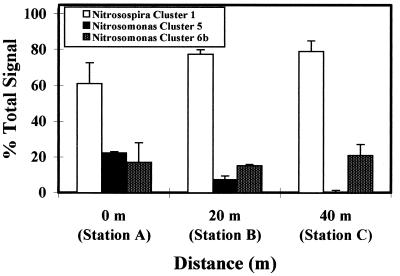
FIG. 6.
Relative abundance of β-subgroup ammonia-oxidizing bacteria from 0- to 5-mm-depth-horizon sediment samples obtained directly beneath the fish cage (0 m) and at distances of 20 and 40 m. Ammonia oxidizer clusters were resolved by DGGE, and relative band intensities were quantified by densitometry after probing with β-AMO233, which is specific for all β-subgroup ammonia-oxidizing bacteria. Error bars represent standard errors for duplicate sediment samples.
Quantification of the relative abundance of different clusters was carried out by densitometry. Figure 6 indicates the relative band intensities obtained when probe β-AO223, which detects all clusters of proteobacterial β-subgroup ammonia oxidizers, was used. Nitrosospira cluster 1 is the most abundant group at all three sites, comprising 53 to 83% of the total signal in each lane. This is in agreement with the original finding of Stephen et al. (37) that cluster 1 Nitrosospira spp. are found, and are potentially abundant, in marine environments. The relative quantity of signal from the cluster 5 bands, determined by using the β-AO223 probe (Fig. 5B), was significantly higher at site A than at site B or C (P < 0.01). Similarly, the signal from site B was significantly higher than that at site C (P < 0.01). Although in Fig. 5B to E there appears to be an increase in both clusters 1 and 6 with increasing distance from the fish cage, no significant differences in the relative abundances of these groups at any of the sites were observed. Quantification of the total signal for each lane on the ethidium bromide-stained gel, and after probing with β-AO233, indicates an increase in probe hybridization with distance from the fish cage, despite approximately equal DNA loadings (data not shown). This may be explained by amplification of nontarget sequences during the PCR or the presence within polluted sediments of ammonia oxidizers with rDNA sequences which are not complementary to oligonucleotides used in this study. Quantification of the relative abundance of Nitrosomonas clusters 5 and 6, when probed with the Nitrosomonas probe Nmo254, confirms that the cluster 5 signal significantly decreased, relative to the total signal within each lane, with increasing distance from the fish cage (data not shown). It was not possible in this study, however, to compare quantification of the relative abundance of cluster 5 by using both the Nitrosomonas probe Nmo254 and β-AMO233. A long exposure of the autoradiograph for Nmo254 (Fig. 5E) was necessary to visualize cluster 5 bands in lanes representing station B samples, but this resulted in saturation of cluster 5 and 6 controls, thus preventing normalization of the data for the two probes. However, comparison of the ratios of each cluster within the environmental samples, using both probes, indicates that the probing efficiencies of the two oligonucleotides were similar.
DISCUSSION
The aim of this study was to investigate the relationship between biological and chemical data on nitrogen cycling and community structure of proteobacterial β-subgroup ammonia-oxidizing bacteria in polluted and unpolluted sediments, combining traditional and molecular biology-based methods. In addition, 16S rDNA sequence analysis (37) and DGGE analysis were compared by analyzing ammonia oxidizers from the same sediment samples with specific PCR primers targeted to different parts of the 16S rRNA gene. Only members of the β-subgroup ammonia oxidizers were targeted for molecular analysis, and γ-subgroup ammonia oxidizers and unknown nitrifying organisms may have a role in nitrogen flux within polluted and unpolluted sediments.
Chemical and biological analyses indicated that nitrogen cycling beneath the fish cage had been severely disrupted by sedimentation of nitrogen-rich organic material from the cage. This detritus accumulation led to the formation of anaerobic sediment within the vicinity of the cage and undetectable levels of nitrification and denitrification. This is consistent with studies of model systems artificially enriched with organic material (8, 36) and other fish farm locations (11, 21). It is possible that nitrification was inhibited by the low oxygen penetration, which generally decreases from 2 to 6 mm, in normal sediments, to less than 1 mm in areas of high organic loading (8, 18).
The largest populations of ammonia-oxidizing bacteria and the highest nitrification rates were observed at station S, 10 m from the fish cage, while lower but detectable levels of both were observed at the 20- and 40-m stations. From these data it can be concluded that organic deposition from the fish cage did not affect process measurements for nitrification at a distance of 20 m, and the relative stimulation of ammonia oxidation at station S can be explained by the presence of elevated, but noninhibitory, concentrations of ammonium and organic material. Molecular analysis, however, did indicate differences in the compositions of the ammonia oxidizer communities directly beneath the fish cage and at 20 m, as shown by the presence of novel sequences within clone libraries (37) and the detection of additional DGGE bands. The MPN method for enumeration of ammonia oxidizers indicated that populations of less than 7 × 104 bacteria m−2 (5-mm sampling depth) were present in the sediment underlying the fish cage, since no growth was shown in any replicates at any dilution factor tested. Stephen et al. (37), however, enriched ammonia-oxidizing bacteria from all of the same sediment samples by using longer incubation periods. The appearance of novel bands in those samples from polluted sites suggests that the changes in environmental conditions in the vicinity of the fish cage led to selection for particular ammonia-oxidizing bacteria through the growth of these organisms and/or a decline of other members of the community. The relative importance of each of these factors is unknown, although pure-culture studies indicate that ammonia oxidizer cell numbers remain stable throughout a range of environmental stresses (15, 17).
Cell concentrations of ammonia and nitrite oxidizers and nitrification rates over the sampling transect were not closely correlated. It is highly likely that the standard culture conditions used for enumeration were not suitable for growth of all sediment ammonia oxidizers, particularly since a single ammonium concentration was used in the growth medium while significantly different levels were measured in the sediment samples at each station. In addition, it is possible that inhibitory substances present at the highly polluted site adversely affected enumeration. Stephen et al. (37) demonstrated a bias toward Nitrosomonas spp. in enrichment cultures from the same sediment samples, while Nitrosospira spp. were more abundant within clone libraries, suggesting that only a proportion of the total population will be enumerated. Similarly, the standard laboratory conditions used for process measurements may have been suboptimal for measurement of nitrification and denitrification in the sediments obtained during this study. For example, the chlorate method for measurement of nitrification may have been adversely affected within the polluted samples via inhibition of ammonia oxidation through reduction of chlorate to chlorite by nitrite oxidizers (13). The application of 15N-based techniques might have provided more-accurate flux data but would have required considerably larger samples.
Although nitrification and denitrification were not detected, nitrous oxide production was measured at the fish cage site and could have resulted from internal cycling of nitrogen within an oxygen gradient. Ammonia and nitrite oxidation may have occurred within aerobic regions, providing substrate for the reduction of nitrate to ammonium, via nitrite and nitrous oxide, within anaerobic regions. The resulting accumulation of nitrate and nitrous oxide beneath the fish cage may, therefore, have occurred because of an imbalance in the flux of nitrogen transformations within the system due to the particularly high levels of ammonium, i.e., higher rates of ammonia oxidation than of ammonium recycling. It is also possible that nitrous oxide was produced as a by-product of ammonia oxidation, while Schmidt and Bock (34) recently demonstrated simultaneous nitrification and denitrification by Nitrosomonas eutropha. Reduction of nitrate to ammonium may have been carried out by the nitrite-oxidizing bacteria (28, 39) rather than by heterotrophic nitrate-respiring bacteria. This is supported by high MPN counts for nitrite oxidizers and detection of only small populations of nitrate respirers at the polluted station. In a similar study, Kaspar et al. (21) showed that potential denitrification in sediment core slices collected from beneath a salmon fish cage was undetectable to a depth of 6 cm, while other studies have demonstrated that ammonium recycling is favored over denitrification when nitrate levels are low and the level of organic matter is high (25).
In this study, DGGE analysis demonstrated a significant change in the community structure of ammonia oxidizers, the appearance of a novel subgroup of Nitrosomonas spp., within the polluted-sediment samples, confirming preliminary results based on sequence analysis of 16S rDNA gene libraries of ammonia oxidizers prepared from the same sediment samples with different PCR primers. These strains may have increased tolerance to low oxygen tensions, to increased ammonium concentrations, or to the presence of inhibitory substances or may have been introduced from external sources, such as fish feed or fecal pellets. Cluster 5 sequences showing 98.5 to 99.5% sequence similarity to near-full-length (1.1-kb) cluster 5 sequences from the polluted sediments have also been detected in an unpolluted, seaward sand dune site (23). Information on the relative abundance of this novel subgroup in the sand dune site is not available, but in contrast to the polluted sediments, the nitrogen content of the sand dune site was low. The physiological and environmental significance of this group, therefore, remains unclear in the absence of physiological analysis of pure-culture representatives.
The presence of cluster 5 Nitrosomonas-like ammonia oxidizer strains within sediment collected at the 20-m site (Fig. 5A and 5E) corresponds with visual observations of minimal organic impact at this site but not with any chemical indication of nitrogen cycling disruption. This suggests that the community structure of the β-subgroup ammonia oxidizers is more sensitive to environmental changes than the physiological process carried out by these organisms, although it is important to note that community structure was assessed by rDNA analysis alone in the present study. Since nitrification was also detected at the 10-m distance, it can be concluded that the detrimental impact of pollution from the fish farm was limited to sediments in the immediate vicinity of the fish cage.
In conclusion, this study demonstrates clearly that DGGE analysis, in combination with Southern hybridization, represents an extremely elegant ecological tool for the analysis of bacterial diversity and is particularly applicable to the investigation of proteobacterial β-subgroup ammonia oxidizers, a phylogenetically and physiologically constrained group of microorganisms. DGGE is a relatively inexpensive and rapid method of assessing population diversity, and while this technology cannot supersede sequence analysis, which is comparatively expensive, repetitive, and time-consuming, it has been shown to complement analysis of rDNA clone libraries. In this investigation, it was possible to link gross process and biological measurements with more-subtle variation detected in the community structure. Inclusion of data from a broad range of environmental parameters—physical, chemical, and biological—proved invaluable in the interpretation of results from the molecular analyses, highlighting the necessity for comprehensive, multidisciplinary approaches to the study of microbial ecology. This study has confirmed the selection of a novel group of β-subgroup ammonia-oxidizing bacteria in sediments underlying and in proximity to fish farm cages. Further analysis of this group is necessary to understand fully its physiological and ecological significance in natural environments.
ACKNOWLEDGMENTS
We thank the Coastal Impact Research Group at Dunstaffnage Marine Laboratory (Oban, United Kingdom) for collection of the samples.
This work was funded by NERC grant GST/02/568 to J.I.P. and T.M.E. as part of the NERC GAMES initiative and by MAFF grant AE 0511 to S.M.H.
REFERENCES
- 1.Alexander M. Denitrifying bacteria. In: Black C A, editor. Methods of soil analysis. Vol. 9. Madison, Wis: American Society of Agronomy; 1965. pp. 1484–1486. [Google Scholar]
- 2.Alexander M, Clark F E. Nitrifying bacteria. In: Black C A, editor. Methods of soil analysis. Vol. 9. Madison, Wis: American Society of Agronomy; 1965. pp. 1477–1483. [Google Scholar]
- 3.Bédard C, Knowles R. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol Rev. 1989;53:68–84. doi: 10.1128/mr.53.1.68-84.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianchi M, Bonin P, Feliatra F. Bacterial nitrification and denitrification rates in the Rhone River plume (northwestern Mediterranean Sea) Mar Ecol Prog Ser. 1994;103:197–202. [Google Scholar]
- 5.Blackburn T H, Blackburn N D. Model of nitrification and denitrification in marine sediments. FEMS Microbiol Lett. 1992;100:517–522. [Google Scholar]
- 6.Brosius J, Dull T L, Sleeter D D, Noller H F. Gene organisation and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;14:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 7.Byers S C, Mills E L, Stewart P L. Measurements of the physical and chemical environments. In: Holme N A, McIntyre A D, editors. Methods for the study of marine benthos. Oxford, United Kingdom: Blackwell Scientific; 1978. pp. 30–58. [Google Scholar]
- 8.Caffrey J M, Sloth N P, Kaspar H F, Blackburn T H. Effect of organic loading on nitrification and denitrification in a marine sediment microcosm. FEMS Microbiol Ecol. 1993;12:159–167. [Google Scholar]
- 9.Dugdale R C. Nutrient limitation in the sea: dynamics, identification and significance. Limnol Oceanogr. 1967;12:685–695. [Google Scholar]
- 10.Felsenstein J. PHYLIP: phylogeny inference package. Seattle: University of Washington; 1993. [Google Scholar]
- 11.Hall P O J, Holby O, Kollberg S, Samuelsson M-O. Chemical fluxes and mass balances in a marine fish cage farm. IV. Nitrogen Mar Ecol Prog Ser. 1992;89:81–91. [Google Scholar]
- 12.Head I M, Hiorns W D, Embley T M, McCarthy A J, Saunders J R. The phylogeny of autotrophic ammonia-oxidizing bacteria as determined by analysis of 16S ribosomal RNA gene sequences. J Gen Microbiol. 1993;139:1147–1153. doi: 10.1099/00221287-139-6-1147. [DOI] [PubMed] [Google Scholar]
- 13.Hynes R K, Knowles R. Inhibition of chemoautotrophic nitrification by sodium chlorate and sodium chlorite: a reexamination. Appl Environ Microbiol. 1983;45:1178–1182. doi: 10.1128/aem.45.4.1178-1182.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnsen R I, Grahl-Nielsen O, Lunestad B T. Environmental distribution of organic waste from a marine fish farm. Aquaculture. 1993;118:229–244. [Google Scholar]
- 15.Johnstone B H, Jones R D. Effects of light and CO on the survival of a marine ammonia-oxidizing bacterium during energy source deprivation. Appl Environ Microbiol. 1988;54:2890–2893. doi: 10.1128/aem.54.12.2890-2893.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones R D, Hood M A. Effects of temperature, pH, salinity, and inorganic nitrogen on the rate of ammonium oxidation by nitrifiers isolated from wetland environments. Microb Ecol. 1980;6:339–347. doi: 10.1007/BF02010496. [DOI] [PubMed] [Google Scholar]
- 17.Jones R D, Morita R Y. Survival of a marine ammonium-oxidiser under energy-source deprivation. Mar Ecol Prog Ser. 1985;26:175–179. [Google Scholar]
- 18.Jorgensen K S, Revsbech N-P. Diffusive boundary layers and the oxygen uptake of sediments and detritus. Limnol Oceanogr. 1985;30:111–122. [Google Scholar]
- 19.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 20.Juliette L Y, Hyman M R, Arp D J. Inhibition of ammonia oxidation in Nitrosomonas europaea by sulfur compounds: thioethers are oxidized to sulfoxides by ammonia monooxygenase. Appl Environ Microbiol. 1993;59:3718–3727. doi: 10.1128/aem.59.11.3718-3727.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaspar H F, Hall G H, Holland A J. Effects of sea cage salmon farming on sediment nitrification and dissimilatory nitrate reductions. Aquaculture. 1988;70:333–344. [Google Scholar]
- 22.Keener W K, Arp D J. Transformations of aromatic compounds by Nitrosomonas europaea. Appl Environ Microbiol. 1994;60:1914–1920. doi: 10.1128/aem.60.6.1914-1920.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kowalchuk G A, Stephen J R, De Boer W, Prosser J I, Embley T M, Woldendorf J W. Analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl Environ Microbiol. 1997;63:1489–1497. doi: 10.1128/aem.63.4.1489-1497.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laanbroek H J, Bodelier P L E, Gerards S. Oxygen consumption kinetics of Nitrosomonas europaea and Nitrobacter hamburgensis in mixed continuous cultures at different oxygen concentrations. Arch Microbiol. 1994;161:156–162. [Google Scholar]
- 25.MacFarlane G T, Herbert R A. Dissimilatory nitrate reduction and nitrification in estuarine sediments. J Gen Microbiol. 1984;130:2301–2308. [Google Scholar]
- 26.Maidak B L, Larsen N, McCaughey J, Overbeek R, Olsen G J, Fogel K, Blandy J, Woese C R. The ribosomal database project. Nucleic Acids Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCaig A E, Embley T M, Prosser J I. Molecular analysis of enrichment cultures of marine ammonia oxidisers. FEMS Microbiol Lett. 1994;120:363–368. doi: 10.1111/j.1574-6968.1994.tb07059.x. [DOI] [PubMed] [Google Scholar]
- 28.Meincke M, Bock E, Kastrauand D, Kroneck P M H. Nitrite oxidoreductase from Nitrobacter hamburgensis—redox centers and their catalytic role. Arch Microbiol. 1992;158:127–131. [Google Scholar]
- 29.Muyzer G, De Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parsons T R, Maita Y, Lalli C M. A manual of chemical and biological methods for seawater analysis. Oxford, United Kingdom: Pergamon Press; 1984. [Google Scholar]
- 31.Raymond N, Bonin P, Bertrand J-C. Comparison of methods for measuring denitrifying activity in marine sediments from the western Mediterranean coast. Oceanol Acta. 1992;15:137–143. [Google Scholar]
- 32.Rowe R, Todd R, Waide J. Microtechnique for most-probable-number analysis. Appl Environ Microbiol. 1977;33:675–680. doi: 10.1128/aem.33.3.675-680.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt I, Bock E. Anaerobic ammonia oxidation with nitrogen dioxide by Nitrosomonas eutropha. Arch Microbiol. 1997;167:106–111. [PubMed] [Google Scholar]
- 35.Sheffield V C, Cox D R, Lerman L S, Myers R M. Attachment of a 40-base pair G+C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc Natl Acad Sci USA. 1989;86:232–236. doi: 10.1073/pnas.86.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sloth N P, Blackburn H, Hansen L H, Risgaard-Petersen N, Lomstein B A. Nitrogen cycling in sediments with different organic loading. Mar Ecol Prog Ser. 1995;116:163–170. [Google Scholar]
- 37.Stephen J R, McCaig A E, Smith Z, Prosser J I, Embley T M. Molecular diversity of soil and marine 16S rRNA gene sequences related to β-subgroup ammonia-oxidizing bacteria. Appl Environ Microbiol. 1996;62:4147–4154. doi: 10.1128/aem.62.11.4147-4154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stephen J R, Kowalchuk G A, Bruns M-A V, McCaig A E, Phillips C J, Embley T M, Prosser J I. Analysis of β-subgroup proteobacterial ammonia oxidizer populations in soil by denaturing gradient gel electrophoresis analysis and hierarchical phylogenetic probing. Appl Environ Microbiol. 1998;64:2958–2965. doi: 10.1128/aem.64.8.2958-2965.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sundermeyer-Klinger H, Meyer W, Warninghoff B, Bock E. Membrane-bound nitrite oxidoreductase of Nitrobacter—evidence for a nitrate reductase system. Arch Microbiol. 1984;140:153–158. [Google Scholar]
- 40.Utåker J B, Bakken L, Jiang Q Q, Nes I F. Phylogenetic analysis of seven new isolates of ammonia-oxidising bacteria based on 16S rRNA gene sequences. Syst Appl Microbiol. 1996;18:549–559. [Google Scholar]
- 41.Warner G F, Domaniewski J C J. Fish farming and the environment. Biologist. 1993;40:202–205. [Google Scholar]
- 42.Watson S W, Bock E, Harms H, Koops H-P, Hooper A B. Nitrifying bacteria. In: Staley J T, Bryant M P, Pfennig N, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 3. Baltimore, Md: Williams and Wilkins Co.; 1989. pp. 1808–1834. [Google Scholar]
- 43.Woese C R, Weisburg W G, Paster B J, Hahn C M, Tanner R S, Krieg N R, Koops H-P, Harms H, Stackebrandt E. The phylogeny of the purple bacteria: the beta-subdivision. Syst Appl Microbiol. 1984;5:327–336. doi: 10.1016/s0723-2020(84)80034-x. [DOI] [PubMed] [Google Scholar]
- 44.Woese C R, Weisburg W G, Hahn C M, Paster B J, Zablen L B, Lewis B J, Macke T J, Ludwig W, Stackebrandt E. The phylogeny of the purple bacteria: the gamma-subdivision. Syst Appl Microbiol. 1985;6:25–33. [Google Scholar]