Abstract
With the ever-increasing use of 3D cell models toward studying bio-nano interactions and offering alternatives to traditional 2D in vitro and in vivo experiments, methods to image biological tissue in real time and with high spatial resolution have become a must. A suitable technique therefore is surface-enhanced Raman scattering (SERS)-based microscopy, which additionally features reduced photocytotoxicity and improved light penetration. However, optimization of imaging and postprocessing parameters is still required. Herein we present a method to monitor cell proliferation over time in 3D, using multifunctional 3D-printed scaffolds composed of biologically inert poly(lactic-co-glycolic acid) (PLGA) as the base material, in which fluorescent labels and SERS-active gold nanoparticles (AuNPs) can be embedded. The combination of imaging techniques allows optimization of SERS imaging parameters for cell monitoring. The scaffolds provide anchoring points for cell adhesion, so that cell growth can be observed in a suspended 3D matrix, with multiple reference points for confocal fluorescence and SERS imaging. By prelabeling cells with SERS-encoded AuNPs and fluorophores, cell proliferation and migration can be simultaneously monitored through confocal Raman and fluorescence microscopy. These scaffolds provide a simple method to follow cell dynamics in 4D, with minimal disturbance to the tissue model.
Keywords: SERS imaging, 3D bioprinting, scaffolds, SERS tags, multimodal imaging
Introduction
The use of 3D-printed scaffolds for in vitro cell culture has become an important element in current biomedical research, in addition to offering an alternative route to in vivo studies.1,2 By 3D printing the scaffold, both the physical and biological properties can be tailored, obtaining high degrees of spatial resolution and reproducibility.3−5 These 3D-printed scaffolds can be used in various ways, such as supporting cell growth,6,7 inducing cell differentiation,8 acting as an imaging reference point,9 or as a sensing substrate to analyze molecular changes over time.10 Moreover, 3D printing has become an excellent fabrication tool for polymeric scaffolds,11,12 as it provides numerous possibilities to customize the final design toward reproducible fabrication at a relatively low cost.13,14 The production of biocompatible scaffolds with micron-scale resolution, yet combining large footprint areas, has been achieved by means of 3D jet writing of polymer inks.15−18 This modified electrospinning process can produce, for example, a honeycomb tessellated structure featuring a highly organized open structure, which can support the long-term growth of a wide variety of cell types, including fibroblasts, endothelial cells, cancer cells, and even mesenchymal stem cells, maintaining the mechanical integrity and even allowing in vivo implantation.15,16 Indeed, gold nanoparticles (AuNPs), and/or molecules such as drugs or fluorophores, can also be included to form multicompartment particles capable of controlled drug release and multimodal imaging.19−21
An important condition toward the development of new materials for sophisticated 3D models is the ability to accurately image and characterize them in situ, hence providing real-time information relating to aspects such as cell proliferation, migration, and differentiation, over time.22−24 For this purpose, multimodal imaging techniques that provide complementary information can be explored. However, the translation of imaging techniques from 2D to 3D cell models is, in general, not trivial,25 and the lack of standardization in emerging characterization techniques26 is also slowing down the development of new substrates for 3D culture and their translation into real clinical protocols.27
We thus identified a need to explore alternative methods and optimize various live cell imaging parameters, toward achieving accurate characterization of 3D cell models with time. To this aim, we have identified surface-enhanced Raman scattering (SERS) imaging as a convenient technique for the interrogation of cells in situ, which furthermore can be combined with fluorescence imaging.28,29 SERS imparts both high sensitivity and high selectivity, thus being an excellent option for imaging live cell-containing 3D structures.30 For SERS imaging, cells are labeled with SERS tags comprising AuNPs encoded by adsorbing Raman-active molecules (Raman reporters) with strong affinity for the AuNP surface and well-defined Raman fingerprints.31
The biocompatible nature and optical stability of purposely designed SERS tags allows long-term imaging in a noninvasive manner, and the wide choice of available Raman reporters provides extensive opportunities for SERS multiplexing, using a single irradiation wavelength.30,32−34 Compared with fluorescence microscopy, SERS tags do not suffer from probe photobleaching,19 and they can be excited by a near-infrared (NIR) laser, resulting in improved penetration of light through tissue.35 It should however be recognized that 3D SERS bioimaging is still in a developmental phase, and not only sophisticated microscopes but also optimization of measurement parameters and complex data analysis are required.
Herein, we show that the optimization of 3D SERS live imaging parameters allowed us to achieve the imaging of SERS-labeled live cells, grown within 3D-printed scaffolds carrying SERS and fluorescent labels. We show that the presence of SERS tags in the scaffold or in the cells does not hinder fluorescence imaging, nor vice versa, and therefore, both SERS and fluorescence imaging techniques can be combined to achieve simultaneous multimodal imaging.36,37 Using the 3D jet writing technique, we fabricated PLGA tessellated scaffolds containing fluorescent polymers and/or SERS-encoded gold nanostars (AuNSs). We demonstrate that such hybrid scaffolds offer a suitable support frame for the long-term growth of human dermal fibroblasts (HDFs); cells grow in a suspended 3D form which offers sufficient optical transparency for both fluorescence and SERS imaging. Indeed, cell–scaffold contact is kept to a minimum, thus promoting the cell–cell interactions that are found in the natural in vivo microenvironment. The relevant parameters for both imaging techniques were optimized taking into consideration factors such as imaging time, resolution, and signal intensity. To achieve multiplexing, we prelabeled separate HDF populations with SERS tags of different Raman fingerprints, which can all be excited at the same laser wavelength.34 By including fluorescence and/or SERS tags, the scaffold could be used not only as a support but also as an imaging reference point to study the migration of different HDF populations. Our results demonstrate multimodal 3D imaging of complex living cell systems over long periods of time, with no need for fixing the cells, thereby opening the way toward 3D live cell imaging in complex systems, using 3D SERS imaging as a complementary bioimaging technique.
Results and Discussion
PLGA Scaffolds Labeled for SERS and Fluorescence Imaging
The preparation of SERS tags responsive to NIR irradiation was achieved by first synthesizing AuNSs with a localized surface plasmon resonance (LSPR) maximum around 800 nm, in resonance with the 785 nm laser wavelength usually available in Raman microscopes.31,38 AuNSs are ideal SERS substrates because of the high electromagnetic field enhancements at their multiple tips,39 as well as their biocompatibility and flexibility regarding the choice of Raman-active molecular reporters that can be incorporated.34 AuNSs were synthesized in aqueous solution40 and transferred to chloroform (CHCl3)34,41 upon adsorption of biphenyl-4-thiol (BPT), which provides a well-defined Raman signature and hydrophobicity. The resulting particles (AuNS-BPT) had an average size of 50 nm, as confirmed by transmission electron microscopy (TEM), and a main extinction band around 785 nm (Figure S1A).42 The SERS spectrum of AuNS-BPT was found to match the characteristic molecular fingerprint of BPT (Figure S1B).
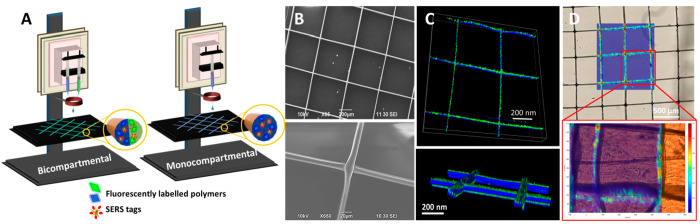
The scaffolds fabricated with the 3D jet writing technique were composed of PLGA fibers of 10 μm diameter, each of which can also be divided into different compartments if desired. As such, we fabricated scaffolds composed of monocompartmental and bicompartmental fibers including SERS tags and different fluorophores. For their fabrication, AuNS-BPT ([Au0] = 3 × 10–3 M) SERS tags, dispersed in chloroform (CHCl3), were subsequently mixed with the fluorescently labeled polymers (poly[(mphenylenevinylene)-alt-(2,5-dihexyloxy-p-phenylenevinylene] and poly[tris(2,5-bis(hexyloxy)-1,4-phenylenevinylene)-alt-(1,3-phenylenevinylene)]) and 50–75 kDaA PLGA (30 wt/vol %), all dissolved in CHCl3. N,N-Dimethylformamide (DMF) was then added to the solution, resulting in a solvent mixture of CHCl3 and DMF at ca. 93:7 vol/vol %16 (see details in the Experimental Section). To fabricate the scaffolds, the different SERS tag PLGA solutions were loaded in individual syringes and flown in a laminar regime through parallel mounted metallic 20-gauge needles (Figure 1A). Hybrid PLGA scaffolds were printed with either 5 or 10 aligned fibers of 10 μm in diameter (Figure S2), resulting in a 10 × 10 mm2 mesh with individual 500 × 500 μm2 squares, as observed by scanning electron microscopy (SEM) (Figure 1B). The bicompartmental composition of the fibers allows the incorporation of either one or two fluorophores (green and blue), as well as AuNSs-BPT (or other SERS tags). Shown in Figure 1C and Figure S2C–E are confocal fluorescence microscopy images of the structure containing different fluorophores and AuNS-BPT, resulting in strong SERS and fluorescence signals, readily detectable in the whole scaffold (Figure 1D).
Figure 1.
Characterization of PLGA hybrid scaffolds. (A) Schematic illustration of the 3D printing system for bicompartmental and monocompartmental scaffolds. (B) Representative SEM images of the hybrid scaffolds. (C) Fluorescence confocal microscopy images showing emission from blue and green labeled compartments. (D) SERS mapping of the signal from AuNS-BPT SERS tags, distributed throughout the PLGA fibers.
SERS Tag Labeling of HDF Cells
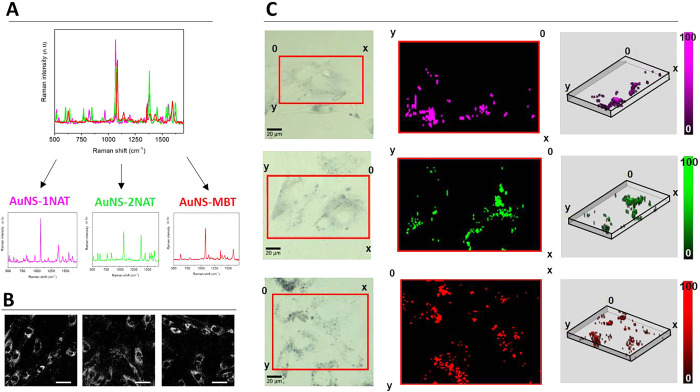
Primary human dermal fibroblasts (HDF) were selected as a model cell system to test the ability of hybrid scaffolds to support cell growth in 3D over time. In their natural setting, HDFs are the main cellular component of the dermis layer of the skin and play a crucial role in maintaining the structural integrity of this connective tissue. We have previously shown that HDF cells avidly uptake positively charged SERS tags and can retain them for long time periods (ca. 2 weeks).30 We therefore incubated HDF cells with different SERS tags, to obtain a mixed group of cells which could be monitored over time. We chose SERS tags composed of AuNSs labeled with a Raman reporter (1-naphthalenethiol (1NAT), 2-naphthalenethiol (2NAT), or 4-methylbenzenethiol (MBT)), and stabilized with dodecylamine-modified polyisobutylene-alt-maleic anhydride (PMA) and further coated with poly-l-arginine hydrochloride (PA) to achieve a positive surface charge (Figure S3). We denoted these SERS tags as AuNS-1NAT, AuNS-2NAT, and AuNS-MBT. The choice of 1NAT, 2NAT, and MBT as the Raman reporter molecules allowed multiplexing through their spectral features, which could be distinguished using postprocessing tools such as multiple linear regression analysis (MLRA)38 or True Component Analysis (Figure 2A).
Figure 2.
Characterization and imaging of SERS tags for cell labeling. (A) SERS spectra of AuNS-1NAT, AuNS-2NAT, and AuNS-MBT SERS tags, measured in water. (B) SERS tag uptake into HDF, as detected via two-photon confocal microscopy (780 nm excitation, 570–610 nm emission). Scale bars: 50 μm. (C) Left: brightfield images of cells selected for SERS imaging (red box); middle: maximum intensity projection SERS maps showing distribution of signals corresponding to separate SERS tags; right: 3D volume reconstruction of z-stack SERS maps. SERS imaging was conducted using 0.02 s integration time with 5 mW laser power and 1 × 1 × 4 μm step size (XYZ).
Indeed, exposure of HDF cells to all three SERS tag formulations for 24 h resulted in high levels of uptake (ca. 10–30% of added NP), which was verified via two-photon and SERS confocal microscopy, showing SERS signals located in peri-nuclear vesicles (Figure 2B,C). In both cases, SERS tags were added at a concentration of 0.1 mM (in FBS-containing media) to previously serum starved cells, thereby promoting endocytosis of SERS tags by cells, at a similar rate.43 Previous studies with similar NPs showed no cytotoxicity, thanks to the protective PMA coating and the inherent biocompatible nature of gold.30,34 Verification was performed by analyzing cell viability via propidium iodide (PI) uptake as a marker of damaged cell membranes. As shown in Figure S4, no significant changes in overall cell number, as well as the lack of PI stained cells in the entire cell population, confirm that SERS tags are not cytotoxic to HDF, even at relatively high concentrations (0.1 mM). We thus explored the labeling of HDF cells with SERS for long-term SERS imaging. First, we studied the Raman signal intensity over time, using HDF cells prelabeled with AuNS-2NAT as an example (Figure 3). The initial average signal of the Raman spectra collected between 600 and 1600 cm–1 (4 days in vitro, 4 “DIV”) showed little change in intensity, while after a further 4 days (8 DIV), the intensity reduced by approximately half. To conduct SERS imaging, we used a confocal Raman microscope (Alpha300 R, Witec) with a 20× immersion long working distance objective, and an XYZ step size of 1 × 1 × 4 μm3. Whereas for initial cell uptake characterization (Figure 2C), we used a 0.02 s integration time, working with 5 mW laser power, for interrogation of the cell culture with time, a longer integration time (0.05 s vs 0.02 s) and higher laser power (10 mW vs 5 mW) were needed to obtain clear signals. The 1 × 1 × 4 μm3 step size in XYZ coordinates was sufficient to spatially resolve the SERS tags in a 2D cell culture over time. Even higher SERS signal intensities could be achieved by further increasing the excitation laser power intensity, while avoiding damage to cells (Figure S5). As a result, cells can be imaged for longer periods if needed, much in the same way as laser power and detector gain can be increased in fluorescence imaging.
Figure 3.

Monitoring HDF cells labeled with AuNS-2NAT over time in 3D. (A) Volume reconstructions from SERS confocal imaging (left) of areas containing HDF cells, framed by colored boxes in the brightfield optical images (right). Mapping was performed on day 1 (DIV 1, blue), DIV 4 (orange), and DIV 8 (yellow). (B) Average SERS spectra from DIV 1, DIV 4, and DIV 8, calculated from the entire imaging volume. SERS imaging was conducted using a 0.05 s integration time with 10 mW laser power and 1 × 1 × 4 μm step size (XYZ).
SERS Imaging of Cells within Tessellated Scaffolds
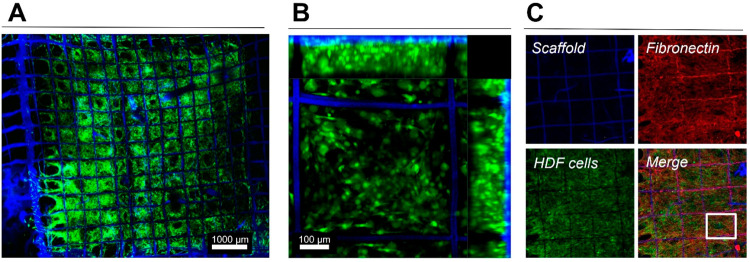
In addition to the improved signal stability, as compared with fluorescence confocal microscopy, SERS microscopy offers the ability to image deeper in a biological sample by using a NIR laser with deeper penetration length.30,44 This feature is of special interest in the characterization of in vitro and in vivo 3D models, which require noninvasive imaging over time and may intrinsically present high absorption and scattering of visible light. SERS imaging conditions and parameters were thus optimized to establish both the limitations and the advantages of SERS for 3D cell model imaging. We first analyzed the growth kinetics of HDF cells, seeded onto the hybrid multilabeled scaffolds, using GFP-transfected HDF cells (Figure S6). HDF growth could be clearly observed to progressively fill the spaces of the scaffold, eventually resulting in the formation of a continuous HDF film, growing intertwined with the scaffold fibers (Figures 4a, S7). Within 14 days of cell growth, a complete HDF film with a thickness larger than 100 μm was identified within the printed scaffold. Immunocytochemical staining revealed the presence of fibronectin in the suspended imaging windows, colocalizing with the presence of HDF cells. This suggests that, aside from the fibronectin used initially to coat the scaffolds to aid cell adherence, HDF cells were producing their own extracellular matrix to support their growth in 3D (Figure 4b,c).
Figure 4.
HDF cell network formed in 3D jet printed scaffolds, 14 days after seeding. The scaffold shows blue fluorescence, HDF cells were transfected to express GFP (green emission), and immunocytochemical staining after fixing allowed the detection of scaffold associated and HDF secreted fibronectin (red). (A) Tile image showing the entire scaffold (1 cm2) with HDF cells filling most of the squares. (B) Maximum intensity projection (MIP) of a Z-stack (ca. 150 μm thick) showing HDF 3D organization in the scaffold. (C) MIPs of Z-stacks (ca. 150 μm thick) showing individual components and a merged image. The square in (C, Merge) depicts the area shown at higher magnification in Figure S7.
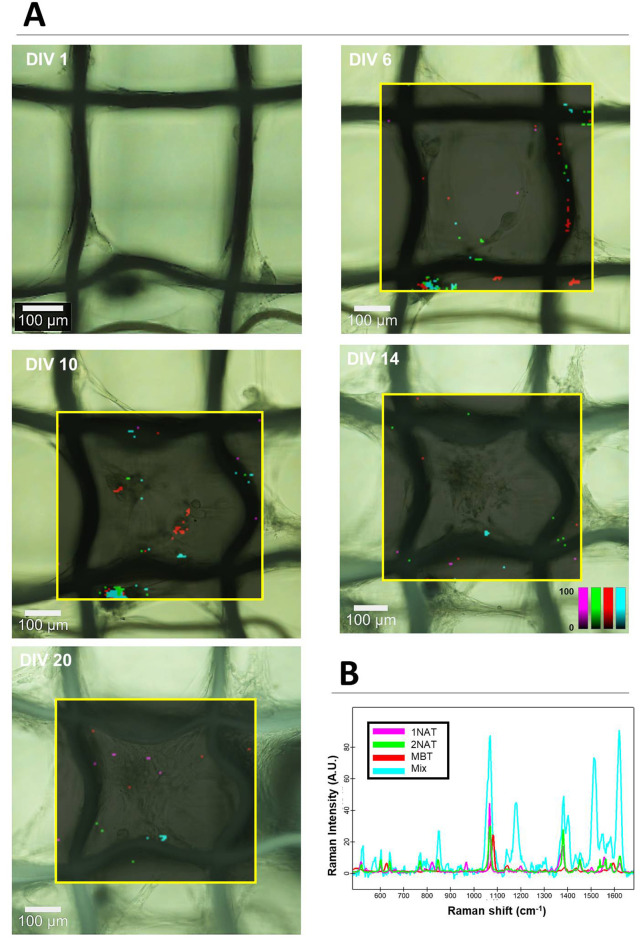
To study the kinetics of HDF cell growth via SERS, nontransfected HDF cells prelabeled with the three SERS tags were seeded onto the scaffold and SERS spectra collected every 3–5 days for a period of ca. 3 weeks. The scaffold geometry, which supports cell growth yet with minimum cell–scaffold contact, was ideal for SERS imaging, as cells were essentially in a suspended state but growing in a 3D extracellular matrix (Figure 5). This provided an imaging “window” that allowed continued monitoring of cells in a noninvasive manner. SERS imaging in 2D was conducted, and signals corresponding to the three SERS tags were plotted and overlaid on a brightfield image of the same area (Figure 5, Figure S8). Whereas we did observe changes in the distribution of the SERS tags within the field of view, the 2D nature of this mapping led to a significant loss of information. We could confirm the presence of HDF cells in the field of view via brightfield imaging, but few SERS tag-positive cells were observed by SERS at a fixed viewing plane. On the basis of the expected dilution of SERS tags with time, and knowing that HDF cells most likely occupy the thickness of ca. 100 μm, the 3D distribution of SERS tags in the scaffold was studied using 30 mW laser power with 0.05 s integration time (Figure 6, Figure S9). Indeed, the measurements conducted at 25 DIV clearly show the presence of many HDF cells labeled with all three SERS tags, with a homogeneous distribution throughout the field of view. The z-dimension is also in agreement with that determined via fluorescence (ca. 100 μm, Figure 4b).
Figure 5.
HDF proliferation and migration. HDF cells prelabeled with three different SERS tags were used: AuNS-1NAT (magenta), AuNS-2NAT (green), and AuNSs-MBT (red). HDFs were allowed to grow in a tessellated scaffold; SERS mapping of the area framed in yellow was conducted every 3–5 days. Cells containing all three SERS tags are shown in cyan. (A) Brightfield images with overlaid SERS maps at different time points. (B) Graph showing the average SERS spectra for all three SERS tags, as well as the intensity of pixels with positive match for all three tags, as analyzed by True Component Analysis (Witec). SERS imaging was conducted using a 0.05 s integration time, 10 mW laser power, and a 5 × 5 μm2 step size (XY).
Figure 6.
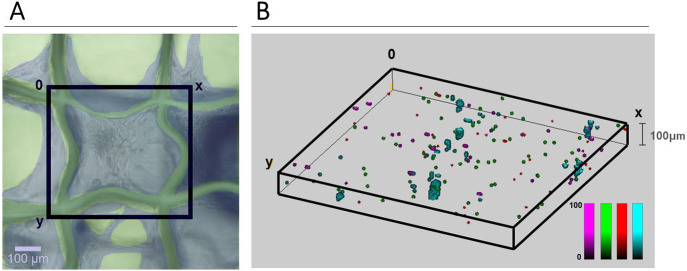
3D SERS mapping. HDF cell distribution after 25 DIV, as viewed with brightfield microscopy (A) and 3D SERS imaging (B). (A) HDF cells are false-colored in gray blue, the cell-filled imaging window (black square box) was subsequently mapped by SERS imaging. (B) SERS mapping of HDF cells, prelabeled with AuNS-1NAT (magenta), AuNS-2NAT (green), and AuNS-MBT (red). Cells containing all three SERS tags are displayed in cyan. SERS maps were analyzed using True Component Analysis (Witec). SERS imaging was conducted using 0.05 s integration, 30 mW laser power, and 5 × 5 × 5 μm3 step size (XYZ).
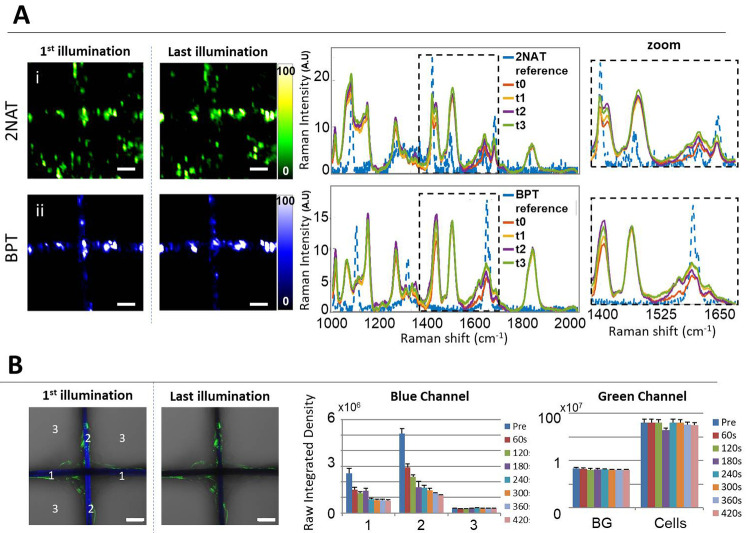
With regards to the targeted imaging spatial resolution and time required to achieve it, it should be noted that 3D cell imaging was carried out using cubic voxels, measuring 5 μm3 in XYZ. Therefore, to complete an image measuring 600 × 600 × 100 μm3 (in XYZ) with this level of imaging resolution (5 μm3), 12 h were needed. Although, admittedly, this could be considered as a long time, we rely on the absence of photoinduced cytotoxicity and probe bleaching during SERS (opposite to fluorescence) imaging. As an additional evidence, we conducted a bleaching experiment in which HDF cells, labeled with either SERS tags or AF488-labeled actin fluorophores, were imaged by SERS and fluorescence microscopy, respectively. The cells were grown on a scaffold that was itself labeled with either AuNS-MBT (for SERS imaging) or poly[(mphenylenevinylene)-alt-(2,5-dihexyloxy-p-phenylenevinylene] light-emitting polymer (for fluorescence imaging). For the SERS bleaching test, we conducted a mock imaging experiment in which four sequential 2D images over an area of 250 × 250 μm2, with 5 μm resolution (XY), were conducted using a 785 nm laser–in resonance with both SERS tags (each of these runs lasted for 1 h). For the fluorescence bleaching test, an image size of 354 × 354 μm2 was selected, and seven sequential bleaching/imaging cycles were undertaken using 405 nm laser excitation, in resonance for the scaffold fluorophore. The pinhole was opened to 24 μm, and a 60 s bleaching cycle followed by imaging acquisition was used. Intentionally, only 405 nm irradiation was used to excite the scaffold fluorophore, so that the green fluorescence from AF488-actin labeled cells would remain unaffected and provide an internal control. The results are shown in Figure 7. No changes were revealed in SERS mapping over time as it relates to the signal intensity of either BPT (from the scaffold) or 2NAT (from cells). Oppositely, a significant decrease was recorded in the fluorescence intensity of the blue channel (scaffold) while the green fluorescence remained unchanged. These results confirm that, unlike fluorescence microscopy, SERS can be used for repetitive measurements without any changes in signal intensity due to photon-induced chemical changes in the Raman molecules. It should be noted that, in all cases, the BPT signal was recorded at the well-defined area of the scaffold and showed negligible fluctuations over time. This result is in agreement with a high stability of AuNS-BPT SERS tags inside the PLGA scaffold structure, with no leaching of SERS tags and no Raman molecules leaving the NP surface, which are essential aspects to be considered when long irradiation times are required.
Figure 7.
Comparison of SERS and fluorescence stability upon sequential bleaching. (A) Bleaching test using cell-internalized AuNR-2NAT and AuNS-BPT NPs inside PLGA scaffolds. For SERS imaging, the selected area (250 × 250 μm2) was repeatedly illuminated for 1 h four times using a 785 nm laser, resonant with all SERS tags used. For fluorescence imaging, a different area (354 × 354 μm2) was irradiated (in scanning mode) for 60 s seven times using a 405 nm laser. Images show the first and last illumination SERS maps, as well as the corresponding average spectra. Zoomed spectra highlight one of the main representative Raman peaks for each label. A 50× (NA 0.5) long working distance objective was used. Scale bars: 50 μm. (B) Fluorescence imaging bleaching test. Images in the left panel show the blue and green channels overlaid with corresponding optical images, for the first and last illuminations. Scale bars: 50 μm. The graphs to the right show the evolution of fluorescence intensity for both fluorophores.
Using Other NP Shapes for SERS Imaging
While in this study we focused on the use of AuNS, it should be noted that AuNPs with different morphologies can also be used, as long as their LSPR is in resonance with the Raman excitation wavelength used for imaging. Gold nanorods (AuNRs) are an obvious example; they feature a longitudinal LSPR that can be tuned into the NIR during synthesis. We conducted a supplementary study in which two SERS tags, composed of AuNR-2NAT and AuNS-MBT both coated with PMA and PA, were used to label HDF cells. The absorbance spectrum of AuNR, after functionalization with 2NAT and surface coating with PMA and PA, shows a main peak at 780 nm (Figure S10A). Incubation of HDF cells with AuNR-2NAT and AuNS-MBT resulted in SERS tag endocytosis, as verified using both TEM and SERS microscopy (Figure S11). Therefore, we monitored HDF cell growth with time (Figure S12) and obtained a 3D reconstruction, which confirmed that two different NP morphologies with two different Raman molecules can indeed be used for cell imaging in 3D (Figure 8). Apart from the additional flexibility regarding the synthesis of SERS tags using NRs and NSs, the use of NPs with significantly different morphologies may additionally provide a method to unmistakably distinguish them in TEM.
Figure 8.
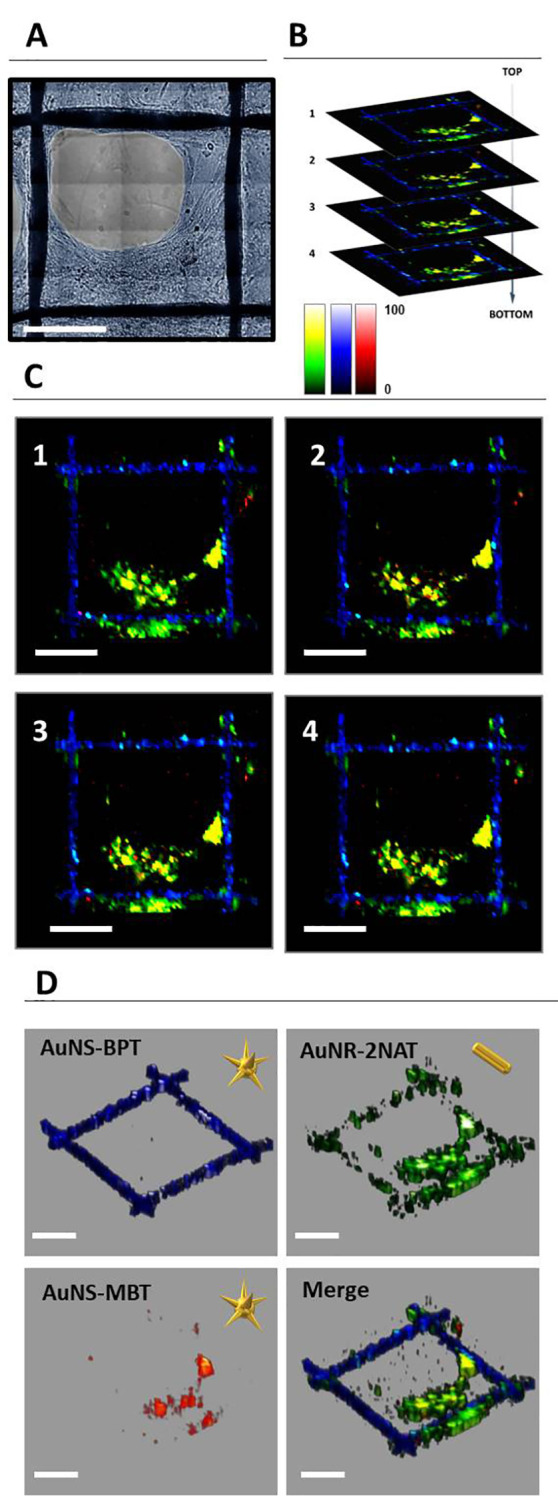
(A) Optical image of the area selected for full 3D SERS mapping. (B, C) Four different layers from different z-stack measurements, on which pixels containing AuNS-BPT (blue), AuNR-2NAT (green), and AuNS-MBT (red) were highlighted. AuNS-BPT corresponds to signal from the scaffold, whereas AuNR-2NAT and AuNS-MBT corresponds to a signal from HDF cells. (D) Individual and merged 3D reconstructions of (C). A 50× (NA 0.5) long working distance objective was used with a 1s integration time, at 29.55 mW laser power, and a step size of 8 × 8 × 20 μm3 (XYZ). Scale bars: 200 μm.
3. Conclusions
By use of the complementary confocal imaging techniques SERS and fluorescence microscopy, we have been able to follow the proliferation and migration of human fibroblasts in 3D and to determine appropriate imaging parameters to better characterize such systems in live conditions (i.e., not chemically fixed). We fabricated hybrid scaffolds, labeled with SERS tags and/or fluorophores, that allow the proliferation of fibroblasts in 3D, reaching thicknesses of ca. 100 μm in a suspended matrix. Fibroblasts were selected because they can be easily labeled with SERS tags and form an interwoven 3D film whose growth can be supported by the multimodal labeled scaffold, which in turn provides a reference framework for imaging. As expected, prelabeling of fibroblasts with SERS tags allowed us to follow their growth in 2D with relative ease, achieving a relatively high level of resolution along the XY dimension. However, distinguishing cells with sufficient resolution along the Z-axis required optimization of the imaging system, mainly increasing the laser excitation power and slowing down the integration time. By using a high-resolution Raman confocal microscope, we improved the cell imaging resolution along the Z-axis to 5 μm, thereby obtaining a more realistic view of the true cell density within the scaffold. In addition, our results showed that continuous irradiation during SERS imaging had little-to-no effect on the acquired signal, whereas similar experiments using fluorescence confocal microscopy caused significant photobleaching. This is especially important if we consider the long irradiation times (in the scale of many hours for large samples) required to achieve high resolution 3D SERS maps. For the data analysis, we took advantage of the True Component Analysis tool, a postprocessing function provided in the Witec Raman microscope software, or MLRA38 analysis when using our Renishaw Raman microscope. Both methods are suitable for dealing with large data sets with multiple components, allowing their separation in a fast and convenient way. This is particularly important when large 3D areas are to be scanned and when working with multiple SERS tags. While in this study we only labeled a single cell type (HDF), but with three SERS tags, the lessons learned are certainly applicable to imaging of multiple cell types, with even more SERS tags. We have previously shown that the 2D imaging of up to five different cell types, labeled with five different SERS tags, was possible, albeit with poor resolution.34 We were able to perform the current work with an upgraded Raman microscope with confocal imaging capability, thus significantly improving the spatial resolution. We propose that all these improvements contribute to the characterization of live cellular 3D structures and will help establish new protocols to understand cell behavior in real time.
4. Experimental Section
Chemicals
Tetrachloroauric acid trihydrate (HAuCl4·3H2O, ≥ 99%), citric acid (≥99.5%), sodium borohydride (NaBH4, 99%), l-ascorbic acid (≥99%), silver nitrate (AgNO3, ≥ 99%), hexadecyltrimethylammonium bromide (CTAB, ≥ 99%), O-[2-(3-mercaptopropionylamino)ethyl]-O′-methylpolyethylene glycol (PEG, MW 5000 g mol–1), 2-naphthalenethiol (2NAT, 99%), 4-methylbenzenethiol (MBT, 98%), biphenyl-4-thiol (BPT, 97%), poly-l-arginine hydrochloride (PA, Aldrich no. 26982-20-7 > 70 000 Da), poly(isobutylene-alt-maleic anhydride) (average MW ∼ 6000 g mol–1), dodecylamine (98%),1-decanol, tetrahydrofuran (THF, 99.85%, extra dry), chloroform (CHCl3, ≥ 99.8%), and sodium hydroxide (NaOH, > 97%) were purchased from Sigma-Aldrich. Hydrochloric acid solution (37 wt%) was purchased from Fisher Chemical. All chemicals were used without further purification. Milli-Q water (resistivity 18.2 MΩ cm at 25 °C) was used in all experiments. All glassware was washed with aqua regia, rinsed with Milli-Q water, and dried prior to use.
AuNP Synthesis
AuNSs were prepared following a seed-mediated growth method.40 The seed solution was prepared by adding 5 mL of 34 mM citrate solution to 95 mL of boiling 0.5 mM HAuCl4 solution under vigorous stirring. After 15 min of boiling, the solution was cooled down and stored at 4 °C. For the synthesis of 70 nm AuNSs with LSPR maximum at 780 nm, 2.5 mL of the citrate-stabilized seed solution was added to 50 mL of 0.25 mM HAuCl4 solution (with 50 μL of 1 M HCl) in a 100 mL glass vial at room temperature under moderate stirring. Quickly, 500 μL of 2 mM AgNO3 and 250 μL of 100 mM ascorbic acid were added simultaneously. The solution rapidly turns from light red to greenish, indicating AuNSs formation. The resulting solution was mixed with 410 μL of 0.1 mM PEG-SH and stirred for 15 min, washed by centrifugation at 1190g, for 25 min, at 10 °C, and redispersed in water. AuNRs with LSPR maximum at 750 nm were prepared by a seeded-growth method. In brief, 1–2 nm gold seeds were grown in the presence of CTAB to form small anisotropic seeds (21 nm length, 8 nm width). These seeds provided the base for the synthesis of larger NRs with LSPR maximum at 750 nm. The exact protocol has been reported by González-Rubio et al.45
AuNP RaR Labeling
Both AuNSs and AuNRs were labeled with Raman active molecules following a previously developed method,34 including wrapping with an amphiphilic polymer to make them biocompatible and coating with poly-l-arginine hydrochloride to make them positively charged and thereby enhance cell uptake, as previously reported.30
NP Characterization
TEM images were collected with a JEOL JEM-1400PLUS transmission electron microscope operating at 120 kV, using carbon-coated 400 square mesh copper grids. UV–vis optical extinction spectra were recorded using an Agilent 8453 UV–vis diode array spectrophotometer.
Tessellated Scaffold Fabrication
Using a 50–75 kDaA PLGA (no. 430471 Aldrich) and a solvent 97:3 volume ratio of CHCl3 and DMF, 10 μm diameter fibers which can be printed forming 3D scaffolds can be obtained. Briefly, a chloroform dispersion of AuNS-BPT was subsequently mixed with 50–75 kDaA PLGA at a 30 wt/vol % solution, also in chloroform. Different fluorescently labeled polymers dissolved in CHCl3 were also included in the mixture to allow the fluorescence microscopy of the scaffolds. Specifically, poly[(mphenylenevinylene)-alt-(2,5-dihexyloxy-p-phenylenevinylene] with excitation/emission wavelengths at λex: 404 nm/ λem: 451 nm, resulting in blue fluorescence; or poly[tris(2,5-bis(hexyloxy)-1,4-phenylenevinylene)-alt-(1,3-phenylenevinylene)] with excitation/emission wavelengths at λex: 448 nm; λem: 518 nm, resulting in green fluorescence were used. Then the corresponding DMF amount was added to the solution. The mentioned final solvent mixture correspond to ca. 93:7 CHCl3:DMF volume ratio.16 The exact composition mixtures are detailed below:
Composition of monocompartmental scaffolds: 0.15 g of PLGA in 55.45 μL of CHCl3 was mixed with 54.55 μL of ((poly[(mphenylenevinylene)-alt-(2,5-dihexyloxy-p-phenylenevinylene) blue polymer (1 mg mL–1 diluted in CHCl3) and 350 μL of SERS tag (AuNS-BPT) ([Au0] = 3 × 10–3 M) in CHCl3 and 35 μL of DMF.
Composition of bicompartmental scaffolds: In the compartment (I) 0.15 g of PLGA in 55.45 μL of CHCl3 was mixed with 54.55 μL of blue polymer (1 mg mL–1 diluted in CHCl3) and 350 μL of SERS tag (AuNS-4BPT) ([Au0] = 3 × 10–3 M) in CHCl3 and 35 μL of DMF. Compartment (II) was prepared by mixing 0.15 g of PLGA in 55.45 μL of CHCl3 with 54.55 μL of green polymer (1 mg mL–1 diluted in CHCl3) and 350 μL of SERS tag (AuNS-4BPT) ([Au0] = 3 × 10–3 M) in CHCl3 and 35 μL of DMF.
Then for the fabrication of the scaffolds, the solution mixtures with or without AuNS-BPT and the two different fluorophores (one for each compartment in the case of bicompartmental scaffolds) were loaded in individual syringes and flown in a laminar regime through a 20-gauge needle for monocompartmental fiber fabrication or parallel mounted metallic 20-gauge needles for bicompartmental fiber ones, at 40 μL h–1, with an applied voltage of 15 kV. As the fluid jet descends to the ground electrode, it passes through a copper ring at 10 kV. The jetted fibers was collected on the ground electrode, a stainless-steel plate, which is translated through X–Y coordinates by computer-controlled motions to stack the depositing fiber onto itself in a desired pattern.
Confocal Imaging of PLGA Scaffolds
NP-loaded scaffolds were sandwiched between two Quartz glass coverslips with the use of mounting media (Dako). This ensured that the scaffold did not move or dry out. All confocal images were taken using a Zeiss LSM 880 confocal laser scanning microscope equipped with 405 nm (blue fluorophore excitation) and 488 nm (green fluorophore excitation), and Plan-Apochromat 10× objective (0.45 N.A.) and Plan-Apochromat 20× objective (0.8 N.A.) objectives. A postimaging 3-pixel median filter was generally applied to remove noise pixels. In the case of 3D characterization of scaffolds, Z-stacks of approximately 70 μm in thickness were obtained and postimaging 3-pixel median filter applied prior to 3D rendering, to obtain images from different angles. Finally, Z-stacks of higher resolution were obtained by imaging scaffolds composed solely of blue fluorophore, again with the same 20× objective but with increased pixel resolution and Z-depth. In this case, we obtained Z-stacks of approximately 200 μm in thickness with XYZ pixel resolution of 0.3 × 0.3 × 0.5 μm3.
NP-Incubation with Cells for Growth on PLGA Scaffolds
Human dermal fibroblasts (HDF) were purchased from Invitrogen and grown in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (PS) (herein termed complete DMEM, cDMEM). To expose cells to NPs for uptake, HDF were seeded in 24 plates (3 × 104 cells per well) and allowed to adhere before replacing media with a solution of NPs diluted in cDMEM. NPs were added at a final concentration of 0.1 mM. After 24 h, nonuptaken NPs were removed by washing the adhered HDF cell monolayer, and the adhered cells were detached using trypsin. After recounting, cells were added to the scaffold as described below.
Quantification of SERS Tag Uptake and Viability
SERS tag uptake by HDF cells was characterized via ICP-MS and via two-photon confocal microscopy. For inductively coupled plasma-mass spectrometry (ICP-MS) measurements, trypsinized HDF cells were digested using aqua regia and analyzed using standard protocols. For two-photon microscopy and PI staining, GFP-expressing HDF cells were seeded in optically transparent 96-well plates (Ibidi) and exposed to SERS tags for 24 h. Samples were washed to remove nonendocytosed NPs and imaged using two-photon confocal microscopy with a 780 nm laser excitation with a BP emission filter of 570–610 nm (880 Zeiss Confocal microscope). A three-pixel mean filter was applied for postprocessing. For PI staining, cells were stained with 5 μg/mL PI for 20 min, followed by detection of PI expressing cells (dead) using a Cell Observer Zeiss Microscope.
Scaffold Preparation for In Vitro Studies
A modified previously reported protocol16 for preparation of NP-loaded scaffolds was used for imaging experiments. Scaffolds were sandwiched between two metallic windows (0.5 × 1 cm2) and placed in a 2 mL sterile Eppendorf tube. 50 μL of human fibronectin (50 μg) was placed on top of the scaffold which lay suspended in the center of the Eppendorf tube. The fibronectin was allowed to adhere to the scaffold fibers at 37 °C for 30 min. HDF cells, previously incubated with NPs as described above, were washed, trypsinized, counted, and readjusted to a concentration of 4–8 × 105 cells mL–1. A concentrated drop of SERS tag-incubated cells (100 μL) was then added to the scaffold and allowed to incubate for approximately 30 min before transferring the scaffold to a 24-well plate and adding cDMEM. Either mixed SERS tag-cell populations or single SERS tag populations were used. The scaffolds were stored for approximately 2 weeks to allow cells to form a 3D mesh around the scaffold. cDMEM was carefully replaced every 2–3 days without disturbing the scaffold.
Multimodal Cell Imaging
For live SERS imaging of the scaffold, the metal frames were picked up with tweezers, and the whole component (scaffold and frames) was transferred to a quartz slide with an adhered in-house 3D printed well,30 which allowed long-term cell growth and insertion of an immersion objective without disturbing the cells. The metal holder lays flat on the base allowing upright and inverted imaging. For live fluorescence imaging, HDF cells were transfected to express eGFP using a multiplicity of infection (MOI) of 10 and hygromycin for selection, allowing imaging using 488 nm excitation. For fixed cell imaging, cell-containing scaffolds were fixed in situ with the metal holder in the 24-well plate. To do so, a 4% solution of formaldehyde was used at RT for 20 min, followed by washing with PBS. Immunocytochemical staining using antifibronectin (F3648 clone) with AF633 labeled antirabbit secondary antibody was conducted to image fibronectin.
SERS Imaging
For high resolution SERS imaging of AuNS-1NAT, AuNS-2NAT, and AuNS-MBT NPs incubated with HDF cells, measurements were performed with a confocal Raman microscope (Alpha300 R – Confocal Raman Imaging Microscope, Witec) equipped with 1024 × 512 CCD detectors. A 785 nm laser excitation source (maximum output 79 mW) and a 300 l/mm diffraction grating were used. Measurements were recorded in static mode (center of scattered wavenumber 1450 cm–1) using a 20× immersion long working distance objective (numerical aperture, NA = 0.5; Zeiss, Jena, Germany). The integration time and laser power varied between 0.02–0.05 s and 5–30 mW, respectively. Exact values are given in the legend of each figure. SERS data were analyzed using True Component Analysis provided by the Witec microscope software. This is a nonopen access script which is described by Witec as “The unique post-processing function for confocal Raman imaging measurements automatically establishes the number of components in a sample, locates them in the image, and differentiates their individual spectra. It delivers meaningful results in a fast and convenient way.”
SERS measurements of HDF cells incubated with AuNR-2NAT and AuNS-MBT SERS tags were performed with a confocal Raman microscope (Renishaw inVia) equipped with 1024 × 512 CCD detectors, using a 785 nm laser excitation source (maximum output 270 mW) and a 1200 l/mm diffraction grating. SERS maps were recorded in static mode (center of scattered wavenumber 1450 cm–1). For overtime imaging, a 40× immersion objective (numerical aperture, NA = 0.8; Nikon, Tokyo, Japan) with 0.8s integration time, at 3.1 mW laser power was used. The map of one selected area (325 × 225 μm) was acquired with a resolution of 5 μm in X and Y, and required approximately 1 h and 10 min to be completed. For 3D imaging, a 50× long distance objective (N.A. = 0.5; Leica Microsystems, Wetzlar, Germany), with 1s integration time, at 29.55 mW laser power was used. The map of one selected area (584 × 584 × 60 μm) was acquired with a resolution of 8 μm in X and Y, and 20 μm in Z, and the total four layers required approximately 2 h and 30 min to be completed. SERS data were analyzed using the MLRA method, as described in previous work.
SERS reference spectra were collected from a 5 μL drop of the SERS tags ([Au]0 = 0.5 mM) on top of a quartz slide. We used a 50× long working distance objective (NA = 0.5; Leica Microsystems, Wetzlar, Germany) in expanded scan mode with an integration time of 10 s, at a laser power of 1.11 mW and five accumulative scans. SERS data were analyzed using Multiple Linear Regression Analysis (MLRA) (regress function of Matlab).38
SERS Bleaching Test Measurements
Maps of a fixed area–different from that tested by fluorescence–were acquired four times with a resolution of 5 μm in X and Y using a 50× long distance objective (N.A. = 0.5; Leica Microsystems, Wetzlar, Germany), a full-1200 lines mm–1 diffraction grating and a 785 nm HeNe laser. Each point was exposed to 1.11 mW of laser power for 1s. Each map has a dimension of 250 × 250 μm and took approximately 1 h to complete. Reference spectra: Raman measurements of colloidal SERS tags in solution were performed with a 50× long distance objective (N.A. = 0.5; Leica Microsystems, Wetzlar, Germany), a full-1200 lines mm–1 diffraction grating and a 785 nm HeNe laser. The volume of the sample was exposed for 10 s in total during scanning; the PLGA Raman spectrum was obtained by measuring a scaffold without SERS tags, with a 50× long distance objective (N.A. = 0.5; Leica Microsystems, Wetzlar, Germany), a full-1200 l/mm diffraction grating and a 785 nm HeNe laser. SERS data were analyzed using the MLRA method, as described in previous work.38 The average spectra are represented in Figure 7.
Fluorescence Scaffold Bleaching Experiments
To determine the stability of fluorescence signal in the scaffolds after repetitive confocal imaging, we devised an experimental setup in which the scaffold and cells were irradiated with a defined laser wavelength and the mean intensity of scaffold, cellular, and background regions were analyzed using regions of interest (ROI). Importantly, we did not change the laser power or pinhole for bleaching and imaging, as we were interested in seeing the effect that repetitive imaging has on the fluorescence properties. In detail, HDF cells were grown on scaffolds containing blue fluorophore, using the method described above to seed cells. Cells were fixed with formaldehyde and then stained with Actin 488 ReadyProbes fluorophore (Invitrogen). The sample was sandwiched between two quartz coverslips using mounting media to provide physical and chemical stability. To conduct the experiment, the sample was alternatively irradiated with a 405 nm laser for 60 s, and then imaged using the same 405 nm laser and a 488 nm laser, thereby exciting the fluorophores in the scaffold and in the cells, respectively. A Plan-Apochromat 20× objective (0.8 N.A.) was used throughout, and the pinhole was set to 24 μm for both bleaching and imaging. Images were transferred to ImageJ and, by working with the blue and the green channels separately, ROIs were drawn to represent the scaffold (blue channel), the cells (green channel), and their corresponding backgrounds. In the case of the ROIs representing signal from the blue channel, they were 10 × 10 μm2. In the case of the ROIs representing signal from the green channel, they measured 200 × 100 μm, to capture a greener signal (which comes from the actin cytoskeleton and is therefore sparser). Using ImageJ, the Raw Integrated Density of each ROI was calculated (that is, the sum of the pixel intensities in that ROI), and the average of each area was plotted. Images for visual understanding were postprocessed with a three-pixel median filter for clarity.
Acknowledgments
Dr. Beatriz Molina is thanked for performing the transfection of HDF cells with eGFP. Dr. Judith Langer is thanked for help with 3D SERS imaging.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsami.2c02615.
Additional TEM images, UV–vis and SERS spectra of SERS tags; SEM and fluorescence microscopy images of scaffolds; cytotoxicity studies of SERS tags in HDF cells at different concentrations; SERS measurements in cells at different laser power intensities; evolution of HDF cell growth and scaffold structure over 14 days; SERS imaging of labeled cells in 2D; 3D distribution of SERS tags in different scaffold areas; TEM images and SERS mappings of endocytosed SERS tags in HDF cells (PDF)
Author Contributions
∥ D.J.d.A. and M.H.-L. contributed equally to this work. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
This work has been funded by the European Research Council (ERC Advanced Grant 787510, 4DbioSERS), the Spanish State Research Agency (Project PID2019-108854RA-I00 and MDM-2017-0720) and the National Science Foundation (Grant No. EEC-1647837 and Grant No. DGE 1256260). P. Piñeiro acknowledges an FPU fellowship (PRE2020-094237) from the Spanish Ministry of Universities.
The authors declare no competing financial interest.
Supplementary Material
References
- Alhaque S.; Themis M.; Rashidi H. Three-Dimensional Cell Culture: From Evolution to Revolution. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170216. 10.1098/rstb.2017.0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derakhshanfar S.; Mbeleck R.; Xu K.; Zhang X.; Zhong W.; Xing M. 3D Bioprinting for Biomedical Devices and Tissue Engineering: A Review of Recent Trends and Advances. Bioactive Mater. 2018, 3, 144–156. 10.1016/j.bioactmat.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skylar-Scott M. A.; Mueller J.; Visser C. W.; Lewis J. A. Voxelated Soft Matter via Multimaterial Multinozzle 3D Printing. Nature 2019, 575, 330–335. 10.1038/s41586-019-1736-8. [DOI] [PubMed] [Google Scholar]
- Zhang Q.; Lee Y. H.; Phang I. Y.; Lee C. K.; Ling X. Y. Hierarchical 3D SERS Substrates Fabricated by Integrating Photolithographic Microstructures and Self-Assembly of Silver Nanoparticles. Small 2014, 10, 2703–2711. 10.1002/smll.201303773. [DOI] [PubMed] [Google Scholar]
- Wang X.; Zhang M.; Ma J.; Xu M.; Chang J.; Gelinsky M.; Wu C. 3D Printing of Cell-Container-Like Scaffolds for Multicell Tissue Engineering. Engineering 2020, 6, 1276–1284. 10.1016/j.eng.2020.08.001. [DOI] [Google Scholar]
- Zhou D.; Chen L.; Ding J.; Zhang X.; Nie Z.; Li X.; Yang B.; Xu T. A 3D Engineered Scaffold for Hematopoietic Progenitor/Stem Cell Co-Culture in Vitro. Sci. Rep. 2020, 10, 11485. 10.1038/s41598-020-68250-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova M. P.; Chavali M. S. Recent Advances in Biomaterials for 3D Scaffolds: A Review. Bioact. Mater. 2019, 4, 271–292. 10.1016/j.bioactmat.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetzke R.; Keijdener H.; Franzen J.; Ostrowska A.; Nüchtern S.; Mela P.; Wagner W. Differentiation of Induced Pluripotent Stem Cells towards Mesenchymal Stromal Cells Is Hampered by Culture in 3D Hydrogels. Sci. Rep. 2019, 9, 1–12. 10.1038/s41598-019-51911-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf B. W.; Boppart S. A. Imaging and Analysis of Three-Dimensional Cell Culture Models. Methods Mol. Biol. 2010, 591, 211. 10.1007/978-1-60761-404-3_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Astrain C.; Lenzi E.; Jimenez de Aberasturi D.; Henriksen-Lacey M.; Binelli M. R.; Liz-Marzán L. M. 3D-Printed Biocompatible Scaffolds with Built-In Nanoplasmonic Sensors. Adv. Funct. Mater. 2020, 30, 2005407. 10.1002/adfm.202005407. [DOI] [Google Scholar]
- Wang X.; Jiang M.; Zhou Z.; Gou J.; Hui D. 3D Printing of Polymer Matrix Composites: A Review and Prospective. Composites Part B: Engineering 2017, 110, 442–458. 10.1016/j.compositesb.2016.11.034. [DOI] [Google Scholar]
- Tamay D. G.; Usal T. D.; Alagoz A. S.; Yucel D.; Hasirci N.; Hasirci V. 3D and 4D Printing of Polymers for Tissue Engineering Applications. Front. Bioeng. Biotechnol. 2019, 7, 164. 10.3389/fbioe.2019.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman B. 3-D Printing: The New Industrial Revolution. Business Horiz. 2012, 55, 155–162. 10.1016/j.bushor.2011.11.003. [DOI] [Google Scholar]
- Murphy S. V.; Atala A. 3D Bioprinting of Tissues and Organs. Nat. Biotechnol. 2014, 32, 773–785. 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- Jordahl J. H.; Solorio L.; Sun H.; Ramcharan S.; Teeple C. B.; Haley H. R.; Lee K. J.; Eyster T. W.; Luker G. D.; Krebsbach P. H.; Lahann J. 3D Jet Writing: Functional Microtissues Based on Tessellated Scaffold Architectures. Adv. Mater. 2018, 30, 1707196. 10.1002/adma.201707196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordahl S.; Solorio L.; Neale D. B.; McDermott S.; Jordahl J. H.; Fox A.; Dunlay C.; Xiao A.; Brown M.; Wicha M.; Luker G. D.; Lahann J. Engineered Fibrillar Fibronectin Networks as Three-Dimensional Tissue Scaffolds. Adv. Mater. 2019, 31, 1904580. 10.1002/adma.201904580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steier A.; Schmieg B.; Irtel von Brenndorff Y.; Meier M.; Nirschl H.; Franzreb M.; Lahann J. Enzyme Scaffolds with Hierarchically Defined Properties via 3D Jet Writing. Macromol. Biosci. 2020, 20, 2000154. 10.1002/mabi.202000154. [DOI] [PubMed] [Google Scholar]
- Moon S.; Jones M. S.; Seo E.; Lee J.; Lahann L.; Jordahl J. H.; Lee K. J.; Lahann J. 3D Jet Writing of Mechanically Actuated Tandem Scaffolds. Sci. Adv. 2021, 7, 5289–5303. 10.1126/sciadv.abf5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strozyk M. S.; Jimenez de Aberasturi D.; Gregory J. V.; Brust M.; Lahann J.; Liz-Marzán L. M. Spatial Analysis of Metal-PLGA Hybrid Microstructures Using 3D SERS Imaging. Adv. Funct. Mater. 2017, 27, 1701626. 10.1002/adfm.201701626. [DOI] [Google Scholar]
- Gregory J. V.; Vogus D. R.; Barajas A.; Cadena M. A.; Mitragotri S.; Lahann J. Programmable Delivery of Synergistic Cancer Drug Combinations Using Bicompartmental Nanoparticles. Adv. Healthc. Mater. 2020, 9, 2000564. 10.1002/adhm.202000564. [DOI] [PubMed] [Google Scholar]
- Strozyk M. S.; Jimenez de Aberasturi D.; Gregory J. V.; Brust M.; Lahann J.; Liz-Marzán L. M. Spatial Analysis of Metal-PLGA Hybrid Microstructures Using 3D SERS Imaging. Adv. Funct. Mater. 2017, 27, 1701626. 10.1002/adfm.201701626. [DOI] [Google Scholar]
- Riss T.; Trask O. J. Factors to Consider When Interrogating 3D Culture Models with Plate Readers or Automated Microscopes. In Vitro Cell. Developm. Biol. Animal 2021, 57, 238–256. 10.1007/s11626-020-00537-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allam M.; Cai S.; Coskun A. F. Multiplex Bioimaging of Single-Cell Spatial Profiles for Precision Cancer Diagnostics and Therapeutics. NPJ. Precis. Oncol. 2020, 4, 11. 10.1038/s41698-020-0114-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J.-W.; Son S. U.; Lim E.-K.. Recent Advances in Bioimaging for Cancer Research. In: State Art Nano-bioimaging 2018. 10.5772/intechopen.72725. [DOI] [Google Scholar]
- Charwat V.; Schütze K.; Holnthoner W.; Lavrentieva A.; Gangnus R.; Hofbauer P.; Hoffmann C.; Angres B.; Kasper C. Potential and Limitations of Microscopy and Raman Spectroscopy for Live-Cell Analysis of 3D Cell Cultures. J. Biotechnol. 2015, 205, 70–81. 10.1016/j.jbiotec.2015.02.007. [DOI] [PubMed] [Google Scholar]
- Placone J. K.; Mahadik B.; Fisher J. P. Addressing Present Pitfalls in 3D Printing for Tissue Engineering to Enhance Future Potential. APL Bioeng. 2020, 4, 010901. 10.1063/1.5127860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prina-Mello A.; Bonacina L.; Staedler D.; Movia D. Editorial: Use of 3D Models in Drug Development and Precision Medicine - Advances and Outlook. Front. Bioeng. Biotechnol. 2021, 9, 658941. 10.3389/fbioe.2021.658941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabris L. Gold-Based SERS Tags for Biomedical Imaging. J. Opt. 2015, 17, 114002. 10.1088/2040-8978/17/11/114002. [DOI] [Google Scholar]
- Wang Y.; Yan B.; Chen L. SERS Tags: Novel Optical Nanoprobes for Bioanalysis. Chem. Rev. 2013, 113, 1391–1428. 10.1021/cr300120g. [DOI] [PubMed] [Google Scholar]
- Jimenez de Aberasturi D.; Henriksen-Lacey M.; Litti L.; Langer J.; Liz-Marzán L. M. Using SERS Tags to Image the Three-Dimensional Structure of Complex Cell Models. Adv. Funct. Mater. 2020, 30, 1909655. 10.1002/adfm.201909655. [DOI] [Google Scholar]
- Lenzi E.; Jimenez de Aberasturi D.; Liz-Marzán L. M. Surface-Enhanced Raman Scattering Tags for Three-Dimensional Bioimaging and Biomarker Detection. ACS Sens. 2019, 4, 1126–1137. 10.1021/acssensors.9b00321. [DOI] [PubMed] [Google Scholar]
- Zhuo X.; Henriksen-Lacey M.; Jimenez De Aberasturi D.; Sánchez-Iglesias A.; Liz-Marzán L. M. Shielded Silver Nanorods for Bioapplications. Chem. Mater. 2020, 32, 5879–5889. 10.1021/acs.chemmater.0c01995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Jimenez de Aberasturi D.; Henriksen-Lacey M.; Langer J.; Liz-Marzán L. M. Live-Cell Surface-Enhanced Raman Spectroscopy Imaging of Intracellular PH: From Two Dimensions to Three Dimensions. ACS Sens. 2020, 5, 3194–3206. 10.1021/acssensors.0c01487. [DOI] [PubMed] [Google Scholar]
- Jimenez De Aberasturi D.; Serrano-Montes A. B.; Langer J.; Henriksen-Lacey M.; Parak W. J.; Liz-Marzán L. M. Surface Enhanced Raman Scattering Encoded Gold Nanostars for Multiplexed Cell Discrimination. Chem. Mater. 2016, 28, 6779–6790. 10.1021/acs.chemmater.6b03349. [DOI] [Google Scholar]
- Langer J.; Jimenez de Aberasturi D.; Aizpurua J.; Alvarez-Puebla R. A.; Auguié B.; Baumberg J. J.; Bazan G. C.; Bell S. E.; Boisen A.; Brolo A.; Choo J.; Cialla-May D.; Deckert V.; Fabris L.; Faulds K.; García de Abajo F. J.; Goodacre R.; Graham D.; Haes A. J.; Haynes C. L.; Huck C.; Itoh T.; Käll M.; Kneipp J.; Kotov N. A.; Kuang H.; Le Ru E. C.; Lee H. K.; Li J.-F.; Ling X. Y.; Maier S. A.; Mayerhöfer T.; Moskovits M.; Murakoshi K.; Nam J.-M.; Nie S.; Ozaki Y.; Pastoriza-Santos I.; Pérez-Juste J.; Popp J.; Pucci A.; Reich S.; Ren B.; Schatz G. C.; Shegai T.; Schlücker S.; Tay L.-L.; Thomas K. G.; Tian Z.-Q.; Van Duyne R. P.; Vo-Dinh T.; Wang Y.; Willets K. A.; Xu C.; Xu H. X.; Xu Y.; Yamamoto Y. S.; Zhao B.; Liz-Marzán L. M. Present and Future of Surface Enhanced Raman Scattering. ACS Nano 2020, 14, 28–117. 10.1021/acsnano.9b04224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. L.; SoRelle E. D.; Ilovich O.; Liba O.; James M. L.; Qiu Z.; Perez V.; Chan C. T.; de la Zerda A.; Zavaleta C. Multimodal Assessment of SERS Nanoparticle Biodistribution Post Ingestion Reveals New Potential for Clinical Translation of Raman Imaging. Biomaterials 2017, 135, 42–52. 10.1016/j.biomaterials.2017.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter A.; Paul-Gilloteaux P.; Plochberger B.; Sefc L.; Verkade P.; Mannheim J. G.; Slezak P.; Unterhuber A.; Marchetti-Deschmann M.; Ogris M.; Bühler K.; Fixler D.; Geyer S. H.; Weninger W. J.; Glösmann M.; Handschuh S.; Wanek T. Correlated Multimodal Imaging in Life Sciences: Expanding the Biomedical Horizon. Front. Phys. 2020, 8, 47. 10.3389/fphy.2020.00047. [DOI] [Google Scholar]
- Lenzi E.; Litti L.; Jimenez de Aberasturi D.; Henriksen-Lacey M.; Liz-Marzán L. M. SERSTEM: An App for the Statistical Analysis of Correlative SERS and TEM Imaging and Evaluation of SERS Tags Performance. J. Raman Spectrosc. 2021, 52, 355–365. 10.1002/jrs.6043. [DOI] [Google Scholar]
- Guerrero-Martínez A.; Barbosa S.; Pastoriza-Santos I.; Liz-Marzán L. M. Nanostars Shine Bright for You: Colloidal Synthesis, Properties and Applications of Branched Metallic Nanoparticles. Curr. Opin. Colloid Interface Sci. 2011, 16, 118–127. 10.1016/j.cocis.2010.12.007. [DOI] [Google Scholar]
- Yuan H.; Khoury C. G.; Hwang H.; Wilson C. M.; Grant G. A.; Vo-Dinh T. Gold Nanostars: Surfactant-Free Synthesis, 3D Modelling, and Two-Photon Photoluminescence Imaging. Nanotechnology 2012, 23, 075102. 10.1088/0957-4484/23/7/075102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Montes A. B.; Jimenez de Aberasturi D.; Langer J.; Giner-Casares J. J.; Scarabelli L.; Herrero A.; Liz-Marzán L. M. A General Method for Solvent Exchange of Plasmonic Nanoparticles and Self-Assembly into SERS-Active Monolayers. Langmuir 2015, 31, 9205–9213. 10.1021/acs.langmuir.5b01838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Yuan H.; Kersey F.; Register J.; Parrott M.; Vo-Dinh T. Plasmonic Gold Nanostars for Multi-Modality Sensing and Diagnostics. Sensors 2015, 15, 3706–3720. 10.3390/s150203706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.; Huang J.; Yang X.; Liu B.; Zhang W.; Huang L.; Deng F.; Ma J.; Bai Y.; Lu R.; Huang B.; Gao Q.; Zhuo Y.; Ge J. Serum Starvation Induced Cell Cycle Synchronization Facilitates Human Somatic Cells Reprogramming. PLoS One 2012, 7, e28203 10.1371/journal.pone.0028203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbora A.; Bohar O.; Sivan A. A.; Magory E.; Nause A.; Minnes R. Higher Pulse Frequency of Near-Infrared Laser Irradiation Increases Penetration Depth for Novel Biomedical Applications. PLoS One 2021, 16, e0245350 10.1371/journal.pone.0245350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Rubio G.; Kumar V.; Llombart P.; Díaz-Núñez P.; Bladt E.; Altantzis T.; Bals S.; Peña-Rodríguez O.; Noya E. G.; MacDowell L. G.; Guerrero-Martínez A.; Liz-Marzán L. M. Disconnecting Symmetry Breaking from Seeded Growth for the Reproducible Synthesis of High Quality Gold Nanorods. ACS Nano 2019, 13, 4424–4435. 10.1021/acsnano.8b09658. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.