Abstract
Recognition of viruses as the most abundant component of aquatic microbial communities has stimulated investigations of the impact of viruses on bacterio- and phytoplankton host communities. From results of field studies to date, it is concluded that in most aquatic environments, a reduction in the number of bacteria on a daily basis is caused by viral infection. However, the modest amount of in situ virus-mediated mortality may be less significant than viral infection serving to maintain clonal diversity in the host communities directly, through gene transmission (i.e., transduction), and indirectly, by elimination of numerically dominant host species. If the latter mechanism for controlling community diversity prevails, then the overall structure of aquatic viral communities would be expected to change as well over short seasonal and spatial scales. To determine whether this occurs, pulsed-field gel electrophoresis (PFGE) was used to monitor the population dynamics of Chesapeake Bay virioplankton for an annual cycle (1 year). Virioplankton in water samples collected at six stations along a transect running the length of the bay were concentrated 100-fold by ultrafiltration. Viruses were further concentrated by ultracentrifugation, and the concentrated samples were embedded in agarose. PFGE analysis of virus DNA in the agarose plugs yielded several distinct bands, ranging from 50 to 300 kb. Principal-component and cluster analyses of the virus PFGE fingerprints indicated that changes in virioplankton community structure were correlated with time, geographical location, and extent of water column stratification. From the results of this study, it is concluded that, based on the dynamic nature of the Chesapeake Bay virioplankton community structure, the clonal diversity of bacterio- and phytoplankton host communities is an important component of the virus community.
Interactions among planktonic organisms may be critical to maintaining homeostatic conditions. Furthermore, these interactions may influence the flow of nutrients and energy through aquatic ecosystems. In ecological studies, planktonic organisms, for practical purposes, are divided into groups based on size, trophism, and/or taxon. Until recently, the bacteria were considered to be numerically the most abundant component of the total plankton community. However, during the past decade, a number of studies have shown that the virioplankton are numerically the most abundant component of the plankton. Direct counting methods have been employed, with results showing that within the water column, viruses frequently outnumber potential hosts by approximately 10-fold (3, 6, 8, 29, 37, 46, 50). The abundance of viruses in natural waters is now generally accepted, but it is not yet clear whether viruses and virus infection are fundamental to planktonic community function. In the years following discovery of large numbers of viruses in marine and estuarine waters, field studies have shown virioplankton to be a dynamic component of aquatic microbial communities and ecosystems. Virioplankton abundance appears to be seasonal (20, 50) and related to physiochemical factors associated with depth (12, 35) and trophic gradients (46). In general, the number of viruses in water samples decreases with increasing distance from shore (29) and is generally correlated with trophic conditions in the water column (46). However, in shallow (<50-m) coastal and estuarine water, the number of viruses does not appear to vary with depth (12, 50).
The dynamic nature of virioplankton abundance and the broad geographical distribution of viruses in marine and estuarine water leads to interesting hypotheses, one of which is that, through lysis, viruses directly limit productivity of the host community. Also, viruses are thereby an important mechanism for maintaining clonal and genetic diversity of host populations (18, 41). That is, genetic diversity within host populations occurs by viral elimination of specific numerically dominant hosts and by transduction (32, 45), which are not mutually exclusive. However, the relative importance of each in describing interactions of virioplankton and host plankton communities is subject to debate. Research on the ecology of viruses has focused largely on determining the effect of viruses on numbers of bacteria and phytoplankton in ecosystems. Because it is not possible to measure in situ rates of virus-induced mortality, a variety of indirect approaches have been employed (see references 18, 36, and 48 for a review of the literature). While estimates vary significantly, the early indications are that ca. 10 to 20% of the marine heterotrophic bacterial standing stock is lost per day to viral infection (36). Despite variability in results, the studies demonstrate that a certain percentage of primary and secondary production is lost because of virus infection. Yet, it is difficult to conceptualize viruses as being regulators of bacterial production, mainly because it is known that bacterial hosts quickly acquire resistance to cooccurring viruses (24).
The incidence of phage resistance within a natural bacterial host population was reported by Waterbury and Valois (45). Their results, combined with field data on Synechococcus and cyanophage abundances, led to the conclusion that marine Synechococcus populations and their cooccurring cyanophages have evolved a relationship in which cyanophages “scavenge rare sensitive cells” and exist at concentrations an order of magnitude lower than that of the host populations (45). Therefore, Waterbury and Valois (45) concluded that viral lysis was not significant in controlling host densities and was more likely to influence, and perhaps determine, the clonal composition of Synechococcus populations in Woods Hole harbor.
High cyanophage titers, exceeding those obtained by Waterbury and Valois (45), were recorded by Suttle and Chan (37) in the Gulf of Mexico. In this case, the number of cyanophage frequently exceeded that of the Synechococcus hosts. Nevertheless, the most conservative estimates of cyanophage-induced mortality reported by Suttle and Chan (37) are similar to those of Waterbury and Valois (45). Thus, a slight loss in productivity may be incidental to the more important outcome of viral infection and lysis, namely, virus-mediated control of host community structure. If clonal diversity describes the impact of viral infection on host communities accurately, then virioplankton populations should be unstable in species composition over short temporal and spatial scales.
Morphology data obtained by transmission electron microscopy of naturally occurring viruses suggest that the majority of viruses in estuarine (50), marine (12), and fresh (15) waters are bacteriophages. Despite the lack of direct evidence, this assumption is supported by the fact that bacteria are the most abundant planktonic hosts. Previous studies examining the compositions of aquatic virus populations have documented the morphological types of naturally occurring viruses, from direct counts, and purified phage-host systems isolated from a marine environment. Sediment samples collected from Lake Plußsee included 39 distinct morphotypes (15), with the number of morphotypes in water samples ranging from 2 in samples collected during January 1990 to 10 in samples collected during April 1990. However, differences in the compositions of the phage populations could not be correlated with environmental parameters. Similarly, virus-like particles observed in direct counts have been classified into groups based on approximate capsid diameter (6, 12, 50). The small number of studies on virioplankton morphological diversity does not permit definitive conclusions with respect to distribution of capsid size that could be related to environmental factors.
The morphology of bacteriophages isolated from seawater is significantly diverse (5, 17, 30). On the basis of morphotypes, bacteriophage isolates from the western and eastern north Atlantic Ocean could also be separated into distinct bacteriophage populations (17), supporting the assertion that genetically distinct bacterioplankton populations exist in the eastern and western north Atlantic Ocean. Based on biology and morphology, marine bacteriophages are concluded to be similar to other phages that have been described (27). Unfortunately, morphological diversity of the virioplankton, i.e., the marine phages, cannot be used to assess virus population dynamics (1, 31). Studies of genetic diversity of viruses have demonstrated the utility and analytical power of molecular genetic methods, such as a combination of restriction fragment length polymorphism (RFLP) and hybridization analysis, as was employed to classify 67 Vibrio parahaemolyticus phages isolated from southern Florida and Hawaii seawater samples into six genetic groups (21).
A powerful method for determining microbial diversity is a combination of PCR amplification of a conserved genetic element or gene, principally 16S rRNA genes, and RFLP or sequence analysis of the amplification products. Chen and Suttle (9) used a highly degenerate primer set to amplify selectively viral DNA polymerase (pol) genes from viruses infecting members of three microalga genera. Within water samples from an onshore-to-offshore transect in the Gulf of Mexico, a diverse community of algal viruses was detected and could be divided into five operational taxonomic groups based on RFLP patterns of PCR products. Subsequent phylogenetic analysis established a close relationship between environmental pol amplicons and demonstrated the high diversity of algal viruses (10). Another approach, namely, pulsed-field gel electrophoresis (PFGE), enables size fractionation of intact large DNAs and has proven useful in separating viruses in a natural community of ruminant bacteriophages according to genome size (22), as well as in detecting differences between bacteriophage populations in ruminal fluid samples (39).
The objective of the study reported here was to determine the population dynamics of Chesapeake Bay virioplankton communities by employing PFGE fingerprinting.
MATERIALS AND METHODS
Virioplankton concentration.
Water samples were collected in 10 liter Niskin bottles mounted on an instrument rosette. The samples were collected from the mid-stem of the Chesapeake Bay at six stations (50). Sampling stations traversed the bay, from the Patapsco River to the York River. Station designations and locations are 908 (39°08′N, 76°20′W), 858 (38°58′N, 76°23′W), 845 (38°45′N, 76°26′W), 818 (38°18′N, 76°26′W), 744 (37°44′N, 76°11′W), and 724 (37°24′N, 76°05′W). A total of 23 water samples were collected on four cruises during 21 to 23 August 1995, 9 to 11 May 1996, 31 May to 2 June 1996, and 5 to 9 July 1996.
Sample processing was conducted aboard ship immediately following collection. On the 1996 research cruises, Chesapeake Bay virioplankton were concentrated by using a spiral-cartridge filtration method described by Suttle et al. (38). Removal of bacteria and larger plankton was accomplished by two-stage filtration, using 142-mm-diameter filters mounted in stainless steel filter holders. Each 50-liter water sample, under low vacuum (<300 mm Hg), was first passed through a glass fiber filter (GF-D; nominal pore size, 1.2 μm; Gelman, Ann Arbor, Mich.), collected, and passed through a 0.2-μm-pore-size polycarbonate filter (Poretics, Livermore, Calif.). Viruses in the 50-liter filtrates were concentrated by using the CH 2 system and S1Y30 filter (Amicon, Bedford, Mass.) from 50 liters to 250 ml) (38). Final water sample concentrates contained particulates of between 30,000 Da (approximately 2 nm) and 0.22 μm in size.
On the August 1995 cruise, virioplankton were concentrated from 0.22-μm-pore-size-filtered water samples to a 10× final concentration by using immersible-CX ultrafilters (Millipore, Burlington, Mass.) and vacuum (49). The August 1995 virioplankton concentrates contained particulates of between 10,000 Da (approximately 1 nm) and 0.22 μm in size. Volume measurements were recorded to ensure accurate calculation of the sample concentration.
Preparation of viral concentrates for PFGE.
Thirty milliliters of virioplankton concentrate in a 32-ml ultracentrifuge tube was centrifuged for 3.5 h at 100,000 × g. The supernatant was gently decanted, and the virus pellet was resuspended and incubated overnight at 4°C in 450 μl of SM buffer (0.1 M NaCl, 8 mM MgSO4 · 7H2O, 50 mM Tris-HCl, and 0.005% [wt/vol] glycerol) with gentle shaking. Equal volumes of the viral concentrate and molten (50°C) 1.5% InCert agarose (FMC, Rockland, Maine) were mixed, vortexed, and dispensed into plug molds. After solidification of the gel, plugs were punched out from the molds into a small volume of buffer (250 mM EDTA, 1% sodium dodecyl sulfate) containing 1 mg of proteinase K per ml. The plugs were incubated in the dark at room temperature overnight. The proteinase K digestion buffer was decanted, and the plugs were washed three times for 30 min each in 10 mM Tris–1 mM EDTA, pH 8.0. The virioplankton agarose plugs were stored at 4°C in 20 mM Tris–50 mM EDTA, pH 8.0.
PFGE.
Optimal conditions for electrophoresis were determined empirically. Virioplankton plugs and plugs containing phage lambda concatamers (Promega, Madison, Wis.), serving as molecular size markers, were placed into wells of a 1% SeaKem GTG agarose (FMC) gel with an overlay of molten 1% agarose. PFGE for samples collected in 1996 was performed with the contour-clamped homogeneous electric field DR-II Cell (Bio-Rad, Richmond, Calif.) under the following conditions: 1× TBE gel buffer (90 mM Tris-borate and 1 mM EDTA, pH 8.0), 0.5× TBE tank buffer, 1- to 15-s pulse ramp, 200 V, 14°C, and 22 h. Virioplankton preparations of the August 1995 samples were analyzed under identical electrophoretic conditions for 24 h. After electrophoresis, the gels were stained for 30 min in SYBR Green I (Molecular Probes, Eugene, Oreg.) according to the manufacturer’s instructions and digitally scanned for fluorescence by using a laser fluoroimager (FluorImager; Molecular Dynamics, Sunnyvale, Calif.).
Densitometric analysis of virioplankton PFGE fingerprints.
Each virioplankton PFGE fingerprint was scanned, and the densitometric profiles (relative fluorescence) were recorded. The staining intensity (relative fluorescence units) for entire molecular size regions was used as a direct measure of the concentration of virioplankton genomic DNA within each region. The ratio of staining intensity to genome size and viral abundance was calculated, utilizing a standard of known viral abundance. Standards consisted of PFGE plugs containing known titers of bacteriophage processed under conditions identical to those for the viral concentrates and loaded onto PFGE fingerprint gels. The ratio was used to calculate the viral abundance within each molecular size region. In the calculation of viral abundance, V represents the number of viruses within a single band or collection of bands in a molecular size region, and G represents the genome size in kilobase pairs of DNA visualized on the virioplankton PFGE fingerprint. A represents the area under all peaks within a region, u represents the unknown viruses, and k represents the known virus standard. The calculation was as follows: VuGu/Au = VkGk/Ak and Vu = AuVkGk/GuAk.
Two standards, CB 7Φ and CB 45Φ, of known titer, were used independently in the calculation of viral abundances within the molecular size regions. The estimates of viral abundance obtained with the two standards were averaged to obtain a final estimate of virioplankton titer within each molecular size region.
Analysis of virioplankton PFGE fingerprints.
Digital gel images, recorded by using a FluorImager (Molecular Dynamics), were imported as TIFF files into a Power Macintosh 7600/132 computer (Apple Computer, Cupertino, Calif.). Densitometry data were obtained from pulsed-field gel images, using an image analysis program (ImageQuant; Molecular Dynamics).
Four virioplankton PFGE fingerprint gels were analyzed by using the Molecular MA fingerprinting program (Bio-Rad). Bands within the virioplankton PFGE fingerprints were normalized according to a single molecular size standard of phage lambda concatamers and identified according to a background intensity setting of 71 to 72%. A schematic representation of the banding patterns, based on the interpretation of the Molecular MA software, was prepared. Among the 23 water samples, a total of 35 bands, ranging from 314 to 12 kb, were identified. A binary matrix identifying the presence or absence of each band within each sample was constructed, and a similarity index for the virioplankton populations within each water sample was calculated, using the Dice coincidence index (16).
The similarity value (Sab) for water samples a and b is equal to the number of common bands in their virioplankton PFGE fingerprints (nab) divided by the average number of fragments in both water samples [Sab = 2nab/(na + nb)]. A dendrogram, based on the matrix of Sab values, was obtained by using the unweighted pair group method with arithmetic averages (UPGMA) (34).
Considering that the objective was to compare virioplankton community structures between water samples and to analyze the complexity within the samples, the multiaxis ordination principal-component analysis was applied to the binary matrix. Eigenvalues and coordinates for water sample ordination were calculated from the binary matrix. Cluster and principal-component analyses were performed with NTSYS (Exeter Software, New York, N.Y.).
RESULTS
Virioplankton PFGE fingerprinting.
Virioplankton PFGE fingerprints were obtained (Fig. 1; see Fig. 3). All virioplankton PFGE fingerprints examined in this study contained DNAs with molecular sizes of between 12 and 314 kb. The band intensity within each virioplankton PFGE fingerprint is related to the number of viruses within the water sample, while the overall banding pattern reflects viral genomic diversity within the virioplankton. To determine whether DNA in the virioplankton PFGE fingerprints was encapsulated viral DNA, the viral concentrates were treated with DNase prior to embedding in agarose. Virioplankton PFGE fingerprints obtained from DNase-treated samples and untreated samples were identical (47), leading to the conclusion that the virioplankton PFGE fingerprints represent encapsulated viral genomic DNA, i.e., DNA protected from DNase digestion.
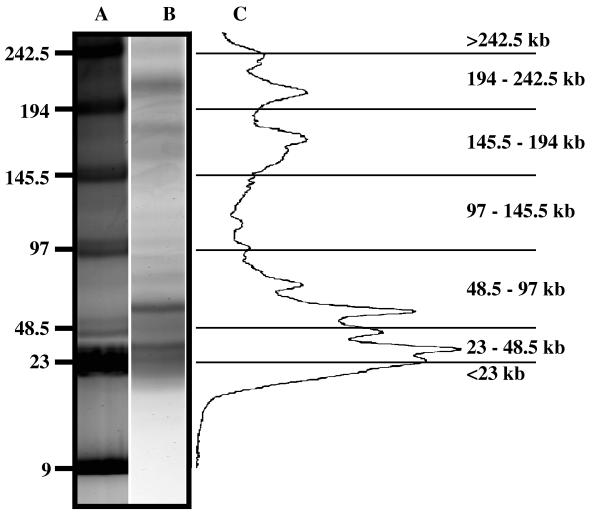
FIG. 1.
Contour-clamped homogeneous electric field PFGE of Chesapeake Bay virioplankton and densitometric scan of virioplankton PFGE fingerprints. Lane A, molecular size markers; lane B, virioplankton PFGE fingerprint from a water sample collected at station 744 on the August 1995 cruise. (C) Densitometric scan of fluorescence intensity from the lane B virioplankton PFGE fingerprint. The densitometric scan data were divided into seven molecular size classes, shown at the far right. Molecular size markers are in kilobases.
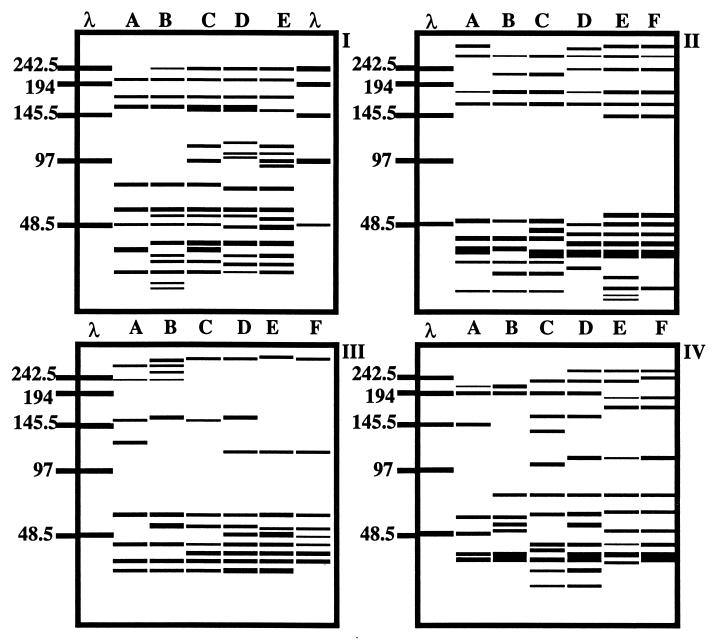
FIG. 3.
Computer-generated banding patterns based on virioplankton PFGE fingerprints shown in Fig. 2. Gel and lane designations are identical to those for Fig. 2. Banding patterns were used to calculate a similarity matrix (data not shown). Band positions were standardized to a single marker lane. Molecular size markers are in kilobases.
Virus fractions concentrated from replicate water samples collected from stations in the lower Chesapeake Bay (station 744) and in the upper Chesapeake Bay (station 845) in June 1996 yielded identical virioplankton PFGE fingerprints (47). Moreover, banding patterns of samples collected from two of the lower-bay stations (744 and 724) were similar, and they were identical in July 1996 (see Fig. 3 and 4).
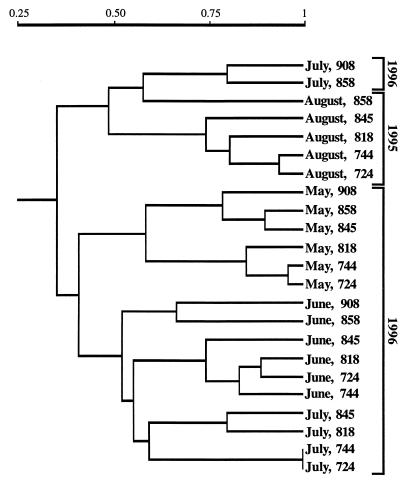
FIG. 4.
Dendrogram based on a similarity matrix of virioplankton PFGE fingerprint banding patterns from all water samples.
The sensitivity of the method for detecting viruses was tested by using three bacteriophages isolated from Chesapeake Bay. Plugs containing mixtures of the three bacteriophages at different titers, after analysis by PFGE, gave results indicating that the detection limit for CB 7Φ and CB 45Φ was 106 virus particles within a single band, while the detection limit for CB 908Φ was 105 virus particles/band (data shown in reference 47). If the detection limit is indeed 106 virus particles/band, virioplankton PFGE fingerprinting should detect a virus which comprises 1% of the total virioplankton abundance in the original water sample.
Densitometric analysis of virioplankton PFGE fingerprints.
A study of ruminal fluid (22) showed that band staining intensity on PFGE gels can be used to estimate the abundance of viruses of a given genome size. In this study the staining intensity of virioplankton PFGE fingerprints was used to characterize Chesapeake Bay virioplankton populations. As shown in Fig. 1, virioplankton PFGE fingerprints were divided into seven molecular size groups: <23, 23 to 48.5, 48.5 to 97, 97 to 145.5, 145.5 to 194, 194 to 242.5, and >242.5 kb. Results of virioplankton abundance analyses are shown in Table 1. The majority of viruses were in the 23- to 48.5- and 48.5- to 97-kb molecular size groups, with the former estimated to be more numerous. Viruses with genome sizes of 23 to 97 kb comprised 66, 90, 93, and 86% of the total virus abundances in the August 1995 and May, June, and July 1996 water samples, respectively. Based on PFGE fingerprint data, ca. 75% of the Chesapeake Bay virioplankton in the water samples analyzed in this study comprised viruses with genome sizes of less than 97 kb. In fact, the 23- to 48.5-kb viruses account for ca. 30 to 60% of the total virioplankton.
TABLE 1.
Composition of Chesapeake Bay virioplankton according to molecular size of the virus genome
| Cruise date | Molecular size region (kb) | % of total virioplankton at station:
|
|||||
|---|---|---|---|---|---|---|---|
| 908 | 858 | 845 | 818 | 744 | 724 | ||
| August 1995 | >242.5 | ||||||
| 197–242.5 | 0.4 | 1.6 | 3.0 | 2.5 | 2.2 | ||
| 145.5–194 | 1.5 | 1.9 | 2.7 | 1.9 | 2.5 | ||
| 97–145.5 | 0.3 | 0.2 | |||||
| 48.5–97 | 16.3 | 24.0 | 28.5 | 28.5 | 29.8 | ||
| 23–48.5 | 52.4 | 47.7 | 46.1 | 50.3 | 52.1 | ||
| <23 | 29.5 | 24.8 | 19.7 | 16.5 | 13.2 | ||
| May 1996 | >242.5 | 0.5 | 0.1 | 0.4 | 0.4 | 0.5 | 0.4 |
| 197–242.5 | 0.1 | 0.2 | 0.1 | ||||
| 145.5–194 | 1.7 | 2.0 | 3.9 | 1.5 | 1.5 | 1.2 | |
| 97–145.5 | |||||||
| 48.5–97 | 34.1 | 30.4 | 26.9 | 22.1 | 18.0 | 20.4 | |
| 23–48.5 | 61.2 | 64.7 | 64.2 | 74.1 | 78.0 | 76.1 | |
| <23 | 2.6 | 2.8 | 4.6 | 1.8 | 2.0 | 1.9 | |
| June 1996 | >242.5 | 0.2 | 0.4 | 0.2 | 0.2 | 0.4 | 0.4 |
| 197–242.5 | 0.1 | 0.1 | |||||
| 145.5–194 | 0.3 | 0.5 | 0.2 | 0.3 | 0.3 | 0.2 | |
| 97–145.5 | 0.6 | 0.1 | 0.6 | 0.8 | 0.8 | 1.3 | |
| 48.5–97 | 24.5 | 22.4 | 18.9 | 14.2 | 17.5 | 13.7 | |
| 23–48.5 | 74.4 | 76.6 | 80.2 | 84.5 | 81.0 | 84.4 | |
| <23 | |||||||
| July 1996 | >242.5 | ||||||
| 197–242.5 | 1.0 | 0.5 | 1.2 | 0.8 | 0.4 | 0.8 | |
| 145.5–194 | 1.7 | 0.6 | 0.6 | 0.7 | 1.1 | 0.8 | |
| 97–145.5 | 0.1 | 0.8 | 0.7 | 0.8 | 0.7 | ||
| 48.5–97 | 12.4 | 19.0 | 20.2 | 19.2 | 22.3 | 25.1 | |
| 23–48.5 | 75.6 | 71.6 | 67.8 | 69.4 | 67.2 | 64.6 | |
| <23 | 9.4 | 8.3 | 9.3 | 9.2 | 8.2 | 8.0 | |
Virus titers within the molecular size groups were 105 to 108 virus particles per PFGE plug. The virus abundance data presented in Table 1 showed temporal differences in frequency distribution. All water samples except those collected in June 1996 contained <23-kb viruses. August 1995 water samples did not contain viruses of >242 kb. Viruses with a genome size of 97 to 145.5 kb were present in water samples collected from the lower-bay stations (818, 744, and 724) and during the summer months.
Similarity analysis of Chesapeake Bay virioplankton populations.
Four pulsed-field gels, each containing virioplankton PFGE fingerprints of water samples collected on each of the cruises, are shown in Fig. 2. All samples were analyzed under identical electrophoretic conditions, with the exception of the August 1995 samples, which were run for 24 h. In addition to the virioplankton concentrate plugs, each gel contained molecular size markers (lambda phage concatamers) and a plug containing a known titer of CB 7Φ and CB 45Φ bacteriophages. Reconstructed gels, shown in Fig. 3, were normalized to these single molecular size standards.
FIG. 2.
Virioplankton PFGE fingerprints of water samples analyzed in the study. (I) August 1995 water samples from stations 858, 845, 818, 744, and 724 (lanes A to E, respectively). (II to IV) May 1996 (II), June 1996 (III), and July 1996 (IV) water samples from stations 908, 858, 845, 818, 744, and 724 (lanes A to F, respectively). Lanes λ, molecular size markers (kilobases) specific for each pulsed-field gel.
A total of 35 bands were identified, ranging from 314 to 12 kb. Individual virioplankton PFGE fingerprints contained an average of 11 (±3 [standard error]) bands and ranged from 7 bands (July, station 908) to 16 bands (August, stations 744 and 724). The most common band (28.9 kb) was detected in 20 samples. Only five bands were identified as being unique to a given sample. Similarity analysis of water samples was based on the presence or absence of a band in a virioplankton PFGE fingerprint.
The similarity of viral populations within a water sample is shown in a dendrogram (Fig. 4) constructed from a matrix of similarity values (Dice’s correlation coefficients), using the UPGMA clustering algorithm. The correlation coefficient (r) between the matrix of Sab values (data not shown) and the cophenetic matrix (calculated from UPGMA), on which the dendrogram is based, was 0.84. Therefore, the dendrogram was a good representation of the similarity between virioplankton populations in the water samples. Overall, the water samples clustered into groups according to cruise date, with a few exceptions. Water samples collected in May 1996 clustered most tightly, with the lowest Sab value (0.45) for stations 744 and 858.
The lower-bay stations 724 and 744 yielded similar fingerprints, with similarity values ranging from 0.82 (June 1996) to 1.0 (July 1996). Virioplankton populations in water samples collected from neighboring stations in the upper bay, 908 and 858, were the least similar, while samples collected at neighboring stations in the lower bay were the most similar. Middle-bay stations, 845 and 818, yielded results that were variable.
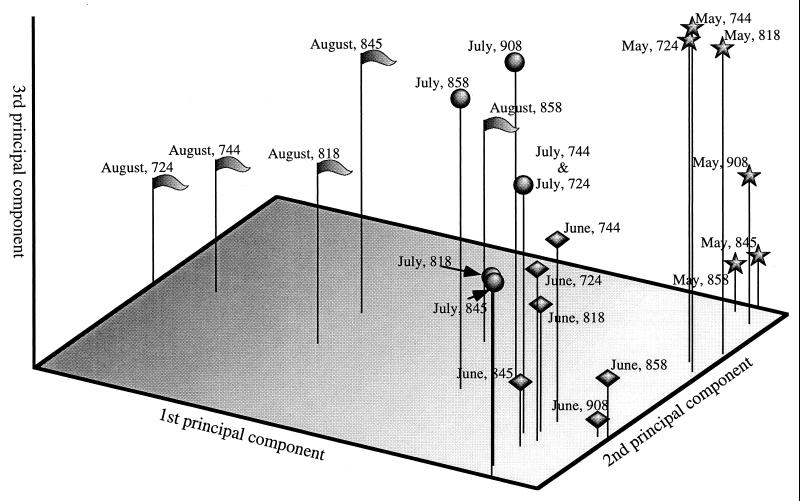
To gain insight into environmental factors influencing virioplankton community structure, the community ordination technique principal-component analysis was applied to the data set. Results of this analysis are shown in Fig. 5. The first three axes, to which sample positions are mapped, accounted for 22, 15, and 10% of the variation between samples, respectively. Closer examination in light of environmental data (Table 2) allows formation of hypotheses with respect to environmental variables correlating with each of the principal-component axes.
FIG. 5.
Three-dimensional plot generated from principal-component analysis of virioplankton PFGE fingerprint data. Positions of samples are marked according to cruise date (August [ ], May [★], June [⧫], or July [•]) and station.
], May [★], June [⧫], or July [•]) and station.
TABLE 2.
Environmental data for Chesapeake Bay stations
| Station | Depth (m) | August 1995
|
May 1996
|
June 1996
|
July 1996
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Temp (°C) | O2a | PSUb | Temp (°C) | O2 | PSU | Temp (°C) | O2 | PSU | Temp (°C) | O2 | PSU | ||
| 908 | Surface | 15.4 | 7.0 | 1.6 | 18.3 | 8.1 | 1.3 | 25.0 | 8.2 | 4.7 | |||
| 6 | 14.7 | 6.6 | 4.3 | 15.4 | 5.7 | 8.6 | 21.0 | 3.4 | 10.6 | ||||
| 858 | Surface | 26.9 | 6.8 | 11.9 | 15.1 | 6.0 | 3.9 | 17.0 | 8.7 | 3.9 | 24.1 | 7.4 | 6.5 |
| 19 | 26.2 | 0.4 | 23.3 | 12.2 | 3.0 | 11.2 | 15.1 | 0.2 | 14.5 | 19.3 | 0.2 | 16.1 | |
| 845 | Surface | 18.1 | 9.1 | 5.5 | |||||||||
| 21 | 15.8 | 0.2 | 17.7 | ||||||||||
| 818 | Surface | 27.2 | 5.8 | 14.5 | 15.1 | 7.0 | 6.0 | 17.5 | 8.7 | 6.3 | 24.6 | 5.9 | 9.9 |
| 30 | 25.7 | 1.9 | 25.0 | 9.8 | 2.5 | 14.5 | 16.1 | 0.7 | 21.6 | 22.7 | 0.1 | 19.7 | |
| 744 | Surface | 28.4 | 6.0 | 16.4 | 14.9 | 7.9 | 12.1 | 19.4 | 7.2 | 9.7 | 25.1 | 5.2 | 12.2 |
| 23 | 25.9 | 3.4 | 27.2 | 15.1 | 6.0 | 17.0 | 16.7 | 3.0 | 22.0 | 23.6 | 1.4 | 19.9 | |
| 724 | Surface | 26.5 | 5.8 | 21.1 | 16.1 | 9.2 | 13.5 | 19.0 | 7.4 | 11.9 | |||
| 18 | 25.8 | 2.8 | 29.7 | 14.3 | 6.4 | 23.0 | 17.3 | 4.8 | 21.9 | ||||
Oxygen concentration in milliliters of O2 per liter of water.
PSU, practical salinity units.
The position of virioplankton populations along the first principal-component axis correlates approximately to the time of sample collection. All samples collected in late spring and early summer of 1996 grouped along this axis. The August 1995 samples, while not as tightly grouped along the first axis, were notably different from the 1996 samples. Thus, a seasonal trend in the Chesapeake Bay virioplankton community structure is suggested. Positions of virioplankton populations along the second principal-component axis correlate with the degree of water column stratification during sampling. Virioplankton populations of samples collected in June and July 1996, were in the upper 1 m of a highly stratified water column. Conversely, virioplankton populations in the May 1996 surface water samples derived from a well-mixed water column. August 1995 virioplankton were collected from surface waters of a moderately stratified water column. Within a cruise, the locations of virioplankton samples along the third axis indicate the station from which the water sample was collected. From their positions along the third axis, virioplankton grouped into three subpopulations, i.e., upper (908 and 858), middle (845 and 818), and lower (744 and 724) bay (July 1996 samples), or into two subpopulations, i.e., upper (908, 858, and 845) and lower (818, 744, and 724) bay (August 1995 and May and June 1996).
DISCUSSION
Chesapeake Bay virioplankton populations are dynamic, not only in abundance (50) but also in composition. Changes in virioplankton community structure were observed for the samples examined in this study, both within a cruise and between cruises. Differences observed in virioplankton PFGE fingerprints suggest that unique virioplankton populations exist in the upper and lower Chesapeake Bay regions. Changes observed in the virioplankton are concluded to reflect corresponding differences in the compositions of phyto-/zoo- and bacterioplankton host populations. In a broader interpretation, three conclusions can be drawn. First, viruses are an active and dynamic component of plankton and are responsive to environmental factors. Second, Chesapeake Bay bacterioplankton host populations do not comprise a single group of cosmopolitan species capable of existing in a diverse range of physiochemical environments but instead constitute specialized communities, separated by time and space. Third, the dynamic nature of Chesapeake Bay virioplankton is indicative of a virus population which, through infection and lysis of numerically dominant hosts, is capable of influencing the clonal diversity of its host population.
The technique used in this study to monitor Chesapeake Bay virioplankton population dynamics, i.e., PFGE, has been employed as a tool for molecular fingerprinting (strain typing) and molecular phylogenetic studies (33, 40, 42). However, prior to this study, the application of PFGE to analyses of complex microbial communities has been demonstrated only in studies of sheep ruminal bacteriophage populations (22, 39). This study is the first to demonstrate the utility of PFGE in cataloging and comparing community structures of the virioplankton. Furthermore, because this procedure does not require selective methods, except for filter fractionation, or DNA amplification steps in preparation of virioplankton concentrates for PFGE, the band staining intensity within a virioplankton PFGE fingerprint provides a direct estimate of viral abundance.
The single selective component of the method is that only viruses containing double-stranded DNA (dsDNA) are detected by the SYBR Green I fluorescent stain used to label DNA separated on PFGE gels. Accurate estimation of viral genome sizes from virioplankton PFGE fingerprints is possible only for dsDNA viruses with linear, nonsegmented chromosomes. Despite these restrictions, genomic DNA from five of the nine described bacteriophage families would be visualized on virioplankton PFGE fingerprints. Bacteriophage genomes containing cohesive ends, which enable the formation of circular DNAs or concatamers consisting of two to several single genomes linked together, could be represented at several molecular size locations. Treatment to eliminate concatamers from virioplankton PFGE plugs (heating to 95°C followed by rapid cooling) did not alter the banding patterns of the virioplankton PFGE fingerprints (data not shown), thus indicating that the large viral genomic DNAs observed in this study were not artifacts.
Because only dsDNA viruses were detected, it could be concluded that virioplankton PFGE fingerprinting overlooks a large portion of viral diversity. However, of the approximately 3,500 known and purified bacteriophage strains, 3,300 belong to the three families of tailed phages (Myoviridae, Podoviridae, and Siphoviridae), all of which contain dsDNA (2). Among the tailed phages, the genome size varies from 17 to 590 kb but most commonly is between 30 and 60 kb (1), values that correlate with the observation in this study that, in all water samples, DNAs of 23 to 97 kb accounted for the majority of virioplankton DNA and with the fact that the majority of viruses in the Chesapeake Bay virioplankton are bacteriophages (50).
After bacteria, eukaryotic algae are the second most abundant host group within the Chesapeake Bay plankton. Algal viruses should be detected in virioplankton PFGE fingerprints, since large concentrations of viruses infecting unicellular algae (>103/ml) have been reported in marine (13, 14) and freshwater (44) systems. Genome size, topology, and composition have been reported for only a few of the algal viruses (43). Of those reported, most contain dsDNA genomes of ≥200 kb in size, with a few examples of algal viral genomes as small as 77 kb (14). It is interesting that neither the August nor July water samples contained viruses with genomes of >242.5 kb in size (Fig. 3). A possible interpretation is that the abundance of algal viruses is low during mid- and late summer in the Chesapeake Bay and that the mid- and late summer viruses may be associated with zooplankton, i.e., zooplankton commensal and/or symbiotic bacteria.
Maruyama and coauthors (26) used DNase to quantify the proportions of free and coated (non-DNase-digestible) DNA within estuarine waters of Tokyo Bay. They found that nearly 90% of DNA contained within the <0.2-μm size fraction was not DNase digestible. Most of the coated DNA was 20 to 30 kb in size, from which the authors concluded that coated DNA originated from viruses within the <0.2-μm size fraction. It is interesting that the 20- to 30-kb size of coated DNA in the Tokyo Bay water samples corroborates the finding in this study that in all water samples the single most abundant DNA size class is 23 to 48.5 kb (Table 1). Thus, it is highly probable that the DNA separated and visualized by PFGE consists entirely of viral genomic DNA. If dissolved DNA was concentrated in the water samples analyzed in this study, it was either too small (<9 kb) or too dilute to be detected by the method employed.
Microbial ecologists, recognizing the inadequacies of traditional culturing methods for characterizing microbial communities, have increasingly turned to molecular techniques for examining population dynamics of bacterial consortia, including total-community DNA hybridization (23), low-molecular-weight DNA profiles (4, 19), and denaturing gradient gel electrophoresis of PCR-amplified 16S rRNA genes (28). An ideal method for examining bacterioplankton community structure would offer a combination of species-level resolution and unbiased, quantifiable sampling of each species within a consortium. Because of genetic homology among closely related bacterial species and difficulties in quantitative lysis of bacterial consortia, none of the methods used for direct examination of the bacterioplankton possess these attributes. The PFGE-based method, as used in this study, can claim a measure of both species-level resolution and unbiased sampling for examining virioplankton composition. This is because even closely related viruses sometimes lack significant genetic homology (7). Viral capsids, in comparison, consist mostly of protein and are relatively easy to lyse by using standard enzymatic methods. Thus, virioplankton PFGE fingerprints provide the best measure of host community dynamics to date.
PFGE was utilized by Klieve et al. (22) and Swain and coworkers (39) to estimate the abundance of viruses of a specific genome size in the ruminal fluid of sheep and to observe ruminal bacteriophage population dynamics. Comparison of Chesapeake Bay virus populations with those of the sheep rumen suggests that ruminal populations are more diverse in composition. In contrast to Chesapeake Bay virioplankton PFGE fingerprints, viral PFGE fingerprints from ruminal fluid contained a broad smear of DNA between 23 and 450 kb in size. Within this wide molecular size range, the individual DNA bands could not be resolved, perhaps because of a larger number of viruses in ruminal fluid (ca. 1010/ml) than in estuarine water samples (ca. 107/ml). In trials conducted with larger numbers of Chesapeake Bay viruses, band smearing did occur (data not shown). By utilizing the direct relationship between staining intensity and numbers of viruses, it was estimated that phages with genome sizes of between 23 and 100 kb were the most common in ruminal fluid (22) and in Chesapeake Bay water samples (Table 1). The dynamics of bacteriophage consortia in sheep rumen indicated that there were differences in the virus populations between animals, varying significantly over a diurnal period. The diverse and dynamic nature of rumen bacteriophage populations led Swain et al. to suggest that phage infection serves to “…maintain bacterial population diversity and balance…” (39).
Because individual bands were readily discernible in Chesapeake Bay virioplankton PFGE fingerprints, cluster analysis could be used to group the samples (25). Hierarchical clustering methods, such as UPGMA, impose a one-dimensional structure on similarity data, which can distort relationships between samples (11). For these reasons, we chose to apply also the ordination method principal-component analysis to the virioplankton community data. Unlike cluster analysis, ordination does not divide samples into groups but determines the coordinates of a sample within a hyperspace defined by several axes (one axis for each species). Therefore, the principal benefit of a community ordination approach is the ability to view the relationship between virioplankton populations in terms of “species hyperspace” (25). Ultimately, principal-component analysis should provide insight into environmental factors involved in shaping the structure of an ecological community (25). Conclusions drawn from examination of Chesapeake Bay virioplankton population dynamics by principal-component analysis (Fig. 5) were in agreement with those from cluster analyses (Fig. 4). In both analyses, the May 1996 virioplankton samples formed the most cohesive of the groups and the virioplankton from the lower-bay stations, 744 and 724, were highly similar. Principal-component analysis results showed clustering of June and July samples, as well as groupings of virioplankton populations into upper, middle, and lower Chesapeake Bay populations.
The spatial distributions of virioplankton communities within species hyperspace indicated that the physiochemical environmental changes associated with seasonality, the degree of stratification of the water column, and other factors related to geographical location in the Chesapeake Bay are important in determining virioplankton composition and structure. In fact, the degree of stratification of the water column may account for more differences observed in the virioplankton populations than geographical location. Since the abundance of a virus is, of course, dependent on its specific host, these factors are significant in shaping the compositions of host communities. From the study reported here and the results of other investigations, it is clear that the viruses in estuarine and marine environments are an interesting and important component of these ecosystems.
ACKNOWLEDGMENTS
We acknowledge the excellent cooperation of Wayne Coates and Diane Stoecker, permitting K.E.W. to participate in research cruises. The crew of the R/V Cape Henlopen provided valuable assistance during sampling cruises.
Footnotes
Contribution no. 315 from the Center of Marine Biotechnology; Contribution no. 912 from the Australian Institute of Marine Science.
REFERENCES
- 1.Ackermann H W, DuBow M S. Viruses of prokaryotes: natural groups of bacteriophages. Boca Raton, Fla: CRC Press, Inc.; 1987. [Google Scholar]
- 2.Ackermann H W, DuBow M S, Jarvis A W, Jones L A, Krylov V N, Maniloff J, Rocourt J, Safferman R S, Schneider J, Seldin L. The species concept and its application to tailed phages. Arch Virol. 1992;124:69–82. doi: 10.1007/BF01314626. [DOI] [PubMed] [Google Scholar]
- 3.Bergh O, Borsheim K Y, Bratbak G, Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–468. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- 4.Bidle K D, Fletcher M. Comparison of free-living and particle-associated bacterial communities in the Chesapeake Bay by stable low-molecular-weight RNA analysis. Appl Environ Microbiol. 1995;61:944–952. doi: 10.1128/aem.61.3.944-952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Børsheim K Y. Native marine bacteriophages. FEMS Microbiol Ecol. 1993;102:141–159. [Google Scholar]
- 6.Børsheim K Y, Bratbak G, Heldal M. Enumeration and biomass estimation of planktonic bacteria and viruses by transmission electron microscopy. Appl Environ Microbiol. 1990;56:352–356. doi: 10.1128/aem.56.2.352-356.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Botstein D. A theory of modular evolution for bacteriophages. Ann N Y Acad Sci. 1980;354:484–491. doi: 10.1111/j.1749-6632.1980.tb27987.x. [DOI] [PubMed] [Google Scholar]
- 8.Bratbak G, Heldal M, Norland S, Thingstad T F. Viruses as partners in spring bloom microbial trophodynamics. Appl Environ Microbiol. 1990;56:1400–1405. doi: 10.1128/aem.56.5.1400-1405.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen F, Suttle C A. Amplification of DNA polymerase gene fragments from viruses infecting microalgae. Appl Environ Microbiol. 1995;61:1274–1278. doi: 10.1128/aem.61.4.1274-1278.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen F, Suttle C A, Short S M. Genetic diversity in marine algal virus communities as revealed by sequence analysis of DNA polymerase genes. Appl Environ Microbiol. 1996;62:2869–2874. doi: 10.1128/aem.62.8.2869-2874.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chun J. Computer assisted classification and identification of actinomycetes. Ph.D. dissertation. Newcastle upon Tyne, United Kingdom: University of Newcastle upon Tyne; 1996. [Google Scholar]
- 12.Cochlan W P, Wikner J, Steward G F, Smith D C, Azam F. Spatial distribution of viruses, bacteria, and chlorophyll a in neritic, oceanic and estuarine environments. Mar Ecol Prog Ser. 1993;92:77–87. [Google Scholar]
- 13.Cottrell M T, Suttle C A. Dynamics of a lytic virus infecting the photosynthetic marine picoflagellate Micromonas pusilla. Limnol Oceanogr. 1995;40:730–739. [Google Scholar]
- 14.Cottrell M T, Suttle C A. Wide-spread occurance and clonal variation in viruses which cause lysis of a cosmopolitan, eukaryotic marine phytoplankter, Micromonas pusilla. Mar Ecol Prog Ser. 1991;78:1–9. [Google Scholar]
- 15.Demuth J, Neve H, Witzel K P. Direct electron microscopy study on the morphological diversity of bacteriophage populations in Lake Plußsee. Appl Environ Microbiol. 1993;59:3378–3384. doi: 10.1128/aem.59.10.3378-3384.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dice L R. Measures of the amount of ecological association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 17.Frank H, Moebus K. An electron microscopic study of bacteriophages from marine waters. Helgoländer Meeresuntersuchungen. 1987;41:385–414. [Google Scholar]
- 18.Fuhrman J A, Suttle C A. Viruses in marine planktonic systems. Oceanography. 1993;6:50–62. [Google Scholar]
- 19.Hofle M G. Bacterioplankton community structure and dynamics after large-scale release of nonindigenous bacteria as revealed by low-molecular-weight-RNA analysis. Appl Environ Microbiol. 1992;58:3387–3394. doi: 10.1128/aem.58.10.3387-3394.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang S C, Paul J H. Seasonal and diel abundance of viruses and occurence of lysogeny/bacteriocinogeny in the marine environment. Mar Ecol Prog Ser. 1994;104:163–172. [Google Scholar]
- 21.Kellogg C A, Rose J B, Jiang S C, Thurmond J M, Paul J H. Genetic diversity of related vibriophages isolated from marine environments around Florida and Hawaii, USA. Mar Ecol Prog Ser. 1995;120:89–98. [Google Scholar]
- 22.Klieve A V, Swain R A. Estimation of ruminal bacteriophage numbers by pulsed-field gel electrophoresis and laser densitometry. Appl Environ Microbiol. 1993;59:2299–2303. doi: 10.1128/aem.59.7.2299-2303.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S H, Fuhrman J A. Spatial and temporal variation of natural bacterioplankton assemblages studied by total genomic DNA cross-hybridization. Limnol Oceanogr. 1991;36:1277–1287. [Google Scholar]
- 24.Lenski R E. Dynamics of interactions between bacteria and virulent bacteriophage. Adv Microb Ecol. 1988;10:1–44. [Google Scholar]
- 25.Ludwig J A, Reynolds J F. Statistical ecology: a primer on methods and computing. New York, N.Y: John Wiley and Sons; 1988. [Google Scholar]
- 26.Maruyama A, Oda M, Higashihara T. Abundance of virus-sized non-DNase-digestible DNA (coated DNA) in eutrophic seawater. Appl Environ Microbiol. 1993;59:712–717. doi: 10.1128/aem.59.3.712-717.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moebus K. Ecology of marine bacteriophages. In: Goyal S M, Gerba C P, Britton G, editors. Phage ecology. New York, N.Y: John Wiley and Sons; 1987. pp. 137–155. [Google Scholar]
- 28.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paul J H, Rose J B, Jiang S C, Kellogg C A, Dickson L. Distribution of viral abundance in the reef environment of Key Largo, Florida. Appl Environ Microbiol. 1993;59:718–724. doi: 10.1128/aem.59.3.718-724.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Proctor L M. Advances in the study of marine viruses. Microsc Res Tech. 1997;37:136–161. doi: 10.1002/(SICI)1097-0029(19970415)37:2<136::AID-JEMT3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 31.Reanney D C, Ackermann H W. Comparative biology and evolution of bacteriophages. Adv Virus Res. 1982;27:205–280. doi: 10.1016/s0065-3527(08)60436-4. [DOI] [PubMed] [Google Scholar]
- 32.Ripp S, Ogunseitan O A, Miller R V. Transduction of a freshwater microbial community by a new Pseudomonas aeruginosa generalized transducing phage, UT1. Mol Ecol. 1994;3:121–126. doi: 10.1111/j.1365-294x.1994.tb00112.x. [DOI] [PubMed] [Google Scholar]
- 33.Roux V, Raoult D. Inter- and intraspecies identification of Bartonella (Rochalimaea) species. J Clin Microbiol. 1995;33:1573–1579. doi: 10.1128/jcm.33.6.1573-1579.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sokal R R, Michener C D. A statistical method for evaluating systematic relationships. Kansas Univ Sci Bull. 1958;38:1409–1438. [Google Scholar]
- 35.Steward G F, Smith D C, Azam F. Abundance and production of bacteria and viruses in the Bering and Chukchi seas. Mar Ecol Prog Ser. 1996;131:287–300. [Google Scholar]
- 36.Suttle C A. The significance of viruses to mortality in aquatic microbial communities. Microb Ecol. 1994;28:237–243. doi: 10.1007/BF00166813. [DOI] [PubMed] [Google Scholar]
- 37.Suttle C A, Chan A M. Dynamics and distribution of cyanophages and their effect on marine Synechococcus spp. Appl Environ Microbiol. 1994;60:3167–3174. doi: 10.1128/aem.60.9.3167-3174.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suttle C A, Chan A M, Cottrell M T. Use of ultrafiltration to isolate viruses from seawater which are pathogens of marine phytoplankton. Appl Environ Microbiol. 1991;57:721–726. doi: 10.1128/aem.57.3.721-726.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swain R A, Nolan J V, Klieve A V. Natural variability and diurnal fluctuations within the bacteriophage population of the rumen. Appl Environ Microbiol. 1996;62:994–997. doi: 10.1128/aem.62.3.994-997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamplin M L, Jackson J K, Buchrieser C, Murphree R L, Portier K M, Gangar V, Miller L G, Kaspar C W. Pulsed-field gel electrophoresis and ribotype profiles of clinical and environmental Vibrio vulnificus isolates. Appl Environ Microbiol. 1996;62:3572–3580. doi: 10.1128/aem.62.10.3572-3580.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thingstad T F, Heldal M, Bratbak G, Dundas I. Are viruses important partners in pelagic food webs? Trends Ecol Evol. 1993;8:209–212. doi: 10.1016/0169-5347(93)90101-T. [DOI] [PubMed] [Google Scholar]
- 42.Townen K J, Cockayne A. Molecular methods for microbial identification and typing. London, United Kingdom: Chapman and Hall; 1993. [Google Scholar]
- 43.Van Etten J L, Lane L C, Meints R H. Viruses and viruslike particles of eukaryotic algae. Microbiol Rev. 1991;55:586–620. doi: 10.1128/mr.55.4.586-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Etten J L, Van Etten C H, Johnson J K, Burbank D E. A survey for viruses from fresh water that infect a eucaryotic chlorella-like green alga. Appl Environ Microbiol. 1985;49:1326–1328. doi: 10.1128/aem.49.5.1326-1328.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waterbury J B, Valois F W. Resistance to cooccuring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl Environ Microbiol. 1993;59:3393–3399. doi: 10.1128/aem.59.10.3393-3399.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weinbauer M G, Fuks D, Peuzzi P. Distribution of viruses and dissolved DNA along a coastal trophic gradient in the northern Adriatic Sea. Appl Environ Microbiol. 1993;59:4074–4082. doi: 10.1128/aem.59.12.4074-4082.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wommack K E. Aspects of the ecological role of bacteriophages. Ph.D. dissertation. College Park: University of Maryland; 1998. [Google Scholar]
- 48.Wommack, K. E., and R. R. Colwell. Unpublished data.
- 49.Wommack K E, Hill R T, Colwell R R. A simple method for the concentration of viruses from natural water samples. J Microbiol Methods. 1995;22:57–67. [Google Scholar]
- 50.Wommack K E, Hill R T, Kessel M, Russek-Cohen E, Colwell R R. Distribution of viruses in the Chesapeake Bay. Appl Environ Microbiol. 1992;58:2965–2970. doi: 10.1128/aem.58.9.2965-2970.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]